Abstract
Microtubule plus-end depolymerization rate is a potentially important target of physiological regulation, but it has been challenging to measure, so its role in spatial organization is poorly understood. Here we apply a method for tracking plus ends based on time difference imaging to measure depolymerization rates in large interphase asters growing in Xenopus egg extract. We observed strong spatial regulation of depolymerization rates, which were higher in the aster interior compared with the periphery, and much less regulation of polymerization or catastrophe rates. We interpret these data in terms of a limiting component model, where aster growth results in lower levels of soluble tubulin and microtubule-associated proteins (MAPs) in the interior cytosol compared with that at the periphery. The steady-state polymer fraction of tubulin was ∼30%, so tubulin is not strongly depleted in the aster interior. We propose that the limiting component for microtubule assembly is a MAP that inhibits depolymerization, and that egg asters are tuned to low microtubule density.
INTRODUCTION
Microtubules undergo rapid polymerization dynamics in many cell types, as first revealed by polarization microscopy (Inoué and Sato, 1967). Polymerization dynamics occur mostly on plus ends and are well-approximated by a two-state dynamic model of dynamic instability (Dogterom and Leibler, 1993). Plus-end behavior is described by four parameters in this model: polymerization, depolymerization, catastrophe, and rescue rates. These parameters are regulated in both space and time to promote robust organization of intracellular structures and allow for rapid responses to internal and external cues. Most recent biology and mechanism studies focused on polymerization and catastrophe rates, in part because multiple factors are known to regulate these parameters, and in part because they are the easiest to measure in cells. Tracking of the plus-tip binding proteins EB1/3 allows measurement of polymerization rate with high reliability, and inference of catastrophe rate from comet lifetime with somewhat lower reliability. In contrast, depolymerization rates are harder to measure. Most studies reporting depolymerization rates focused on pure tubulin systems or on manual measurements of small numbers of microtubules at the periphery of living cells (Desai and Mitchison, 1997; Brouhard and Rice, 2018). As a result, we know rather little about regulation of depolymerization rates in complex microtubule structures such as the mitotic spindle or microtubule asters. Here, we apply imaging-based methods to systematically measure microtubule depolymerization rates in Xenopus egg extract, and report spatial regulation of depolymerization for the first time.
One important emergent property from the four dynamic instability factors, combined with nucleation, is the fraction of polymerized tubulin at steady state, which we will call the “polymer fraction.” In a closed system like a cell, an increase in microtubule mass leads to depletion of soluble subunits, which limits further assembly giving rise to a negative feedback of microtubule growth. When continuously supplied with GTP, the system comes to a steady state where the polymer fraction is approximately constant over time, and sufficient tubulin remains in solution to power robust polymerization of GTP-capped plus ends. The steady-state polymer fraction in interphase tissue culture cells was found to be 60–80% (Zhai and Borisy, 1994; Kim et al., 2012). What determines this value, and its significance for regulation, are largely unknown. Negative feedback from microtubule mass on further assembly in a closed system was conceptualized in the “component limitation” model for mitotic spindle size (Mitchison et al., 2015). A feedback of this kind is inevitable, but its precise mechanism is nonobvious. For pure tubulin, polymerization rate is a linear function of soluble dimer concentration (Desai and Mitchison, 1997; Brouhard and Rice, 2018). Thus, increased microtubule mass in a closed system decreases polymerization rate by subunit consumption. Catastrophe rate should increase in parallel, assuming it negatively correlates with polymerization rate as in most models (Walker et al., 1988; Gardner et al., 2008). This assumption has been questioned for growing ends (Gardner et al., 2011), but not for shrinking ends to our knowledge. In physiological contexts, tubulin need not be the only limiting component in a closed system. Microtubule-associated proteins (MAPs) that regulate dynamics are also depleted when microtubule mass increases, by binding to microtubules. MAP limitation could provide a negative feedback from polymer fraction to microtubule growth. How individual dynamic instability parameters would respond to MAP limitation is unknown, because different MAPs regulate different parameters.
Xenopus egg extract provides undiluted, metabolically active cytoplasm that reconstitutes physiological polymerization dynamics and is useful for investigating regulatory mechanisms (Field and Mitchison, 2018). It also allows microscopy-based scoring of large numbers of dynamic microtubules in a relatively homogeneous environment that is ideal for quantitative analysis. Here, we focus on interphase asters, which model the egg-spanning asters that position centrosomes, nuclei, and cleavage furrows during early divisions (Wühr et al., 2010; Field et al., 2015). These asters grow to hundreds of microns in radius at a rate of ∼20 µm/min. EB1 tracking showed that the microtubules within them are relatively short, ∼16 µm on average. Their plus-end dynamics are in the bounded regime of dynamic instability, so they polymerize transiently, but are biased toward eventual depolymerization with a half-life of ∼1 min (Ishihara et al., 2016). The aster as a whole grows because each microtubule nucleates more than one daughter microtubule during its lifetime by a poorly understood branching process. The density of plus ends is approximately homogeneous within interphase asters, and there are two interesting boundaries: the stationary microtubule organizing center (MTOC) and the growing aster periphery. The MTOC nucleates continuously and is thought to provide the information that directs microtubule polarity. The periphery controls the rate of aster expansion and determines whether expansion is bounded or unbounded. Plus ends at the periphery polymerize into a microtubule-free environment, while those in the aster interior polymerize into an environment that is dense in microtubules, and as we will show below, partially depleted of soluble subunits. Many questions remain about these asters, including the polymer fraction, minus-end behavior, and whether dynamics differ between the interior and periphery. These dynamical properties likely have important implications for aster mechanics and function. Here, we report that plus-end depolymerization rate is subject to strong spatial regulation and infer that this parameter is important in feedback regulation of polymer fraction. Mechanistically, we interpret this observation in terms of a model where the depolymerization rate is regulated by MAP depletion.
RESULTS
Polymerization rate increases slightly at the aster periphery
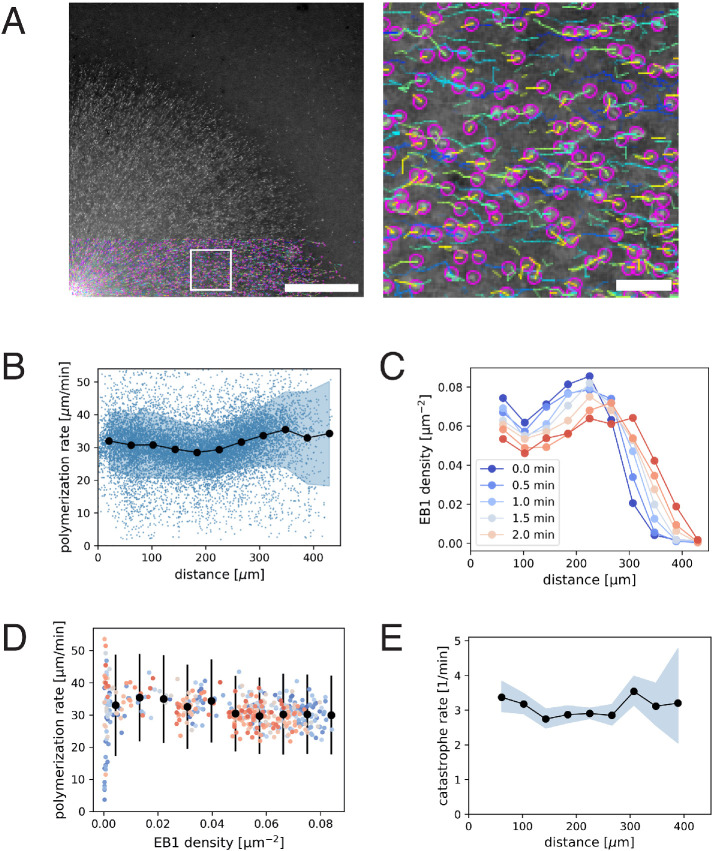
We first investigated the spatial variation of plus-end polymerization and catastrophe rates using standard EB1 comet tracking methods (Figure 1A and Supplemental Movie 1). We found that the polymerization rate remained relatively constant at 30.9 µm/min (N = 9348 tracks, SD 5.52, SEM 0.06) as a function of distance from the center of asters (Figure 1B). We detected an ∼10% increase in polymerization rate at the aster periphery 33.7 ± 10.9 µm/min (mean, std) compared with the interior 30.0 ± 9.5 µm/min (mean, std). The plus-end density is lower at the periphery compared with the aster center (Figure 1C), and resulted in a slight anti-correlation between plus-end density and polymerization rate (Figure 1D). This anti-correlation was smaller than reported in an independent study (Geisterfer et al., 2020).
FIGURE 1:
EB1 tracking-based measurements of microtubule plus-end polymerization rate and catastrophe rate during aster growth. (A) Time-lapse movies of EB1-GFP comets are analyzed by particle tracking. Region in the white box is magnified on the right. Purple spots show the location of individual EB1 comets in this frame, while the trailing lines show the trajectory of the corresponding plus end for the preceding 10 s. The velocity of EB1 comets report microtubule polymerization rate. (B) Polymerization rate as a function radial distance from the center of the aster. N = 9348 EB1 tracks are represented as blue dots. Filled black circles and the shaded region indicate the mean and the SD of the polymerization rate for each spatial bin. The estimated mean difference in the periphery (distance > 300 µm) vs. interior (50 µm < distance < 280 µm) for t = 1.5 min was 3.7 µm/min (95% CI = [2.4, 4.9]). (C) EB1 comet density as a function radial distance. (D) Polymerization rate vs. EB1 comet density. Filled black circles and error bars indicate the mean and the SD of polymerization rate for each EB1 density bin. The different colors correspond to the same time points as in the previous panel. The estimated mean difference in the dense (0.066 1/µm2) vs. sparse (0.022 1/µm2) was 3.8 µm/min (95% CI = [2.6, 5.0]). (E) Catastrophe rate is calculated from the duration of EB1 tracks and plotted over radial distance. Filled black circles and the shaded region indicate the mean and the corresponding 95% confidence interval for the catastrophe rate for each spatial bin. Original data used for this analysis is Supplemental Movie 1. Scale bars, 100 µm and 10 µm.
Movie S1.
Timelapse imaging of EB1-GFP comets during aster growth.
Next, we asked whether the plus-end catastrophe rate showed spatiotemporal variation using EB1 comet lifetimes as a proxy. The average EB1 comet persisted for 19 s, which corresponds to a catastrophe rate of 3.1 per min. This value did not show much variation as a function of location or plus-end density (Figure 1E). These measurements performed with wide-field imaging were consistent with our previous measurements with spinning disk confocal microscopy (Ishihara et al., 2016). Therefore, we expect our measurements to be sensitive to spatial variation in catastrophe rate. In summary, our analysis of EB1 comet imaging demonstrated a modest 10% increase in polymerization rate at the aster periphery, where local plus-end density is relatively low, and no measurable differences in catastrophe rate.
Spatial regulation of depolymerization rates
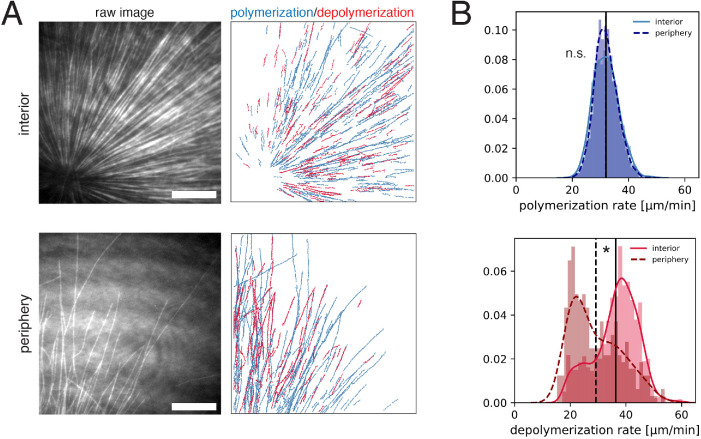
To measure microtubule depolymerization rates, we applied tubulin intensity difference analysis. This method relies on collecting high-quality tubulin images at frequent intervals using total internal reflection fluorescence (TIRF) microscopy, subtracting the intensity of sequential frames to generate time difference images, then applying a tracking algorithm. It allows tracking of polymerizing and depolymerizing ends even when most microtubules are in small bundles as is the case in egg extract asters. Our previous analysis of growing and shrinking ends used manual kymograph analysis of difference images and was restricted to the interior of the interphase asters (Ishihara et al., 2014). Here, we applied a newly developed automated workflow (Caldas et al., 2019, 2020), and systematically compared spatial differences in polymerization dynamics (Figure 2A). In brief, this method applies a low pass spatial filter to the difference images, followed by particle-based tracking to quantify many microtubule ends. Using this workflow, the polymerization rate was found to have a mean of 32.0 ± 4.5 µm/min (mean, std) in the aster interior, and 32.0 ± 4.0 µm/min (mean, std) in the periphery (Figure 2B, upper panel). Polymerization rates exhibited a sharp, unimodal histogram, which overlapped for the interior and periphery with no statistical difference. In contrast, the depolymerization rate showed a striking difference in the aster interior versus periphery. The depolymerization rate was 36.3 ± 7.9 µm/min (mean, std) in the aster interior, compared with 29.2 ± 8.9 µm/min (mean, std) at the aster periphery. The distribution of the depolymerization rates was spread out and had a positive skew in the aster interior, and a negative skew in the aster periphery. In summary, our analysis based on an improved intensity difference analysis showed that polymerization rates are similar in the aster periphery relative to the interior, while depolymerization rates show strong spatial variation and are faster inside asters.
FIGURE 2:
Intensity difference–based measurement of polymerization and depolymerization rates in the interior and the periphery of microtubule asters. (A) TIRF microscopy movies of fluorescent tubulin were subjected to intensity difference analysis, which revealed a spatial map of polymerization and depolymerization for the interior (<100 µm away from the MTOC) and peripheral (>200 µm from the MTOC with few microtubules) region of microtubule asters. These images correspond to Supplemental Movies 2 and 3. (B) The distributions of polymerization and depolymerization rates measured from N = 6 interior movies and N = 6 peripheral movies. The polymerization rate (median 32.0 vs. 32.0 µm/min) showed no statistical difference in the interior vs. periphery (p-value = 0.819, t test). The depolymerization rate (median 37.6 vs. 26.7 µm/min) was higher in the interior (p-value = 4.44e-16, two sample Kolmogorov-Smirnov tests) with estimated difference of mean and median depolymerization rates of 7.1 µm/min (95% CI = [5.9, 8.3]) and 11.0 µm/min (95% CI = [8.4, 12.4]). Vertical lines indicate the mean for each distribution. See Supplemental Figure 1 for analysis of individual movies. Scale bar, 20 µm.
Movie S2.
TIRF microscopy-based intensity difference imaging of the aster interior. Blue correspond to positive intensity difference interpreted as polymerization, while red correspond to negative intensity difference interpreted as depolymerization.
Movie S3.
TIRF microscopy-based intensity difference imaging of the aster periphery.
Tubulin difference imaging revealed other useful information on polymerization dynamics. Plus-end polymerization and depolymerization rates were both variable, but over fairly narrow ranges, so the data supported a two-state model of dynamic instability over alternatives such as a biased random walk (Needleman et al., 2010). Our time sampling was not frequent enough to score rate fluctuations within bouts of depolymerization. We observed few outward-moving depolymerization events (<5%, all restricted to the periphery). We also saw very few inward-moving polymerization events. These data suggest minus ends neither polymerize nor depolymerize on the timescale of our imaging, and are perhaps capped.
Laser ablation–induced depolymerization rates
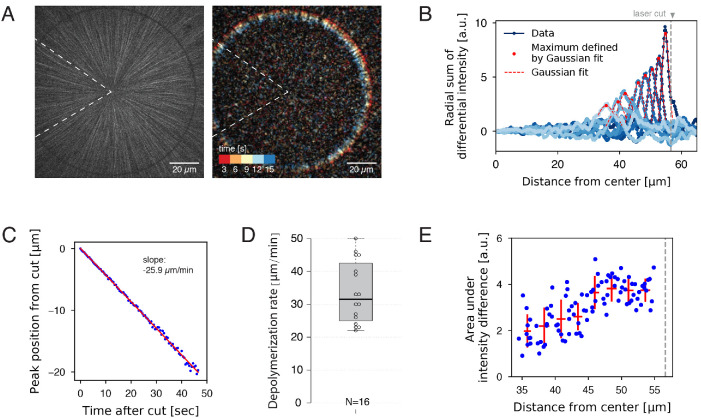
To gain additional information on aster dynamics, we used femtosecond laser ablation to artificially induce depolymerization. Ablation along a line normal to the microtubule axis triggers a spatially defined, synchronous wave of depolymerization (Decker and Brugués, 2015; Decker et al., 2018). Using circular patterns, we ablated microtubules at a fixed distance from the center of asters (Figure 3A and Supplemental Movie 4). We were not able to reliably cut and measure at the aster periphery due to the low microtubule density. Intensity difference analysis (Figure 3B) revealed a resulting wave of depolymerization that moved inward at a constant rate (Figure 3C). No outward-moving wave was detected. This single inward wave is consistent with a model in which (a) the majority of microtubules are oriented with plus ends outward, (b) cut plus ends immediately depolymerize, and (c) newly formed minus ends are stable. By applying this laser ablation assay to multiple asters, we measured an average depolymerization wave velocity of 33.5 ± 9.4 µm/min (mean, std) with a 31.5 µm/min median value, similar to the rate of spontaneous depolymerization in the aster interior (Figure 2B). The average depolymerization rate was surprisingly variable between asters and extract batches (Figure 3D). This variability suggests that the plus-end depolymerization rate is not governed by microtubule structure alone, which should be constant, but also by the precise concentration of MAPs and motors that regulate depolymerization, which are likely to vary between extract batches. We found no correlation between the rate of depolymerization wave and the distance at which the ablation was induced (15–100 µm; Supplemental Figure 2). Overall, these observations confirm that depolymerization rates are variable, and we find no statistical distinction of rates between plus ends following a natural catastrophe versus ablation.
FIGURE 3:
Measurement of depolymerization rates following laser ablation. (A) An interphase aster was subjected to a circular laser cut with radius 56 µm (left), which induced a wave of microtubule depolymerization (Supplemental Movie 4). The resulting movie was used to construct a differential intensity movie (right and Supplemental Movie 5). Dotted wedge region indicates the region excluded from image analysis. (B) Radial sum of differential intensities at different time points (from dark to light blue) of the same laser cut experiment. The area under each curve equals the mass of microtubules depolymerized per time interval of 5 s. Vertical dotted line indicates the location of the laser cut. (C) For each cut, the peak position of the differential intensity travels at constant speed. (D) Depolymerization rates measured from N = 16 laser cuts positioned at 15–56 µm from the center of the aster. (E) The area under differential intensity curves decreases as the depolymerization wave travels inward. This example corresponds to the cut depicted in panels A–C. Vertical dotted line indicates the location of the laser cut.
Movie S4.
A circular laser ablation induces an inward wave of depolymerization.
Careful analysis of microtubules density versus time following laser ablation offers insights into the organization of microtubule assemblies (Brugués et al., 2012; Decker et al., 2018). We found that the strength of the depolymerization wave (i.e., the area under the curves of the summed differential intensities) decreased with progression (Figure 3E). This is explained by the fact that depolymerization is halted when individual plus ends reach their corresponding minus ends. Thus, this observation provides evidence that minus ends exist throughout the aster, and confirms our previous proposal that asters are built of short microtubules (Ishihara et al., 2016).
Aster growth results in spatial gradients of soluble tubulin and MAPs
Spatial regulation of depolymerization rate could be due to complex biochemical schemes, such as activation of a dynamics-regulating kinase in the aster interior. We cannot rule out this kind of hypothesis, but prefer an interpretation based on component limitation, in part because this is a mechanistically simpler hypothesis, and in part because component limitation is inevitable in a closed system. Component limitation is a relevant consideration when comparing microtubule dynamics between the inside and periphery of large asters because aster cytosol is not well mixed by diffusion on relevant time and length scales. Specifically, the aster grows ballistically into fresh cytosol outside the aster much faster than its interior can diffusively equilibrate with that fresh cytosol. A simple Péclet number calculation serves to make this point. Consider the dimensionless Péclet number:  . While a typical protein molecule is transported in a diffusive process with cytoplasmic diffusion coefficient
. While a typical protein molecule is transported in a diffusive process with cytoplasmic diffusion coefficient 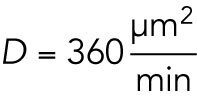 (Salmon et al., 1984), aster growth is a ballistic or advective process, that is, radius increases linearly with time, with a velocity
(Salmon et al., 1984), aster growth is a ballistic or advective process, that is, radius increases linearly with time, with a velocity 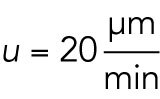 (Ishihara et al., 2014). Given a relevant length scale L, we evaluate the magnitude of the Péclet number. The extremes are as follows: Pe≫1, peripheral microtubules grow so fast that they will always grow into fresh cytoplasm with plenty of available material. Pe≪1, aster growth is limited by the diffusive transport of material to the aster periphery. At a depth of L = 200 μm from the aster periphery, which is typical for our internal measurements, Pe = 11, so diffusion makes little contribution to supplying subunits. Thus, we expect the aster interior to effectively constitute a closed system, shut off from fresh cytosol supplied by diffusion from the outwardly growing aster boundary.
(Ishihara et al., 2014). Given a relevant length scale L, we evaluate the magnitude of the Péclet number. The extremes are as follows: Pe≫1, peripheral microtubules grow so fast that they will always grow into fresh cytoplasm with plenty of available material. Pe≪1, aster growth is limited by the diffusive transport of material to the aster periphery. At a depth of L = 200 μm from the aster periphery, which is typical for our internal measurements, Pe = 11, so diffusion makes little contribution to supplying subunits. Thus, we expect the aster interior to effectively constitute a closed system, shut off from fresh cytosol supplied by diffusion from the outwardly growing aster boundary.
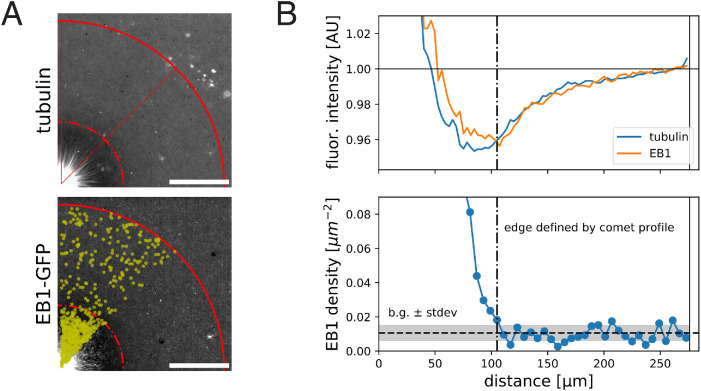
To seek evidence for depletion of soluble subunits by growing asters, we imaged fluorescently labeled tubulin and analyzed its intensity profile as a proxy of local tubulin concentration, summing both polymer and soluble forms (Figure 4A). The center of asters showed the highest signal, reflecting the highest microtubule polymer density. In addition, we noticed an annular region at the aster periphery with ∼4% lower value of fluorescence signal relative to the background cytosol levels (Figure 4B). We interpret this as evidence for depletion of soluble tubulin by aster growth. Considering the potential contribution of background signal, the 4% decrease of tubulin intensity is a lower bound for the degree of depletion. The intensity profiles of EB1-GFP (Figure 4B) and Tau-mCherry (Supplemental Figure 3) showed a similar spatial pattern, suggesting that MAPs may also be depleted to some degree. Finally, we compared the relative position of tubulin/EB1 intensity profiles to that of the EB1 comet density profile (Figure 4B). This suggested that even the most peripheral plus ends exist in a cytosolic environment that is 4% (or more) depleted of tubulin and EB1 compared with unconsumed cytosol. The half-width of the depleted zone extended ∼50 µm beyond the growing aster periphery, which is smaller than the typical aster radius. This analysis indicated that soluble protein levels may vary across subregions of growing asters due to subunit consumption.
FIGURE 4:
Fluorescent intensity profiles of tubulin and EB1 indicate spatial variation of these species within growing interphase asters. (A) Wide-field images of a growing aster visualized with Alexa 647–labeled tubulin and EB1-GFP. For both images, the contrast is adjusted to emphasize the zone of low fluorescence intensity. (B) Corresponding quantification of the fluorescence intensity profiles averaged over the quadrant and normalized to the intensity outside the aster (top) and EB1 comet density profile (bottom). We define the aster as the region that has EB1 density higher than the background level + SD. Scale bar, 100 µm.
DISCUSSION
Systematic measurement of dynamic instability parameters over space
The initial motivation of this study was to test our previous assumption that microtubule dynamics was spatially homogenous during aster growth (Ishihara et al., 2016). EB1 tracking experiments showed that the catastrophe rate was largely constant over hundreds of microns (Figure 1), and the polymerization rate was ∼10% higher in the periphery than the interior. Intensity difference imaging showed that the depolymerization rate varied from modal values of ∼20 µm/min at the aster periphery to ∼40 µm/min in the interior (Figure 2). Average values were closer, 29 versus 36 µm/min. Following laser ablation, plus ends depolymerized at 22–48 µm/min (Figure 3). This may be the first report of systematic spatial variation of depolymerization rate. We also found that individual depolymerization rates were more variable within individual asters than polymerization rates, and also that average depolymerization rates tended to vary more between extract batches.
The use of multiple methods to measure microtubule dynamics highlighted their relative strengths as well as some discrepancies. Notably, we detected a 10% increase in polymerization rate at the periphery with the EB1 tracking method but not the TIRF intensity difference method despite larger measurement variation (SD) in the former. This may be because the EB1 tracking method (i) reports a large number of observations across a large 600 × 600 µm field of view providing statistical power and (ii) allows unequivocal determination of plus-end locations with respect to the center of the aster. In contrast, the TIRF method is limited to a small 80 × 80 µm field of view, which must be fixed in one position for 2 min of the time-lapse acquisition before moving to another position. During this time, the peripheral region is moving outward due to aster growth. Thus, our definition of the periphery in the TIRF method is less precise, possibly explaining the high position-to-position variability in the peripheral depolymerization rate (Supplemental Figure 1) and the failure to detect the difference in polymerization rate in the interior versus periphery. Despite these limitations in the TIRF intensity difference method, we found evidence of spatial regulation for depolymerization rate, suggesting that the effect size on depolymerization is much larger than for polymerization. We anticipate future studies to develop novel probes and methods that provide better measurements of depolymerization, which is clearly understudied in the field.
Limiting component hypothesis for the spatial regulation of depolymerization rate
Multiple hypotheses could be considered to account for the observed spatial regulation of depolymerization rates, including chemical activity gradients or timer mechanisms coupled to the nucleotide state in the microtubule lattice (Bollinger et al., 2020; Roostalu et al., 2020). We will focus on component depletion models because this is the simplest hypothesis; the growth of structures in confined cytoplasm necessarily consumes the available building blocks and limits further growth (see Introduction). Consistent with such a model, we found evidence for depletion of soluble tubulin and EB1 inside asters by imaging (Figure 4), and a Péclet number calculation showed that the cytosol inside and outside growing asters does not mix by diffusion once the distance between them is larger than ∼50 µm. We considered three candidate limiting factors: tubulin, MAPs, and GTP. GTP is our least favored candidate, though we cannot rule it out. It diffuses faster than proteins and the concentration and turnover rate of tubulin are probably insufficient to significantly perturb the GTP/GDP ratio in the aster interior relative to the periphery.
Tubulin is the most obvious limiting component in a microtubule system, and is assumed to be limiting in previous models. As tubulin concentration decreases, the literature predicts that plus ends should grow more slowly and catastrophe more often (see Introduction). However, we observed little spatial variation in these parameters, though EB1-measured growth rates trended in the expected direction (Figure 1). One reason for a lack of effect of depletion on polymerization rate may be that the steady-state polymer fraction in our system is relatively low, ∼30% (Materials and Methods, Tubulin polymer fraction estimate), which is the upper bound for difference in soluble tubulin concentration inside and outside asters. Given this small difference and lack of literature on how soluble tubulin levels regulate depolymerization, we explore alternative candidates for the limiting component that is responsible for the spatial regulation of depolymerization rates.
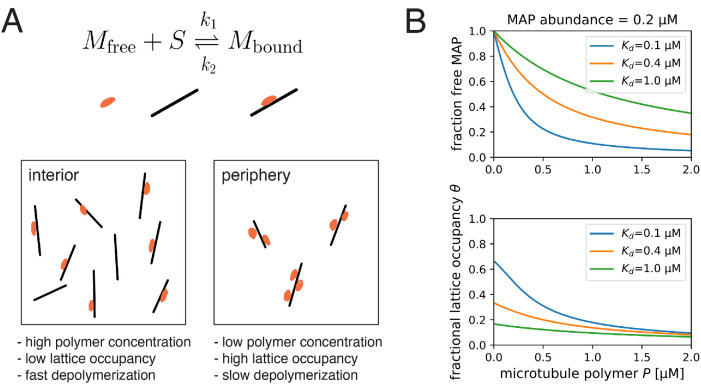
One or more MAPs that regulate depolymerization are our preferred candidate for the limiting factor. Several MAPs have been shown to slow down depolymerization rates (Drechsel et al., 1992; Andersen and Karsenti, 1997). We propose that one or more MAPs that are present at low concentrations relative to microtubule polymer binds to microtubules at lower density in the interior compared with the periphery, resulting in faster depolymerization in the interior (Figure 5A). The key parameters that determine whether a given MAP can cause spatial regulation in this way are concentration in cytosol and affinity to the microtubule lattice, which together determine whether the MAP is significantly depleted by microtubule binding. We present a toy model to illustrate how these parameters may help us predict which MAPs regulate depolymerization (see Materials and Methods, A model of single MAP species binding to microtubule lattice). The model predicts the fraction of freely diffusing MAP molecules 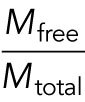 and the site occupancy of the microtubule lattice
and the site occupancy of the microtubule lattice 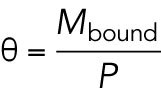 , both of which decrease with microtubule polymer concentration P as expected (see Figure 5B). For a hypothetical MAP whose abundance is 0.2 µM and associates with microtubules with a relatively tight dissociation constant of 100 nM, the fractional occupancy of the microtubule lattice decreases from 0.67 to 0.12 when the polymer concentration increases from 0 to 1.6 µM. We argue that such 5.6-fold difference in microtubule lattice occupancy may be sufficient to cause differences in depolymerization rate that account for spatial regulation. Soluble tubulin, in contrast, only decreases by 0.7-fold. In Table 1, we summarize how different MAPs present in the frog egg are predicted to change their lattice occupancies. Based on the predicted fold change lattice occupancies, we favor EML4, MAP7, and its homologues MAP7D1, MAP7D2, and MAP1B as candidate regulators of depolymerization that are subject to component limitation effects in large interphase asters, leading to faster depolymerization in the interior. Given this long list of candidate factors, it was not practical to deplete them in this study.
, both of which decrease with microtubule polymer concentration P as expected (see Figure 5B). For a hypothetical MAP whose abundance is 0.2 µM and associates with microtubules with a relatively tight dissociation constant of 100 nM, the fractional occupancy of the microtubule lattice decreases from 0.67 to 0.12 when the polymer concentration increases from 0 to 1.6 µM. We argue that such 5.6-fold difference in microtubule lattice occupancy may be sufficient to cause differences in depolymerization rate that account for spatial regulation. Soluble tubulin, in contrast, only decreases by 0.7-fold. In Table 1, we summarize how different MAPs present in the frog egg are predicted to change their lattice occupancies. Based on the predicted fold change lattice occupancies, we favor EML4, MAP7, and its homologues MAP7D1, MAP7D2, and MAP1B as candidate regulators of depolymerization that are subject to component limitation effects in large interphase asters, leading to faster depolymerization in the interior. Given this long list of candidate factors, it was not practical to deplete them in this study.
FIGURE 5:
Proposed model for the regulation of depolymerization rate by a MAP species as the limiting component. (A) We consider the equilibrium of a single MAP species that associates with the microtubule lattice with dissociation constant 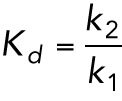 . We hypothesize that the degree at which a microtubule is bound by this MAP species slows down the depolymerization rate. This effect is greater in the aster periphery, where the microtubule density is lower. (B) Fraction of free MAP and occupied microtubule lattice as a function of microtubule concentration for a hypothetical MAP present at 0.2 µM.
. We hypothesize that the degree at which a microtubule is bound by this MAP species slows down the depolymerization rate. This effect is greater in the aster periphery, where the microtubule density is lower. (B) Fraction of free MAP and occupied microtubule lattice as a function of microtubule concentration for a hypothetical MAP present at 0.2 µM.
TABLE 1:
Summary of microtubule-associated proteins (MAPs) that may slow down plus-end depolymerization rate.
| Gene symbol | Protein description | Abundance (nM) | Kd (nM) | Reference | θ at low P | θ at P = 1.6 µM | θ fold change |
|---|---|---|---|---|---|---|---|
| MAP4 | Microtubule-associated protein, XMAP230 | 964.5 | 300 | Tokuraku et al., 2003; Andersen et al., 1994 | 0.76 | 0.45 | 1.69 |
| EML2 | Echinoderm microtubule-associated protein-like 2 | 578.18 | ? | ||||
| EML4 | EML4 protein | 504.2 | 180 | Eichenmuller et al., 2001 | 0.74 | 0.27 | 2.74 |
| EML1 | Echinoderm microtubule-associated protein-like 1 | 135.67 | ? | ||||
| MAP7 | Ensconsin | 91.3 | 460 | Monroy et al., 2020 | 0.17 | 0.04 | 4.25 |
| MAP1B | Microtubule-associated protein 1B, XMAP310 | 84.68 | 200 | Andersen and Karsenti, 1997 | 0.3 | 0.05 | 6.0 |
| MAP1S | MAP1S light chain | 66.04 | ? | ||||
| MAP7D1 | MAP7 domain-containing protein 1 | 46.97 | ? | ||||
| MAP7D2 | MAP7 domain-containing 2 protein variant 2 (fragment) | 8.55 | ? |
Protein abundance in the frog egg is from proteome data in Wühr et al. (2014). For those with reported dissociation constants (Andersen et al., 1994; Andersen and Karsenti, 1997; Eichenmüller et al., 2001; Tokuraku et al., 2003; Monroy et al. 2020), we provide the estimated value of θ, the fractional occupancy of the microtubule lattice at low (i.e., excess MAP) and high polymer concentrations (P = 1.6 µM). See Supplemental Table 1 for an extended list of MAPs.
Our proposal that a spatially varying MAP:microtubule ratio regulates depolymerization comes with several limitations. First, while a single, low-abundance, tight-binding MAP was considered in our model, multiple MAPs may regulate the depolymerization rate in eggs. The total abundance of all MAP species (see Supplemental Table 1 for an expanded list of MAPs in the frog egg) easily exceeds 2 µM. Thus, in a more realistic scenario, multiple MAP species may compete for microtubule binding and their order of association is governed by their abundances and affinities. Kinases such as Cdk1, MARK, and Nek may regulate the affinity of these MAPs (Drewes et al., 1992; Illenberger et al., 1996; Drewes et al., 1997; Ookata et al., 1997; Chang et al., 2001; Adib et al., 2018). Our current model is a useful starting point for future studies that consider complex multispecies dynamics of cytoskeletal self-organization including the mitotic spindle (Good et al., 2013; Hazel et al., 2013; Rieckhoff et al., 2019, 2020). Second, while most MAPs that slow down depolymerization have been reported to accelerate polymerization in minimal conditions (Drechsel et al., 1992; Andersen et al., 1994; Andersen and Karsenti, 1997), we observed little regulation of polymerization rate in our aster reactions. We speculate that, in a cellular context where the synergistic effects of XMAP215 and EB1 play a prominent role in modulating plus-end dynamics (Kinoshita et al., 2001; Zanic et al., 2013), our hypothetical MAP may not further accelerate polymerization. It will be interesting to carefully revisit depolymerization rates in other cellular and reconstitution systems.
Biological implications
Our findings have both general and egg-specific implications. In all cells with dynamic microtubules, a negative feedback from growth to further assembly must exist to prevent all the tubulin from polymerizing and leave enough in the soluble form to support rapid polymerization of growing ends at steady state. Since the original formulation of the dynamic instability model, this feedback was assumed to occur by component limitation of polymerization and catastrophe rates, where the limiting component was tubulin itself or a polymerization-promoting MAP (see citations in Introduction). Component limitation of depolymerization rates was not considered, mainly because this reaction is insensitive to tubulin concentration. Here, we propose that depletion of a limiting MAP can provide the necessary negative feedback by increasing depolymerization rate. This finding could be frog egg–specific or much more general. We do not expect spatial regulation of depolymerization rate in small cells because the soluble components of cytoplasm are well mixed by diffusion. One way to test how component limitation regulates microtubule dynamics in small cells might be to carefully measure changes in dynamics as cells recover from a cold shock or nocodazole washout.
In large eggs cells, our findings have implications for the morphology and mechanics of the asters that position centrosomes and cleavage furrows during cleavage divisions. Previously, we modeled egg aster growth as a combination of polymerization dynamics and autocatalytic nucleation at the periphery (Ishihara et al., 2016). We assumed spatially unform dynamics for simplicity. To prevent the uncontrolled exponential growth expected from an autocatalytic process, we had to assume negative feedback from growth to further assembly, which we implemented as a logistic function that made no mechanistic assumptions. Our current findings suggest that this feedback occurs mainly by increased depolymerization rate in the aster interior, and that it leads to a steady state with a relatively low polymer fraction, estimated as ∼30% tubulin polymerized. Higher microtubule stability at the periphery might account for the higher density of microtubules often observed at the periphery of growing asters in fixed images (Mitchison et al., 2012). Low polymer density may be important for how centrosomes move in response to force from dynein distributed through the cytoplasm (Wühr et al., 2010). Egg asters are gels comprising a network of relatively short microtubules entangled with F-actin and organelles (Pelletier et al., 2020). Microtubules are the stiffest component of this composite gel, so the bulk stiffness of the aster is likely to depend strongly on the tubulin polymer fraction. We speculate that a low polymer fraction allows the aster gel to deform in response to forces from dynein and the ingressing furrow while maintaining sufficient connectivity to transmit forces across large length scales.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Aster reconstitution in Xenopus egg extract
Interphase microtubule asters were reconstituted in Xenopus egg extract with anti-AurkA–coated beads as microtubule organizing centers (Field et al., 2014, 2017). All reactions were supplemented with a 0.04 mg/ml p150-CC1 fragment of dynactin to inhibit dynein motor activity.
Measurement of polymerization and catastrophe rates from EB1 comets
Wide-field images were acquired on a Nikon Eclipse Ni-E upright microscope equipped with a CFI Plan Apochromat Lambda 20× 0.75 NA objective lens (Nikon), Nikon motorized XY stage, and Hamamatsu ORCA-Flash4.0 LT scientific CMOS camera, driven by NIS-Elements. EB1-GFP was supplemented to the reactions at a final concentration of 200–400 nM. EB1 comets were imaged at 2 s intervals, and analyzed with TrackMate (Tinevez et al., 2017). For identifying spots, the LoG detector was used with 2.0 µm spot diameter, threshold 10.0, no median filter, subpixel localization enabled. For linking tracks, the LAP tracker was applied with a max search radius of 3 µm without gap closing. The results were further analyzed by a custom script written in Python to calculate the polymerization velocity (average of the frame-to-frame velocity for each track) and the catastrophe rate (fitting an exponential function to the comet duration distribution). The DABEST-Python package was used for statistics and effect size estimation (Ho et al., 2019).
Measurement of polymerization and depolymerization rates of microtubule ends
Physiological microtubule polymerization and depolymerization rates were measured by TIRF microscopy and tubulin intensity difference analysis. Briefly, asters labeled with Alexa 647–labeled bovine tubulin were assembled under K-casein–coated coverslips and imaged with TIRF microscopy (Ishihara et al., 2014). The Nikon Ti-E motorized inverted microscope was equipped with a Nikon motorized TIRF illuminator, Perfect Focus, a Prior Proscan II motorized stage, Agilent MLC400B laser launch (488 nm, 561 nm, 647 nm), and an Andor DU-897 EM-CCD camera. Alexa 647–labeled bovine tubulin was imaged with a 100× CFI Apo 1.49 NA TIRF objective lens (Nikon) with or without 1.5× Optovar. Stream acquisition of images at 500-ms intervals was performed using the RAM capture mode in NIS-Elements software (Nikon Instruments). We defined the aster interior as fields of view (FOVs) with the artificial MTOC in the corner (= center of the FOV less than 100 µm from the center of the aster), while peripheral regions were chosen as the leading edge of the growing aster with very few microtubules in the FOV (= center of the FOV at least 200 µm from the center of the aster).
To obtain spatial information regarding polymerization and depolymerization rates, we applied a recently developed workflow used to quantify treadmilling dynamics in bacterial cytoskeletal filaments (Caldas et al., 2019, 2020). This method allows one to track hundreds of growing and shrinking microtubule ends in an automated manner, overcoming the limitations of a standard manual kymograph analysis. We first constructed differential time-lapse movies by subtracting the intensity differences between each of the two consecutive frames. This image-processing step removes static objects from the movie while regions containing intensity fluctuations give rise to either a positive or negative signal (speckles), which corresponds to growth (polymerization) or shrinkage (depolymerization) rates, respectively. Next, the particle-tracking software for ImageJ, TrackMate (Tinevez et al., 2017), was used to automatically detect and follow the resultant moving speckles to rebuild trajectories for analysis. Finally, a custom Python script is used to quantify densities and rates of all the detect spots along their trajectories. For TrackMate, we used the LoG (Laplacian Gaussian) detector with an estimated diameter of 1 µm to detect the moving spots. To discard potential false positives, we considered only particles with a signal-to-noise ratio lower than 0.8 and a track displacement distance larger than 1 µm. To build the final trajectories, we used the “Simple LAP tracker” with a “Max linking distance” of 1 µm, a “Maximal gap-closing distance” of 1 µm, and a “Max frame gap” of five frames. Later, we only considered for analysis trajectories longer than 5 s. To construct the distributions in Figure 2B, we pooled the frame-to-frame velocities for each track without averaging. The DABEST-Python package was used for statistics and effect size estimation (Ho et al., 2019).
Measurement of laser cutting–induced depolymerization rates
During the cutting experiments, interphase asters were labeled with Atto565 porcine tubulin and imaged using a Nikon microscope (Ti Eclipse) with a Yokogawa CSU-X1 confocal spinning disk, an EMCCD camera (Andor iXon DU-888), a 60× 1.2 NA water immersion objective, and the software AndorIQ for image acquisition. Laser cutting and image analysis were done as described in Decker et al. (2018) with the only difference that the laser pulse repetition rate was reduced by a pulse picker (APE pulseSelect) from 80 MHz to 20 kHz for some of the cuts, which reduced the probability of destroying the asters. In brief, the sample stage was moved according to the desired cut shape in multiple z planes to reach a cut depth of ∼2 µm around the focal plane. The resulting inward traveling microtubule depolymerization wave was recorded at two to five frames/s. The measurement of the polymerization speed involved calculating differential intensities from the raw images with a time difference of 2–3 s (see Figure 3 and Supplemental Movies 4 and 5). These differential intensities were integrated with respect to the radius leading to a Gaussian-shaped integrated intensity profile plotted over the radius for each time point. Using the peak position of fitted Gaussians to these profiles, we determined the distance that the depolymerization wave traveled over time. The slope of this traveled distance over time plot equals the depolymerization speed of the cut microtubules, which was constant over the entire time that the wave was visible (in agreement with depolymerization in metaphase spindles and monopoles; Decker et al., 2018).
Movie S5.
Intensity difference movie corresponding to the laser ablation in Movie 4.
Fluorescence intensity–based estimation of local depletion of tubulin and MAPs
Wide-field images were acquired on a Nikon Eclipse Ni-E upright microscope equipped with a CFI Plan Apochromat Lambda 20× 0.75 NA objective lens (Nikon), Nikon motorized XY stage, and Andor Zyla 4.2 Plus scientific CMOS camera, driven by NIS-Elements. Alexa 647–labeled bovine tubulin, EB1-GFP (110 nM final), Tau-mCherry (20 nM final; Mooney et al., 2017), and/or Alexa 568–labeled 10 kDa dextran were supplemented to the reactions. Dark current subtraction and flat field correction were applied to both channels. A custom script written in MATLAB was used to quantify the fluorescent intensity as a function of radial distance from the MTOC (Pelletier et al., 2020). Alexa 568–labeled dextran showed a flat intensity profile with respect to the aster as expected for a molecule that shows no spatial variation (Supplemental Figure 4). Spot detection of EB1 comets was performed with TrackMate as described above.
Tubulin polymer fraction estimate
To estimate the fraction of tubulin that is in polymer state, we performed a simple calculation based on our measurements. Specifically, we calculated the total concentration of microtubule polymer in the interior of our aster reactions:
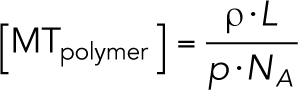 |
where
ρ: number of microtubules per volume [#/µm3]
L: average length of a microtubule—16 µm (Ishihara et al., 2016)
p: pitch of tubulin dimers—8 nm/13 heterodimers
NA: Avogadro’s number— 6*1023 heterodimers/mol
By analyzing a z-stack of EB1-GFP images, we measured the thickness of a reaction prepared by squashing 4 µl of extract under an 18 × 18 mm coverslip as ∼10 µm. Further, we found that a single focal plane of a 20 × 0.75 NA lens captures as much as 30–40% of all EB1 comets that are axially distributed in such reaction. Thus, we estimate the microtubule density from 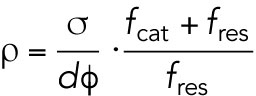 .
.
σ: number of microtubules per area in a single focal plane—0.08 per µm2
d: thickness of the coverslip reaction—10 µm
φ: fraction of EB1 comet detected in a single focal plane—0.35
fcat: catastrophe rate—3.3 [1/min] (Ishihara et al., 2016)
fres: rescue rate—2.0 [1/min] (Ishihara et al., 2016)
The ratio of catastrophe and rescue rates allows us to account for the shrinking plus ends, which are not detected by EB1 comets. We estimate that asters contain [polymer] = 2.65 µM of polymerized tubulin. Comparing this value to the abundance of tubulin heterodimers in the frog egg, 8.6 µM (Wühr et al., 2015), we estimate that ∼31% of tubulin is in the polymer form.
A model of single MAP species binding to microtubule lattice
We consider a single MAP species M (total concentration Mtotal) that binds and unbinds from the microtubule lattice (total concentration P). Let S denote the concentration of free lattice sites and k1 and k2 denote the binding and unbinding rates, respectively.
Conservation of MAP species: Mtotal = Mfree + Mbound
Conservation of microtubule lattice sites: P = S + Mbound
The equation for mass action kinetics is:
 |
Solving for the steady-state solution 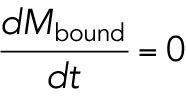 , we find
, we find
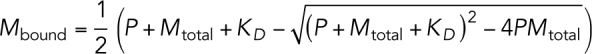 |
This allows us to calculate 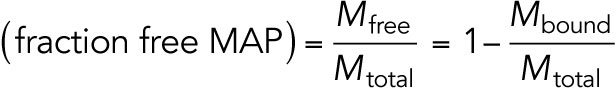
and  .
.
Animal experimentation
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All animals at Harvard Medical School were handled according to approved institutional animal care and use committee (IACUC) protocols (IS00000519) of Harvard Medical School. The protocol was approved by the Harvard Medical Area Standing Committee on Animals (SIRIUS/Procurement no. 2762). All animals at MPI-CBG were handled according to the directive 2010/63/EU on the protection of animals used for scientific purposes, and the German animal welfare law under the license document no. DD24-5131/367/9 from the Landesdirektion Sachsen (Dresden)–Section 24D.
Supplementary Material
Acknowledgments
The authors thank the members of Mitchison, Brugués, and Jay Gatlin groups (University of Wyoming) for discussions. We thank Heino Andreas (MPI-CBG) for frog maintenance. We thank Nikon for microscopy support at Marine Biological Laboratory (MBL). K.I. was supported by fellowships from the Honjo International Scholarship Foundation and Center of Systems Biology Dresden. F.D. was supported by the DIGGS-BB fellowship provided by the German Research Foundation (DFG). P.C. is supported by a Boehringer Ingelheim Fonds PhD fellowship. J.F.P. was supported by a fellowship from the Fannie and John Hertz Foundation. M.L.’s research is supported by European Research Council (ERC) Grant no. ERC-2015-StG-679239. J.B.’s research is supported by the Human Frontiers Science Program (CDA00074/2014). T.J.M.’s research is supported by National Institutes of Health Grant no. R35GM131753.
Abbreviations used:
- GTP
Guanosine triphosphate
- MAP
microtubule associated protein
- MTOC
microtubule organizing center
- TIRF
total internal reflection fluorescence.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-11-0723) on January 13, 2021.
REFERENCES
- Adib R, Montgomery JM, Atherton J, O’Regan L, Richards MW, Straatman KR, Roth D, Straube A, Bayliss R, Moores CA, et al. (2019). Mitotic phosphorylation by Nek6 and Nek7 reduces microtubule affinity of EML4 to alter spindle dynamics and promote chromosome congression. Sci Signaling 12, eaaw2939. [DOI] [PubMed] [Google Scholar]
- Andersen SS, Buendia B, Domínguez JE, Sawyer A, Karsenti E (1994). Effect on microtubule dynamics of XMAP230, a microtubule-associated protein present in Xenopuslaevis eggs and dividing cells. J Cell Biol 127, 1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SS, Karsenti E (1997). XMAP310: a Xenopus rescue-promoting factor localized to the mitotic spindle. J Cell Biol 139, 975–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger JA, Imam ZI, Stevens MJ, Bachand GD (2020). Tubulin islands containing slowly hydrolyzable GTP analogs regulate the mechanism and kinetics of microtubule depolymerization. Sci Rep 10, 13661–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard GJ, Rice LM (2018). Microtubule dynamics: an interplay of biochemistry and mechanics. Nat Rev Mol Cell Biol 19, 451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugués J, Nuzzo V, Mazur E, Needleman DJ (2012). Nucleation and transport organize microtubules in metaphase spindles. Cell 149, 554–564. [DOI] [PubMed] [Google Scholar]
- Caldas P, López-Pelegrín M, Pearce DJG, Budanur NB, Brugués J, Loose M (2019). Cooperative ordering of treadmilling filaments in cytoskeletal networks of FtsZ and its crosslinker ZapA. Nat Commun 10, 5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldas P, Radler P, Sommer C, Loose M (2020). Computational analysis of filament polymerization dynamics in cytoskeletal networks. Methods Cell Biol 158, 145–161. [DOI] [PubMed] [Google Scholar]
- Chang W, Gruber D, Chari S, Kitazawa H, Hamazumi Y, Hisanaga S, Bulinski JC (2001). Phosphorylation of MAP4 affects microtubule properties and cell cycle progression. J Cell Sci 114, 2879–2887. [DOI] [PubMed] [Google Scholar]
- Decker F, Brugués J (2015). Dissecting microtubule structures by laser ablation. Methods Cell Biol 125, 61–75. [DOI] [PubMed] [Google Scholar]
- Decker F, Oriola D, Dalton B, Brugués J (2018). Autocatalytic microtubule nucleation determines the size and mass of Xenopus laevis egg extract spindles. Elife 7 e31149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ (1997). Microtubule polymerization dynamics. Annu Rev Cell Dev Biol 13, 83–117. [DOI] [PubMed] [Google Scholar]
- Dogterom M, Leibler S (1993). Physical aspects of the growth and regulation of microtubule structures. Phys Rev Lett 70, 1347–1350. [DOI] [PubMed] [Google Scholar]
- Drechsel DN, Hyman AA, Cobb MH, Kirschner MW (1992). Modulation of the dynamic instability of tubulin assembly by the microtubule-associated protein tau. Mol Biol Cell 3, 1141–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E (1997). MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell 89, 297–308. [DOI] [PubMed] [Google Scholar]
- Drewes G, Lichtenberg-Kraag B, Döring F, Mandelkow EM, Biernat J, Goris J, Dorée M, Mandelkow E (1992). Mitogen activated protein (MAP) kinase transforms tau protein into an Alzheimer-like state. EMBO J 11, 2131–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenmüller B, Ahrens DP, Li Q, Suprenant KA (2001). Saturable binding of the echinoderm microtubule-associated protein (EMAP) on microtubules, but not filamentous actin or vimentin filaments. Cell Motil Cytoskeleton 50, 161–172. [DOI] [PubMed] [Google Scholar]
- Field CM, Groen AC, Nguyen PA, Mitchison TJ (2015). Spindle-to-cortex communication in cleaving, polyspermic Xenopus eggs. Mol Biol Cell 26, 3628–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field CM, Mitchison TJ (2018). Assembly of spindles and asters in Xenopus egg extracts. Cold Spring Harb Protoc 2018, 5984692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field CM, Nguyen PA, Ishihara K, Groen AC, Mitchison TJ (2014). Xenopus egg cytoplasm with intact actin. Methods Enzymol 540, 399–415. [DOI] [PubMed] [Google Scholar]
- Field CM, Pelletier JF, Mitchison TJ (2017). Xenopus extract approaches to studying microtubule organization and signaling in cytokinesis. Methods Cell Biol 137, 395–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, Charlebois BD, Jánosi IM, Howard J, Hunt AJ, Odde DJ (2011). Rapid microtubule self-assembly kinetics. Cell 146, 582–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, Hunt AJ, Goodson HV, Odde DJ (2008). Microtubule assembly dynamics: new insights at the nanoscale. Curr Opin Cell Biol 20, 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisterfer ZM, Zhu DY, Mitchison TJ, Oakey J, Gatlin JC (2020). Microtubule growth rates are sensitive to global and local changes in microtubule plus-end density. Curr Biol 30, 3016–3023.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good MC, Vahey MD, Skandarajah A, Fletcher DA, Heald R (2013). Cytoplasmic volume modulates spindle size during embryogenesis. Science 342, 856–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazel J, Krutkramelis K, Mooney P, Tomschik M, Gerow K, Oakey J, Gatlin JC (2013). Changes in cytoplasmic volume are sufficient to drive spindle scaling. Science 342, 853–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J, Tumkaya T, Aryal S, Choi H, Claridge-Chang A (2019). Moving beyond P values: data analysis with estimation graphics. Nat Methods 16, 565–566. [DOI] [PubMed] [Google Scholar]
- Illenberger S, Drewes G, Trinczek B, Biernat J, Meyer HE, Olmsted JB, Mandelkow EM, Mandelkow E (1996). Phosphorylation of microtubule-associated proteins MAP2 and MAP4 by the protein kinase p110mark. Phosphorylation sites and regulation of microtubule dynamics. J Biol Chem 271, 10834–10843. [DOI] [PubMed] [Google Scholar]
- Inoué S, Sato H (1967). Cell motility by labile association of molecules. The nature of mitotic spindle fibers and their role in chromosome movement. J Gen Physiol 50 (Suppl), 259–292. [PMC free article] [PubMed] [Google Scholar]
- Ishihara K, Korolev KS, Mitchison TJ (2016). Physical basis of large microtubule aster growth. Elife 5, e19145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara K, Nguyen PA, Groen AC, Field CM, Mitchison TJ (2014). Microtubule nucleation remote from centrosomes may explain how asters span large cells. Proc Natl Acad Sci USA 111, 17715–17722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Peshkin L, Mitchison TJ (2012). Vascular disrupting agent drug classes differ in effects on the cytoskeleton. PLoS One 7, e40177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K, Arnal I, Desai A, Drechsel DN, Hyman, AA (2001). Reconstitution of physiological microtubule dynamics using purified components. Science 294, 1340–1343. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Wühr M, Nguyen P, Ishihara K, Groen A, Field CM (2012). Growth, interaction, and positioning of microtubule asters in extremely large vertebrate embryo cells. Cytoskeleton (Hoboken) 69, 738–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Ishihara K, Nguyen P, Wühr M (2015). Size scaling of microtubule assemblies in early Xenopus embryos. Cold Spring Harb Perspect Biol 7, a019182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroy BY, Tan TC, Oclaman JM, Han JS, Simó S, Niwa S, Nowakowski DW, McKenney RJ, Ori-McKenney KM (2020). A combinatorial MAP code dictates polarized microtubule transport. Dev Cell 53, 60–72.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney P, Sulerud T, Pelletier JF, Dilsaver MR, Tomschik M, Geisler C, Gatlin JC (2017). Tau-based fluorescent protein fusions to visualize microtubules. Cytoskeleton (Hoboken) 74, 221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman DJ, Groen A, Ohi R, Maresca T, Mirny L, Mitchison T (2010). Fast microtubule dynamics in meiotic spindles measured by single molecule imaging: evidence that the spindle environment does not stabilize microtubules. Mol Biol Cell 21, 323–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ookata K, Hisanaga S, Sugita M, Okuyama A, Murofushi H, Kitazawa H, Chari S, Bulinski JC, Kishimoto T (1997). MAP4 is the in vivo substrate for CDC2 kinase in HeLa cells: identification of an M-phase specific and a cell cycle-independent phosphorylation site in MAP4. Biochemistry 36, 15873–15883. [DOI] [PubMed] [Google Scholar]
- Pelletier JF, Field CM, Fürthauer S, Sonnett M, Mitchison TJ (2020). Co-movement of astral microtubules organelles and F-actin suggests aster positioning by surface forces in frog eggs. Elife 7, e60047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckhoff EM, Berndt F, Golfier S, Decker F, Elsner M, Ishihara K, Brugués J (2020). Spindle scaling is governed by cell boundary regulation of microtubule nucleation. Curr Biol 30, P4973–P4983.eE10. [DOI] [PubMed] [Google Scholar]
- Rieckhoff EM, Ishihara K, Brugués J (2019). How to tune spindle size relative to cell size? Curr Opin Cell Biol 60, 139–144. [DOI] [PubMed] [Google Scholar]
- Roostalu J, Thomas C, Cade NI, Kunzelmann S, Taylor IA, Surrey T (2020). The speed of GTP hydrolysis determines GTP cap size and controls microtubule stability. Elife 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon ED, Saxton WM, Leslie RJ, Karow ML, McIntosh JR (1984). Diffusion coefficient of fluorescein-labeled tubulin in the cytoplasm of embryonic cells of a sea urchin: video image analysis of fluorescence redistribution after photobleaching. J Cell Biol 99, 2157–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez JY, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, Eliceiri KW (2017). TrackMate: an open and extensible platform for single-particle tracking. Methods 115, 80–90. [DOI] [PubMed] [Google Scholar]
- Tokuraku K, Matsushima K, Matui T, Nakagawa H, Katsuki M, Majima R, Kotani S (2003). The number of repeat sequences in microtubule-associated protein 4 affects the microtubule surface properties. J Biol Chem 278, 29609–29618. [DOI] [PubMed] [Google Scholar]
- Walker RA, O’Brien ET, Pryer NK, Soboeiro MF, Voter, WA, Erickson HP, Salmon ED (1988). Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J Cell Biol 107, 1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wühr M, Freeman RMJ, Presler M, Horb ME, Peshkin L, Gygi S, Kirschner MW (2014). Deep proteomics of the Xenopuslaevis egg using an mRNA-derived reference database. Curr Biol 24, 1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wühr M, Güttler T, Peshkin L, McAlister GC, Sonnett M, Ishihara K, Groen AC, Presler M, Erickson BK, Mitchison TJ, et al. (2015). The nuclear proteome of a vertebrate. Curr Biol 25, 2663–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wühr M, Tan ES, Parker SK, Detrich HW 3rd, Mitchison TJ (2010). A model for cleavage plane determination in early amphibian and fish embryos. Curr Biol 20, 2040–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanic M, Widlund PO, Hyman AA, Howard J (2013). Synergy between XMAP215 and EB1 increases microtubule growth rates to physiological levels. Nat Cell Biol 15, 688–693. [DOI] [PubMed] [Google Scholar]
- Zhai Y, Borisy GG (1994). Quantitative determination of the proportion of microtubule polymer present during the mitosis-interphase transition. J Cell Sci 107 (Pt 4), 881–890. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.