Abstract
In the spring of 2020, we and others hypothesized that T cells in COVID-19 patients may recognize identical protein fragments shared between the coronaviruses of the common cold and COVID-19 and thereby confer cross-virus immune memory. Here, we look at this issue by screening studies that, since that time, have experimentally addressed COVID-19 associated T cell specificities. Currently, the identical T cell epitope shared between COVID-19 and common cold coronaviruses most convincingly identified as immunogenic is the CD8 + T cell epitope VYIGDPAQL if presented by the MHC class I allele HLA-A*24:02. The HLA-A*24:02 allele is found in the majority of Japanese individuals and several indigenous populations in Asia, Oceania, and the Americas. In combination with histories of common cold infections, HLA-A*24:02 may affect their protection from COVID-19.
Keywords: COVID-19, T cell, MHC, HLA, peptide, epitope, Japanese, VYIGDPAQL, SPRWYFYYL
Introduction
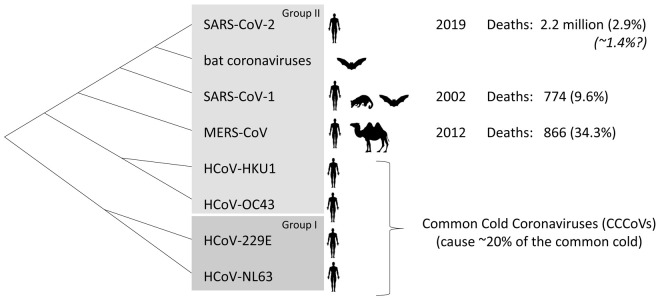
The virus causing the COVID-19 pandemic is severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) ( Wu et al., 2020; Zhou et al., 2020). SARS-CoV-2 is one of the seven coronaviruses that are known to infect humans, the others being SARS-CoV-1 (causing SARS), Middle East respiratory syndrome coronavirus (MERS-CoV), and the common cold coronaviruses (CCCoVs): human coronavirus OC43 (HCoV-OC43), HCoV-HKU1, HCoV-229E, and HCoV-NL63. These coronaviruses belong to the serological/phylogenetic clades designated as group I (alphacoronaviruses) and group II (betacoronaviruses) ( Figure 1). SARS-CoV-1 and MERS-CoV infected relatively few people and therefore should have little effect on global cross-virus immune memory. On the other hand, the CCCoVs cause ~20% of common cold cases, are globally distributed, and all adults have probably been infected with them multiple times in their lives ( Hirsch et al., 2013; Mäkelä et al., 1998; Zhou et al., 2013).
Figure 1. Cladogram of the phylogeny of coronaviruses infecting humans ( Ceraolo & Giorgi, 2020; Forni et al., 2017).
Viruses closely related to SARS-CoV-2 are found in bats. Bats and civets are the probable sources of SARS-CoV-1, and camels are alternative hosts for MERS-CoV. The first reported outbreaks in people by infections with SARS-CoV-1, MERS-CoV, and SARS-CoV-2 occurred in 2002, 2012, and 2019, respectively, with differences in number of deaths and case fatality ratios among registered cases (percentages indicated in regular font between parentheses) ( https://www.who.int/publications/m/item/summary-of-probable-sars-cases-with-onset-of-illness-from-1-november-2002-to-31-july-2003; https://www.who.int/health-topics/middle-east-respiratory-syndrome-coronavirus-mers; https://coronavirus.jhu.edu/map.html). SARS-CoV-2 infections and deaths are not always registered and based on data from New York it was estimated that the true fatality rates may be ~1.4% (Italic font) ( Yang et al., 2021).
The immune defense against viruses includes both innate and adaptive immune responses. Cell types that specialize in adaptive immunity (immune memory) are B cells, CD4 + T cells, and CD8 + T cells. B cells can secrete antibodies, but there is probably little or no protective cross-virus anti-SARS-CoV-2 humoral immunity deriving from infections by CCCoVs (e.g., Amanat et al., 2020; Shrock et al., 2020). CD8 + T cells recognize peptides presented by major histocompatibility complex class I (MHC-I) cells and can kill the presenting cell; the peptides bound by MHC-I are approximately 8-13 amino acids (aa) length, and mostly are 9 aa (“9-mers”) ( Rammensee et al., 1995; Schellens et al., 2015). CD4 + T cells recognize peptides if presented by MHC class II (MHC-II) molecules and help regulate immune responses; the peptides bound to MHC-II typically are 12-25 aa ( Rammensee et al., 1995), although the part binding within the MHC-II groove is only 9 aa as commonly found for MHC-I ( Stern & Wiley, 1994). In the case of MHC-I alleles, available computational software provides a powerful in silico tool in predicting which peptides are presented. For a broader discussion on T cell functions in COVID-19 patients, including whether T cell responses are always beneficial or could also be detrimental, we refer to other studies ( Altmann & Boyton, 2020; Bacher et al., 2020b; Jarjour et al., 2020).
In the spring of 2020, Nguyen et al. (2020) and Dijkstra & Hashimoto (2020) reported on possible MHC-I binding epitopes shared between CCCoVs and SARS-CoV-2 based on in silico analyses and speculated on cross-virus T cell immune memory. Whereas Nguyen et al. (2020) made a comprehensive analysis of possible MHC epitopes of SARS-CoV-2, Dijkstra & Hashimoto (2020) concentrated on identical 9-mers shared between SARS-CoV-2 and at least one of the CCCoVs. Probably for that reason, Dijkstra & Hashimoto (2020) were more accurate in identifying such 9-mers, as they found >230 whereas Nguyen et al. (2020) only listed 144. These identical 9-mers, or even 8-mers, were mostly found in several well-conserved nonstructural proteins that are expressed as part of the ORF1ab polyprotein, while they are absent or nearly absent in the structural proteins S, M, N, and E ( Dijkstra & Hashimoto, 2020; Lee et al., 2020; Nguyen et al., 2020). Since then, it has been shown indeed that a recent history of CCCoV infections reduces the severity of COVID-19 infections ( Sagar et al., 2021), and a considerable number of studies have investigated SARS-CoV-2 T cell epitopes (summarized in this article). The present study screened the recent literature to investigate—in line with the hypothesis of our earlier report ( Dijkstra & Hashimoto, 2020)—which of the identical peptides shared between SARS-CoV-2 and any of the CCCoVs have been confirmed experimentally to bind to MHC molecules and/or to stimulate T cells. Arguably, the only one of such peptides convincingly reported as immunogenic by independent research groups is the helicase-derived peptide VYIGDPAQL which binds MHC-I allele HLA-A*24:02 and then can stimulate CD8 + T cells. In some populations, such as the Japanese, HLA-A*24:02 is found in >50% of the individuals and the allele may affect their resistance against COVID-19.
Methods
As we did before ( Dijkstra & Hashimoto, 2020), proteins encoded by a reported genomic sequence for SARS-CoV-2 (GenBank MN908947; Wu et al., 2020) were compared with those for HCoV-OC43 (NC_005147; Vijgen et al., 2005), HCoV-HKU1 (NC_006577; Woo et al., 2005), HCoV-229E (NC_002645; Thiel et al., 2001), and HCoV-NL63 (NC_005831; van der Hoek et al., 2004) by performing BLAST homology searches at the NCBI database ( https://blast.ncbi.nlm.nih.gov/Blast.cgi) and by making multiple sequence alignments using CLUSTALW software ( https://www.genome.jp/tools-bin/clustalw); continuous stretches of 9 aa acids identical between SARS-CoV-2 and one of the other viruses were identified manually. A complete list of the detected 9-mers, minus a very few that we had missed at that time, are shown in Dijkstra & Hashimoto (2020). Furthermore, from published reports found by Google and PubMed searches, sequences of SARS-CoV-2 peptides reported to activate T cells were compared with the above listed CCCoV proteomes using tblastn (align) function at the NCBI database, in search for identical sequences in the case of 8-mers (which we did not find) or stretches of ≥9 consecutive identical aa in the case of 9-mers or larger peptides. The peptide sequences collected by either method were screened against the Immune Epitope Database (IEDB; http://www.iedb.org/; Dhanda et al., 2019) for reports in the human species context, and, unless these database reports represented the same studies that also appeared in article form, their IEDB information was added to Table 2 (available here). The collected peptide sequences were also analyzed by ANN 4.0 software at IEDB Analysis Resource ( http://tools.immuneepitope.org/mhci/) for prediction of their affinity to a set of representative human MHC-I alleles, which were chosen because of their global abundancy, their relevance for the presented data, or as representatives of MHC-I supertypes ( Lund et al., 2004). Identical 9-mers for which no MHC binding was found or predicted, for which no labeling or activation of T cells was reported, and which were not part of larger immunogenic T cell epitopes, were not included in Table 2.
The HLA allele frequencies as shown in Table 3 are based on data as summarized in the Allele Frequency Net Database http://www.allelefrequencies.net/ (settings: HLA > HLA classical allele freq search) ( Gonzalez-Galarza et al., 2020).
Results and discussion
The first two experimental studies on CCCoV-derived anti-SARS-CoV-2 cross-virus T cell memory
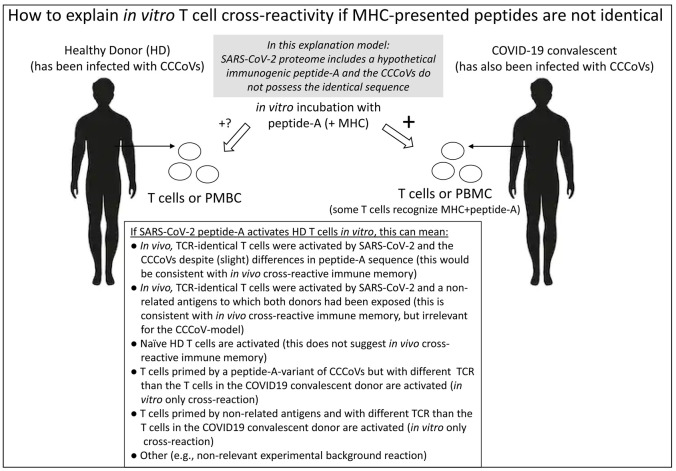
The first two studies that experimentally investigated possible CCCoV-induced T-cell memory against SARS-CoV-2 were Grifoni et al. (2020) and Braun et al. (2020) ( Leslie, 2020). Braun et al. (2020) tested CD4 + T cell activation using S (spike) protein derived peptide pools, whereas Grifoni et al. (2020) investigated both CD4 + T and CD8 + T cell activations using peptide pools derived from various SARS-CoV-2 proteins. Both groups found SARS-CoV-2 peptide pools to activate T cells from healthy donors (HD), proposedly representing memory T cells primed by similar peptides during earlier CCCoV infections. Notably, T cells of HD were also activated if their peripheral blood mononuclear cells (PBMC) were incubated with pools of peptides derived from not very well conserved SARS-CoV-2 proteins such as S ( Braun et al., 2020; Grifoni et al., 2020), although—depending on the virus isolates—SARS-CoV-2 S protein does not share identical 9-mers with any of the CCCoVs but may share two identical 8-mer sequences ( Dijkstra & Hashimoto, 2020). Braun et al. (2020) assumed that even a <50% identity between the corresponding SARS-CoV-2 and CCCoV peptides might explain the assumed cross-virus CD4 + T cell memory. Similarly, the authors of the Grifoni et al. (2020) study—answering our question on how S-derived peptides could activate cross-virus CD4 + and CD8 + T cell memory despite not sharing identical 9-mers or (presumably) immunogenic identical 8-mers—explained that CD8 + T cell activation in their type of in vitro assay could be found for a substantial part of any peptides sharing only ≥70% identity and that for CD4 + T cell activation the requirements for peptide similarity were even lower (see comments section below the Grifoni et al., 2020 article at https://www.cell.com/cell/fulltext/S0092-8674(20)30610-3). Although it has been well established that T cells can be promiscuous in recognizing pMHC (MHC + peptide) complexes, the extents to which this is relevant for in vivo T cell memory and does or does not tend to involve peptides with very similar sequences are being debated (e.g., Grant et al., 2016; Petrova et al., 2012). As explained by Peng et al. (2020), in vitro observations that suggested the existence of CCCoV-primed anti-SARS-CoV-2 “cross-reactive” T cell memory might alternatively be caused by the activation of naive T cells or of T cells primed by not-so-similar peptides of non-related pathogens. Possibilities for explaining in vitro observations suggesting cross-virus T cell memory in the case of non-identical peptides are summarized in Figure 2. Naturally, the proportions of false negative and false positive outcomes depend on the sensitivity of the assay. The best chance for in vitro results truly representing stimulation of the same T cells (TCR-identical T cells) activated during CCCoV and subsequent SARS-CoV-2 infections—hence representing functional memory—is if the MHC-presented peptides are identical. Hence, the present article predominantly focuses on such identical peptides.
Figure 2. Theoretical possibilities for explaining in vitro activation of T cells from healthy donors (HD) by a hypothetical immunogenic SARS-CoV-2 peptide (“peptide-A”) that has no perfect sequence match in the CCCoVs.
Even if the CCCoVs do possess a very similar sequence, the in vitro activation does not need to be indicative for peptide-A being an epitope for in vivo cross-virus T cell memory. In the model, peptide-A is either used directly or as part of pMHC complexes, and the T cells are stimulated after their isolation or as part of PBMC.
Summary, from the perspective of possible cross-virus immune memory, of studies that identified SARS-CoV-2 T cell epitopes
Table 1 summarizes the Grifoni et al. (2020) and Braun et al. (2020) studies, as well as later studies that experimentally investigated potential SARS-CoV-2 T cell epitopes. Section A of the table summarizes studies that only investigated peptide pools and/or intact proteins, and section B summarizes the studies that (also) investigated individual peptides. The table explains whether indications for CD4 + and/or CD8 + T cell activation were observed, whether MHC association was addressed and/or MHC binding tested, and whether indications for SARS-CoV-2 and CCCoV cross-virus T cell memory were investigated and observed. Importantly, the consensus was that anti-SARS-CoV-2 T cell memory in individuals that had only been infected with CCCoVs was weak and only detectable in a subset of individuals ( Sette & Crotty, 2020). It should be noted that none of the studies listed in Table 1 had specifically selected individuals with a known recent history of CCCoV infection, although presumably in such group a stronger T cell response against SARS-CoV-2 antigens would be found ( Sagar et al., 2021).
Table 1. Summary of experimental studies on SARS-CoV-2 proteins/peptides in relation to T cell activation.
|
(A) Studies that only used peptide pools or intact
proteins |
||||
|---|---|---|---|---|
| Indications for | Indications for | |||
| SARS-CoV-2- | cross-virus | MHC | ||
| Reference | specific T cells a | T cell memory b | alleles c | The investigated peptides (and positive findings for
cross-virus shared 9-mer sequences) d, e |
| Anft et al., 2020 | CD4, CD8 | CD4, CD8 | n.d. | peptide pools derived from S |
| Bacher et al., 2020a | CD4 | CD4 | n.d. | peptides pools derived from S, M, N, E, NS6, NS7a, NS7b,
NS8, ORF3a, ORF9B, ORF10, and ORF14 |
| Braun et al., 2020 | CD4 | CD4 | n.d. | peptide pools derived from S |
| Dan et al., 2021 | CD4, CD8 | n.d. | n.d. | peptide pools derived from throughout the SARS-CoV-2
proteome |
| Grifoni et al., 2020 | CD4, CD8 | CD4, CD8 | n.d. | peptide pools derived from throughout the SARS-CoV-2
proteome |
| Meckiff et al., 2020 | CD4 | CD4 | n.d. | peptide pools derived from S and M |
| Ni et al., 2020 | Yes, not
specified |
Yes | n.d. | S, N, and NSP5 proteins |
| Rydyznski Moderbacher et al., 2020 | CD4, CD8 | CD4, CD8 | n.d. | peptide pools derived from throughout the SARS-CoV-2
proteome |
| Steiner et al., 2020 | CD4, CD8 | CD4, CD8 | n.d. | peptide pools derived from S and N |
| Thieme et al., 2020 | CD4, CD8 | CD4, CD8 | n.d. | peptide pools derived from S, M, and N |
| Weiskopf et al., 2020 | CD4, CD8 | CD4, CD8 | n.d. | peptide pools derived from throughout the SARS-CoV-2
proteome |
| (B) Studies that (also) investigated individual peptides | ||||
| Ferretti et al., 2020 | CD8 | CD8 | a, b | peptides derived from throughout the SARS-CoV-2
proteome; activation of CD8+ T cells by VYI and SPR peptides |
| Gangaev et al., 2020 | CD8 | CD8 | a, b | peptides throughout the SARS-CoV-2 proteome but the
preprint does not provide all details |
| Habel et al., 2020 | CD4, CD8 | maybe | a, b | peptides derived from S, M, N, NSP3, NSP4, NSP6, and
NSP12 |
| Kared et al., 2020 | CD8 | No | a, b | peptides derived from throughout the SARS-CoV-2
proteome; activation of CD8+ T cells by VYI and SPR peptides |
| Keller et al., 2020 | CD4, CD8 | Yes, not
specified |
a | peptides derived from S, M, N, and E |
| Le Bert et al., 2020 | CD4, CD8 | CD4, CD8 | a | peptides derived from N, NSP7, and NSP13; activation of
CD4+ T cells by SPR encompassing peptide |
| Mateus et al., 2020 | CD4, CD8 | CD4, CD8 | n.d. | peptides derived from throughout the SARS-CoV-2
proteome; activation of CD4+ T cells by LKS, YLR (+ LRK, RKH), IER (+ ERF, RFV, FVS, VSL), and NVN (+ VNR, NRF, RFN, FNV) encompassing peptides |
| Nelde et al., 2021 | CD4, CD8 | CD4, CD8 | a | peptides derived from throughout the SARS-CoV-2
proteome; activation of PBMC (probably CD8+ T cells) by VYI peptide; activation of CD4+ T cells by peptide that partially overlaps SPR peptide |
| Peng et al., 2020 | CD4, CD8 | No | a, b | peptides derived from S, M, N, E, ORF3a, ORF6, ORF7a,
and ORF8; binding of CD8+ T cells by HLA-B*07:02/ SPR pentamers; activation of CD4+ and CD8+ T cells by peptide encompassing SPR peptide |
| Poluektov et al., 2020 | n.d. | n.d. | b | peptides derived from throughout the SARS-CoV-2 proteome |
| Prachar et al., 2020 | n.d. | n.d. | b | peptides derived from throughout the SARS-CoV-2
proteome; SLA peptide bound to HLA-A*02:01; KYT, AYA, and VYI peptide bound to HLA-A*24:02; HRF peptide bound to HLA-B*40:01; peptide encompassing RFY (+ FYR, YRL, RLA) bound to HLA-DR4 |
| Schulien et al., 2021 | CD8 | CD8 | a, b | peptides derived from throughout the SARS-CoV-2
proteome; activation of CD8+ T cells by SPR peptide; binding of CD8+ T cells by HLA-B*07:02/ SPR tetramers |
| Sekine et al., 2020 | CD4, CD8 | CD4, CD8 | (a?), b | peptides derived from S, M, N, E, ORF3a, and ORF6;
activation of CD8+ T cells by SPR peptide |
| Shomuradova et al., 2020 | CD4, CD8 | CD4, CD8 | a, b | peptides derived from S, M, and N |
| Snyder et al., 2020 | CD8 | n.d. | a | peptides derived from throughout the SARS-CoV-2
proteome; activation of CD8+ T cells by FVD, RIL, AIM, and IVD peptides and by a combined set of SPR peptide plus an SPR-overlapping peptide |
| Takagi & Matsui, 2020 | CD8 (in
HLA-A*02+ mice) |
n.d. | a, b | peptides derived from NSP1-to-10 |
| Tarke et al., 2020 | CD4, CD8 | CD4, CD8 | a, b | peptides derived from throughout the SARS-CoV-2
proteome; activation of CD4 + T cells by peptides that encompass or partially overlap LKS, YPK (+ PKC), RFY (+ FYR, RLA, LAN), FNI (+ NIC, ICQ), IER (+ ERF, RFV, FVS, VSL, SLA), SPR (+ PRW, RWY, WYF, YFY), or RAK (+AKH); activation of CD8 + T cells by QTV encompassing 10-mer, YAI (+AIS) encompassing 10-mer, DLT encompassing 12-mer, and by 10-13 mers that encompassed DYV, YVY, VYL, YLP, LPY, and/or PYP |
| Woldemeskel et al., 2020 | CD4
(presumably) |
CD4
(presumably) |
(a?) | peptides derived from S, M, and N |
(a) In most of the listed studies experimental evidence was obtained for the existence of SARS-CoV-2-specific CD4 + and/or CD8 + T cells in COVID-19 convalescent donors
(b) In many of the listed studies experimental evidence was obtained suggesting that CCCoV infections induced, or could induce, anti-SARS-CoV-2 T cell memory. Naturally, no samples were used of healthy donors without CCCoV infection history, and for this table, as done in the majority of the listed studies, all positive reactions in healthy donors that indicated SARS-CoV-2-specific T cell activation were interpreted as indications for possible cross-virus T cell memory. In the Habel et al. (2020) study, for T cells from healthy donors activations of similar extent were found for SARS-CoV-2 peptides and peptides from other pathogens for which the donors did not have an infection history.
(c) Some of the listed studies determined the association (a) of T cell responses with MHC alleles or found binding (b) of peptides to MHC alleles
(d) This column lists the proteins or peptides that were investigated. In most cases, though not all, there had been a preselection of peptides based on software predictions for MHC binding. In addition, positive findings for identical 9-mers shared between SARS-CoV-2 and at least one of the CCCoVs are summarized, with VYI and SPR peptides highlighted in bold.
(e) The 3-letter names for peptides here only refer to the 9-mers "Not specified" indicates that it was not determined whether reacting cells were CD4+ or CD8+ T cells.
A question mark is added if we are uncertain about what the authors did.
n.d. = not determined
Table 3. Frequency of HLA-A*24:02 in different populations.
| % of individuals
that have the allele |
Allele
frequency |
Sample
size |
|
|---|---|---|---|
| Taiwan Paiwan | 96 | 0.86 | 51 |
| Taiwan Tsou | 98 | 0.78 | 51 |
| Taiwan Rukai | 96 | 0.76 | 50 |
| Papua New Guinea Eastern Highlands Goroka Asaro | 0.74 | 57 | |
| Papua New Guinea Karimui Plateau Pawaia | 0.74 | 80 | |
| Taiwan Puyuma | 88 | 0.64 | 50 |
| Taiwan Ami | 85 | 0.63 | 98 |
| Papua New Guinea Wanigela Keapara | 0.63 | 66 | |
| Taiwan Atayal | 82 | 0.62 | 106 |
| Ecuador Cayapa | 0.61 | 183 | |
| New Caledonia | 0.61 | 65 | |
| Venezuela Perja Mountain Bari | 0.60 | 55 | |
| Taiwan Thao | 90 | 0.60 | 30 |
| Colombia Waunana NA-DHS_20 | 85 | 0.60 | 20 |
| Taiwan Bunun | 84 | 0.58 | 101 |
| USA Alaska Yupik | 0.58 | 252 | |
| Taiwan Saisiat | 86 | 0.57 | 51 |
| Taiwan Tao | 78 | 0.54 | 50 |
| Colombia Embera NA-DHS_19 | 93 | 0.54 | 14 |
| Colombia/Brazil Ticuna Tarapaca NA-DHS_22 | 74 | 0.53 | 19 |
| Papua New Guinea Wosera Abelam | 0.51 | 131 | |
| Colombia/Brazil Ticuna Arara NA-DHS_21 | 67 | 0.50 | 17 |
| Taiwan Siraya | 78 | 0.47 | 51 |
| Colombia North Chimila Amerindians | 0.46 | 47 | |
| Taiwan Taroko | 73 | 0.45 | 55 |
| Colombia Arhuaco NA-DHS_16 | 65 | 0.44 | 17 |
| Colombia Kogi NA-DHS_17 | 67 | 0.43 | 15 |
| Colombia North Wiwa El Encanto | 0.43 | 52 | |
| Colombia Zenu NA-DHS_18 | 75 | 0.42 | 16 |
| New Zealand Maori with Full Ancestry | 65 | 0.38 | 46 |
| Japan Central | 0.38 | 371 | |
| Mexico Chihuahua Tarahumara | 0.38 | 44 | |
| Colombia Inga NA-DHS_11 | 53 | 0.37 | 16 |
| Japan pop 16 | 0.36 | 18604 | |
| Japan pop 3 | 0.36 | 1018 | |
| Costa Rica Guaymi NA-DHS_10 | 72 | 0.36 | 18 |
| USA Arizona Pima | 0.36 | 100 | |
| Chile Easter Island | 0.36 | 21 | |
| USA Arizona Gila River Pima | 0.36 | 3000 | |
| USA NMDP Japanese | 0.35 | 24582 | |
| Costa Rica Amerindians | 57 | 0.35 | 125 |
| USA Hawaii Okinawa | 0.34 | 106 | |
| Costa Rica Cabecar NA-DHS_9 | 53 | 0.34 | 19 |
| USA Arizona Gila River Amerindian | 0.34 | 492 | |
| Taiwan Pazeh | 58 | 0.34 | 55 |
| Japan Okinawa Ryukyuan | 0.34 | 143 | |
| New Zealand Polynesians with Admixed History | 59 | 0.33 | 27 |
| American Samoa | 0.33 | 51 | |
| Japan pop 5 | 0.33 | 117 | |
| Philippines Ivatan | 58 | 0.32 | 50 |
| Papua New Guinea East New Britain Rabaul | 0.32 | 60 | |
| New Zealand Polynesians with Full Ancestry | 57 | 0.31 | 21 |
| USA New Mexico Canoncito Navajo | 0.31 | 42 | |
| Australia Yuendumu Aborigine | 0.30 | 191 | |
| Australia Groote Eylandt Aborigine | 0.29 | 75 | |
| New Zealand Maori with Admixed History | 51 | 0.29 | 105 |
| 9 other populations with HLA-A*24:02 frequencies between 0.24 and 0.29 (not shown) | |||
| Japan Hokkaido Ainu | 0.24 | 50 | |
Data, and also the nomenclature, were retrieved from the Allele Frequency Net Database.
Only populations with HLA-A*24:02 frequencies ≥0.29 and Japanese Ainu are listed.
Table 2 lists the subset of the 9-mers that are identical between SARS-CoV-2 and at least one of the CCCoVs ( Dijkstra & Hashimoto, 2020) and additionally were predicted to bind representative MHC-I alleles or experimentally found to bind MHC-I, be part of MHC-II binding peptides, or to activate T cells. It also lists >9-mers for which evidence of T cell activation was reported. The experimental results summarized within Table 2 were collected from the articles listed in Table 1, from articles on SARS-CoV-1, and from reports uniquely deposited to the Immune Epitope Database and Analysis Resource (IEDB; http://www.iedb.org/; Dhanda et al., 2019). The 9-mer peptides listed in Table 2 are referred to in Table 1 by using the letter code of their N-terminal three amino acids.
The only identical T cell epitope repeatedly found to be immunogenic by independent research groups was peptide “VYI” (VYIGDPAQL) (highlighted in bold in Table 1; details in Table 2). The VYI peptide is shared between SARS-CoV-2, HCoV-HKU1, and HCoV-OC43 viruses and part of a larger identical stretch AKHYVYIGDPAQLPAPR in their helicase protein (aka nonstructural protein 13 or NSP13). Prachar et al. (2020) showed that the VYI peptide bound to HLA-A*24:02, although not very stable. To our knowledge, all studies that investigated the VYI peptide—three in total—found it to stimulate T cells in an HLA-A*24:02 context ( Ferretti et al., 2020; Kared et al., 2020; Nelde et al., 2021). Ferretti et al. (2020) investigated the entire SARS-CoV-2 proteome by a “T-scan” assay measuring activation of CD8 + memory T cells from COVID-19 convalescent donors after incubation with HEK293 cells engineered to express a single HLA allele and one of a set of overlapping 61 aa stretches. By this method, Ferretti et al. (2020) detected only three SARS-CoV-2 stretches with “dominant” epitopes that activated CD8 + T cells from multiple HLA-A*24:02 + COVID-19 convalescent donors; one of these three stretches encompassed the VYI peptide and stimulated two of five investigated samples. Involvement of the VYI peptide was confirmed by activation of the HLA-A*24:02 + T cells upon coculturing with HLA-A*24:02 + target cells pulsed with VYI peptide. If for the T-scan screen 61 aa stretches of CCCoV proteomes were used instead, Ferretti et al. (2020) appear to have found a weak but noticeable response by HLA-A*24:02 + memory CD8 + T cells in the cases of HCoV-HKU1 and HCoV-OC43 (our interpretation of their Figure 5A). Kared et al. (2020) performed a binding assay, testing 94 peptides from across the SARS-CoV-2 proteome—predicted by software to bind HLA-A*24:02— using pHLA-A*24:02 tetramers for labeling of CD8 + T cells from five HLA-A*24:02 + COVID-19 convalescent donors. They found positive reactions for only eight of the 94 peptides. One of these eight peptides was the VIY peptide, which detectably labeled T cells of only one of the five donors. Regarding possible CCCoV-induced anti-SARS-CoV-2 T cell memory, Kared et al. (2020) mentioned “Notably, SARS-CoV-2 specific CD8 + T cells were not detected in any of the healthy donors recruited before the official SARSCoV-2 pandemic;” their number of HD controls, however, was low, and for their HLA-A*24:02-matched experiments seems to have been between one and four. Nelde et al. (2021) tested ten predicted HLA-A*24:02 SARS-CoV-2 epitopes and found the VYI peptide to be one of the three “dominant T cell epitopes” as it elicited activation of CD8 + T cells from seven of ten HLA-A*24:02 + COVID-19 convalescent donors upon incubation with their PBMC. On the other hand, for PBMC of 17 healthy HLA-A*24:02 + donors a stimulation of CD8 + T cells could not be observed. To summarize these three studies, the VYI peptide is among the most immunogenic SARS-CoV-2 T cell epitopes in HLA-A*24:02 + individuals, although there is no evidence yet that this was primed by previous CCCoV infections. Presumably, because of the latter, none of the three studies mentioned that VYI peptide is shared between SARS-CoV-2, HCoV-HKU1, and HCoV-OC43 ( Ferretti et al., 2020; Kared et al., 2020; Nelde et al., 2021). We assume that not finding anti-VYI T cells in HLA-A*24:02 + HD was only a matter of assay sensitivity, because CCCoV-induced T cell memory is expected as the VYI peptide is embedded in a longer identical AKHYVYIGDPAQLPAPR stretch shared between SARS-CoV-2, HCoV-HKU1, and HCoV-OC43; thus, the MHC-I pathway processing of the peptide is expected to be similar in each viral background, and software predicts that the immunoproteasome efficiently generates the VYI peptide from all three viruses ( https://imed.med.ucm.es/Tools/pcps/; Gomez-Perosanz et al., 2020). An additional reason for assuming the involvement of CCCoV-induced T cell memory is that the helicase is not one of the more abundant (structural) viral proteins ( Davidson et al., 2020) whereas nevertheless VYI is consistently found as one of the dominant T cell epitopes. Future experiments selectively investigating HLA-A*24:02 donors with a recent HCoV-HKU1 or HCoV-OC43 infection will probably be more sensitive in detecting anti-VYI CD8 + T cells primed by CCCoV infection.
HLA-A*23:01 belongs to the same “supertype” as HLA-A*24:02 ( Lund et al., 2004) meaning that they tend to bind similar peptides, and in the IEDB database it is reported that HLA-A*23:01 also binds VYI peptide (IEDB Reference:1000425). It has not been described yet whether VYI peptide can stimulate T cells in an HLA-A*23:01 context.
Although not identical throughout the sequence, as an exception, Table 2 also lists the 9-mer SPRWYFYYL (“SPR”) for SARS-CoV-2 and its matching LPRWYFYYL (“LPR”) for HCoV-HKU1 and HCoV-OC43. Several independent studies ( Table 1; SPR indicated in bold) suggest that SPR is highly immunogenic and involved in cross-virus immune memory (summarized in Table 2). The only amino acid difference between the SPR and LPR peptides is at the P1 position, which is not necessarily important for peptide conformation in pMHC-I complexes or for binding T cell receptors (TCRs) (e.g., Sewell, 2012). Thus, it seems plausible that a fraction of the T cells primed by pMHC/LPRWYFYYL may also recognize pMHC/SPRWYFYYL. It was convincingly shown that SPR peptide binds HLA-B*07:02 alleles and in that context can stimulate CD8 + T cells from COVID-19 convalescent donors ( Ferretti et al., 2020; Kared et al., 2020; Peng et al., 2020; Schulien et al., 2021; Snyder et al., 2020). CD8 + T cells from HLA-B*07:02 + HD could also be stimulated by SPR, suggesting cross-virus CD8 + T cell memory induced by the LPR peptide of HCoV-HKU1 or HCoV-OC43 ( Schulien et al., 2021), which is consistent with in vitro results showing that CD8 + memory T cells from HLA-B*07:02 + COVID-19 convalescent donors were activated by HLA-matched cells expressing 61 aa fragments of HCoV-HKU1 or HCoV-OC43 encompassing the LPR peptide ( Ferretti et al., 2020). However, SARS-CoV-2 peptides encompassing or even only partially overlapping the SPR peptide also induced responses—remarkably strong in some cases—of CD4 + T cells from COVID-19 convalescent donors and, to a lesser extent, from HD (also summarized in Table 2; Le Bert et al., 2020; Nelde et al., 2021; Peng et al., 2020; Tarke et al., 2020). The combined results suggest an overlap of highly immunogenic MHC-I and MHC-II epitopes, and—although such overlap is not impossible—some caution for the possibility that SPR peptide might (additionally) cause non-MHC-restricted immune stimulation seems to be warranted. Except for HLA-B*07:02, SPR peptide was also found to bind the MHC-I alleles HLA-B*51:01, -*53:01, and -*54:01 (IEDB database Reference:1000425).
Other than VYI, we did not find any other identical peptides shared between SARS-CoV-2 and CCCoVs for which current publications convincingly indicate a high immunogenicity. However, the 9-mers FVDGVPFVV (“FVD”) and LPYPDPSRI (“LPY”) could be promising, although they were only reported as immunogenic CD8 + T cell epitopes in single publications. Snyder et al. (2020) appear to have identified FVD peptide as an immunodominant SARS-CoV-2 T cell epitope in the HLA-A*02:01 context, although their descriptions of this matter could be more detailed. Tarke et al. (2020) found that CD8 + T cells from a few HLA-B*51:01 + COVID-19 convalescent donors could be stimulated by several peptides that encompassed the LPY 9-mer peptide sequence, namely peptide DYVYLPYPDPSRI (a 13-mer shared between SARS-CoV-2 and HCoV-HKU1), its shorter versions VYLPYPDPSRI (an 11-mer) or YLPYPDPSRI (a 10-mer), or peptide LPYPDPSRIL (a 10-mer); software predicts that the encompassed LPYPDPSRI (a 9-mer) is the best HLA-B*51:01 binder and that of the other lengths only the 10-mers YLPYPDPSRI and LPYPDPSRIL are expected to bind this allele ( Table 2), suggesting that some processing of the longer peptides may explain the combined results. Future analysis of the immunogenicity of the LPY 9-mer peptide would be interesting.
In addition to the above, Mateus et al. (2020) and Tarke et al. (2020) found some other SARS-CoV-2 peptides that encompassed ≥9-mers shared with CCCoVs and stimulated CD4 + T cells ( Table 2), but whether those identical stretches formed the immunogenic epitope has not been determined yet.
Approximately 60% of the Japanese population carries the MHC class I allele HLA-A*24:02
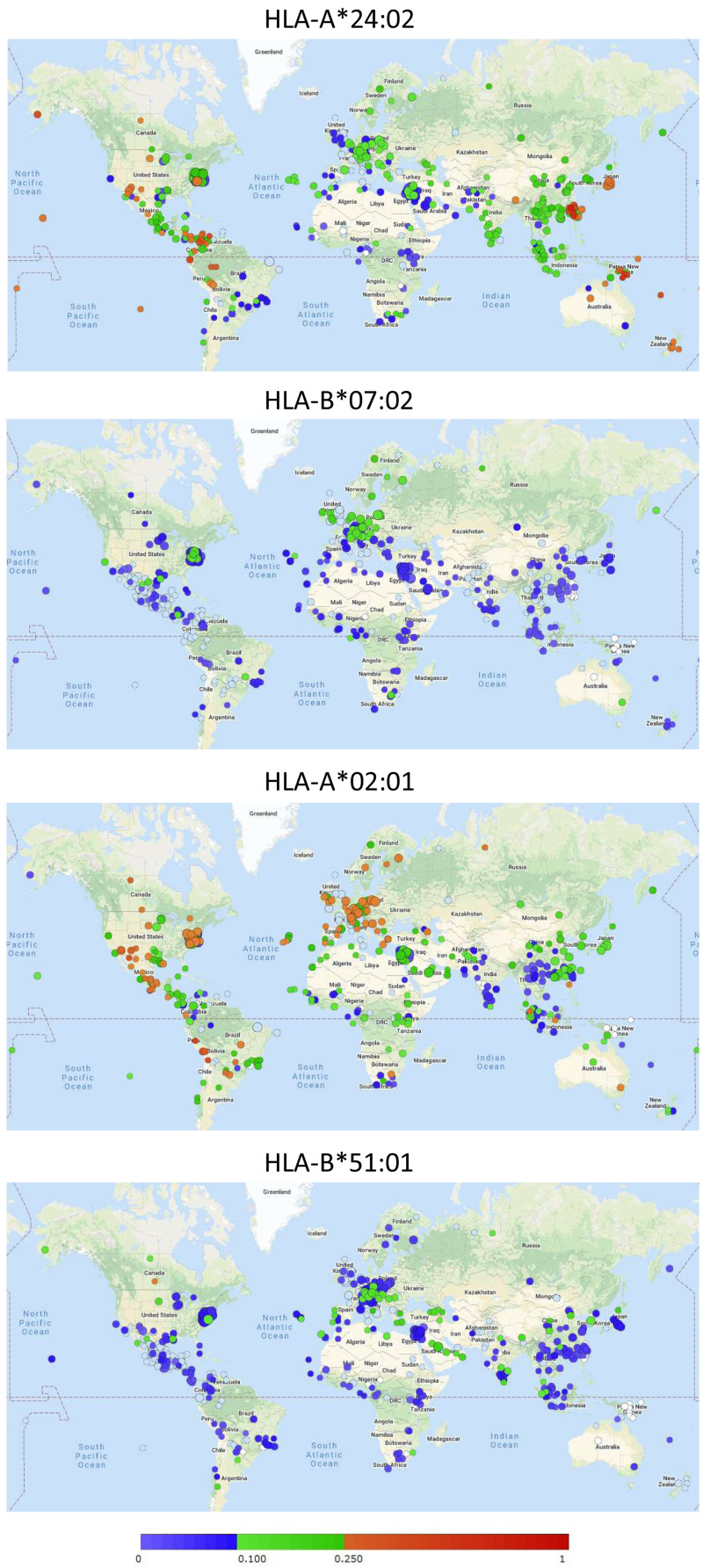
For speculation on the global implications of the ability of different MHC-I alleles to bind conserved peptides, their global distributions must be appreciated. The online Allele Frequency Net Database (AFND; http://allelefrequencies.net/; Gonzalez-Galarza et al., 2020) comprises information of MHC-I allele frequencies in different populations, and Figure 3 includes their visual summaries of global distributions of the alleles HLA-A*24:02 (binder of VYI peptide), HLA-B*07:02 (binder of SPR peptide), HLA-A*02:01 (binder of FVD peptide), and HLA-B*51:01 (predicted binder of LPY peptide). Table 3 lists populations with the highest frequencies of HLA-A*24:02 according to AFND. For all studies summarized in Table 3 the allele frequencies and for a subset also the percentage of individuals carrying the allele had been determined (note that if randomly distributed, an allele frequency of >0.29 would correspond to a prevalence of >50%). Data for populations in Japan and for Japanese in the USA are highlighted in shades of gray ( Table 3). In Japanese populations the HLA-A*24:02 allele frequencies are ≥0.33 except for in the Ainu which are a racial minority indigenous to Hokkaido in Northern Japan. Ikeda et al. (2015; the “Japan pop 16” study in Table 3) investigated 18604 individuals from “all parts of Japan” and the authors concluded that the HLA-A*24:02 allele is distributed in ~60% of the Japanese population. Other populations in which HLA-A*24:02 is present in >50% of the investigated members are various indigenous populations in Asia, Oceania, and the Americas ( Table 3). Those non-Japanese populations are not further discussed in this study as it is harder to collect their data relevant to COVID-19.
Figure 3. Global distribution of HLA-A*24:02, HLA-B*07:02, HLA-A*02:01, and HLA-B*51:01 allele frequencies as visually summarized by the Allele Frequency Net Database ( Gonzalez-Galarza et al., 2020).
Circles refer to individual studies with the color indicating the detected allele frequency following the color bar. See http://www.allelefrequencies.net/ for more detailed information on those studies. Permission to reproduce this image was obtained from AFND.
HLA-B*07:02 is common in Northern Europe ( Figure 3) and is carried by approximately a third of the Irish population (reports at AFND). HLA-A*02:01 is common in Europe, and, for example, found in 49% of the Polish population (report at AFND). HLA-B*51:01 is common in Southern Europe and the Middle East ( Figure 3) and was found at an allele frequency of 0.19 in Saudi Arabia (report at AFND).
COVID-19 and CCCoV infections in Japan
A year into the pandemic, the accumulated number of COVID-19 deaths per million inhabitants in Japan is 45, which is >30 times fewer than in countries such as Italy (1465), the UK (1559), and the USA (1362) (on February 1st, 2021, according to https://www.worldometers.info/coronavirus/). At least for the initial wave of the disease in the first half of 2020 the apparent low prevalence of COVID-19 was supported by finding specific antibodies in only ~0.1% of citizens of Tokyo in June 2020 (governmental report https://www.mhlw.go.jp/content/000648706.pdf) and minimal or even negative excess mortality rates until at least July 2020 ( Yorifuji et al., 2021). These low numbers are quite surprising since the stringency of behavioral regulations to protect against COVID-19 have been less severe in Japan than in most Western countries ( https://www.bsg.ox.ac.uk/research/research-projects/coronavirus-government-response-tracker; Hale et al., 2020). The surprise about the relatively low incidence of COVID-19 in Japan was captured well in the title of a BBC article on July 2020 “Coronavirus: Japan's mysteriously low virus death rate” ( https://www.bbc.com/news/world-asia-53188847). Most of the difference with Western countries can probably be explained by voluntary behaviors such as the willingness of the Japanese to wear masks (despite absence of obligation) and by better individual health status such as a relatively low prevalence of obesity. As a note, however, the Japanese are not per se better protected against respiratory viruses, as mortalities per capita resulting from the 2009 influenza pandemic were higher than in European countries ( Simonsen et al., 2013).
In Japan, as in other countries, CCCoV infections are poorly monitored, but available data indicate that from the winter 2014–2015 until November 2019 (we were not able to find more recent data) the most common CCCoV species in Japan has been HCoV-OC43. Especially during the winters of 2014–2015 and 2018–2019, this virus species appears to have been prevalent ( Komabayashi et al., 2020; Kubota-Koketsu et al., 2020). Thus, many Japanese individuals probably received a relatively recent immune boost with the VYI peptide.
Currently, in the winter 2020–2021, there has been a surge in the number of COVID-19 cases and deaths in Japan ( https://www.worldometers.info/coronavirus/; https://www.aljazeera.com/news/2021/1/4/japan-weighs-state-of-emergency-amid-severe-covid-19-surge), and it is unclear whether the factors that during the first half of 2020 protected the Japanese better than the populations of many other countries are still in place. Possibly, the warm winter season of 2019–2020 (Japan Meteorological Agency reports https://www.data.jma.go.jp/obd/stats/data/en/smp/index.html), and the associated early hay fever season ( https://global.weathernews.com/news/13178/; http://kafun.taiki.go.jp/), may have helped to protect the Japanese population during the first half of 2020. In Japan, largely because of aging monoculture coniferous forests, depending on the definition approximately half of the population may be considered to suffer from pollen-induced allergic rhinitis, which has been called a national affliction (e.g., Minami et al., 2019; Nakamura et al., 2019; Yamada et al., 2014). Although speculative, the associated inflammation of the respiratory tract might non-specifically elevate both innate and specific (possibly anti-VYI T cells) immunity against COVID-19. Although each of these individual biological factors probably is not very protective against COVID-19 (see also the below paragraph), at the population scale a combination of such factors might significantly impact the virus reproduction number (R).
In short, (i) most Japanese individuals possess the MHC-I allele HLA-A*24:02 that presents a highly immunogenic SARS-CoV-2 T cell epitope VYI, (ii) in recent years many of them have been exposed to this epitope by HCoV-OC43 infection, and (iii) at the time of the first COVID-19 wave many of them had an elevated immune status of their respiratory tracts because of pollen allergy.
How might MHC polymorphism affect anti-COVID-19 immunity?
MHC polymorphism is believed to be driven by differences in immune responses conferred by the different alleles, but actual evidence for this to cause differences in disease resistance is close to absent ( Kelly & Trowsdale, 2017; Yamaguchi & Dijkstra, 2019). Presumably, this is caused by each set of MHC alleles having enough choice within a pathogen proteome for presenting some peptides efficiently to the immune system. Theoretically, the MHC allelic effect on differences in disease resistance should become larger if the choice of possible immunogenic epitopes becomes more limited. Hence, the effect of MHC polymorphism on immune memory induced against a related virus should be larger than on immune memory against the same virus. However, several studies have investigated the association of MHC polymorphism with differences in COVID-19 resistance, and—arguably—no convincing associations have yet been presented ( Iturrieta-Zuazo et al., 2020; Littera et al., 2020; Lorente et al., 2021; Wang et al., 2020). From these combined within population studies, however, it probably follows that HLA-A*24:02 does not necessarily have a detectable positive or negative impact on the two commonly analyzed COVID-19 parameters, which are the frequency of contracting COVID-19 and the severity of the disease. As far as we know, there has not been a thorough investigation yet of a possible association between MHC polymorphism and the virus titers in the upper respiratory tract. The severity of COVID-19 is predominantly determined by whether the virus infects the lower respiratory tract ( Cyranoski, 2020), which, curiously, has been described as largely disconnected from the level of virus replication in the upper respiratory tract ( He et al., 2020; Lavezzo et al., 2020). He et al. (2020), for example, stated “There was no obvious difference in viral loads across sex, age groups and disease severity.” Therefore, if cross-virus memory T cells would not be sufficient to help block an infection but instead help to expedite the end of upper respiratory tract infections, the HLA-A*24:02 allele might give protection at the population level by limiting spread without having an impact on the COVID-19 parameters typically studied in the HLA-association studies (contraction and severity at the level of individuals). We speculate that HLA-*24:02-restricted anti-VYI CD8+ T cell immune memory—especially if recently boosted by HCoV-OC43 or HCoV-HKU1 infections or nonspecific immune stimulations—can reduce the total number of virus particles secreted by COVID-19 patients and so the replication number (R) of the virus at the population level.
Concluding remarks
The only CD8 + T cell epitope shared between CCCoVs and SARS-CoV-2 for which the immunogenicity was convincingly proven is the VYIGDPAQL peptide if presented by HLA-A*24:02. This MHC-I allele is found in the majority of the Japanese population and may help explain their surprising resistance to the virus. More studies on T cells activated during CCCoV infections and on possible associations of MHC alleles with COVID-19 parameters are necessary. Considering that for the S protein there may not be meaningful CCCoV-induced anti-SARS-CoV-2 cross-virus immune memory (depending on how one interprets the various publications)—related to the weak conservative pressure on this protein—it is feasible that SARS-CoV-2 will mutate its S proteins and escape the immune protection induced by the current generation of S-only vaccines. Potentiating such vaccines by adding immunogenic peptides from better conserved parts of the proteome, for example VYI peptide in the case of HLA-A*24:02 + individuals, may be a viable option to help prevent such immune escape.
Data availability
Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome, Accession number MN908947: https://www.ncbi.nlm.nih.gov/nuccore/MN908947
Human coronavirus OC43, complete genome, Accession number NC_005147.1: https://www.ncbi.nlm.nih.gov/nuccore/NC_005147.1
Human coronavirus HKU1, complete genome, Accession number NC_006577: https://www.ncbi.nlm.nih.gov/nuccore/NC_006577
Human coronavirus 229E, complete genome, Accession number NC_002645: https://www.ncbi.nlm.nih.gov/nuccore/NC_002645
Human Coronavirus NL63, complete genome, Accession number NC_005831: https://www.ncbi.nlm.nih.gov/nuccore/NC_005831
Funding Statement
The author(s) declared that no grants were involved in supporting this work.
[version 1; peer review: 2 approved
References
- Altmann DM, Boyton RJ: SARS-CoV-2 T cell immunity: Specificity, function, durability, and role in protection. Sci Immunol. 2020;5(49):eabd6160. 10.1126/sciimmunol.abd6160 [DOI] [PubMed] [Google Scholar]
- Amanat F, Stadlbauer D, Strohmeier S, et al. : A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020;26(7):1033–1036. 10.1038/s41591-020-0913-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anft M, Paniskaki K, Blazquez-Navarro A, et al. : COVID-19-Induced ARDS Is Associated with Decreased Frequency of Activated Memory/Effector T Cells Expressing CD11a +. Mol Ther. 2020;28(12):2691–2702. 10.1016/j.ymthe.2020.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher P, Rosati E, Esser D, et al. : Low-Avidity CD4 + T Cell Responses to SARS-CoV-2 in Unexposed Individuals and Humans with Severe COVID-19. Immunity. 2020a;53(6):1258–1271.e5. 10.1016/j.immuni.2020.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher P, Rosati E, Esser D, et al. : Pre-existing T cell memory as a risk factor for severe 1 COVID-19 in the elderly. medRxiv. 2020b; 2020.09.15.20188896. 10.1101/2020.09.15.20188896 [DOI] [Google Scholar]
- Braun J, Loyal L, Frentsch M, et al. : SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587(7833):270–274. 10.1038/s41586-020-2598-9 [DOI] [PubMed] [Google Scholar]
- Ceraolo C, Giorgi FM: Genomic variance of the 2019-nCoV coronavirus. J Med Virol. 2020;92(5):522–528. 10.1002/jmv.25700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyranoski D: Profile of a killer: the complex biology powering the coronavirus pandemic. Nature. 2020;581(7806):22–26. 10.1038/d41586-020-01315-7 [DOI] [PubMed] [Google Scholar]
- Dan JM, Mateus J, Kato Y, et al. : Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371(6529):eabf4063. 10.1126/science.abf4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AD, Williamson MK, Lewis S, et al. : Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020;12(1):68. 10.1186/s13073-020-00763-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda SK, Mahajan S, Paul S, et al. : IEDB-AR: immune epitope database-analysis resource in 2019. Nucleic Acids Res. 2019;47(W1):W502–W506. 10.1093/nar/gkz452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra JM, Hashimoto K: Expected immune recognition of COVID-19 virus by memory from earlier infections with common coronaviruses in a large part of the world population [version 2; peer review: 2 approved]. F1000Res. 2020;9:285. 10.12688/f1000research.23458.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti AP, Kula T, Wang Y, et al. : Unbiased Screens Show CD8 + T Cells of COVID-19 Patients Recognize Shared Epitopes in SARS-CoV-2 that Largely Reside outside the Spike Protein. Immunity. 2020;53(5):1095–1107.e3. 10.1016/j.immuni.2020.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D, Cagliani R, Clerici M, et al. : Molecular Evolution of Human Coronavirus Genomes. Trends Microbiol. 2017;25(1):35–48. 10.1016/j.tim.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaev A, Ketelaars SLC, Isaeva OI, et al. : Identification and characterization of an immunodominant SARS-CoV-2-specific CD8 T cell response. Researchsquare. 2020. 10.21203/rs.3.rs-33197/v2 [DOI] [Google Scholar]
- Grant EJ, Josephs TM, Valkenburg SA, et al. : Lack of Heterologous Cross-reactivity toward HLA-A*02: 01 Restricted Viral Epitopes Is Underpinned by Distinct alphabetaT Cell Receptor Signatures. J Biol Chem. 2016;291(47):24335–24351. 10.1074/jbc.M116.753988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A, Weiskopf D, Ramirez SI, et al. : Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181(7):1489–1501.e15. 10.1016/j.cell.2020.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Perosanz M, Ras-Carmona A, Lafuente EM, et al. : Identification of CD8+ T cell epitopes through proteasome cleavage site predictions. BMC Bioinformatics. 2020;21(Suppl 17):484. 10.1186/s12859-020-03782-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Galarza FF, McCabe A, Dos Santos EJM, et al. : Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res. 2020;48(D1):D783–D788. 10.1093/nar/gkz1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habel JR, Nguyen THO, van de Sandt CE, et al. : Suboptimal SARS-CoV-2-specific CD8 + T cell response associated with the prominent HLA-A*02: 01 phenotype. Proc Natl Acad Sci U S A. 2020;117(39):24384–24391. 10.1073/pnas.2015486117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale T, Petherick A, Phillips T, et al. : Variation in government responses to COVID-19. Blavatnik school of government working paper 31. 2020. Reference Source [Google Scholar]
- He X, Lau EHY, Wu P, et al. : Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26(5):672–675. 10.1038/s41591-020-0869-5 [DOI] [PubMed] [Google Scholar]
- Hirsch HH, Martino R, Ward KN, et al. : Fourth European Conference on Infections in Leukaemia (ECIL-4): guidelines for diagnosis and treatment of human respiratory syncytial virus, parainfluenza virus, metapneumovirus, rhinovirus, and coronavirus. Clin Infect Dis. 2013;56(2):258–266. 10.1093/cid/cis844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda N, Kojima H, Nishikawa M, et al. : Determination of HLA-A, -C, -B, -DRB1 allele and haplotype frequency in Japanese population based on family study. Tissue Antigens. 2015;85(4):252–259. 10.1111/tan.12536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka J, Grebe K, Shenderov E, et al. : Quantitating T cell cross-reactivity for unrelated peptide antigens. J Immunol. 2009;183(7):4337–4345. 10.4049/jimmunol.0901607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturrieta-Zuazo I, Rita CG, García-Soidán A, et al. : Possible role of HLA class-I genotype in SARS-CoV-2 infection and progression: A pilot study in a cohort of Covid-19 Spanish patients. Clin Immunol. 2020;219:108572. 10.1016/j.clim.2020.108572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarjour NN, Masopust D, Jameson SC: T Cell Memory: Understanding COVID-19. Immunity. 2021;54(1):14–18. 10.1016/j.immuni.2020.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kared H, Redd AD, Bloch EM, et al. : CD8+ T cell responses in convalescent COVID-19 individuals target epitopes from the entire SARS-CoV-2 proteome and show kinetics of early differentiation. bioRxiv. 2020;2020.10.08.330688. 10.1101/2020.10.08.330688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A, Trowsdale J: Introduction: MHC/KIR and governance of specificity. Immunogenetics. 2017;69(8–9):481–488. 10.1007/s00251-017-0986-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MD, Harris KM, Jensen-Wachspress MA, et al. : SARS-CoV-2 specific T-cells Are Rapidly Expanded for Therapeutic Use and Target Conserved Regions of Membrane Protein. Blood. 2020;136(25):2905–2917. 10.1182/blood.2020008488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komabayashi K, Seto J, Matoba Y, et al. : Seasonality of Human Coronavirus OC43, NL63, HKU1, and 229E Infection in Yamagata, Japan, 2010-2019. Jpn J Infect Dis. 2020;73(5):394–397. 10.7883/yoken.JJID.2020.525 [DOI] [PubMed] [Google Scholar]
- Kubota-Koketsu R, Terada Y, Yunoki M, et al. : Neutralizing and binding activities against SARS-CoV-1/2, MERS-CoV, and human coronaviruses 229E and OC43 by normal human intravenous immunoglobulin derived from healthy donors in Japan. Transfusion. 2020;61(2):356–360. 10.1111/trf.16161 [DOI] [PubMed] [Google Scholar]
- Lavezzo E, Franchin E, Ciavarella C, et al. : Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo'. Nature. 2020;584(7821):425–429. 10.1038/s41586-020-2488-1 [DOI] [PubMed] [Google Scholar]
- Le Bert N, Tan AT, Kunasegaran K, et al. : SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584(7821):457–462. 10.1038/s41586-020-2550-z [DOI] [PubMed] [Google Scholar]
- Lee CH, Pinho MP, Buckley PR, et al. : Potential CD8+ T Cell Cross-Reactivity Against SARS-CoV-2 Conferred by Other Coronavirus Strains. Front Immunol. 2020;11:579480. 10.3389/fimmu.2020.579480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie M: T cells found in coronavirus patients 'bode well' for long-term immunity. Science. 2020;368(6493):809–810. 10.1126/science.368.6493.809 [DOI] [PubMed] [Google Scholar]
- Littera R, Campagna M, Deidda S, et al. : Human Leukocyte Antigen Complex and Other Immunogenetic and Clinical Factors Influence Susceptibility or Protection to SARS-CoV-2 Infection and Severity of the Disease Course. The Sardinian Experience. Front Immunol. 2020;11:605688. 10.3389/fimmu.2020.605688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorente L, Martín MM, Franco A, et al. : HLA genetic polymorphisms and prognosis of patients with COVID-19. Med Intensiva. 2021;45(2):96–103. 10.1016/j.medin.2020.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund O, Nielsen M, Kesmir C, et al. : Definition of supertypes for HLA molecules using clustering of specificity matrices. Immunogenetics. 2004;55(12):797–810. 10.1007/s00251-004-0647-4 [DOI] [PubMed] [Google Scholar]
- Mäkelä MJ, Puhakka T, Ruuskanen O, et al. : Viruses and bacteria in the etiology of the common cold. J Clin Microbiol. 1998;36(2):539–42. 10.1128/JCM.36.2.539-542.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J, Grifoni A, Tarke A, et al. : Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370(6512):89–94. 10.1126/science.abd3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckiff BJ, Ramírez-Suástegui C, Fajardo V, et al. : Imbalance of Regulatory and Cytotoxic SARS-CoV-2-Reactive CD4 + T Cells in COVID-19. Cell. 2020;183(5):1340–1353.e16. 10.1016/j.cell.2020.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami T, Fukutomi Y, Inada R, et al. : Regional differences in the prevalence of sensitization to environmental allergens: Analysis on IgE antibody testing conducted at major clinical testing laboratories throughout Japan from 2002 to 2011. Allergol Int. 2019;68(4):440–449. 10.1016/j.alit.2019.03.008 [DOI] [PubMed] [Google Scholar]
- Moderbacher CR, Ramirez SI, Dan JM, et al. : Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell. 2020;183(4):996–1012.e19. 10.1016/j.cell.2020.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Tsunoda S, Sakaida H, et al. : Analysis of factors associated with cedar pollen sensitization and development of pollinosis in a young Japanese adult population. Allergol Int. 2019;68(1):39–45. 10.1016/j.alit.2018.05.006 [DOI] [PubMed] [Google Scholar]
- Nelde A, Bilich T, Heitmann JS, et al. : SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat Immunol. 2021;22(1):74–85. 10.1038/s41590-020-00808-x [DOI] [PubMed] [Google Scholar]
- Nguyen A, David JK, Maden SK, et al. : Human Leukocyte Antigen Susceptibility Map for Severe Acute Respiratory Syndrome Coronavirus 2. J Virol. 2020;94(13):e00510–20. 10.1128/JVI.00510-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L, Ye F, Cheng ML, et al. : Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity. 2020;52(6):971–977.e3. 10.1016/j.immuni.2020.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Mentzer AJ, Liu G, et al. : Broad and strong memory CD4 + and CD8 + T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol. 2020;21(11):1336–1345. 10.1038/s41590-020-0782-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova G, Ferrante A, Gorski J: Cross-reactivity of T cells and its role in the immune system. Crit Rev Immunol. 2012;32(4):349–372. 10.1615/critrevimmunol.v32.i4.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poluektov Y, Daftarian P, Delcommenne MC: Assessment of SARS-CoV-2 Specific CD4(+) and CD8 (+) T Cell Responses Using MHC Class I and II Tetramers. bioRxiv. 2020; 2020.07.08.194209. 10.1101/2020.07.08.194209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prachar M, Justesen S, Steen-Jensen DB, et al. : Identification and validation of 174 COVID-19 vaccine candidate epitopes reveals low performance of common epitope prediction tools. Sci Rep. 2020;10(1):20465. 10.1038/s41598-020-77466-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rammensee HG, Friede T, Stevanoviíc S: MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41(4):178–228. 10.1007/BF00172063 [DOI] [PubMed] [Google Scholar]
- Sagar M, Reifler K, Rossi M, et al. : Recent endemic coronavirus infection is associated with less-severe COVID-19. J Clin Invest. 2021;131(1):e143380. 10.1172/JCI143380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellens IMM, Hoof I, Meiring HD, et al. : Comprehensive Analysis of the Naturally Processed Peptide Repertoire: Differences between HLA-A and B in the Immunopeptidome. PLoS One. 2015;10(9):e0136417. 10.1371/journal.pone.0136417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulien I, Kemming J, Oberhardt V, et al. : Characterization of pre-existing and induced SARS-CoV-2-specific CD8 + T cells. Nat Med. 2021;27(1):78–85. 10.1038/s41591-020-01143-2 [DOI] [PubMed] [Google Scholar]
- Sekine T, Perez-Potti A, Rivera-Ballesteros O, et al. : Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell. 2020;183(1):158–168.e14. 10.1016/j.cell.2020.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A, Crotty S: Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat Rev Immunol. 2020;20(8):457–458. 10.1038/s41577-020-0389-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewell AK: Why must T cells be cross-reactive? Nat Rev Immunol. 2012;12(9):669–677. 10.1038/nri3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomuradova AS, Vagida MS, Sheetikov SA, et al. : SARS-CoV-2 Epitopes Are Recognized by a Public and Diverse Repertoire of Human T Cell Receptors. Immunity. 2020;53(6):1245–1257.e5. 10.1016/j.immuni.2020.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrock E, Fujimura E, Kula T, et al. : Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370(6520):eabd4250. 10.1126/science.abd4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen L, Spreeuwenberg P, Lustig R, et al. : Global mortality estimates for the 2009 Influenza Pandemic from the GLaMOR project: a modeling study. PLoS Med. 2013;10(11):e1001558. 10.1371/journal.pmed.1001558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder TM, Gittelman RM, Klinger M, et al. : Magnitude and Dynamics of the T-Cell Response to SARS-CoV-2 Infection at Both Individual and Population Levels. medRxiv. 2020;2020.07.31.20165647. 10.1101/2020.07.31.20165647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner S, Sotzny F, Bauer S, et al. : HCoV- and SARS-CoV-2 Cross-Reactive T Cells in CVID Patients. Front Immunol. 2020;11:607918. 10.3389/fimmu.2020.607918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern LJ, Wiley DC: Antigenic peptide binding by class I and class II histocompatibility proteins. Structure. 1994;2(4):245–251. 10.1016/s0969-2126(00)00026-5 [DOI] [PubMed] [Google Scholar]
- Sylvester-Hvid C, Nielsen M, Lamberth K, et al. : SARS CTL vaccine candidates; HLA supertype-, genome-wide scanning and biochemical validation. Tissue Antigens. 2004;63(5):395–400. 10.1111/j.0001-2815.2004.00221.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi A, Matsui M: Identification of HLA-A *02:01-restricted candidate epitopes derived from the non-structural polyprotein 1a of SARS-CoV-2 that may be natural targets of CD8 + T cell recognition in vivo. J Virol. 2020;JVI.01837–20. 10.1128/JVI.01837-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A, Sidney J, Kidd CK, et al. : Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. bioRxiv. 2020;2020.12.08.416750. 10.1101/2020.12.08.416750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V, Herold J, Schelle B, et al. : Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J Gen Virol. 2001;82(Pt 6):1273–1281. 10.1099/0022-1317-82-6-1273 [DOI] [PubMed] [Google Scholar]
- Thieme CJ, Anft M, Paniskaki K, et al. : Robust T Cell Response Toward Spike, Membrane, and Nucleocapsid SARS-CoV-2 Proteins Is Not Associated with Recovery in Critical COVID-19 Patients. Cell Rep Med. 2020;1(6):100092. 10.1016/j.xcrm.2020.100092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L, Pyrc K, Jebbink MF, et al. : Identification of a new human coronavirus. Nat Med. 2004;10(4):368–73. 10.1038/nm1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L, Keyaerts E, Moës E, et al. : Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol. 2005;79(3):1595–1604. 10.1128/JVI.79.3.1595-1604.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhang W, Zhang J, et al. : Distribution of HLA allele frequencies in 82 Chinese individuals with coronavirus disease-2019 (COVID-19). HLA. 2020;96(2):194–196. 10.1111/tan.13941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Schmitz KS, Raadsen MP, et al. : Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol. 2020;5(48):eabd2071. 10.1126/sciimmunol.abd2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldemeskel BA, Kwaa AK, Garliss CC, et al. : Healthy donor T cell responses to common cold coronaviruses and SARS-CoV-2. J Clin Invest. 2020;130(12):6631–6638. 10.1172/JCI143120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PCY, Lau SKP, Chu CM, et al. : Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79(2):884–895. 10.1128/JVI.79.2.884-895.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, et al. : A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Saito H, Fujieda S: Present state of Japanese cedar pollinosis: the national affliction. J Allergy Clin Immunol. 2014;133(3):632–9.e5. 10.1016/j.jaci.2013.11.002 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Dijkstra JM: Major Histocompatibility Complex (MHC) Genes and Disease Resistance in Fish. Cells. 2019;8(4):378. 10.3390/cells8040378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Kandula S, Huynh M, et al. : Estimating the infection-fatality risk of SARS-CoV-2 in New York City during the spring 2020 pandemic wave: a model-based analysis. Lancet Infect Dis. 2021;21(2):203–212. 10.1016/S1473-3099(20)30769-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorifuji T, Matsumoto N, Takao S: Excess All-Cause Mortality During the COVID-19 Outbreak in Japan. J Epidemiol. 2021;31(1):90–92. 10.2188/jea.JE20200492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Wang W, Wang H, et al. : First infection by all four non-severe acute respiratory syndrome human coronaviruses takes place during childhood. BMC Infect Dis. 2013;13:433. 10.1186/1471-2334-13-433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yang XL, Wang XG, et al. : A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]