Abstract
In fluctuating environmental conditions, organisms must modulate their bioenergetic production in order to maintain cellular homeostasis for optimal fitness. Mitochondria are hubs for metabolite and energy generation. Mitochondria are also highly dynamic in their function: modulating their composition, size, density, and the network-like architecture in relation to the metabolic demands of the cell. Here, we review the recent research on the post-transcriptional regulation of mitochondrial composition focusing on mRNA localization, mRNA translation, protein import, and the role that dynamic mitochondrial structure may have on these gene expression processes. As mitochondrial structure and function has been shown to be very important for age-related processes, including cancer, metabolic disorders, and neurodegeneration, understanding how mitochondrial composition can be affected in fluctuating conditions can lead to new therapeutic directions to pursue.
Keywords: Mitochondria, mRNA, Mitochondrial morphology, mRNA localization, protein import
Subjects: Cell Biology, Gene Expression & Regulation
Introduction
Mitochondria are eukaryotic organelles of symbiotic origin that are important for many cellular functions including oxidative phosphorylation, ROS generation, the biosynthesis of amino acids, iron-sulfur clusters and heme. Over the approximately 2 billion years since the initial symbiosis, mitochondria have lost the majority of their genes to the nuclear-genome, yet a small number are still encoded by mitochondrial DNA (1). While mitochondria are colloquially referred to as “the powerhouse of the cell”, because of their ability to produce the energy currency ATP through respiration, a more apt description may be the “powerhouse of certain cell types under certain conditions”, as oxidative phosphorylation is not the primary source of ATP generation in many conditions and cell types. For example, many cancer cells repress mitochondrial oxidative phosphorylation and generate ATP almost exclusively through glycolytic flux (2). Similarly many yeast, including Sacchacromyces cerevisiae, are Crabtree-positive in that they will use fermentation even in the presence of oxygen as the primary source of ATP generation (3). In these cells, while mitochondria are essential organelles in all conditions, oxidative phosphorylation is non-essential in certain conditions as cells are able to grow in glucose media even if they have lost their mitochondrial DNA, because they are able to obtain ATP through aerobic glycolysis (4). In other conditions such as non-fermentable carbon sources, the ability to respire is a requirement for optimal growth, so cells and mitochondria must rapidly adjust their bioenergetic capabilities to fit the current environment.
Proteomic studies show that there are large shifts in mitochondrial composition as the structure and function of mitochondria change. While the importance of transcriptional control in these mitochondrial functional changes have been explored and reviewed (5–7), post-transcriptional regulation is another important layer of regulation in the how mitochondria properly adjust their composition to the metabolic needs of the cell. Intriguingly, changing mitochondrial structure may also directly impact post-transcriptional steps of gene regulation. As abnormalities in mitochondrial structure and function have been identified as key pathological components of aging and disease (8–10), it is imperative to fully understand the mechanisms of mitochondrial control (Figure 1). In this review we discuss the impact of dynamic conditions on the structure, function, and composition of mitochondria, focusing on the impact of post-transcriptional regulation.
Figure 1. Interrelationship between cellular conditions, gene expression, and mitochondrial morphology with each other on the mitochondrial activity of the cell.
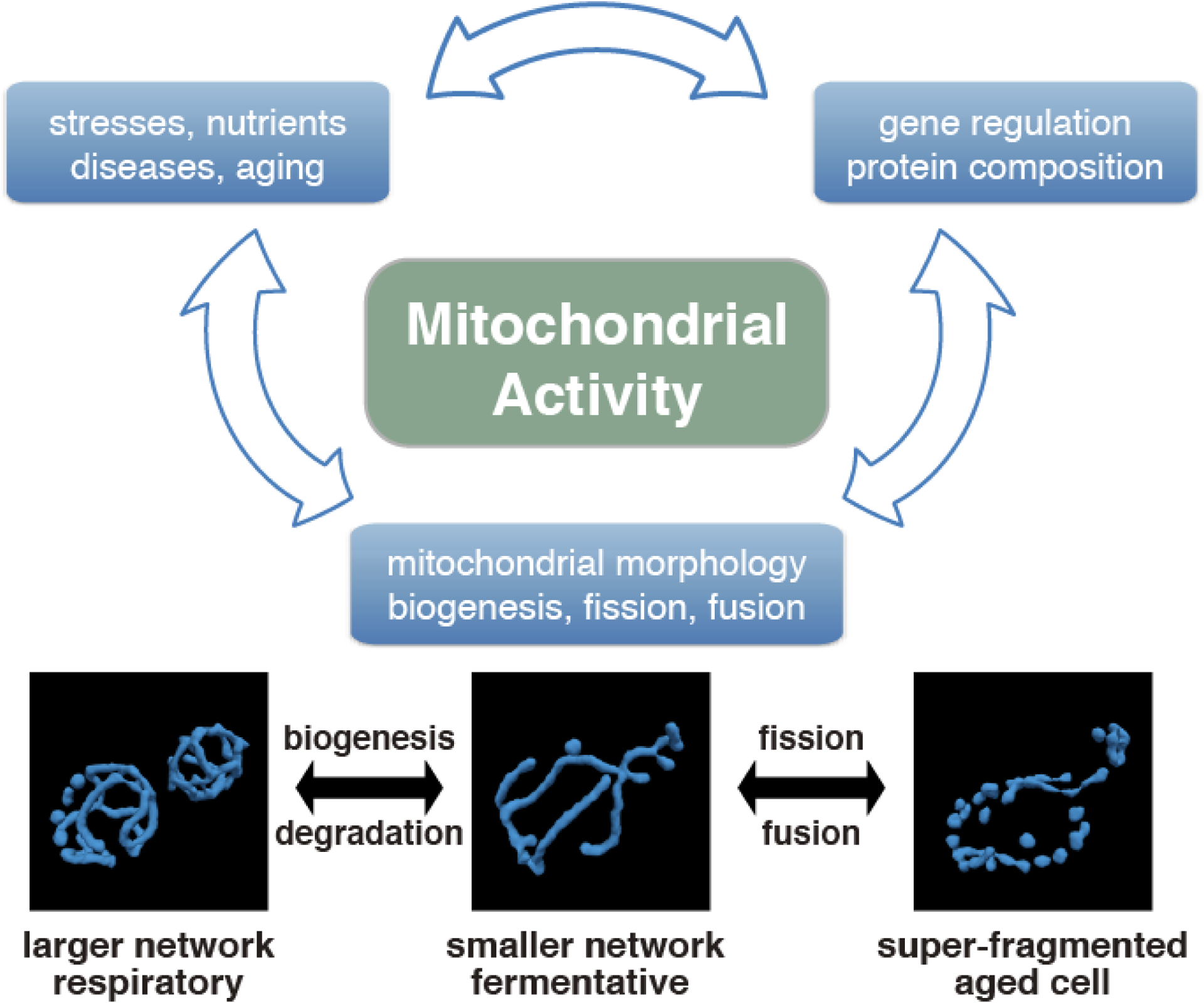
The reconstructed mitochondrial morphology in fermentative, respiratory condition, and aged yeast cells are shown below. These active morphological changes are regulated by gene regulatory networks in different cellular states such as metabolic function of the cell and disease phenotypes and has a close relationship with mitochondrial activity.
Cells control mitochondrial size and morphology according to cellular environment
Mitochondrial architecture is highly dynamic as these organelles can modulate their morphology through fission and fusion as well as their overall size through biogenesis and mitophagy. Mitochondria size and morphology are known to correlate with the metabolic function of the cell and disease phenotypes (Figure 1) with fusion and biogenesis known to be important for OXPHOS activity.(Reviewed in (8–11). Morphological control is governed by fission and fusion proteins, and once this balanced regulation is altered by external or internal signals, the morphology changes to either a hyperfused or super-fragmented structure (8,10,12). For example when yeast cells are grown in non-fermentable carbon sources they have a large expansion of their mitochondria leading to a highly branched mitochondrial network (13,14). Similarly, forcing cancer cells to switch their metabolism towards respiration through growth in galactose also drives elongated mitochondria in human cells (15). It has been proposed that elongated mitochondria lead to increased bioenergetics efficiency of oxidative phosphorylation (16). This fits with the observation that during nutrient starvation, when efficient metabolism is essential for survival, mitochondrial also become highly interconnected, forming long tubular networks (17,18). Along with potentially driving more efficient oxidative phosphorylation, expansion of a tubular network of mitochondria has also been found to protect mitochondria from nutrient deprivation induced autophagic degradation (17,18).
While elongated mitochondrial morphology is associated with increased ATP production and efficient oxidative phosphorylation, mitochondrial fragmentation is associated with variety of pathological states including aging, neurodegenerative dieseases, and metabolic diseases such as diabetes and insulin resistance (19–22). In S. cerevisiae mitochondrial fragmentation is an early event in the aging process (23) and inhibiting fission has been found to extend lifespan (24). Similarly, in C. elegans and D. melanogaster increased mitochondrial fusion is associated with extended lifespan (25,26). In mammalian cells high levels of mitochondrial fission activity are associated with high proliferation and invasiveness in some cancer cells (27). Also mutations in fusion proteins such as Mfn1/2 and Opa1 in mice lead to neurodegeneration, developmental delay, and further embryonic lethality (10,28). Future studies will be needed to help elucidate the drivers of these changes in mitochondrial morphology, whether it is changing metabolic regulation during aging or a general loss of homeostasis, as well as the direct pathological impact these changes in morphology have.
While mitochondrial fission and fusion are important to mitochondrial dynamics, mitochondria can also change their overall size/volume, dominated by alterations in mitochondrial biogenesis. In yeast, the HAP complex is known to play an important role in the transcriptional upregulation of mitochondrial biogenesis upon a shift to non-fermentable carbon sources (29). In mammalian cells PGC-1α is a key co-transcriptional regulatory factor in the regulation of mitochondrial biogenesis (Reviewed in (30)). This size control is well quantified in yeast cells (13,31) and also observed in human cell culture supplemented with ethanol (32), or cells grown in galactose (15). When it comes to estimating the mitochondrial activity of a cell or tissue, it is important to take into consideration what fraction of the cell or tissue volume can actually be occupied by enzymes and mitochondria. While mitochondrial volume fraction differs depending on the definition in each study, mitochondrial volume fraction varies from 1% to 14% during fermentative and respiratory conditions in S. cerevisiae (31), and varies from 5% to 25% for human tissues (Table 1) (33–36). This volume fraction is regulated through the changing of both cellular and mitochondrial volume such that in respiratory conditions, cellular volume decreases 1.5-fold and mitochondrial volume increases 1.4-fold, relative to fermentative conditions in yeast. These changes lead to a roughly 2-fold increase in mitochondrial volume fraction (31). It is also reported that there are unconventional morphological changes which do not involve fission and fusion proteins (37). Together, mitochondrial size and morphology have the potential to help regulate cell metabolism to adapt to the cellular environment (Reviewed in (11)).
Table 1. Mitochondrial volume fraction varies in different cells, tissues, and conditions.
Measurements of mitochondrial volume fraction in cells of different organism and tissues.
Mitochondria show large compositional changes across conditions
Structural changes to mitochondria in response to stimuli are accompanied by changes in the function and composition of mitochondria. Mitochondria contain over 1,000 distinct proteins that encode for a still expanding number of functions (38–40). While the ability to produce ATP through oxidative phosphorylation is a key function of mitochondria, respiration is actually repressed in many conditions and cell types (2,3). Other mitochondrial functions include maintaining ion homeostasis and biosynthetic production of lipids, proteins, nucleotides, and Fe-S(iron-sulfur) clusters. Mitochondria also play active roles integrating signaling pathways in response to nutrients and a variety of stressors (41).
There have been a number of studies that have systematically explored the quantity of mitochondrial proteins present across shifting nutrient conditions (39,40,42). In yeast, shifting cells from glucose (fermentation) to glycerol (respiration) causes an expansion of the mitochondria, but all proteins did not change proportionally. While overall mitochondrial mass increases about 2-fold in respiratory conditions, overall energy-related processes increase more than 3-fold, while respiratory complexes increase almost 4-fold (31,39). Interestingly, essential processes such as the biosynthesis of Fe-S clusters and protein import have little change in abundance from glucose to glycerol when the overall mitochondrial protein content doubles (39). The impact this may have on protein import is discussed further below. Similar changes in mitochondrial composition were found as yeast naturally transition from fermenting glucose, to the diauxic shift after glucose supplies are exhausted and cells need to alter their metabolism, to the respiratory phase where yeast consume the ethanol previously produced as a byproduct of fermentation. During the diauxic shift there is a transition in mitochondrial composition from biosynthetic processes to energy-related processes, with only a slight further upregulation of energy-related processes once the cell has completely shifted to respiratory metabolism (42). There are also significant changes in structural proteins as the metabolic state shifts, driven by a decrease in fission proteins which may underlie some of the previous structural changes that take place in different metabolic states. Overall, these findings support a changing function for mitochondria depending on the metabolic needs of the cell. During times of fast growth, mitochondria are used for the biosynthesis of important building blocks for proliferation as well as common processes necessary in all conditions like Fe-S cluster generation. But mitochondria can also transition their composition to be the primary source for energy production within the cell.
Mitochondrial proteins can be imported post- and co-translationally
Of the approximately 1000 proteins found in mitochondria, only 8 in yeast and 13 in humans are encoded from mitochondrial DNA(1). Therefore, the vast majority of necessary nuclear-encoded proteins must be translated in the cytosol and imported to the mitochondria. There are currently two methods proposed for the creation, transportation, and subsequent import of mitochondrial proteins – a co-translational pathway and a non-co-translational pathway. Until fairly recently, the most popular theory is that mitochondrial proteins are translated in the cytosol and then translocated and imported to the inner membrane, intermembrane space, or outer membrane to serve their intended purpose.
While post-translational import is a well characterized and widely accepted mechanism, it is a complex sequence of events that we aim to briefly summarize. In order for these proteins to enter the mitochondria, they must first interact with the Translocase of the Outer Membrane (TOM) complex. This complex includes protein receptors Tom20 which recognizes an N-terminal mitochondrial targeting signal (MTS) of pre-proteins, and Tom70 which recognizes hydrophobic pre-proteins containing internal signals. Cytosolic chaperones, such as Hsp70 (heat shock protein 70), play a role in stabilizing the pre-protein and guiding it toward the TOM complex (43). Recently, it has been shown that DnaJ-like protein, Djp1, acts as a chaperone for MTSs in the initial step of localizing proteins to import machinery. (44,45). After recognition, the pre-protein travels through the outer membrane-spanning translocase pore and begins interaction with various Translocase of the Inner Membrane (TIM) complexes. Mitochondrial membrane potential is important for inner membrane import, as a negative matrix-side potential drives pre-proteins with positively charged MTSs toward the voltage-mediated TIM22 complex. Mitochondrial membrane potential is sufficient to sort proteins to the mitochondrial inner membrane via the TIM22 complex, but pre-proteins destined for the mitochondrial matrix via a second translocon, the TIM23 complex, require further aid.Mitochondrial Hsp70p (encoded by SSC1 in yeast, HSPA9 in humans) binds to matrix side Tim44p to form an ATP driven motor that can shuttle the pre-protein into the matrix, where it undergoes pre-sequence cleavage and further folding in order to reach maturity (Reviewed in (46–48)).
One surprising finding from proteomics measurements under different metabolic states was that the import proteins do not increase in abundance as yeast transition to respiratory metabolism, and the mitochondrial surface area expands ~1.6 fold (from 48μm2 to 79μm2) (Figure 2A, B). This will lead to a decrease in the density of import machinery proteins as the cell is actively importing more mitochondrial proteins (39). It is possible that conditional changes may cause an alteration of protein import machinery architecture, thereby altering the function and efficiency of the import complex in different conditions. The TOM complex structure has been analyzed through cryo-EM and it has been reported that the complex shows dimeric to tetrameric structures (49–55). Most of the complex purification was conducted in fermentative conditions (for Neurospora crassa, 2% sucrose, for S. cerevisiae 2% glucose) in which a trimeric complex was seen. Interestingly, when the complex was purified from respiratory conditions (S. cerevisiae 2% ethanol and 3% glycerol), the TOM complex was observed as dimeric structure (49) (Figure 2C).
Figure 2. Metabolic state affects mitochondrial surface area and may contribute to TOM complex polymer composition.
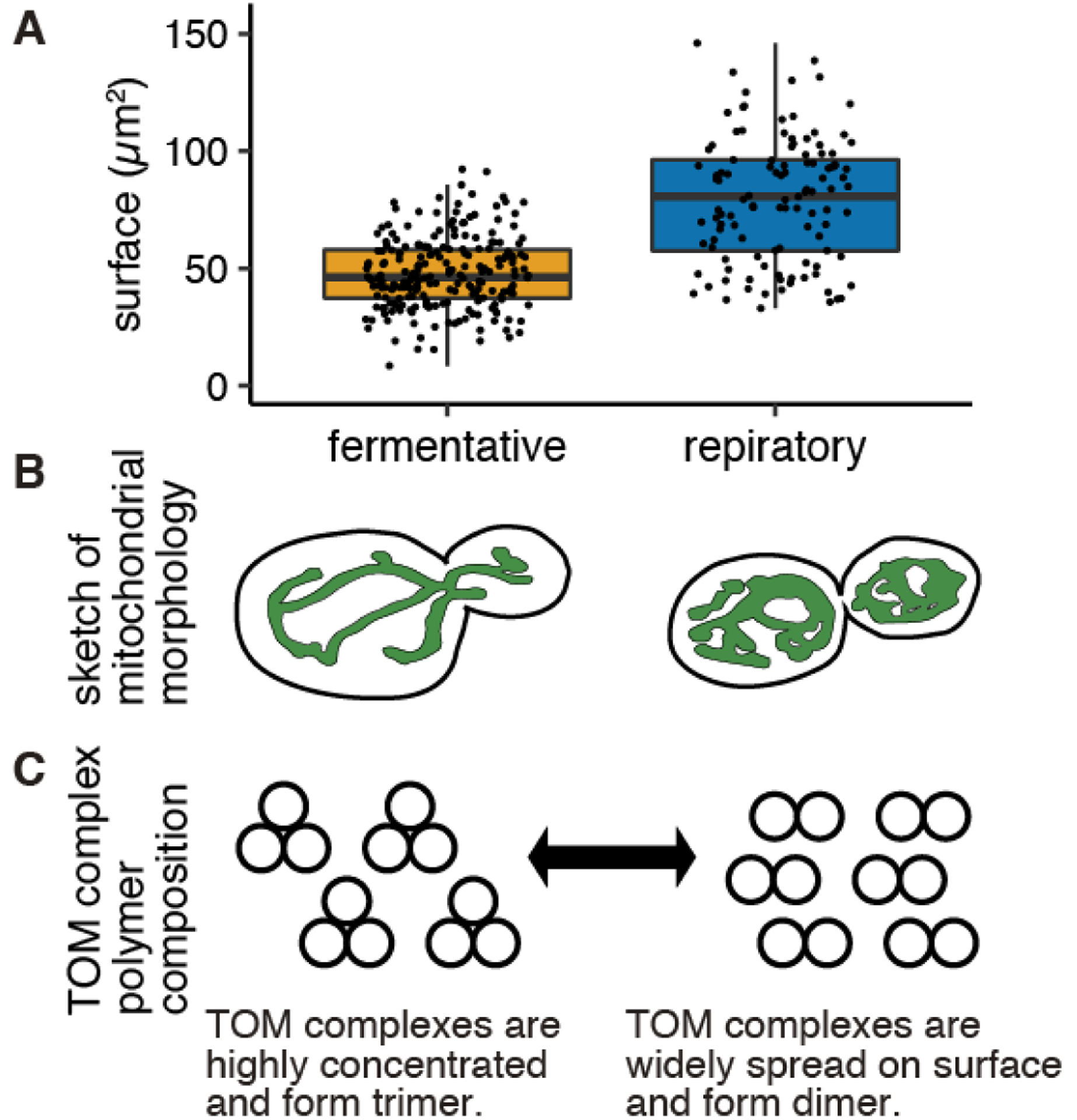
(A) Surface area of yeast mitochondria in fermentative and respiratory condition was measured from fluorescent microscopy image processed by MitoGraph (N>112). Mean of the area for fermentative and respiratory was 48μm2 and 79μm2 respectively. (B) Cartoon of the typical mitochondrial morphology in fermentative (left) and respiratory (right) condition. (C) Cartoon of potential TOM complex polymer composition in fermentative (left) and respiratory (right) condition. Conditional changes may cause an alteration of protein import machinery architecture.
In contrast to the well-defined model of post-translational import, the co-translational model of importing a protein that is being synthesized at the outer mitochondrial membrane is a relatively new idea. This process utilizes the nascent chain associated complex (NAC), a heterodimeric protein which associates with ribosome nascent chain complexes (RNCs), consisting of a nascent polypeptide chain with its C terminus still attached to its translating ribosome. NAC binds to ribosomes near the protein exit tunnel and has been shown to target ribosomes to the mitochondria. Eliminating NAC and Mft52p, a cytosolic targeting factor that further helps it localize to the mitochondria, is shown to lead to mitochondrial defects, loss of mitochondrial DNA, and disturbance of organelle morphology, suggesting that NAC is essential to mitochondria biogenesis (56), While the receptor for NAC was unknown for quite a while, a 2014 study identified outer mitochondrial membrane protein OM14 as a NAC receptor, as well as an important factor for co-translational import (57) (Figure 3).
Figure 3. Nuclear-encoded mRNA localization to mitochondria and potential regulatory mechanism in changing environmental condition.
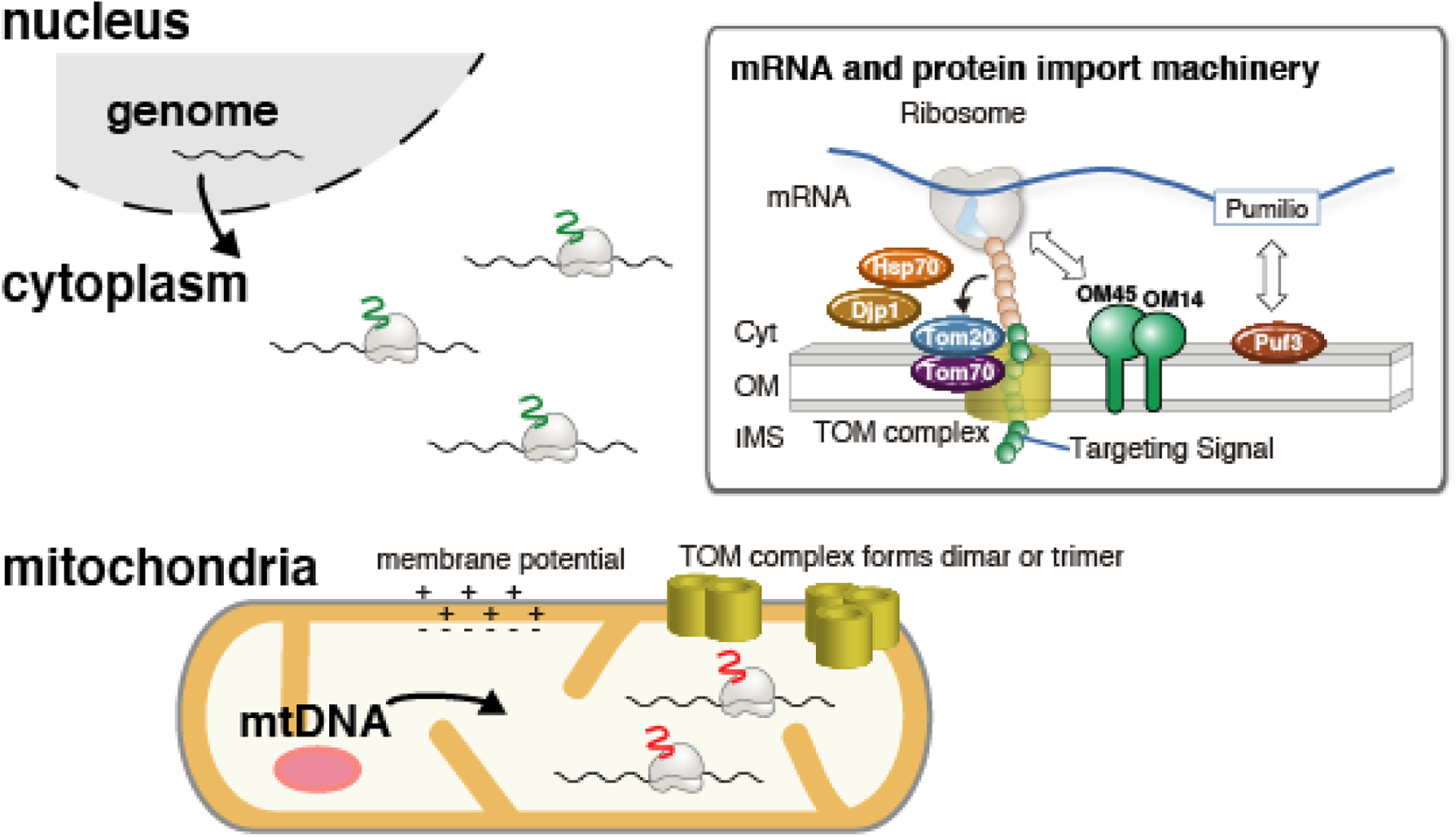
After the export form nuclear pore, nuclear-encoded mitochondrial mRNAs are diffusively localized to mitochondrial surface by binding to mitochondrial protein import machinery, TOM complex. The interaction between mRNA and Protein import machinery are summarized in the right box. Briefly, mRNAs bind mitochondria through a mitochondrial targeting signal in the nascent polypeptide chain. Targeting signal associates with Tom20 and Tom70 with the help of chaperone proteins Hsp70 and Djp1. Pumilio family protein (Puf3 in yeast and CLUH in mammalian cells) directly bind mRNA in a sequence-dependent manner. Association of ribosomes to OM14 and OM45 are also reported.
An even more recent study, in which yeast mitochondria were purified from cells treated with magnesium ions and cycloheximide (CHX)-- in order to stabilize RNCs, and stall the ribosomes, respectively-- was able to visualize ribosomes bound to the outer mitochondrial membrane through electron cryo-tomography (cryoET). Furthermore, these ribosomes were oriented such that the polypeptide exit tunnel points toward the outer membrane, suggesting that the nascent chains will be imported to the mitochondria as they emerge from the ribosome. The association between ribosomes and the TOM complex was seen to be reversible with release of nascent chains from the ribosome, strongly suggesting that mitochondrial imports are occurring as these ribosomes locally synthesize proteins (58).
mRNA localization to the mitochondria
Asymmetric localization of mRNAs within the cell is a means of regulating gene expression in space and time used by various organisms (59). Functions of mRNA localization to the targeted area of the encoded protein include the regulation of further protein synthesis, aid in the assembly of organelle complexes, and specific localized function of proteins. It is also speculated that by restricting translation to the target site, the cell is also able to save energy that would otherwise be spent on the active transport of large proteins along the cytoskeleton (60). Localization of mRNAs can also allow for a quickened production of proteins that are necessary to keep up with the constantly changing conditions of the cell, and local translation reduces the risks of unwanted interactions or folding as the protein travels from the cytosol to its final destination.
Localization that occurs before an mRNA is translated into a protein is dependent on RNA binding proteins (RBPs) which recognize and bind to a specific portion of the mRNA’s sequence, known as a zip code. In many examples the mRNA is held in a translationally inactive state until it reaches its target destination, in which case it is able to begin translation of its encoded protein (61). Alternatively, mRNA localization can be dependent on nascent peptide tethering the entire mRNA-ribosome-nascent protein chain complex to the region of localization (Figure 3). Different translational inhibitors can be used to distinguish between these two mechanisms of localization. If the localization is driven by RBPs then the translational inhibitor puromycin, which dissociates the nascent peptide from the mRNA, should not affect localization, while it should inhibit localization of co-translationally localized mRNAs. CHX, which stalls translation elongation and preserves the mRNA-ribosome-nascent protein chain complex, can increase the localization of co-translationally localized mRNAs.
A number of experiments have investigated nuclear-encoded mRNA localization to the mitochondrial outer membrane at both a global and gene specific level across diverse organisms (62–68). These experiments have found that 100s of mRNAs localize to the OMM and that this localization can proceed through both a puromycin insensitive and sensitive manner in both yeast and mammals (63,66). It has been found that mitochondrially localized mRNAs are more likely to be conserved mRNAs and mRNAs whose proteins reside in the inner membrane (68,69). Translation duration has also been found to be an important factor in localizing mRNAs to the mitochondrial membrane, as longer ORFs are more likely to localize to the OMM and ribosome stalling can increase mitochondrial mRNA localization (31,69). In both yeast and mammalian cells, CHX can increase the mitochondrial localization of a subset of mRNAs (Figure 4) (66,68). Translation duration is thought to be important because nascent polypeptide chains contribute to mRNA localization to the mitochondria. Certain polypeptides have been found to contain an N-terminal MTS that is sufficient to direct the newly synthesized protein to the mitochondria, and is cleaved once localization is complete, similar to that of secreted proteins. This signal is recognized by TOM complex, which, as previously described, is the initial contact point for mitochondrial protein import. It is suggested that the association of nascent polypeptides with these receptors leads to an enrichment of mRNAs encoding for mitochondrial proteins at the outer membrane. Deletion of Tom20, a key subunit of the TOM complex, has been shown to decrease mitochondrial association of many mRNAs that are known to normally localize to the mitochondria, suggesting that this protein receptor plays an important role in anchoring mRNAs as they are being translated (70).
Figure 4. Treatment with the translation elongation inhibitor cycloheximide (CHX) can increase the mitochondrial localization of mitochondrial protein coding mRNAs.
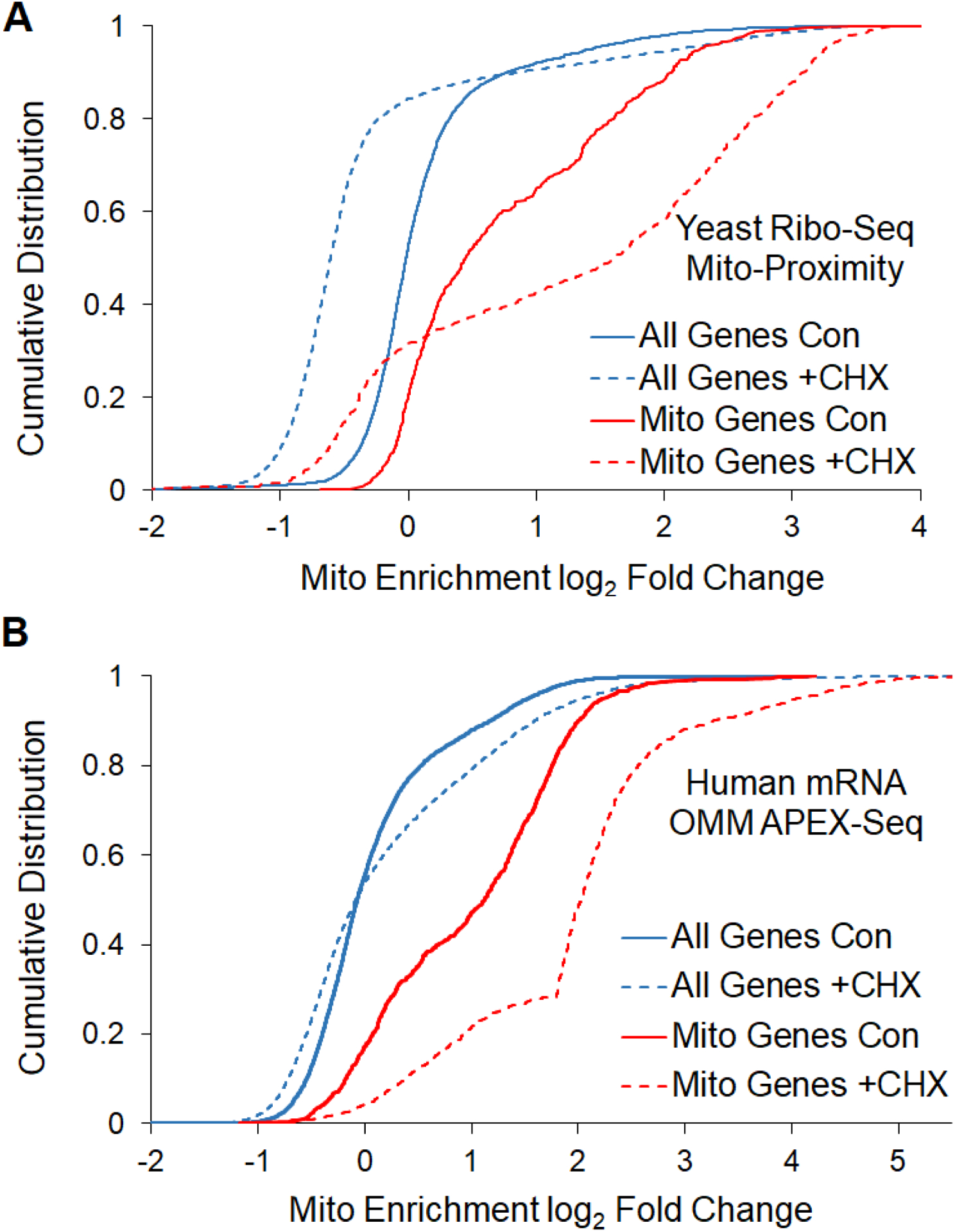
Cumulative distribution of mRNA enrichment (ribosome reads) to mitochondria of mitochondrial genes and all genes of (A) yeast cells measured by proximity specific ribosome profiling (68) and (B) HEK293T cells measured by APEX-seq (66) were depicted with or without addition of CHX.
Mitochondrial mRNA localization promotes translation
Translation is important for the localization of many mRNAs to the mitochondria, but recent research has pointed to localization to the mitochondria potentially reciprocally impacting translation. In Drosophila and mammalian cells it was shown that certain oxidative phosphorylation mRNAs are repressed translationally until they are localized to OMM where they are translationally activated by PINK1/Parkin (67). Disruption of components of the TIM/TOM complex further amplified the effects of PINK1 mutation on mRNA localization to the mitochondria, showing that localization is linked to co-translational import of proteins. It has also been found in flies that the human AKAP1 homolog MDI (mitochondrial DNA insufficient) can recruit the La-related protein (Larp), an RNA-binding protein, to the OMM and that this complex promotes the selective translation of nuclear-encoded mitochondrial proteins at the OMM (71). Interestingly in yeast the Larp homologs, Slf1 and Sro9, have been found to bind large numbers of nuclear-encoded mitochondrial mRNAs (72) and overexpression of SLF1 leads to increased respiration and extended lifespan (73). In yeast it has been found that forced localization of a cytoplasmic reporter to the OMM through MS2 tethering leads to large increases in protein synthesis as well as increased ribosome engagement (31). The mechanism of this increased protein synthesis from mRNA localization to the OMM will be an important future direction to understand.
mRNA decay impacts mitochondrial gene expression
While mRNA level changes are generally ascribed to transcriptional regulation, mRNA decay also plays an important and understudied role in modulating the transcriptome. Mitochondria may rely on mRNA decay regulation more than other cellular functions during changing environmental conditions. In yeast, it was found that mRNA levels for respiration and mitochondria-related genes are inversely correlated to growth rate across many diverse perturbations and that this is primarily impacted by changes in mRNA stability (74). This differed from functions such as translational control, which is primarily regulated at the level of transcription. Changes in mRNA decay have been shown to be directly important for altering mRNA levels of the oxidative phosphorylation mRNAs SDH1 and SDH2 in changing nutrient conditions (75). When yeast cells switch from a non-fermentable carbon source to glucose, SDH1 and SDH2 have a dramatic reduction in their mRNA half-lives, with the 5’UTR playing an important role in SDH2’s glucose-dependent mRNA decay. RBPs that bind a large number of nuclear-encoded mitochondrial proteins also impact mRNA decay (76,77). Interestingly, mRNA decay can also be directly affected by the proximity of mRNAs to the mitochondria. A significant increase in mRNA levels was found when mRNAs were tethered to the OMM, relative to mRNAs under identical promoters that were untethered to the mitochondria (31). Similarly removing a translation elongation stall, which decreases mRNA localization to the mitochondria, also drove a decrease in mRNA levels relative to an otherwise identical reporter mRNA (31). These data point to the importance of post-transcriptional regulation on impacting mitochondrial composition across growth rates and nutrient conditions.
Multifunctional RNA-binding proteins
RBPs have been found to have roles that link the interconnected processes of mRNA decay, translation, and mRNA localization with the metabolic state of the cell for nuclear-encoded mitochondrial mRNAs. The RBP, Puf3 is a well-known pumilio family RBP, conserved from yeast to humans, that binds to hundreds of mRNAs, mostly encoding proteins that support mitochondrial protein import, mitochondrial transcription and translation, and oxidative phosphorylation assembly(78,79). Puf3 is notable in that different carbon sources will have varying effects on Puf3’s ability to stimulate mRNA decay or promote translation of its target transcripts. Wild type yeast cells grown in ethanol or raffinose based medias exhibited stabilization of nuclear-encoded mitochondrial protein mRNAs that were similar to a puf3Δ strain grown in respective media, indicating that these non-fermentable sugar conditions fully inhibit Puf3’s ability to decay mRNA. Galactose-based media showed a significant inhibition of Puf3 mediated mRNA decay, but not complete inhibition (80). Furthermore, it was found that switching cells from a non-fermentable carbon source to a media containing glucose, and vice versa, could rapidly activate or inactivate Puf3 activity within 2 minutes. This indicates that Puf3’s mRNA decay capabilities are strongly based on available carbon sources, and likely stabilizes mRNA of mitochondrial proteins during respiratory conditions, as this is when functional mitochondria are most essential for cell growth (80). Further studies showed that upon glucose depletion, Puf3 is phosphorylated and begins to associate with actively translating ribosomes, promoting translation of its target mRNAs instead of deadenylation in order to support the need for increased mitochondrial biogenesis (81)
The condition dependent action of Puf3 was further explored in a study that looked at yeast cells in which both Tom20 and Puf3 were deleted, as well as strains with single deletions of either gene (70). Under fermentative conditions, all strains grew well, with no significant defects. When switched to respiratory conditions, single deletion strains grew normally, but the double deletion strain suffered severe growth defects. Adding back endogenously expressed Puf3 and Tom20 proteins to the double deletion strain led to rescue of growth, suggesting that both of these proteins are necessary for cells under respiratory conditions. This suggests that both post translational and pre-translation localization of mRNAs are essential for cell function under conditions that require highly functioning mitochondria (70).
While Puf3p is a key regulator of mitochondrial gene expression in yeast, the function of Pumilio family members does not seem to be conserved to mammalian cells. Instead, similar to Puf3, a conserved RBP clustered mitochondria homolog (CLUH) protein shows selective nuclear-encoded mitochondrial mRNA binding in mammalian cells. Loss of CLUH leads a clustered mitochondrial network close to the nucleus in several organisms (82). In mammals it was shown that ~80% of its mRNA targets encode for mitochondrial proteins (83) and upon loss of CLUH its target mRNAs show decreased stability and lower translation (77). CLUH function has been linked to cellular control of the metabolic state of the cell, as loss of CLUH leads to a decrease in respiratory function and a metabolic shift towards glucose dependency (77,84). As CLUH impacts mRNA decay and translation of proteins targeted to the mitochondria, it will be interesting to explore the role of CLUH on mRNA localization to the mitochondria, especially in relation to the large impact it has on the structure of the mitochondria.
Condition-dependent mRNA localization
Even though there have been obvious links between metabolism and RBPs impacting post-transcriptional gene expression, surprisingly, there has been little exploration into how mRNA localization to the mitochondria may be altered under different metabolic states when there are large changes in both mitochondrial morphology and gene expression. In yeast, most of the global localization experiments have been performed under non-fermentable carbon sources. It was recently found that the β and γ ATP synthase subunit mRNAs (ATP2, ATP3) can be localized to the mitochondria in a condition dependent manner. Under fermentative conditions these mRNAs have minimal mitochondrial localization, but under respiratory conditions they become strongly localized (31). Through mutant analysis and computational modeling, this localization was shown to be related to the changing geometric constraints of the cell as mitochondrial volume fraction increased from fermentation to respiration. It was also shown that increasing mitochondrial volume fraction effectively shrinks the cytoplasm, favoring random interactions with the mitochondrial membrane, which would result in altered association based on the competency of an mRNA for binding. Therefore, translation elongation and mitochondrial volume fraction were interrelated variables on impacting mRNA localization as competency for binding and sensitivity to mitochondrial volume fraction were strongly impacted by the duration that an mRNA resides as an mRNA-ribosome-nascent protein chain complex (31). One way to increase this competency is to stabilize the mRNA-ribosome-nascent protein chain complex through CHX administration.
While none of the global mRNA localization experiments have been concurrently performed in different metabolic states, a mitochondrial proximity specific ribosome profiling was performed in glucose media (fermentative) and the localization of previously characterized glycerol (respiratory) localized mRNAs can be compared (68,85). In glucose conditions, the majority of non-localized Class III mRNAs (98%) were also non-localized. For many of the Class I (Puf3-targets) and Class II (Puf3-independent) mRNAs that were localized to the mitochondria in respiratory conditions, they were not found to be localized in glucose conditions. This was especially true for the mRNAs that are most sensitive to Puf3 for their localization, Class I-1, in which only 6% of the mRNAs were categorized as mitochondrially localized in glucose conditions. This may be related to Puf3’s changing role in gene regulation, from inducing mRNA decay and translational repression in glucose conditions, to potentially being a translational activator in respiratory conditions (Table 2). For many mitochondrial mRNAs there was a strong increase in localization upon CHX treatment, including the mitochondrial volume fraction sensitive Class II mRNAs ATP2 and ATP3 (Figure 4A). Though more experiments are needed to confirm, the previous connections between mitochondrial volume fraction and translation duration imply that many of the conditionally localized Class II mRNAs would be sensitive to mitochondrial volume fraction in their localization to the mitochondria. Overall, nuclear-encoded mitochondrial mRNAs showed a robust shift in their enrichment to the mitochondria upon CHX treatment, and this was especially true for mRNAs that were localized to the mitochondria under respiratory conditions. Similarly, mammalian mitochondrial mRNAs show a robust shift in enrichment to the OMM upon CHX treatment (Figure 4B) (66). From the current limited cellular measurements, transformed cells appear to have relatively low mitochondrial volume fraction (Table 1) and are known to switch their metabolism away from respiration and towards aerobic glycolysis. It will be interesting to explore if mammalian mitochondrial mRNAs become more localized to the mitochondria under alternative metabolic states and whether changing mitochondrial volume fraction contributes to mRNA localization changes in mammalian cells.
Table 2. Localization of previously characterized respiratory localized mRNAs are compared with the localization in fermentative condition.
Mitochondrial proximity specific ribosome profiling was performed in fermentative conditions (68) and the localization of previously characterized respiratory localized mRNAs are compared (85). Class I-1 were mRNA targets that were completely dependent on Puf3 for their localization to the mitochondria. Class I-2 are Puf3 targets that could be localized independent on Puf3. Class II mRNAs were non-Puf3 targets that localized to the mitochondria in respiratory conditions. Class III mRNAs were non-mitochondrially localized mRNAs. The percentage of mitochondrial localized mRNAs with or without CHX addition was indicated. n is a number of mRNAs in each Class.
| % Mito Localized mRNAs | ||||
|---|---|---|---|---|
| Class | n | Fermentative−CHX | Fermentative+CHX | Respiratory−CHX |
| Class I-1 | 82 | 6% | 21% | 100% |
| Class I-2 | 173 | 58% | 87% | 100% |
| Class II | 224 | 34% | 59% | 100% |
| Class III | 278 | 2% | 14% | 0% |
Future directions
Many post-transcriptional processes are interconnected. Translation can impact mRNA localization and mRNA decay, but recent work has also found that mRNA localization to the mitochondria can impact mRNA decay and translation. Further mechanistic detail will be needed to disentangle these post-transcriptional mechanisms especially in relation to the impact of changing mitochondrial structure on these processes.
The Endoplasmic Reticulum (ER) is an organelle that interacts with the mitochondria through both signal transduction pathways, as well as physical membrane contact sites to help the cell regulate energy metabolism in shifting conditions (The relationship between ER and mitochondria is well described in (86–88)). Localization of mRNAs to the ER is a well-studied phenomenon that draws many parallels to that of the mitochondria. Similar to mitochondria, translation duration has been found to be important for substrate recognition and mRNA targeting to the ER (89,90). It was even proposed that global reductions in translation elongation could lead to increased targeting of suboptimal substrate proteins and that stressful conditions could lead to changes in the proteins targeted to the ER (90). Also similar to the mitochondria there are a number of mRNAs that show CHX-dependent enrichment to the ER by proximity specific ribosome profiling (91). Targeting mRNAs to the ER was also found to strongly upregulate protein synthesis of reporter mRNAs (31). Similar to the large condition-dependent changes in mitochondrial structure, the ER can also undergo extensive membrane expansion during times of ER stress or differentiation, when B lymphocytes differentiate into plasma cells (92,93). This membrane expansion is essential for maximal fitness during ER stress (92). It is unclear if this expansion of the ER changes mRNA localization to this organelle? Do these ER structural changes lead to direct effects in protein synthesis? It will be interesting to explore how the structure of organelles can directly impact the post-transcriptional regulation of mRNAs.
As previously mentioned changes in mitochondrial morphology are associated with a broad set of diseases from diabetes, to cancer, to neurodegeneration and even aging (9,94). Yet it is unclear the impact that these disease related changes in mitochondrial structure have on mRNA localization, gene expression, and mitochondrial composition. It will be important to further explore the mechanisms of mRNA localization and post-transcriptional control under alternative metabolic states and mitochondrial structures in order to better understand mitochondrial function in health and disease.
Perspectives.
Importance of the field: In fluctuating environmental conditions organisms must modulate their bioenergetic production in order to maintain cellular homeostasis for optimal fitness. While the importance of transcriptional control in these mitochondrial functional changes have been explored, post-transcriptional regulation is another important layer of regulation in the how mitochondria properly adjust their composition to the metabolic needs of the cell.
Current thinking: Intriguingly, changing mitochondrial structure may also directly impact post-transcriptional steps of gene regulation. As abnormalities in mitochondrial structure and function have been identified as key pathological components of aging and disease, it is imperative to fully understand the mechanisms of mitochondrial control.
Future directions: As mitochondrial structure and function has been shown to be very important for age-related processes, including cancer, metabolic disorders, and neurodegeneration, understanding how mitochondrial composition can be affected in fluctuating conditions can lead to new therapeutic directions to pursue.
Funding
This work was supported by startup funds from UCSD and from the National Institutes of Health R35GM128798 (to B.M.Z.). T.T. acknowledges support from a Japan Society for the Promotion of Science (JSPS) for a postdoctoral fellowship (18J00995).
Footnotes
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Reference
- 1.Scheffler IE. A century of mitochondrial research: Achievements and perspectives. Mitochondrion. 2001;1(1):3–31. [DOI] [PubMed] [Google Scholar]
- 2.Warburg O. On Respiratory Impairment in Cancer Cells. Adv Sci. 1956;124(3215):267–72. [PubMed] [Google Scholar]
- 3.De Deken RH. The Crabtree Effect: A Regulatory System in Yeast. J Gen Microbiol. 1966;44(2):149–56. [DOI] [PubMed] [Google Scholar]
- 4.Shadel GS. Yeast as a model for human mtDNA replication. Am J Hum Genet. 1999;65(5):1230–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hock MB, Kralli A. Transcriptional Control of Mitochondrial Biogenesis and Function. Annu Rev Physiol. 2009; [DOI] [PubMed] [Google Scholar]
- 6.Scarpulla RC, Vega RB, Kelly DP. Transcriptional integration of mitochondrial biogenesis. Trends in Endocrinology and Metabolism. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turcotte B, Liang XB, Robert F, Soontorngun N. Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res. 2010;10(1):2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Publ Gr [Internet]. 2010;11(12):872–84. Available from: 10.1038/nrm3013 [DOI] [PubMed] [Google Scholar]
- 9.Spinelli JB, Haigis MC. The multifaceted contributions of mitochondria to cellular metabolism. Nat Cell Biol [Internet]. 2018;20(7):745–54. Available from: 10.1038/s41556-018-0124-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisner V, Picard M, Hajnóczky G. Mitochondrial dynamics in adaptive and maladaptive cellular stress responses. Nat Cell Biol [Internet]. 2018;20(7):755–65. Available from: 10.1038/s41556-018-0133-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mishra P, Chan DC. Metabolic regulation of mitochondrial dynamics. J Cell Biol. 2016;212(4):379–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Youle RJ, Van Der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egner A, Jakobs S, Hell SW. Fast 100-nm resolution three-dimensional microscope reveals structural plasticity of mitochondria in live yeast. Proc Natl Acad Sci U S A. 2002. March;99(6):3370–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jakobs S, Martini N, Schauss AC, Egner A, Westermann B, Hell SW. Spatial and temporal dynamics of budding yeast mitochondria lacking the division component Fis1p. J Cell Sci. 2003;116(10):2005–14. [DOI] [PubMed] [Google Scholar]
- 15.Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, Capaldi RA. Energy Substrate Modulates Mitochondrial Structure and Oxidative Capacity in Cancer Cells. Cancer Res. 2004; [DOI] [PubMed] [Google Scholar]
- 16.Skulachev VP. Mitochondrial filaments and clusters as intracellular power-transmitting cables. Trends Biochem Sci. 2001;26(1):23–9. [DOI] [PubMed] [Google Scholar]
- 17.Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci U S A. 2011;108(25):10190–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomes LC, Benedetto G Di, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13(5):589–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon Y, Galloway CA, Jhun BS, Yu T. Mitochondrial dynamics in diabetes. Antioxidants and Redox Signaling. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galloway CA, Yoon Y. Mitochondrial morphology in metabolic diseases. Antioxidants Redox Signal. 2013;19(4):415–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao J, Wang L, Liu J, Xie F, Su B, Wang X. Abnormalities of Mitochondrial Dynamics in Neurodegenerative Diseases. 2017; [DOI] [PMC free article] [PubMed]
- 22.Liu YJ, McIntyre RL, Janssens GE, Houtkooper RH. Mitochondrial fission and fusion: A dynamic role in aging and potential target for age-related disease. Mech Ageing Dev [Internet]. 2020;186(January):111212. Available from: 10.1016/j.mad.2020.111212 [DOI] [PubMed] [Google Scholar]
- 23.Hughes AL, Gottschling DE. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature [Internet]. 2012. November 21 [cited 2012 Nov 22];492(7428):261–5. Available from: http://www.nature.com/doifinder/10.1038/nature11654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheckhuber CQ, Erjavec N, Tinazli A, Hamann A, Nyström T, Osiewacz HD. Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat Cell Biol [Internet]. 2007;9(1):99–105. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17173038 [DOI] [PubMed] [Google Scholar]
- 25.Chaudhari SN, Kipreos ET. Increased mitochondrial fusion allows the survival of older animals in diverse C. Elegans longevity pathways. Nat Commun [Internet]. 2017;8(1). Available from: 10.1038/s41467-017-00274-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rana A, Oliveira MP, Khamoui AV., Aparicio R, Rera M, Rossiter HB, et al. Promoting Drp1-mediated mitochondrial fission in midlife prolongs healthy lifespan of Drosophila melanogaster. Nat Commun [Internet]. 2017;8(1). Available from: 10.1038/s41467-017-00525-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H, Chan DC. Mitochondrial Dynamics in Regulating the Unique Phenotypes of Cancer and Stem Cells. Cell Metabolism. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knott AB, Perkins G, Schwarzenbacher R, Bossy-Wetzel E. Mitochondrial fragmentation in neurodegeneration. Nature Reviews Neuroscience. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buschlen S, Amillet JM, Guiard B, Fournier A, Marcireau C, Bolotin-Fukuhara M. The S. cerevisiae HAP complex, a key regulator of mitochondrial function, coordinates nuclear and mitochondrial gene expression. Comp Funct Genomics. 2003;4(1):37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jornayvaz FR, Shulman GI. Regulation of mitochondrial biogenesis. Essays Biochem. 2010;47:69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsuboi T, Viana MP, Xu F, Yu J, Chanchani R, Arceo XG, et al. Mitochondrial volume fraction and translation duration impact mitochondrial mRNA localization and protein synthesis. Elife. 2020;9:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madesh M, Zong W-X, Hawkins BJ, Ramasamy S, Venkatachalam T, Mukhopadhyay P, et al. Execution of Superoxide-Induced Cell Death by the Proapoptotic Bcl-2-Related Proteins Bid and Bak. Mol Cell Biol. 2009. June;29(11):3099 LP–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh I, Tsang KY, Ludwig GD. Alterations in the Mitochondria of Human Osteosarcoma Cells with Glucocorticoids. Cancer Res. 1974. November;34(11):2946 LP–2952. [PubMed] [Google Scholar]
- 34.Posakony JW, England JM, Attardi G. Mitochondrial growth and division during the cell cycle in HeLa cells. J Cell Biol. 1977; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barth E, Stämmler G, Speiser B, Schaper J. Ultrastructural quantitation of mitochondria and myofilaments in cardiac muscle from 10 different animal species including man. J Mol Cell Cardiol. 1992; [DOI] [PubMed] [Google Scholar]
- 36.Sullivan SM, Pittman RN. Relationship between mitochondrial volume density and capillarity in hamster cells. Am J Physiol - Hear Circ Physiol. 1987; [DOI] [PubMed] [Google Scholar]
- 37.Helle SCJ, Feng Q, Aebersold MJ, Hirt L, Grüter RR, Vahid A, et al. Mechanical force induces mitochondrial fission. Elife. 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: An updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44(D1):D1251–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgenstern M, Stiller SB, Lübbert P, Peikert CD, Dannenmaier S, Drepper F, et al. Definition of a High-Confidence Mitochondrial Proteome at Quantitative Scale. Cell Rep. 2017;19(13):2836–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paulo JA, O’Connell JD, Everley RA, O’Brien J, Gygi MA, Gygi SP. Quantitative mass spectrometry-based multiplexing compares the abundance of 5000 S. cerevisiae proteins across 10 carbon sources. J Proteomics [Internet]. 2016;148:85–93. Available from: 10.1016/j.jprot.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tait SWG, Green DR. Mitochondria and cell signalling. J Cell Sci. 2012;125(4):807–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Di Bartolomeo F, Malina C, Campbell K, Mormino M, Fuchs J, Vorontsov E, et al. Absolute yeast mitochondrial proteome quantification reveals trade-off between biosynthesis and energy generation during diauxic shift. Proc Natl Acad Sci U S A. 2020;117(13):7524–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stan T Recognition of preproteins by the isolated TOM complex of mitochondria. EMBO J. 2000; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papic D, Elbaz-Alon Y, Koerdt SN, Leopold K, Worm D, Jung M, et al. The Role of Djp1 in Import of the Mitochondrial Protein Mim1 Demonstrates Specificity between a Cochaperone and Its Substrate Protein. Mol Cell Biol. 2013; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hansen KG, Aviram N, Laborenz J, Bibi C, Meyer M, Spang A, et al. An ER surface retrieval pathway safeguards the import of mitochondrial membrane proteins in yeast. Science (80-). 2018; [DOI] [PubMed] [Google Scholar]
- 46.Wiedemann N, Pfanner N. Mitochondrial Machineries for Protein Import and Assembly. Annu Rev Biochem. 2017; [DOI] [PubMed] [Google Scholar]
- 47.Mokranjac D, Neupert W. The many faces of the mitochondrial TIM23 complex. Biochimica et Biophysica Acta - Bioenergetics. 2010. [DOI] [PubMed] [Google Scholar]
- 48.Jensen RE, Dunn CD. Protein import into and across the mitochondrial inner membrane: Role of the TIM23 and TIM22 translocons. Biochimica et Biophysica Acta - Molecular Cell Research. 2002. [DOI] [PubMed] [Google Scholar]
- 49.Tucker K, Park E. Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution. Nat Struct Mol Biol. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bausewein T, Mills DJ, Langer JD, Nitschke B, Nussberger S, Kühlbrandt W. Cryo-EM Structure of the TOM Core Complex from Neurospora crassa. Cell. 2017;170(4):693–700.e7. [DOI] [PubMed] [Google Scholar]
- 51.Shiota T, Imai K, Qiu J, Hewitt VL, Tan K, Shen HH, et al. Molecular architecture of the active mitochondrial protein gate. Science (80-). 2015; [DOI] [PubMed] [Google Scholar]
- 52.Model K, Prinz T, Ruiz T, Radermacher M, Krimmer T, Kühlbrandt W, et al. Protein translocase of the outer mitochondrial membrane: Role of import receptors in the structural organization of the TOM complex. J Mol Biol. 2002; [DOI] [PubMed] [Google Scholar]
- 53.Model K, Meisinger C, Kühlbrandt W. Cryo-Electron Microscopy Structure of a Yeast Mitochondrial Preprotein Translocase. J Mol Biol. 2008; [DOI] [PubMed] [Google Scholar]
- 54.Künkele KP, Heins S, Dembowski M, Nargang FE, Benz R, Thieffry M, et al. The preprotein translocation channel of the outer membrane of mitochondria. Cell. 1998; [DOI] [PubMed] [Google Scholar]
- 55.Araiso Y, Tsutsumi A, Qiu J, Imai K, Shiota T, Song J, et al. Structure of the mitochondrial import gate reveals distinct preprotein paths. Nature. 2019; [DOI] [PubMed] [Google Scholar]
- 56.George R, Beddoe T, Landl K, Lithgow T. The yeast nascent polypeptide-associated complex initiates protein targeting to mitochondria in vivo. Proc Natl Acad Sci U S A. 1998;95(5):2296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lesnik C, Cohen Y, Atir-Lande A, Schuldiner M, Arava Y. OM14 is a mitochondrial receptor for cytosolic ribosomes that supports co-translational import into mitochondria. Nat Commun [Internet]. 2014;5:1–10. Available from: 10.1038/ncomms6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gold VA, Chroscicki P, Bragoszewski P, Chacinska A. Visualization of cytosolic ribosomes on the surface of mitochondria by electron cryo-tomography. EMBO Rep [Internet]. 2017;18(10):e201744261. Available from: http://embor.embopress.org/lookup/doi/10.15252/embr.201744261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buxbaum AR, Haimovich G, Singer RH. In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol [Internet]. 2014. December 30 [cited 2014 Dec 31];(December). Available from: http://www.nature.com/doifinder/10.1038/nrm3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Besse F, Ephrussi A. Translational control of localized mRNAs: Restricting protein synthesis in space and time. Nature Reviews Molecular Cell Biology. 2008. [DOI] [PubMed] [Google Scholar]
- 62.Marc P, Margeot A, Devaux F, Blugeon C, Corral-Debrinski M, Jacq C. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep [Internet]. 2002. February;3(2):159–64. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1083966&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saint-Georges Y, Garcia M, Delaveau T, Jourdren L, Le Crom S, Lemoine S, et al. Yeast mitochondrial biogenesis: a role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS One [Internet]. 2008. January;3(6):e2293. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18523582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garcia M, Darzacq X, Delaveau T, Jourdren L, Singer RH, Jacq C. Mitochondria-associated Yeast mRNAs and the Biogenesis of Molecular Complexes. Mol Biol Cell. 2007;18(February):362–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gadir N, Haim-vilmovsky L, Kraut-cohen J, Gerst JE. Localization of mRNAs coding for mitochondrial proteins in the yeast Saccharomyces cerevisiae. RNA. 2011; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fazal FM, Han S, Parker KR, Kaewsapsak P, Xu J, Boettiger AN, et al. Atlas of Subcellular RNA Localization Revealed by APEX-Seq. Cell [Internet]. 2019;11:1–18. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0092867419305550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gehrke S, Wu Z, Klinkenberg M, Sun Y, Auburger G, Guo S, et al. PINK1 and parkin control localized translation of respiratory chain component mRNAs on mitochondria outer membrane. Cell Metab [Internet]. 2015;21(1):95–108. Available from: 10.1016/j.cmet.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Williams CC, Jan CH, Weissman JS. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014;748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sylvestre, Vialette Stéphane MCD and CJ, Jacq C. Long mRNAs coding for yeast mitochondrial proteins of prokaryotic origin preferentially localize to the vicinity of mitochondria Julien Sylvestre, Stéphane Vialette, Marisol Corral Debrinski and. Genome Biol. 2003;1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eliyahu E, Pnueli L, Melamed D, Scherrer T, Gerber AP, Pines O, et al. Tom20 Mediates Localization of mRNAs to Mitochondria in a Translation-Dependent Manner. Mol Cell Biol [Internet]. 2010;30(1):284–94. Available from: http://mcb.asm.org/cgi/doi/10.1128/MCB.00651-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y, Chen Y, Gucek M, Xu H. The mitochondrial outer membrane protein MDI promotes local protein synthesis and mtDNA replication. EMBO J. 2016;35(10):1045–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kershaw CJ, Costello JL, Castelli LM, Talavera D, Rowe W, Sims PFG, et al. The Yeast La Related Protein Slf1p Is a Key Activator of Translation during the Oxidative Stress Response. PLoS Genet [Internet]. 2015. January [cited 2015 Jan 12];11(1):e1004903. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25569619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chatenay-Lapointe M, Shadel GS. Repression of mitochondrial translation, respiration and a metabolic cycle-regulated gene, SLF1, by the yeast Pumilio-family protein Puf3p. PLoS One. 2011;6(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.García-Martínez J, Delgado-Ramos L, Ayala G, Pelechano V, Medina DA, Carrasco F, et al. The cellular growth rate controls overall mRNA turnover, and modulates either transcription or degradation rates of particular gene regulons. Nucleic Acids Res. 2016;44(8):3643–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de la Cruz BJ, Prieto S, Scheffler IE. The role of the 5’ untranslated region (UTR) in glucose-dependent mRNA decay. Yeast [Internet]. 2002. July;19(10):887–902. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12112242 [DOI] [PubMed] [Google Scholar]
- 76.Olivas W The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 2000;19(23):6602–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schatton D, Pla-Martin D, Marx MC, Hansen H, Mourier A, Nemazanyy I, et al. CLUH regulates mitochondrial metabolism by controlling translation and decay of target mRNAs. J Cell Biol. 2017;216(3):675–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lapointe CP, Stefely JA, Jochem A, Hutchins PD, Wilson GM, Kwiecien NW, et al. Multi-omics Reveal Specific Targets of the RNA-Binding Protein Puf3p and Its Orchestration of Mitochondrial Biogenesis. Cell Syst [Internet]. 2018;6(1):125–135.e6. Available from: 10.1016/j.cels.2017.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller MA, Russo J, Fischer AD, Leban FAL, Olivas WM. Carbon source-dependent alteration of Puf3p activity mediates rapid changes in the stabilities of mRNAs involved in mitochondrial function. Nucleic Acids Res. 2014;42(6):3954–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Der Lee C, Tu BP. Glucose-Regulated Phosphorylation of the PUF Protein Puf3 Regulates the Translational Fate of Its Bound mRNAs and Association with RNA Granules. Cell Rep [Internet]. 2015;11(10):1638–50. Available from: 10.1016/j.celrep.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schatton D, Rugarli EI. A concert of RNA-binding proteins coordinates mitochondrial function. Crit Rev Biochem Mol Biol [Internet]. 2018;53(6):652–66. Available from: 10.1080/10409238.2018.1553927 [DOI] [PubMed] [Google Scholar]
- 83.Gao J, Schatton D, Martinelli P, Hansen H, Pla-Martin D, Barth E, et al. CLUH regulates mitochondrial biogenesis by binding mRNAs of nuclear-encoded mitochondrial proteins. J Cell Biol. 2014;207(2):213–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wakim J, Goudenege D, Perrot R, Gueguen N, Desquiret-Dumas V, de la Barca JMC, et al. CLUH couples mitochondrial distribution to the energetic and metabolic status. J Cell Sci. 2017;130(11):1940–51. [DOI] [PubMed] [Google Scholar]
- 85.García-Rodríguez LJ, Gay AC, Pon LA. Puf3p, a Pumilio family RNA binding protein, localizes to mitochondria and regulates mitochondrial biogenesis and motility in budding yeast. J Cell Biol. 2007;176:197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Phillips MJ, Voeltz GK. Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol. 2016;17(2):69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xia MF, Zhang YZ, Jin K, Lu ZT, Zeng Z, Xiong W. Communication between mitochondria and other organelles: a brand-new perspective on mitochondria in cancer. Cell Biosci [Internet]. 2019;9(1):1–19. Available from: 10.1186/s13578-019-0289-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stacchiotti A, Favero G, Lavazza A, Garcia-Gomez R, Monsalve M, Rezzani R. Perspective: Mitochondria-ER Contacts in Metabolic Cellular Stress Assessed by Microscopy. Cells. 2018;8(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pechmann S, Chartron JW, Frydman J. Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo. Nat Publ Gr [Internet]. 2014;21(12):1100–5. Available from: 10.1038/nsmb.2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang D, Shan SO. Translation elongation regulates substrate selection by the signal recognition particle. J Biol Chem. 2012;287(10):7652–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jan CH, Williams CC, Weissman JS. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science (80-) [Internet]. 2014. November 6 [cited 2014 Nov 6];346(6210):1257521–1257521. Available from: http://www.sciencemag.org/cgi/doi/10.1126/science.1257521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schuck S, Prinz WA, Thorn KS, Voss C, Walter P. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J Cell Biol. 2009;187(4):525–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wiest DL, Burkhardt JK, Hester S, Hortsch M, Meyer DI, Argon Y. Membrane biogenesis during B cell differentiation: Most endoplasmic reticulum proteins are expressed coordinately. J Cell Biol. 1990;110(5):1501–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu T, Chen J, Wang Q, Shao W, Qi B. Modulation of Mitochondrial Dynamics in Neurodegenerative Diseases : An Insight Into Prion Diseases. Front Aging Neurosci. 2018;10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]


