Version Changes
Revised. Amendments from Version 1
Revisions have been made to this manuscript to address comments raised by the reviewers. We have specifically - Added a table (table 4) to summarise the characteristics of the test methods being evaluated here - Added specific details on Salmonella stool culture and Taqman Array Card assay - Added more background information on the specificity of ttr and InvA - Added information on the genomic prevalence of ttr and InvA gene in different Salmonella serotypes - Provided more information on the statistical analysis used -Proofread the manuscript and revised accordingly -Clearly stated the limitations and conclusion
Abstract
Background: The relationship between asymptomatic Salmonella exposure within the gastrointestinal tract and Salmonella bacteraemia is poorly understood, in part due to the low sensitivity of stool culture and the lack of validated molecular diagnostic tests for the detection of Salmonella in the stool. The study aimed to determine a reliable molecular diagnostic test for Salmonella in stool specimens.
Methods: We optimised an in-house monoplex real-time polymerase chain reaction (PCR) for the detection of Salmonella ttr and InvA genes in stool by including a selenite broth pre-culture step for Salmonella before DNA extraction and validated their specificity against other local common pathogens. Then we assessed their performance against a well-validated multiplex PCR targeting the same ttr and InvA genes and against stool culture using clinical stool specimens collected from a cohort of 50 asymptomatic healthy Malawian children that were sampled at 1-month intervals over 12 months. We employed a latent Markov model to estimate the specificities and sensitivities of PCR methods.
Results: Ttr and InvA primers were both able to detect all the different Salmonella serovars tested and had superior limits of detection when DNA was extracted after selenite pre-culture. T tr sensitivity and specificity for monoplex-PCR were (99.53%, 95.46%) and for multiplex-PCR (90.30%, 99.30%) respectively. InvA specificity and specificity for using monoplex-PCR was (95.06%, 90.31%) and multiplex-PCRs (89.41%, 98.00%) respectively. Sensitivity and specificity for standard stool culture were 62.88% and 99.99%, respectively. Culture showed the highest PPV (99.73%), and monoplex- ttr had the highest NPV (99.67%).
Conclusion: Test methods demonstrated high concordance, although stool culture and monoplexed ttr primers had superior specificity and sensitivity, respectively. The use of selenite pre-enrichment step increased Salmonella detection rate. Taken together, molecular detection methods used here could be used to reveal the true extent of both asymptomatic and symptomatic Salmonella exposure events.
Keywords: Salmonella Typhi, nontyphoidal Salmonella, bacteremia, gastrointestinal tract, diagnostics, stool culture, polymerase chain reaction
Introduction
Salmonellae cause a huge global burden of morbidity and mortality. They are globally estimated to be responsible for 300,000 deaths 1– 4. Salmonella enterica serovars Typhi and Paratyphi A are the predominant cause of invasive Salmonella infections in south and southeast Asia and cause between 129,000 to 223,000 global deaths per year 1, 3, 5. In contrast, non-Typhoidal Salmonella (NTS) serovars, principally S. Typhimurium and S. Enteritidis, are a common cause of invasive disease in sub-Saharan Africa (SSA) 4, 6. In 2017, NTS caused an estimated 535,000 cases, with SSA having the highest incidence 7. Risk factors for invasive NTS (iNTS) disease include young age, recent malaria, and advanced HIV disease. Case fatality rates for iNTS in young children, people infected with HIV, and living in the SSA region were estimated at 13.5%, 41.8%, and 15.8%, respectively 4. This is in marked contrast to the presentation of Salmonella disease in high-income countries, where NTS typically cause a self-limiting diarrhoeal disease in healthy individuals, while bloodstream or focal infections are rare and mainly occur in individuals with specific risk factors such as diabetes, neoplastic and autoimmune disease, or immunosuppressive therapy 8. However, it is notable in both settings that invasive NTS disease in adults and children is not always associated with diarrhoea 9.
We previously described in under-five-year-old children the sequential development of cellular and humoral immunity against the Salmonella serovars causing iNTS disease, and that acquisition of this immunity is associated with decreasing incidence of disease 10, 11, suggesting that this immunity is protective. Previous studies have reported that healthy young children experience transient asymptomatic episodes of gastrointestinal infection with non-typhoidal Salmonella 12, 13, and we, therefore, hypothesise that episodes of asymptomatic Salmonella exposure in the healthy gastrointestinal tract during early childhood may facilitate the development of protective immunity. Balanced against this beneficial effect of exposure, diarrhoeal disease results from enteric Salmonella exposure, and invasive NTS disease also follows episodes of asymptomatic gastrointestinal exposure in susceptible children, including those with malaria or malnutrition, or immunocompromised individuals.
Elucidating the relationship between Salmonella exposure events within the gastrointestinal tract and resultant Salmonella immunity or Salmonella disease is critical for understanding iNTS disease pathogenesis. Lack of affordable and rapid diagnostic tools for the detection of bloodstream and intestinal Salmonella disease hampers our understanding of Salmonella disease epidemiology and pathogenesis. Blood culture is considered the gold standard diagnostic test for Salmonella bacteremia and is highly specific but has a number of drawbacks; poor turnaround time of between 2 to 7 days, and low sensitivity of about 20% – 30% for samples collected 7 days post-infection 14– 16. Molecular detection of Salmonella in blood also has limited apparent sensitivity, and various assays are in development 13, 17.
Stool culture is similarly considered the gold standard test for the detection of Salmonella in the intestinal tract. However, stool culture, even for diarrhoeal disease when the bacterial load is likely to be high, has poor sensitivity (<50%), and is labour and time-consuming 18. Real-time PCR has a short turnaround time and is potentially highly sensitive compared to standard culture, and has the capacity for automation and testing for multiple targets 19. However, stool PCR test performance is hindered by PCR inhibitors and a large number of genetically closely related enteric bacteria. These pose a challenge in generating highly specific and sensitive primers for real-time PCR (qPCR) for Salmonella. Furthermore, a lower infective load of Salmonella colonisation during asymptomatic infection may further limit detection by PCR.
With this background, we validated an in-house monoplex qPCR method for the detection of Salmonella in stool specimens and compared them with a validated multiplex-based qPCR and standard stool culture. Both qPCR assays used primers and probes based on the Salmonella tetrathionate respiration gene ( ttr) and the Salmonella invasion gene A ( InvA). Stool specimens were collected from healthy, mainly asymptomatic healthy Malawian children aged 6–18 months. Assessing a diagnostic test's performance is challenging when the existing “gold standard” test being used has known low sensitivity or specificity. Statistical methods, such as the Latent Markov model, are used to assess diagnostic tests' performance without assigning a gold standard test. Since the current reference standard is known to lack sensitivity, we employed a latent Markov model to estimate the specificities and sensitivities of PCR methods without assigning a gold standard.
Methods
Description of study participants and specimens
Stool specimens collected from a longitudinal cohort of children aged 6 – 18 months recruited from Zingwangwa Health Centre (ZHC) in Blantyre, Malawi, were used to compare the performance of molecular and standard culture for detection of Salmonella in stool. The primary study started recruitment in August 2013, and follow-up was concluded in December 2014 17. Group sensitisation of the study by well-trained study nurses was done to parents or guardians of six-month-old children attending a vaccination clinic at ZHC. Individual sensitisation was also done to parents or guardian that were interested in joining the study. Children who met the inclusion criteria of being healthy were recruited into the study after obtaining consent. Children born preterm (less than 38 weeks gestation), HIV positive or HIV exposed, and those with fever >38°C or any acute illness were excluded from the study 20.
Stool samples were collected monthly until they were aged 18 months. Stool specimens were collected in sterile and clean containers and transported to the laboratory on the same day. From 60 children recruited at 6 months of age, 10 children withdrew from the study, and 600 stool specimens were collected and tested by culture on the day of sample collection at the College of Medicine and Malawi Liverpool Wellcome Laboratory. Molecular tests were done on frozen samples that were available at the time the tests were done.
Salmonella stool culture
A matchstick head-size sample of stool was inoculated in 10 ml of selenite F broth (Oxoid, UK, catalog number: 2300631) and aerobically incubated overnight at 37 °C for 18–24 hours. The top layer (1 ml) of an overnight culture was spun at 20,000 g for 5 minutes. This is a method that has been developed in our laboratory. A 1 ul loop was used to subculture Salmonella from the pellet by spreading on Xylose Lysine Deoxycholate (XLD) agar (Oxoid, UK, catalog number: 2547703) to achieve single colonies. Careful plate spreading prevents overcrowding of colonies. A single colony was picked and cultured on sheep blood agar. An aliquot of the selenite broth was also frozen for molecular detection. A single colony of presumptive Salmonella was cultured onto sheep blood agar (Oxoid, UK, catalog number: 2910831) and MacConkey agar plates (Oxoid, UK, catalog number: 2529552) and incubated aerobically at 37°C for 18–24 hours. Salmonella colonies were then distinguished from other enteric bacteria (i.e., Citrobacter and Serratia) using triple sugar iron agar (Oxoid, UK, catalog number: 1882283) and Urea agar (Oxoid, UK, catalog number: 1779617) biochemical tests. Further Salmonella identification was determined using API® 10S (bioMérieux, France, catalog number: 1007181060) according to the manufacturer’s instructions.
Monoplex-qPCR ttr and InvA assay
Validation of the monoplex- qPCR ttr and InvA assay. For the monoplex-qPCR, the ttr primers and probe were designed and validated by Federal Institute for Risk Assessment, Berlin, Germany, according to the published DNA sequence of the S. enterica serotype Typhimurium ttr locus for Salmonella detection (GenBank accession no. AF282268). The use of ttr was based on 21. In this study, the ttr gene's specificity was assessed using 110 Salmonella strains representing 31 serotypes, and it demonstrated 100% specificity. The InvA gene has been widely studied and used as a pan Salmonella marker. In 22, the specificity of the InvA gene was assessed using 242 Salmonella strains representing 43 serotypes. It also demonstrated 100% specificity. InvA has shown 100% specificity in several other studies. Both primers required optimisation for use in stool specimens. The DNA sequences of all the primers and probes used in this study are listed in Table 1.
Table 1. List of primers and probes sequences used in this study.
Primers and probes sequences used in this study include in-house designed InvA, ttr previously validated for Salmonella detection in food, and TAC- InvA and TAC- ttr used on a well-validated TAC assay as pan Salmonella primers.
| Primer name | Primer direction | Primer code/Probe description | |
|---|---|---|---|
| 1 | INVA | Forward | 5’-AGCGTACTGGAAAGGGAAAG-3’ |
| 2 | INVA | Reverse | 5’-CACCGAAATACCGCCAATAAAG-3’ |
| 3 | INVA | Probe | FAM-TTACGGTTCCTTTGACGGTGCGAT-BHQ1 |
| 4 | ttr | Forward | 5’-CTCACCAGGAGATTACAACATGG-3’ |
| 5 | ttr | Reverse | 5’-AGCTCAGACCAAAAGTGACCATC-3’ |
| 6 | ttr | Probe | FAM-CACCGACGGCGAGACCGACTTT-BHQ1 |
| 7 | InvA-TAC | Forward | 5’-GGCAATTCGTTATTGGCGATA-3’ |
| 8 | InvA-TAC | Reverse | 5’-CACGGTGACAATAGAGAAGACAACA-3’ |
| 9 | InvA-TAC | Probe | FAM-CCTGGCGGTGGGTT-MGB |
| 10 | ttr-TAC | Forward | 5’-CTCACCAGGAGATTACAACATGG-3’ |
| 11 | ttr-TAC | Reverse | 5’-AGCTCAGACCAAAAGTGACCATC-3’ |
| 12 | ttr-TAC | Probe | FAM-CACCGACGGCGAGACCGACTTT-MGB |
Specificity of ttr and InvA primer/probe set for Salmonella compared to other local pathogens
To determine the prevalence of ttr and InvA in the genomes of Salmonella serotypes, NCBI nucleotide blast was conducted (10 February 2021). Ttr and InvA nucleotide sequences were used as the query sequence against Salmonella enterica subspecies enterica (taxid:59201) genomes with a maximum target sequence set at 1000. Both primer sequences demonstrated 100% identity for most commonly isolated Salmonella serotypes (supplementary material). To determine the specificity of the primers in vitro, 9 different locally isolated and whole genome sequenced Salmonella strains and 26 pure isolates of non- Salmonella bacterial strains locally isolated from blood culture were tested using ttr and InvA primer/probe sets ( Table 2). These 9 Salmonella strains represented all known Salmonella serotypes in the MLW isolate archive. The non- Salmonella strains were chosen because they are genetically closely related to Salmonella or because their growing conditions are similar to Salmonella. These strains were collected from MLW bacterial blood culture repository. Overnight cultures of the frozen samples were made on SBA or LB agar. One colony was then cultured in liquid media. After reaching stationary growth phase, a known and matched concentration of about 10 6 CFU was used for DNA extraction using QIAamp Fast DNA Stool Mini Kit (QIAGEN, Netherlands, catalog number: 51604) but without the bead beating step. Miles and Misra technique was used for bacteria quantification.
Table 2. Bacterial organisms tested for the specificity of ttr and InvA primer/probe sets.
Bacterial organisms used in this study to test for the specificity of ttr and InvA primer/ probe sets. Nine Salmonella and 26 non- Salmonella isolates previously isolated at MLW laboratory were retrieved and tested either as direct or selenite sub-cultured isolates.
| Bacteria isolates | Number
tested |
Direct | Selenite sub-
cultured |
||
|---|---|---|---|---|---|
|
ttr
Positive |
InvA
Positive |
ttr
Positive |
InvA
Positive |
||
| Morganella morgana | 1 | 0 | 0 | 0 | 0 |
| Streptococcus pneumonia | 1 | 0 | 0 | 0 | 0 |
| Staphylococcus aureus | 1 | 0 | 0 | 0 | 0 |
| Citrobacter | 1 | 0 | 0 | 0 | 0 |
| Klebsiella | 1 | 0 | 0 | 0 | 0 |
| Enterobacter | 1 | 0 | 0 | 0 | 0 |
| Acinetobacter | 1 | 0 | 0 | 0 | 0 |
| Enterobacter intermedius | 1 | 0 | 0 | 0 | 0 |
| Enterococcus feacium | 1 | 0 | 0 | 0 | 0 |
| E. coli | 17 | 0 | 0 | 0 | 0 |
| S. Typhi | 1 | 1 | 1 | 1 | 1 |
| S. Typhimurium | 1 | 1 | 1 | 1 | 1 |
| S. Enteritidis | 1 | 1 | 1 | 1 | 1 |
| S. Braenderup | 1 | 1 | 1 | 1 | 1 |
| S. Virchow | 1 | 1 | 1 | 1 | 1 |
| S. Bonn/Fann | 1 | 1 | 1 | 1 | 1 |
| S. Oesterbro/Zanzibar | 1 | 1 | 1 | 1 | 1 |
| S. Heidelberg | 1 | 1 | 1 | 1 | 1 |
| S. Dublin | 1 | 1 | 1 | 1 | 1 |
Limits of detection in different conditions A well-characterised invasive S. Typhimurium ST313 strain (D23580), isolated from an HIV-negative child in Malawi, and representative of our commonest invasive bloodstream infections, was used as a reference strain for determining limits of detection in varying kinds of sample 23, 24. Three types of Salmonella samples were prepared for comparison using RT-PCR; 1) pure Salmonella isolates picked from a blood agar plate, 2) Salmonella cultured in selenite broth, and 3) Salmonella spiked into stool. Salmonella stool spiking was done to determine the inhibitory effect that stool may have on the assay, affecting the limit of detection. For this, a stool sample was collected from a healthy individual and confirmed as Salmonella negative by culture. The stool sample was thereafter diluted with PBS (50% w/v) and then spiked with S. Typhimurium, D23580, at varying doses of viable bacteria . The viable dose of Salmonella was adjusted across a range from 10 0–10 6 CFU/ml and quantified using Miles and Misra technique. DNA was extracted for RT-PCR, as above. All experiments were repeated three times on different days by the same operator.
Detection of Salmonella in clinical samples using monoplex-qPCR ttr and InvA assay. The primer/probe sets were then used to detect Salmonella in clinical stool samples collected from the longitudinal cohort study of healthy asymptomatic children. For the monoplex qPCR, approximately 200μl top layer of frozen Selenite F broth overnight stool culture, or 200 mg of stool, was suspended in 500 μl of PBS. DNA was extracted using QIAamp Fast DNA Stool Mini Kit (QIAGEN, Netherlands, catalog number: 51604) according to the manufacturer’s instructions, with an added bead-beating step. Eluted DNA was stored at –20°C.
A previously-optimised in-house PCR protocol was used 17. Briefly, the master mix for RT-PCR was prepared using pre-defined quantities. A total of 20μl master-mix for each sample was comprised of the following: 12.5μl Platinum ® Quantitative PCR Super Mix-UDG (Life Technologies, USA, Catalog number: 11730025), 0.10μl specific forward primer, 0.10 specific reverse primer, 0.10 specific probe (all primers and probes at 200nM), 0.05μl ROX reference dye (Life Technologies, USA, Catalog number: 12223012) at 50nM final concentration, and 7.15μl nuclease-free water. This mixture was transferred to 96-well plate PCR wells. 5μl of test DNA, positive controls DNA (DNA from D23580), technical extraction negative control, and assay negative control (UV treated water) were added in triplicates to appropriate wells containing 20ul of master-mix. The qPCR was run for 40 cycles using Applied Biosystems® 7500 Real-Time PCR Systems (Life Technologies, USA). The following cycling conditions were used; initial denaturation at 95°C for 1 minute, denaturation at 95°C for 15 seconds, annealing/extension at 60°C for 30 seconds, final extension: 12°C. The threshold was set in the lag phase. An assay was considered to have passed when the positive controls were positive, and both the technical extraction negative and assay negative controls were negative. Test sample cycle threshold (Ct) values were evaluated after subtracting the baseline value. Samples with cycle threshold (Ct) values of less than or equal to 35 were considered positive.
Detection of Salmonella using multiplex qPCR assay
As a comparator, we used a well-validated TAC assay on DNA extracted from stool samples, according to the manufacturer’s protocol. The customised Taqman Array Card assay developed and validated at the University of Virginia was used. The performance of the TAC method has been previously described and has now been widely used 12, 25– 27. It is used to detect multiple enteric pathogens, including bacteria, viruses, protozoa, and helminths. Targets included on the TAC card for pan Salmonella detection are InvA and ttr 28. Phocine Herpesvirus (PHhv) and MS2 targets are included as internal positive controls.
To extract total nucleic acid (TNA) from the clinical samples for TAC assay, we used QIAamp Fast DNA Stool Mini Kit (QIAGEN, Netherlands, catalog number: 51604) - the same DNA extraction kit and protocol that were used to extract whole-stool DNA for the monoplex qPCR assay, with the addition of internal extraction positive controls. For TNA extraction, each sample was extracted together with internal positive controls, Phocine Herpesvirus (PHhv), and MS2. PHhv and MS2 were added to the inhibitX buffer before being added to each sample, as previously described 28. An assay was considered to have passed when both MS2 and PhHv internal positive (amplification crossing the threshold) and negative controls (no amplification crossing the threshold line) passed and when the sample had a sigmoid curve that crossed the threshold line. A sample was classified as pathogen positive at a Ct value of <35. Only results for Salmonella are reported here.
Statistical analysis
Data were recorded and analysed in MS Excel (version 16.14.1 (18061302)). Sensitivities and specificities of the different PCR methods were estimated using a latent Markov model (LMM) 29. We have previously described the LMM and various extensions that we considered for modeling longitudinal diagnostic test data 30. We implemented the LMM within a Bayesian framework using R (version v3.5.1) and JAGS (version 4.3.0) via the rjags (version 4.6) R package 31. LMMs have been extensively used for discrete-time longitudinal data in the absence of a gold standard diagnostic procedure 32, 33. LMMs consists of a process model for a latent condition (in our case, the unobserved true infection status) evolving over time and a measurement model for the observed outputs (in our case, the results from the 5 diagnostic test methods) conditional on the latent state. We considered several LMMs, with and without mixed effects and with either time-homogeneous or time-heterogeneous transition matrices 30. Convergence and identifiability of the LMM were checked by inspecting trace plots and computing Gelman-Rubin potential scale reduction factors 34. The more complex models exhibited poor mixing or convergence of MCMC chains (most likely due to the sparse number of positive samples). As a result, the LMM we used for this dataset is a basic LMM with no random effects and a time-homogeneous transition matrix. To report positive predictive values (PPV) and negative predictive values (NPV), we calculated an estimate of the infection prevalence. For the Bayesian LMM, we report maximum a posteriori (MAP) parameter estimates together with 95% credible intervals (Crl), specifically the highest posterior density intervals (HDI) with 95% coverage. All other analyses report (frequentist) parameter estimates and corresponding 95% confidence estimates (CI).
Ethical considerations
Ethical approval for this work was granted by the University of Malawi, College of Medicine Research Ethics Committee (P.01/13/1327). Written informed consent was obtained from the parent or guardian of each participating child.
Results
Ttr and InvA primers for Salmonella do not cross-react with closely related enteric micro-organisms
We first validated the ttr and InvA primers that were used in the monoplex-qPCR assay by assessing the sensitivity and specificity of the primers for Salmonella, using a standardised number of 10 0–10 6 CFU/ml of 9 different locally-relevant Salmonella strains and 26 non- Salmonella bacterial strains as indicated in Table 2. We included 17 strains of E. coli because of the close genomic relatedness of Salmonella and E. coli. Bacterial isolates enriched in Selenite F broth (referred here as selenite sub-cultured) or not (referred here as direct culture) were used in this evaluation. We found that ttr and InvA assays both achieved 100% sensitivity and specificity either as direct isolates or selenite sub-cultured isolates. Table 2 demonstrates that all Salmonella strains tested positive with both monoplexed primer pairs, and all other bacterial strains were negative, confirming a lack of cross-reactivity. Additionally, the NCBI nucleotide blast demonstrated that the genomic prevalence of ttr and InvA genes is high. Both primer sequences demonstrated 100% identity for most commonly isolated Salmonella serotypes.
Selenite broth culture enhances the detection of Salmonella in stool using either ttr or InvA primers
The limits of detection (LOD) of qPCR for Salmonella were then determined using S. Typhimurium strain D23580 serially diluted and tested as direct isolates, selenite broth cultured samples, or isolates spiked into a culture-negative stool specimen. We found that limits of detection for ttr were 1, 10, and 100 CFU/ml, and for InvA were 1, 100, and 100 CFU/ml for selenite sub-cultured broth, direct isolates, and stool-spiked isolates, respectively, with 98.5% qPCR efficiency for ttr and 97.2% qPCR efficiency for InvA. No statistically significant difference was observed in the LOD when ttr was compared with InvA in either direct isolates (p = 0.3212), selenite sub-cultured samples (P = 0.2534), or salmonella spiked stool samples (P = 0.2361). Importantly, we found that the ttr assay was significantly different when direct isolates (LOD = 10 CFU/ml) were compared with selenite sub-cultured samples (LOD = 1 CFU/ml) (p<0.0001), and when selenite sub-cultured isolates were compared to Salmonella spiked stool ( p <0.0001), and there was no significant difference when direct isolates were compared to Salmonella spiked stool ( p=0.2965).
Similarly, we found that detection in InvA qPCR assay direct isolates was significantly different compared to selenite broth cultures isolates ( p < 0.0001), and selenite subculture isolates were also significantly different from Salmonella spiked stool ( p < 0.0001). In contrast, there was no significant difference between direct isolates compared to Salmonella spiked stool samples ( p = 0.2862). In summary, we found that selenite broth overnight liquid culture of stool samples enhanced the molecular detection of Salmonella using either ttr or InvA primers, even if culture of the broth remained negative.
Ttr and InvA primers had both high specificity and sensitivity rates, while stool culture had high specificity but low sensitivity
The samples from healthy children were used to determine the performance of stool culture, monoplex ttr, monoplex InvA, multiplex TAC ttr, and multiplex TAC InvA. Standard stool culture was performed on a total of 600 specimens at different time points. Molecular tests were used to detect Salmonella in the available 421 stool DNA specimens. We detected Salmonella in 23, 40, 29, 56, and 47 of 421 stool specimens, using standard stool culture, ttr, InvA, TAC- ttr, and TAC- InvA respectively. Of the 23 Salmonella stool culture-positive samples, 21 samples were also positive with either one or more molecular tests, while 2 were molecular tests negative.
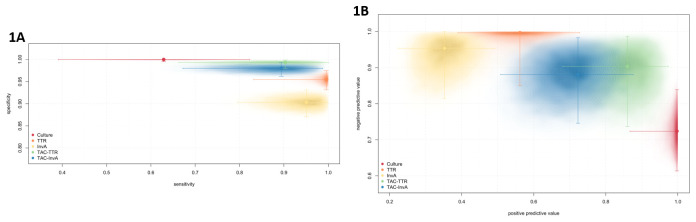
Based on a time-homogeneous LMM without random effects ( Table 3 and Figure 1A), we report the specificities and sensitivities of the detection methods with their 95% credible intervals (Bayesian confidence intervals). The observed specificity rates from highest to lowest were for stool culture (99.99%), TAC- ttr (99.30%), TAC- InvA (98.00%), monoplex ttr (95.46%), and monoplex InvA (90.31%), respectively. The observed sensitivity rates from highest to lowest were monoplex ttr (99.53%), monoplex InvA (95.06%), TAC- ttr (90.30%), TAC- InvA (89.41%,) and stool culture (62.88%) respectively ( Table 3 and Figure 1A). While stool culture achieved the highest specificity and monoplex ttr the highest sensitivity, monoplex ttr achieved arguably the best sensitivity-specificity trade-off: very high sensitivity (99.53%) at a relatively small drop in specificity (95.46) compared to stool culture.
Figure 1.
Maximum a posteriori probability estimates of the specificities and sensitivities (Figure 1A), positive and negative predictive values (Figure 1B) together with 95% highest density credible intervals (segments) and posterior density estimates (contours) for stool culture, ttr, InvA, TAC- ttr, and TAC. Big dots and error bars represent the median values and 25 and 75 percentile.
Table 3. Probability estimates of the specificities and the sensitivities, PPV, and NPV of the diagnostic tests.
Maximum a posterior probability estimates of the specificities and the sensitivities, PPV, and NPV of the diagnostic tests. Also reported are the 95% highest density credible intervals for each parameter.
| Sensitivity | Specificity | Positive predictive
value |
Negative predictive
value |
|||||
|---|---|---|---|---|---|---|---|---|
| MAP | (95% Crl) | MAP | 95% Crl | MAP | 95% Crl | MAP | 95% Crl | |
| Stool culture | 0.6288 | (0.3916,0.8223) | 0.9999 | (0.9949,10000) | 0.9973 | (0.8668,10000) | 0.7238 | (0.6135,0.8389) |
| ttr | 0.9953 | (0.8315,1.0000) | 0.9546 | (0.9317,0.9749) | 0.5615 | (0.3897,07275) | 0.9967 | (0.8501,10000) |
| InvA | 0.9506 | (0.7950,10000) | 0.9031 | (0.8702,0.9311) | 0.3521 | (0.2233,0.4915) | 0.9536 | (0.8147,10000) |
| TAC- ttr | 0.903 | (0.6628,10000) | 0.993 | (0.9797,0.9987) | 0.8597 | (0.6798,0.9736) | 0.9033 | (0.7367,0.9869) |
| TAC- InvA | 0.8941 | (0.6721,0.9869) | 0.98 | (0.9618,0.9928) | 0.7228 | (0.5079,0.8757) | 0.8807 | (0.7459,0.9828) |
High negative and positive concordance for stool culture, monoplex ttr, monoplex InvA, Multiplex ttr, and multiplex InvA
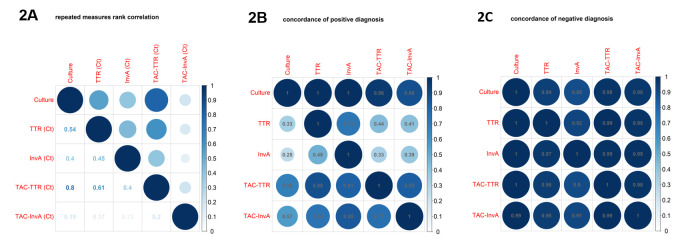
Next, we explored correlations between stool culture, monoplex ttr, monoplex InvA, Multiplex ttr, and multiplex InvA. In this exploration, we considered all test results, whether positive or negative. To account for both censored observations and the longitudinal nature of the data, we calculated repeated measures of correlation coefficients 35 using the ranks of observations for each test (akin to a repeated-measures Spearman correlation coefficient) for measuring the correlation between the Ct values for the four molecular tests and point bi-serial correlation coefficients based on ranks for measuring correlations between standard stool culture and each of the qPCR tests ( Figure 2A). The correlation coefficients vary quite widely from 0.12 (monoplex InvA and TAC- InvA) to 0.8 (stool culture and TAC- ttr). Given that for truly negative samples, the Ct values are effectively randomly distributed near the threshold used to discriminate between positive and negative samples and that most samples were negative in most tests, the somewhat weak correlations we observe can be driven by the random Ct values for negative samples. For this reason, using only the binary negative / positive outcomes for each test, we computed positive ( Figure 2B) and negative ( Figure 2C) concordance: for example, in Figure 2B, the intersection of the row labeled ‘ ttr,’ and the column labeled ‘ InvA’ lists the proportion of positive test results for the ttr test that are also positive for the InvA test. Unexpectedly (given that most samples were negative), negative concordance ( Figure 2C) was very high, with the lowest negative concordance being 89%. Results for positive concordance ( Figure 2B) are also relatively high, though there is more variation, ranging from 25% (for positive InvA results confirmed by positive stool cultures) to 100% (positive stool cultures confirmed by positive monoplex ttr or positive monoplex InvA).
Figure 2.
Correlation coefficients for the four molecular tests (using Ct values) and stool culture using positive or negative (Figure 2A). Concordance coefficients for positive (Figure 2B) and negative (Figure 2C) diagnosis obtained using binary negative or positive outcomes for each test. For example, in Figure 2B, the intersection of the row labeled ‘Culture’ and the column labeled ‘ ttr’ lists the proportion of positive test results for the Culture test that are also positive for the ttr test. Both the size and colour depth represent the magnitude of correlation.
Stool culture had high positive predictive value while molecular tests methods had high negative predictive values
To report PPV and NPV, for an estimate of prevalence, we use the model-estimated stationary (time-homogeneous model) probability of being infected (MAP 5.25%, 95% credible interval [3.27%, 8.14%]). From highest to lowest, the estimated PPVs were culture (99.73%), TAC- ttr (85.97%), TAC- InvA (72.28%), mono- ttr (56.15%), mono- InvA (35.21%). From highest to lowest, the estimated NPVs were mono- ttr (99.67%), mono- InvA (95.36%), TAC- ttr (90.33%), TAC- InvA (88.07%), and culture (72.38%) as indicated in Table 3 and Figure 1B. While stool culture has the highest PPV (99.73%) and mono- ttr the largest NPV (99.67%), a good trade-off between PPV and NPV is achieved by TAC- ttr (both high PPV, 85.97%, and high NPV, 90.33%). We note that these PPV and NPV estimates are for an asymptomatic population of children in urban Blantyre, Malawi, and are not directly generalisable to different contexts and populations where prevalence may be higher or lower.
Discussion
The burden of asymptomatic gastrointestinal exposure to Salmonella, which could be linked to either the development of immunity or, conversely, to bloodstream infection, is not known due to lack of robust Salmonella detection methods for stool specimens. This study aimed to optimise detection methods and to validate and compare the performance of monoplex ttr and InvA qPCR assays ( ttr and InvA) against ttr and InvA qPCR assays on a validated multiplex qPCR platform (TAC- ttr and TAC- InvA), and compare all molecular methods to standard Salmonella stool culture. Validation of the monoplex ttr and InvA primers showed that the primers do not cross-react with other enteric pathogens, and LOD testing showed that selenite pre-culture promotes molecular detection, even when culture is negative. Stool culture demonstrated the highest specificity but low sensitivity than all the molecular tests. Stool culture, despite having low sensitivity, still remains important in Salmonella diagnosis. Culture allows for antimicrobial susceptibility testing and strain typing. Ttr detected on the monoplex platform demonstrated superior sensitivity to stool culture, InvA, TAC- ttr, and TAC- InvA. All the test methods, however, displayed high concordance to each other.
Several studies have developed Salmonella detection methods based on antigen detection or nucleic acid amplification 16, 18, 36, 37. Both monoplex and multiplex nucleic acid amplification-based detection methods have been developed 38– 41. Most of these have, however, focused on Salmonella detection in blood as opposed to stool specimens. Some multiplex qPCRs to specifically detect Salmonella and its serovars or for the detection of multiple enteric pathogens in stool specimen (including Salmonella) have recently been developed 28, 42– 44. The advantage of multiplex qPCR is that it is fast in determining the primary etiological agent in cases where multiple pathogens or different serovars cause the outcome, but it is expensive if one is interested in detecting only one particular pathogen ( Table 4). By contrast, the advantage of a monoplex test is that it is economical than the multiplex, faster than stool culture, even with the addition of the selenite enrichment step and allows for batch-processing of samples which increases the efficiency of the test method. In this study, the same primer/ probe sets were tested using both the monoplex and multiplex qPCR platforms. The monoplex qPCR maximised sensitivity, while the multiplex panel provided a balanced pay-off between sensitivity and specificity ( Table 4). The high sensitivities of the monoplex qPCR could be attributed to the use of selenite pre-cultured stool as opposed to extraction of DNA from neat stool samples, which is used in the multiplex qPCR. Selenite sub-cultured stool samples were not used on the multiplex platform because the manufacturer’s protocol was followed. Other studies have, however, also demonstrated superior performance of monoplex qPCR when compared with multiplex qPCR. The monoplex qPCR is therefore ideal for studies that are only interested in determining the presence or absence of Salmonella while capitalising on the sensitivity of the test, while multiplex qPCR will have an added advantage if a study wants to detect multiple pathogens while having a pay-off between sensitivity and specificity.
Table 4. Characteristics of Salmonella stool culture, Monoplex -qPCR ttr, Monoplex -qPCR InvA, Multiplex -qPCR ttr and Multiplex -qPCR InvA.
A summary of the characteristics of the test methods. Specific values are indicated for sensitivity and specificity. Estimates were made for time to assay result, person-labour time requirement, cost of the test, specialist equipment cost by ranking them from Low to very high. Comparisons were also made for follow-up tests, antibiotic sensitivity and serotyping, and efficiency in batching.
|
Salmonella stool
culture |
Monoplex -qPCR ttr | Monoplex -qPCR InvA | Multiplex -qPCR ttr | Multiplex -qPCR InvA | |
|---|---|---|---|---|---|
| Sensitivity | Moderate (62.88%) | Very high (99.53%) | Very High (95.06%) | High (90.30%) | High (89.41%) |
| Specificity | Very high (99.99%) | Very High (95.46%) | High (90.31%) | Very high (99.30%) | Very high (98.00%) |
|
Estimated time to
assay result (10 samples) |
3 –7 days | 1.5 days including
overnight Selenite F broth enrichment |
1.5 days including overnight
Selenite F broth enrichment |
0.5 day | 0.5 day |
|
Person-labour time
requirement |
Very high | High | High | high | high |
| Cost estimate | Low | High | High | Very high | Very high |
|
Specialist
equipment cost |
Low: can be done
in a standard microbiology laboratory |
High: qPCR machine | High: qPCR machine | High: TAC-compatible qPCR
machine Compatible centrifuge buckets TAC plate sealer |
High: TAC-compatible qPCR machine
Compatible centrifuge Buckets TAC plate sealer |
|
Follow-up antibiotic
sensitivity |
Possible | Not possible | Not possible | Not possible | Not possible |
| Serotyping | Possible | Possible using serotype-
specific primers |
Possible using serotype-
specific primers |
Possible using serotype-specific
primers |
Possible using serotype-specific
primers |
|
Efficiency in
batching |
Low: working on
more samples at once reduces efficiency |
Very High: batched
samples can be efficiently extracted and tested 96 samples and controls can be tested using 1 plate |
Very High: batched samples
can be efficiently extracted and tested 96 samples and controls can be tested using 1 plate |
Very High: batched samples can
be efficiently extracted and tested 8 samples and multiple pathogens can be tested using 1 plate |
Very High: batched samples can be
efficiently extracted and tested 8 samples and multiple pathogens can be tested using 1 plate |
The ttr primer/ probe set used in the monoplex qPCR was previously validated for use in food samples and required validation in stool specimens. Our in-house developed InvA primer/ probe set also needed validation. Both assays demonstrated that they could detect all the different Salmonella strains, including S. Enteritidis, S. Typhimurium, and S. Typhi strains which are the commonly isolated strains in Malawi and SSA 45. Comparing the limits of detection of different Salmonella isolate conditions demonstrated that selenite pre-culture achieves a significantly lower limit of detection (1 CFU/ml) as opposed to direct isolates (10 CFU/ml) and Salmonella-spiked stool (10 CFU/ml). Selenite F broth is a selective broth that suppresses fecal coliforms and streptococci growth to optimise Salmonella growth 46. The LOD achieved after sub-culturing samples in Selenite enrichment broth agrees with results demonstrated by other studies, including a study done by Boer et al., who showed that sub-culturing samples in Selenite F broth promotes the recovery of Salmonella in stool samples and improves sensitivity if samples are subsequently tested using molecular methods like PCR 46, 47.
Given the lack of a true gold standard diagnostic test, we took a model-based approach and used an LMM to estimate the specificities and sensitivities of the 5 Salmonella detection methods. Stool culture demonstrated the highest specificity but had the lowest sensitivity. All molecular assays; TAC- ttr, TAC- InvA, ttr, and InvA, demonstrated high specificity and sensitivity rates. Compared to the other methods, the monoplex based qPCR ttr achieved, in our opinion, the best sensitivity-specificity trade-off as it demonstrates near-perfect sensitivity (99.53%) and still achieves high specificity (95.43%). While monoplex ttr has the best overall performance, depending on the research context or clinical purpose, practitioners may still prefer tests with higher specificity such as stool culture, TAC- ttr, or TAC- InvA. All molecular test methods had significantly higher sensitivities than stool culture. High specificity and low sensitivity rates for culture have been widely reported 18. Such low sensitivity rates should be taken into consideration when evaluating diagnostic tests. It is clear that a reference test with poor sensitivity is not adequate to evaluate alternative test methods. In such a situation, alternative means of evaluating the assays should be used, such as the LMM that has been used here. LMMs, and their counterpart for cross-sectional data, latent class models (LCMs), have been used to evaluate diagnostic tests for various pathogens, including Salmonella 48.
PPV and NPV vary depending on the prevalence of the condition being tested in any particular population. Our samples were collected from a population that was considered healthy and asymptomatic at the time of recruitment. Using the model-estimated stationary probability of being infected, we estimated the Salmonella infection prevalence of 5.25% in this population. With this prevalence estimate, stool culture demonstrated a high PPV (99.73%) when compared to molecular tests that had high NPVs, with the highest NPV recorded by mono- ttr (99.67%). As a trade-off between PPV and NPV, in a population of asymptomatic children in urban Blantyre, TAC- ttr achieved high PPV (85.97%) and high NPV (90.33%). Whether a test with high PPV or NPV is to be preferred depends on the research and/or clinical context. When prevalence is low, a slight change in specificity will have significant effects on the PPV. Higher PPVs could be observed in a situation where prevalence is high such as when using a cohort of hospitalised diarrheal cases or during a diarrhoeal outbreak.
Molecular methods had higher sensitivity but lower specificity relative to stool culture. The loss in specificity is slight compared to the gain in sensitivity, and in the case of Salmonella, the public health cost of false-negative results could be higher if the infection becomes potentially life-threatening due to withholding or delay of treatment. With the high sensitivity, molecular methods were able to detect asymptomatic Salmonella events, critical for the research questions we hoped to pose in this cohort. All the events that were detected here were asymptomatic in healthy children, which are potentially very important in transmission or the development of immunity. The detection of low bacterial burden events could also be relevant in settings like Malawi where unprescribed over-the-counter antibiotic procurement and use is common. Studies that have reported on risk factors of having a culture-negative result have indicated that antibiotic usage before sample collection is the main risk factor. Using molecular techniques such as PCR could overcome this challenge because it detects bacterial DNA regardless of pathogen viability. This might increase the probability of identifying the infection and reduce sample processing time, leading to proper patient management and treatment if needed.
Our study has several limitations. One main limitation is the use of different sample types for the two qPCR platforms. The use of selenite sub-cultured stool samples in monoplex qPCR may have contributed to the superior performance when compared with the multiplex qPCR. We used neat stool samples for multiplex qPCR to comply with the manufacturer’s protocol. Other studies have, however, demonstrated that testing primer/ probe sets in the monoplex platform perform better than in the multiplex qPCR platform. Clinical samples used to test the performance of the test are a limitation, especially in determining the PPV and NPV. Clinical samples used in the study were collected from a cohort of children that were asymptomatic to Salmonella and remained healthy for most of the one-year study period. Using samples from participants with a clinical diagnosis of Salmonella or diarrhoea would have resulted in PPV and NPV estimates more directly relevant in a clinical setting. Another important limitation is the model-based nature of our approach. This was a necessity given the lack of a gold standard diagnostic test but does mean that our estimates depend on the validity of the model’s assumptions, in particular, i) the assumption that the latent infection state at a given time only depends on the immediate previous timepoint, the so-called Markov assumption, and ii) the conditional independence assumption of the basic LMM. While our modelling framework had been extended to relax the latter assumption, a better fit was achieved for the basic model.
Conclusion
The data presented here demonstrate that the addition of selenite F broth pre-enrichment step increases Salmonella detection in stool samples and that ttr and InvA primer and probe sets used can detect different Salmonella strains. The ability of ttr to detect Salmonella with such high levels of specificity and sensitivity when tested using clinical samples collected from a mostly healthy cohort makes it a promising assay that could be used for research surveillance studies. The assays could be very useful in studying the transmission of Salmonella infections. This method may perform with different sensitivity and specificity in a chronic carriage, diarrhoeal or invasive Salmonella disease state, since the load and culturability of the pathogen within the stool may be different, and further validation studies would be needed
We established that selenite pre-culture increased diagnostic yield for molecular detection and identified ttr primers as molecular tools that could best help reveal the true extent of Salmonella exposure events within the gastrointestinal tract. This will allow us to understand their importance to diarrhoeal and invasive disease pathogenesis and epidemiology in the future.
Data availability
Underlying data
Figshare: Data and software code for Bayesian mixed latent Markov models for binary diagnostic data, https://doi.org/10.6084/m9.figshare.12911870.
-
1.
gitMarcH-Bayesian-mixed-latent-Markov-models-for-binary-diagnostic-data.zip (software code for Latent Markov Model used in this study)
-
2.
Data files used by the uploaded software code:
-
-
salexpoLIMSDataSetComplete.csv (Date of sample collection and follow-up visit number)
-
-
TACResults_4Mar TAC ttr TAC InvA Ct For Correlation.csv (Ct values for TAC_ ttr and TAC_ InvA)
-
-
ttr & InvA master file Ct for correlation.csv (Ct values for monoplex TTR and InvA)
-
-
TtrInvASensitivity20170724_corrected.csv (Combined binary results for stool culture, ttr, InvA, TAC_ ttr, and TAC_ InvA used to calculate sensitivity, specificity and correction of the test methods)
-
-
-
3.
Raw data:
-
-
TAC Results_TAC- ttr_TAC- InvA_I_Ct ValuesTAC Results_TAC- ttr_TAC- InvA_IC_Ct-values.csv (raw Taqman array card assay results for test and control sample)
-
-
Salmonella_Detection_Stool_ ttr_ InvA_raw_data.xlsx (raw data for the monoplex qPCR assay. Includes results for test and control sample)
-
-
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
Acknowledgements
The authors are very thankful to Sr. Rose Nkhata who helped with recruitment and following up of the study participants.
Funding Statement
This work was supported by the Wellcome Trust through core funding to the Malawi Liverpool Welcome Trust Clinical Research Programme (MLW) which funds a Training Fellowship award to AC [206545], and funding to the Consortium for Advanced Research Training in Africa (CARTA) which provides a Post-Doctoral Training Fellowship to TSN [087547]. CARTA is jointly led by African Population and Health Research Centre and the University of the Witwatersrand and funded by Wellcome Trust (UK) (Grant: 087547/Z/08/Z), the Carnegie Corporation of New York (Grant No-B 8606.R02), SIDA (Swedish International Development Aid Agency) (Grant No: 54100029). The study was also supported by Bill and Melinda Gates Foundation (Grant: OPP1128435).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; peer review: 2 approved]
References
- 1. Crump JA: Progress in Typhoid Fever Epidemiology. Clin Infect Dis. 2019;68(Suppl 1):S4–s9. 10.1093/cid/ciy846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ao TT, Feasey NA, Gordon MA, et al. : Global burden of invasive nontyphoidal Salmonella disease, 2010(1). Emerg Infect Dis. 2015;21(6);941–949. 10.3201/eid2106.140999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. GBD 2017 Typhoid and Paratyphoid Collaborators: The global burden of typhoid and paratyphoid fevers: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect Dis. 2019;19(4);369–381. 10.1016/S1473-3099(18)30685-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. GBD 2017 Non-Typhoidal Salmonella Invasive Disease Collaborators: The global burden of non-typhoidal salmonella invasive disease: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect Dis. 2019;19(12);1312–1324. 10.1016/S1473-3099(19)30418-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Crump JA, Luby SP, Mintz ED: The global burden of typhoid fever. Bull World Health Organ. 2004;82(5):346–53. [PMC free article] [PubMed] [Google Scholar]
- 6. WHO: Salmonella (non-typhoidal). Fact sheets -detail.2018[cited 2019 October 4]. Reference Source [Google Scholar]
- 7. GBD 2017 Non-Typhoidal Salmonella Invasive Disease Collaborators: The global burden of non-typhoidal salmonella invasive disease: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect Dis. 2019;19(12):1312– 1324. 10.1016/S1473-3099(19)30418-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gordon MA: Salmonella infections in immunocompromised adults. J Infect. 2008;56(6):413–422. 10.1016/j.jinf.2008.03.012 [DOI] [PubMed] [Google Scholar]
- 9. Feasey NA, Dougan G, Kingsley RA, et al. : Invasive non-typhoidal salmonella disease: an emerging and neglected tropical disease in Africa. Lancet. 2012;379(9835):2489–2499. 10.1016/S0140-6736(11)61752-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nyirenda TS, Gilchrist JJ, Feasey NA, et al. : Sequential acquisition of T cells and antibodies to nontyphoidal Salmonella in Malawian children. J Infect Dis. 2014;210(1):56–64. 10.1093/infdis/jiu045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. MacLennan CA, Gondwe EN, Msefula CL, et al. : The neglected role of antibody in protection against bacteremia caused by nontyphoidal strains of Salmonella in African children. J Clin Invest. 2008;118(4):1553–1562. 10.1172/JCI33998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kotloff KL: The Burden and Etiology of Diarrheal Illness in Developing Countries. Pediatr Clin North Am. 2017;64(4):799–814. 10.1016/j.pcl.2017.03.006 [DOI] [PubMed] [Google Scholar]
- 13. Darton TC, Zhou L, Blohmke CJ, et al. : Blood culture-PCR to optimise typhoid fever diagnosis after controlled human infection identifies frequent asymptomatic cases and evidence of primary bacteraemia. J Infect. 2017;74(4):358–366. 10.1016/j.jinf.2017.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mogasale V, Ramani E, Mogasale VV, et al. : What proportion of Salmonella Typhi cases are detected by blood culture? A systematic literature review. Ann Clin Microbiol Antimicrob. 2016;15(1):32. 10.1186/s12941-016-0147-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Antillon M, Saad NJ, Baker S, et al. : The Relationship Between Blood Sample Volume and Diagnostic Sensitivity of Blood Culture for Typhoid and Paratyphoid Fever: A Systematic Review and Meta-Analysis. J Infect Dis. 2018;218(suppl_4):S255–S267. 10.1093/infdis/jiy471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tennant SM, Toema D, Qamar F, et al. : Detection of Typhoidal and Paratyphoidal Salmonella in Blood by Real-time Polymerase Chain Reaction. Clin Infect Dis. 2015;61 Suppl 4(Suppl 4):S241–50. 10.1093/cid/civ726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Msefula CL, Olgemoeller F, Jambo N, et al. : Ascertaining the burden of invasive Salmonella disease in hospitalised febrile children aged under four years in Blantyre, Malawi. PLoS Negl Trop Dis. 2019;13(7):e0007539. 10.1371/journal.pntd.0007539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Andrews JR, Ryan ET: Diagnostics for invasive Salmonella infections: Current challenges and future directions. Vaccine. 2015;33 Suppl 3(03):C8–15. 10.1016/j.vaccine.2015.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Trafford G, Ratnaraja N, Wickramasinghe N: Molecular diagnostic testing for common stool pathogens. J Hosp Infect. 2015;90(3):196–8. 10.1016/j.jhin.2015.01.019 [DOI] [PubMed] [Google Scholar]
- 20. Nyirenda TS: Natural Immunity to Salmonella in Humans. In: Institute of Infection and Global Health.University of Liverpool.2015. [Google Scholar]
- 21. Malorny B, Paccassoni E, Fach P, et al. : Diagnostic real-time PCR for detection of Salmonella in food. Appl Environ Microbiol. 2004;70(12):7046–52. 10.1128/AEM.70.12.7046-7052.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Malorny B, Hoorfar J, Bunge C, et al. : Multicenter validation of the analytical accuracy of Salmonella PCR: towards an international standard. Appl Environ Microbiol. 2003;69(1):290–6. 10.1128/aem.69.1.290-296.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. MacLennan CA, Gondwe EN, Msefula CL, et al. : The neglected role of antibody in protection against bacteremia caused by nontyphoidal strains of Salmonella in African children. J Clin Invest. 2008;118(4):1553–1562. 10.1172/JCI33998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kingsley RA, Msefula CL, Thomson NR, et al. : Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 2009;19(12):2279–87. 10.1101/gr.091017.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lertsethtakarn P, Silapong S, Sakpaisal P, et al. : Travelers' Diarrhea in Thailand: A Quantitative Analysis Using TaqMan® Array Card. Clin Infect Dis. 2018;67(1):120–127. 10.1093/cid/ciy040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Platts-Mills JA, Liu J, Rogawski ET, et al. : Use of quantitative molecular diagnostic methods to assess the aetiology, burden, and clinical characteristics of diarrhoea in children in low-resource settings: a reanalysis of the MAL-ED cohort study. Lancet Glob Health. 2018;6(12):e1309–e1318. 10.1016/S2214-109X(18)30351-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu J, Kabir F, Manneh J, et al. : Development and assessment of molecular diagnostic tests for 15 enteropathogens causing childhood diarrhoea: a multicentre study. Lancet Infect Dis. 2014;14(8):716–724. 10.1016/S1473-3099(14)70808-4 [DOI] [PubMed] [Google Scholar]
- 28. Liu J, Gratz J, Amour C, et al. : A laboratory-developed TaqMan Array Card for simultaneous detection of 19 enteropathogens. J Clin Microbiol. 2013;51(2):472–80. 10.1128/JCM.02658-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bartolucci F, Farcomeni A, Pennoni F: Latent Markov Models for Longitudinal Data. Chapman & Hall, CRC Press, Boca Raton, FL.2013. [Google Scholar]
- 30. Henrion MYR, Chirambo A, Nyirenda TS, et al. : Mixed latent Markov models for longitudinal multiple diagnostics data with an application to salmonella in Malawi. JSM Proceedings Vancouver Canada. 2018. Reference Source [Google Scholar]
- 31. Plummer M: rjags: Bayesian Graphical Models using MCMC. R package version 4-6.2016. Reference Source [Google Scholar]
- 32. Koukounari A, Donnelly CA, Moustaki I, et al. : A latent Markov modelling approach to the evaluation of circulating cathodic antigen strips for schistosomiasis diagnosis pre- and post-praziquantel treatment in Uganda. PLoS Comput Biol. 2013;9(12):e1003402. 10.1371/journal.pcbi.1003402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koukounari A, Moustaki I, Grassly NC, et al. : Using a nonparametric multilevel latent Markov model to evaluate diagnostics for trachoma. Am J Epidemiol. 2013;177(9):913–922. 10.1093/aje/kws345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gelman SA, Rubin DB, Rubin DB: Inference from Iterative Simulation Using Multiple Sequences. Stat Sci. 1992;7:457–472. 10.1214/ss/1177011136 [DOI] [Google Scholar]
- 35. Bakdash JZ, Marusich LR: Repeated Measures Correlation. Front Psychol. 2017;8:456. 10.3389/fpsyg.2017.00456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuijpers LMF, Chung P, Peeters M, et al. : Diagnostic accuracy of antigen-based immunochromatographic rapid diagnostic tests for the detection of Salmonella in blood culture broth. PLoS One. 2018;13(3):e0194024. 10.1371/journal.pone.0194024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bugarel M, den Bakker H, Grout J, et al. : CRISPR-based assay for the molecular identification of highly prevalent Salmonella serotypes. Food Microbiol. 2018;71:8–16. 10.1016/j.fm.2017.03.016 [DOI] [PubMed] [Google Scholar]
- 38. Pouzol S, Tanmoy AM, Pouzol DA, et al. : Clinical Evaluation of a Multiplex PCR for the Detection of Salmonella enterica Serovars Typhi and Paratyphi A from Blood Specimens in a High-Endemic Setting. Am J Trop Med Hyg. 2019;101(3):513–520. 10.4269/ajtmh.18-0992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heymans R, Vila A, van Heerwaarden CAM, et al. : Rapid detection and differentiation of Salmonella species, Salmonella Typhimurium and Salmonella Enteritidis by multiplex quantitative PCR. PLoS One. 2018;13(10):e0206316. 10.1371/journal.pone.0206316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Scopes E, Screen J, Evans K, et al. : Evaluation of the Thermo Scientific RapidFinder™ Salmonella Species, Typhimurium, and Enteritidis Multiplex PCR Kit. J AOAC Int. 2018;101(4):1059–1100. 10.5740/jaoacint.17-0344 [DOI] [PubMed] [Google Scholar]
- 41. Park SH, Ricke SC: Development of multiplex PCR assay for simultaneous detection of Salmonella genus, Salmonella subspecies I, Salm. Enteritidis, Salm. Heidelberg and Salm. Typhimurium. J Appl Microbiol. 2015;118(1):152–60. 10.1111/jam.12678 [DOI] [PubMed] [Google Scholar]
- 42. Hannet I, Engsbro AL, Pareja J, et al. : Multicenter evaluation of the new QIAstat Gastrointestinal Panel for the rapid syndromic testing of acute gastroenteritis. Eur J Clin Microbiol Infect Dis. 2019;38(11):2103–2112. 10.1007/s10096-019-03646-4 [DOI] [PubMed] [Google Scholar]
- 43. Martín A, Pérez-Ayala A, Chaves F, et al. : Evaluation of the multiplex PCR Allplex-GI assay in the detection of bacterial pathogens in diarrheic stool samples. J Microbiol Methods. 2018;144:33–36. 10.1016/j.mimet.2017.10.016 [DOI] [PubMed] [Google Scholar]
- 44. Beckman AK, Ferrieri P: Prospective Investigation of an Automated PCR/Nucleic Acid Microarray-Based Platform for Enteric Pathogen Testing. Lab Med. 2019;50(4):390–395. 10.1093/labmed/lmz022 [DOI] [PubMed] [Google Scholar]
- 45. Gilchrist JJ, MacLennan CA: Invasive Nontyphoidal Salmonella Disease in Africa. EcoSal Plus. 2019;8(2). 10.1128/ecosalplus.ESP-0007-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Boer MD, de Boer RF, Lameijer A, et al. : Selenite enrichment broth to improve the sensitivity in molecular diagnostics of Salmonella. J Microbiol Methods. 2018;157:59–64. 10.1016/j.mimet.2018.12.018 [DOI] [PubMed] [Google Scholar]
- 47. Goneau LW, Mazzulli A, Trimi X, et al. : Evaluating the preservation and isolation of stool pathogens using the COPAN FecalSwab™ Transport System and Walk-Away Specimen Processor. Diagn Microbiol Infect Dis. 2019;94(1):15–21. 10.1016/j.diagmicrobio.2018.11.020 [DOI] [PubMed] [Google Scholar]
- 48. Moore CE, Pan-Ngum W, Wijedoru LPM, et al. : Evaluation of the diagnostic accuracy of a typhoid IgM flow assay for the diagnosis of typhoid fever in Cambodian children using a Bayesian latent class model assuming an imperfect gold standard. Am J Trop Med Hyg. 2014;90(1):114–120. 10.4269/ajtmh.13-0384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Henrion MYR: Data and software code for Bayesian mixed latent Markov models for binary diagnostic data. figshare.Dataset.2020. 10.6084/m9.figshare.12911870.v3 [DOI] [Google Scholar]