It has been hoped that rapid SARS-CoV-2 antigen testing could reduce the tragic toll of COVID-19 in nursing homes. The performance of the BinaxNOW antigen test in a nursing home during an ongoing SARS-CoV-2 outbreak was evaluated.
Visual Abstract. Serial SARS-CoV-2 Rapid Antigen Testing During a Nursing Home Outbreak.
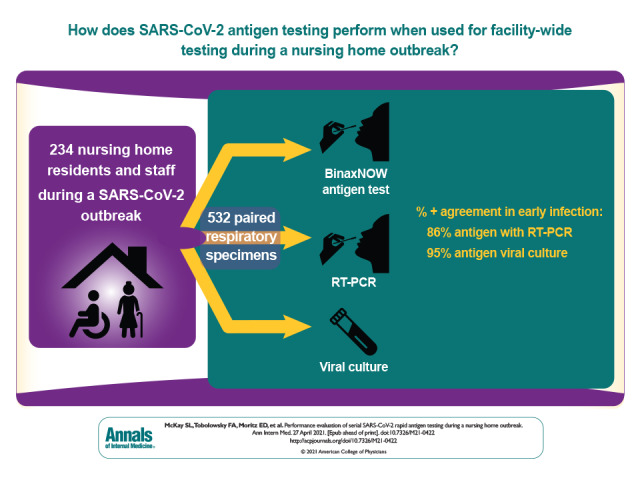
It has been hoped that rapid SARS-CoV-2 antigen testing could reduce the tragic toll of COVID-19 in nursing homes. The performance of the BinaxNOW antigen test in a nursing home during an ongoing SARS-CoV-2 outbreak was evaluated.
Abstract
Background:
To address high COVID-19 burden in U.S. nursing homes, rapid SARS-CoV-2 antigen tests have been widely distributed in those facilities. However, performance data are lacking, especially in asymptomatic people.
Objective:
To evaluate the performance of SARS-CoV-2 antigen testing when used for facility-wide testing during a nursing home outbreak.
Design:
A prospective evaluation involving 3 facility-wide rounds of testing where paired respiratory specimens were collected to evaluate the performance of the BinaxNOW antigen test compared with virus culture and real-time reverse transcription polymerase chain reaction (RT-PCR). Early and late infection were defined using changes in RT-PCR cycle threshold values and prior test results.
Setting:
A nursing home with an ongoing SARS-CoV-2 outbreak.
Participants:
532 paired specimens collected from 234 available residents and staff.
Measurements:
Percentage of positive agreement (PPA) and percentage of negative agreement (PNA) for BinaxNOW compared with RT-PCR and virus culture.
Results:
BinaxNOW PPA with virus culture, used for detection of replication-competent virus, was 95%. However, the overall PPA of antigen testing with RT-PCR was 69%, and PNA was 98%. When only the first positive test result was analyzed for each participant, PPA of antigen testing with RT-PCR was 82% among 45 symptomatic people and 52% among 343 asymptomatic people. Compared with RT-PCR and virus culture, the BinaxNOW test performed well in early infection (86% and 95%, respectively) and poorly in late infection (51% and no recovered virus, respectively).
Limitation:
Accurate symptom ascertainment was challenging in nursing home residents; test performance may not be representative of testing done by nonlaboratory staff.
Conclusion:
Despite lower positive agreement compared with RT-PCR, antigen test positivity had higher agreement with shedding of replication-competent virus. These results suggest that antigen testing could be a useful tool to rapidly identify contagious people at risk for transmitting SARS-CoV-2 during nascent outbreaks and help reduce COVID-19 burden in nursing homes.
Primary Funding Source:
None.
As of 10 January 2021, in the United States, 1 022 297 nursing home residents and staff have tested positive for SARS-CoV-2, the virus that causes COVID-19, and 108 447 have died (1). Nursing home residents might be asymptomatic, have atypical symptoms, or be unable to verbalize their symptoms, making diagnosis using symptom-based screening alone inadequate (2, 3). Serial, facility-wide testing for SARS-CoV-2 can help identify cases in outbreak settings, allowing for rapid implementation of transmission-based precautions and infection prevention and control strategies (3, 4). Although real-time reverse transcription polymerase chain reaction (RT-PCR) testing performed in a laboratory has the highest sensitivity, its prolonged turnaround time can delay quarantine and isolation implementation (5, 6). Furthermore, RT-PCR can be a poor indicator for infectiousness because people might shed measurable amounts of viral RNA despite the absence of infectious virus (7–10). Conversely, the ability to culture virus from clinical specimens is a better indication of contagiousness than RT-PCR (11). Positive virus culture is most often detected within 10 days after onset or when viral loads are high (>7.0 log10 copies/mL) (12, 13).
Antigen tests are easy to use and produce results in minutes, facilitating rapid action, particularly during outbreaks in congregate settings (4, 14, 15). In 2020, the U.S. Food and Drug Administration granted emergency use authorization (EUA) to 11 rapid antigen tests. The U.S. Department of Health and Human Services sent 3 of these, including the Abbott BinaxNOW COVID-19 Ag Card, to nursing homes nationwide (16). According to the 3 products' EUAs, among symptomatic people tested 5 to 7 days from symptom onset, the percentage of positive agreement (PPA) of antigen tests with RT-PCR is 84% to 99% and the percentage of negative agreement (PNA) remains close to 100% (16). However, antigen test performance in asymptomatic people and those with longer time to symptom onset than defined in the EUAs is not well characterized, with mixed reports on performance and concerns about false-positive results (16–19). Although mathematical models have suggested potential benefits from frequent, rapid-turnaround testing even with lower-PPA tests, limited data exist on antigen test performance in capturing early SARS-CoV-2 infections when people are most likely to be contagious (20–22).
On 7 October 2020, a 149-bed nursing home in Georgia identified its index COVID-19 case in a resident using the BinaxNOW antigen test, which prompted additional antigen testing in the facility. Despite attempts to implement mitigation measures, including cohorting, 43 residents and 5 staff had tested positive for SARS-CoV-2 by 21 October. The Centers for Disease Control and Prevention (CDC) worked with the Georgia Department of Public Health to evaluate the performance of the BinaxNOW antigen test compared with RT-PCR and virus culture. This report describes test characteristics of the BinaxNOW antigen test platform when used for symptomatic and asymptomatic people tested serially every 5 or 6 days during a nursing home outbreak.
Methods
Study Design and Data Sources
Between 22 October and 3 November 2020, serial, facility-wide testing of all residents and staff was done 3 times over a 13-day period during an ongoing SARS-CoV-2 outbreak. Specimens were collected from all available and assenting residents and staff present on days of testing, including people identified as SARS-CoV-2–positive before 21 October. During the first round of facility-wide testing, trained project personnel collected paired bilateral swabs from the anterior nares (AN) of residents for antigen testing and RT-PCR and, from nursing home staff, an AN swab for antigen testing and a nasopharyngeal swab from a single naris for RT-PCR. Because of patient intolerance, nasopharyngeal swabbing was discontinued during the second and third testing rounds and paired bilateral AN swabs were collected from both residents and staff (Appendix 2). All specimens were collected in accordance with CDC guidelines for specimen collection and handling (4). Trained laboratory scientists tested 1 AN swab onsite using the BinaxNOW COVID-19 Ag Cards per manufacturer instructions for use (23). The other was sent to the CDC for RT-PCR and virus culture reference testing.
The facility provided demographic characteristics and prior antigen testing results for residents and staff. During 7 to 21 October, the facility exclusively used BinaxNOW testing, and prior antigen positivity was defined as any positive result on a SARS-CoV-2 test during this time. At each visit, project personnel administered a standardized questionnaire assessing COVID-19–like symptoms (24). Able residents and staff self-reported symptoms at the time of testing. For residents who could not self-report, symptom information was obtained from nursing staff and electronic medical records and confirmed by residents, if possible. A symptomatic participant was defined as a resident or staff member who, at the time of collection, reported any new or worsening symptoms similar to those of COVID-19 (24) in the 14 days before that round of testing.
Participant specimens were tested for SARS-CoV-2 RNA by RT-PCR using the CDC Influenza SARS-CoV-2 (Flu SC2) Multiplex Assay (25) on the Applied Biosystems 7500 Fast Dx Real-Time PCR Instrument (Thermo Fisher Scientific). Nucleic acid was extracted by either the QIAGEN EZ1 or the Roche MagNA Pure 96 extraction platforms. Cycle threshold (Ct) values were reported for the SARS-CoV-2 viral nucleocapsid protein gene target. Values less than 40 indicated that a specimen was positive for SARS-CoV-2 RNA. Previous experience showed an inability to detect culture-positive virus in samples with a Ct greater than 34. Therefore, virus culture was attempted on RT-PCR–positive specimens with Ct values of 34 or less and RT-PCR–negative, antigen-positive specimens. Culture was done using Vero CCL-81 cells, as previously described (26). Cells showing cytopathic effect up to 8 days after culture inoculation were tested for the presence of SARS-CoV-2 by RT-PCR to confirm virus isolation and growth in culture (Appendix 2).
Specimens were categorized into stages of infection using prior test results and Ct values. Stages were defined as early (low or decreasing Ct values), late (increasing or sustained high Ct values), resolved (negative test result in a person with a prior positive result), or uninfected (consecutive negative results in specimens from a person with no prior positive result). Table 1 gives full definitions.
Table 1.
Defining Stages of Infection With RT-PCR Ct Values

Statistical Analysis
Descriptive analyses were done using SAS, version 9.4 (SAS Institute). We determined PPA and PNA by comparing antigen test results with reference tests. Paired specimens with at least 1 invalid test result were excluded from analysis. We calculated PPA and PNA for all participants and stratified by resident or staff, symptom status, previous positivity by any test, specimen type, and stage of infection (27). Clopper–Pearson exact binomial methods were used to calculate CIs.
Role of the Funding Source
This activity was reviewed by CDC, and its conduct was consistent with applicable federal law and CDC policy (28–32). This work did not receive any non-CDC funding support.
Results
Demographic Characteristics and Test Results, by Resident Versus Staff
A total of 107 staff members participated in at least 1 round of paired testing; the median age was 39 years (range, 21 to 72 years), 81% (n = 87) were female, and 75% (n = 80) were Black. A total of 127 residents participated in at least 1 round of paired testing; the median age was 75 years (range, 35 to 101 years), 43% (n = 55) were female, and 60% (n = 76) were Black (Appendix Table 1). Among 234 participants, 54% of residents (68 of 127) and 11% of staff (12 of 107) had at least 1 positive result on antigen or RT-PCR testing, including 43 of 68 residents and 5 of 12 staff who had tested positive at the facility between 7 and 21 October 2020.
Appendix Table 1.
Demographic Characteristics and Test Results of Residents and Staff Participating in ≥1 Round of Paired Testing (n = 234)

During 3 facility-wide testing events between 22 October and 3 November 2020, a total of 532 paired specimens were collected, including 388 from people who had not previously tested positive (Appendix Table 2) and 144 from those who had tested positive at least once since 7 October 2020. Details on the number of people tested during each facility-wide testing event are in Table 2. No specimens tested positive for influenza.
Appendix Table 2.
BinaxNOW COVID-19 Ag Card Performance Compared With Reference Standard RT-PCR, by Symptom Status

Table 2.
Participant Flow, by First Round of Testing, Further Stratified by Completed Rounds of Facility-wide Testing

Antigen Testing Results Compared With RT-PCR
Overall, 113 of 532 paired specimens (21%) were positive by antigen or RT-PCR testing. Of those that tested positive, 64% (72 of 113) were positive for both antigen and RT-PCR, 29% (33 of 113) were discordant RT-PCR–positive and antigen-negative, and 7% (8 of 113) were discordant RT-PCR–negative and antigen-positive (Appendix Table 2). The 8 discordant paired specimens that were RT-PCR–negative and antigen-positive were collected from 7 people who had previously tested positive, and 6 occurred 2 weeks or longer after the first positive test result (median, 18 days [range, 6 to 20 days]). Across all 532 paired specimens, PPA between antigen test and RT-PCR was 69% (95% CI, 59% to 77%) and PNA was 98% (CI, 96% to 99%) (Figure 1, top). Among 388 specimens from people without a prior positive result, PPA between antigen test and RT-PCR was 63% (CI, 44% to 79%). Antigen test performance was similar to the overall results when limited to 1 test per person (at first test, and when stratified by round of facility-wide testing) (Appendix Table 3). When stratified by symptom reports, PPA between antigen test and RT-PCR was 82% (CI, 48% to 98%) among specimens from symptomatic participants and 52% (CI, 30% to 74%) among those from asymptomatic participants. Between antigen test and RT-PCR, PNA remained close to 100% across all categories (Figure 1 [top] and Appendix Table 2). Antigen test performance (that is, PPA and PNA) compared with RT-PCR was also similar for staff and residents (Appendix Table 4) overall and stratified by symptom status. Antigen test performance compared with RT-PCR was also similar for nasopharyngeal and AN swabs (Appendix Table 5).
Figure 1. PPA and PNA between antigen test and RT-PCR and virus culture.
PNA = percentage of negative agreement; PPA = percentage of positive agreement; RT-PCR = reverse transcription polymerase chain reaction. Top. PPA and PNA between antigen test and reference standard RT-PCR for all paired specimens and paired specimens collected from people without a prior positive test result; those without a prior positive result were further stratified by symptomatic versus asymptomatic individuals. Detailed data for these PPA and PNA figures are provided in Appendix Table 2. Bars indicate 95% CIs. Bottom. PPA between antigen test and alternative reference standard virus culture for all paired specimens and paired specimens collected from people without a prior positive test result; those without a prior positive result were further stratified by symptomatic versus asymptomatic individuals. Bars indicate 95% CIs. Note: Virus culture was attempted only for RT-PCR–positive specimens with a cycle threshold value ≤34. Because specimens not likely to harbor infectious virus were not assessed for virus culturing, PNAs were not calculated. Detailed data for these PPA and PNA figures are provided in Appendix Table 6. Antigen test: BinaxNOW COVID-19 Ag Card.
Appendix Table 3.
BinaxNOW COVID-19 Ag Card Performance Compared With Reference Standard RT-PCR, by Testing Round

Appendix Table 4.
BinaxNOW COVID-19 Ag Card Performance Among Staff and Residents Compared With Reference Standard RT-PCR, Stratified by Symptom Status

Appendix Table 5.
BinaxNOW COVID-19 Ag Card Performance Compared With Reference Standard RT-PCR From Nasopharyngeal Swabs and AN Swabs

Antigen Testing Results Compared With Virus Culture
Virus was recovered from 21% of positive specimens (21 of 101) where virus culture was attempted (Appendix Table 6), including 29% (20 of 69) of concordant RT-PCR–positive and antigen-positive specimens and 4% (1 of 24) of specimens that were RT-PCR–positive and antigen-negative (Appendix Figure). Virus was not recovered from the 8 discordant paired specimens that were RT-PCR–negative and antigen-positive (Appendix Figure). Using virus culture as the reference standard, PPA with antigen testing was 95% (CI, 86% to 100%; note that negative agreement with virus culture was not applicable because only specimens most likely to harbor infectious virus, including those with Ct ≤34 and antigen-positive specimens, were subjected to virus culturing) (Figure 1, bottom). Antigen test performance was similar to the overall results when limited to 1 test per person (Appendix Table 7). The majority of culture-positive specimens (15 of 21 [71%]) were collected 0 to 5 days from the first positive test result; 1 specimen was culture-positive at 13 days (Appendix Figure). In the subset of 31 paired specimens from people without a prior positive result, PPA between antigen test and virus culture was 92% (CI, 62% to 100%) (Appendix Table 6).
Appendix Table 6.
BinaxNOW COVID-19 Ag Card Performance Compared With Alternative Reference Standard Virus Culture, by Symptom Status

Appendix Figure. Agreement between antigen testing and RT-PCR and VC among 101 paired specimens with VC result, over time.
Ag, RT-PCR, and VC results for 101 specimens from 63 individuals. VC was attempted for specimens that were Ag-positive and RT-PCR–negative (circle with X) but not attempted for RT-PCR–positive specimens with Ct values ≥35 (black squares). Ag = antigen; Ct = cycle threshold; RT-PCR = reverse transcription polymerase chain reaction; VC = virus culture.
Appendix Table 7.
BinaxNOW COVID-19 Ag Card Performance Compared With Alternative Reference Standard Virus Culture, by Testing Round

Antigen Testing and Virus Culture Results Compared With RT-PCR Ct Values
Among 105 RT-PCR–positive specimens, we compared Ct values in relation to antigen test result and virus culture (Figure 2). The median Ct value was significantly lower for antigen-positive paired specimens (median, 28.0 [range, 15.4 to 36.4]) than for antigen-negative paired specimens (median, 33.2 [range, 21.3 to 38.7]) (Wilcoxon P < 0.001) (Figure 2). Similarly, among the 93 paired specimens that were RT-PCR–positive and had a Ct value of 34 or less for which virus culture was attempted, the median Ct that resulted in positive virus culture was significantly lower (median, 21.3 [range, 15.4 to 26.7]) than that for culture-negative specimens (median, 30.2 [range, 22.5 to 35.0]; Wilcoxon P < 0.001).
Figure 2. Antigen test result, by SARS-CoV-2 nucleocapsid Ct values and virus culture result.

Ct values of all RT-PCR–positive respiratory specimens (n = 105). Shown are nucleocapsid Ct values for all paired RT-PCR–positive specimens stratified by antigen-positive and antigen-negative results and further by virus culture results. Median Ct for each category of antigen results is noted by the black bar. Virus culture was attempted for all RT-PCR–positive specimens with Ct ≤34. Culture was attempted for 8 additional specimens that were antigen-positive and RT-PCR–negative; all were culture-negative. Randomized jitter of 0.5 was added to x-axis values to improve visibility. Antigen test: BinaxNOW COVID-19 Ag Card. Ct = cycle threshold; RT-PCR = reverse transcription polymerase chain reaction.
Consensus Test Performance From Serial Testing
Among 173 people who were tested in more than 1 round of testing between 22 October and 3 November, 56 (32%) tested positive by RT-PCR at least once. Of these, 49 had at least 1 positive result on an antigen test during the evaluation (PPA, 88% [CI, 76% to 95%]). Among 30 RT-PCR–positive people who had more than 1 paired test and symptom information, antigen test performance at the lowest Ct value among all tests for an individual was similar between symptomatic people (16 of 20; PPA, 80%) and asymptomatic people (8 of 10; PPA, 80%).
Antigen Test Performance, by Stage of Infection
Of the 532 paired specimens analyzed, 356 (67%) were collected from people with a negative test result and no previous positive test result during the outbreak period and were categorized as uninfected. Of the remaining 176 paired specimens (33%), 56 (32%) were categorized as early infection, 88 (50%) as late infection, and 30 (17%) as resolved infection; we could not categorize the infection stage for 2 paired specimens (1%) (Appendix Table 8).
Appendix Table 8.
BinaxNOW COVID-19 Ag Card Performance, by Stage of Infection, Compared With Reference Standard RT-PCR

Among specimens categorized as early infection, PPA was 86% (CI, 74% to 94%) with RT-PCR (Appendix Table 8) and 95% (CI, 76% to 100%) with virus culture (Appendix Table 9). Among specimens categorized as late infection, PPA was 51% (CI, 36% to 66%) with RT-PCR and none were positive by virus culture. Among paired specimens categorized as early infection, the median Ct value was significantly lower for antigen-positive pairs (median, 25.1 [range, 15.4 to 36.4]) than for antigen-negative pairs (median, 28.6 [range, 15.4 to 36.4]) (Wilcoxon P = 0.049) (Figure 3). The median Ct value among paired specimens categorized as late infection was significantly lower for antigen-positive pairs (median, 31.7 [range, 30.1 to 36.4]) than for antigen-negative pairs (median, 34.8 [range, 30.1 to 38.7]) (Wilcoxon P = 0.006) (Figure 3).
Appendix Table 9.
BinaxNOW COVID-19 Ag Card Performance, by Stage of Infection, Compared With Alternative Reference Standard Virus Culture

Figure 3. Antigen test result during early and late infection, by SARS-CoV-2 nucleocapsid Ct values and virus culture result.

Shown are nucleocapsid Ct values for all paired RT-PCR–positive specimens with evidence of early (n = 56) or late (n = 47) infection by antigen and virus culture results. Median Ct for each category of antigen results is noted by the black bar. Virus culture was attempted for all RT-PCR–positive specimens with Ct ≤34. Culture was attempted for an additional 6 late-infection and 2 unknown-stage specimens that were antigen-positive and RT-PCR–negative; all were culture-negative. Randomized jitter of 0.5 was added to x-axis values to improve visibility. Antigen test: BinaxNOW COVID-19 Ag Card. Ct = cycle threshold; RT-PCR = reverse transcription polymerase chain reaction.
Discussion
Although highly sensitive RT-PCR can be an effective tool for thorough case finding during a nursing home outbreak, using RT-PCR to provide actionable results requires rapid turnaround and is likely to identify noninfectious people in addition to infectious ones. Despite low overall PPA compared with RT-PCR, in this evaluation antigen testing performed well in identifying early infections and specimens with replication-competent virus (that is, culture-positive). Further, consensus test analysis of test-positive individuals with more than 1 test result suggested that repeated testing produced similar PPA for antigen testing compared with RT-PCR regardless of the presence of symptoms. Our data suggest that early and frequent antigen testing during a SARS-CoV-2 outbreak can effectively identify infectious people with the greatest potential to transmit the virus.
Previous studies have shown that people with asymptomatic and presymptomatic SARS-CoV-2 infections can harbor high viral loads and contribute to widespread transmission within a nursing home (3, 33, 34). Rapid identification of these people is essential, and frequent facility-wide testing is recommended, particularly in outbreak settings (3). Although our data suggest that nearly a third of RT-PCR–positive infections were missed overall, the antigen test was able to identify 86% of infections when testing was done during early infection when people are more likely to be infectious. Previous work has shown that people can continue to test positive for SARS-CoV-2 by RT-PCR for weeks after they are no longer infectious (7, 9). Thus, comparisons of antigen testing with virus culture might provide a more accurate measure of antigen test performance for identifying infectious people. In this evaluation, PPA was very high (95%) among participants who had replication-competent virus in their specimen, suggesting that rapid antigen tests might be more useful for detecting people who are infectious. Pekosz and colleagues (35) found similar agreement (96%) between antigen testing and virus culture using the BD Veritor System for Rapid Detection of SARS-CoV-2, a lateral flow antigen detection test, on a convenience sample of RT-PCR–positive specimens. Of note, we found that 1 participant who had virus culture–positive specimens from 2 consecutive rounds of testing done 6 days apart had corresponding specimens that were also antigen-positive.
The antigen test was effective for identifying SARS-CoV-2 during early infection when viral RNA load might be high but was less effective during late infection. Kissler and colleagues (36) used serial RT-PCR testing to define and characterize infection stage dynamics (proliferation, clearance, and persistence) for symptomatic and asymptomatic infections. In their analysis, the average proliferation stage lasted 2 to 4 days and was similar regardless of symptoms. Thus, by doing point prevalence surveys every 5 to 6 days, we might have identified additional infections but potentially missed the proliferation stage of some new infections. Taken together, these data suggest that frequent antigen testing during an initial outbreak response might be an effective strategy for screening and identifying new SARS-CoV-2 infections that are in the early, proliferative stage—that is, the highly infectious period.
Despite previous concern about false-positive test results (17), only 8 false positives occurred during this evaluation (PNA, 98%), similar to rates reported in the EUA (99%) (37). Even with a high PNA because of a large volume of testing in nursing homes with a low percentage of positivity, there are concerns that frequent false positives may be problematic within a facility. Of note, all 8 specimens were collected from people with previously positive results (6 of 7 of whom tested positive by RT-PCR during the evaluation), suggesting that these false positives had some association with true infection and were not solely attributable to user error.
Our findings are subject to several limitations. Antigen test performance for asymptomatic infections might have been overestimated because of challenges in symptom ascertainment that led to misclassification of symptomatic people as asymptomatic. In addition, although virus culture was used to identify replication-competent virus, the inability to culture virus from a given specimen does not mean that replication-competent virus was not present in that specimen or person. Further, antigen testing was done by CDC laboratory staff, and test performance by nonlaboratory staff may not be equivalent (38). Finally, although Kissler and colleagues also showed that repeated quantitative testing with RT-PCR can be used to infer infection stages (36), the categorizations described for this evaluation might not be generalizable to other populations, test protocols, or testing frequencies and do not account for host factors, including antibody development (36).
Many antigen tests are inexpensive, fast, and relatively easy to perform and can be used to augment the testing capacity of clinical and public health laboratories. Despite the overall lower PPA compared with the reported EUA data, these findings show that the BinaxNOW antigen test performed well for identifying people who are infectious and will likely perform well when used serially as a screening tool for nascent and emerging COVID-19 outbreaks. Further, the generally high PNA between antigen testing and RT-PCR supports not doing confirmatory testing on antigen-positive individuals when the pretest probability is high, as in a large nursing home outbreak (4). Taken together, these data suggest that serial antigen testing early and often could be an effective testing strategy to support infection control in nursing homes having a SARS-CoV-2 outbreak. These findings merit further evaluation in other congregate settings, such as university campuses, hospitals, and detention centers.
Appendix 1: Members of the CDC Infection Prevention and Control Team and the CDC COVID-19 Surge Laboratory Group
Members of the CDC Infection Prevention and Control Team who authored this work: Amelia Bhatnagar, BS; Allison C. Brown, PhD, MPH; Jonathan Bryant-Genevier, PhD; Davina Campbell, MS, MPH; Brandi Freeman, PhD, MPH; Sarah E. Gilbert, MPH; Kelly M. Hatfield, MSPH; David A. Jackson, MD, MPH; John A. Jernigan, MD, MS; Preeta K. Kutty, MD, MPH; Stephen P. LaVoie, PhD; K. Danielle Lecy, RN; L. Clifford McDonald, MD; Susannah L. McKay, PhD, MPH; Erin D. Moritz, PhD, MS; Sujan C. Reddy, MD, MSc; and Farrell A. Tobolowsky, DO, MS.
Members of the CDC Infection Prevention and Control Team who contributed to this work but did not author it: Laura Green Brown, PhD; Dustin W. Currie, PhD, MPH; Juliana Carvalho DaSilva, MA; James L. Dawson, PhD; Matthew J. Hudson, MD, MPH; Kahaliah Joseph, MSc; Michelle A. Waltenburg, DVM, MPH; and Malania M. Wilson, MS, MBA.
Members of the CDC COVID-19 Surge Laboratory Group as part of the Laboratory and Testing Task Force who authored this work: Raydel Anderson, MS; Bettina Bankamp, PhD; Michael Bowen, PhD; Jennifer M. Folster, PhD; Magdalena Medrzycki, PhD; Kay W. Radford, BS; and Patricia L. Shewmaker, PhD.
Members of the CDC COVID-19 Surge Laboratory Group as part of the Laboratory and Testing Task Force who contributed to this work but did not author it: Leslie Barclay, MPH; Theresa K. Bessey, PhD; Caitlin Bohannon, PhD; Hannah E. Browne, BS; Heather Colley; Min-hsin Chen, PhD; Preeti Chhabra, PhD; Ebenzer David, PhD; Richard Ebai, MS; Brian Emery, BS; Matthew D. Esona, PhD; Lalitha Gade, MPharm; Renee Galloway, MPH; Rashi Gautam, PhD; Claire Hartloge, BS; Amy L. Hopkins, BS; Shilpi Jain, PhD; Brent Jenkins, BS; Baoming Jiang, PhD; Eric M. Katz, MS; Gimin Kim; Benton Lawson, MS; John M. Metz, MS; Congrong Miao, MS; Slavica Mijatovic-Rustempasic, MSc; Sung-Sil Moon, PhD; Kenny Nguyen, BS; Marla E. Petway, MPH; Shannon Rogers, MS; Kashif Iqbal Sahibzada, MS; Sarah L. Smart, BS; Jonathan Spencer, BS; Suganthi Suppiah, PhD; Alexandra Tejada-Strop, MS; Dexter Thompson, MCS; Srinivasan Velusamy, PhD; Jan Vinje; Houping Wang, PhD; Adam K. Wharton, MBA; Phili Wong, MS; Nhien Tran Wynn, MS; and HaoQiang Zheng, MD.
Appendix 2: Additional Methods and Results
Methods
Specimen Collection
Bilateral AN swab collection was done in 2 steps: First, the RT-PCR swab was inserted into 1 naris and the antigen swab into the other; then, each swab was removed and used to sample the opposite naris. When paired specimens included a nasopharyngeal swab for RT-PCR and AN swab for antigen testing, the bilateral AN swab was collected first followed by the nasopharyngeal swab collected from a single naris.
Virus Culture
To perform virus culture, 100 µl of clinical specimen was diluted 2-fold across a 96-well plate in serum-free Dulbecco's Modified Eagle Medium supplemented with 2 × penicillin–streptomycin and 2 × amphotericin B (Sigma). Vero CCL-81 cells were trypsinized and resuspended in Dulbecco's Modified Eagle Medium plus 10% fetal bovine serum plus 2 × penicillin–streptomycin plus 2 × amphotericin B at 2.5 × 105 cells/mL. A 100-µl cell suspension was added directly to the clinical specimen dilutions and mixed gently by pipetting. The inoculated cultures were grown in a humidified 37°C incubator with 5% CO2 and observed for cytopathic effect daily. When cytopathic effect was observed, presence of SARS-CoV-2 was confirmed by RT–PCR.
Results
Among 69 concordant antigen-positive and RT-PCR–positive specimens that were assessed for virus culture, 20 were virus culture–positive. These concordant specimens had a lower median Ct value (median, 21.1 [range, 15.4 to 26.7]) than 49 that were virus culture–negative (median, 29.7 [range, 22.5 to 35.0]) (P < 0.001). The remaining culture-positive specimen was discordant antigen-negative and RT-PCR–positive; this specimen was cultured from a nasopharyngeal swab.
Footnotes
This article was published at Annals.org on 27 April 2021.
* For members of the CDC Infection Prevention and Control Team and the CDC COVID-19 Surge Laboratory Group, see Appendix 1.
References
- 1. Centers for Medicare & Medicaid Services. COVID-19 nursing home data. 2020. Accessed at https://data.cms.gov/stories/s/COVID-19-Nursing-Home-Data/bkwz-xpvg on 10 January 2021.
- 2. Shi SM , Bakaev I , Chen H , et al. Risk factors, presentation, and course of coronavirus disease 2019 in a large, academic long-term care facility. J Am Med Dir Assoc. 2020;21:1378-1383.e1. [PMID: 32981664] doi: 10.1016/j.jamda.2020.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arons MM , Hatfield KM , Reddy SC , et al. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med. 2020;382:2081-2090. [PMID: 32329971] doi: 10.1056/NEJMoa2008457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. CDC. SARS-CoV-2 antigen testing in long term care facilities. Updated 7 January 2020. Accessed at www.cdc.gov/coronavirus/2019-ncov/hcp/nursing-homes-antigen-testing.html on 10 January 2021.
- 5. McGarry BE , SteelFisher GK , Grabowski DC , et al. COVID-19 test result turnaround time for residents and staff in US nursing homes. JAMA Intern Med. 2020. [PMID: 33125044] doi: 10.1001/jamainternmed.2020.7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Taylor J , Carter RJ , Lehnertz N , et al. Serial testing for SARS-CoV-2 and virus whole genome sequencing inform infection risk at two skilled nursing facilities with COVID-19 outbreaks — Minnesota, April–June 2020. MMWR Morb Mortal Wkly Rep. 2020;69:1288-1295. [PMID: 32966272] doi: 10.15585/mmwr.mm6937a3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bullard J , Dust K , Funk D , et al. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis. 2020;71:2663-2666. [PMID: 32442256] doi: 10.1093/cid/ciaa638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. La Scola B, Le Bideau M, Andreani J, et al.. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059-1061. [PMID: 32342252] doi: 10.1007/s10096-020-03913-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wölfel R , Corman VM , Guggemos W , et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465-469. [PMID: 32235945] doi: 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
- 10. Gniazdowski V , Morris CP , Wohl S , et al. Repeat COVID-19 molecular testing: correlation of SARS-CoV-2 culture with molecular assays and cycle thresholds. Clin Infect Dis. 2020. [PMID: 33104776] doi: 10.1093/cid/ciaa1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sia SF , Yan LM , Chin AWH , et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834-838. [PMID: 32408338] doi: 10.1038/s41586-020-2342-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Perera RAPM , Tso E , Tsang OTY , et al. SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg Infect Dis. 2020;26:2701-2704. [PMID: 32749957] doi: 10.3201/eid2611.203219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. CDC. Interim guidance on duration of isolation and precautions for adults with COVID-19. 2020. Accessed at www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html on 10 January 2021.
- 14. CDC. Interim guidance for antigen testing for SARS-CoV-2. Updated 16 December 2020. Accessed at www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines.html on 10 January 2021.
- 15. CDC. Testing guidelines for nursing homes. Updated 7 January 2020. Accessed at www.cdc.gov/coronavirus/2019-ncov/hcp/nursing-homes-testing.html on 10 January 2021.
- 16. U.S. Food and Drug Administration. Individual EUAs for antigen diagnostic tests for SARS-CoV-2. 2020. Accessed at www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/vitro-diagnostics-euas#individual-antigen on 10 January 2021.
- 17. U.S. Food and Drug Administration. Potential for false positive results with antigen tests for rapid detection of SARS-CoV-2 - letter to clinical laboratory staff and health care providers. 3 November 200. Accessed at www.fda.gov/medical-devices/letters-health-care-providers/potential-false-positive-results-antigen-tests-rapid-detection-sars-cov-2-letter-clinical-laboratory on 10 January 2021.
- 18. Pray IW , Ford L , Cole D , et al. Performance of an antigen-based test for asymptomatic and symptomatic SARS-CoV-2 testing at two university campuses — Wisconsin, September–October 2020. MMWR Morb Mortal Wkly Rep. 2021;69:1642-1647. [PMID: 33382679] doi: 10.15585/mmwr.mm695152a3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pilarowski G , Marquez C , Rubio L , et al. Field performance and public health response using the BinaxNOW TM Rapid SARS-CoV-2 antigen detection assay during community-based testing. Clin Infect Dis. 2020. [PMID: 33367619] doi: 10.1093/cid/ciaa1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Larremore DB , Wilder B , Lester E , et al. Test sensitivity is secondary to frequency and turnaround time for COVID-19 surveillance. med. Rxiv. 2020. [PMID: 32607516] doi: 10.1101/2020.06.22.20136309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mina MJ , Parker R , Larremore DB . Rethinking covid-19 test sensitivity — a strategy for containment. N Engl J Med. 2020;383:e120. [PMID: 32997903] doi: 10.1056/NEJMp2025631 [DOI] [PubMed] [Google Scholar]
- 22. See I , Paul P , Slayton RB , et al. Modeling effectiveness of testing strategies to prevent COVID-19 in nursing homes —United States, 2020. Clin Infect Dis. 2021. [PMID: 33564862] doi: 10.1093/cid/ciab110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. U.S. Department of Health & Human Services. Coronavirus (COVID-19) testing. 2020. Accessed at www.hhs.gov/sites/default/files/abbott-binaxnow-fact-sheet.pdf on 25 February 2021.
- 24. CDC. Symptoms of coronavirus. 2020. Accessed at www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html on 10 January 2021.
- 25. U.S. Food and Drug Administration. Individual EUAs for molecular diagnostic tests for SARS-CoV-2. 2020. Accessed at www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas-molecular-diagnostic-tests-sars-cov-2#individual-molecular on 10 January 2021.
- 26. COVID-19 Investigation Team. Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat Med. 2020;26:861-868. [PMID: 32327757] doi: 10.1038/s41591-020-0877-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. U.S. Food and Drug Administration. Statistical guidance on reporting results from studies evaluating diagnostic tests: guidance for industry and FDA staff. 13 March 2007. FDA docket no. 2003D-0044.
- 28.Protection of Human Subjects. 45 C.F.R. part 46.102(1)(1) (2018).
- 29.Institutional Review Boards. 21 C.F.R. part 56 (1981).
- 30.Records Maintained on Individuals. 5 U.S.C. §552a (2018).
- 31.Purposes. 44 U.S.C. §3501 (2018).
- 32.Research and Investigations Generally. 42 U.S.C. §241(d) (2011).
- 33. Kimball A , Hatfield KM , Arons M , et al. Asymptomatic and presymptomatic SARS-CoV-2 infections in residents of a long-term care skilled nursing facility — King County, Washington, March 2020. MMWR Morb Mortal Wkly Rep. 2020;69:377-381. [PMID: 32240128] doi: 10.15585/mmwr.mm6913e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. White EM , Santostefano CM , Feifer RA , et al. Asymptomatic and presymptomatic severe acute respiratory syndrome coronavirus 2 infection rates in a multistate sample of skilled nursing facilities. JAMA Intern Med. 2020;180:1709-1711. [PMID: 33074318] doi: 10.1001/jamainternmed.2020.5664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pekosz A, Cooper CK, Parvu V, et al. Antigen-based testing but not real-time PCR correlates with SARS-CoV-2 virus culture. medRxiv. Preprint posted online 5 October 2020. doi:10.1101/2020.10.02.20205708
- 36. Kissler SM, Fauver JR, Mack C, et al. SARS-CoV-2 viral dynamics in acute infections. medRxiv. Preprint posted online 1 December 2020. doi:10.1101/2020.10.21.20217042
- 37. Abbott. BinaxNOW COVID-19 Ag Card – instructions for use. U.S. Food and Drug Administration; 2020.
- 38. Peto T; UK COVID-19 Lateral Flow Oversight Team. COVID-19: rapid antigen detection for SARS-CoV-2 by lateral flow assay:a national systematic evaluation for mass-testing. medRxiv. Preprint posted online 26 January 2021. doi:10.1101/2021.01.13.21249563




