Introduction
The recently published original article by Mortaji et al. (2020) characterized for the first time the function of a type I toxin-antitoxin (TA) system in the gastric pathogen Helicobacter pylori. It was found that the high expression of an AapA1 toxin, which is part of this system, causes a drastic decrease in the amount of culturable H. pylori cells and their transformation from a spiral to a coccoid morphotype. It was also established that AapA1 is a hydrophobic peptide disrupting cell division and that oxidative stress is an inducer of the toxin expression.
The development of genomics and bioinformatics in recent years has contributed to the discovery of a high frequency of TA systems in microorganisms, which was a strong stimulus for the intensification of research on their structure and function (Lee and Lee, 2016; Yang and Walsh, 2017). Prokaryotic TA modules are genetic elements that encode information about a toxin involved in inhibiting growth of the bacterial producer and an antitoxin that counteracts the activity of the former. Toxins belonging to TA systems restrict microbial replication by targeting key processes for cell physiology, including replication, transcription, translation and/or cell wall synthesis (Harms et al., 2018). Attention is being paid increasingly to the participation of these systems in suppressing the microbial multiplication and the stimulatory effect on adaptation to stressful conditions, i.e., nutritional starvation, exposure to antimicrobial substances or immune system cells’ attack (Lee and Lee, 2016; Yang and Walsh, 2017).
To date, five type II TA systems of H. pylori have been identified. These include chromosomally encoded HP0892-HP0893 (Han et al., 2013), HP0894-HP0895 (Han et al., 2011), HP0315-HP0316 (Kwon et al., 2012), and HP0967-HP0968 (Cárdenas-Mondragón et al., 2016), and the newly identified TfiT-TfiA (Boampong et al., 2020), which is encoded on mobile genetic fragments. The expression of toxins belonging to the above modules arrest the growth of bacterial producers and cause the reduction of their number (expressed in CFU/mL). Similar observations were made in 2017 by Arnion et al. (2017), who first identified the existence of the type I TA system in H. pylori (called AapA1-IsoA1), and noted that the expression of the toxin significantly decreases the amount of culturable H. pylori cells. At this point it is worth mentioning that Mortaji et al. (2020) deepened the knowledge related to the above phenomenon. They proved in their next original article that this decline was caused by a reduction in the culturability (observed as the optical density of the culture) but not the viability of H. pylori (preserved cell membrane integrity and a stable ATP level), and was accompanied by the transition of morphology from spiral to coccoidal (Mortaji et al., 2020). This observation is very valuable from the scientific point of view and confirms the postulates presented by our research group, pointing to difficulties in the correct interpretation of the H. pylori viability (understood as the sum of various cell parameters suggesting its physiological activity) and frequent mistakes made by scientists taking the culturability (detected by culture optical density or CFU/mL) as the only determinant of the viability of this pathogen (Krzyżek and Grande, 2020).
An additional valuable cognitive element shown by Mortaji et al. (2020) was a proof that oxidative stress was an inducer of the aapA1 expression in H. pylori, and thus a trigger for the spiral-to-coccoid transition. Exposure to high concentrations of oxygen, understood here as oxidative stress, is a well-known stress factor for H. pylori determining its intensive transformation into spherical forms (Chuang et al., 2005; Zeng et al., 2008). Thus, Mortaji et al. (2020) neatly revealed a possible molecular mechanism governing this process. In regard to this, it is also worth paying attention to the results presented by many research teams that have shown that bactericidal antibiotics, unlike bacteriostatic ones, stimulate the formation of oxygen free radicals and oxidative stress in bacterial cells, regardless of their target site (Kohanski et al., 2007; Brynildsen et al., 2013; Dwyer et al., 2014; Belenky et al., 2015; Lobritz et al., 2015; Li et al., 2017). According to Lobritz et al. (2015), this effect was particularly visible with the use of antibiotics acting on the microbial cell wall and DNA, but neither translation nor transcription. The above information, in conjunction with the results provided by Mortaji et al. (2020), seem to be extremely interesting, as they may explain why bactericidal antibiotics (amoxicillin, levofloxacin or metronidazole) induce morphological transformation into spherical forms in H. pylori significantly faster than bacteriostatic antibiotics (Sörberg et al., 1997; Sörberg et al., 1998; Akada et al., 1999; Faghri et al., 2014; Krzyżek et al., 2019a; Krzyżek et al., 2019b). Still, it should be remembered that the process of cell death and/or formation of coccoids by H. pylori during the exposure to bactericidal antibiotics may depend on many factors simultaneously or be independent of oxidative stress.
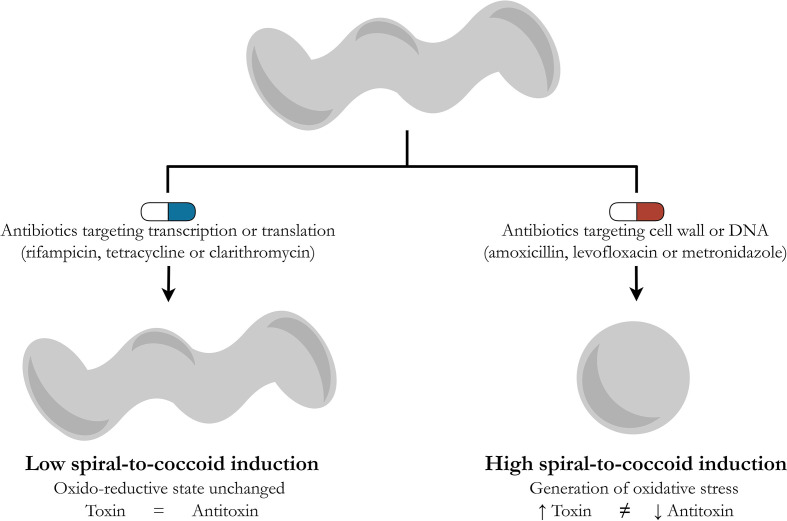
In the original article by Mortaji et al. (2020), H. pylori was exposed to one of two antibiotics: rifampicin or tetracycline targeting transcription or translation, respectively. The authors did not observe any significant increase in the aapA1 expression in rifampicin- or tetracycline-treated cells, concluding that exposure of H. pylori to antibiotics did not affect the expression of this toxin. In the light of the above presented deduction, however, it seems that divergent results may arise for other antibiotics used in the therapy of H. pylori, especially those with a strong bactericidal activity, e.g., amoxicillin, levofloxacin or metronidazole. Extending research to include these antibiotics would allow it to be established whether the hypothesis presented by an author of this commentary about the inducing effect of bactericidal antibiotics and their oxidative stress-dependent generation of morphological transition into spherical forms by H. pylori is correct ( Figure 1 ).
Figure 1.
Schematic drawing presenting a hypothetical model describing differences in the potential of antibiotics to generate the spiral-to-coccoid transformation in H. pylori. In the routine therapy of H. pylori, the following antibiotics are used: rifampicin (transcription), tetracycline and clarithromycin (translation), metronidazole and levofloxacin (the DNA structure or replication), and amoxicillin (the cell wall) (Jones et al., 2008; Francesco, 2011; Nishizawa and Suzuki, 2014). Based on reports showing the ability of bactericidal antibiotics to stimulate oxidative stress in microbial cells (Kohanski et al., 2007; Brynildsen et al., 2013; Dwyer et al., 2014; Belenky et al., 2015; Lobritz et al., 2015; Li et al., 2017) and the results of Mortaji et al. (2020), demonstrating the oxidative stress-dependent induction of the toxin-antitoxin system in H. pylori, a hypothetical model integrating the above observations has been proposed. Antibiotics acting on transcription and translation (rifampicin, tetracycline or clarithromycin) have a marginal effect on the oxido-reductive state of bacterial cells and therefore do not significantly affect the toxin-antitoxin balance. The opposite situation is suggested for antibiotics targeting the cell wall or DNA (amoxicillin, levofloxacin or metronidazole), all of which stimulate the accumulation of reactive oxygen species in bacterial cells and the oxidative stress-related disturbance of the toxin-antitoxin balance in favor of the former. The increased production of this toxin is accompanied by the conversion of H. pylori into spherical forms.
Finally, it is worth noting that the results presented by Mortaji et al. (2020) may have clinically significant implications, especially in the context of the eradication of difficult-to-treat, recurrent H. pylori infections. Recently, Morales-Espinosa et al. (2020) showed that the expression of HP0315, one of the components of the type II TA systems, is expressed significantly higher in intracellular H. pylori subpopulations and that the expression of this gene was accompanied by the formation of coccoid forms by these bacteria. Therefore, it seems very interesting to determine whether this type of relationship can also be demonstrated for other TA modules, including AapA1-IsoA1, and whether lowering the expression of the toxin or increasing the expression of the antitoxin would positively influence the frequency of H. pylori eradication.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
The study was supported by the Wroclaw Medical University grant No: SUB.A130.21.031. The funder had no role in the preparation of the manuscript.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Akada J. K., Shirai M., Fujii K., Okita K., Nakazawa T. (1999). In Vitro Anti-Helicobacter pylori Activities of New Rifamycin Derivatives, KRM-1648 and KRM-1657. Antimicrob. Agents Chemother. 43, 1072–1076. 10.1128/aac.43.5.1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnion H., Korkut D. N., Gelo S. M., Chabas S., Reignier J., Iost I., et al. (2017). Mechanistic Insights Into Type I Toxin Antitoxin Systems in Helicobacter pylori: The Importance of mRNA Folding in Controlling Toxin Expression. Nucleic Acids Res. 45, 4782–4795. 10.1093/nar/gkw1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenky P., Ye J. D., Porter C. B. M., Cohen N. R., Lobritz M. A., Ferrante T., et al. (2015). Bactericidal Antibiotics Induce Toxic Metabolic Perturbations That Lead to Cellular Damage. Cell Rep. 13, 968–980. 10.1016/j.celrep.2015.09.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boampong K., Smith S. L., Delahay R. M. (2020). Rapid Growth Inhibitory Activity of a YafQ-Family Endonuclease Toxin of the Helicobacter pylori Tfs4 Integrative and Conjugative Element. Sci. Rep. 10, 18171. 10.1038/s41598-020-72063-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brynildsen M. P., Winkler J. A., Spina C. S., MacDonald I. C., Collins J. J. (2013). Potentiating Antibacterial Activity by Predictably Enhancing Endogenous Microbial ROS Production. Nat. Biotechnol. 31, 160–165. 10.1038/nbt.2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas-Mondragón M. G., Ares M. A., Panunzi L. G., Pacheco S., Camorlinga-Ponce M., Girón J. A., et al. (2016). Transcriptional Profiling of Type II Toxin-Antitoxin Genes of Helicobacter pylori Under Different Environmental Conditions: Identification of HP0967-HP0968 System. Front. Microbiol. 7, 1872. 10.3389/fmicb.2016.01872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang M.-H., Wu M.-S., Lin J.-T., Chiou S.-H. (2005). Proteomic Analysis of Proteins Expressed by Helicobacter pylori Under Oxidative Stress. Proteomics 5, 3895–3901. 10.1002/pmic.200401232 [DOI] [PubMed] [Google Scholar]
- Dwyer D. J., Belenky P. A., Yang J. H., Cody MacDonald I., Martell J. D., Takahashi N., et al. (2014). Antibiotics Induce Redox-Related Physiological Alterations as Part of Their Lethality. Proc. Natl. Acad. Sci. U. S. A. 111, E2100–E2109. 10.1073/pnas.1401876111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghri J., Poursina F., Moghim S., Zarkesh Esfahani H., Nasr Esfahani B., Fazeli H., et al. (2014). Morphological and Bactericidal Effects of Different Antibiotics on Helicobacter pylori . Jundishapur. J. Microbiol. 7, e8704. 10.5812/jjm.8704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francesco V. (2011). Mechanisms of Helicobacter pylori Antibiotic Resistance: An Updated Appraisal. World J. Gastrointest. Pathophysiol. 2, 41. 10.4291/wjgp.v2.i3.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K. D., Ahn D. H., Lee S. A., Min Y. H., Kwon A. R., Ahn H. C., et al. (2013). Identification of Chromosomal HP0892-HP0893 Toxin-Antitoxin Proteins in Helicobacter pylori and Structural Elucidation of Their Protein-Protein Interaction. J. Biol. Chem. 288, 6004–6013. 10.1074/jbc.M111.322784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K. D., Matsuura A., Ahn H. C., Kwon A. R., Min Y. H., Park H. J., et al. (2011). Functional Identification of Toxin-Antitoxin Molecules From Helicobacter pylori 26695 and Structural Elucidation of the Molecular Interactions. J. Biol. Chem. 286, 4842–4853. 10.1074/jbc.M109.097840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms A., Brodersen D. E., Mitarai N., Gerdes K. (2018). Toxins, Targets, and Triggers: An Overview of Toxin-Antitoxin Biology. Mol. Cell 70, 768–784. 10.1016/j.molcel.2018.01.003 [DOI] [PubMed] [Google Scholar]
- Jones K. R., Cha J.-H., Merrell D. S. (2008). Who’s Winning the War? Molecular Mechanisms of Antibiotic Resistance in Helicobacter pylori . Curr. Drug Ther. 3, 190–203. 10.2174/157488508785747899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski M. A., Dwyer D. J., Hayete B., Lawrence C. A., Collins J. J. (2007). A Common Mechanism of Cellular Death Induced by Bactericidal Antibiotics. Cell 130, 797–810. 10.1016/j.cell.2007.06.049 [DOI] [PubMed] [Google Scholar]
- Krzyżek P., Franiczek R., Krzyżanowska B., Łaczmański Ł., Migdał P., Gościniak G. (2019. a). In Vitro Activity of 3-Bromopyruvate, an Anti-Cancer Compound, Against Antibiotic-Susceptible and Antibiotic-Resistant Helicobacter pylori Strains. Cancers (Basel) 11, 229. 10.3390/cancers11020229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyżek P., Franiczek R., Krzyżanowska B., Łaczmański Ł., Migdał P., Gościniak G. (2019. b). In Vitro Activity of Sertraline, an Antidepressant, Against Antibiotic-Susceptible and Antibiotic-Resistant Helicobacter pylori Strains. Pathogens 8, 228. 10.3390/pathogens8040228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyżek P., Grande R. (2020). Transformation of Helicobacter pylori Into Coccoid Forms as a Challenge for Research Determining Activity of Antimicrobial Substances. Pathogens 9, 184. 10.3390/pathogens9030184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon A. R., Kim J. H., Park S. J., Lee K. Y., Min Y. H., Im H., et al. (2012). Structural and Biochemical Characterization of HP0315 From Helicobacter pylori as a VapD Protein With an Endoribonuclease Activity. Nucleic Acids Res. 40, 4216–4228. 10.1093/nar/gkr1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. Y., Lee B. J. (2016). Structure, Biology, and Therapeutic Application of Toxin-Antitoxin Systems in Pathogenic Bacteria. Toxins (Basel) 8, 305. 10.3390/toxins8100305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Tan J., Shao L., Dong X., Ye R. D., Chen D. (2017). Selenium-Mediated Protection in Reversing the Sensitivity of Bacterium to the Bactericidal Antibiotics. J. Trace Elem. Med. Biol. 41, 23–31. 10.1016/j.jtemb.2017.02.007 [DOI] [PubMed] [Google Scholar]
- Lobritz M. A., Belenky P., Porter C. B. M., Gutierrez A., Yang J. H., Schwarz E. G., et al. (2015). Antibiotic Efficacy is Linked to Bacterial Cellular Respiration. Proc. Natl. Acad. Sci. U. S. A. 112, 8173–8180. 10.1073/pnas.1509743112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Espinosa R., Delgado G., Serrano L. R., Castillo E., Santiago C. A., Hernández-Castro R., et al. (2020). High Expression of Helicobacter pylori VapD in Both the Intracellular Environment and Biopsies From Gastric Patients With Severity. PLoS One 15, e0230220. 10.1371/journal.pone.0230220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortaji L., Tejada-Arranz A., Rifflet A., Boneca I. G., Pehau-Arnaudet G., Radicella J. P., et al. (2020). A Peptide of a Type I Toxin-Antitoxin System Induces Helicobacter pylori Morphological Transformation From Spiral Shape to Coccoids. Proc. Natl. Acad. Sci. U. S. A. 117, 31398–31409. 10.1073/pnas.2016195117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa T., Suzuki H. (2014). Mechanisms of Helicobacter pylori Antibiotic Resistance and Molecular Testing. Front. Mol. Biosci. 1:19. 10.3389/fmolb.2014.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sörberg M., Hanberger H., Nilsson M., Björkman A., Nilsson L. E. (1998). Risk of Development of In Vitro Resistance to Amoxicillin, Clarithromycin, and Metronidazole in Helicobacter pylori . Antimicrob. Agents Chemother. 42, 1228. 10.1128/AAC.42.5.1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sörberg M., Hanberger H., Nilsson M., Nilsson L. E. (1997). Pharmacodynamic Effects of Antibiotics and Acid Pump Inhibitors on Helicobacter pylori . Antimicrob. Agents Chemother. 41, 2218–2223. 10.1128/aac.41.10.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q. E., Walsh T. R. (2017). Toxin-Antitoxin Systems and Their Role in Disseminating and Maintaining Antimicrobial Resistance. FEMS Microbiol. Rev. 41, 343–353. 10.1093/femsre/fux006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H., Guo G., Mao X. H., De Tong W., Zou Q. M. (2008). Proteomic Insights Into Helicobacter pylori Coccoid Forms Under Oxidative Stress. Curr. Microbiol. 57, 281–286. 10.1007/s00284-008-9190-0 [DOI] [PubMed] [Google Scholar]