SUMMARY
Naked mole-rats are highly vocal, eusocial, subterranean rodents with, counterintuitively, poor hearing. The causes underlying their altered hearing are unknown. Moreover, whether altered hearing is degenerate or adaptive to their unique lifestyles is controversial. We used various methods to identify the factors contributing to altered hearing in naked and the related Damaraland mole-rats and to examine whether these alterations result from relaxed or adaptive selection. Remarkably, we found that cochlear amplification was absent from both species despite normal prestin function in outer hair cells isolated from naked mole-rats. Instead, loss of cochlear amplification appears to result from abnormal hair bundle morphologies observed in both species. By exploiting a well-curated deafness phenotype-genotype database, we identified amino acid substitutions consistent with abnormal hair bundle morphology and reduced hearing sensitivity. Amino acid substitutions were found in unique groups of six hair bundle link proteins. Molecular evolutionary analyses revealed shifts in selection pressure at both the gene and the codon level for five of these six hair bundle link proteins. Substitutions in three of these proteins are associated exclusively with altered hearing. Altogether, our findings identify the likely mechanism of altered hearing in African mole-rats, making them the only identified mammals naturally lacking cochlear amplification. Moreover, our findings suggest that altered hearing in African mole-rats is adaptive, perhaps tailoring hearing to eusocial and subterranean lifestyles. Finally, our work reveals multiple, unique evolutionary trajectories in African mole-rat hearing and establishes species members as naturally occurring disease models to investigate human hearing loss.
Graphical Abstract
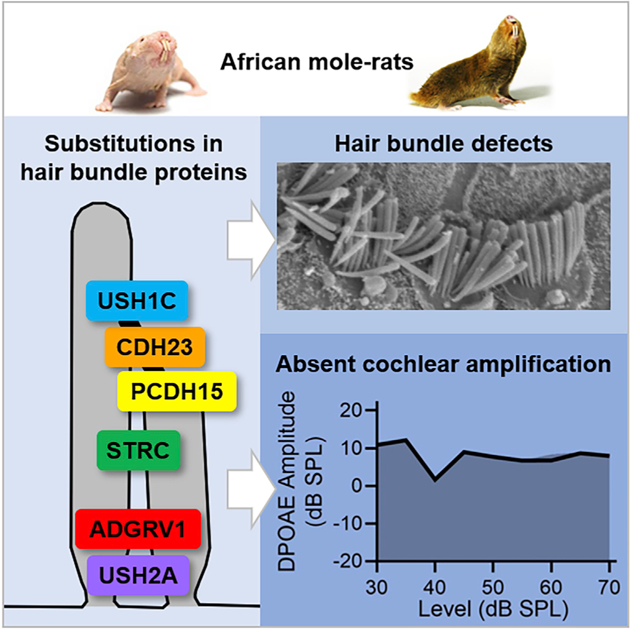
In Brief
Pyott et al. attribute comparatively poor hearing in African naked and Damaraland mole-rats to lack of cochlear amplification, disrupted hair bundles, and hair bundle proteins bearing deafness-associated amino acid substitutions. Positive selection in some bundle proteins suggests altered hearing is adaptive to subterranean and eusocial lifestyles.
INTRODUCTION
Most mammals hear with astounding sensitivity over broad frequency ranges [1]. Evolutionary adaptations within the cochlea—the auditory portion of the mammalian inner ear—underlie this feat. The most remarkable adaptation is prestin-mediated outer hair cell (OHC) electromotility [2], which enables cochlear amplification necessary for greater hearing sensitivity and high-frequency hearing [3]. Within mammals, rodents collectively exhibit an impressive sensitivity and range of hearing [4]. For example, gerbils (Meriones unguiculatus) hear frequencies over an impressive nine octaves, from 125 Hz to 60 kHz, with the most sensitive frequency at thresholds equivalent to that of humans, who hear over a narrower frequency range [5].
Curious then are the subterranean rodents. Most subterranean rodents are placed within four families: Bathyergidae (African mole-rats), Ctenomyidae (tuco-tucos), Geomyidae (pocket gophers), and Spalacidae (e.g., blind mole-rats) [6, 7]. Representatives from these families show elevated auditory thresholds and restricted frequency ranges of hearing compared with other surface-dwelling rodents [4, 8] (not known in Ctenomyidae). Members of the Bathyergidae family are among the best studied. Naked mole-rats (Heterocephalus glaber) hear within a narrow frequency range, from 125 cHz to 8 kHz, with only slightly more sensitive hearing at 4 kHz [9, 10]. Species in the genus Fukomys also hear within a narrow frequency range, between 125 cHz and 4 kHz, with the best hearing around 1 kHz [11]. Even at their best frequencies, audiograms indicate thresholds are elevated by 25 to 35 dB in these mole-rat species compared with mouse, rat, gerbil, guinea pig, and chinchilla [4].
The factors contributing to the comparatively reduced sensitivity and narrower frequency range of hearing in bathyergid mole-rats have not been identified. Most conspicuously, African mole-rats have no pinnae (ear lobes). Although the lack of pinnae may contribute to their poor sound localization (reported previously in naked mole-rats [9]), it cannot account for their elevated auditory thresholds at low frequencies (because pinnae in rodents are responsible for acoustical gain at frequencies above 8 kHz [12, 13]). A study using microcomputed tomography concluded that the gross dimensions of the middle and inner ear, including cochlear length, are not sufficient to explain altered hearing in bathyergids [14]. Another study reported alterations in the patterns of innervation of the cochlear hair cells in naked and Damaraland mole-rats (Fukomys damarensis) compared with mice and gerbils, but these alterations are again not sufficient to explain the decreased hearing sensitivity in mole-rats [15]. Finally, the most comprehensive examination of auditory function has been performed in Fukomys anselli. This species shows altered cochlear amplification [16] and expansion of low-frequency representation in the cochlear tonotopic map [17]. It is not known whether these features are shared by other bathyergid mole-rats.
The restricted frequency range and elevated auditory thresholds of naked mole-rats have been interpreted as degenerate hearing [9]. Loss of hearing acuity may result from loss of selective pressures imposed by above-ground acoustic environments, analogous to reduced visual acuity in mole-rats living in darkness [18]. A competing argument suggests that changes may be adaptive [19], arguing that auditory thresholds of subterranean rodents have not been assessed ecologically and do not account for the stethoscope effect [20]—the amplification of sound in subterranean burrows [21]. Indeed, measurements in natural burrows of various Fukomys species [21] and blind mole-rats [22] indicate amplification of low-frequency sounds (200–800 Hz) within the audible frequency ranges in these species [8, 11]. Reduced hearing sensitivity in subterranean species may have evolved to avoid acoustic overexposure. Moreover, the frequency and intensity range of hearing and vocalizations in naked mole-rats are well matched [10] and may support eusociality.
Recent genomic and transcriptomic analyses have been used to examine whether biological processes, including those related broadly to cochlear function, have undergone relaxed or adaptive selection in bathyergids. One study, which examined the genome from naked mole-rats and transcriptomes from all bathyergid genera, identified a cluster of genes relevant to the inner ear that underwent positive selection in the ancestral bathyergid [23]. However, examination of the genes within this cluster does not immediately identify the factors contributing to elevated thresholds in naked mole-rats. Another study identified convergent adaptive substitutions in genes related to cochlear function in both echolocating bats and cetaceans [24]. Although subterranean mammals were not included in this particular analysis, a separate analysis in this study reported both relaxed and adaptive selection in genes involved in hearing in three subterranean species, including naked mole-rats.
In this work, we used various methods to identify the factors contributing to altered hearing in bathyergid mole-rats and to examine whether these alterations result from relaxed or adaptive selection. By taking advantage of a well-curated hearing genotype-phenotype database [25], we identified amino acid substitutions matching pathogenic mutations in humans in the hair bundle link proteins from both naked and Damaraland mole-rats. These substitutions are consistent with abnormal hair bundle morphology observed by scanning electron microscopy and reduced cochlear amplification measured in vivo in both naked and Damaraland mole-rats despite normal prestin function observed in OHCs isolated from naked mole-rats. Therefore, our findings suggest that previously unrecognized amino acid substitutions in hair bundle proteins contribute to the elevated auditory thresholds and likely the narrower range of hearing in bathyergid mole-rats. Additional phylogenetic and molecular evolutionary analyses indicate that some of these substitutions accumulated independently in bathyergid mole-rats, evolved non-neutrally, and are likely adaptive. Our work, together with previous work identifying positive selection in genes regulating cochlear function, suggests that modifications to cochlear function and hearing in African mole-rats are adaptive.
RESULTS
Auditory Thresholds Are Elevated and the Cochlear Amplifier Is Absent from Bathyergid Mole-Rats
To assess cochlear function in mature naked and Damaraland mole-rats, we measured wave I auditory brainstem responses (ABRs) to pure tones ranging from 0.25 to 6 kHz (Figure 1A, red and blue traces, respectively). For comparison, we also measured wave I ABRs to pure tones ranging from 6 to 32 kHz in mice, a commonly used model system (Figure 1A, black traces). Wave I ABRs provide a measure of neural activity in the cochlea (arising from activation of the auditory nerve). In mice (n = 13), the hearing range (over which absolute thresholds were ≤60 dB SPL) extended from 6 to about 32 kHz, with the mean most sensitive frequency (lowest thresholds) between 12 and 16 kHz. In naked mole-rats (n = 2), the hearing range extended from 0.5 to 4 kHz (with thresholds ≤ 60 dB SPL). In Damaraland mole-rats (n = 4), the hearing range extended from 0.5 to 2 kHz (with thresholds ≈ 60 dB SPL). These findings agree with previously obtained audiograms from naked [9, 10] and Fukomys [11] mole-rats. Importantly, mole-rats show elevated auditory thresholds at their frequencies of highest sensitivity compared with many other surface-dwelling rodents [4]. Even at equivalent low frequencies (i.e., 1 kHz), thresholds are elevated by approximately 25 to 35 dB in naked and Damaraland mole-rats in comparison to other well-studied surface-dwelling rodents, including gerbils [5] and the hystricognath guinea pigs [26] and chinchillas [27]. In gerbils, guinea pigs, and chinchillas, sensitive low-frequency hearing does not come at the expense of reduced sensitivity to higher frequencies: in each of these three species, auditory thresholds are below 60 dB SPL within the range of 63 Hz to 32 kHz [5, 26, 27].
Figure 1. Auditory Thresholds Are Elevated and DPOAEs Are Absent from Bathyergid Mole-Rats.
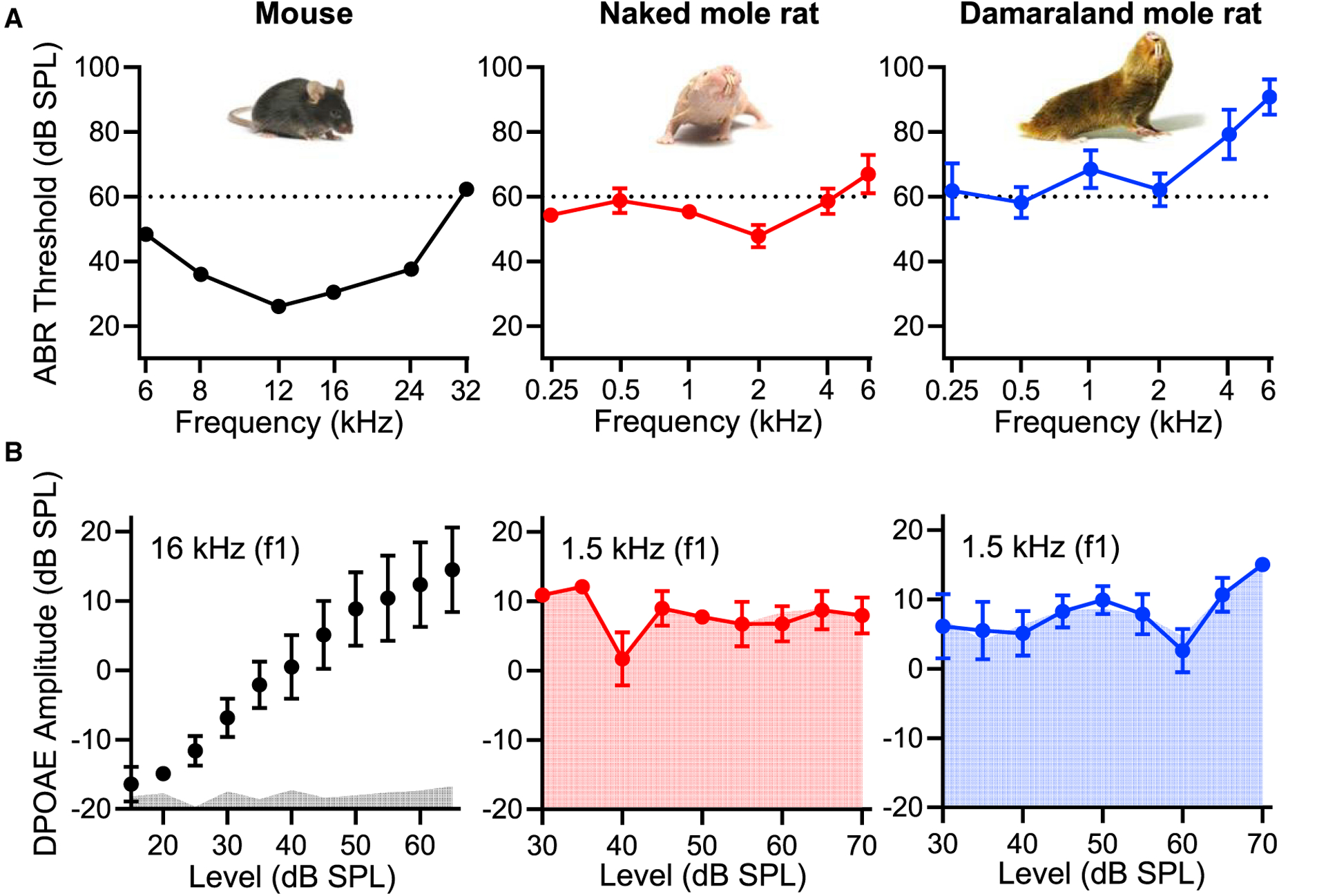
(A) In comparison to mice (black traces), auditory thresholds measured as the wave I ABR thresholds to pure tones (frequency) are elevated in naked mole-rats (red traces) and Damaraland mole-rats (blue traces). Mean ± SEM, n = 2–13.
(B) In contrast to mice (black traces and gray fills), 2f2-f1 DPOAE levels (DPOAE amplitude) as a function of the f1 level (indicated by the curves) are not significantly above the noise floor (indicated by the fills) in naked mole-rats (red traces) and Damaraland mole-rats (blue traces). Mean ± SEM, n = 3–11.
Elevated auditory thresholds in naked and Damaraland mole-rats could arise for many reasons. To examine specifically the integrity of the cochlear amplifier, we measured distortion product otoacoustic emissions (DPOAEs) (Figure 1B). Otoacoustic emissions are sounds emitted from the ear due to electromotility of the OHCs necessary for cochlear amplification. Thus, DPOAEs measure the robustness of cochlear amplification. In mice (n = 11), DPOAE response levels (measured near their most sensitive frequency) increased with stimulus level and were well above the noise floor (Figure 1B, black traces). Gerbils, guinea pigs, and chinchillas show robust DPOAEs over lower frequency ranges [28–30]. In contrast, in both naked mole-rats (n = 4) and Damaraland mole-rats (n = 3), DPOAE responses did not increase with the stimulus level. They were not significantly detectable above the noise floor (Wilcoxon matched-pairs signed-rank test; Figure 1B, red and blue traces, respectively). Absent DPOAEs in naked and Damaraland mole-rats indicate reduced (absent) cochlear amplification, consistent with their elevated thresholds measured by ABRs [9].
Prestin Localization and Function Are Normal in OHCs from Bathyergid Mole-Rats
In mammals, the membrane protein prestin is necessary for OHC electromotility and cochlear amplification [31]. Prestin knockout mice show wave I ABR threshold shifts of approximately 40 dB [32], similar to the elevations in auditory thresholds observed in both naked and Damaraland mole-rats at their frequencies of highest sensitivity (Figure 1). Moreover, variations in the amino acid sequence of prestin have been associated with nonsyndromic deafness in humans [33]. Therefore, we suspected that naked and Damaraland mole-rats might show differences in prestin expression compared with mice and/or that substitutions in the prestin protein sequence might give rise to loss of prestin function and consequently loss of cochlear amplification.
To test this prediction, we examined prestin expression and localization using immunofluorescence in the cochlear sensory epithelia (organs of Corti) isolated from mature mice and naked and Damaraland mole-rats (Figure 2A). Comparable to mice, prestin was expressed in the OHCs and localized to the basolateral cell membrane in both naked and Damaraland mole-rats. Qualitative differences in prestin abundance were not observed between individual OHCs, between rows of OHCs, or among these three species. Nevertheless, examination of prestin expression revealed differences in the morphology and organization of the OHCs (Figures 2A and 2B). Median cross-sectional areas of OHCs were larger in both naked mole-rats (38.4 μm2, n = 89 OHCs from 3 individuals; Figure 2B, red traces) and Damaraland mole-rats (57.3 μm2, n = 197 OHCs from 3 individuals; Figure 2B, blue traces) compared with mice (27.8 μm2, n = 199 OHCs from 3 individuals; Figure 2B, black traces). Based on previous observations [34] and the results of this study, the change in OHC diameter may relate allometrically to body weight. In mice and naked mole-rats, OHCs were relatively homogeneous in diameter and organized into three discrete rows. In contrast, OHCs in Damaraland mole-rats varied in size and were not organized into three discrete rows.
Figure 2. Prestin Localization and Function in OHCs Are Normal in Bathyergid Mole-Rats.
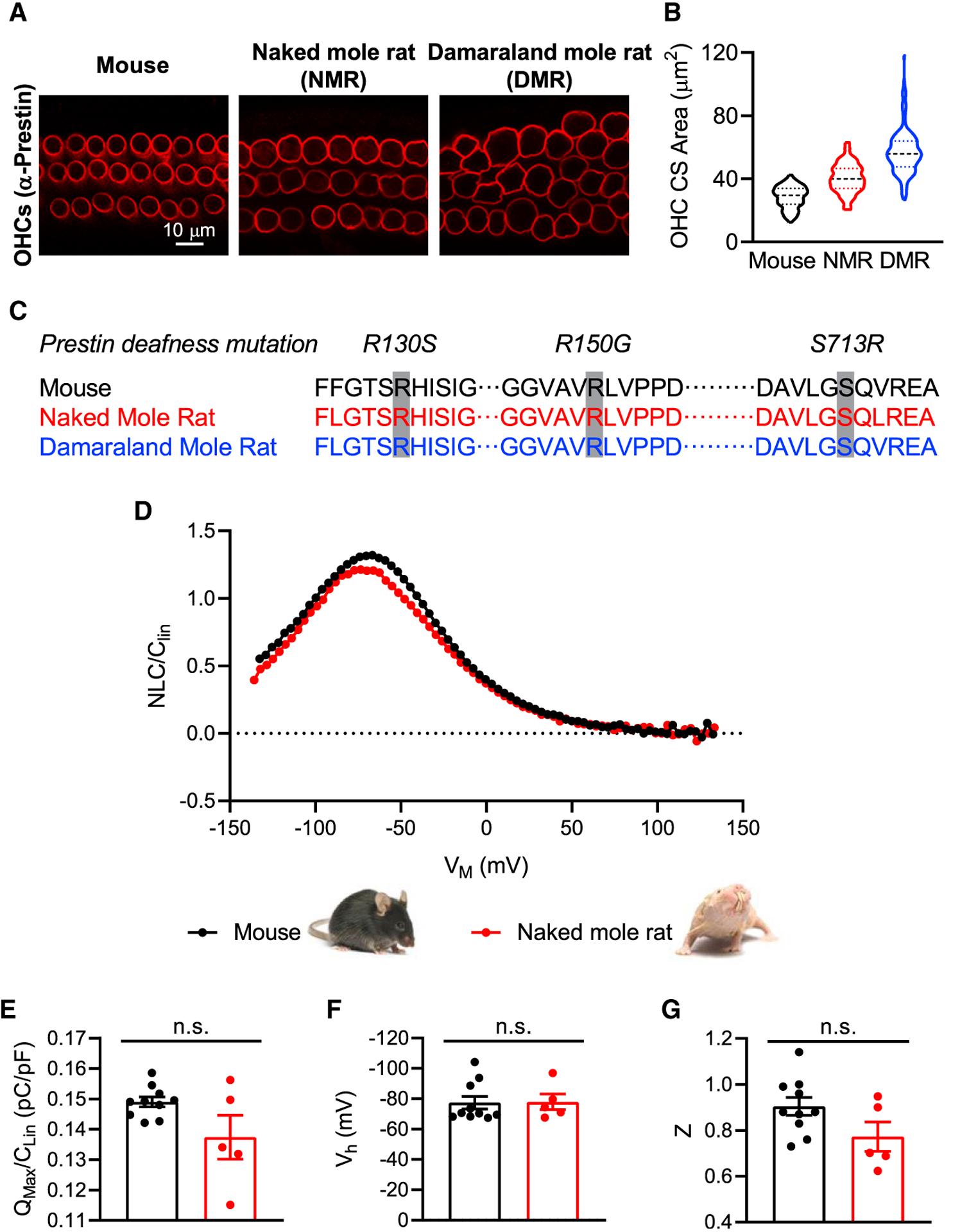
(A) Prestin expression identified in OHCs by immunofluorescence microscopy is distributed comparably in the lateral walls of OHCs from mice and naked and Damaraland mole-rats.
(B) OHC cross-sectional areas (OHC CS area), shown as violin plots (with median and quartiles) for mice (black), naked mole-rats (NMR, red), and Damaraland mole-rats (DMR, blue), are larger in mole-rats in comparison to mice. Violin plots showing distribution, median, and quartiles, n = 89–199.
(C) Prestin amino acid sequence in naked and Damaraland mole-rats in comparison to mouse shows no substitutions at sites associated with prestin loss of function and deafness in humans (R130S, R150G, and S713R).
(D) Measurement of the normalized NLC (NLC/CLin) as a function of the membrane voltage (VM) revealed no qualitative differences in prestin function in OHCs isolated from naked mole-rats (red trace) in comparison to mice (black trace).
(E–G) No significant differences of QMax/CLin (E), Vh (F), or Z (G) were observed between naked mole-rats and mice. Mean ± SEM, n = 5–10; n.s indicates p > 0.05, Mann-Whitney test.
To gain insight into the functional status of prestin in OHCs from these mole-rats, we first examined the prestin protein sequence (Figure 2C). Specifically, we examined three pathogenic amino acid mutations (R130S, R150G, and S713R) associated with prestin loss of function [25] and deafness in humans [35]. We found no substitutions in these three positions in either naked or Damaraland mole-rats compared with gerbil (data not shown), mouse, or human. We did observe amino acid substitutions in other positions in both naked and Damaraland mole-rat prestin sequences, but the functional consequences of these substitutions are not known.
We also assessed prestin function in OHCs isolated from naked mole-rats (Figures 2D–2G). Prestin-mediated OHC electromotility is accompanied by charge movement measured as a bell-shaped nonlinear capacitance (NLC) function [36]. The measurement of this NLC indicates prestin function. The normalized NLC function recorded from OHCs isolated from naked mole-rats (n = 5, red trace) was indistinguishable from those recorded from OHCs isolated from mature mice (n = 10; Figure 2D, blue trace). We found no significant differences in the normalized maximum nonlinear charge (QMax/CLin) moved between mice (0.149 ± 0.0102 pC/pF) and naked mole-rats (0.137 ± 0.007 pC/pF; Mann-Whitney test, p = 0.3097; Figure 2E). Comparable values have been reported previously for gerbils [37]. The variance in these measures was greater in naked mole-rats compared with mice and may reflect the greater variance in OHC diameter and non-prestin-containing membrane (which contributes to QMax/CLin) in naked mole-rats compared with mice (Figure 2B). There were also no significant differences in the half-activation voltage (Vh) between mice (−77.5 ± 4.2 mV) and naked mole-rats (−78.0 ± 5.1 mV; Mann-Whitney test, p = 0.5941; Figure 2F). Finally, there were no significant differences in the unitary charge valence (z) between mice (0.90 ± 0.04) and naked mole-rats (0.77 ± 0.06; Mann-Whitney test, p = 0.1215; Figure 2G).
These results show that reduced cochlear amplification in naked and Damaraland mole-rats does not result from the absence or altered function of prestin in the OHC. These findings, together with previous work showing that OHCs from both naked and Damaraland mole-rats have both afferent and efferent innervation [15], indicate that reduced cochlear sensitivity in these mole-rats is most likely the consequence of events preceding prestin-mediated OHC electromotility.
Hair Bundle Morphology Is Abnormal in Bathyergid Mole-Rats
Depolarization of the OHCs is required for prestin-mediated OHC electromotility and cochlear amplification. Depolarization occurs following sound-evoked, positive deflection of the hair bundles. The precise arrangement and orientation of hair bundles across the sensory epithelium is essential for efficient deflection of the OHC hair bundles, depolarization of the OHCs, and prestin activation. Not surprisingly, abnormal morphology of the hair bundles can cause hearing loss and deafness [38]. We used SEM to examine whether abnormal morphology of the hair bundles in naked and Damaraland mole-rats might contribute to their reduced cochlear amplification. We examined apical, apical/middle, middle/basal, and basal cochlear regions. Although cochlear frequency place maps are not known for these species, progressively more basal regions are expected to encode progressively higher frequencies [16].
SEMs revealed intact hair bundles from inner hair cells (IHCs) in all regions in both naked mole-rats (n = 4) and Damaraland mole-rats (n = 3), with a supernumerary row observed in apical turns in some naked mole-rats (Figure 3A). In contrast, hair bundles from OHCs were absent from or disorganized in the apical and apical/middle turns of both naked and Damaraland mole-rats (Figure 3A). Hair bundles were absent from the most apical regions of the naked mole-rat cochleae. In both naked and Damaraland mole-rats, abnormal hair bundles were observed in regions in which prestin immunoreactivity in the OHCs was observed (Figure 2A), suggesting that although OHCs were present, their hair bundles were missing or disorganized. Qualitatively, the morphology of the hair bundles became increasingly organized in progressively more basal regions in both naked and Damaraland mole-rats. Nonetheless, the orientations of the v-shaped hair bundles showed considerable variability. We quantified relative hair bundle orientation in naked and Damaraland mole-rats compared with mice (Figure 3B). In naked and Damaraland mole-rats, relative orientation angles ranged from −30.5° to 34.5° (n = 53 bundles, red bars) and −39.0° to 37.0° (n = 61 bundles, blue bars), respectively. In contrast, the relative orientation angles in mice were restricted to within −6.5° and 3.5° (n = 14 bundles, black bars). Previous examination of hair bundles from gerbils revealed an organized arrangement and orientation in the low-frequency apical turns comparable to our observations in mice [39]. Absent or disrupted alignment of the hair bundles would be expected to reduce the efficiency of sound-evoked OHC depolarization and, in turn, cochlear amplification.
Figure 3. Hair Bundle Morphology and Orientation Is Disrupted in Bathyergid Mole-Rats.
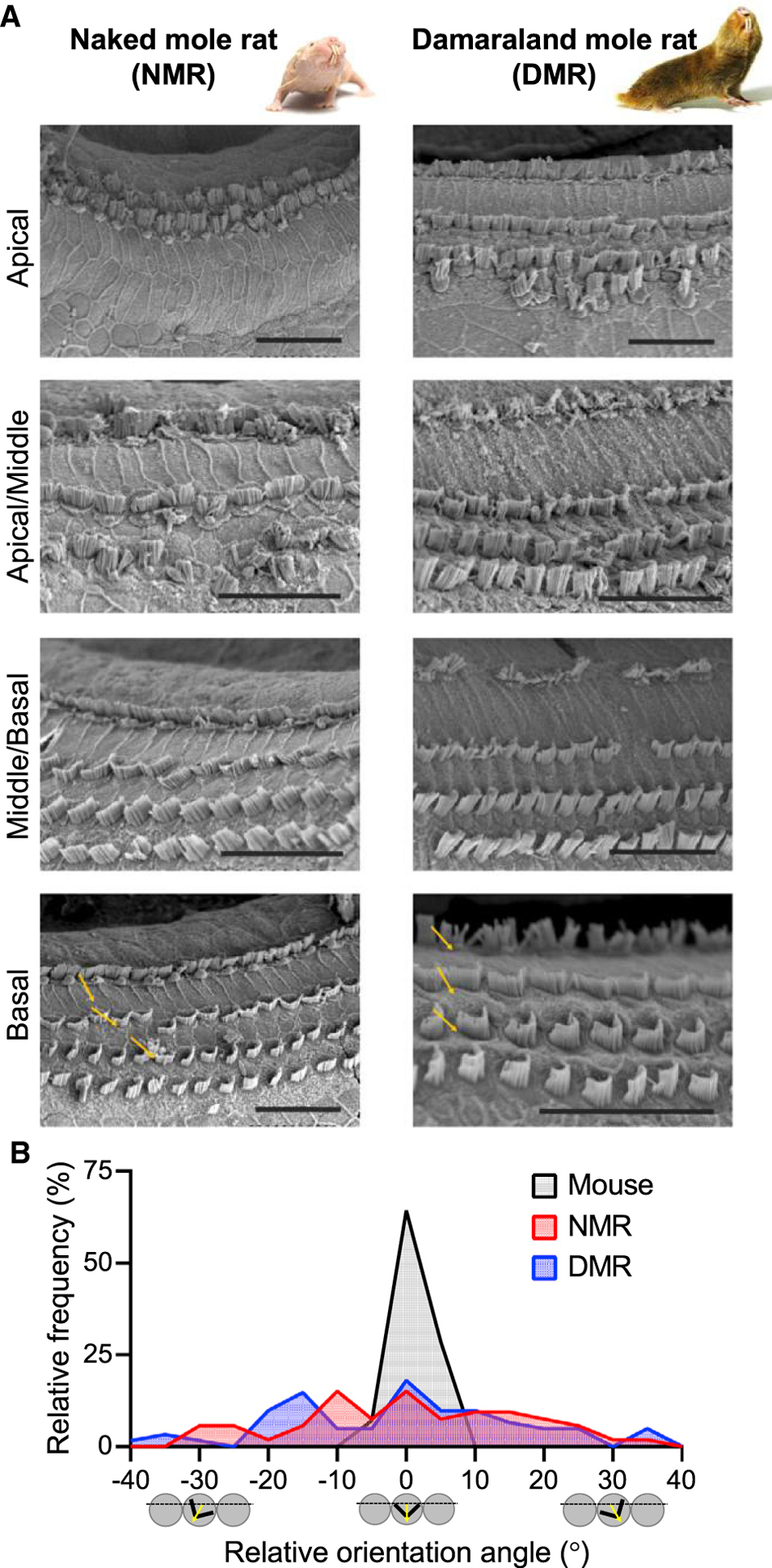
(A) Scanning electron micrographs show absent and/or disorganized OHC hair bundles in apical cochlear regions in both naked and Damaraland mole-rats. Hair bundle morphology improves in progressively more basal cochlear regions but nevertheless shows considerable variation in orientation (yellow arrows). Scale bars, 25 μm.
(B) OHC hair bundle orientation angles from basal cochlear regions show broader distributions in both naked mole-rats (red) and Damaraland mole-rats (blue) compared with mice (black; shown here for comparison).
Hair Bundle Link Proteins in Bathyergid Mole-Rats Contain Amino Acid Substitutions Matching Pathogenic Mutations in Humans
Not surprisingly, given the importance of hair bundle morphology to cochlear amplification, hearing loss and deafness can result from mutations in hair bundle proteins [25, 38]. Of the 66 genes known to encode hair bundle proteins, amino acid mutations in 29 of these genes are associated with deafness and hearing loss in humans [25]. To determine whether the abnormal hair bundle morphology and loss of cochlear amplification in naked and Damaraland mole-rats might result from the presence of amino acid substitutions that match pathogenic mutations identified in humans, we examined the amino acid sequences of these 29 hair bundle proteins in both naked and Damaraland mole-rats (Figure 4). Only variants associated with deafness and/or hearing loss were included to exclude the possibly confounding associations with nonauditory phenotypes.
Figure 4. Amino Acid Substitutions Are Present in Stereocilia Proteins in Bathyergid Mole-Rats.
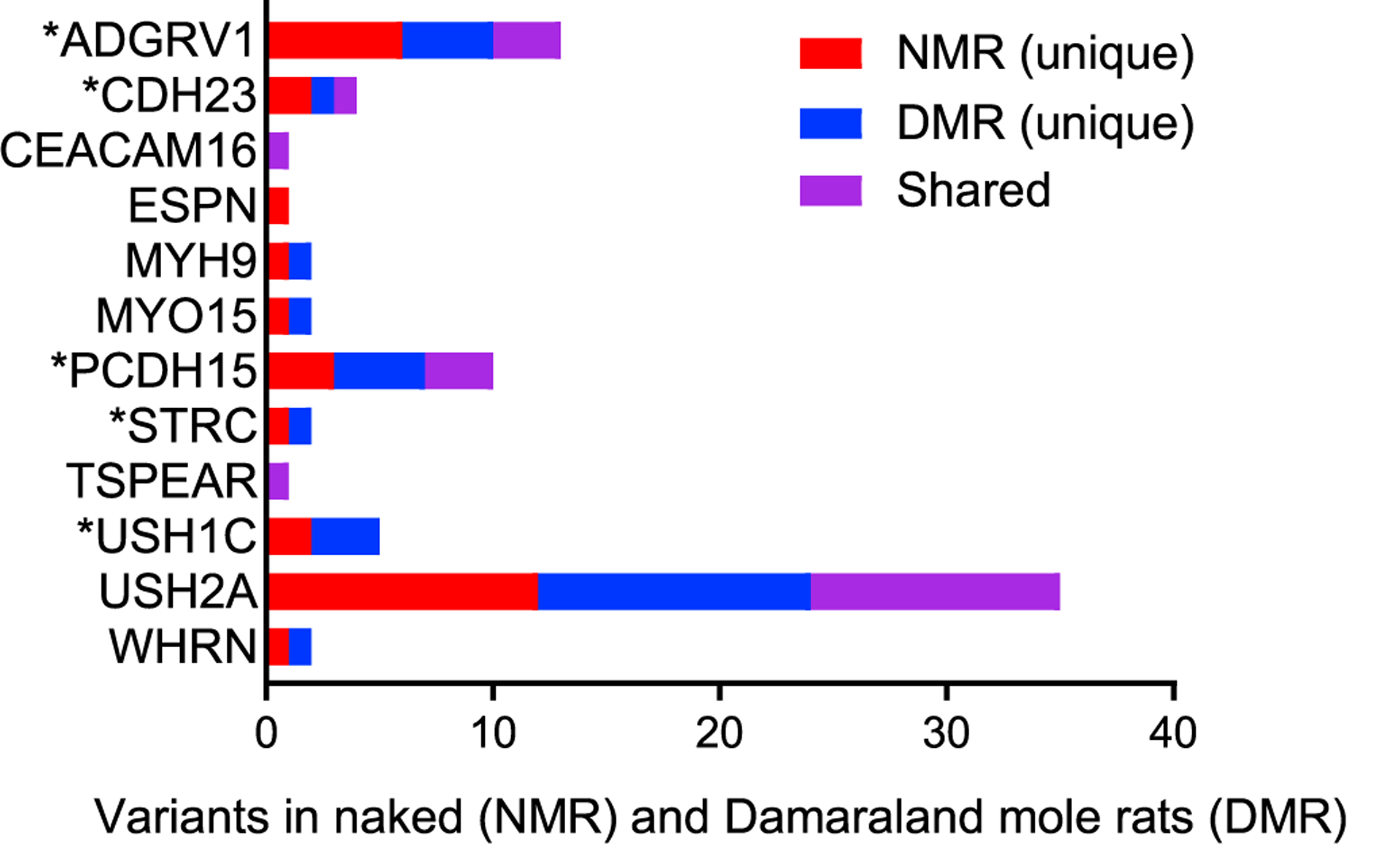
Twelve of the 29 identified stereocilia proteins have amino acid substitutions at positions corresponding to known sites of pathogenic mutations associated with deafness in humans. A total of 78 substitutions were identified: 30 unique substitutions in naked mole-rats (red bars), 28 unique substitutions in Damaraland mole-rats (blue bars), and 20 shared substitutions (purple bars). Seven substitutions in five hair bundle (stereociliar) proteins (marked with asterisks) match pathogenic variations associated with deafness in humans. These are CDH23 (V1090I) and STRC (R1541Q) in naked mole-rats and ADGRV1 (L4112W), CDH23 (E2438K), PCDH15 (V1622A), and USH1C (S190L and R339Q) in Damaraland mole-rats.
From both mole-rats, we observed a combined total of 78 amino acid substitutions at positions corresponding to known sites of pathogenic mutations in 12 (of the 29) hair bundle (stereociliar) proteins examined: ADGRV1 (adhesion G-coupled receptor V1), CDH23 (cadherin 23), CEACAM16, ESPN, MYH9, MYO15, PCDH15 (protocadherin 15), STRC (stereocilin), TSPEAR, USH1C (harmonin), USH2A (usherin), and WHRN (whirlin) (Figure 4). Although some substitutions were shared between species (Figure 4, purple bars), substitutions were mostly unique to either naked mole-rats (Figure 4, red bars) or Damaraland mole-rats (Figure 4, blue bars). Substitutions that match known pathogenic mutations in humans were found uniquely in naked and Damaraland mole-rats. There were seven substitutions in five proteins, including five substitutions in Damaraland mole-rats (ADGRV1, CDH23, PCDH15, and two in USH1C) and two substitutions in two proteins in naked mole-rats (CDH23 and STRC). Interestingly, substitutions that match known pathogenic mutations were restricted to hair bundle link proteins, proteins that interconnect hair bundle stereocilia [40].
The unique pattern of substitutions between naked and Damaraland mole-rats prompted us to examine the phylogenetic distribution of these substitutions among the larger group of bathyergid species, as well as the infraorder of hystricognath rodents that contains bathyergid mole-rats (Figure 5). Pathogenic substitutions were additionally identified in the common mole-rat (Cryptomys hottentotus), the silvery mole-rat (Heliophobius argenteocinereus), the greater cane-rat (Thryonomys swinderianus), and the social tuco-tuco (Ctenomys sociabilis). Moreover, substitutions matching pathogenic mutations in humans were found more frequently on fossorial (burrowing) branches (thick gray lines) and most frequently in subterranean lineages (thick black lines) compared with surface-dwelling rodents (thin black lines). Phylogenetic analysis also indicated that three pathogenic substitutions were modified further in naked, but not Damaraland, mole-rats (ADGRV1, PCDH15, and USH2A, indicated by the prime and double-prime symbols). Most amino acid substitutions (substitutions listed in Figure 5) are expected to result in altered chemical properties. None of the genes encoding these six proteins (ADGRV1, CDH23, PCDH15, STRC, USH1C, and USH2A) are pseudogenes. Three of the seven identified pathogenic substitutions (ADGRV1-2, CDH23-3, and STRC-1) are associated with nonsyndromic hearing loss (and thus not additionally associated with reported visual or vestibular deficits).
Figure 5. Phylogenetic Distribution of Amino Acid Substitutions Matching Pathogenic Variations in Hair Bundle Link Proteins in Bathyergid Mole-Rats and Hystricognath Rodents Indicates Independent Evolutionary Accumulation of Substitutions.
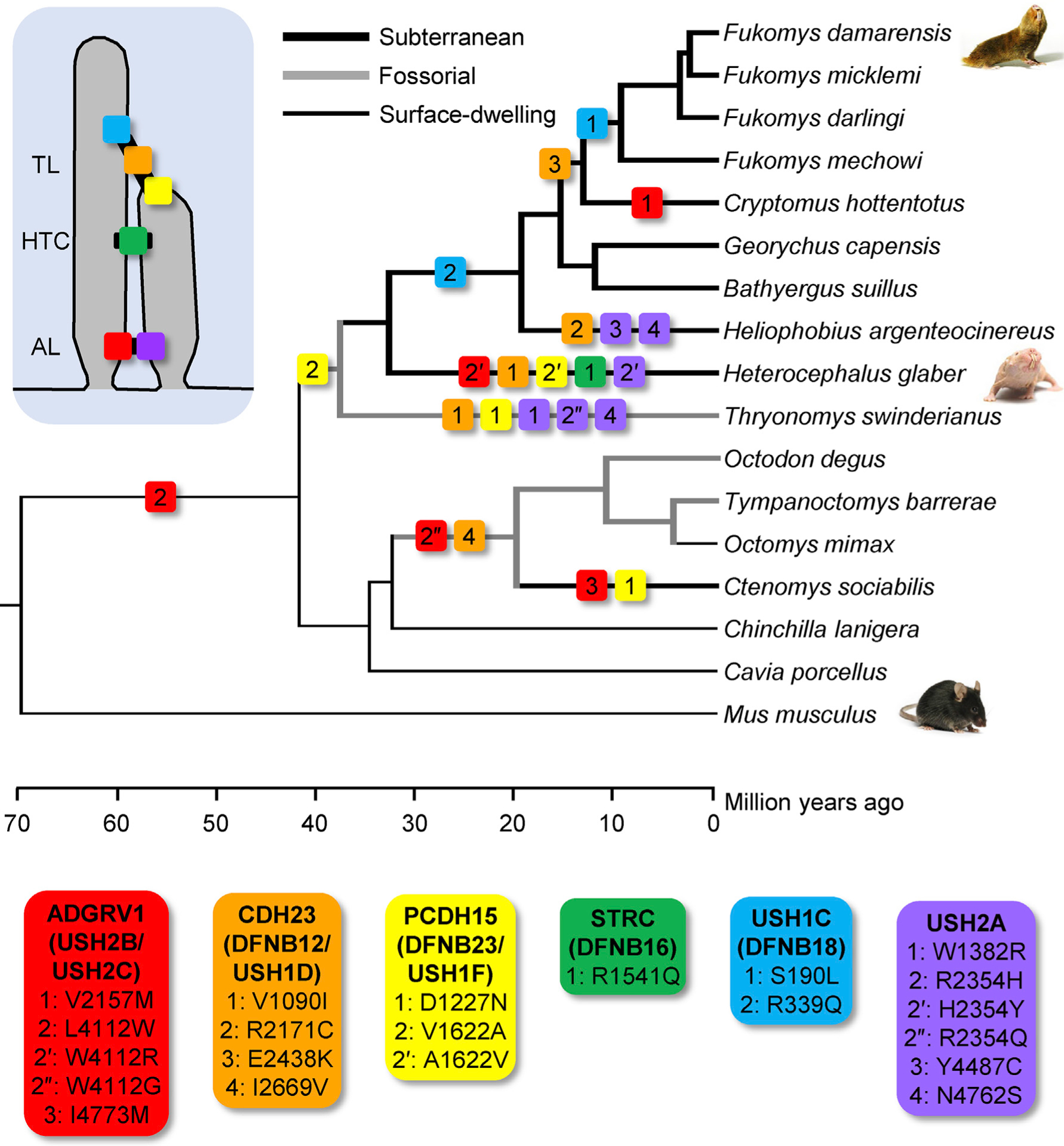
Phylogenetic distribution of the substitutions identified in six link proteins (ADGRV1, CDH23, PCDH15, STRC, USH1C, and USH2A; see insets) shows that these substitutions are found more frequently on fossorial branches (thick gray lines) and most frequently in subterranean lineages (thick black lines) compared with surface-dwelling rodents (thin black lines). Three substitutions were modified further in naked mole-rats (indicated by prime and double-prime symbols). In an equally parsimonious scenario, the CDH23-1 and PCDH15-2 substitutions could have alternatively arisen ancestral to the Thryonomys-bathyergid split, with reversions occurring ancestral to the branch leading to Fukomys species. In either scenario, naked and Damaraland mole-rats show unique substitutions. Divergence times are based on published estimates summarized in TimeTree [41] and should be considered approximate. TL, tip link; HTC, horizontal top connectors; AL, ankle link.
Molecular Evolutionary Analyses Suggest Shifting Selective Pressures and Potentially Adaptive Substitutions in Hair Bundle Link Proteins Harboring Substitutions
We performed several analyses to investigate changes in selection in the genes encoding these six hair bundle link proteins. First, we estimated the ratio of the numbers of nonsynonymous substitutions per nonsynonymous site to the number of synonymous substitutions per synonymous site (dN/dS) in the bathyergid and nonbathyergid (NB) portions of the phylogeny. For most genes, selective constraint results in the number of synonymous substitutions vastly outnumbering the number of nonsynonymous substitutions. Thus, increases in the dN/dS ratio between lineages indicate shifting selection. This analysis revealed higher average dN/dS ratios for all six hair bundle link proteins in the bathyergid portion of the phylogeny compared with the NB portion, indicating shifting selection in bathyergids (Table 1).
Table 1.
Molecular Evolutionary Analyses Suggest Non-neutral and Adaptive Evolution in Hair Bundle Link Proteins
| Average dN/dS (Gene Wide) | dN-dS (Codon Specific) | EP | Overall Evolutionary Signala | ||||
|---|---|---|---|---|---|---|---|
| B | NB | B+NB | NB | derived | ancestral | ||
| ADGRV1-2 | 0.357 | 0.221 | 1.13 | 1.58 | 0.655 | 0.174 | selection shifting/adaptive |
| ADGRV1-2′ | 0.006 | 0.655 | adaptive | ||||
| CDH23-1 | 0.086 | 0.043 | 0.50 | 0.00 | 0.012 | 0.971 | adaptive |
| CDH23-3 | 0.46 | 0.00 | 0.002 | 0.975 | adaptive | ||
| PCDH15-2 | 0.144 | 0.099 | 0.00 | 0.00 | 0.084 | 0.811 | neutral |
| PCDH15-2′ | 0.811 | 0.084 | neutral | ||||
| STRC-1 | 0.352 | 0.178 | −0.12 | −1.73 | 0.008 | 0.941 | selection shifting/adaptive |
| USH1C-1 | 0.378 | 0.097 | −0.89 | −3.00 | 0.002 | 0.908 | selection shifting/adaptive |
| USH1C-2 | 0.55 | −0.91 | 0.007 | 0.946 | adaptive | ||
| USH2A-2′ | 0.388 | 0.302 | 0.42 | 0.42 | 0.014 | 0.775 | adaptive |
Greater gene-wide dN/dS and codon-specific dN-dS values in columns including bathyergid (B) species compared with values in columns including non-bathyergid (NB) species indicate shifts toward greater numbers of nonsynonymous substitutions in B species compared with NB species and therefore shifting selection in B species. If dN-dS is additionally greater than zero in columns including B species, then shifting selection is considered likely adaptive in B species. EP values below 0.05 in columns including B species support adaptive shifts in selection. dN-dS values for ADGRV1 and USH2A-2′ are greater than zero for B+NB species, consistent with the occurrence of these substitutions before the ancestral divergence of Bathyergidae.
Because selective pressures can vary at different positions within a gene, we also investigated codon-specific dN-dS values for each of the seven pathogenic substitutions identified in naked and Damaraland mole-rats, as well as the three reversals/substitutions identified in naked mole-rats (ADGRV1-2′, PCDH15-2′, and USH2A-2′). Under strictly neutral evolution, dN-dS values will equal zero. In contrast, adaptive selection results in dN-dS values that are positive, and negative selection results in dN-dS values that are negative. Because the number of changes at each position is small, statistical tests estimating the likelihood that dN > dS at a single site have rather low power [42]. Instead, codon-specific dN-dS values determined from a sampling of bathyergid and nonbathyergid (B+NB) species were compared with the codon-specific dN-dS values determined from a control sampling of only NB species. For codons harboring the CDH23-1, CDH23-3, and USH1C-2 pathogenic substitutions, the dN-dS values exceeded zero when bathyergid mole-rats were included and were zero or less than zero when bathyergid mole-rats were excluded, indicating adaptive selection. Moreover, the codons harboring the STRC-1 and USH1C-1 pathogenic substitutions had more positive dN-dS values when bathyergid mole-rats were included compared with when they were excluded, consistent with shifting selection (Table 1).
Finally, we calculated the neutral evolutionary probability (EP) for these seven pathogenic substitutions and three reversals/substitutions. The EP quantifies the probability of observing an amino acid residue at a given site and is higher when this residue occurs frequently across related species. A high EP suggests neutral evolution or constraint. In contrast, function-altering alleles have low EPs and are candidate adaptive polymorphisms when found with a high frequency in a population [43]. When compared with the ancestral EP values (which averaged 0.88), the derived EP values for the bathyergid acid substitutions were lower (EP < 0.05) for ADGRV1-2′, CDH23-1/3, STRC-1, USH1C-1/2, and USH2A-2′ proteins. Therefore, amino acid substitutions in these positions are not expected in naked and Damaraland mole-rats based on the long-term evolutionary patterns observed at these positions. Thus, these results support shifting and likely adaptive selection in these genes. EP values are shown in Table 1.
In summary, molecular evolutionary analyses suggest non-neutral evolution and signs of positive selection in at least five of the hair bundle link proteins: ADGRV1, CDH23, STRC, USH1C, and USH2A. These patterns, coupled with the finding that three pathogenic substitutions (ADGRV1-2′, CDH23-3, and STRC-1) are associated with nonsyndromic hearing loss, indicate adaptive selection specifically on auditory function in bathyergids.
DISCUSSION
In this work, we provide insight into the functional and evolutionary mechanisms responsible for altered hearing in bathyergid mole-rats compared with surface-dwelling rodents. Our findings suggest that hair bundle defects, likely resulting from amino acid substitutions in hair bundle link proteins, diminish cochlear amplification and result in elevated auditory thresholds in naked and Damaraland mole-rats. Moreover, because the ancestor to naked and Damaraland mole-rats comprises the earliest divergence in the bathyergid family—covering approximately 30 million years of evolutionary history [23]—the data presented here also provide insight into the evolution of bathyergid hearing. Importantly, amino acid substitutions in hair bundle link proteins accumulated uniquely, independently, and possibly adaptively in the evolutionary history of these two species, suggesting that auditory function may be tailored to species-specific differences in vocalizations [44–46], eusociality [47], and subterranean burrow architecture [48, 49].
In naked and Damaraland mole-rats and related hystricognath rodents, amino acid substitutions matching pathogenic mutations in humans were found in five hair bundle link proteins— ADGRV1, CDH23, PCDH15, STRC, and USH1C—with an additional modification identified in USH2A. Although the effect of these amino acid substitutions in mole-rats is not known, there is remarkable conservation between the pathology of hearing loss observed in humans and that observed in rodents (mice) [50]. The shared pathology between humans and rodents likely reflects the conserved function of these proteins in mammalian hair bundles [51].
To summarize these effects, CDH23, PCDH15, and USH1C are tip link proteins [40]. In humans, mutations in CDH23 result in Usher syndrome type 1D (USH1D [52, 53]) and nonsyndromic autosomal recessive deafness DFNB12 [53–55]. Similarly, mice with null and missense mutations in CDH23 [56, 57] show, respectively, congenital syndromic and progressive nonsyndromic hearing loss associated with disrupted hair bundles. PCDH15 is a kinocilial and tip link protein that interacts with CDH23 [40]. Various mutations in PCDH15 give rise to nonsyndromic deafness DFNB23 and Usher syndrome type 1F [58, 59]. Spontaneous and engineered mutations in PCDH15 in mice cause hair bundle defects and degeneration of the cochlear neuroepithelium [60–64]. Mutations in USH1C cause Usher syndrome type 1C (also associated with nonsyndromic recessive deafness DFNB18 [65]). Again, mouse models show hair bundle defects and subsequent degeneration of the cochlear neuroepithelium [61, 66, 67]. Our analyses indicate adaptive selection in CDH23 in both naked and Damaraland mole-rats and USH1C in Damaraland mole-rats. The CDH23-3 pathogenic substitution is specifically associated with nonsyndromic hearing loss. Our analyses do not indicate adaptive selection for the PCDH15-2/2′ pathogenic substitution and reversal.
STRC is a horizontal top connector [40]. In humans, mutations in STRC lead to nonsyndromic autosomal recessive deafness DFNB16 [68]. In mice, STRC connects OHC stereocilia to one another and to the tectorial membrane [69], and STRC knockout mice show marked OHC dysfunction, absent DPOAEs [70], and hearing loss [69, 71]. Our analyses indicate adaptive selection (based on dN/dS and EP values) in STRC in naked mole-rats. The STRC-1 pathogenic substitution is specifically associated with nonsyndromic hearing loss.
Finally, ADGRV1 and USH2A are both ankle link proteins [40]. ADGRV1 (also known as very large G-protein receptor 1 or G-protein coupled receptor 98) is clinically associated with Usher syndrome type 2 [72]. ADGRV1 knockout mice show absent DPOAEs, elevated auditory thresholds, and progressively disorganized OHC hair bundles [73, 74]. Mutations in USH2A causes Usher syndrome type 2A, the most common subtype of Usher syndrome [75]. Null mutant mice show elevated DPOAEs and hair cell loss in the high-frequency regions of the cochlea [76]. Maximum parsimony suggests that both genes underwent substitutions before the common ancestor of Bathyergidae, with further modification/reversal in naked mole-rats. Our analyses further indicate that both reversals (ADGRV1-2′ and USH2A-2′) are adaptive. The ADGRV1-2 pathogenic substitution is specifically associated with nonsyndromic hearing loss.
The presence of hair bundle defects and reduced cochlear amplification in mole-rats harboring amino acid substitutions matching pathogenic mutations in humans is therefore consistent with the pathology observed in humans and rodents (mice). Future work should particularly examine tip link integrity in naked and Damaraland mole-rats. Nevertheless, there is an unexpected difference in the phenotype observed in mole-rats compared with humans and mice. Pathology typically progresses from basal (high frequency) to apical (low frequency) cochlear regions in humans and mice [77]. In contrast, in naked and Damaraland mole-rats, hair bundles were more disorganized in apical compared with basal cochlear regions, a pattern not immediately consistent with their poor high-frequency hearing. Various factors may contribute to this discrepancy. In general, hearing loss associated with these proteins shows considerable variability and progression depending on the specific mutations and is influenced by other factors, including genetic background [78], transcriptional variants [79], digenic inheritance [80, 81], and protein interactions [82]. Moreover, attenuation of high-frequency sounds in underground burrows [21] may limit sound exposure to lower frequencies that preferentially damage only the low-frequency regions of the cochlea. Previous examination of the blind mole-rat (Spalax ehrenbergi), which belongs to the family Spalacidae, also reported worse hair bundle defects in apical compared with basal cochlear regions [83]. Poor high-frequency hearing in these three mole-rat species may result from off-frequency hearing: the low-frequency regions of the cochlea may function so poorly that sound is detected off-place (in higher-frequency regions) with sufficiently greater sound intensities [84].
Our findings of adaptive rather than degenerate changes in hearing in bathyergid mole-rats are supported by independent lines of evidence and may extend to other aspects of cochlear function in additional groups of subterranean mammals. First, previous genomic analyses identified STRC and CDH23 as having undergone adaptive selection on the branch leading to naked mole-rats [85] and Damaraland mole-rats [86], respectively. Similar work identified additional hair bundle proteins that have undergone adaptive selection, including LOXHD1 on the branch leading to naked mole-rats [87] and MYO3A and CEACAM16 on the branch ancestral to hystricognath rodents [86]. Mutations in each of these hair bundle proteins are associated with nonsyndromic hearing loss in humans. Moreover, various studies have failed to find support for the loss of selection (indicated by pseudogenization and/or gene loss) in stereocilia proteins in bathyergid mole-rats [85, 87, 88]. Second, adaptive selection has been previously identified in genes encoding proteins essential for other aspects of cochlear function, including SLC12A2, an ion transporter essential for the maintenance of the endocochlear potential and associated with deafness in knockout mice [89, 90], in naked mole-rats [87]; TECTA, a component of the tectorial membrane-associated with DFNA12 [91], in Damaraland mole-rats [87]; and SLC4A7, another ion transporter associated with deafness in mice [92, 93], in both naked and Damaraland mole-rats [23]. Moreover, adaptive selection has been identified in ADGRV1 and USH1C in talpid moles and MYO15A in golden moles [94], suggesting that adaptive changes we observed in Bathyergidae and likely in Ctenomyidae and Thryonomidae extend to other mammalian orders with members that have independently obtained the subterranean lifestyles. One of the ADGRV1 amino acid substitutions (ADGRV1-2: L4112W) is found ancestral to all hystricognath rodents (Figure 5). Some members of this group, notably guinea pigs and chinchillas, have sensitive low-frequency hearing [4], suggesting the intriguing possibility that this substitution associates with low-frequency specializations that evolved commonly in all hystricognath rodents as an adaptation for long-distance communication in open, arid environments [95].
In summary, our findings indicate that adaptations in hearing and cochlear function in bathyergid mole-rats followed distinct evolutionary trajectories, similar to adaptations to sociality [96] and physiology [97]. Therefore, researchers should be wary of collapsing members to a single bathyergid representative when examining adaptations to subterranean lifestyles. These results also establish members of this fascinating group as naturally occurring disease models to investigate human hearing loss.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sonja J. Pyott (s.pyott@umcg.nl).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Original/source data for figures in the paper are available from the corresponding author on request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal protocols conformed to the respective legislation at the University of Illinois at Chicago (Chicago, IL, USA), Johns Hopkins University School of Medicine (Baltimore, MD, USA), Yale University (New Haven, CT, USA), and the University Medical Center Groningen (Groningen, the Netherlands). Naked mole-rats (Heterocephalus glaber) and Damaraland mole-rats (Fukomys damarensis) were obtained from colonies maintained at the University of Illinois at Chicago. Mice (C57BL6, Mus musculus) were obtained from colonies maintained at the University of Illinois at Chicago, the University Medical Center Groningen and Yale University. A total of 38 mice, 18 naked mole-rats, and 13 Damaraland mole-rats were used. To avoid examination of age-related pathology of the inner ear, all animals were mature but not aged. Mice were used at 6 weeks of age for all experiments. Naked mole-rats were 1 year old for in vivo auditory assessments (Figure 1) and in vitro hair bundle examination by SEM (Figure 3) and 9 months old for in vitro examination of the OHC (Figure 2). Damaraland mole-rats were 5 years old for all experiments. Considering the longevity of naked and Damaraland mole-rats, these animals would be considered mature but certainly not aged (see also [108]).
METHOD DETAILS
Auditory brainstem response (ABR) and distortion product otoacoustic emission (DPOAE) measurements
Naked and Damaraland mole-rats were anesthetized with a ketamine/xylazine mixture (80 mg/kg ketamine and 20 mg/kg xylazine, i.p.) and placed on a gauze-covered heating pad in a sound attenuated booth (IAC Acoustics, Inc., Naperville, IL). Animals were maintained at 37°C. ABR measurements were performed as described previously [109]. Briefly, electrodes were placed at the vertex of the skull and behind the pinna. A ground electrode was placed in the ipsilateral hind leg. Animals were presented a 5-ms monaural click stimuli (0.1-ms square wave pulse of alternating polarity) or a 5-ms tone at frequencies of 250, 500, 1000, 2000, 4000, and 6000 Hz (0.5-ms onset/offset) at a rate of 20 Hz. Stimuli were generated and presented using custom MATLAB software, Tucker-Davis Technology (TDT, RX6, PA5), dome tweeter speaker (Fostex model FT28D), and a desktop computer. Stimuli intensity was calibrated using a quarter-inch free-field microphone (type 4939, Brüel and Kjӕr) at the location of the animals’ head (30 cm from the speaker). Click stimuli were tested first to ensure proper electrode placement. Responses were recorded first to supra-threshold levels for all stimuli (85 to 105 dB, depending on the stimulus), and intensity was decreased in 10 dB SPL steps until threshold was reached. Each intensity level was repeated 300 times to generate an average waveform before moving to the next intensity level. Responses were amplified using a World Precision Instruments ISO-80 biological amplifier and filtered between 30 and 3000 Hz using a Krohn-Hite bandpass filter. Threshold was determined by the absence of any wave above the noise. Thresholds were additionally verified by statistical determination that estimates the level of the response that is two standard deviations above the noise.
Animals were anesthetized and prepared for measurements as described for ABR measurements. DPOAE measurements were performed as described previously [110]. Custom MATLAB software (generously provided by Michael E. Smith (University of Maryland, MD, USA) and TDT RX6 processor were used to control stimulus presentation and response recording. The processor was connected to an Etymotic Research ER-10C low noise, battery-powered DPOAE system, which included a sound source and microphone. A 20 μL small pipette tip was cut and placed onto the end of the ER-10C probe and then fitted into the animal’s ear canal. The probe was inserted approximately 2 to 4 mm into the ear canal. No visible exudate was observed before inserting the probe tip or stuck to the probe tip after removal from the ear canal. Attempts to clear the ear canal were not performed since these procedures are invasive and risk damaging the structures of the outer and middle ear. The acoustic seal of the microphone in the ear canal was tested before the beginning of each test to ensure proper placement of the probe by measuring the ear canal sound pressure levels of tone sweeps emanating from each speaker to confirm that neither speaker was occluded and that background noise was as low as possible. The ear canal response to tone sweeps was tested for each speaker to ensure proper probe placement before the beginning of the DPOAE test. The cubic (2f1-f2, where f2/f1 = 1.2 and f1 is 10 dB SPL lower than f2) DPOAE thresholds were determined across a range of frequencies (between 0.25 and12 kHz) and evoked using f1 stimuli with randomly presented intensity levels (up to 65 dB SPL in mouse and 70 dB SPL in naked and Damaraland mole-rats) in 5 to 10 dB interval increments to establish thresholds. In vivo DPOAE amplitudes vary substantially across species and are often very small at low frequencies, even in the healthy cochlea (sometimes only 5 dB above noise floor [111]), partly because the microphone noise floor is higher at low frequencies. More intense stimuli cannot be used since the system itself (speaker/probe/tube) produces distortion above the levels examined in this study.
Previous work detected DPOAEs in a species, Fukomys anselli, recently reclassified to the same genus as the Damaraland mole-rat [16]. These authors indicated that measurement of DPOAEs were methodologically difficult due to the unusual external ear anatomy of mole-rats. In our experiments, DPOAEs could not be detected in either naked or Damaraland mole-rats despite careful positioning (and repositioning) of the recording microphone in the ear canal. Importantly, Fukomys anselli shows slightly more sensitive hearing [lower ABR wave I thresholds: [11]] and a region of increased frequency representation in the cochlea [termed an acoustic fovea: [17]]. Differences in the cochlear function between Fukomys anselli and naked and Damaraland mole-rats might account for the presence of DPOAEs in Fukomys anselli and their absence in naked and Damaraland mole-rats. Unfortunately, Fukomys anselli was not available for parallel testing in our facilities.
Immunofluorescence, confocal microscopy, and image analysis
To obtain cochleae, animals were anaesthetized by isoflurane inhalation and then rapidly decapitated. Procedures published previously were used to isolate intact organs of Corti from cochleae for immunofluorescence and confocal microscopy [112, 113]. In these experiments, a rabbit polyclonal antibody against-prestin was generously provided by Dr. Mary Ann Cheatham (Northwestern University, IL, USA) and used diluted 1:1000. The secondary antibody, AlexaFluor 594 goat anti-rabbit, was purchased from Invitrogen and used diluted 1:500. High magnification confocal micrographs of middle turns of the organs of Corti were acquired using a Leica SP8 confocal microscope with a 60 × Olympus PlanApo oil-immersion lens (NA 1.42) under the control of the Leica Application Suite (LAS) software. Single optical sections were acquired in a 1024 × 1024 pixel raster at a sampling speed of 100 Hz. The laser power and PMT voltage, gain and offset adjustments were adjusted to optimize the dynamic range of the detected intensity values. To quantify outer hair cell cross-sectional areas, images containing clearly defined and intact outer hair cell membranes (visualized by prestin immunoreactivity) were converted to binary (black and white) images to allow measurement of the cross-sectional areas. Average outer hair cell cross-sectional areas are presented in the text as the grand average of the average outer hair cell cross-sectional areas from each individual.
Measurements of non-linear capacitance in isolated outer hair cells
Naked mole-rats and C57BL/6J mice were anesthetized with 100 mg/kg ketamine and 20 mg/kg xylazine (i.p.) and then rapidly decapitated. Apical turns of the organs of Corti were isolated for whole cell voltage clamp recordings performed as described previously [114]. Briefly, the extracellular solution contained (in mM): 100 NaCl, 20 tetraethylammonium (TEA)-Cl, 20 CsCl, 2 CoCl2, 1 MgCl2, 1 CaCl2, 10 HEPES, pH 7.2. The intracellular solution contained (in mM): 140 CsCl, 2 MgCl2, 10 HEPES, and 10 EGTA, pH 7.2. Pipettes had resistances of 3–5 MΩ. Gigaohm seals were made and stray capacitance was balanced out with amplifier circuitry prior to establishing whole-cell conditions. A Nikon Eclipse E600-FN microscope with 40 × water immersion lens was used to observe cells during voltage clamp. Outer hair cells from all three rows were recorded at room temperature using jClamp software and an Axopatch 200B amplifier. Data were low pass-filtered at 10 kHz and digitized at 100 kHz with a Digidata 1320A.
To extract Boltzmann parameters, capacitance-voltage data were fit to the first derivative of a two-state Boltzmann function:
where
Qmax is the maximum non-linear charge moved, Vh is the voltage at peak capacitance or equivalently at half-maximum charge transfer, Vm is the Rs-corrected membrane potential, z is the valence, Clin is the linear membrane capacitance, e is the electron charge, kB is the Boltzmann’s constant, and T is the absolute temperature. Csa is a component of capacitance that characterizes sigmoidal changes in specific membrane capacitance. ΔCsa is the total sum of unitary changes per prestin motor protein.
Scanning electron microscopy (SEM) of isolated cochleae
Animals were anesthetized with 100 mg/kg ketamine and 20 mg/kg xylazine (i.p.) and transcardially perfused with 2% paraformal-dehyde (PFA) and 2.5% glutaraldehyde in sodium cacodylate buffer (0.1 M sodium cacodylate with 3 mM CaCl2, pH 7.4). After perfusion, temporal bones were removed from the skull and post-fixed for 24h at 4°C overnight. The temporal bone was placed in SEM buffer (0.05 mM HEPES Buffer pH 7.4, 10 mM CaCl2, 5 mM MgCl2, and 0.9% NaCl), and the bony cochlear shell, tectorial membrane and Reissner’s membrane were removed, leaving the organ of Corti exposed on the modiolus. Samples were washed in SEM buffer before being post-fixed in 1% OsO4 using the OTOTO method (alternating osmium tetroxide and thiocarbohydrazide immersion [115]). The sample was rinsed in 0.1% cacodylate buffer and then dehydrated in an ethanol series (50, 70, 80, 90, 95, then two 100% EtOH) for 10 min each at room temperature. Samples were dried using a chemical drying agent, hexamethyldisilizane (HMDS), for 30 min in 50% HMDS/50% EtOH and then 30 min in 100% HMDS. Afterward samples were placed in a vacuum desiccator to evaporate the HMDS. Samples were then mounted on stubs with double-sided adhesive carbon disks and sputter coated with 10 nm platinum. Apical, middle, and basal sections of the organ of Corti were viewed and images obtained with a Hitachi S-3000N scanning electron microscope (RRC, University of Illinois Chicago, Chicago, IL).
Hair bundle orientation was quantified as the angular deviation of the hair bundle axis and the mediolateral axis, similar to previously published methodology [79]. Specifically, for each micrograph, a line was drawn through the axes of all complete hair bundles (yellow arrows in Figure 3). The angle corresponding to the mediolateral axis for that micrograph was determined as the median of the individual angles drawn through the hair bundle axes. The angle of the mediolateral axis was determined in this way because of the variability in the orientation of the rows of outer hair cells between micrographs and species. An angular deviation of 0° indicates the expected orientation of the hair bundle along the mediolateral axis. The observation of both positive and negative angular deviations indicate that hair bundles are sometimes oriented in opposite directions.
Hair bundle (stereociliar) protein sequence analysis
Hair bundle (stereociliar) proteins were identified from the list of genes retrieved when searching the gene ontology (GO) term “stereo-cilium” (specifying mouse) in AmiGO v2.0 (accessed on November 15, 2017) [103, 104]. Additional proteins were added to this list by including proteins localized to stereocilia in the Hereditary Hearing Loss Homepage (hereditaryhearingloss.org, accessed on November 15, 2017) [105]. Independent literature searches confirmed localization of each of these proteins to the stereocilia. Of the 66 hair bundle proteins identified, 29 are encoded by deafness genes associated with known pathogenic missense variants in humans identified in the Deafness Variation Database (deafnessvariationdatabase.org, accessed on January 6, 2019, version 8.1) [25]. Only variants associated with Usher syndrome, deafness, and/or hearing loss were included to exclude possibly confounding associations with non-auditory phenotypes. Orthologous amino acid sequences for these 29 stereocilia proteins were retrieved from naked and Damaraland mole-rats, related bathyergid and hystricognath species, and mouse using Blastp against annotated genomes and tblastn against mammal whole genome sequence (WGS) and sequence read archive (SRA) data (when annotated sequences were not available) and screened for the presence of these pathogenic missense variants. Prior to screening, possible pseudogenization was assessed via published genomic scans of pseudogenes [85, 87, 116], and gene expression was verified via The Naked Mole-Rat Genome Resource (naked-mole-rat.org, accessed on March 5, 2020) [106, 107]. Pathogenic substitutions were mapped onto a well-established phylogeny of hystricognath rodents [117] using maximum parsimony.
Molecular evolutionary analyses
Protein-coding nucleotide sequences were retrieved from the NCBI RefSeq non-redundant protein database (https://www.ncbi.nlm.nih.gov/refseq/about/nonredundantproteins). Non-human sequences for each gene were selected by using the NCBI BLASTn utility (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to query each of the target human sequences, and selecting the top BLAST hit for each of the six non-human species. After selecting the protein coding portion of each transcript, the sequences for each gene were aligned using the MEGA X software [101]. Genes were aligned by codon using the ClustalW algorithm with default settings. Estimation of gene-wide dN/dS ratios for bathyergid and non-bathyergid portions of the phylogeny were computed online at the Datamonkey web-site using the Fixed Effects Likelihood (FELL) method [100]. Taxa included naked and Damaraland mole rats, guinea pig, chinchilla, degu, mouse, and human. Per-codon estimates of dN-dS were obtained by using the Single-Likelihood Ancestor Counting (SLAC) method [100]. Taxa for the bathyergid and non-bathyergid sampling included naked and Damaraland mole rats, guinea pig, chinchilla, and degu. Taxa for the non-bathyergid only sampling included guinea pig, chinchilla, degu, mouse, and human. Evolutionary probability of variants was estimated using the evolutionary probability (EP) method in MEGA X software [43, 118], which estimates the probability of observing an amino acid residue at a protein position in a given species using a multispecies sequence alignment and phylogenetic relationships among sequences. EP is independent of population-level information on the frequency of alleles at the focal position in the species of interest. EP was estimated using the amino acid sequences derived from the same seven-species alignments used to estimate dN/dS ratios, with either naked or Damaraland mole rats as the focal species depending on which species the target variant was found in.
QUANTIFICATION AND STATISTICAL ANALYSIS
All data are presented as mean ± standard error unless otherwise noted. Tests for statistical significance are provided in the text. In general, data were not assumed to be normally distributed and non-parametric tests for significance were used. These tests included the Wilcoxon signed-rank test for paired data or the Kruskal Wallis with Dunn’s correction for multiple comparisons. P values ≤ 0.05 were used to establish significance.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-prestin | Dr. Mary Ann Cheatham (Northwestern University, IL, USA) [98] | N/A |
| Experimental Models: Organisms/Strains | ||
| Heterocephalus glaber | Dr. Thomas J. Park [15] | N/A |
| Fukomys damarensis | Dr. Thomas J. Park [15] | N/A |
| Mouse: CBA/CaJ mice | The Jackson Laboratory | JAX:000654 |
| Software and Algorithms | ||
| ImageJ | [99] | https://imagej.nih.gov/ij/ |
| jClamp Software | https://scisoftco.com | |
| Adobe Suite | N/A | https://adobe.com |
| FELL | [100] | https://datamonkey.org |
| GraphPad Prism8 | N/A | https://graphpad.com/scientific-software/prism |
| Microsoft Suite | N/A | https://microsoft.com |
| NCBI GenBank/BLAST | N/A | https://ncbi.nlm.nih.gov |
| SLAC | [100] | https://datamonkey.org |
| MEGA X | [101] | https://megasoftware.net |
| ClustalW | [102] | https://megasoftware.net |
| Other | ||
| AmiGO v2.0 | [103, 104] | http://amigo.geneontology.org/amigo |
| Hereditary Hearing Loss Homepage | [105] | https://hereditaryhearingloss.org |
| Deafness Variation Database | [25] | http://www.deafnessvariationdatabase.org |
| The Naked Mole-Rat Genome Resource | [106, 107] | http://www.naked-mole-rat.org |
Highlights.
Hearing is comparatively poor in African naked and Damaraland mole-rats
These mole-rats lack cochlear amplification and have disrupted hair bundles
Hair bundle proteins bear deafness-associated amino acid substitutions
Positive selection in some bundle proteins suggests altered hearing is adaptive
ACKNOWLEDGMENTS
The project was supported by grants from the University of Groningen (the Netherlands) to S.J.P.; the NSF (USA) to T.J.P. (grant 1655494) and to K.M.S. and L.A.S. (NRSA postdoctoral fellowships); the NIH (USA) to S.K. (R01-HG008146 and GM0126567), to J.S.-S. (R01-DC016318 and R01-DC008130), to A.M.L. (R01-DC017620 and RO1-DC012347); and the David M. Rubenstein Fund for Hearing Research (USA) to A.M.L. We acknowledge Paul A. Fuchs for training support (NIH T32-DC000032) and Dr. Elizabeth Brittan-Powell for technical support.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Köppl C, and Manley GA (2019). A Functional Perspective on the Evolution of the Cochlea. Cold Spring Harb. Perspect. Med 9, a033241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Franchini LF, and Elgoyhen AB (2006). Adaptive evolution in mammalian proteins involved in cochlear outer hair cell electromotility. Mol. Phylogenet. Evol 41, 622–635. [DOI] [PubMed] [Google Scholar]
- 3.Ashmore J, Avan P, Brownell WE, Dallos P, Dierkes K, Fettiplace R, Grosh K, Hackney CM, Hudspeth AJ, Jülicher F, et al. (2010). The remarkable cochlear amplifier. Hear. Res 266, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dent ML, Screven LA, and Kobrina A (2013). Rodent Bioacoustics. In Springer Handbook of Auditory Research, Volume 67, Dent ML, Popper AN, and Fay RR, eds. (Springer; ), pp. 71–105. [Google Scholar]
- 5.Ryan A (1976). Hearing sensitivity of the Mongolian gerbil, Meriones un-guiculatis. J. Acoust. Soc. Am 59, 1222–1226. [DOI] [PubMed] [Google Scholar]
- 6.Lacey EA, Patton JL, and Cameron GN (2000). Life Underground: The Biology of Subterranean Rodents (University of Chicago Press; ). [Google Scholar]
- 7.Nevo E (1999). Mosaic Evolution of Subterranean Mammals—Regression, Progression and Global Convergence (Oxford University Press; ). [Google Scholar]
- 8.Heffner RS, and Heffner HE (1992). Hearing and sound localization in blind mole rats (Spalax ehrenbergi). Hear. Res 62, 206–216. [DOI] [PubMed] [Google Scholar]
- 9.Heffner RS, and Heffner HE (1993). Degenerate hearing and sound localization in naked mole rats (Heterocephalus glaber), with an overview of central auditory structures. J. Comp. Neurol 331, 418–433. [DOI] [PubMed] [Google Scholar]
- 10.Okanoya K, Yosida S, Barone CM, Applegate DT, Brittan-Powell EF, Dooling RJ, and Park TJ (2018). Auditory-vocal coupling in the naked mole-rat, a mammal with poor auditory thresholds. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol 204, 905–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerhardt P, Henning Y, Begall S, and Malkemper EP (2017). Correction: Audiograms of three subterranean rodent species (genus Fukomys) determined by auditory brainstem responses reveal extremely poor high-frequency cut-offs. J. Exp. Biol 220, 4747. [DOI] [PubMed] [Google Scholar]
- 12.Koka K, Jones HG, Thornton JL, Lupo JE, and Tollin DJ (2011). Sound pressure transformations by the head and pinnae of the adult chinchilla (Chinchilla lanigera). Hear. Res 272, 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lauer AM, Slee SJ, and May BJ (2011). Acoustic basis of directional acuity in laboratory mice. J. Assoc. Res. Otolaryngol 12, 633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mason MJ, Cornwall HL, and Smith ES (2016). Ear structures of the naked mole-rat, Heterocephalus glaber, and its relatives (Rodentia: Bathyergidae). PLoS ONE 11, e0167079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barone CM, Douma S, Reijntjes DOJ, Browe BM, Köppl C, Klump G, Park TJ, and Pyott SJ (2019). Altered cochlear innervation in developing and mature naked and Damaraland mole rats. J. Comp. Neurol 527, 2302–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kössl M, Frank G, Burda H, and Müller M (1996). Acoustic distortion products from the cochlea of the blind African mole rat, Cryptomys spec. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol 178, 427–434. [DOI] [PubMed] [Google Scholar]
- 17.Müller M, Laube B, Burda H, and Bruns V (1992). Structure and function of the cochlea in the African mole rat (Cryptomys hottentotus): evidence for a low frequency acoustic fovea. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol 171, 469–476. [DOI] [PubMed] [Google Scholar]
- 18.Partha R, Chauhan BK, Ferreira Z, Robinson JD, Lathrop K, Nischal KK, Chikina M, and Clark NL (2017). Subterranean mammals show convergent regression in ocular genes and enhancers, along with adaptation to tunneling. eLife 6, e25884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burda H (2006). Ear and eye in subterranean mole-rats, Fukomys anselli (Bathyergidae) and Spalax ehrenbergi (Spalacidae): progressive specialisation or regressive degeneration? Anim. Biol. Leiden Neth 56, 475–486. [Google Scholar]
- 20.Quilliam TA (1966). The mole’s sensory apparatus. J. Zool 149, 76–88. [Google Scholar]
- 21.Lange S, Burda H, Wegner RE, Dammann P, Begall S, and Kawalika M (2007). Living in a “stethoscope”: burrow-acoustics promote auditory specializations in subterranean rodents. Naturwissenschaften 94, 134–138. [DOI] [PubMed] [Google Scholar]
- 22.Heth G, Frankenberg E, and Nevo E (1986). Adaptive optimal sound for vocal communication in tunnels of a subterranean mammal (Spalax ehrenbergi). Experientia 42, 1287–1289. [DOI] [PubMed] [Google Scholar]
- 23.Davies KT, Bennett NC, Tsagkogeorga G, Rossiter SJ, and Faulkes CG (2015). Family wide molecular adaptations to underground life in African mole-rats revealed by phylogenomic analysis. Mol. Biol. Evol 32, 3089–3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcovitz A, Turakhia Y, Chen HI, Gloudemans M, Braun BA, Wang H, and Bejerano G (2019). A functional enrichment test for molecular convergent evolution finds a clear protein-coding signal in echolocating bats and whales. Proc. Natl. Acad. Sci. USA 116, 21094–21103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azaiez H, Booth KT, Ephraim SS, Crone B, Black-Ziegelbein EA, Marini RJ, et al. (2018). Genomic landscape and mutational signatures of deafness-associated genes. Am. J. Hum. Genet 103, 484–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heffner R, Heffner H, and Masterton B (1971). Behavioral measurements of absolute and frequency-difference thresholds in guinea pig. J. Acoust. Soc. Am 49, 1888–1895. [DOI] [PubMed] [Google Scholar]
- 27.Heffner RS, and Heffner HE (1991). Behavioral hearing range of the chinchilla. Hear. Res 52, 13–16. [DOI] [PubMed] [Google Scholar]
- 28.Boettcher FA, and Schmiedt RA (1995). Distortion-product otoacoustic emissions in Mongolian gerbils with resistance to noise-induced hearing loss. J. Acoust. Soc. Am 98, 3215–3222. [DOI] [PubMed] [Google Scholar]
- 29.Eddins AC, Zuskov M, and Salvi RJ (1999). Changes in distortion product otoacoustic emissions during prolonged noise exposure. Hear. Res 127, 119–128. [DOI] [PubMed] [Google Scholar]
- 30.Peng JH, Tao ZZ, and Huang ZW (2007). Long-term sound conditioning increases distortion product otoacoustic emission amplitudes and decreases olivocochlear efferent reflex strength. Neuroreport 18, 1167–1170. [DOI] [PubMed] [Google Scholar]
- 31.Dallos P (2008). Cochlear amplification, outer hair cells and prestin. Curr. Opin. Neurobiol 18, 370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liberman MC, Gao J, He DZ, Wu X, Jia S, and Zuo J (2002). Prestin is required for electromotility of the outer hair cell and for the cochlear amplifier. Nature 419, 300–304. [DOI] [PubMed] [Google Scholar]
- 33.Liu XZ, Ouyang XM, Xia XJ, Zheng J, Pandya A, Li F, Du LL, Welch KO, Petit C, Smith RJ, et al. (2003). Prestin, a cochlear motor protein, is defective in non-syndromic hearing loss. Hum. Mol. Genet 12, 1155–1162. [DOI] [PubMed] [Google Scholar]
- 34.Zajic G, and Schacht J (1987). Comparison of isolated outer hair cells from five mammalian species. Hear. Res 26, 249–256. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi S, Cheatham MA, Zheng J, and Homma K (2016). The R130S mutation significantly affects the function of prestin, the outer hair cell motor protein. J. Mol. Med. (Berl) 94, 1053–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos-Sacchi J (1991). Reversible inhibition of voltage-dependent outer hair cell motility and capacitance. J. Neurosci 11, 3096–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Yang S, Jia S, and He DZ (2010). Prestin forms oligomer with four mechanically independent subunits. Brain Res 1333, 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barr-Gillespie PG (2015). Assembly of hair bundles, an amazing problem for cell biology. Mol. Biol. Cell 26, 2727–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davies S, and Forge A (1987). Preparation of the mammalian organ of Corti for scanning electron microscopy. J. Microsc 147, 89–101. [DOI] [PubMed] [Google Scholar]
- 40.Richardson GP, and Petit C (2019). Hair-Bundle Links: Genetics as the Gateway to Function. Cold Spring Harb. Perspect. Med 9, a033142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar S, Stecher G, Suleski M, and Hedges SB (2017). TimeTree: A resource for timelines, timetrees, and divergence times. Mol. Biol. Evol 34, 1812–1819. [DOI] [PubMed] [Google Scholar]
- 42.Kumar S, Filipski AJ, Battistuzzi FU, Kosakovsky Pond SL, and Tamura K (2012). Statistics and truth in phylogenomics. Mol. Biol. Evol 29, 457–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patel R, and Kumar S (2019). On estimating evolutionary probabilities of population variants. BMC Evol. Biol 19, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bednářová R, Hrouzková-Knotková E, Burda H, Sedláček F, and Sumbera R (2013). Vocalizations of the giant mole-rat (Fukomys mecho-wii), a subterranean rodent with the richest vocal repertoire. Bioacoustics 22, 87–107. [Google Scholar]
- 45.Dvořáková V, Hrouzková E, and Šumbera R (2016). Vocal repertoire of the social Mashona mole-rat (Fukomys darlingi) and how it compares with other mole-rats. Bioacoustics 25, 253–266. [Google Scholar]
- 46.Pepper JW, Braude SH, Lacey EA, and Sherman PW (1991). Vocalizations of the naked mole-rat. In The Biology of the Naked Mole-Rat, Sherman PW, Jarvis JUM, and Alexander RD, eds. (Princeton University Press; ), pp. 243–274. [Google Scholar]
- 47.Bennett NC, and Faulkes CG (2000). African mole-rats: ecology and eusociality (Cambridge University Press; ). [Google Scholar]
- 48.Thomas HG, Swanepoel D, and Bennett NC (2016). Burrow architecture of the Damaraland mole-rat (Fukomys damarensis) from South Africa. African Zoology 51, 29–36. [Google Scholar]
- 49.Brett RA (1991). The ecology of naked-mole-rat colonies: burrowing, food, and limiting factors. In The Biology of the Naked Mole-Rat, Sherman PW, Jarvis JUM, and Alexander RD, eds. (Princeton University Press; ), pp. 137–148. [Google Scholar]
- 50.Brown SD, Hardisty-Hughes RE, and Mburu P (2008). Quiet as a mouse: dissecting the molecular and genetic basis of hearing. Nat. Rev. Genet 9, 277–290. [DOI] [PubMed] [Google Scholar]
- 51.Hosoya M, Fujioka M, Ogawa K, and Okano H (2016). Distinct expression patterns of causative genes responsible for hereditary progressive hearing loss in non-human primate cochlea. Sci. Rep 6, 22250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolz H, von Brederlow B, Ramírez A, Bryda EC, Kutsche K, Nothwang HG, Seeliger M, del C-Salcedó Cabrera M, Vila MC, Molina OP, et al. (2001). Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat. Genet 27, 108–112. [DOI] [PubMed] [Google Scholar]
- 53.Schultz JM, Bhatti R, Madeo AC, Turriff A, Muskett JA, Zalewski CK, King KA, Ahmed ZM, Riazuddin S, Ahmad N, et al. (2011). Allelic hierarchy of CDH23 mutations causing non-syndromic deafness DFNB12 or Usher syndrome USH1D in compound heterozygotes. J. Med. Genet 48, 767–775. [DOI] [PubMed] [Google Scholar]
- 54.Bork JM, Peters LM, Riazuddin S, Bernstein SL, Ahmed ZM, Ness SL, Polomeno R, Ramesh A, Schloss M, Srisailpathy CR, et al. (2001). Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am. J. Hum. Genet 68, 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagatsuma M, Kitoh R, Suzuki H, Fukuoka H, Takumi Y, and Usami S (2007). Distribution and frequencies of CDH23 mutations in Japanese patients with non-syndromic hearing loss. Clin. Genet 72, 339–344. [DOI] [PubMed] [Google Scholar]
- 56.Di Palma F, Holme RH, Bryda EC, Belyantseva IA, Pellegrino R, Kachar B, Steel KP, and Noben-Trauth K (2001). Mutations in Cdh23, encoding a new type of cadherin, cause stereocilia disorganization in waltzer, the mouse model for Usher syndrome type 1D. Nat. Genet 27, 103–107. [DOI] [PubMed] [Google Scholar]
- 57.Schwander M, Xiong W, Tokita J, Lelli A, Elledge HM, Kazmierczak P, Sczaniecka A, Kolatkar A, Wiltshire T, Kuhn P, et al. (2009). A mouse model for nonsyndromic deafness (DFNB12) links hearing loss to defects in tip links of mechanosensory hair cells. Proc. Natl. Acad. Sci. USA 106, 5252–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ahmed ZM, Riazuddin S, Ahmad J, Bernstein SL, Guo Y, Sabar MF, Sieving P, Riazuddin S, Griffith AJ, Friedman TB, et al. (2003). PCDH15 is expressed in the neurosensory epithelium of the eye and ear and mutant alleles are responsible for both USH1F and DFNB23. Hum. Mol. Genet 12, 3215–3223. [DOI] [PubMed] [Google Scholar]
- 59.Doucette L, Merner ND, Cooke S, Ives E, Galutira D, Walsh V, Walsh T, MacLaren L, Cater T, Fernandez B, et al. (2009). Profound, prelingual nonsyndromic deafness maps to chromosome 10q21 and is caused by a novel missense mutation in the Usher syndrome type IF gene PCDH15. Eur. J. Hum. Genet 17, 554–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alagramam KN, Murcia CL, Kwon HY, Pawlowski KS, Wright CG, and Woychik RP (2001). The mouse Ames waltzer hearing-loss mutant is caused by mutation of Pcdh15, a novel protocadherin gene. Nat. Genet 27, 99–102. [DOI] [PubMed] [Google Scholar]
- 61.Lefèvre G, Michel V, Weil D, Lepelletier L, Bizard E, Wolfrum U, Hardelin JP, and Petit C (2008). A core cochlear phenotype in USH1 mouse mutants implicates fibrous links of the hair bundle in its cohesion, orientation and differential growth. Development 135, 1427–1437. [DOI] [PubMed] [Google Scholar]
- 62.Raphael Y, Kobayashi KN, Dootz GA, Beyer LA, Dolan DF, and Burmeister M (2001). Severe vestibular and auditory impairment in three alleles of Ames waltzer (av) mice. Hear. Res 151, 237–249. [DOI] [PubMed] [Google Scholar]
- 63.Zheng QY, Yu H, Washington JL 3rd, Kisley LB, Kikkawa YS, Pawlowski KS, Wright CG, and Alagramam KN (2006). A new spontaneous mutation in the mouse protocadherin 15 gene. Hear. Res 219, 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Washington JL 3rd, Pitts D, Wright CG, Erway LC, Davis RR, and Alagramam K (2005). Characterization of a new allele of Ames waltzer generated by ENU mutagenesis. Hear. Res 202, 161–169. [DOI] [PubMed] [Google Scholar]
- 65.Ahmed ZM, Smith TN, Riazuddin S, Makishima T, Ghosh M, Bokhari S, Menon PS, Deshmukh D, Griffith AJ, Riazuddin S, et al. (2002). Nonsyndromic recessive deafness DFNB18 and Usher syndrome type IC are allelic mutations of USHIC. Hum. Genet 110, 527–531. [DOI] [PubMed] [Google Scholar]
- 66.Grillet N, Xiong W, Reynolds A, Kazmierczak P, Sato T, Lillo C, Dumont RA, Hintermann E, Sczaniecka A, Schwander M, et al. (2009). Harmonin mutations cause mechanotransduction defects in cochlear hair cells. Neuron 62, 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lentz JJ, Gordon WC, Farris HE, MacDonald GH, Cunningham DE, Robbins CA, Tempel BL, Bazan NG, Rubel EW, Oesterle EC, and Keats BJ (2010). Deafness and retinal degeneration in a novel USH1C knock-in mouse model. Dev. Neurobiol 70, 253–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Verpy E, Masmoudi S, Zwaenepoel I, Leibovici M, Hutchin TP, Del Castillo I, Nouaille S, Blanchard S, Lainé S, Popot JL, et al. (2001). Mutations in a new gene encoding a protein of the hair bundle cause nonsyndromic deafness at the DFNB16 locus. Nat. Genet 29, 345–349. [DOI] [PubMed] [Google Scholar]
- 69.Verpy E, Leibovici M, Michalski N, Goodyear RJ, Houdon C, Weil D, Richardson GP, and Petit C (2011). Stereocilin connects outer hair cell stereocilia to one another and to the tectorial membrane. J. Comp. Neurol 519, 194–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Verpy E, Weil D, Leibovici M, Goodyear RJ, Hamard G, Houdon C, Lefèvre GM, Hardelin JP, Richardson GP, Avan P, and Petit C (2008). Stereocilin-deficient mice reveal the origin of cochlear waveform distortions. Nature 456, 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Avan P, Le Gal S, Michel V, Dupont T, Hardelin JP, Petit C, and Verpy E (2019). Otogelin, otogelin-like, and stereocilin form links connecting outer hair cell stereocilia to each other and the tectorial membrane. Proc. Natl. Acad. Sci. USA 116, 25948–25957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weston MD, Luijendijk MW, Humphrey KD, Möller C, and Kimberling WJ (2004). Mutations in the VLGR1 gene implicate G-protein signaling in the pathogenesis of Usher syndrome type II. Am. J. Hum. Genet 74, 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yagi H, Tokano H, Maeda M, Takabayashi T, Nagano T, Kiyama H, Fujieda S, Kitamura K, and Sato M (2007). Vlgr1 is required for proper stereocilia maturation of cochlear hair cells. Genes Cells 12, 235–250. [DOI] [PubMed] [Google Scholar]
- 74.Yan W, Long P, Chen T, Liu W, Yao L, Ren Z, et al. (2018). A natural occurring mouse model with ADGRV1 mutation of Usher Syndrome 2C and characterization of its recombinant inbred strains. Cell. Physiol. Biochem 47, 1883–1897. [DOI] [PubMed] [Google Scholar]
- 75.Eudy JD, Weston MD, Yao S, Hoover DM, Rehm HL, Ma-Edmonds M, Yan D, Ahmad I, Cheng JJ, Ayuso C, et al. (1998). Mutation of a gene encoding a protein with extracellular matrix motifs in Usher syndrome type IIa. Science 280, 1753–1757. [DOI] [PubMed] [Google Scholar]
- 76.Liu X, Bulgakov OV, Darrow KN, Pawlyk B, Adamian M, Liberman MC, and Li T (2007). Usherin is required for maintenance of retinal photoreceptors and normal development of cochlear hair cells. Proc. Natl. Acad. Sci. USA 104, 4413–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tsang SH, Aycinena ARP, and Sharma T (2018). Ciliopathy: Usher Syndrome. Adv. Exp. Med. Biol 1085, 167–170. [DOI] [PubMed] [Google Scholar]
- 78.Kane KL, Longo-Guess CM, Gagnon LH, Ding D, Salvi RJ, and Johnson KR (2012). Genetic background effects on age-related hearing loss associated with Cdh23 variants in mice. Hear. Res 283, 80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Webb SW, Grillet N, Andrade LR, Xiong W, Swarthout L, Della Santina CC, Kachar B, and Müller U (2011). Regulation of PCDH15 function in mechanosensory hair cells by alternative splicing of the cytoplasmic domain. Development 138, 1607–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zheng QY, Scarborough JD, Zheng Y, Yu H, Choi D, and Gillespie PG (2012). Digenic inheritance of deafness caused by 8J allele of myosin-VIIA and mutations in other Usher I genes. Hum. Mol. Genet 21, 2588–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zheng QY, Yan D, Ouyang XM, Du LL, Yu H, Chang B, Johnson KR, and Liu XZ (2005). Digenic inheritance of deafness caused by mutations in genes encoding cadherin 23 and protocadherin 15 in mice and humans. Hum. Mol. Genet 14, 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kremer H, van Wijk E, Märker T, Wolfrum U, and Roepman R (2006). Usher syndrome: molecular links of pathogenesis, proteins and pathways. Hum. Mol. Genet 15, R262–R270. [DOI] [PubMed] [Google Scholar]
- 83.Raphael Y, Lenoir M, Wroblewski R, and Pujol R (1991). The sensory epithelium and its innervation in the mole rat cochlea. J. Comp. Neurol 314, 367–382. [DOI] [PubMed] [Google Scholar]
- 84.Moore BC (2001). Dead regions in the cochlea: diagnosis, perceptual consequences, and implications for the fitting of hearing aids. Trends Amplif 5, 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim EB, Fang X, Fushan AA, Huang Z, Lobanov AV, Han L, Marino SM, Sun X, Turanov AA, Yang P, et al. (2011). Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature 479, 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pisciottano F, Cinalli AR, Stopiello JM, Castagna VC, Elgoyhen AB, Rubinstein M, et al. (2019). Inner ear genes underwent positive selection and adaptation in the mammalian lineage. Mol. Biol. Evol 36, 1653–1670. [DOI] [PubMed] [Google Scholar]
- 87.Fang X, Seim I, Huang Z, Gerashchenko MV, Xiong Z, Turanov AA, Zhu Y, Lobanov AV, Fan D, Yim SH, et al. (2014). Adaptations to a subterranean environment and longevity revealed by the analysis of mole rat genomes. Cell Rep 8, 1354–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Emerling CA (2018). Regressed but not gone: patterns of vision gene loss and retention in subterranean mammals. Integr. Comp. Biol 58, 441–451. [DOI] [PubMed] [Google Scholar]
- 89.Delpire E, Lu J, England R, Dull C, and Thorne T (1999). Deafness and imbalance associated with inactivation of the secretory Na-K-2Cl co-transporter. Nat. Genet 22, 192–195. [DOI] [PubMed] [Google Scholar]
- 90.Dixon MJ, Gazzard J, Chaudhry SS, Sampson N, Schulte BA, and Steel KP (1999). Mutation of the Na-K-Cl co-transporter gene Slc12a2 results in deafness in mice. Hum. Mol. Genet 8, 1579–1584. [DOI] [PubMed] [Google Scholar]
- 91.Verhoeven K, Van Laer L, Kirschhofer K, Legan PK, Hughes DC, Schatteman I, Verstreken M, Van Hauwe P, Coucke P, Chen A, et al. (1998). Mutations in the human alpha-tectorin gene cause autosomal dominant non-syndromic hearing impairment. Nat. Genet 19, 60–62. [DOI] [PubMed] [Google Scholar]
- 92.Bok D, Galbraith G, Lopez I, Woodruff M, Nusinowitz S, BeltrandelRio H, Huang W, Zhao S, Geske R, Montgomery C, et al. (2003). Blindness and auditory impairment caused by loss of the sodium bicarbonate cotransporter NBC3. Nat. Genet 34, 313–319. [DOI] [PubMed] [Google Scholar]
- 93.Lopez IA, Acuna D, Galbraith G, Bok D, Ishiyama A, Liu W, and Kurtz I (2005). Time course of auditory impairment in mice lacking the electroneutral sodium bicarbonate cotransporter NBC3 (slc4a7). Brain Res. Dev. Brain Res 160, 63–77. [DOI] [PubMed] [Google Scholar]
- 94.Davies KTJ, Bennett NC, Faulkes CG, and Rossiter SJ (2018). Limited evidence for parallel molecular adaptations associated with the subterranean niche in mammals: a comparative study of three superorders. Mol. Biol. Evol 35, 2544–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Argyle EC, and Mason MJ (2008). Middle ear structures of Octodon degus (Rodentia: Octodontidae), in comparison with those of subterranean Caviomorphs. J. Mammal 89, 1447–1455. [Google Scholar]
- 96.Faulkes C, and Bennett N (2007). African mole-rats: social and ecological diversity. Rodent Societies, 427–437. [Google Scholar]
- 97.Eigenbrod O, Debus KY, Reznick J, Bennett NC, Sánchez-Carranza O, Omerbašić D, Hart DW, Barker AJ, Zhong W, Lutermann H, et al. (2019). Rapid molecular evolution of pain insensitivity in multiple African rodents. Science 364, 852–859. [DOI] [PubMed] [Google Scholar]
- 98.Zheng J, Du GG, Anderson CT, Keller JP, Orem A, Dallos P, and Cheatham M (2006). Analysis of the oligomeric structure of the motor protein prestin. J. Biol. Chem 281, 19916–19924. [DOI] [PubMed] [Google Scholar]
- 99.Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kosakovsky Pond SL, and Frost SD (2005). Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol 22, 1208–1222. [DOI] [PubMed] [Google Scholar]
- 101.Kumar S, Stecher G, Li M, Knyaz C, and Tamura K (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol 35, 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- 103.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. ; The Gene Ontology Consortium (2000). Gene ontology: tool for the unification of biology. Nat. Genet 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.The Gene Ontology Consortium (2019). The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 47 (D1), D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Van Camp G, and Smith RJH (2020). Hereditary Hearing Loss Homepage https://hereditaryhearingloss.org/.
- 106.Keane M, Craig T, Alföldi J, Berlin AM, Johnson J, Seluanov A, Gorbunova V, Di Palma F, Lindblad-Toh K, Church GM, and de Magalhães JP (2014). The Naked Mole Rat Genome Resource: facilitating analyses of cancer and longevity-related adaptations. Bioinformatics 30, 3558–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yu C, Li Y, Holmes A, Szafranski K, Faulkes CG, Coen CW, Buffenstein R, Platzer M, de Magalhães JP, and Church GM (2011). RNA sequencing reveals differential expression of mitochondrial and oxidation reduction genes in the long-lived naked mole-rat when compared to mice. PLoS ONE 6, e26729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yu C, Wang S, Yang G, Zhao S, Lin L, Yang W, et al. (2017). Breeding and rearing naked mole-rats (Heterocephalus glaber) under laboratory conditions. J. Am. Assoc. Lab. Anim. Sci 56, 98–101. [PMC free article] [PubMed] [Google Scholar]
- 109.Schrode KM, Muniak MA, Kim YH, and Lauer AM (2018). Central compensation in auditory brainstem after damaging noise exposure. eNeuro 5, ENEURO.0250–18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.May BJ, Lauer AM, and Roos MJ (2011). Impairments of the medial olivocochlear system increase the risk of noise-induced auditory neuropathy in laboratory mice. Otol. Neurotol 32, 1568–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shaffer LA, and Long GR (2004). Low-frequency distortion product otoacoustic emissions in two species of kangaroo rats: implications for auditory sensitivity. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol 190, 55–60. [DOI] [PubMed] [Google Scholar]
- 112.Braude JP, Vijayakumar S, Baumgarner K, Laurine R, Jones TA, Jones SM, and Pyott SJ (2015). Deletion of Shank1 has minimal effects on the molecular composition and function of glutamatergic afferent postsynapses in the mouse inner ear. Hear. Res 321, 52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McLean WJ, Smith KA, Glowatzki E, and Pyott SJ (2009). Distribution of the Na,K-ATPase alpha subunit in the rat spiral ganglion and organ of Corti. J. Assoc. Res. Otolaryngol 10, 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bai JP, Navaratnam D, and Santos-Sacchi J (2019). Prestin kinetics and corresponding frequency dependence augment during early development of the outer hair cell within the mouse organ of Corti. Sci. Rep 9, 16460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Malick LE, and Wilson RB (1975). Modified thiocarbohydrazide procedure for scanning electron microscopy: routine use for normal, patho-logical, or experimental tissues. Stain Technol 50, 265–269. [DOI] [PubMed] [Google Scholar]
- 116.Emerling CA, Widjaja AD, Nguyen NN, and Springer MS (2017). Their loss is our gain: regressive evolution in vertebrates provides genomic models for uncovering human disease loci. J. Med. Genet 54, 787–794. [DOI] [PubMed] [Google Scholar]
- 117.Fabre PH, Hautier L, Dimitrov D, and Douzery EJ (2012). A glimpse on the pattern of rodent diversification: a phylogenetic approach. BMC Evol. Biol 12, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu L, Tamura K, Sanderford M, Gray VE, and Kumar S (2016). A molecular evolutionary reference for the human variome. Mol. Biol. Evol 33, 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Original/source data for figures in the paper are available from the corresponding author on request.


