Abstract
In transplantation, the ever-increasing number of organ’s demand and long-term graft dysfunction constitute some of the major problems. Therefore, alternative solutions to increase the quantity and quality of the organ supply for transplantation are desired. On this subject, revolutionary CRISPR technology holds enormous potential for the scientific community with its expanding toolbox. In this minireview, we summarize the history and mechanism of CRISPR/Cas9 systems and explore its potential applications at cellular and organ level transplantation. The last part of this review includes future opportunities as well as the challenges in the transplantation field.
Keywords: Transplantation, Genome-Editing, CRISPR/Cas9, Genomics, Translational Research
1. Introduction
Transplantation represents the most successful curative treatment for acute and chronic organ failure. Unfortunately, the gap between the availability of organs for transplantation and the increasing number of patients in the national waiting lists continues. Nowadays, more than 110,000 patients in the United States are waiting for a transplant, and unfortunately around half of them would receive an organ, with around 20 listed patients losing their lives every day ( optn.transplant.hrsa.gov)1. This issue motivated surgeons and scientists to explore innovative ways to find a solution for high organ demand. In this regard, while some researchers are focusing on organoid generation from stem cells or induced pluripotent cells (iPS), others are trying to modify genomes of cells or the entire organ of the donor to make the graft more resistant to injury or tolerant for allo- and xenotransplantation.
While moving to a new decade, we witnessed many awe-inspiring scientific discoveries in the last decade. Some of the major mind-boggling breakthroughs in the last 10 years include improvements in next-generation sequencing technology which allow us to sequence an entire human genome for a few hundred dollars rather than billions of dollars, evaluation of multi-omics data at single-cell resolution, producing organoids as a miniaturized organ and genome editing by revolutionary CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)-technology. State-of-the-art technologies have been exploited to understand the molecular and cellular mechanisms of major conditions during transplantation, such as ischemia-reperfusion injury, transplant tolerance, and rejection, among other issues. In this minireview, we summarize the successes of CRISPR technologies and its combinatorial enrollment with the aforementioned innovations since 2013, and we further address potential future CRISPR applications in solid organ transplantation (SOT).
2. Genome Editing
H.W Boyer and S.N. Cohen’s studies for the recombinant DNA technology in the early 1970s paved the foundation of modern genome editing technologies2. Scientific efforts over the following two decades demonstrated the importance of double-strand DNA break near the target site to achieve precise and efficient genome editing 3, 4. To take advantage of this observation, artificial restriction enzymes have been proposed to eliminate the unwanted consequences of random DNA integration as well as inefficiency. To do this, first, ZFNs (zinc finger nucleases) were engineered to alter genetic material precisely by designing a special peptide for each codon of 18-24 nucleotide long target site 5, 6. Second, TALEN (Transcription activator-like effector nuclease) was developed for the same purpose by generating a DNA binding domain that can recognize single specific nucleotide 7, 8. Despite their great advantages in genome engineering, both systems need expensive and time-consuming de-novo protein synthesis for each target site. Finally, the CRISPR system adds versatility to genome engineering by recognizing the target DNA on an RNA dependent “molecular scissors” rather than protein-DNA dependent mechanism. Due to low feasibility and higher cost of the first two methods, recent clinical trials have been performed by the recent CRISPR technologies (Table 1).
Table 1.
Summary of the main gene editing technologies.
| Method | Recognition mechanism |
Complexity | Specificity | Cost | Scalability | Overall Feasibility |
Clinical Trial (numbers; field) |
|---|---|---|---|---|---|---|---|
| ZFN (since 1996) | protein-DNA | high | low | high | no | low | 15; mainly for HIV |
| TALEN (since 2009) | protein-DNA | moderate | high | high | no | medium | 6; for blood cancer |
| CRISPR (since 2013) | RNA-DNA | low | high | low | yes | high | 33; sickle cell, blood cancer, CAR-T, diagnostic |
2.1. The Rise of CRISPR:
CRISPR stands for Clustered Regularly Interspaced Short Palindromic Repeats. These palindromic repeats and spacer sequences in between were first discovered in E.Coli in 1987, long before the establishment of ZFN and TALEN system9. Subsequent studies further showed that half of the bacteria and >90% of archaea carry those DNA fragments in their genome10, 11, however, researchers had to wait for almost 20 years to understand their natural function as an adaptive immune system of bacteria and archaea against bacteriophages12-14. In 2012, E. Charpentier and J.A. Doudna repurposed the type II CRISPR/Cas9 system as a new genome-editing tool, much like ZFN and TALEN, in their in-vitro study15. Three groups further applied this RNA-programmed genome editing to the human cells 16-18. Initial findings demonstrate CRISPR/Cas9 as a facile, robust, and multiplexable system compared to other gene-editing tools.
2.2. Mechanism of CRISPR/Cas9 System:
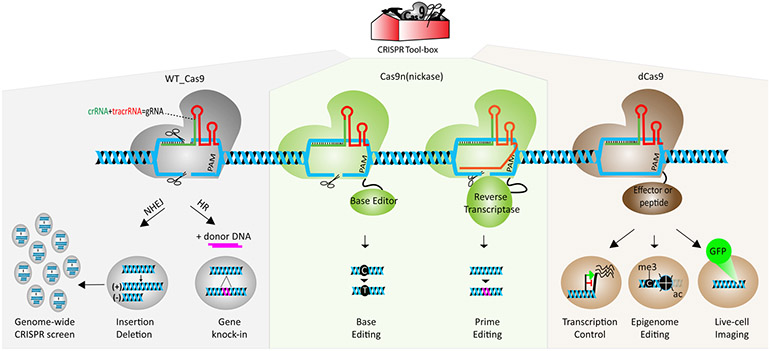
The most widely used CRISPR/Cas9 system, derived from Streptococcus pyogenes(sp), requires one single protein called spCas9(CRISPR associated protein), two natural non-coding RNA molecules, crRNA (CRISPR RNA) and tracrRNA (trans-activating crRNA) and ribonuclease III15, 19. By taking advantage of RNA engineering, two non-coding RNA molecules are fused to produce single guide RNA (sgRNA) which has two critical features18: 20 nucleotides at the 5’ end determine the DNA target site by Watson-Crick base pairing if target DNA has a PAM (protospacer adjacent motif) sequence, and the scaffold structure at the 3’ end interacts with Cas9. In other words, the first 20 nucleotides (originally crRNA) of the sgRNA helps Cas9 to inspect 3 billion bases in the human genome to find the exact match like a Google search engine. Once the sgRNA/Cas9 complex finds the right site, Cas9 performs its endonuclease function like a “molecular surgeon” to produce a double-strand cut that is corrected mainly by two different DNA repair mechanisms: error-prone “non-homologous end-joining”(NHEJ) and error-free “homologous repair”(HR). If the donor template is supplied to the target cells, the HR system gets a chance to edit the target site by integrating the given DNA fragment during cell division; however, NHEJ generally repairs the double-strand break by random insertion and deletion of nucleotides in both dividing and non-dividing cells16, 18. These mechanisms comprise the canonical Cas9-mediated cleavage activity. It is also noteworthy that researchers have gone further and developed many other tools by modifying the wild type Cas9 endonuclease (Figure 1).
Figure 1: Overview of Cas9-based CRISPR mechanisms.
CRISPR toolbox expands by relying on three versions of Cas9 molecule. (WT: wild type, n: nickase, d: dead (inactive) Cas9, gRNA: guide RNA, PAM: Protospacer Adjacent Motif, me3: methylation of DNA at cytosine, ac: acetylation mark on histones, GFP: green fluorescent protein).
2.3. CRISPR-tool box is expanding:
Due to ease of use and higher editing efficiency, CRISPR/Cas9 became a popular gene-editing tool to edit the genome of different cell types (cell lines, primary cells, iPSC) and to generate transgenic animals in a short amount of time20, 21. These signs of progress inspire researchers to increase their efforts to find novel applications of this system across a variety of fields. The scientific community challenged traditional approaches by utilizing inactive or death Cas9 (dCas9), which lacks nuclease activity on both domains but generates a higher affinity to the target DNA. Therefore, de-novo dCas9-fused complex allows us to dock effector protein or peptides on the desired location wherein researchers can change gene expression, alter the epigenome, image the chromatin in live cells and modify the chromatin architecture22.Cas9 nickase which has only one active nuclease domain initiated the second-generation CRISPR system in which “base editing” and “prime editing” allow us to edit the genome without donor DNA and double-strand DNA cut23, 24 (Table 2). We further repurposed base editing to silence genes in a safer method called CRISPR-STOP 25. One of the major advantages of the CRISPR system is its scalability or multiplex targeting due to an RNA-based editing mechanism. By designing a genome-wide CRISPR library containing 80-100k gRNA to target the entire genome, CRISPR screens have become a tremendous tool for functional genomics where one can test the effect of loss-of-function or gain-of-function at the genomic level in the presence of selectable phenotype or outcome. Studies extrapolating the CRISPR library demonstrate the ease with which researchers can carry out parallel high-throughput screening 26, 27. Collectively, these findings have helped expand the CRISPR toolbox in less than a decade, and each of these tools holds great promise for future translational research (Figure 1).
Table 2:
Summary of the different Cas9 variants and their roles for the improvement of this technology.
| Date | Cas variants | Purpose | Improvements | Reference | |
|---|---|---|---|---|---|
| 2013 | WT Cas9 | native Cas9(spCas9) | genome editing | specific genome editing in eukaryotic cell | 16-18 |
| 2016 | e-SpCas9 | increase the specificity of Cas9 | mutations in non-catalytic domain and reduced off-target effects | 70 | |
| 2016 | HF1-SpCas9 | 71 | |||
| 2017 | hypa-spCas9 | 72 | |||
| 2018 | xCas9 | expand the PAM recognition | broad range of PAM sequence and greater DNA sensitivity | 73 | |
| 2015 | saCas9 | Cas9 alternative in other organisms | smaller Cas9 with different PAM | 74 | |
| 2015 | other major Cas molecules | Cas12a(Cpf1) | finding Cas9 alternative | smaller Cas9, easy to pack in virus | 75 |
| 2016 | Cas13a(C2c2) | ability to modify RNA sequence | 76, 77 | ||
| 2018 | Cas14 | ability to target ss DNA | 78 | ||
| mutated spCas9 |
fused domains | ||||
| 2013 | Cas9n(nickease) (one mutation) | - | genome editing | reduce the chance of random double strand cut | 79 |
| 2016 | Cytidine Deaminase | base editing (C to T) | genome editing without ds DNA cut and donor DNA | 23 | |
| 2017 | Deoxyadenosine deaminase | base editing (A to G) | 80 | ||
| 2019 | Reverse Transcriptase | prime editing | 24 | ||
| 2013 | dCas9 (death or inactive Cas9) (double mutation) | KRAB | gene regulation | site specific gene silencing | 81 |
| 2013 | VP64,VPR | site specific gene activation | 82, 83 | ||
| 2014 | Fokl | reduce off-target effect | less off-target activities | 84 | |
| 2015 | p300 | epigenome editing | site specific acetylation of H3K27 | 85 | |
| 2016 | Tet | epigenome editing | site specific removal of methyl group | 86 | |
| 2017 | DNMT3a | epigenome editing | site specific addition of methyl group | 87 | |
| 2017 | GFP | live-cell imaging | specific loci labeling in vivo | 88 | |
| 2017 | PYL1,ABI1 | reorganizing chromatin architecture | locus specific chromatin looping | 89 | |
| 2019 | EZH2 | epigenome editing | site specific methylation of H3K27 | 90 | |
3. Transplantation studies in the age of gene editing
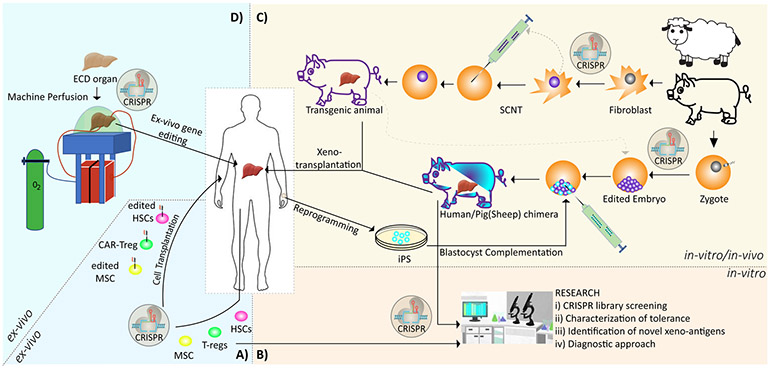
Compared to the current applications of CRISPR technology in medical fields such as oncology, neuroscience or developmental biology, there is a limited number of applications in the transplantation. However, promising improvements are continually developing, particularly those related to xenotransplantation. In this section, the on-going and potential future applications of this revolutionary system from cell to organ level for transplantation are discussed and summarized (Figure 2).
Figure 2: On-going and future applications of CRISPR technology for solid organ transplantation.
A) Applications of CRISPR-based technologies at cellular level for tolerance induction. B) CRISPR mediated research strategies. C) Xenotransplantation from CRISPRed transgenic animals and CRISPRed chimeric animal for solid organ. D) Potential ex-vivo CRISPR application for solid organs inside the pump. (ECD: Expanded Criteria Donor, SCNT: Somatic Cell Nuclear Transfer, HSC: hematopoietic stem cell, MSC: Mesenchymal Stem Cell).
3.1). In-vivo and ex-vivo gene editing at cellular level
3.1.1). Gene editing of universal human cells for transplantation
Cas9 and gRNA can be efficiently delivered to target cells by using viral (AAV, lentiviral) and non-viral (electroporation, liposome, nanoparticles, RNP) methods to exert a therapeutic effect 28,29. Delivering the CRISPR system into ex-vivo isolated cells is relatively more feasible compared to in-vivo targeting of solid tissue30, 31. Therefore, initial CRISPR clinical trials (33 total phase I/II studies) have been performed to cure cancer and blood cell-related diseases by manipulating T-cells and hematopoietic stem cells (HSC) ex-vivo31. Studies regarding pluripotent stem cells (PSC) and induced stem cells (iPSC) combined with CRISPR gene editing opened tremendous opportunities to understand regenerative medicine and disease settings. Researchers are not yet able to generate an organ from stem cells on a dish in the laboratory with the current technology. However, researchers have engineered allogeneic stem cells by removing MHC I and II molecules and inserting CD47 protein by CRISPR to produce universal stem cells, which are termed “hypoimmunogenic” and evade immune rejection. This strategy might be used in future projects for organ development in the lab32, 33. On the other hand, in hereditary tyrosinemia type 1 (HT1) disease, allogeneic liver/hepatocyte cell transplantation has potential therapeutic benefits, but the availability of enough donor cells and GvHD (graft versus host disease) represent major limitations that are common in cell-based transplantation. In a mouse model of this disease, both in-vivo and ex-vivo CRISPR mediated gene editing in hepatocytes improved liver metabolism and demonstrated the potential of the CRISPR system as an alternative therapy for cell/organ transplantation34, 35. Another potential target for cell-based therapy is a mesenchymal stem/stromal cell (MSC) due to its anti-inflammatory, immunomodulatory and tissue-repair properties that can play critical roles during solid organ transplantation36, 37. CRISPR manipulation of those cells will demonstrate potential therapeutic approaches in the near future38.
3.1.2). Gene Editing and Tolerance
Tolerance, the specific absence of a harmful alloimmune response to a graft tissue in the absence of immunosuppression, is the main step for successful complete immunosuppressive drug (IS) withdrawal following solid organ transplantation. Dendritic and Regulatory T cells (Tregs) have been studied for a while to understand their roles in immune tolerance39, 40. Therefore, these cells became the natural target of the CRISPR gene editing with the potential to provide further insights into the mechanisms behind their protective role in immunotolerance and improve their activity. Scientists can engineer both cells to induce tolerance41, 42. In a recent study, Cas9 and gRNA targeting CD40 were encapsulated into nanoparticles and successfully disrupted CD40 in dendritic cells both in-vivo and in-vitro41. Currently, approximately 50 clinical trials are using Tregs and 15 of them have been conducted for solid organ transplant (SOT) (clinicaltrials.gov) 43. Adoptive cell therapy (ACT) of polyclonal T-reg cells have been clinically tested in living donor liver and kidney transplantation and demonstrated the efficacy in IS cessation44, 45. Alloantigen-specific T-reg cells are more potent inhibitors than the polyclonal T-reg cells; however, producing a high yield of these cells is very challenging 46. Researchers can generate these antigen-specific T-reg cells using CRISPR technology by i) engineered TCR, ii) chimeric antigen receptors (CAR) like CAR-T cells in immune-oncology or iii) overexpression of FoxP3 in antigen-specific effector CD4+ cells. CRISPR-dCas9 mediated epigenome editing has been shown to change FoxP3 expression significantly47, 48. As of today, there is no clinical trial using CRISPR engineered T-reg cells. Collectively, alloantigen-specific CRISPR gene editing will improve the specificity, stability, and efficiency of T-reg cells, and it might allow scientists to produce off-the-shelf T-reg cells with advanced engineering. T-reg adoptive cell therapy in which RNA-guided nuclease technology and T-reg are merged will be one of the most effective treatments for solid organ transplants. Furthermore, genome-wide CRISPR-screening studies shed lights on details of molecular mechanisms behind the T-reg differentiation like revealing the activation and differentiation of helper Th2 cells49.
3.2). Gene editing at organ level
The increasing need of organ donors for end-stage patients forces researchers and clinicians to evaluate alternative options (i.e., xenotransplantation, organoids, 3D bioprinting). These alternative methods are taking advantage of developments from CRISPR technology to shorten the needed time for transferring applications from bench to bedside.
3.2.1). Xenotransplantation and humanized pig liver
Farm animals and non-human primates (NHP) have been considered alternative sources for organ supply since the 1900s together with some medical, legal and ethical issues50. Chimpanzees and baboons have been used in initial trials, but in the modern era of xenotransplantation, porcine are currently preferred due to their similar organ size, ease of breeding, high offspring number, and less disease transmission risk compared to NHP. However, there are still three major barriers preventing successful pig-to-primate(human) organ transplants: i) Glycans on the surface of porcine endothelial cells act like xenoantigens (α-Gal, Neu5Gc, SDa) and cause hyperacute rejection (HAR), ii) dysregulated coagulation due to the disagreement between the pig and human coagulation system (THBD, TFPI, CD39) and iii) porcine endogenous retroviruses (PERV) in porcine genome can transfer vertically into human cells and cause xenosis. Due to the efficiency and simplicity of CRISPR gene editing, researchers have tested around 40 different combinations of pig-gene knock-out and human-gene knock-in into the porcine genome with great efficiency in less than a decade51, 52. Recently, G. Church and colleagues established the most advanced transgenic pig by using CRISPR technology in which they successfully deleted 3 pig genes (GGTA1, CMAH, B4GALNT2), and specifically inserted 9 human transgenes (CD46, CD55, CD59, B2M, HLA-E, CD47, THBD, TFB1, CD39 ) in a single locus in addition to the inactivation of 25 PERV loci in pig cells 53. Biotech companies like eGenesis (Boston), Qihan (China) and United Therapeutics (MD) have accelerated their progress to begin the first human trial. Further improvements can be anticipated in these transgenic animals after evaluating the results of the first trials leading to a better understanding of xeno-immune response mechanisms. One possible way to improve the xenotransplantation is to identify novel non-gal xenoantigens and powerful genome-wide CRISPR screening might be a suitable method to assess the role of pig genes involved in immune rejection54. As of today, there is only one study conducted with the pig CRISPR library55. Noteworthy, CRISPR technology with future gadgets will likely improve the feasibility of genome project (GP)-write aiming to write a virus-free genome with the desired modifications56.
3.2.2). Chimeric Organs and Blastocyst Complementation
With current technologies, it is not possible to generate human organs with in-vitro conditions; however, an animal can serve as a biological incubator to produce human organs at least partially. In theory, patient-specific and immune-matched chimeric organs can be generated by injecting human pluripotent/stem cells into an animal blastocyst (blastocyst complementation) or a targeted organ of utero (utero transplantation)57. Rat to mice or mice to rat chimeric animals have been generated with great success. However, the ratio of chimerism is very low for interspecies chimeras such as human/pig or human/sheep58-60. CRISPR gene editing increases the efficiency of human/animal chimerism by disabling particular organ development in the host zygote that can be filled by human cells61. In future chimeric organ generation studies, scientists will utilize the most advanced CRISPR mediated transgenic pigs or sheep as a source of zygote and surrogate animals as well.
4. Potential future applications of CRISPR technology in transplantation
In several clinical trials, normothermic pulsatile machine perfusion has been used for organ procurement up to 24 hours with increasing success and progress62, 63. In a recent study, Clavien and his group were able to increase the viability of poor-quality livers up to 7 days by modifying the regular pump with multiple core physiological units64. These improvements in machine perfusion and enhancement in delivery methods of CRISPR can surpass the limitation of in-vivo organ editing by employing ex-vivo gene editing of donor organs inside the pump. We speculate that future studies might focus on re-evaluating the ex-vivo approaches associated to normothermic organ perfusions in 1- discarded organ and making them viable for transplantation, 2- poor quality organs (e.x Fatty livers) and treat them before implantation and 3- inducing transplant tolerance making grafts less immunogenic. Another critical factor during organ transplants is the ischemia-reperfusion injury to the organ. In our unpublished study, we exploited the genome-wide CRISPR screen to identify the genes whose depletion induce resistance against the oxygen deprivation in an in-vitro model. This powerful functional genomic screening will also be a very useful tool to answer diverse problems of transplantation, such as identifying novel xenoantigens and novel molecules of immunotolerance. After the transplant procedure, patient monitoring against rejection is important. Recent studies illustrate CRISPR mediated diagnostic tools for virus detection in a shorter period. SHARLOCK and DETECTR are two CRISPR-based identification methods that can be tailored to detect biomarkers of rejections from the patient’s plasma or urine 65, 66. CRISPR-Chip which can sense unamplified DNA samples on graphene within 15 minutes will also be used for the same purpose 67.
5. Limitations of CRISPR technology:
The off-target effect, efficient delivery methods of CRISPR molecules for in-vitro and particularly for in-vivo applications and immunogenicity of Cas9 protein against the human immunity system are major barriers for CRISPR technology. Since Cas9 molecules can tolerate some mismatches especially at the distal gRNA/DNA dimer, intense work should be considered for the design of gRNA and the concentration of Cas9 molecules. Any undesirable change which is caused by off-target in the human (or animal for xenotransplant) genome might cause serious unwanted effects, such as activation of oncogenes,or any other de novo SNPs changing the behaviour of the cells. Improved Cas proteins, redesigned gRNA molecules, and new strategies like prime-editing might eliminate some off-target problems. Transfering Cas9 and gRNA molecules into cells especially for in-vivo treatment associate with major limitations since there is no chance to choose the edited cells as in the case of in-vitro or ex-vivo treatment. Depending on the organ and disease, researcher might need different levels of editing, partial or full edited organ which is not possible with current delivery method. Additionally, it has been shown that >60% of humans have a pre-existing humoral and cell-mediated adaptive response against major molecules of the CRISPR system, i.e. Cas968. Therefore, if a persistent expression of Cas9 is required during the treatment, the immunologic response of Cas9 protein needs to be taken into consideration. A possible solution to this issue might be to modify the epitope of Cas9 protein without altering its function and specificity69.
6. Conclusion
CRISPR is an adaptive immune system of bacteria and repurposing CRISPR technology in the development of cell/organ for transplantation will be sententious. So far, the scientific community has benefited from broad applications of CRISPR technology. In the era of genomics and super-computers, the next-generation researchers may discover more from “simple” organisms such as bacteria, archaea, and even viruses. It will be very exciting to follow the future progress of these revolutionary technologies for medical research.
Acknowledgment:
This research was funded through generous support from James D. Eason Transplant Institute and R01DK080074(VM), R01DK109581(VM) and R01DK117183(AB). We would like to thank members of the Transplant Research Institute, especially C. Watkins and M. McKay for their critical reading of the manuscript.
References:
- 1.OPTN/SRTR 2018 Annual Data Report: Introduction. Am J Transplant 20 Suppl s1, 11–19 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Cohen SN, Chang AC, Boyer HW & Helling RB Construction of biologically functional bacterial plasmids in vitro. Proc Natl Acad Sci U S A 70, 3240–3244 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rudin N, Sugarman E & Haber JE Genetic and physical analysis of double-strand break repair and recombination in Saccharomyces cerevisiae. Genetics 122, 519–534 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rouet P, Smih F & Jasin M Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol 14, 8096–8106 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bibikova M, Beumer K, Trautman JK & Carroll D Enhancing gene targeting with designed zinc finger nucleases. Science 300, 764 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Kim YG, Cha J & Chandrasegaran S Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A 93, 1156–1160 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boch J et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Moscou MJ & Bogdanove AJ A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Ishino Y, Shinagawa H, Makino K, Amemura M & Nakata A Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 169, 5429–5433 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mojica FJ, Diez-Villasenor C, Soria E & Juez G Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol Microbiol 36, 244–246 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Grissa I, Vergnaud G & Pourcel C The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8, 172 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrangou R et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Makarova KS, Grishin NV, Shabalina SA, Wolf YI & Koonin EV A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct 1, 7 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pourcel C, Salvignol G & Vergnaud G CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151, 653–663 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Jinek M et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cong L et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jinek M et al. RNA-programmed genome editing in human cells. Elife 2, e00471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mali P et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deltcheva E et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou Z et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci U S A 110, 15644–15649 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adli M The CRISPR tool kit for genome editing and beyond. Nat Commun 9, 1911 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anzalone AV et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuscu C et al. CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations. Nat Methods 14, 710–712 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Szlachta K et al. CRISPR knockout screening identifies combinatorial drug targets in pancreatic cancer and models cellular drug response. Nat Commun 9, 4275 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shalem O et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glass Z, Lee M, Li Y & Xu Q Engineering the Delivery System for CRISPR-Based Genome Editing. Trends Biotechnol 36, 173–185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lino CA, Harper JC, Carney JP & Timlin JA Delivering CRISPR: a review of the challenges and approaches. Drug Deliv 25, 1234–1257 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilbie D, Walther J & Mastrobattista E Delivery Aspects of CRISPR/Cas for in Vivo Genome Editing. Acc Chem Res 52, 1555–1564 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Glass Z, Huang M, Chen ZY & Xu Q Ex vivo cell-based CRISPR/Cas9 genome editing for therapeutic applications. Biomaterials 234, 119711 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deuse T et al. Hypoimmunogenic derivatives of induced pluripotent stem cells evade immune rejection in fully immunocompetent allogeneic recipients. Nat Biotechnol 37, 252–258 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malik NN, Jenkins AM, Mellon J & Bailey G Engineering strategies for generating hypoimmunogenic cells with high clinical and commercial value. Regen Med 14, 983–989 (2019). [DOI] [PubMed] [Google Scholar]
- 34.VanLith CJ et al. Ex Vivo Hepatocyte Reprograming Promotes Homology-Directed DNA Repair to Correct Metabolic Disease in Mice After Transplantation. Hepatol Commun 3, 558–573 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin H et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol 32, 551–553 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vandermeulen M et al. Mesenchymal Stromal Cells in Solid Organ Transplantation. Transplantation (2019). [DOI] [PubMed] [Google Scholar]
- 37.Kaundal U, Bagai U & Rakha A Immunomodulatory plasticity of mesenchymal stem cells: a potential key to successful solid organ transplantation. J Transl Med 16, 31 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Golchin A, Shams F & Karami F Advancing Mesenchymal Stem Cell Therapy with CRISPR/Cas9 for Clinical Trial Studies. Adv Exp Med Biol (2020). [DOI] [PubMed] [Google Scholar]
- 39.Hasegawa H & Matsumoto T Mechanisms of Tolerance Induction by Dendritic Cells In Vivo. Front Immunol 9, 350 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaban E et al. Targeting Regulatory T Cells for Transplant Tolerance: New Insights and Future Perspectives. Kidney Dis (Basel) 4, 205–213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y et al. In situ repurposing of dendritic cells with CRISPR/Cas9-based nanomedicine to induce transplant tolerance. Biomaterials 217, 119302 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Raffin C, Vo LT & Bluestone JA Treg cell-based therapies: challenges and perspectives. Nat Rev Immunol (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Esensten JH, Muller YD, Bluestone JA & Tang Q Regulatory T-cell therapy for autoimmune and autoinflammatory diseases: The next frontier. J Allergy Clin Immunol 142, 1710–1718 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Mathew JM et al. A Phase I Clinical Trial with Ex Vivo Expanded Recipient Regulatory T cells in Living Donor Kidney Transplants. Sci Rep 8, 7428 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Todo S et al. A pilot study of operational tolerance with a regulatory T-cell-based cell therapy in living donor liver transplantation. Hepatology 64, 632–643 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Sagoo P et al. Human regulatory T cells with alloantigen specificity are more potent inhibitors of alloimmune skin graft damage than polyclonal regulatory T cells. Sci Transl Med 3, 83ra42 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okada M, Kanamori M, Someya K, Nakatsukasa H & Yoshimura A Stabilization of Foxp3 expression by CRISPR-dCas9-based epigenome editing in mouse primary T cells. Epigenetics Chromatin 10, 24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forstneric V et al. CRISPRa-mediated FOXP3 gene upregulation in mammalian cells. Cell Biosci 9, 93 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henriksson J et al. Genome-wide CRISPR Screens in T Helper Cells Reveal Pervasive Crosstalk between Activation and Differentiation. Cell 176, 882–896 e818 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reemtsma K Xenotransplantation: A Historical Perspective. ILAR J 37, 9–12 (1995). [DOI] [PubMed] [Google Scholar]
- 51.Cowan PJ, Hawthorne WJ & Nottle MB Xenogeneic transplantation and tolerance in the era of CRISPR-Cas9. Curr Opin Organ Transplant 24, 5–11 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Thomas A, Hawthorne WJ & Burlak C Xenotransplantation literature update, November/December 2019. Xenotransplantation 27, e12582 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Yue Y et al. Extensive Mammalian Germline Genome Engineering. bioRxiv, 2019.2012.2017.876862 (2019). [Google Scholar]
- 54.Li P, Skill JN, Kubal CA, Fridell JA & Ekser B Identification of novel xenoreactive non-gal antigens: tetraspanin CD37 and CD81. Xenotransplantation 24 (2017). [Google Scholar]
- 55.Zhao C et al. CRISPR screening of porcine sgRNA library identified host factors essential for Japanese encephalitis virus replication. bioRxiv, 840835 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Servick K Genome writing project confronts technology hurdles. Science 356, 673–674 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Lu Y, Zhou Y, Ju R & Chen J Human-animal chimeras for autologous organ transplantation: technological advances and future perspectives. Ann Transl Med 7, 576 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kobayashi T et al. Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell 142, 787–799 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Wu J et al. Interspecies Chimerism with Mammalian Pluripotent Stem Cells. Cell 168, 473–486 e415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamaguchi T et al. Interspecies organogenesis generates autologous functional islets. Nature 542, 191–196 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Vilarino M et al. CRISPR/Cas9 microinjection in oocytes disables pancreas development in sheep. Sci Rep 7, 17472 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Summers DM et al. Cold pulsatile machine perfusion versus static cold storage for kidneys donated after circulatory death: a multicenter randomized controlled trial. Transplantation (2019). [DOI] [PubMed] [Google Scholar]
- 63.Weissenbacher A, Vrakas G, Nasralla D & Ceresa CDL The future of organ perfusion and re-conditioning. Transpl Int 32, 586–597 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eshmuminov D et al. An integrated perfusion machine preserves injured human livers for 1 week. Nat Biotechnol 38, 189–198 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen JS et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gootenberg JS et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360, 439–444 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hajian R et al. Detection of unamplified target genes via CRISPR-Cas9 immobilized on a graphene field-effect transistor. Nat Biomed Eng 3, 427–437 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Charlesworth CT et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat Med 25, 249–254 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ferdosi SR et al. Multifunctional CRISPR-Cas9 with engineered immunosilenced human T cell epitopes. Nat Commun 10, 1842 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Slaymaker IM et al. Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kleinstiver BP et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen JS et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550, 407–410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hu JH et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ran FA et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zetsche B et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Abudayyeh OO et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.East-Seletsky A et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Harrington LB et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 362, 839–842 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ran FA et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gaudelli NM et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gilbert LA et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maeder ML et al. CRISPR RNA-guided activation of endogenous human genes. Nat Methods 10, 977–979 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chavez A et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods 12, 326–328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tsai SQ et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol 32, 569–576 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hilton IB et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33, 510–517 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morita S et al. Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions. Nat Biotechnol 34, 1060–1065 (2016). [DOI] [PubMed] [Google Scholar]
- 87.Stepper P et al. Efficient targeted DNA methylation with chimeric dCas9-Dnmt3a-Dnmt3L methyltransferase. Nucleic Acids Res 45, 1703–1713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qin P et al. Live cell imaging of low- and non-repetitive chromosome loci using CRISPR-Cas9. Nat Commun 8, 14725 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Morgan SL et al. Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat Commun 8, 15993 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fukushima HS, Takeda H & Nakamura R Targeted in vivo epigenome editing of H3K27me3. Epigenetics Chromatin 12, 17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]