Abstract
Background:
The regeneration of larvae zebrafish fin emerged as a new model of regeneration in the last decade. In contrast to genetic tools to study fin regeneration, chemical probes to modulate and interrogate regeneration processes are not well developed.
Results:
We set up a zebrafish larvae fin regeneration assay system and tested activities of natural product compounds and extracts, prepared from various microbes. Colomitide C, a recently isolated product from a fungus obtained from Antarctica, inhibited larvae fin regeneration. Using fluorescent reporter transgenic lines, we show that colomitide C inhibited fibroblast growth factor (FGF) signaling and WNT/β-catenin signaling, which were activated after larvae fin amputation. By using the endothelial cell reporter line and immunofluorescence, we showed that colomitide C did not affect migration of the blood vessel and nerve into the injured larvae fin. Colomitide C did not show any cytotoxic activities when tested against FGF receptor-amplified human cancer cell lines.
Conclusion:
Colomitide C, a natural product, modulated larvae fin regeneration likely acting upstream of FGF and WNT signaling. Colomitide C may serve as a template for developing new chemical probes to study regeneration and other biological processes.
Keywords: Colomitide C, natural product, zebrafish larvae, fin regeneration, Fibroblast growth factor signaling, WNT signaling
INTRODUCTION
The zebrafish (Danio rerio) is a freshwater fish native to south Asia, and is a long-established model organism that shares genetics and cell biology with humans1,2. Zebrafish regenerate a variety of tissues and organs, such as the fin, heart, retina, kidney, liver and the central nervous system3,4. Knowledge of tissue/organ regeneration in zebrafish contributes to understanding of this remarkable biological process. Moreover, the study of regeneration in zebrafish is relevant to the field of regenerative medicine with the potential of applying the knowledge and tools to enhance regenerative capacity in humans. Fin regeneration in zebrafish, in particular, is an amenable experimental system and has been extensively studied5,6.
Signaling pathways regulate a variety of biological processes from embryonic development and tissue/organ regeneration to cancer7. The pathways include those initiated by fibroblast growth factors (FGFs), WNTs, bone morphogenic proteins, hedgehog, insulin-like growth factor, Notch and retinoic acid8. Various studies have demonstrated that FGF signaling is critical for fin regeneration in adult zebrafish. Multiple fgf ligand genes (fgf3, fgf10a, fgf20a, fgf20b) and fgf receptor 1 (fgfr1) are expressed in the regenerating caudal fin. Inhibition of FGF signaling by dominant negative fgfr1 expression or SU5402, a small molecule inhibitor for Fgfr1 tyrosine kinase, has a significant inhibitory effect on fin regeneration9–12. WNT signaling is also critical for the regulation of adult fin regeneration. Inhibiting WNT/ß-catenin signaling in adult zebrafish blocks fin regeneration13–15.
Zebrafish embryos hatch at 48–72 hours post fertilization (hpf)16. At this stage, most of morphogenesis is complete, and larvae mainly increase in size17. In the last decade, fin fold regeneration in zebrafish embryos (amputation at 48 hpf)18–21 and larvae (amputation at 72 hpf)22,23 has emerged as an alternative model of regeneration. The embryonic fin fold, amputated at 48 hpf, may fully regenerate within 72 hours after amputation, compared to two weeks in the adult. In addition, fin fold regeneration shares mechanisms with adult fin regeneration. For example, inhibition of Fgfr signaling by SU5402 blocks fin fold regeneration18, similar to the adult fin. Contrary to this report, a molecular difference has also been reported in regeneration of fins between adults and larvae. For instance, inhibition of WNT signaling in adult zebrafish causes failure to regenerate the fin13–15. In contrast, during fin fold regeneration, activating WNT signaling by 6-bromoindirubin-3’-oxime (BIO), a small molecule that blocks GSK3ß and hence activates ß-catenin signaling, inhibits fin regeneration24. While the larvae fin is a suitable system for regeneration studies, the difference from adult fin regeneration described above suggests that more characterization of larvae fin regeneration is needed.
In the past two decades, various genetic methods have been developed to visualize and/or modulate activities of signaling pathways. Transgenic reporters are one of the most widely used approaches25. Tools to manipulate activities of specific components of signaling pathways have also been adopted such as recombinase-driven cassette insertion26 and CRISPR/Cas9 targeted knockouts27. Such tools have contributed to uncovering mechanisms of fin regeneration and the roles of the signaling pathways. In addition to genetic methods, development and use of chemical tools that regulate specific components of signaling pathways is an important approach. Although some chemical tools have been developed, further discovery and development of additional molecular probes that modulate activities of signaling pathways would extend fin regeneration research.
Natural products are compounds produced by living organisms including bacteria, fungi, plants, and animals. More than 200,000 diverse secondary metabolites have been isolated and identified, and many have demonstrated significant biological activities. More than 70% of clinically used drugs for infectious disease and cancer are derived from or inspired by natural products and their active pharmacophores. Because these chemicals are “biologically pre-validated”, they also hold great potential to be adapted as chemical tools and probes for a diverse array of complex biological processes, such as cancer treatment and immune modulation28.
Zebrafish are an ideal model organism to use in phenotypical screening of natural products for effects on endogenous processes such as regeneration27,29. Whole animal assays take into account both membrane permeability and overall toxicity which can be limitations with in vitro target-based or cellular screens, respectively. Due to the relatively low yields of natural products, the available material for screening is also frequently a limiting factor for conducting vertebrate assays. However, due to their small body size, zebrafish larvae have been a suitable model for testing chemicals and even for performing large scale chemical screening in a multiple-well plate format with small volumes30–34.
We assayed a microbial natural products library to identify compounds that modulate signaling processes involved in larval zebrafish fin regeneration. One active metabolite identified in this screen was colomitide C, a volatile polyketide isolated from an Antarctic fungus35. We found that colomitide C inhibits larval fin regeneration and both FGF and WNT signaling. Furthermore, our in vitro assays demonstrated that colomitide C has no cytotoxic activities to various mammalian cell lines. Colomitide C may serve as a template for development as a nontoxic tool for chemical modulation of signaling pathways.
RESULTS
Testing activities of natural compounds on larvae fin regeneration
Natural products have been used as sources for chemical tool development28. Due to the relative lack of small molecule modulators of regeneration, we chose to screen a broad, diverse library of natural products from bacteria, fungi and plants developed in-house over many years. In addition to screening pure compounds, we also tested crude extracts and fractions to potentially identify additional molecules of interest in complex mixtures, which would then require follow up assay-guided fractionation. As an assay system, we chose zebrafish fin regeneration. Due to their small body size, the larvae fin regeneration system allows for chemical treatments in a small volume. We anesthetized and amputated the caudal fin of larvae at 4 days post fertilization (dpf) at the posterior end of the spinal cord and allowed them to regenerate the fin for 48 hours (Fig. 1A, B). The larvae were treated with various microbial natural products, including both fractionated extracts and purified compounds. As a control, we used DMSO (vehicle) and 0.5 μM BIO which is known to inhibit larvae fin regeneration24. We treated five larvae in a well of a 24-well plate dish with 1 mL of fish water with compounds. We collected regeneration and toxicity data using the following criteria: (1) three or more control larvae survived after 48 hours, which ensures that the specific clutch of larvae were healthy; (2) three or more BIO-treated larvae survived after 48 hours and exhibited inhibition of fin regeneration with p<0.05, which ensures that the specific clutch of larvae responds to a known treatment; and (3) three or more larvae treated with a natural product survived after 48 hours, which allows for examining the effect by t-test. We measured the length of the most regenerated fin tissue from the amputation plane (Fig. 1B) and compared the length of larvae treated by a natural compound to that of the average of DMSO-treated larvae. We then used unpaired t-test to determine whether a relative regeneration length is significant (p<0.05). We performed this screening using fractions of extracts (at 1 – 10 μg/ml) and purified compounds (at 10–100 μM). Several natural products were tested multiple times (Table 1). When a product exhibited both positive (p<0.05 for inhibition or enhancing regeneration) and negative (p ≥ 0.05) results in multiple assays, we considered that the compound had a positive activity.
Fig. 1. Zebrafish larvae fin regeneration assay system.
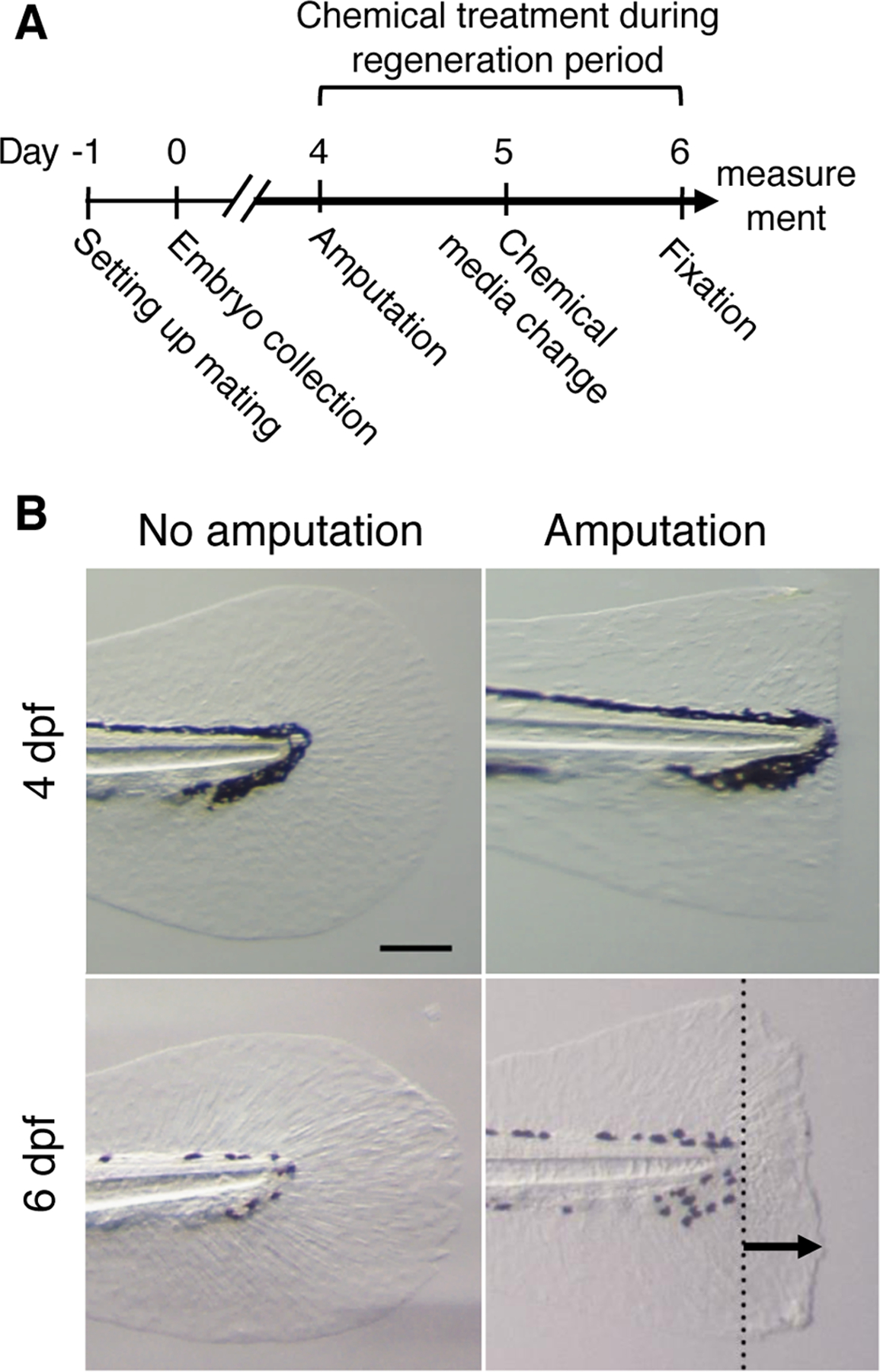
(A) Schematic of experimental design from setting up mating to sample fixation at 6 dpf.
(B) Caudal fin images at 4dpf and 6 dpf with and without amputation at 4dpf. Dotted line in the lower right panel indicates the amputation plane. The arrow in the lower right panel indicates a maximum regenerated length. Scale bar, 150 μm.
Table 1.
Number of assays for natural products
| Positive | Negative | Toxic | |
|---|---|---|---|
| Assayed once | 50 | 162 | 43 |
| Assayed twice | 58 | 34 | 4 |
| Assayed ≥ 3 times | 49 | 17 | 0 |
| Total | 157 | 213 | 47 |
The numbers of natural products that were assayed once, twice or more than three times that exhibited positive (p < 0.05), negative (p ≥ 0.05) or toxic effects are shown.
We screened a total of 417 unique compounds and fractions, which are derived from bacteria, fungi and plants (Table 2). Out of the 417 compounds or fractions, 213 samples did not exhibit significant activities on larval fin regeneration (p≥0.05, Fig. 2). Forty-seven samples were toxic, and more than three larvae died, while control larvae survived. Forty-three compounds and fractions exhibited either inhibitory or stimulatory effects with less than 10% of inhibition or enhancement of regeneration. Among compounds that exhibited <90% regeneration length, 33 and 35 compounds exhibited 80–90% and <80% of regeneration length, respectively. Among compounds that exhibited >110% regeneration length, 31 and 15 compounds exhibited 110–120% and >120% of regeneration length, respectively. Natural compounds that exhibited either positive or negative results included pure compounds, extracts and fractions (Table 3). Most of the toxic samples were crude extracts (Table 2).
Table 2.
Numbers of natural products derived from various sources
| Sources | Positive | Negative | Toxic |
|---|---|---|---|
| Bacteria | 68 | 169 | 44 |
| Fungus | 38 | 22 | 0 |
| Plants | 47 | 18 | 1 |
| Derivatives of purified compounds | 3 | 4 | 2 |
| Purchased compound | 1 | 0 | 0 |
| Total | 157 | 213 | 47 |
The numbers of compounds derived from bacteria, fungus, plants, derivatives of purified compounds or a purchased compound that exhibited positive (p < 0.05), negative (p ≥ 0.05) or toxic effects are shown.
Fig. 2. Summary of larvae fin regeneration screening.
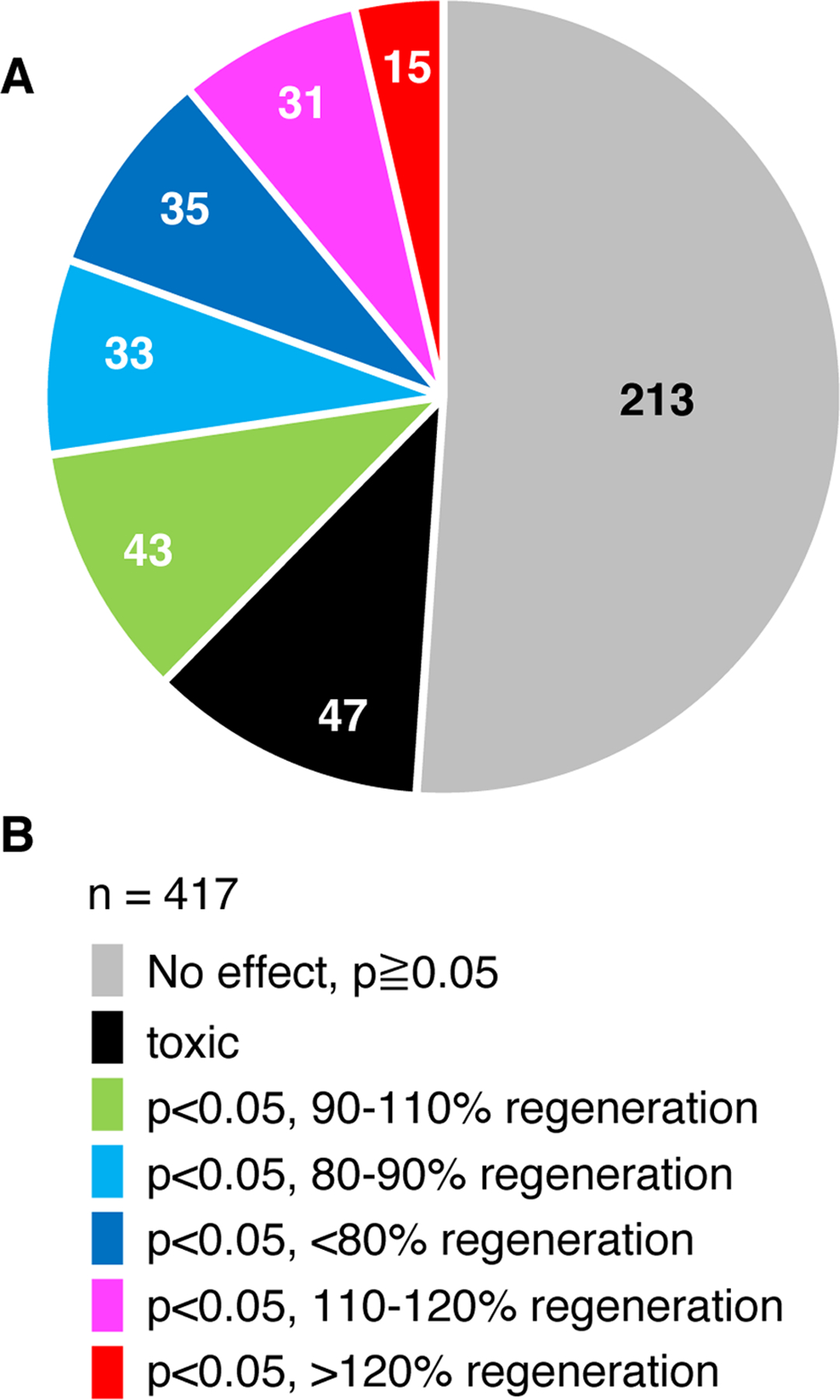
(A) Summary of experiments with 429 unique compounds are shown as a pie chart. The number in each fraction shows the number of fractions or purified compounds.
(B) Color-coding of the results shown in A.
Table 3.
Numbers of pure compounds, extracts and fractions assayed in this study
| Positive | Negative | Toxic | |
|---|---|---|---|
| Pure compounds | 89 | 37 | 3 |
| Extracts | 55 | 62 | 44 |
| Fractions | 13 | 114 | 0 |
| Total | 157 | 213 | 47 |
The numbers of pure compounds, extracts or fractions that exhibited positive (p < 0.05), negative (p ≥ 0.05) or toxic effects are shown.
Colomitide C blocks larvae fin regeneration
We focused our analysis on the polyketide metabolite colomitide C, which we recently characterized and can obtain in high yields (>1g/L) from the producing fungus35. During the initial screening, colomitide C exhibited both positive (n=3) and negative (n=2) results at 50 μM. In the following two assays colomitide C consistently inhibited fin regeneration at 100 μM. Therefore, we further tested colomitide C activity at 100 μM. Given the known function of FGFR signaling and WNT signaling in larvae fin regeneration, we compared the activities of colomitide C with SU5402 (FGFR kinase inhibitor) and BIO (WNT signaling agonist) (Fig. 3) with an increased number of replicates. We measured two parameters with the test samples: one is the maximum length of the regenerated fin from the amputation plane. The other is measuring the total area of regenerated fin. Compared to DMSO-treated controls, BIO and SU5402 inhibited regeneration by both the length (23–26% reduction compared to controls) and area (26–32% reduction compared to controls) (Fig. 3B, C), consistent with previous studies18,24. Colomitide C consistently inhibited fin regeneration by >30% at 100 μM (Fig. 3). Larvae survived and looked healthy after colomitide C treatment with this concentration over 2 days of treatment. The overall morphology and swimming behavior of the colomitide C-treated larvae showed no sign of abnormalities. These results demonstrate that colomitide C inhibits fin regeneration at a high concentration without affecting larvae viability.
Fig. 3. Colomitide C inhibits zebrafish larvae fin regeneration.
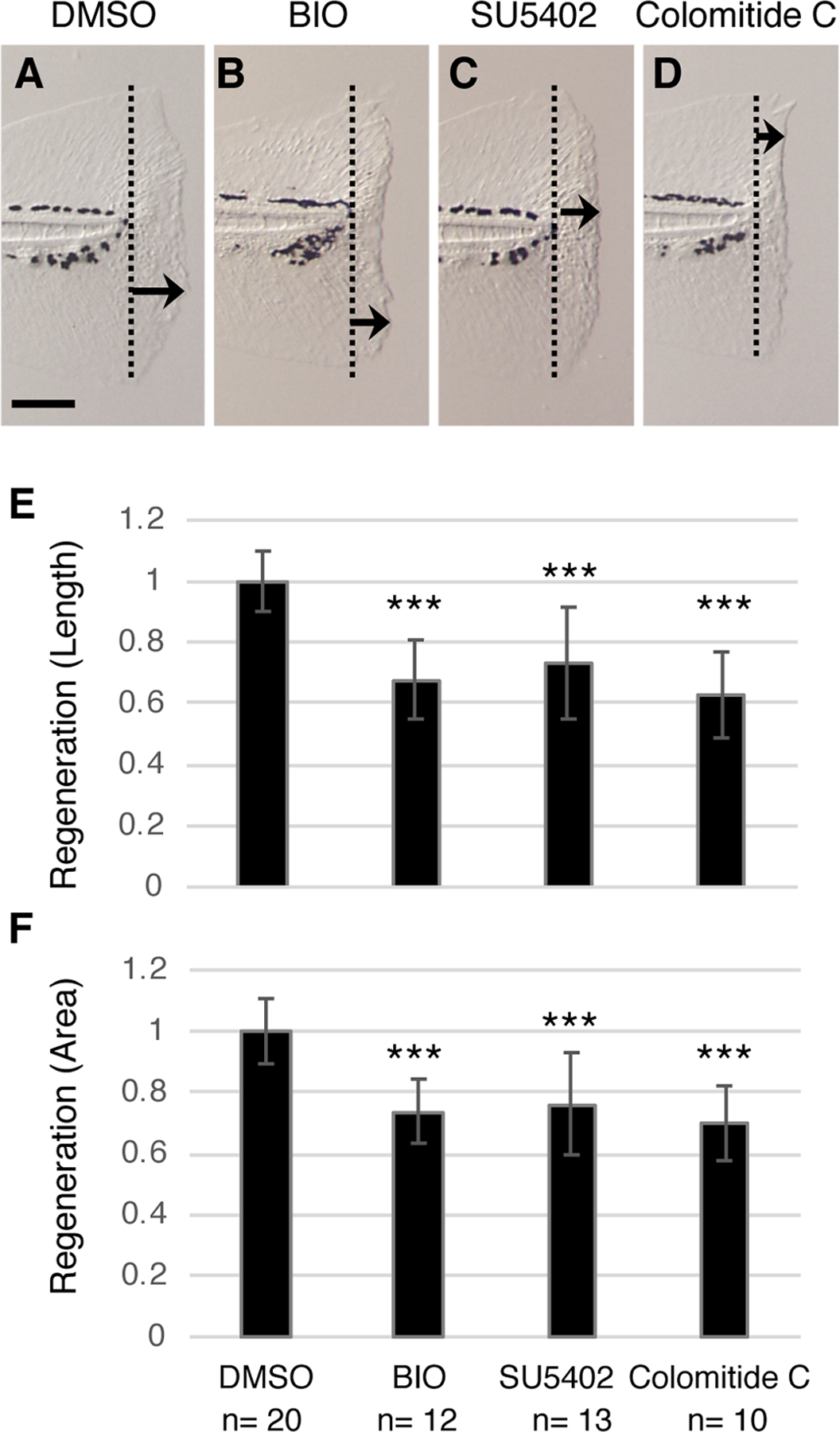
(A-D) Images of larvae fin 2 days after amputation. Larvae were treated with indicated chemicals. Dotted lines indicate the amputation plane. Arrows indicate the maximum regenerated length in each image. Scale bar, 100 μm.
(E,F) Graphic presentation of maximum regenerated length (E) and total area of regenerated fin (F). Values are normalized using the average value of DMSO-treated fin as 1.0. ** p<0.01, *** p<0.001 by t-test.
Colomitide C downregulates FGF signaling and WNT signaling
Inhibition of larvae fin regeneration suggests that colomitide C may influence signaling pathways, such as FGFR signaling known to regulate larvae fin regeneration8. To test this idea, we used a fluorescent reporter fish line. The Et7-EGFP line is an enhancer trap-line, generated by EGFP cassette insertion near the dusp6 locus using the sleeping beauty transposon system36. It has been shown that expression of dusp6 is regulated by FGF signaling37,38, and therefore, the EGFP signal in the Et7-EGFP line reports activation of FGF signaling36. Strong Et7-EGFP signals were detected in the spinal cord without fin amputation (white arrows, Fig. 4A, D, n=5). After fin amputation, the Et7-EGFP signals were upregulated in the regenerating fin mesenchyme (yellow arrows, Fig. 4B, E, n=5), consistent with the requirement of FGF signaling in larvae fin regeneration. Colomitide C treatment significantly downregulated the Et7-EGFP signals in the regenerating fin mesenchyme as well as the endogenous Et7-EGFP signals in the spinal cord (white arrowheads, Fig. 4C, F, n=10). These data indicate that colomitide C inhibits FGF signaling in vivo during larvae fin regeneration.
Fig. 4. Colomitide C modulates FGF and WNT signaling during larvae fin regeneration.
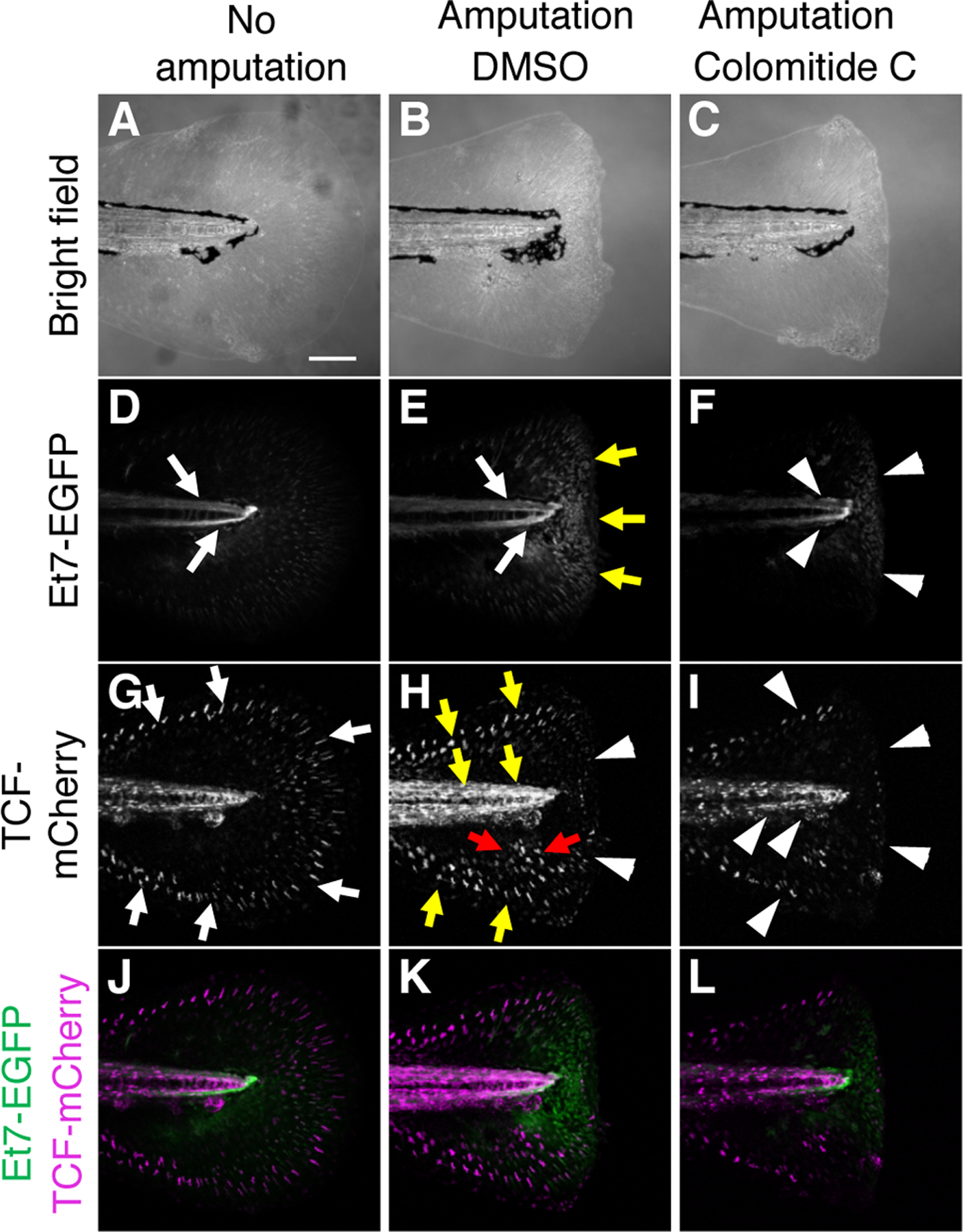
Images of 6 dpf larvae fin without amputation (A, D, G, J). Images of 6 dpf larvae fin, amputated at 4dpf (B, C, E, F, H, I, K, L). Larvae in the middle and right column are treated with DMSO and colomitide C, respectively.
(A-C) Bright filed image. (D-F) Et7-EGFP signals, shown in black/white. (G-I) TCF-mCherry signals, shown in black/white. (J-L) Merged images of Et7-EGFP signals (green) and TCF-mCherry signals (magenta).
The TCF-mCherry line reports activation of the canonical WNT signaling pathway that requires a TCF transcription factor and ß-catenin39. Since activating canonical WNT signaling by BIO inhibits larvae fin regeneration24, we examined endogenous status of the canonical WNT signaling pathway by the TCF-mCherry line. Strong TCF-mCherry signals were detected in the spinal cord and the periphery of the tail fin without amputation (white arrows, Fig. 4G, n=5). After fin amputation, the signals at the fin periphery and the spinal cord became stronger (yellow arrows, Fig. 4H, n=5). In addition, fin mesenchyme also exhibited increase in the TCF-mCherry signals (red arrows, Fig. 4H, n=5). These observations indicate that canonical WNT signaling is upregulated in the regenerating fin and proximal fin mesenchyme. Treating larvae with colomitide C reduced the TCF-mCherry signals in the periphery of the fin and the spinal cord (white arrowheads, Fig. 4I, n=10). These results indicate that colomitide C also inhibits WNT signaling in vivo during larvae fin regeneration.
Colomitide C does not affect migration of the nerve and endothelial cells into the regenerating area
It is known that the nerve plays a crucial role in pectoral fin regeneration in adult zebrafish40. In addition, it has been shown that fin amputation in zebrafish larvae promotes regeneration of nearby axons41. We next investigated whether colomitide C affects innervation of regenerating larvae fins by visualizing axons through immunostaining of acetylated tubulin. Without amputation, the nerve was detected along the spinal cord, which spread to the distal parts of the tail fin (Fig. 5A, D, n=3). Two days after amputation, the nerve was similarly detected in the regenerating fin with DMSO treatment (Fig. 5B, E, n=7) and colomitide C treatment (Fig. 5 C, F, n=7). In order to evaluate whether colomitide C affects innervation more precisely, we assessed the density of axons in the regenerating area by measuring the relative ratio of axon area in the regenerating fin (depicted by red dotted area, Fig. 5E, E’)41. The density of axons is similar between DMSO-treated control and colomitide C-treated larvae (Fig. 5G). This result indicates that nerve migration is not affected by colomitide C, and suggests that the action of colomitide C on larvae fin regeneration does not involve nerves.
Fig. 5. Colomitide C does not affect density of nerve and endothelial cells during larvae fin regeneration.
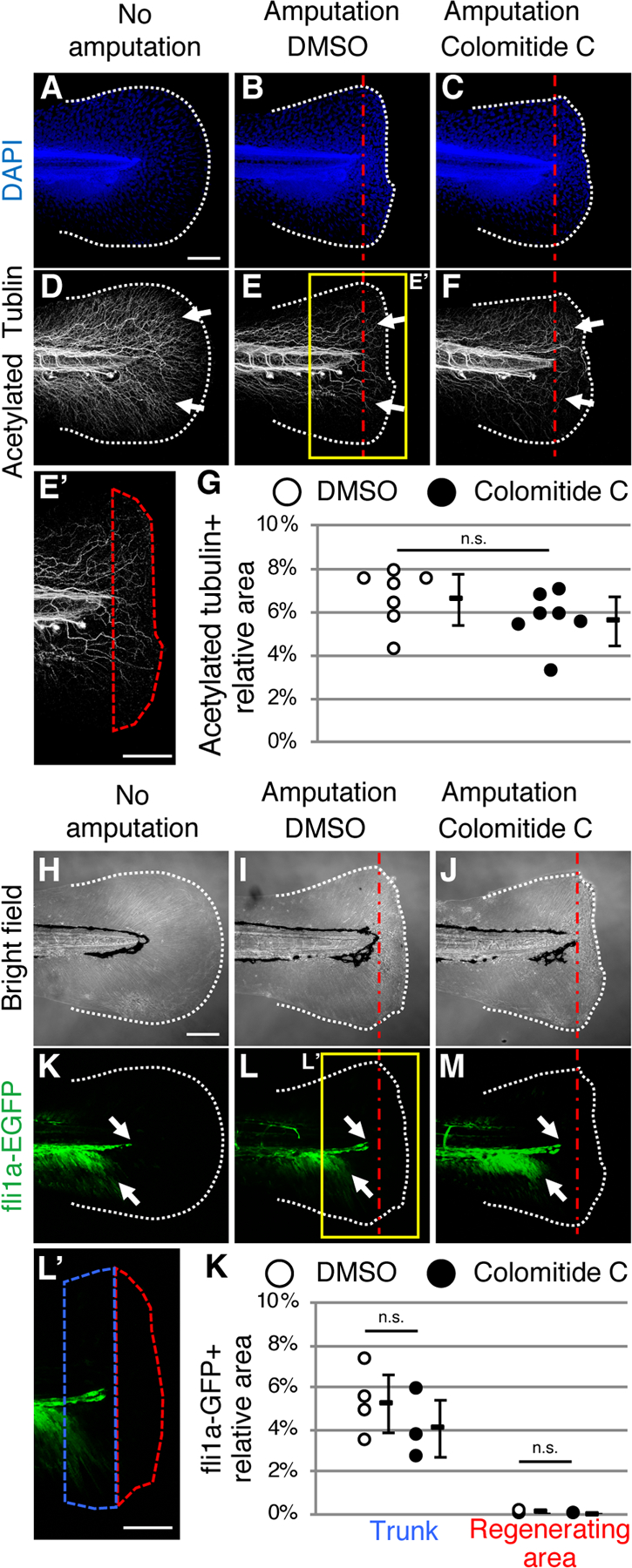
Images of 6 dpf larvae fin without amputation (A, D). Images of 6 dpf larvae fin, amputated at 4dpf (B, C, E, F). Larvae in the middle and right column are treated with DMSO and colomitide C, respectively.
(A-G) DAPI (A-C) and acetylated tubulin (D-F) signals after whole-mount immunostaining. Tail fin without amputation (A, D), with amputation and DMSO-treatment (B, E, E’), and with amputation and colomitide C-treatment (C, F) are shown. E’ shows a closeup indicated in E. Red dotted area in E’ show the regenerating fin. (G) Relative density of the nerve in the regenerating fin. . n.s., not significant by t-test.
(H-K) Bright field images (H-J) and fli1a-GFP signals (K-M) of the tail fin without amputation (H, K), with amputation and DMSO-treatment (I, L, L’), and with amputation and colomitide C-treatment (J, M). L’ shows a closeup indicated in L. Red dotted area and blue dotted area in L’ show the regenerating fin and the trunk area 100 μm from the amputation plane, respectively. (K) Relative density of endothelial cells in the trunk and regenerating fin. . n.s., not significant by t-test.
White dotted lines and red dotted straight lines indicate the amputation plane, respectively. A-F and H-M are at the same scale. Scale bar in A, E’, H and L’, 100 μm
In the amputated adult fin, the blood vessel becomes open at the amputation plane. A previous study showed that after the amputated vessel ends are connected, the truncated vessels regenerate during fin regeneration42. We tested whether vessels in the larvae fin regenerate after amputation, and if colomitide C modulates the blood vessel regeneration process. For this purpose, we used the fli1a-EGFP fish line, which reports endothelial cells43. In 6 dpf fin without amputation, the fli1a-EGFP signals were detected along the spinal cord, which spread ventrally, but do not extend into the fin (Fig. 5H, K, n=2). In regenerating fins, this pattern of fli1a-EGFP signals was similar with both DMSO treatment (Fig. 5I, L, n=4) and colomitide C treatment (Fig 5J, M, n=3). In both DMSO-treated and colomitide C-treated larvae, fli1a-GFP-positive endothelial cells were not detected in the regenerating fin (depicted by red dotted area, Fig. 5L, L’, K). We further assessed whether endothelial cells just proximal to the amputation plane were affected by measuring the ratio of fli1a-GFP-positive area in the trunk within 100 μm from the amputation plane (depicted by blue dotted area, Fig. 5L, L’). The ratio of fli1a-GFP positive area in the trunk was similar with and without colomitide-C-treatment (Fig. 5 K). These results indicate that endothelial cells do not migrate immediately into the regenerating fin in larvae, and that colomitide C does not exhibit activity related to endothelial cell migration.
Colomitide C does not exhibit cytotoxicity to breast cancer cell lines with FGFR amplification
Given the significant inhibition of amputation-induced activation of FGF signaling in zebrafish larvae, we tested whether colomitide C affects other systems where FGF signaling has a critical role. For this purpose, we chose to use several well-characterized cancer cell lines. FGFR1 amplification is observed in ~10% of patients with estrogen receptor (ER)-positive/human epidermal growth factor receptor 2 (HER2, also known as ERBB2)-negative breast cancer, and mutations in FGFR genes have been associated with progression of breast cancer44,45. Inhibition of FGF signaling is considered a potential therapeutic strategy for breast cancer to overcome the resistance to ER-targeting treatment45. With this information, we tested whether colomitide C exhibits cytotoxicity by a standard tetrazolium assay in vitro46. We used three human breast cancer cell lines, CAMA-1, HCC38 and MDA-MB-361, which are known to have FGFR1 amplification and FGFR1 protein overexpression47.
We also compared the activity of colomitide C to known modulators of different components of FGF signaling (AZD4547, brivanib, GSK690693, MK-2206, wortmannin) to identify patterns of activity and potential mechanisms of action. We present full dose-response curves for each compound against each cell line at two time points (Fig. 6, 7) in order to provide a visual indication of both the potency and range of activity (steepness of slope). This presentation provides additional information to supplement the EC50 values (Table 4). AZD4547 is a potent and selective inhibitor of autophosphorylation of FGFR tyrosine kinases 1–348. AZD4547 exhibited moderate cytotoxicity to all three cell lines for the 48 and 72 hour timepoints (EC50 = 8.7 – 20.3 μM, Table 4, Fig. 6B, H, N, Fig. 7B, H, N). Brivanib is an investigational, anti-tumorigenic drug that inhibits VEGFR1 and FGFR 1–349,50. Brivanib was only active towards HCC38 cells after the 72 hour treatment (EC50 = 5.9 μM, Table 4, Figs. 6, 7) and was nontoxic towards CAMA-1 and MDA-MB-361 cell lines (EC50 > 50 μM, Table 4, Figs. 6, 7).
Fig. 6. Cytotoxicity assay results after treating breast cancer cells lines for 24 hours.
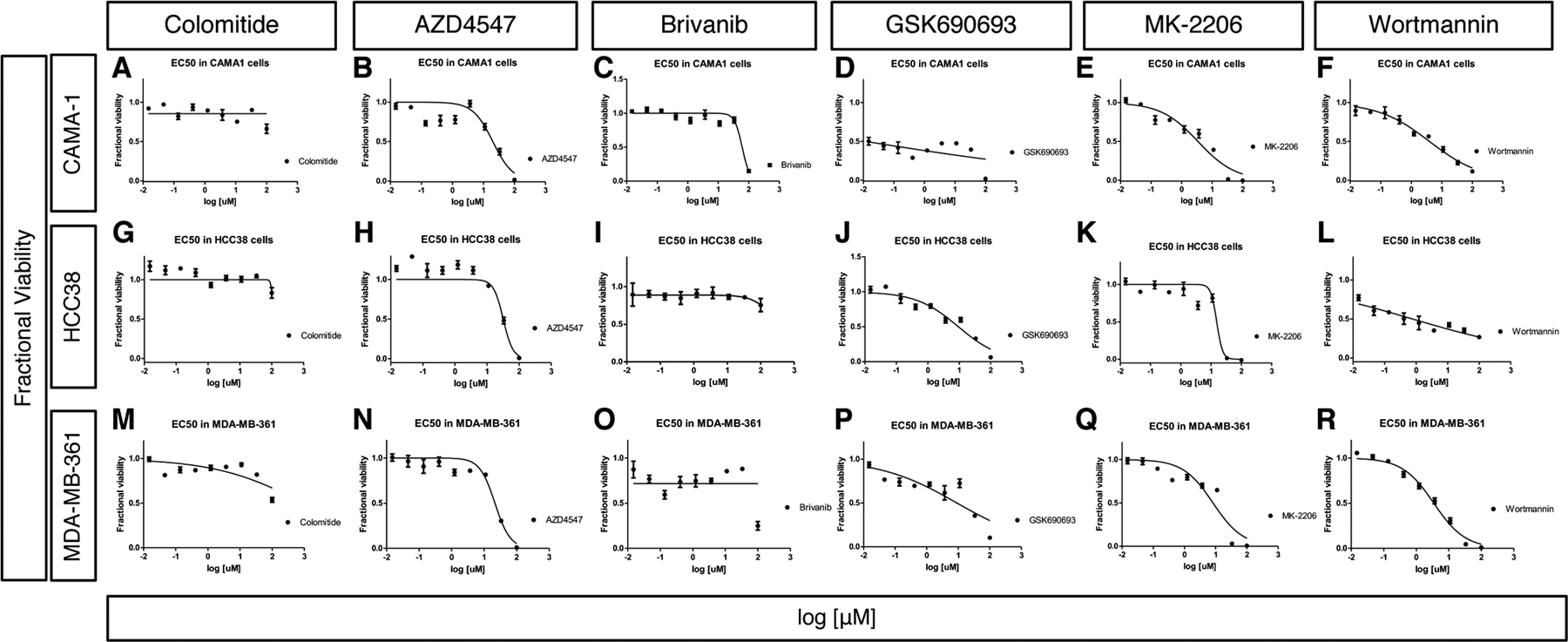
Dose-dependent cytotoxicity assay results after treating CAMA-1 (A-F), HCC38 (G-L) and MDA-MN-361 (M-R) cell lines for 24 hours. Colomitide C (A, G,M), ZAD4547 (B, H, N), Brivanib (C, I, O), GSK690693 (D,J,P), MK-2206 (E,K, Q) and Wortmannin (F, L, R) were used.
The x axis represents concentration of chemicals in log10 scale and the y axis represents fractional viability.
Fig. 7. Cytotoxicity assay results after treating breast cancer cells lines for 48 hours.
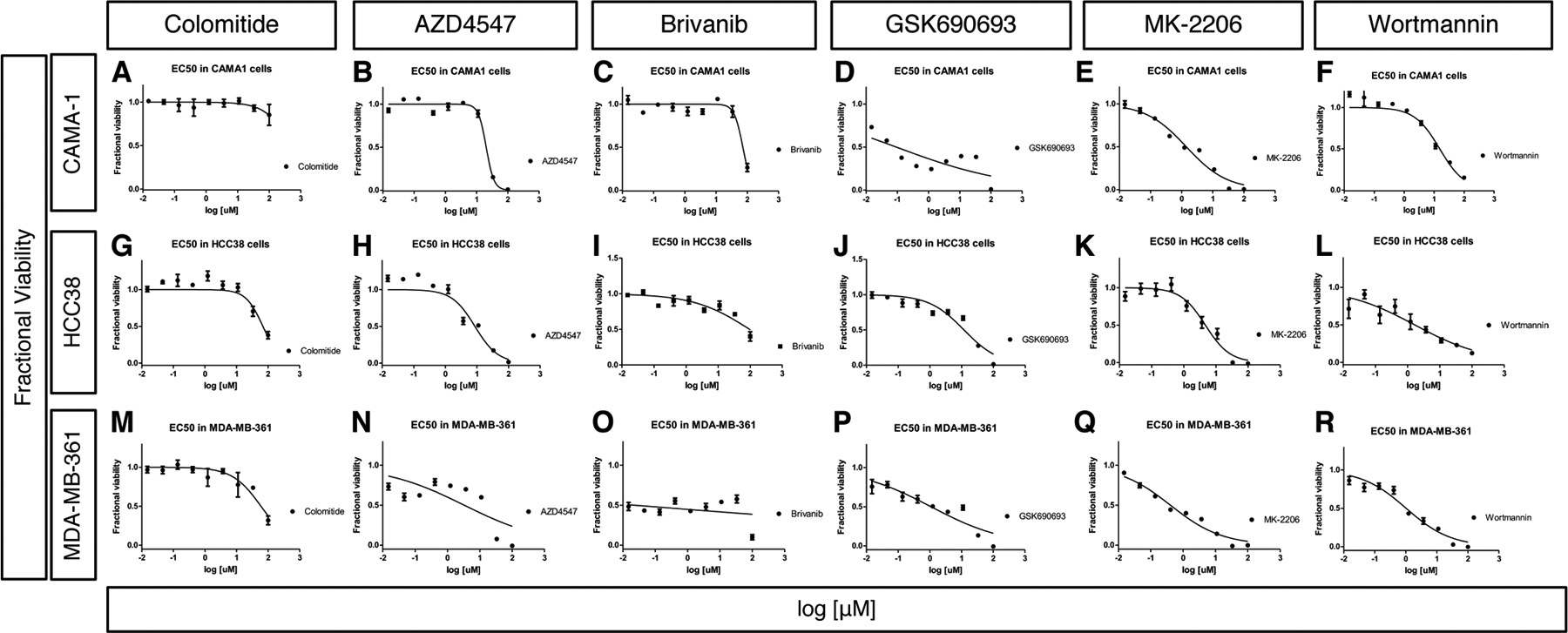
Dose-dependent cytotoxicity assay results after treating CAMA-1 (A-F), HCC38 (G-L) and MDA-MN-361 (M-R) cell lines for 48 hours. Colomitide C (A, G,M), ZAD4547 (B, H, N), Brivanib (C, I, O), GSK690693 (D,J,P), MK-2206 (E,K, Q) and Wortmannin (F, L, R) were used.
The x axis represents concentration of chemicals in log10 scale and the y axis represents fractional viability.
Table 4.
EC50 values of various compounds on cell lines
| Cell line | Compound | 24 hours | 48 hours | 72 hours |
|---|---|---|---|---|
| CAMA-1 | Colomitide | > 100 | > 100 | > 100 |
| CAMA-1 | AZD4547 | 20.13 | 20.25 | 14.75 |
| CAMA-1 | Brivanib | 60.98 | 56.76 | 57.48 |
| CAMA-1 | GSK690693 | 50.84 | 0.0917 | 0.0696 |
| CAMA-1 | MK-2206 | 3.466 | 1.406 | 0.1853 |
| CAMA-1 | Wortmannin | 3.957 | 15.25 | 6.825 |
| HCC38 | Colomitide | > 100 | 68.94 | > 100 |
| HCC38 | AZD4547 | 32.10 | 8.664 | 11.64 |
| HCC38 | Brivanib | > 100 | 95.85 | 5.875 |
| HCC38 | GSK690693 | 9.396 | 11.83 | 3.175 |
| HCC38 | MK-2206 | 15.33 | 4.718 | 13.82 |
| HCC38 | Wortmannin | 0.7717 | 1.671 | 15.89 |
| MDA-MB-361 | Colomitide | > 100 | 59.61 | > 100 |
| MDA-MB-361 | AZD4547 | 21.51 | 16.66 | 15.59 |
| MDA-MB-361 | Brivanib | > 100 | 94.0 | 54.32 |
| MDA-MB-361 | GSK690693 | 9.021 | 4.907 | 0.559 |
| MDA-MB-361 | MK-2206 | 8.205 | 0.5505 | 0.0557 |
| MDA-MB-361 | Wortmannin | 3.474 | 1.692 | 2.909 |
All EC50/IC50 values in μM.
We also tested activities of compounds that target the phosphatidylinositol 3-kinase (PI3K) – protein kinase B (AKT) pathway. GSK690693 is a potent, ATP-competitive, pan-AKT kinase inhibitor51,52. GSK690693 showed potent sub-micromolar inhibition of CAMA-1 cells at the 48 and 72 hour timepoints (EC50 = 0.09 and 0.07 μM, respectively) and MDA-MB-361 cells at 72 hour (EC50 =0.56 μM) (Table 4, Figs. 6, 7). MK-2206 is a selective allosteric inhibitor of AKT 1–353, and exhibited potent cytotoxicity towards CAMA-1 (EC50 = 1.4 and 0.19 μM, 48 and 72 hr, respectively) and MDA-MB361 cells (EC50 = 0.55 and 0.06 μM, 48 and 72 hour, respectively) (Table 4, Figs. 6, 7). Wortmannin is a cell-permeable, fungal metabolite that acts as a potent, selective and irreversible inhibitor of PI3K54. Wortmannin was most cytotoxic towards HCC38 (EC50 =0.77 and 1.67 μM at 24 and 48 hour, respectively) and MDA-MB-361 cell lines (1.69 μM at 48 hour) (Table 4, Figs. 6, 7).
Colomitide C was non-toxic towards CAMA-1, HCC38 and MDA-MB361 cell lines (EC50 >50 μM) for all timepoints (Table 4, Figs. 6, 7). These results suggest that colomitide C downregulated FGFR signaling during larval fin regeneration through other mechanisms distinct from these compounds. Colomitide C was previously tested for antimicrobial activity against a panel of bacterial and fungal pathogens and found to be inactive (minimal inhibitory concentration > 100 μM) (Rusman et al. 2018).
DISCUSSION
Chemical screenings have been successfully performed in several whole organism systems. For example, Drosophila melanogaster55, Caenorhabditis elegans56, mice57 and zebrafish58 have been established as whole organism models for phenotypic screening. These models share physiological similarities and/or pathway homology to humans, so that phenotypes produced by chemicals will translate to the same target phenotypes in humans. Zebrafish and mice are useful as vertebrate model systems, but the small size, a large number of embryos from a mating (~100) and genetic tractability of zebrafish provide for an ideal screening platform. In addition, the zebrafish system offers a unique experimental setting due to the organisms’ exceptional ability to regenerate a variety of tissues and organs59. Chemical screens for modulators of adult fin regeneration have been previously performed, which identified the compound AGN192403 that targets the imidazole receptor as an inhibitor of fin regeneration32. The use of embryos or larval is an ideal way to minimize both the volume of each assay and amount of test material needed and also saves time by reducing the need to grow fish to the adult stage. Accordingly, a previous study showed a chemical screening method for fin regeneration using zebrafish embryos during the hatching period (48–72 hpf), which were amputated at 2 dpf and the results were evaluated at 3 dpf33. Our experiments in larvae were performed by amputating the caudal fin at 4 dpf, after near completion of morphogenesis17. We then evaluated fin regeneration after 2 days of treatment. This assay is an efficient and information-rich system to screen chemical libraries for physiological responses while simultaneously identifying toxicity and membrane permeability.
In addition to assessing fin regeneration by morphological evaluation of length and area, the availability of fluorescent reporter lines further facilitates the testing of compound activity on specific signaling pathways. For example, previous chemical screening using a dusp6-dGFP transgenic reporter fish line led to identification of the compound BCI that modulates FGF signaling60,61. Similarly, the use of the Et7-EGFP enhancer trap line in this study allowed us to assess FGFR signaling status during larvae fin regeneration. Development of other zebrafish lines that report developmental signaling, such as WNT, Hedgehog, bone morphogenetic proteins, transforming growth factor ß and Notch, have been reported25. These reporter lines will be useful tools in screening bioactive compounds and assessing the status of signaling pathways. Moreover, we also utilized the fli1a-EGFP line, which reports endothelial cells and immunofluorescence of the nerve. Various cell-type specific reporter lines have also been established and used for fin regeneration research, which includes those reporting osteoblasts62,63, macrophages64,65 and neutrophils65–67. Such reporter lines in combination with fluorescent imaging are useful tools to identify molecules that modulate signaling pathway activities during complex biological processes, such as fin regeneration.
Our study reported here provides an example of the utility of larval screening for identifying a natural product that modulates fin regeneration. In this study, we observed that colomitide C inhibits fin regeneration and modulates the FGF and WNT signaling pathways, two developmental signaling pathways that have been extensively studied. In contrast, colomitide C was non-toxic to human breast cancer cell lines that are associated with FGFR amplification. Known drugs that target various components of the FGFR signaling pathway exhibited different degrees of cytotoxicity to these cell lines. The difference between in vivo (fin regeneration) and in vitro (cancer cell lines) suggests that colomitide C acts on molecules or mechanisms upstream of FGF and WNT signaling during regeneration. Moreover, the activity of the FGF and WNT reporters in colomitide C-treated regenerating larvae resembles that of non-regenerating fins. This observation indicates that colomitide C affects FGF and WNT signaling related to fin regeneration, but the effect on the endogenous FGF and WNT signaling is minor. Colomitide C may have low affinity to its target molecules, and may modulate activities of putative targets only when their activities are upregulated during regeneration.
A previous study demonstrated that activating WNT signaling through inhibition of GSK3ß by BIO causes inhibition of embryonic fin fold regeneration24. Similarly, BIO inhibited larvae fin regeneration in our study, indicating that the effect of activating WNT signaling has negative impact on regeneration of both embryonic fin fold and larvae fin. In contrast, our analysis using the TCF-mCherry reporter system showed WNT signaling is activated during larvae fin regeneration. Moreover, colomitide C treatment caused downregulation of WNT signaling, which is associated with inhibition of larvae fin regeneration. These results also confirm that activating WNT signaling is associated with larvae fin regeneration. These observations suggest that proper levels of WNT signaling activation is needed for larvae fin regeneration, and modulating WNT signaling above or below certain levels might interfere with proper fin regeneration. Further study will identify the specific molecular targets and mechanisms of action of colomitide C and this work will contribute towards its development as a tool to better understand and manipulate fin regeneration.
For more than a century, secondary metabolites have been central to the drug discovery and development process68,69. For example, a comprehensive and meticulous series of studies of FDA approved drugs revealed that from January1946 – September 2019, 79% of the 259 novel small molecule antitumor drugs were either direct natural products, derivatives of natural products, or synthetic analogues of natural products70–74. These reports clearly indicate that the majority of functional drug categories are mostly comprised of small molecules natural products or inspired by natural product pharmacophores. Colomitide C is a structurally unique secondary metabolite with five chiral centers, and closely related compounds were recently synthesized75. While colomitide C did not exhibit cytotoxic activities to cancer cell lines, synthetic modification of the colomitide C structure and testing the activities of the derivatives may be a useful approach to develop a novel chemical tool that targets cancer cells. Furthermore, identifying target proteins of colomitide C and structural modifications to alter the interaction with components of signaling pathways may offer an opportunity to develop new tools to elucidate mechanisms of fin regeneration.
EXPERIMENTAL PROCEDURES
Zebrafish husbandry
Zebrafish were housed in the University of Minnesota zebrafish core facility with a light cycle of 14:10 at 28.5°C according to approval of the Institutional Animal Care and Use Committee of the University of Minnesota. Adult fish were bred in a 2:2 ratio in non-sloped breeding tanks. Upon embryo collection, embryos were separated into groups of approximately 100 into 10 cm plastic petri dishes with embryo water. Embryos were incubated at 28.5°C, and debris, chorions, disfigured, and dead embryos were removed daily until 4 dpf.
Larval caudal fin amputation assay
At 4 dpf, larvae were anesthetized with MS-222 (also known as Tricaine, pH 7) in embryo medium in a 6 cm petri dish. When they became unresponsive, one larva was moved via a transfer pipette on to a dry 6 cm dish lid. The water around the caudal fin was removed such that the fin adhered to the plastic surface. Making sure not to allow the exposed fin to dry substantially, the larva was aligned such that a vertical amputation could be made perpendicularly and immediately proximal to the posterior end of the spinal cord under stereomicroscope. The amputation was made with a No. 21 surgical blade (Feather). Immediately, a transfer pipette was used to rehydrate the amputated larva and transfer it to a well in a 24-well plate containing 1 mL of embryo water. After five such amputations were successfully performed, each one being moved into the same well, the embryo water in the well was changed to 1 mL of media with the chemical being tested. The 24-well plates, containing a chemical and amputated larvae, were incubated for 24 hours at 28.5°C. At 5 dpf, the treatment media within the wells was replaced with newly prepared chemical treatment media. The plates were incubated for another 24 hours. At 6 dpf, after 48 hours of regeneration with chemical treatment, the larvae were euthanized by cold hypoxia and fixed in 4% paraformaldehyde.
Measurement of regenerated fin area
After fixation, the heads of the larvae were removed with forceps to prevent buoyancy during photography. Larvae were placed onto a translucent agarose gel and photographed by a Zeiss Discovery V8 stereoscope. Pictures were measured for length using iSolutionLite software. Length was measured as the horizontal distance from the amputation plane to the distal-most regenerated tissue. Area was measured with FIJI software. It was measured as the area of regenerated tissue distal to the amputation plane. Length and area values were normalized to the average value of DMSO-treated control larvae in the same set of experiment.
Reporter fish lines and imaging
The mn7Et line (referred to as Et7-EGFP)36, the Tg(7xTCF-Xla.Sia:NLS-mCherry) line (referred to as TCF-mCherry)39 and the Tg(fli1a:EGFP)y1 (referred to as fli1a-EGFP)43 are used in this study. For fluorescent imaging larvae were anesthetized with MS-222, mounted on a glass plate using 1% low melting agarose, and images were taken by Zeiss LSM710 confocal microscope.
Whole-mount immunostaining of acetylated tubulin
Zebrafish larvae were fixed in 4% paraformaldehyde/phosphate buffered saline + 0.1% Tween 20 (PBS-T) at 4°C overnight. The larvae were washed, dehydrated and stored at −20°C. After rehydration, the samples were washed three times with PBS-T at room temperature. The samples were equilibrated in 150mM Tris-HCl (pH9.0) for 5min, heated at 70°C for 15min, and washed with PBT. Then, the samples were blocked with 2% donkey serum + 2% BSA/PBS + 0.1% Triton-X100 for 1 hour at room temperature, and reacted with anti-acetylated tubulin antibody (Sigma, T7451, dilution 1:1000). The larvae were washed with PBT, reacted with Alexa 647-labelled donkey anti-mouse IgG (Invitrogen, A-11091, dilution 1:500), washed with PBT, and stained with 1 μg/ml of 4’,6-diamidino-2-phenylindole, dihydrochloride (DAPI)/PBT. Confocal images were acquired similar to reporter fish lines described above.
Measurement of nerve and endothelial cell density
Confocal images were converted into 8-bit images and binary images were created by FIJI software. The area positive for acetylated tubulin or fli1a-GFP signals was measured as a pixel value, and the ratio of signal-positive area over the area pixel value of regenerating tail fin or the trunk (100 μm from the amputation plane) was determined.
Natural compound libraries
Microbial samples were collected from Minnesota caves, mines, and terrestrial soils and cultured on various media including ISP2, water, nutrient, marine, and tryptone soy broth agars (bacteria) and on Sabouraud dextrose, potato, and yeast malt agars (fungi). Several Cadophora fungi were collected from wooden structures in Antarctica35. Plant extracts were obtained from the National Cancer Institute of the NIH as crude extracts and were fractionated and purified as part of a different project76. Microbial cultures were archived as frozen glycerol stocks at −80 °C in the Center for Drug Design at the University of Minnesota.
For solid production cultures of fungi and bacteria (rice and agar, respectively), the cultures were exhaustively extracted with ethyl acetate and methanol (three times for each solvent). Extracts were combined, concentrated and successively partitioned with ethyl acetate, n-hexane and n-butanol. Each partition sample was dried and subjected to vacuum liquid chromatography with reversed phase C-18 or normal phase silica gel. These fractions were subsequently purified using Sephadex LH-20 size exclusion media, flash chromatography using an ISCO®combiflash instrument, and semipreparative reversed phase C-18 high performance liquid chromatography separations (Agilent 1260). Pure compounds were identified and characterized using nuclear magnetic resonance spectroscopy, mass spectrometry, optical rotation measurements, circular dichroism spectroscopy and semi-synthesis35,77,78.
The mycelia and supernatants from liquid cultures were separated by filtration and separately extracted three times using methanol and ethyl acetate, respectively. Supernatants were also subsequently extracted with n-butanol. Both the liquid culture extracts and plant extracts were further purified as described above for the solid media culture samples.
Cell lines
We used following cell lines. CAMA-1 (ATCC® HTB-21™) is a luminal-type human breast cancer cell line79,80. HCC-38 (ATCC® CRL-2314™) is a human mammary gland-derived breast cancer cell line81. MDA-MB-361 (ATCC® HTB-27™) is a human breast adenocarcinoma-derived cancer cell line82.
CAMA-1 cells were maintained in RPMI 1640 growth media (Invitrogen 11875–119) supplemented with 10% FBS (Invitrogen 16000–044), 1% Penicillin/Streptomycin (Invitrogen 15140–122), 5 μg/ml bovine insulin (Akron Biotech AK8213–0100) and 10 ng/ml human epidermal growth factor (Invitrogen PHG0311L). Cells were plated in 96-well plates at 25 × 104 cells/ml and allowed to grow overnight. After 24 hours, compounds were added at 9, 3x dilutions from 100 μM final concentration in growth media. MDA-MB-361 and HCC-38 cells were maintained in growth media: DMEM/F12 (Invitrogen 11320–033) supplemented with 10% FBS (Invitrogen 16000–044) and 1% Penicillin/Streptomycin (Invitrogen 15140–122). MDA-MB-361 cells were plated in 96-well plates at 50 × 104 cells/ml. HCC38 cells were plated at 25 × 104 cells/ml. After 24 hours, compounds were added at 9, 3x dilutions from 100 μM final concentration in growth media.
Cytotoxicity assay
The cytotoxicity of each compound was determined with a standard tetrazolium assay46. For all cell lines, plates were incubated for 24, 48, and 72 hours at 37°C in a 5% CO2/95% air humidified atmosphere after which time the media was removed and (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was added in RPMI phenol red-free media. The MTT was removed after 3 hours and formazan crystals were solubilized with 200 μL of isopropanol and plates were read on a M5e spectrophotometer (Molecular Devices) at 570 nm for formazan and 690 nm for background subtraction. EC50 (half maximal effective concentration) values were calculated by fitting the data in GraphPad Prism software.
Bullet points.
We performed screening of natural products on zebrafish larvae fin regeneration.
Colomitide C inhibited larvae fin regeneration.
Colomitide C downregulated FGF and WNT signaling, but did not affect endothelial cells and the nerve.
In vitro assays demonstrate that colomitide C has low or no cytotoxicity.
ACKNOWLEDGEMENTS
The authors declare no competing interest. We are grateful to Drs. James Amatruda, Richard Dorsky, Stephen Ekker, Enrico Moro and Angel Raya for sharing zebrafish lines, to Dr. Michael O’Connor for sharing his equipment, to Dr. Anindya Bagchi for helpful discussion, and to the Zebrafish Core Facility at the University of Minnesota for the help with zebrafish husbandry. We also thank Nicole Kilby, Tyler Burgoyne, Tyler Willing, Michael Brush, An Le, Sho Kawakami and Dr. Robyn Leary for their excellent assistance. This study was supported by the University of Minnesota Center for Drug Design Seed Grant to CES (CDD2013) and a grant from the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health to YK (R01AR064195). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors have no conflicts to disclose.
REFERENCES
- 1.Grunwald DJ, Eisen JS. Headwaters of the zebrafish -- emergence of a new model vertebrate. Nat Rev Genet. September 2002;3(9):717–24. 10.1038/nrg892. [DOI] [PubMed] [Google Scholar]
- 2.Kinth P, Mahesh G, Panwar Y. Mapping of zebrafish research: a global outlook. Zebrafish. December 2013;10(4):510–7. 10.1089/zeb.2012.0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gemberling M, Bailey TJ, Hyde DR, Poss KD. The zebrafish as a model for complex tissue regeneration. Trends Genet. November 2013;29(11):611–20. 10.1016/j.tig.2013.07.003S0168-9525(13)00113-3 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tal TL, Franzosa JA, Tanguay RL. Molecular signaling networks that choreograph epimorphic fin regeneration in zebrafish - a mini-review. Gerontology. 2010;56(2):231–40. https://doi.org000259327 [pii] 10.1159/000259327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfefferli C, Jazwinska A. The art of fin regeneration in zebrafish. Regeneration. 2015;2(2):72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sehring IM, Weidinger G. Recent advancements in understanding fin regeneration in zebrafish. Wiley Interdiscip Rev Dev Biol. January 2020;9(1):e367. 10.1002/wdev.367. [DOI] [PubMed] [Google Scholar]
- 7.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. August 2004;10(8):789–99. 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 8.Wehner D, Weidinger G. Signaling networks organizing regenerative growth of the zebrafish fin. Trends Genet. June 2015;31(6):336–43. 10.1016/j.tig.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Poss KD, Shen J, Nechiporuk A, et al. Roles for Fgf signaling during zebrafish fin regeneration. Dev Biol. June 15 2000;222(2):347–58. [DOI] [PubMed] [Google Scholar]
- 10.Lee Y, Grill S, Sanchez A, Murphy-Ryan M, Poss KD. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. December 2005;132(23):5173–83. [DOI] [PubMed] [Google Scholar]
- 11.Whitehead GG, Makino S, Lien CL, Keating MT. fgf20 is essential for initiating zebrafish fin regeneration. Science. December 23 2005;310(5756):1957–60. [DOI] [PubMed] [Google Scholar]
- 12.Shibata E, Yokota Y, Horita N, et al. Fgf signalling controls diverse aspects of fin regeneration. Development. August 15 2016;143(16):2920–9. 10.1242/dev.140699. [DOI] [PubMed] [Google Scholar]
- 13.Kawakami Y, Rodriguez Esteban C, Raya M, et al. Wnt/beta-catenin signaling regulates vertebrate limb regeneration. Genes Dev. December 1 2006;20(23):3232–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoick-Cooper CL, Weidinger G, Riehle KJ, et al. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. February 2007;134(3):479–89. [DOI] [PubMed] [Google Scholar]
- 15.Stoick-Cooper CL, Moon RT, Weidinger G. Advances in signaling in vertebrate regeneration as a prelude to regenerative medicine. Genes Dev. June 1 2007;21(11):1292–315. [DOI] [PubMed] [Google Scholar]
- 16.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. July 1995;203(3):253–310. 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 17.Dahm R Atlas of embryonic stages of development in the zebrafish. In: Christine Nusslein-Volhard RD, ed. Zebrafsih. A Practical Approach Oxford University Press; 2002:219–236. [Google Scholar]
- 18.Kawakami A, Fukazawa T, Takeda H. Early fin primordia of zebrafish larvae regenerate by a similar growth control mechanism with adult regeneration. Dev Dyn. December 2004;231(4):693–9. 10.1002/dvdy.20181. [DOI] [PubMed] [Google Scholar]
- 19.Yoshinari N, Kawakami A. Mature and juvenile tissue models of regeneration in small fish species. Biol Bull. August 2011;221(1):62–78. 10.1086/BBLv221n1p62. [DOI] [PubMed] [Google Scholar]
- 20.Mateus R, Pereira T, Sousa S, et al. In vivo cell and tissue dynamics underlying zebrafish fin fold regeneration. PLoS One. 2012;7(12):e51766. 10.1371/journal.pone.0051766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathew LK, Andreasen EA, Tanguay RL. Aryl hydrocarbon receptor activation inhibits regenerative growth. Molecular pharmacology. January 2006;69(1):257–65. 10.1124/mol.105.018044. [DOI] [PubMed] [Google Scholar]
- 22.Roehl HH. Linking wound response and inflammation to regeneration in the zebrafish larval fin. The International journal of developmental biology. 2018;62(6-7-8):473–477. 10.1387/ijdb.170331hr. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen-Chi M, Laplace-Builhé B, Travnickova J, et al. TNF signaling and macrophages govern fin regeneration in zebrafish larvae. Cell death & disease. August 10 2017;8(8):e2979. 10.1038/cddis.2017.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathew LK, Sengupta SS, Ladu J, Andreasen EA, Tanguay RL. Crosstalk between AHR and Wnt signaling through R-Spondin1 impairs tissue regeneration in zebrafish. FASEB J. August 2008;22(8):3087–96. 10.1096/fj.08-109009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moro E, Vettori A, Porazzi P, et al. Generation and application of signaling pathway reporter lines in zebrafish. Molecular genetics and genomics : MGG. June 2013;288(5–6):231–42. 10.1007/s00438-013-0750-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carney TJ, Mosimann C. Switch and Trace: Recombinase Genetics in Zebrafish. Trends Genet. May 2018;34(5):362–378. 10.1016/j.tig.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Marques IJ, Lupi E, Mercader N. Model systems for regeneration: zebrafish. Development. September 20 2019;146(18). 10.1242/dev.167692. [DOI] [PubMed] [Google Scholar]
- 28.Gunatilaka AA. Natural products from plant-associated microorganisms: distribution, structural diversity, bioactivity, and implications of their occurrence. J Nat Prod. March 2006;69(3):509–26. 10.1021/np058128n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rennekamp AJ, Peterson RT. 15 years of zebrafish chemical screening. Current opinion in chemical biology. February 2015;24:58–70. 10.1016/j.cbpa.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lessman CA. The developing zebrafish (Danio rerio): a vertebrate model for high-throughput screening of chemical libraries. Birth Defects Res C Embryo Today. September 2011;93(3):268–80. 10.1002/bdrc.20212. [DOI] [PubMed] [Google Scholar]
- 31.Kaufman CK, White RM, Zon L. Chemical genetic screening in the zebrafish embryo. Nat Protoc. 2009;4(10):1422–32. 10.1038/nprot.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oppedal D, Goldsmith MI. A chemical screen to identify novel inhibitors of fin regeneration in zebrafish. Zebrafish. March 2010;7(1):53–60. 10.1089/zeb.2009.0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathew LK, Sengupta S, Kawakami A, et al. Unraveling tissue regeneration pathways using chemical genetics. J Biol Chem. November 30 2007;282(48):35202–10. 10.1074/jbc.M706640200. [DOI] [PubMed] [Google Scholar]
- 34.Han Y, Chen A, Umansky KB, et al. Vitamin D Stimulates Cardiomyocyte Proliferation and Controls Organ Size and Regeneration in Zebrafish. Developmental cell. March 25 2019;48(6):853–863.e5. 10.1016/j.devcel.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rusman Y, Held BW, Blanchette RA, He Y, Salomon CE. Cadopherone and colomitide polyketides from Cadophora wood-rot fungi associated with historic expedition huts in Antarctica. Phytochemistry. April 2018;148:1–10. 10.1016/j.phytochem.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 36.Balciunas D, Davidson AE, Sivasubbu S, Hermanson SB, Welle Z, Ekker SC. Enhancer trapping in zebrafish using the Sleeping Beauty transposon. BMC Genomics. September 3 2004;5(1):62. 10.1186/1471-2164-5-621471-2164-5-62 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawakami Y, Rodriguez-Leon J, Koth CM, et al. MKP3 mediates the cellular response to FGF8 signalling in the vertebrate limb. Nat Cell Biol. June 2003;5(6):513–9. [DOI] [PubMed] [Google Scholar]
- 38.Tsang M, Maegawa S, Kiang A, Habas R, Weinberg E, Dawid IB. A role for MKP3 in axial patterning of the zebrafish embryo. Development. June 2004;131(12):2769–79. [DOI] [PubMed] [Google Scholar]
- 39.Moro E, Ozhan-Kizil G, Mongera A, et al. In vivo Wnt signaling tracing through a transgenic biosensor fish reveals novel activity domains. Dev Biol. June 15 2012;366(2):327–40. 10.1016/j.ydbio.2012.03.023. [DOI] [PubMed] [Google Scholar]
- 40.Simoes MG, Bensimon-Brito A, Fonseca M, et al. Denervation impairs regeneration of amputated zebrafish fins. BMC Dev Biol. 2014;14:49. 10.1186/s12861-014-0049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rieger S, Sagasti A. Hydrogen peroxide promotes injury-induced peripheral sensory axon regeneration in the zebrafish skin. PLoS biology. May 2011;9(5):e1000621. 10.1371/journal.pbio.1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang CC, Lawson ND, Weinstein BM, Johnson SL. reg6 is required for branching morphogenesis during blood vessel regeneration in zebrafish caudal fins. Dev Biol. December 1 2003;264(1):263–74. 10.1016/j.ydbio.2003.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. August 15 2002;248(2):307–18. https://doi.orgS0012160602907116 [pii]. [DOI] [PubMed] [Google Scholar]
- 44.Turner N, Pearson A, Sharpe R, et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. March 1 2010;70(5):2085–94. 10.1158/0008-5472.Can-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piasecka D, Braun M, Kitowska K, et al. FGFs/FGFRs-dependent signalling in regulation of steroid hormone receptors - implications for therapy of luminal breast cancer. Journal of experimental & clinical cancer research : CR. May 29 2019;38(1):230. 10.1186/s13046-019-1236-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mosmann T Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. December 16 1983;65(1–2):55–63. 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 47.Shiang CY, Qi Y, Wang B, et al. Amplification of fibroblast growth factor receptor-1 in breast cancer and the effects of brivanib alaninate. Breast cancer research and treatment. October 2010;123(3):747–55. 10.1007/s10549-009-0677-6. [DOI] [PubMed] [Google Scholar]
- 48.Gavine PR, Mooney L, Kilgour E, et al. AZD4547: an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. April 15 2012;72(8):2045–56. 10.1158/0008-5472.Can-11-3034. [DOI] [PubMed] [Google Scholar]
- 49.Hofman J, Sorf A, Vagiannis D, et al. Brivanib Exhibits Potential for Pharmacokinetic Drug-Drug Interactions and the Modulation of Multidrug Resistance through the Inhibition of Human ABCG2 Drug Efflux Transporter and CYP450 Biotransformation Enzymes. Molecular pharmaceutics. November 4 2019;16(11):4436–4450. 10.1021/acs.molpharmaceut.9b00361. [DOI] [PubMed] [Google Scholar]
- 50.Huynh H, Ngo VC, Fargnoli J, et al. Brivanib alaninate, a dual inhibitor of vascular endothelial growth factor receptor and fibroblast growth factor receptor tyrosine kinases, induces growth inhibition in mouse models of human hepatocellular carcinoma. Clin Cancer Res. October 1 2008;14(19):6146–53. 10.1158/1078-0432.Ccr-08-0509. [DOI] [PubMed] [Google Scholar]
- 51.Heerding DA, Rhodes N, Leber JD, et al. Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}−1H- imidazo[4,5-c]pyridin-4-yl)-2-methyl-3-butyn-2-ol (GSK690693), a novel inhibitor of AKT kinase. J Med Chem. September 25 2008;51(18):5663–79. 10.1021/jm8004527. [DOI] [PubMed] [Google Scholar]
- 52.Kumar R, Blakemore SJ, Ellis CE, et al. Causal reasoning identifies mechanisms of sensitivity for a novel AKT kinase inhibitor, GSK690693. BMC Genomics. July 6 2010;11:419. 10.1186/1471-2164-11-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao YY, Tian Y, Zhang J, et al. Effects of an oral allosteric AKT inhibitor (MK-2206) on human nasopharyngeal cancer in vitro and in vivo. Drug design, development and therapy. 2014;8:1827–37. 10.2147/dddt.S67961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arcaro A, Wymann MP. Wortmannin is a potent phosphatidylinositol 3-kinase inhibitor: the role of phosphatidylinositol 3,4,5-trisphosphate in neutrophil responses. The Biochemical journal. December 1 1993;296 (Pt 2):297–301. 10.1042/bj2960297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maitra U, Ciesla L. Using Drosophila as a platform for drug discovery from natural products in Parkinson’s disease. MedChemComm. June 1 2019;10(6):867–879. 10.1039/c9md00099b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu H, Guo M, Xue T, Guan J, Luo L, Zhuang Z. Screening lifespan-extending drugs in Caenorhabditis elegans via label propagation on drug-protein networks. BMC systems biology. December 23 2016;10(Suppl 4):131. 10.1186/s12918-016-0362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Richter L, Kropp S, Proksch P, Scheu S. A mouse model-based screening platform for the identification of immune activating compounds such as natural products for novel cancer immunotherapies. Bioorg Med Chem. December 1 2019;27(23):115145. 10.1016/j.bmc.2019.115145. [DOI] [PubMed] [Google Scholar]
- 58.Wiley DS, Redfield SE, Zon LI. Chemical screening in zebrafish for novel biological and therapeutic discovery. Methods Cell Biol. 2017;138:651–679. 10.1016/bs.mcb.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goessling W, North TE. Repairing quite swimmingly: advances in regenerative medicine using zebrafish. Dis Model Mech. July 2014;7(7):769–76. 10.1242/dmm.016352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Molina G, Watkins S, Tsang M. Generation of FGF reporter transgenic zebrafish and their utility in chemical screens. BMC Developmental Biology. 2007;7:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsang M Zebrafish: A tool for chemical screens. Birth Defects Res C Embryo Today. September 2010;90(3):185–92. 10.1002/bdrc.20183. [DOI] [PubMed] [Google Scholar]
- 62.Knopf F, Hammond C, Chekuru A, et al. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Developmental cell. May 17 2011;20(5):713–24. 10.1016/j.devcel.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 63.Singh SP, Holdway JE, Poss KD. Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Developmental cell. April 17 2012;22(4):879–86. 10.1016/j.devcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ellett F, Pase L, Hayman JW, Andrianopoulos A, Lieschke GJ. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood. January 27 2011;117(4):e49–56. 10.1182/blood-2010-10-314120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petrie TA, Strand NS, Yang CT, Rabinowitz JS, Moon RT. Macrophages modulate adult zebrafish tail fin regeneration. Development. July 2014;141(13):2581–91. 10.1242/dev.098459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mathias JR, Perrin BJ, Liu TX, Kanki J, Look AT, Huttenlocher A. Resolution of inflammation by retrograde chemotaxis of neutrophils in transgenic zebrafish. Journal of leukocyte biology. December 2006;80(6):1281–8. 10.1189/jlb.0506346. [DOI] [PubMed] [Google Scholar]
- 67.Mathias JR, Dodd ME, Walters KB, Yoo SK, Ranheim EA, Huttenlocher A. Characterization of zebrafish larval inflammatory macrophages. Developmental and comparative immunology. November 2009;33(11):1212–7. 10.1016/j.dci.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harvey AL. Natural products in drug discovery. Drug discovery today. October 2008;13(19–20):894–901. 10.1016/j.drudis.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 69.Dias DA, Urban S, Roessner U. A historical overview of natural products in drug discovery. Metabolites. April 16 2012;2(2):303–36. 10.3390/metabo2020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cragg GM, Newman DJ, Snader KM. Natural products in drug discovery and development. J Nat Prod. January 1997;60(1):52–60. 10.1021/np9604893. [DOI] [PubMed] [Google Scholar]
- 71.Newman DJ, Cragg GM, Snader KM. Natural products as sources of new drugs over the period 1981–2002. J Nat Prod. July 2003;66(7):1022–37. 10.1021/np030096l. [DOI] [PubMed] [Google Scholar]
- 72.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. March 23 2012;75(3):311–35. 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Newman DJ, Cragg GM. Natural Products as Sources of New Drugs from 1981 to 2014. J Nat Prod. March 25 2016;79(3):629–61. 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 74.Newman DJ, Cragg GM. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J Nat Prod. March 27 2020;83(3):770–803. 10.1021/acs.jnatprod.9b01285. [DOI] [PubMed] [Google Scholar]
- 75.Yang H, Liu X, Li X, et al. Enantioselective total synthesis of colomitides and their absolute configuration determination and structural revision. Org Biomol Chem. May 3 2017;15(17):3728–3735. 10.1039/c7ob00539c. [DOI] [PubMed] [Google Scholar]
- 76.Hassig CA, Zeng FY, Kung P, et al. Ultra-High-Throughput Screening of Natural Product Extracts to Identify Proapoptotic Inhibitors of Bcl-2 Family Proteins. J Biomol Screen. September 2014;19(8):1201–11. 10.1177/1087057114536227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rusman Y, Held BW, Blanchette RA, Wittlin S, Salomon CE. Soudanones A-G: Antifungal Isochromanones from the Ascomycetous Fungus Cadophora sp. Isolated from an Iron Mine. J Nat Prod. June 26 2015;78(6):1456–60. 10.1021/acs.jnatprod.5b00204. [DOI] [PubMed] [Google Scholar]
- 78.Rusman Y, Oppegard LM, Hiasa H, Gelbmann C, Salomon CE. Solphenazines A-F, glycosylated phenazines from Streptomyces sp. strain DL-93. J Nat Prod. January 25 2013;76(1):91–6. 10.1021/np3007606. [DOI] [PubMed] [Google Scholar]
- 79.Ji H, Stout LE, Zhang Q, Zhang R, Leung HT, Leung BS. Absence of transforming growth factor-beta responsiveness in the tamoxifen growth-inhibited human breast cancer cell line CAMA-1. Journal of cellular biochemistry. March 1994;54(3):332–42. 10.1002/jcb.240540309. [DOI] [PubMed] [Google Scholar]
- 80.Fogh J, Wright WC, Loveless JD. Absence of HeLa cell contamination in 169 cell lines derived from human tumors. Journal of the National Cancer Institute. February 1977;58(2):209–14. 10.1093/jnci/58.2.209. [DOI] [PubMed] [Google Scholar]
- 81.Gazdar AF, Kurvari V, Virmani A, et al. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. Int J Cancer. December 9 1998;78(6):766–74. . [DOI] [PubMed] [Google Scholar]
- 82.Brinkley BR, Beall PT, Wible LJ, Mace ML, Turner DS, Cailleau RM. Variations in cell form and cytoskeleton in human breast carcinoma cells in vitro. Cancer Res. September 1980;40(9):3118–29. [PubMed] [Google Scholar]


