Abstract
Background:
Differences in core or tissue-specific ribosomal protein (Rp) composition within ribosomes contribute to ribosome heterogeneity and functional variability. Yet, the degree to which ribosome heterogeneity modulates development is unknown. The Drosophila melanogaster eRpL22 family contains structurally diverse paralogues, eRpL22 and eRpL22-like. Unlike ubiquitously expressed eRpL22, eRpL22-like expression is tissue-specific, notably within the male germline and the eye. We investigated expression within the developing eye to uncover tissue/cell types where specific paralogue roles might be defined.
Results:
Immunohistochemistry analysis confirms ubiquitous eRpL22 expression throughout eye development. In larvae, eRpL22-like is ubiquitously expressed, but highly enriched in the peripodial epithelium (PE). In early pupae, eRpL22-like is broadly distributed in multiple cell types, but later, is primarily enriched in interommatidial hair cells (IoHC). Adult patterns include the ring of accessory cells around ommatidia. Adult retinae IoHC patterning phenotypes (shown by scanning electron microscopy) may be linked to RNAi-mediated eRpL22-like depletion within larval PE. Immunoblots and polysome profile analyses show multiple variants of eRpL22-like across development, with the variant at the expected molecular mass co-sedimenting with active ribosomes.
Conclusion:
Our data reveal differential patterns of eRpL22-like expression relative to eRpL22 and suggest a specific role for eRpL22-like in developmental patterning of the eye.
Keywords: development, gene expression, interommatidial hair cell, peripodial epithelium, ribosomal proteins
1 ∣. INTRODUCTION
Despite global functioning of the ribosome in all cells, several ribosomal protein (Rp) components of this complex molecular machine are characterized by tissue-specific expression. Defects in ribosomal components would therefore be expected to result in catastrophic effects; however, ribosomal component defects or deficiencies commonly result in a spectrum of tissue-specific or developmental phenotypes.1,2 Among the most common human phenotypes resulting from a ribosomal component defect is the ribosomopathy Diamond Blackfan Anemia (DBA), which can manifest from mutations in one of a host of small or large subunit Rps.1,3,4 Tissue-specific expression of an Rp likely indicates potential for a specific function within those tissues. Thus, exploration of tissue-specific patterns of Rp expression has the potential to provide insights into unique functions of specialized ribosomal components.
Several examples of cell- or tissue-specific Rp functions have highlighted the significance of Rp components in developmental or differentiation processes. In the social slime mold Dictyostelium discoideum, distinct profiles of Rps exist in vegetative cells compared to aggregation-competent cells5,6, supporting a role for specific Rps at specific developmental stages. Mitochondrial Rp, mRpL34, in Drosophila melanogaster is expressed in differentiating photoreceptors, and when depleted, causes a flattened lens and ommatidial alterations.7 Differentiation and cell polarity defects in photoreceptors caused by dystroglycan mutations are exacerbated by mRpL34 defects, demonstrating genetic interactions between mRpL34 and dystroglycan in a neuronal differentiation pathway. It is noteworthy that some Rps function independently from a ribosomal role in processes such as DNA replication and repair, transcription, and splicing.8,9.
Recent advances have revealed that variability in Rp composition within the ribosome can alter translational specificity and define populations of “specialized ribosomes” that may preferentially translate specific mRNAs10,11; reviewed by References 12, 13. Specific mRNA enrichment on a subset of ribosome types was recently demonstrated in mouse embryonic stem cells, showing that mRNAs related to differentiation are enriched on RpS25 ribosomes and mRNAs related to metabolism are enriched on RpL10a ribosomes.11 Segregation of mRNA translational specificity based on functional pathways has clear implications for how developmental pathways may be regulated.
In D melanogaster, the eRpL22 family includes two paralogues, eRpL22 and eRpL22-like - both essential in development.14-16 eRpL22 is ubiquitously expressed17-20; however, eRpL22-like shows tissue-specific expression in adult testes and the adult eye.21 Tissue specificity in this Rp family may underlie specific roles for this paralogue in developing germ cells and in developing eye tissue. In fact, we have recently demonstrated unique patterns of specific translation on eRpL22 ribosomes and eRpL22-like ribosomes in the germline.22 A complete developmental eRpL22-like expression profile within the developing eye is necessary to understand functional contributions of this Rp to developmental and differentiation processes required to build an eye.
Eye development in D melanogaster is a highly characterized and tightly regulated tissue system used as a model to probe changes in various developmental processes.23 Assembly of 750 to 800 ommatidia in the fly eye requires a complex suite of cellular differentiation processes to specify an array of different cell types along with the proper number, orientation, and position of each cell type within the eye disc.24
Here we have used immunohistochemistry (IHC) and confocal microscopy to examine patterns of expression for eRpL22 and eRpL22-like in several stages of eye development: third instar larva, 30-43 hours post pupation (hpp), and adult. While the pattern of expression for eRpL22 is widespread throughout the eye in all developmental stages, eRpL22-like is ubiquitously expressed only at certain times in development, with significant enrichment of eRpL22-like in the peripodial epithelium (PE) of the larval eye disc and in interommatidial hair cells (IoHC) in midpupal eye tissue. A role for the PE in IoHC patterning and/or orientation has previously been proposed.25 RNAi-mediated depletion of eRpL22-like in the PE of larval eye discs may influence downstream IoHC patterning defects in adult retinae, shown by scanning electron microscopy. Immunoblot and polysome profiling analyses reveal multiple eRpL22-like molecular mass variants, with only the predicted molecular mass species co-sedimenting with ribosomal constituents and actively translating ribosomes. Collectively, these data are consistent with a role for eRpL22-like as a specialized Rp in IoHC patterning.
2 ∣. RESULTS
2.1 ∣. eRpL22-like has unique spatial and temporal patterns of expression in eye development
To determine expression patterns for eRpL22 and eRpL22-like, we performed IHC analysis of eye tissues from key stages of eye development (third instar larval, pupal, and adult eye tissue; see diagram in Figure 1A). This approach was pursued for two reasons: to define cell and tissue types where eRpL22 paralogues are either co-expressed or not, and to use localization information as a basis for developing hypotheses about putative functions for eRpL22-like.
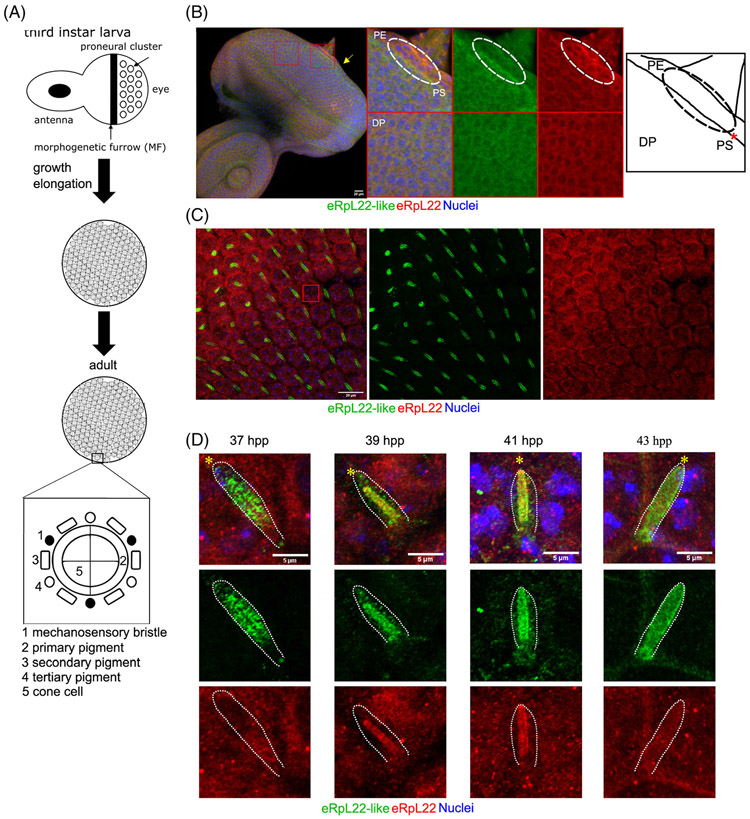
FIGURE 1.
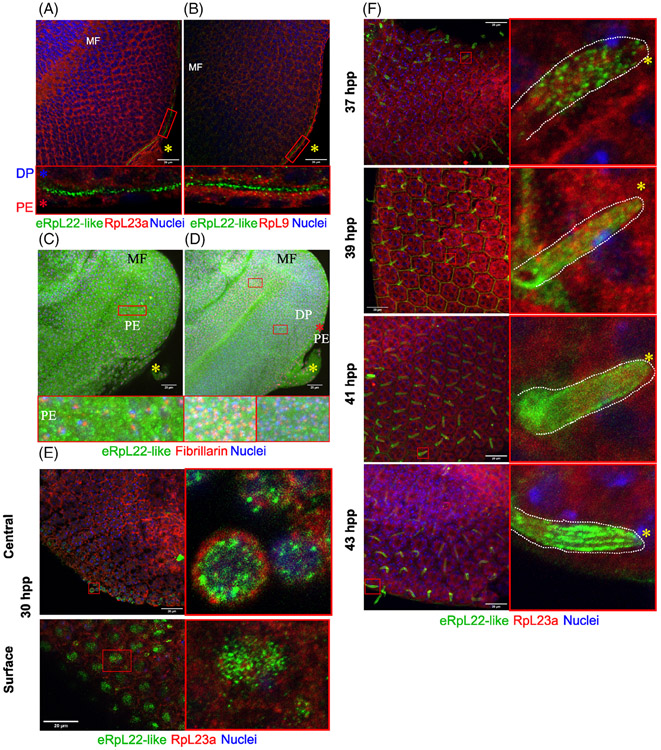
Distribution of eRpL22 family paralogues in larval and pupal eye tissue. A, Diagram of key stages of eye development: third instar larval eye/antennal imaginal disc, pupal retina, adult eye. B, Representative immunohistochemistry (IHC) analysis of a third instar eye/antennal imaginal disc (left) with magnified views of different tissue regions. Yellow arrow shows region of eRpL22-like enrichment on the disc proper (DP) periphery. Top—Central/anterior area of the eye disc. The dotted circle indicates the area of the PE containing either eRpL22-like (green) or eRpL22 (red) enrichment. Bottom—Central area of the DP. Far right—A trace of top middle panel showing tissue structures labeled as PE, PS (marked by a red *), or DP. All tissue was stained with nuclei marker DAPI (blue), eRpL22 (red) and eRpL22-like (green). Scale bar: 20 μm. C, Representative IHC image of 37 hpp pupal retina. Scale bar: 20 μm. D, Magnified view of IoHC from pupal time points (37 hpp, 39 hpp, 41 hpp, 43 hpp). Brightness of both eRpL22 and eRpL22-like stains has been adjusted to sub-threshold levels. A dotted line outlines each IoHC. The distal end of the cell containing eRpL22-like is marked by yellow *. Scale bar: 5 μm
In larval eye/antennal imaginal discs, eRpL22-like is enriched in regions on the perimeter of the disc proper (DP; Figure 1B —left [see arrow]) and is found in low levels in the cytoplasm in all cells across the disc (Figure 1B —left and bottom middle). eRpL22-like enrichment in the space (called the peripodial space [PS]) between the DP and the PE (a structure surrounding one side of the eye disc) is also evident (Figure 1B —top middle). The cognate paralogue, eRpL22, is also detected ubiquitously at low levels; however, it is somewhat enriched in the PE, (Figure 1B —top right). A trace of the eye/antennal imaginal disc PE/PS/DP interface (Figure 1B —far right) shows the specific location of each structure.
In contrast to larval stages of eye development, midpupal eRpL22-like expression is highly enriched primarily in one elongated cell type within each ommatidium (Figure 1C). We have identified this elongated cell as the IoHC within the mechanosensory bristle complex (MBC), as will be described later in further detail (Figure 4). As in larval stages, eRpL22 expression is widespread throughout the retina; however, IHC analysis of midpupal eye tissue from four time points in development (37 hpp, 39 hpp, 41 hpp, and 43 hpp) reveals a degree of spatial asymmetry in paralogue expression (Figure 1D). At 37 hpp, eRpL22 stain is primarily distributed toward the proximal end of the IoHC, whereas the central axis of the IoHC is surrounded by a punctate-like pattern of eRpL22-like stain. At 39 hpp and 41 hpp, both paralogues are dispersed along the central axis. By 43 hpp in midpupal development, paralogue distribution changes, with both paralogues being more equally dispersed throughout the IoHC.
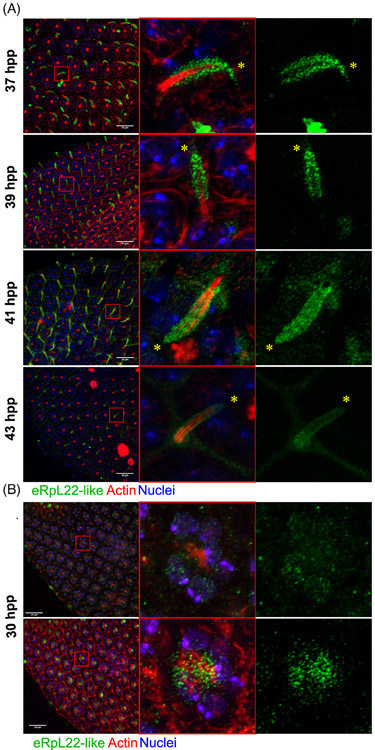
FIGURE 4.
eRpL22-like is expressed within pupal IoHC. A, Left—representative maximum intensity projections (rMIP) of eRpL22-like (green) and actin (red) in a time course between 37 and 43 hpp in 2-hour intervals. Middle —magnification of merged image of IoHC. Right—magnification of image of IoHC—eRpL22-like channel only. Distal end of IoHC is denoted by yellow *. Scale bar: 20 μm. B, Left—rMIP of eRpL22-like (green) and actin (red) at deep (top) and surface (bottom) focal planes. Middle—magnification of merged image of an ommatidium. Right—magnification of image of an ommatidium, eRpL22-like channel only. Scale bar: 20 μm
In adult stages, eRpL22-like expression expands into an array of different cell types (though not all cell types). Both paralogues are co-expressed within the ring of accessory cells (composed of secondary and tertiary pigment cells along with mechanosensory bristles) surrounding each ommatidium (see diagram in Figure 2A). Although paralogues are co-expressed, there is an asymmetrical subcellular distribution within each ommatidium. While eRpL22 is localized in both the cytoplasm and neuronal nuclei, eRpL22-like is detected in the cytoplasm of accessory cells but does not accumulate to detectable levels in neuronal nuclei (Figure 2A). Evaluation of z stack images from multiple eye tissue samples has not uncovered any cell type where eRpL22-like is the only paralogue expressed in that tissue; instead, eRpL22-like is co-expressed along with eRpL22. This observation has implications for proposals related to differential roles for paralogues within these cells.
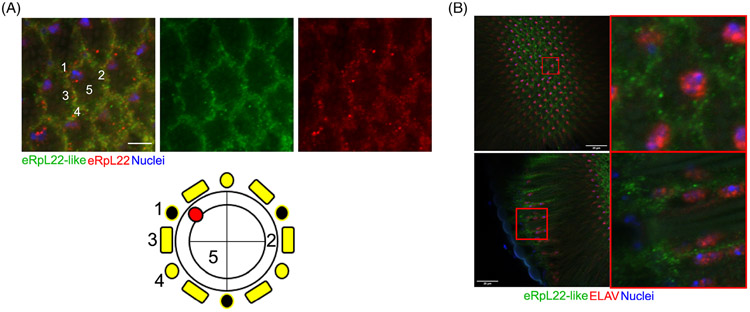
FIGURE 2.
Distribution of eRpL22 family paralogues in adult eye tissue. A, Representative IHC images of adult eyes with corresponding diagram showing paralogue distributions. Tissue was stained with DAPI as a marker for nuclei (blue); eRpL22 (red) and eRpL22-like (green). Paralogue co-expression is indicated by yellow. Scale bar: 5 μm. B, Representative IHC images of ELAV (red), eRpL22-like (green) and nuclei (blue) in adult eyes in a surface focal plane (top) and a longitudinal focal plane (bottom). Scale bar: 20 μm
Each ommatidium is a complex three-dimensional structure; as such, we viewed both apical (Figure 2B — top panels) and longitudinal (Figure 2B —bottom panels) focal planes to completely assess ommatidial distribution of eRpL22-like. eRpL22-like is uniformly distributed throughout an entire ommatidium (Figure 2B —right panels). Neural cell types such as photoreceptors are within the center of each ommatidium, while non-neural cell types, such as pigment cells and mechanosensory bristles, surround each ommatidium (Figures 1A and 2B). We used the pan-neural protein, ELAV26 to mark the position of neural cell nuclei. We focused on two different focal planes (surface and longitudinal) to show photoreceptor nuclear positioning within the complex three-dimensional structure of the adult eye. Based on photoreceptor nuclear staining (positioned internally within an ommatidium) within longitudinal views, eRpL22-like-positive cells were positioned peripherally relative to ELAV-positive neural cell nuclei (Figure 2B). With this pattern of staining, we conclude that eRpL22-like is expressed in non-neural cell types within adult eye tissue. The identity of the cell type with ELAV-positive nuclear staining (in a focal plane at the surface, depicted in Figure 2A) has not been confirmed; however, we propose that it is likely the MBC neuron based on ELAV staining patterns observed by Lai and Orgogozo.27.
2.2 ∣. eRpL22-like is enriched in the peripodial epithelium and interommatidial hair cells
Identification of cells within the eye that express eRpL22-like provides insight into putative functions of this paralogue in a developmental context. We used a combination of cell-specific green fluorescent protein (GFP) expression and cell type-specific markers to determine the identity of cells that express eRpL22-like. Developmental patterning and differentiation of the third instar eye/antennal imaginal disc is influenced by external tissue interactions with the bordering layer of squamous epithelial cells, identified as the PE (reviewed by28).
We used the PE-specific driver C311-Gal429,30 to drive expression of a GFP reporter in the PE and then used IHC to position eRpL22-like relative to GFP expression in the PE of third instar eye/antennal imaginal discs (Figure 3A). GFP and eRpL22-like are widely distributed; however, in an orthogonal view of the tissue (top and right panels, Figure 3A), it is clear that eRpL22-like distribution is enriched between the PE and the DP (see diagram in Figure 3B). Within the PE, eRpL22-like distribution is asymmetric and polarized at the apical end toward the DP (Figure 3C) and is present within the PS. Gibson and Schubiger25 have reported the PS contains microtubule-based “transluminal” extensions (also called cytonemes31) that project from peripodial cells of wing and eye discs across an acellular space to cells of the DP. It is noteworthy that regions within the PE and PS that are enriched for eRpL22-like are depleted of eRpL22 (Figure 3D) in addition to Rps RpL23a and RpL9 (Figure 3E,E'). Computational analysis (using custom image masks) was used to quantify the relative enrichment of Rps within nuclear and cytoplasmic sub-compartments, as well as within the PE and PS (Figure 3F — top). This analysis corroborates the conclusions that regions of eRpL22-like enrichment within the PE and PS are also devoid of the cognate paralogue eRpL22 and core Rps L23a and L9 (Figure 3F —bottom).
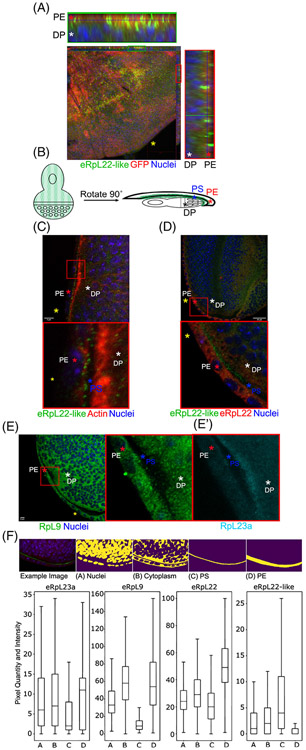
FIGURE 3.
Distribution of eRpL22-like in third instar eye/antennal imaginal discs. A, Representative maximum intensity projection of C311-Gal4 mediated GFP expression in peripodial epithelium (PE); whole tissue (center) and magnified orthogonal views (top and right). GFP (red), eRpL22-like (green), nuclei (blue); Scale bar: 50 μm. B, Diagram of eRpL22-like (green) distribution in third instar eye/antennal imaginal discs. A black * marks the DP, a blue * marks the PS, and a red * marks the PE. C, Representative IHC of wild type third instar eye imaginal discs (top) stained for eRpL22-like (green) and actin (red). Magnified view of top panel (bottom). Yellow * marks disc posterior (A,C,D). White * marks the DP, red * marks the PE, and blue * marks the PS (A, C-E, E'). Scale bar: 5 μm. D, Representative IHC of wild type third instar eye imaginal discs (top) stained for eRpL22-like (green) and eRpL22 (red). Magnified view of top panel (bottom). Both eRpL22 and eRpL22-like stains were adjusted to sub-threshold levels. E, Representative IHC image of wild type third instar eye imaginal discs (left) stained for RpL9 (green). Magnified view of left panel (middle). E', Imaginal disc tissues were stained for RpL23a (cyan/magnified view of left panel). F, Tissue structure-specific masks assigned to tissue/cell structural features (top). A—nuclei, B—cytoplasm, C—peripodial space, D—peripodial membrane. Computational analysis of RpL23a (n = 6), RpL9 (n = 3), eRpL22 (n = 2), and eRpL22-like (n = 6) pixel quantity, intensity and distribution (bottom). n = number of discs. Note that a different Y-axis for each graph is shown to optimize Rp localization for each Rp
In midpupal stages, eRpL22-like expression appears to be confined to a cell(s) present at alternating vertices of each ommatidium (Figure 4A). Based on the cellular composition of an ommatidium, we propose that the identities of cells expressing eRpL22-like are either tertiary pigment cells or cells of the MBC. Four different cell types (tormogen [socket], trichogen [shaft; here referred to as the IoHC27], thecogen [sheath], and sensory neuron) comprise the MBC, but one cell (IoHC) can be distinguished due to its accumulation of complex F-actin bundles.27,32 The actin network provides support as hair cell length increases.32 To distinguish the IoHC from other cell types, we used phalloidin to visualize F-actin. IHC analysis of eye tissue at progressive time points in midpupal development revealed that eRpL22-like expression occurs within cells containing complex F-actin bundles, thereby identifying the growing IoHC as the relevant cell type.
Previous studies have shown that IoHC growth and elongation occurs between 33 and 50 hpp.33 We therefore refined our IHC analysis of eRpL22-like expression within the IoHC to a timeframe consistent with IoHC maturation. As midpupal eye development proceeds, distribution patterns of eRpL22-like change relative to actin organization within the IoHC. At 37 hpp and 39 hpp, eRpL22-like is found in a punctate pattern in proximity to aggregated actin bundles, but by 41 hpp and 43 hpp, eRpL22-like distribution is in a more diffuse pattern (Figure 4A).
From a comprehensive analysis of z stacks of eye tissue from 37 hpp to 39 hpp, eRpL22-like expression is confined to IoHC, and is devoid from other regions of the ommatidium (Figures 1C and 4A). At 41 hpp and 43 hpp, eRpL22-like is present in trace levels throughout the cytoplasm of cells within each ommatidium (Figure 4A).
Given the changes observed in eRpL22-like expression within maturing IoHC, we decided to investigate eRpL22-like expression within IoHC at an earlier point (30 hpp) in pupal development when IoHC are in an immature state.33 When compared to later stages of midpupal eye development, enrichment of eRpL22-like at this earlier stage is dramatically different. At 30 hpp, eRpL22-like is detected in all cell nuclei, as shown in deep focal planes (Figure 4B —left). Additionally, eRpL22-like is highly enriched in the cytoplasm of cells on or near the surface of ommatidia (visualized in focal planes at the surface; Figure 4B —right). Based on their position within the ommatidium (see diagram in Figure 1A), we speculate that these cells may be cone cells and primary pigment cells that collectively secrete the corneal lens reviewed by Reference 34.
2.3 ∣. Ribosomal roles are implicated for eRpL22-like in eye development
Co-expression of eRpL22 along with eRpL22-like within eye tissue cell types is consistent with the hypothesis that eRpL22 and eRpL22-like have functionally distinct roles within these cells. These roles may include distinctive ribosome-related functions, including their assembly into specialized ribosomes (eg,1,11,12,22) or roles that are independent of ribosomal processes (eg, reviewed by9,35). We used IHC and confocal microscopy to assess eRpL22-like localization relative to other large subunit Rps, RpL9 and RpL23a or to the nucleolar marker fibrillarin.36 While it is unclear if RpL9 is a universal component of all cellular ribosomes, its presence as a large subunit and 80S monosome component in mouse embryonic stem cells has been confirmed.11 RpL23a (referred to as uL23 according to Reference 37) is a conserved and essential rRNA binding Rp, required for early assembly events in ribosome biogenesis across all kingdoms of life (eg, 38,39). Thus, L23a is a universal component of all ribosomes. We reasoned that overlap in eRpL22-like localization with other Rps would indicate the presence of ribosomal complexes, although the complete composition of those complexes would not be known. eRpL22-like co-localization with other Rps and fibrillarin was therefore used as a proxy for ribosomal function.
In larval development, some eRpL22-like-enriched regions of the PE are depleted of eRpL22, RpL9, and RpL23a (Figures 3D, E/Figure 5A —right, Figure 3E'/Figure 5A —left, respectively). More striking is the absence of these core Rps from the PS (Figure 3D,E [blue *] and Figure 5A,B) where apical extensions from the PE terminate on cells of the DP.25 Further, there is significant overlap in the distribution patterns for RpL23a and the cognate paralogue eRpL22 in the PE (Figure 3D), but both Rps are absent from the PS where eRpL22-like resides. Taken together, the absence of this subset of Rps (RpL23a, RpL9, and eRpL22) in the PS is suggestive of exclusion of eRpL22-like in the PS from canonical ribosomes. A role for eRpL22-like in ribosome assembly in the PE is supported by co-localization of a fraction of eRpL22-like with fibrillarin (Figure 5C). Although nonribosomal roles for the nucleolus have been confirmed,40 eRpL22-like function as a ribosomal component is also strongly supported by the presence of eRpL22-like within small nucleolar puncta within PE cells (Figure 5C). Both paralogues in the PE are likely ribosomal constituents, creating a heterogeneous pool of paralogue-specific ribosomes within the PE. The origin and trafficking pattern of eRpL22-like into presumptive apical extensions within the PS, however, is unknown. eRpL22-like may indeed have multifunctional roles within the PE and PS.
FIGURE 5.
Ribosomal roles are implicated for eRpL22-like in eye development. A, Representative images of co-localization of eRpL22-like (green) with core ribosomal components RpL23a (red) and B.) RpL9 (red) in larval eye/antennal discs. DAPI staining marks nuclei (blue) in A-F. Red boxes below show magnified areas in the PE/PS/DP interface. Images are from single focal planes. Scale bar: 20 μm (A-F). C,D, Representative images showing co-localization of eRpL22-like (green) and fibrillarin (red). C—Top, Representative maximum intensity projection (rMIP) at surface focal plane including PE. C—Bottom, Magnification of PE tissue. D—Top, rMIP at central focal plane of a disc. Red * marks the PE. D—Bottom, Magnification of tissue anterior (left) and posterior (right) to the morphogenetic furrow (MF). Yellow * marks disc posterior (shown in A-D). E, rMIP of eRpL22-like (green) co-localized with RpL23a (red) at 30 hpp. A central focal plane (top) and surface focal plane (bottom) are shown. Each image is from a different retina. F, rMIP at consecutive stages of pupal retinal development (37, 39, 41, and 43 hpp). Yellow * marks distal end of IoHC. Each magnified IoHC is outlined by a dotted white line. Full images (left) and magnified images right; E,F
Within the developing larval DP, eRpL22-like expression is ubiquitous with a fraction of eRpL22-like co-localizing with fibrillarin in nucleoli of DP cells. Varying degrees of fibrillarin and eRpL22-like overlap (yellow) are evident throughout the DP, with significant overlap occurring in areas anterior of the morphogenetic furrow where significant cellular proliferation occurs (Figure 5D —bottom left) compared to areas in the eye field (Figure 5D —bottom right). Variation in nucleolar size within DP cells in regions of high mitotic activity has previously been reported41 and may reflect differences in translational requirements to support high levels of cellular proliferation in the developing larval DP.
For midpupal stages of development, we evaluated co-localization of eRpL22-like and RpL23a at early (30 hpp) and late (37-43 hpp) stages of IoHC growth and maturation. At 30 hpp, eRpL22-like is detected within nuclei of all cells within developing ommatidia (Figure 5E —top). A ring of RpL23a surrounds the nucleus within these cells, suggestive of ribosomes that may be bound to the endoplasmic reticulum (ER). A pool of eRpL22-like overlaps with RpL23a in the presumptive ER as well as within the cytoplasm, supporting a ribosomal role for eRpL22-like within cells of developing ommatidia. Interestingly, at surface focal planes where eRpL22-like is shown enriched in the center of ommatidia, coincident with the presumptive cone (PC) and primary pigment (PP) cell complex, no overlap with RpL23a is apparent (Figure 5E —bottom), indicating the existence of a pool of eRpL22-like that is excluded from canonical ribosomes. In contrast to early stages where eRpL22-like is broadly distributed in cells across the ommatidium, eRpL22-like is primarily enriched in developing IoHC in later stages (37–43 hpp) of midpupal growth (Figures 1D, 4, and 5F). A majority of eRpL22-like within growing IoHC at 37/39 hpp does not overlap with RpL23a (Figure 5F). At 41/43 hpp, no significant overlap in localization of eRpL22-like and RpL23a was observed (Figure 5F). Overlap of eRpL22-like and RpL23a (would be observed as yellow color if evident) is not observed within IoHC of pupal time points examined. Thus, in both larval and pupal stages of eye development, roles for eRpL22-like as a ribosomal constituent are supported, but in each developmental stage a pool of eRpL22-like appears to be excluded from canonical ribosomes.
2.4 ∣. in vivo eRpL22-like functional depletion studies corroborate eRpL22-like expression patterns and display specific developmental phenotypes
The profile of eRpL22 paralogue expression in the eye over time was constructed using paralogue-specific antibodies previously validated for immunoblots and IHC in the testis.21 As a strategy to corroborate expression patterns found in the eye, we employed several functional in vivo eRpL22-like depletion strategies to corroborate eRpL22-like expression patterns observed in eye tissue at different stages of development. We reasoned that a reduction in the IHC staining pattern for eRpL22-like in response to tissue-specific paralogue depletion would provide strong evidence of authentic eRpL22-like expression in eye tissues, and mitigate concerns for nonspecific antibody binding in tissues.
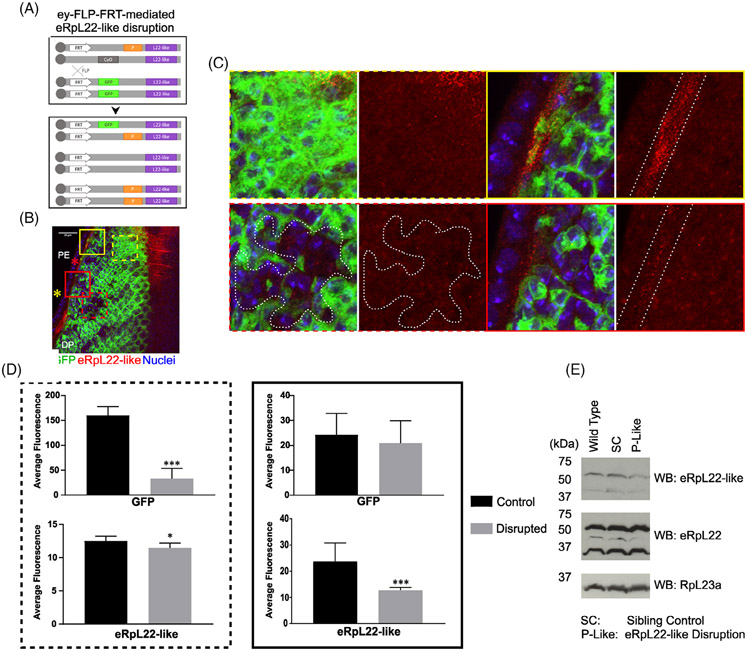
For validation of larval patterns of expression for eRpL22-like in the DP, we used a strain in which eRpL22-like is disrupted by insertion of a P-element (P {lacW}k07136) 156 nucleotides upstream of the transcription start site for the gene (Flybase.org: FB2017_03). This P-element insertion is embryonic lethal when homozy-gous. Therefore, eRpL22-like disruption was induced in a clonal manner through eye-specific somatic recombination, triggered by expression of flippase (FLP) under the control of an eyeless (ey) promoter for expression within embryonic eye primordia and in third instar larval anterior to the morphogenetic furrow at the time of photoreceptor determination.42
Wild type tissue is marked by expression of a GFP reporter whereas eye tissue homozygous for P{lacW} k07136 is marked by the absence of GFP stain (Figure 6A). When recombination occurs, depletion of GFP expression at various points across the eye/antennal imaginal disc is evident (Figure 6B). Significant depletion of eRpL22-like within homozygous P{lacW}k07136 clones in third instar eye/antennal imaginal discs is evident when quantification of eRpL22-like stain within disrupted tissue regions (Figure 6C —red dashed box) is compared to quantification of eRpL22-like stain within control tissue (Figure 6C —yellow dashed box). We have also observed that eRpL22-like staining is significantly depleted within the PE and PS in some regions (Figure 6C —red solid box) while other regions are unaffected (Figure 6C —yellow solid box), likely reflecting the mosaic nature of the disruption pattern. eRpL22-like disruption was quantified by ImageJ analysis of the DP (Figure 6D —left) and the PE and PS (Figure 6D —right). Where GFP disruption is evident within the DP, eRpL22-like expression in flanking regions of the PE and PS was not always diminished. Thus, it is unclear how interactions between the DP and PE or PS influence eRpL22-like expression and function within neighboring cells of third instar eye/antennal imaginal discs. In adult head tissue, eRpL22-like detection through Western blot is also diminished when clones of P{lacW}k07136 are present (Figure 6E). Western blots show a modest depletion of ~15% (as determined by densitometry) of a high molecular mass species of eRpL22-like of unknown function (described further below) relative to levels in wild type and sibling controls (Figure 6E). The lower molecular mass species (at the predicted mass of 34kD) remained relatively unchanged. What role the high molecular mass species of eRpL22-like plays within the developing eye is unknown.
FIGURE 6.
P-element mediated disruption of eRpL22-like diminishes eRpL22-like staining. A, Diagram of P-element induced eRpL22-like disruption through somatic FLP-FRT-mediated recombination (FRT—white box, P-element—orange box, eRpL22-like—purple box, GFP—green box, Cyo—gray box). B, Maximum intensity projection of third instar eye/antennal imaginal discs showing GFP (green), eRpL22-like (red), and nuclei (blue). Yellow * marks disc posterior and red * marks the PE. Scale bar: 20 μm. C, Magnification of image showing P-element disruption of eRpL22-like expression in larval eye/antennal imaginal discs. Magnified images of DP tissue showing eRpL22-like disruption (red dashed box) and control (yellow dashed box), and PE/PS tissue showing eRpL22-like disruption (red solid box) and control (yellow solid box). eRpL22-like disrupted-clone is outlined with a white dotted line. D, Left—Quantification of eRpL22-like and GFP depletion in the DP; Right —eRpL22-like within the PE/PS and GFP within the DP that borders the PE/PS. Number of DP disrupted-clones analyzed: 17; DP control-clones analyzed: 10; PE disrupted-clones analyzed: 6; PE control-clones analyzed: 14. Statistical significance was measured using a Student's t-test. *** = p > .0005, * = P > .05. E, Western blot analysis of eRpL22-like and eRpL22 from adult heads containing P-element disrupted tissue. RpL23a was used as a loading control
We used the pair-rule gene, odd-skipped (Odd) to drive tissue-specific expression of an eRpL22-like.IR construct (previously developed by Mageeney et al16 for expression of inverted repeats (IR) that form short hairpin RNA for eRpL22-like-specific knockdown) for RNAi-mediated depletion of eRpL22-like within the PE of the eye/antennal imaginal disc. Odd is expressed in margin ells of the PE43; margin cells are closest to the DP. Tissue-specific knock-down of eRpL22 (eRpL22.IR) using the Odd-Gal4 driver resulted in complete embryonic lethality, demonstrating a broader requirement for eRpL22 function in embryonic development. Unlike eRpL22 depletion, knock-down of eRpL22-like using Odd-Gal4 did not result in embryonic lethality, but caused eye patterning defects, as indicated below.
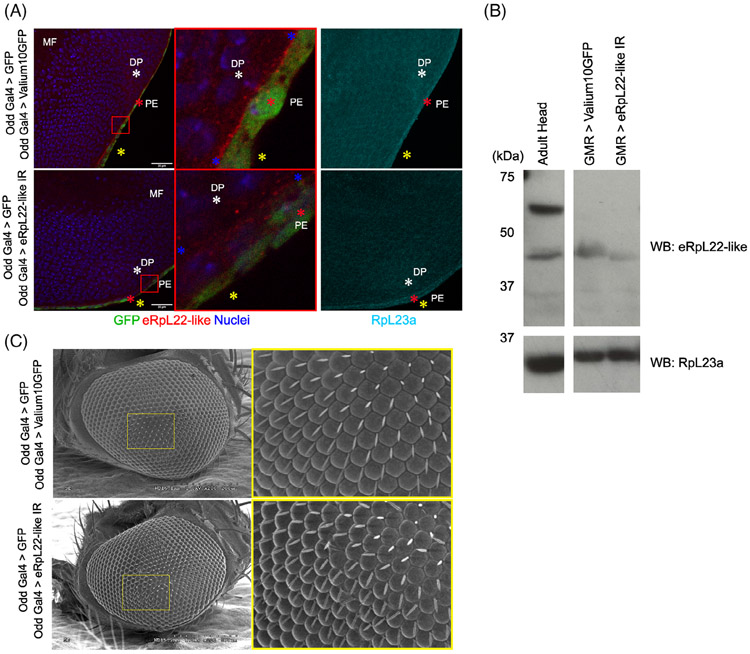
Expression of Odd-Gal4 was confirmed through expression of a GFP reporter (Figure 7A). As previously indicated, significant enrichment of eRpL22-like occurs at the apical surface of the PE and in the PS between the DP and the PE (Figure 3C). RNAi expression within the PE abolishes the eRpL22-like pattern of enrichment within the PE (Figure 7A). Diminished IHC staining in the PE using the RNAi depletion strategy further corroborates the pattern of eRpL22-like expression identified within the PE of wildtype larval eye discs and suggests that staining represents authentic eRpL22-like expression and is not due to nonspecific antibody binding. In addition, diminished IHC staining reveals that RNAi may have depleted eRpL22-like function specific to the PE/PS and may be specific to cytonemes or signaling between the PE and DP. The distribution of core ribosomal component, RpL23a, was uniform throughout the PE and PS and was useful to mark the outline of relevant tissues (Figure 7A —right panels).
FIGURE 7.
RNAi-mediated disruption of eRpL22-like depletes eRpL22-like staining. A, Representative image of a single focal plane from IHC of larval discs showing GFP (green), eRpL22-like (red), RpL23a (cyan), and nuclei (blue) from Odd-Gal4 > UAS-GFP /UAS-eRpL22-like.IR (bottom) and Odd-Gal4 >UAS-GFP/UAS-Valium10GFP as a control (top). Left—full image; middle—magnified image; right—L23a. Yellow * marks disc posterior, white * marks the DP, and red * marks the PE. MF—morphogenetic furrow. In the magnified view, the area of the PE/PS interface containing eRpL22-like enrichment is flanked by two blue *. Scale bar: 20μm. B, Western blot analysis of eRpL22-like in 24 to 50 hpp pupal retina. RpL23a was used as a loading control. C, SEM microscopy of odd-Gal4 > Valium10GFP or odd-Gal4 > eRpL22-like.IR. Yellow boxes show a magnified view of each eye. Scale: 200 μm
Tissue-specific RNAi-mediated depletion of eRpL22-like using a GMR-Gal4 driver was used as an additional strategy to assess the impact on eRpL22-like expression, visualized by Western blot. GMR-mediated expression begins within late stages of third instar larvae in differentiated eye cells and then continues into adult stages. Expression of GMR-Gal4 was confirmed through expression of a GFP reporter (data not shown). RNAi-mediated depletion was assessed by Western blot and IHC analyses. Within 24-50 hpp, the amount of eRpL22-like is reduced by 49% in Western blots compared to controls (Figure 7B).
Elimination of PE tissue through ablation yields severe ommatidia and IoHC patterning defects, indicating that PE tissue is critical to patterning of eye tissue.25 Tissue-specific depletion of eRpL22-like within the third instar eye disc PE results in disruption of IoHC patterns in adult stages, visible through scanning electron microscopy (Figure 7C). Eye defects were evident in 40% (2 of 5 total SEM images) of flies subjected to eRpL22-like depletion with odd-Gal4 > UAS-eRpL22-like.IR. These specific eye defects were not observed in any control images (odd-Gal4 > UAS-Valium10GFP; n = 9). These data support a potential role for eRpL22-like in the regulation of eye development, specifically within the larval eye/antennal imaginal disc PE. Despite the absence of IoHC patterning phenotypic defects in all knockdown flies (likely due to RNAi variability), this experiment revealed the scope of phenotypes that might be expected with more robust perturbation of eRpL22-like expression.
Collectively, P-element disruption and RNAi depletion of eRpL22-like show diminished patterns of eRpL22-like expression within the DP, PE, and IoHC, thereby authenticating IHC patterns of eRpL22-like expression determined for wildtype larval eye discs.
2.5 ∣. Multiple variants of eRpL22-like are expressed during eye development
Previous reports have shown multiple variants of eRpL22 paralogues expressed within adult stage flies. Several molecular mass variants resulting from post-translational modification by SUMOylation and/or phosphorylation of ubiquitously expressed eRpL22 are present in all tissues44 with unique molecular mass variants of eRpL22 being expressed in the male germline. Western blot analysis of adult stage flies demonstrated highly enriched expression of eRpL22-like in the male germline and within adult heads/eyes.21 For eRpL22-like, the most apparent species detected in whole testes extracts was present at its predicted molecular mass of ∣34 kDa, and was determined by co-sedimentation with ribosomes to be a ribosomal component.21 On the other hand, eRpL22-like in adult heads/eyes was detected at a higher molecular mass of >50 kDa.21 Relatively little, if any, eRpL22-like was detected within heads from which eyes were removed, supporting the conclusion that the majority of eRpL22-like expression is found in the adult eye. The molecular mass profile of eRpL22-like in the eye across development was not previously determined; therefore, here we have used immunodetection to interrogate the molecular mass of eRpL22-like in larval eye discs and midpupal eye tissues for comparison to the high molecular mass species previously identified by Kearse et al.21 in adult eye tissue.
We initially confirmed eRpL22-like expression in adult eye tissue using the differentiated eye-specific cell driver GMR45 to drive expression of head involution defective (hid) to promote apoptosis46 in eye tissue. Western blot analysis of genetically ablated head tissue showed that high molecular mass eRpL22-like was eliminated when eye tissue did not develop (Figure 8A). In earlier stages of eye development, eRpL22-like is primarily expressed as lower molecular mass species compared to the predominant variant of >50 kDa found in adult eye tissue (Figure 8A). The variant found in midpupal eye tissue appears identical in mass to that found in the testis and to the less abundant form in the adult eye (Figure 8B,C). On the other hand, a clear difference in mass exists between the larval and midpupal eRpL22-like species.
FIGURE 8.
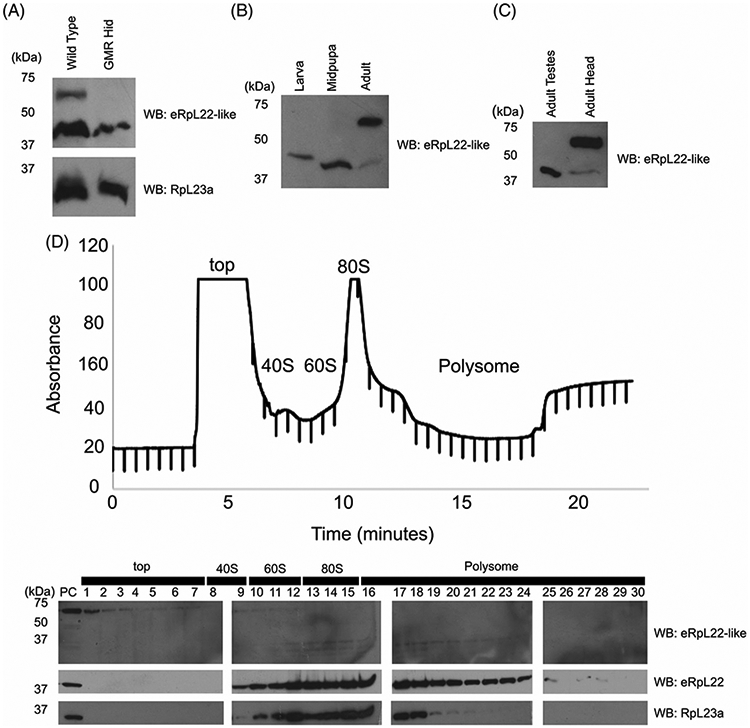
Multiple variants of eRpL22-like are expressed during eye development. A, Western blot analysis of adult head tissue from flies subjected to GMR-Hid-mediated ablation of eye development. eRpL22-like (top) is indicated, with RpL23a (bottom) used as a loading control. B, Western blot analysis of eRpL22-like in eye tissue at key stages of eye development: instar eye/antennal imaginal discs (with brain), pupal retina (with brain), and adult heads. C, Western blot comparison of eRpL22-like in adult testes and heads. D, Top—Polysome profile analysis of adult head tissue extracts on a 10% to 50% sucrose gradient. Bottom—Western blot analysis of fractionated sucrose gradient probed for eRpL22-like and eRpL22. RpL23a was used to mark the position of 60S large ribosomal subunits in the gradient
We have previously shown that high molecular mass species of eRpL22 are post-translationally modified by SUMOylation and phosphorylation and are not components of active ribosomes.44 Exclusion of post-translationally modified variants of eRpL22 from the pool of actively translating ribosomes suggests an alternate role for these eRpL22 variants. Although mechanisms that contribute to the additional size of the adult form of eRpL22-like have not been determined, we probed polysome profiles from adult heads to determine how the sedimentation pattern of this species as well as that of the less predominant form compared to ribosomal components in the gradient (Figure 8D —top). Low molecular mass eRpL22-like is detected in ribosomal fractions (60S, 80S, and polysomes) that contain both eRpL22 and RpL23a while the >50 kDa species sediments at the top of the gradient apart from fractions containing ribosomal components (Figure 8D —bottom). Insufficient tissue quantities for larval discs or midpupal eye tissue prevented polysome analysis for these stages. Yet, based on the polysome profile analysis for the less abundant adult species of eRpL22-like and its co-migration with whole testis eRpL22-like, it is likely that midpupal eRpL22-like is a ribosomal component as well. Whether or not larval eRpL22-like associates with ribosomal components must await additional analyses.
It is important to note that variants represented in each developmental stage shown in Figure 8B,C represent the most abundant species in whole tissues at that stage. Cell and tissue types in low representation within those developmental stages may not be sufficiently represented in this analysis. This is particularly relevant when considering what forms of eRpL22-like are present in the larval PE and PS, and midpupal IoHC.
3 ∣. DISCUSSION
Here we report patterns of eRpL22-like expression and protein processing in the developing fly eye during larval, midpupal and adult stages, as determined by IHC and Western blot analyses. eRpL22-like is widely expressed throughout the developing eye, but is particularly enriched in the PE and PS of the larval eye/antennal imaginal disc and within the IoHC in midpupal eye tissue, as patterning of the eye unfolds. In the adult eye, eRpL22-like expression expands beyond the IoHC into the ring of accessory cells (composed of secondary and tertiary pigment cells along with mechanosensory bristles) surrounding each ommatidium. Disruption of eRpL22-like expression in margin cells of the larval PE by RNAi-mediated knockdown using the odd-Gal4 driver demonstrated a potential link between a role for eRpL22-like in the larval PE and patterning of the adult IoHC. Preliminary biochemical analysis by Western blotting demonstrates that eRpL22-like is not expressed in all stages of eye development as a single species at its predicted molecular mass of ∣34 kDa; instead additional higher molecular mass immunoreactive variants are present in larval and adult stages. Taken together, differential expression and the presence of developmental stage-specific molecular mass variants suggest tissue-specific and/or developmental stage-specific roles for eRpL22-like in the developing eye.
3.1 ∣. eRpL22-like functions as a specialized Rp in the developing eye
Analysis of Rp paralogue functions in yeast has shown that duplicated genes very commonly result in moonlighting activity of the resulting paralogue.47 Moonlighting activities are also more commonly maintained when paralogues are co-expressed, maintaining a stoichiometric balance between each protein. Analysis of eRpL22-like expression and subcellular distribution at each stage of development provides evidence supporting ribosomal functions for eRpL22-like. Co-localization of eRpL22-like with core Rps and fibrillarin suggests ribosomal roles in larval and midpupal stages. Polysome profiling data further support a ribosomal role, as the predominant eRpL22-like variant (at the predicted molecular mass of ∣34kD) found in midpupal eye tissue co-sediments with ribosomal components including actively translating ribosomes.
IHC and microscopy data also suggest that a pool of eRpL22-like is excluded from ribosomal complexes containing core Rps such as RpL23a and RpL9 in the developing eye at each developmental stage. Thus, a fraction of eRpL22-like may function in alternate pathways or be sequestered in noncanonical ribosomal complexes. Within the PE, the distribution pattern for eRpL22-like invokes several interesting questions about how eRpL22-like may be sequestered in the apical domain. Is eRpL22-like derived from de novo synthesis and direct trafficking to the apical domain or is eRpL22-like derived through exchange from within the ribosomal pool? Insights into regulation of eRpL22-like function may be forthcoming from future studies of the PE in larval eye development. Unlike early midpupal stages of eye development, later midpupal stages show a different pattern of eRpL22-like distribution with enrichment in developing IoHC, largely segregated from the distribution pattern for core Rps. Given the relatively small proportion of IoHC compared to other cell types in developing midpupal eye tissue, it is unlikely that an IoHC-specific pool of eRpL22-like with different sedimentation characteristics would be distinguished from the predominant ∣34kD form. Within the ring of accessory cells in adult ommatidia, multifunctional roles for eRpL22-like are implicated, based on the presence of at least two pools of eRpL22-like - a ribosomal pool and a larger pool of a high molecular mass variant that is excluded from actively translating ribosomes.
3.2 ∣. Unique patterns of eRpL22 and eRpL22-like expression may signify paralogue-specific functions
Both eRpL22 paralogues are co-expressed during each stage of eye development. We found no examples of cells within developing eye tissues that were devoid of eRpL22; however, eRpL22 is asymmetrically enriched in peripheral tissue regions, including the PE. It is eRpL22-like expression that changes as eye development proceeds.
Shifts in expression patterns between developmental stages may indicate that the balance between eRpL22 paralogues plays a role in regulation of tissue morphogenesis. Such an impact on tissue morphogenesis has been observed in Arabidopsis thaliana, where the balance between two members of the RpS5 family influences the proliferation or differentiation status of tissues at different times in development.48 One RpS5 member is highly expressed in dividing cells while expression of the other member is correlated with cell differentiation. Co-expression of eRpL22 and eRpL22-like within cells predicts the presence of heterogeneous populations of ribosomes based on paralogue content, if both paralogues are present concurrently within the ribosomal pool. Whether or not paralogue-specific ribosomes within cells of the developing eye regulate translation of specific mRNAs is unknown, but this proposal should be considered in light of recent findings showing differential association of specific mRNAs on eRpL22- or eRpL22-like ribosomes in the male germline of Drosophila.16 Thus, even within the ribosomal pool, eRpL22 and eRpL22-like may have distinctive roles in translation regulation. Alternatively, it is interesting to speculate that some eRpL22-like variants (particularly the high molecular mass species identified) may function outside of the ribosomal pathway in specific cells (as our IHC and Western blot data suggest) in apical regions of the PE and in the midpupal IoHC, reserving ribosomal functions for eRpL22 and the 34kD species of eRpL22-like.
3.3 ∣. eRpL22-like is expressed in apical domains of elongated cell types in the eye
The broad pattern of eRpL22-like expression in the developing eye in both early and later stages of eye development suggests a general role for eRpL22-like in many differentiating cell types. Yet, selective restriction of eRpL22-like expression to the PE and the growing IoHC in midpupal and adult eye tissue suggests a special role for eRpL22-like in key developmental events within these polarized cells. Interestingly, enrichment of eRpL22-like occurs at the apical surface of both the PE and midpupal IoHC. Within the PE (and the PS), eRpL22-like is likely enriched within filopodial extensions or cytonemes that facilitate signaling between the PE and the DP.31 Signaling filopodia are critical regulators of patterning within the developing eye.25 The proximity of eRpL22-like to this region of the PE may connect this paralogue through protein interactions with signaling pathways such as Hedgehog, Decapentaplegic, and Wingless, known to be important in developmental regulation.49 Communication between the PE and the DP is vital for pattern formation and the regulation of polarity within the DP.25 The PE not only contributes signaling molecules to the DP but contributes cells as well (reviewed by28). Within the IoHC, eRpL22-like is found surrounding actin bundles during IoHC growth. Is there a common functional theme for eRpL22-like within the IoHC as this cell type elongates in midpupal stages of eye development?
These studies have uncovered specific cell types and tissue regions in the eye that may be impacted by perturbations of eRpL22-like expression. Future genetic ablation or overexpression strategies will be directed toward assessing phenotypes related to specification, patterning and polarity of tissue within the DP, PE, and midpupal retina as well as morphogenesis and patterning of the IoHC lineage.
4 ∣. EXPERIMENTAL PROCEDURES
4.1 ∣. Fly stocks
All stocks were kept at room temperature on standard cornmeal media. Wild-type Oregon-R D. melanogaster was used for IHC and Western blot experiments. Crosses were maintained at 25°C and performed with a 2:1 (female:male) ratio. The following fly lines were obtained from The Bloomington Drosophila Stock Center: C311-Gal4 driver (BN#5937: y[1]; P{w[+mW.hs]GawB}c311, eRpL22-like P-element line (BN#12195: y[1] w[*]; P{w [+mC] = lacW}k07136, BN#7110 (w[1118] P{ry [+t7.2] = ey-FLP.N}2; P{ry[+t7.2] = neoFRT}42D P{w [+mC] = GMR-myr.GFP}2R, UAS-Valium10GFP (BN#35786: UAS-VALIUM10GFP (y[1] v[1]; p{y[+t7.7] v [+t1.8] = UAS-GFP.VALIUM10}attP2), 7198 (BN#) Balancer line (w[*]; Kr[If-1]/CyO; D[1]/TM3, Ser[1]). The eRpL22-like RNAi line (y[1], v[1]; pVALIUM10{UAS-eRpL22-like.IR} attP2; Chromosomes:1;3) was generated previously16 and is maintained as a homozygous stock. Odd-Gal4 drivers, (Odd-Gal4, UAS-DICER (VDRC 60008)/Sm6::Tm6B), (Odd-Gal4/CyO, 2xTb; UAS-GFP) were a gift from the Pignoni lab (SUNY Upstate Medical University).
4.2 ∣. Generating P{lacW}k07136 recombinant
A fly line containing the P{lacW}k07136 P-element insertion (BN#12195) and crossed with BN#7110, a line containing an FRT site and a GFP marker. Female F1 were collected and then crossed with males from BN#7198. F1 flies were then screened for curly wings and a loss of green fluorescence in eyes. Males and females positive for curly wings and negative for GFP eye fluorescence were mated together. Progeny from this cross were screened for individuals with wild type wing phenotypes. Taking advantage of the homozygous lethality of P{lacW}k07136, absence of wild type wing phenotypes indicated flies were homozygous for the P-element insertion. Presence of the FRT allele was experimentally validated using GFP immunofluorescence. Fly food contained Geneticin (concentration according to the recommended amount for the P{neoFRT}42D allele, Bloomington Stock Center) enriched for flies containing FRT chromosomes.
4.3 ∣. Antibodies and stains
Polyclonal antibodies against eRpL22 and eRpL22-like were generated by Genscript21 (1:1000 Western; 1:100 IHC). A chicken anti-Drosophila peptide polyclonal antibody against RpL23a was also generated by Genscript (22; IHC: 1:1300). HRP-conjugated anti-mouse and anti-rabbit (1:50 000) were used as secondary antibodies for Western blot analysis and anti-mouse Alexa 488 Fluor, anti-rabbit Alexa 488 Fluor, and anti-rabbit Alexa 568 Fluor (all at 1:200) were used as secondary antibodies for IHC analysis. Anti-chicken Alexa 647 Fluor (A21449, Life Technologies; 1:200) was used for IHC. Specific antibodies against core ribosomal components RpL23a (Abgent) (1:1000 Western; 1:100 IHC) and RpL9 (Santa Cruz Biotech) (1:100 IHC) were used as a marker of ribosomes. Rabbit-anti fibrillarin (Ab5821, 1:100) and rabbit-anti GFP (Ab290, 1:200) were obtained from Abcam. Rat-anti ELAV (Elav-9F8A9, 1:50) was obtained from the University of Iowa Developmental Studies Hybridoma Bank. The secondary antibody used to detect ELAV was anti-rat Cy3 (Ab98416, Abcam; 1:200). 4,6-Diamidino-2-phenylindole, dihydrochloride (DAPI) (0.4 μg/mL), and Alexa 568 Fluor-conjugated phalloidin (1:250) were used as counter stains (Life Technologies).
4.4 ∣. Protein extraction, SDS-PAGE, and Western blotting
Drosophila tissue was dissected in 1× PBS after which tissue was immediately frozen on dry ice. Tissue was then homogenized directly in reducing sample buffer (20% β-mercaptoethanol, 10% glycerol, 2% sodium dodecyl sulfate [SDS], 8% stacking gel buffer). Stacking gel buffer contains 2 M Tris, 0.4% SDS, pH 6.8. Samples were then boiled at 95° C three times in 5-minute increments with gentle vortexing between each increment. Whole cell lysate proteins were separated by SDS-PAGE (5% stacking gel; 10% separating gel) at 150 V and electro-transferred onto a 0.2 μm PVDF membrane (Whatman) at 100 V for 1 hour. Membranes were blocked with 5% nonfat dry milk (NFDM) in 1× PBS, 0.1% Tween-20 for hour, after which membranes were incubated in primary antibodies overnight with agitation at 4°C in 3% NFDM, 0.1% Tween-20. Membranes were incubated with secondary antibodies for 1 hour at room temperature with agitation and then visualized with ECL (GE Healthcare) chemiluminescent detection and BioMax Chemiluminescence Film (Kodak).
4.5 ∣. Developmental staging
Flies in crosses were maintained at 25° C in a humidified incubator. Third instar larvae (to be dissected) were characterized by distance traveled from food and low amount of movement, indicating imminent pupation. White prepupae were collected at 0 hours post pupation (hpp), separated into new media vials and aged at 25°C until the desired developmental time point.
4.6 ∣. Immunohistochemistry
Tissue was dissected as demonstrated in Reference 50 with modifications. Eye/antennal imaginal discs were dissected from wandering third instar larva and midpupal retinae were dissected between 37 and 43 hours post pupation. Tissue was dissected in cold 1× PBS then transferred to a 9-well glass spot plate on ice containing fresh 1β PBS. Tissue was maintained in cold PBS until dissection was completed (no longer than 30 minutes). Samples were then fixed for 15 minutes in a 3.7% solution of formaldehyde diluted in 1× PBS. After fixation, tissue was washed with 1× PBS three times and incubated in 1× PBS for 30 minutes at room temperature with agitation. Tissue was washed with 1× PBS, 0.1% triton X-100 three times and incubated for 30 minutes at room temperature with agitation. Primary antibody was diluted in 1× PBS, 0.1% triton X-100, and incubated without agitation overnight at room temperature in a spot plate sealed with parafilm. The following day, after three washes with 1× PBS, 0.1% triton X-100, tissue was incubated for 4 hours at room temperature without agitation. Secondary antibody was diluted in 1× PBS, 0.1% Triton X-100 and incubated at room temperature without agitation overnight in a spot plate sealed with parafilm. After three washes with 1× PBS, 0.1% Triton X-100, the tissue was incubated for 4 hours at room temperature without agitation. Phalloidin (1:250 in 1× PBS, 0.1% Triton X-100) and DAPI (1:500 in 1× PBS, 0.1% Triton X-100) counter stains were performed before mounting tissue for 2 hours and 20 minutes, respectively, with three 1× PBS, 0.1% Triton X-100 washes in between. Tissue was mounted on superfrost slides (VWR) where tissue was overlaid with 10 μL of Slow Fade Diamond Mounting Media (Thermofisher), covered with a cover slip (Corning: 2890-22), and sealed with nail polish. Confocal microscopy was performed on either a Zeiss 710 (Johns Hopkins University) or a Zeiss 880 (Lehigh University) confocal microscope.
4.7 ∣. ImageJ analysis
In P-element disruption experiments, expression levels (mean fluorescence) of eRpL22-like and GFP were measured with the freehand ImageJ tool51 (Figure 6C). Statistically significant differences between P-element-disrupted areas and wild type areas were determined using a Student's t-test of integrated density values.
Quantitative assessment of changes in the relative accumulation (integrated density) of eRpL22-like in Western blots were determined in reference to RpL23a using the ImageJ polygon selection tool51 (Figures 6E and 7B). A ratio was determined between the mean pixel value of eRpL22-like and the mean pixel value of RpL23a within the same sample. Ratios were then compared between control and experimental tissue to establish a percentage decrease from control to experimental samples.
4.8 ∣. Tissue region of interest quantification
The tissue regions of interest (tROI) including nuclei, cytoplasm, PE, and PS were segmented into separate masks per each image through the following methods. Nuclei were identified via DAPI staining and segmented using contrast limited adaptive histogram equalization (CLAHE)52 and secondary thresholding methods from the scikit-image processing algorithm collections.53 Adaptive techniques allowed adjustment for stain intensity within and between images.54 PE, PS, and negative space outside of tissue were identified by phalloidin (actin) staining of cell membranes. Cytoplasmic regions within the DP and PE were identified as the regions left after subtracting the nuclei segmentation mask. The manual segmentation of these regions was constrained so that no delineated tissue structural region overlaps with another (ie, the PS and PE only share borders). The pixel quantity and intensity values for each channel in each masked region in each image analyzed was plotted in box and whisker plots using matplotlib plotting utilities.55 Each plot represents a separate tROI mask (nuclei, cytoplasm PS, and PE) grouped by channel representing a given Rp or paralogue.
4.9 ∣. Sucrose gradient ultracentrifugation
Ribosome extracts were prepared using modified procedures from Qin et al,56 Houmani and Ruf,57 and Pelczar and Filipowicz.58 Newly eclosed wild type (Oregon-R) adult fly heads (1500 male/1500 female) were homogenized in 1.0 mL of lysis buffer (20 mM Tris-HCl, pH 8, 140 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 1% Triton-X 100, 100 μg/mL cycloheximide). Homogenates were clarified by centrifugation in an Eppendorf centrifuge at 10 K rpm for 10 minutes. Approximately 400 μg were loaded onto a 10% to 50% linear sucrose gradient prepared by the horizontal method described in Houmani and Ruf57 and spun at 35 K rpm for 160 minutes in a SW-41 rotor. Gradient preparation, centrifugation conditions, and subsequent protein extraction by TCA precipitation were performed as previously described by Houmani and Ruf.57 Gradients were fractionated using a Brandel syringe pump and Foxy Jr. R1 gradient fractionator along with an Isco UA-6 detector for continuous absorbance readings at 254 nm. Fractions (0.5 mL) were collected at 40 second fraction collection intervals set with 0.75 mL/min pump speed.
4.10 ∣. Scanning electron microscopy
Adult fly heads were dissected at 5 days of age and mounted directly on scanning electron microscope pegs. Images were obtained using a 1 kV setting. Tissue was imaged without sputter coating. Scanning electron microscopy images were captured using a Hitachi S-4300SE Schottky-Emission scanning electron microscopy.
ACKNOWLEDGMENTS
Financial support was provided in part by Lehigh University faculty research funds and NIH 1R15GM132889-01 to Vassie C. Ware. The work described here was in partial fulfillment of the requirements for the PhD degree for Brett W. Gershman, who was funded in part by the Department of Biological Sciences at Lehigh University. We thank members of the fly community (noted in Experimental Procedures) for fly stocks. We thank Dr. Robert Johnston for training on eye tissue dissection techniques and the use of the Zeiss 710 confocal microscope.
Funding information
National Institutes of Health, Grant/Award Number: 1R15GM132889-01; Lehigh University
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Genuth NR, Barna M. The discovery of ribosome heterogeneity and its implications for gene regulation and organismal life. Mol Cell. 2018a;71:364–374. 10.1016/j.molcel.2018.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yelick PC, Trainor PA. Ribosomopathies: global process, tissue specific defects. Rare Dis. 2015;3:e1025185. 10.1080/21675511.2015.1025185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farrar JE, Vlachos A, Atsidaftos E, et al. Ribosomal protein gene deletions in Diamond-Blackfan anemia. Blood. 2011;118: 6943–6951. 10.1182/blood-2011-08-375170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramagopal S Induction of cell-specific ribosomal proteins in aggregation-competent nonmorphogenetic Dictyostelium discoideum. Biochem Cell Biol. 1990;68:1281–1287. 10.1139/o90-190. [DOI] [PubMed] [Google Scholar]
- 6.Ramagopal S, Ennis HL. Regulation of synthesis of cell-specific ribosomal proteins during differentiation of Dictyostelium discoideum. Proc Natl Acad Sci U S A. 1981;78:3083–3087. 10.1073/pnas.78.5.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhan Y, Melian NY, Pantoja M, et al. Dystroglycan and mitochondrial ribosomal protein L34 regulate differentiation in the drosophila eye. PLoS One. 2010;5(5):e10488. 10.1371/journal.pone.0010488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhavsar RB, Makley LN, Tsonis PA. The other lives of ribosomal proteins. Hum Genomics. 2010;4:327–344. 10.1186/1479-7364-4-5-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34:3–11. 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012; 13:355–369. 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Z, Fujii K, Kovary KM, et al. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genomewide. Mol Cell. 2017;67:71–83. 10.1016/j.molcel.2017.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genuth NR, Barna M. Heterogeneity and specialized functions of translation machinery: from genes to organisms. Nat Rev Genet. 2018b;19:431–452. 10.1038/s41576-018-0008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hopes T, Agapiou M, Norris K, McCarthy CGP, O'Connell MJ, Fontana J, Aspden JL. 2020. Specialisation of ribosomes in gonads through paralog-switching. bioRxiv. 913020; doi: 10.1101/2020.01.20.913020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bourbon H, Alin F, Ardourel C, et al. A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech Dev. 2002;110:71–83. [DOI] [PubMed] [Google Scholar]
- 15.Boutros M, Kiger AA, Armknecht S, et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- 16.Mageeney CM, Kearse MG, Gershman BW, Pritchard CE, Colquhoun JM, Ware VC. Functional interplay between ribosomal protein paralogues in the eRpL22 family in Drosophila melanogaster. Fly (Austin). 2018;12:143–163. 10.1080/19336934.2018.1549419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chintapalli VR, Wang J, Dow JAT. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 18.Kai T, Williams D, Spradling AC. The expression profile of purified Drosophila germline stem cells. Dev Biol. 2005;283: 486–502. 10.1016/j.ydbio.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 19.Shigenobu S, Arita K, Kitadate Y, Noda C, Kobayashi S. Isolation of germline cells from Drosophila embryos by flow cytometry. Dev Growth Differ. 2006;48:49–57. 10.1111/j.1440-169X.2006.00845.x. [DOI] [PubMed] [Google Scholar]
- 20.Shigenobu S, Kitadate Y, Noda C, Kobayashi S. Molecular characterization of embryonic gonads by gene expression profiling in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2006; 103:13728–13733. 10.1073/pnas.0603767103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kearse MG, Chen AS, Ware VC. Expression of ribosomal protein L22e family members in Drosophila melanogaster: RpL22-like is differentially expressed and alternatively spliced. Nucleic Acids Res. 2011;39:2701–2716. 10.1093/nar/gkq1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mageeney CM, Ware VC. Specialized eRpL22 paralogue-specific ribosomes regulate specific mRNA translation in spermatogenesis in Drosophila melanogaster. Mol Biol Cell. 2019;30 (17):2240–2253. 10.1091/mbc.E19-02-0086 Epub 2019 Jun 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roignant JY, Treisman JE. Pattern formation in the Drosophila eye disc. Int J Dev Biol. 2009;53:795–804. 10.1387/ijdb.072483jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar JP. Building an Ommatidium one cell at a time. Dev Dyn. 2012;241:136–149. 10.1002/dvdy.23707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson MC, Schubiger G. Peripodial cells regulate proliferation and patterning of drosophila imaginal discs. Cell. 2000;103:343–350. 10.1016/S0092-8674(00)00125-2. [DOI] [PubMed] [Google Scholar]
- 26.Robinow S, White K. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J Neurobiol. 1991;22:443–461. 10.1002/neu.480220503. [DOI] [PubMed] [Google Scholar]
- 27.Lai EC, Orgogozo V. A hidden program in drosophila peripheral neurogenesis revealed: fundamental principles underlying sensory organ diversity. Dev Biol. 2004;269:1–17. 10.1016/j.ydbio.2004.01.032. [DOI] [PubMed] [Google Scholar]
- 28.Atkins M, Mardon G. Signaling in the third dimension: the peripodial epithelium in eye disc development. Dev Dyn. 2009; 238:2139–2148. 10.1002/dvdy.22034.Signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibson MC, Lehman DA, Schubiger G. Lumenal transmission of decapentaplegic in Drosophila imaginal discs. Dev Cell. 2002; 3:451–460. 10.1016/S1534-5807(02)00264-2. [DOI] [PubMed] [Google Scholar]
- 30.Manseau L, Baradaran A, Brower D, et al. GAL4 enhancer traps expressed in the embryo, larval brain, imaginal discs, and ovary of drosophila. Dev Dyn. 1997;209:310–322. . [DOI] [PubMed] [Google Scholar]
- 31.Huang H, Kornberg TB. Cells must express components of the planar cell polarity system and extracellular matrix to support cytonemes. Elife. 2016;5:1–20. 10.7554/eLife.18979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shapira S, Bakhrat A, Bitan A, Abdu U. The Drosophila javelin gene encodes a novel actin-associated protein required for actin assembly in the bristle. Mol Cell Biol. 2011;31:4582–4592. 10.1128/MCB.05730-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bitan A, Rosenbaum I, Abdu U. Stable and dynamic microtubules coordinately determine and maintain Drosophila bristle shape. Development. 2012;139:1987–1996. 10.1242/dev.076893. [DOI] [PubMed] [Google Scholar]
- 34.Charlton-Perkins M, Cook TA. Building a fly eye: terminal differentiation events of the retina, corneal lens, and pigmented epithelia. Curr Topics Dev Biol. 2010;93:129–173. 10.1016/B978-0-12-385044-7.00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou X, Liao WJ, Liao JM, Liao P, Lu H. Ribosomal proteins: functions beyond the ribosome. J Mol Cell Biol. 2015;7:92–104. 10.1093/jmcb/mjv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henras AK, Soudet J, Géras M, et al. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci. 2008; 65:2334–2359. 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ban N, Beckmann R, Cate JHD, et al. A new system for naming ribosomal proteins. Curr Opin Struct Biol. 2014;24:165–169. 10.1016/j.sbi.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ross CLN, Patel RR, Mendelson TC, Ware VC. Functional conservation between structurally diverse ribosomal proteins from Drosophila melanogaster and Saccharomyces cerevisiae: Fly L23a can substitute for yeast L25 in ribosome assembly and function. Nucleic Acids Res. 2007;35:4503–4514. 10.1093/nar/gkm428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Beekvelt CA, de Graaff-Vincent M, Faber AW, van't Riet J, Venema J, Raué HA. All three functional domains of the large ribosomal subunit protein L25 are required for both early and late pre-rRNA processing steps in Saccharomyces cerevisiae. Nucleic Acids Res. 2001;29(24):5001–5008. 10.1093/nar/29.24.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pederson T The nucleolus. Cold Spring Harb Perspect Biol. 2011;3:1–15. 10.1101/cshperspect.a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baker NE. Developmental regulation of nucleolus size during drosophila eye differentiation. PLoS One 2013; 8(3). 10.1371/journal.pone.0058266, e58266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Halder G, Callaerts P, Gehring W. Induction of ectopic eyes by targeted expression of the eyeless gene in drosophila. Science. 1995;267(5205):1788–1792. 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- 43.Bras-Pereira C, Bessa J. Casares F odd-skipped genes specify the signaling center that triggers retinogenesis in Drosophila. Development. 2006;133:4145–4149. 10.1242/dev.02593. [DOI] [PubMed] [Google Scholar]
- 44.Kearse MG, Ireland JA, Prem SM, Chen AS, Ware VC. RpL22e, but not RpL22e-like-PA, is SUMOylated and localizes to the nucleoplasm of Drosophila meiotic spermatocytes. Nucleus. 2013;4(3):241–258. 10.4161/nucl.25261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li WZ, Li SL, Zheng HY, Zhang SP, Xue L. A broad expression profile of the GMR-GAL4 driver in Drosophila melanogaster. Genet Mol Res. 2012;11:1997–2002. 10.4238/2012.August.6.4. [DOI] [PubMed] [Google Scholar]
- 46.Kurada P, White K. Ras promotes cell survival in Drosophila by downregulating hid expression. Cell. 1998;95:319–329. 10.1016/S0092-8674(00)81764-X. [DOI] [PubMed] [Google Scholar]
- 47.Espinosa-Cantú A, Ascencio D, Barona-Gómez F, De Luna A. Gene duplication and the evolution of moonlighting proteins. Front Genet. 2015;6:1–7. 10.3389/fgene.2015.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weijers D, Franke-van Dijk M, Vencken RJ, Quint A, Hooykaas P, Offringa R. An Arabidopsis minute-like phenotype caused by a semi-dominant mutation in a ribosomal protein S5 gene. Development. 2001;128:4289–4299. [DOI] [PubMed] [Google Scholar]
- 49.McClure KD, Schubiger G. Developmental analysis and squamous morphogenesis of the peripodial epithelium in Drosophila imaginal discs. Development. 2005;132:5033–5042. 10.1242/dev.02092. [DOI] [PubMed] [Google Scholar]
- 50.Hsiao H-Y, Johnston RJ, Jukam D, Vasiliauskas D, Desplan C, Rister J. Dissection and immunohistochemistry of larval, pupal and adult Drosophila retinas. J Vis Exp. 2012;69:e4347. 10.3791/4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Fundam Digit Imaging Med. 2010;9: 185–188. 10.1007/978-1-84882-087-6_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pizer SM, Amburn EP, Austin JD, Cromartie R, Geselowitz A, Greer T, ter Haar Romeny B, Zimmerman JB, Zuiderveld K. Adaptive histogram equalization and its variations. Comput Gr Image Process. 1987;39(3):355–368. [Google Scholar]
- 53.Van der Walt S, Schönberger JL, Nunez-Iglesias J, Boulogne F, Warner JD, Yager N, Gouillart E, Yu T. scikit-image: image processing in Python. Peer J. 2014;2:e453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knowles DW, Sudar D, Bator-Kelly C, Bissell MJ, Lelièvre SA. Automated local bright feature image analysis of nuclear protein distribution identifies changes in tissue phenotype. Proceedings of the National Academy of Sciences. 2006;103(12): 4445–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hunter JD. Matplotlib: A 2D graphics environment. Comput Sci Eng. 2007;9(3):90–95. [Google Scholar]
- 56.Qin X, Ahn S, Speed TP, Rubin GM. Global analyses of mRNA translational control during early Drosophila embryogenesis. Genome Biol. 2007;8:R63. 10.1186/gb-2007-8-4-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Houmani JL, Ruf IK. Clusters of basic amino acids contribute to RNA binding and nucleolar localization of ribosomal protein L22. PLoS One. 2009;4(4):e5306. 10.1371/journal.pone.0005306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pelczar P, Filipowicz W. The host gene for intronic U17 small nucleolar RNAs in mammals has no protein-coding potential and is a member of the 5′-terminal oligopyrimidine gene family. Mol Cell Biol. 1998;18:4509–4518. 10.1128/mcb.18.8.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]