Abstract
The recent outbreak of viral infection and its transmission has highlighted the importance of its slowdown for the safeguard of public health, globally. The identification of novel drugs and efficient therapies against these infectious viruses is need of the hour. The eruption of COVID-19 is caused by a novel acute respiratory syndrome virus SARS-CoV-2 which has taken the whole world by storm as it has transformed into a global pandemic. This lethal syndrome is a global health threat to general public which has already affected millions of people. Despite the development of some potential vaccines and repurposed drugs by some Pharma companies, this health emergency needs more attention due to the less efficacy of these vaccines coupled with the emergence of novel and resistant strains of SARS-CoV-2. Due to enormous structural diversity and biological applications, natural products are considered as a wonderful source of drugs for such diseases. Natural product based drugs constitute a substantial proportion of the pharmaceutical market particularly in the therapeutic areas of infectious diseases and oncology. The naturally occurring bioactive antiviral phytochemicals including alkaloids, flavonoids and peptides have been subjected to virtual screening against COVID-19. Since there is no specific medicine available for the treatment of Covid-19, designing new drugs using in silico methods plays an all important role to find that magic bullet which can target this lethal virus. The in silico method is not only quick but economical also when compared to the other conventional methods which are hit and trial methods. Based on this in silico approach, various natural products have been recently identified which might have a potential to inhibit COVID-19 outbreak. These natural products have been shown by these docking studies to interact with the spike protein of the novel coronavirus. This spike protein has been shown to bind to a transmembrane protein called Angiotensin converting enzyme 2 (ACE2), this protein acts as a receptor for the viral spike protein. This comprehensive review article anticipates providing a summary of the authentic and peer reviewed published literature about the potential of natural metabolites that can be developed into possible lead compounds against this new threat of Covid-19. Main focus of the article will be to highlight natural sources of potential anti-coronavirus molecules, mechanism of action, docking studies and the target proteins as well as their toxicity profiles. This review article intends to provide a starting point for the research endeavors that are needed for the design and development of drugs based on pure natural products, their synthetic or semi-synthetic derivatives and standardized plant extracts. This review article will be highly helpful for scientists who are working or intend to work on antiviral drugs from natural sources.
Keywords: Coronaviruses, Natural products, COVID-19, RNA, Spike proteins
Graphical abstract
1. Introduction
Coronaviruses (CoV's) are RNA enveloped family of viruses mostly found in mammals and birds [1]. The virus infection causes fetal respiratory or enteric disorders and in rare cases hosts develop neurological and liver disorders [2]. Species-specific effects of CoV's are observed in hosts like some may cause acute or persistent infections. These viruses are highly communicable and transmission may occur through different modes including respiratory and fecal-oral routes. CoV's show distinctive characteristic features in comparison to other viruses like a large genome size even bigger than those viruses having segmented genome [3]. This huge genome size contributes to wealthy and high variety of genetic expressions shown by CoV's and most importantly these expressions are yet to be fully explored and understood. In the year of 1960, CoV's were assembled under a variant class of viruses [4]. Since 1960, various novel viruses have been discovered which affects the respiratory system of humans in a similar fashion as avian infectious bronchitis virus (IBV) and mouse hepatitis virus (MHV) [5]. Study of CoV's under negative-stained electron microscopy reveals characteristic morphological structures. Their external surface was observed with “fringe” like structures later described as “spikes” or “club like” projections [6,7]. Variant density and morphology of these spikes were observed in MHV's in comparison to myxoviruses. The name “coronaviruses” originated on the bases of structure of these spikes as some of them were similar to solar corona [7]. After almost four decades, a novel pathogen with similar viron morphology appeared to harm human respiratory system. It was the lethal and disastrous outbreak of hazardous SARS-CoV causing severe acute respiratory syndrome (SARS) in the year of 2002–2003 [8]. Its sudden and lethal, emergence and spread led to massive eruption of research towards primary and better understanding of CoV's replication mechanism. Several approaches and targets were investigated to control and prophylaxis the lethal virus. CoV's moves into the host cell via interactions among receptors of host cell and S-protein of virus species [9]. Different species of CoV's involve terminus at S1 site of receptor binding domains like SARS-CoV's involve N-terminus and some others involve C-terminus. MERS-CoV enters the host cell via DPP4 (dipeptidyl-peptidase 4) and CEACAM1 (CEA cell adhesion molecule 1) whereas HCoV-NL63 and SARS use ACE2 (angiotensin-converting enzyme 2) [10]. Thereafter, S-protein undergoes cleavage (acid-dependent proteolytic) through serine 2 (TMPRRS2), transmembrane protease, cathepsin, or another protease. Cleavage is followed by fusion at plasma membrane or acidified endosomes within the cell cytol of host. The cleavage of S20 causes an antiparallel six-helix bundle which permits the collaboration of cellular and viral membranes and leads to the discharge of viral genome into host cell [11]. This review aimed to collect and summarize the published data on coronaviruses and the role of natural products against them.
2. Classification of coronaviruses
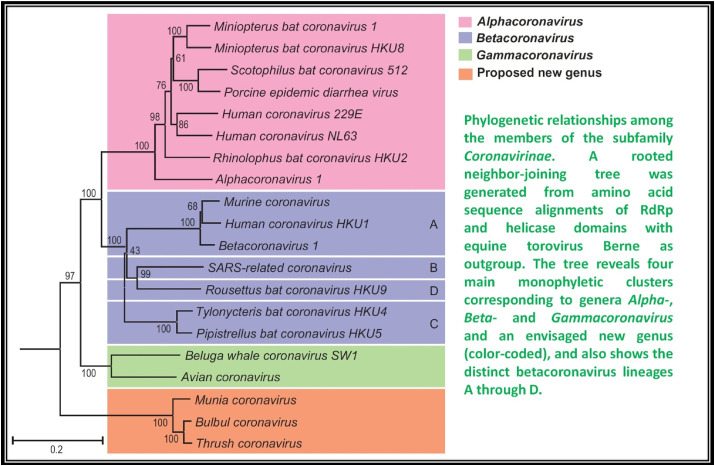
Coronaviruses have been listed under a different sub-family (coronavirinae) of family Coronaviridae [12]. The members of the family Coronaviridae, a monophyletic cluster in the order Nidovirales, are enveloped, positive stranded RNA viruses of three classes of vertebrates: fish (bafiniviruses), birds (coronaviruses) and mammals (toro– and coronaviruses) [13]. On the study of unrooted and rooted phylogenetic trees projected for variant genomic regions, four different CoV clusters are eminent. Three clusters correspond to already found unofficial groups 1, 2 and 3 that are now classified officially as genera, Alpha-, Beta- and Gamma-CoV's, respectively [14]. The fourth cluster is different from the farmer three and found in birds. By all the standards this fourth cluster has been listed under a different genus (unofficial, yet to be approved) tentatively termed as “Delta- CoV's”. Moreover, in case of Beta- CoV's further four divergent linages can be distinguished, assigned as A through D [15], that correspond to former subgroups 2A through D, respectively (Fig. 1 ).
Fig. 1.
Phylogenetic relation among the members of the Coronavirinae [15].
3. Distinctive characteristic features of coronaviruses
Across Coronavirinae, virons are spherical in shape and a size of 120–160 nm, bacilliform, 170–200 × 75–88 nm (Bafinivirus) or both, having a characteristically bent bacilliform into crescents (Torovirus) [16]. These particles are decked with typically large, petal- or club-shaped surface projections (spikes), creating an image of solar corona under electron microscopy. The CoV's bear helical nucleocapsids and can been released from virion on the application of detergents [17]. The nucleocapsids of coronavirus appear to be loosely-wound and in Torovirinae are characteristically tubular. Coronoviridae are the large size RNA viruses discovered so far in terms of genetic complexity and genome size, except rivaled by okaviruses (belonging to the family of Roniviridae, okaviruses are large invertebrate nidoviruses) [18].
The replication mechanism in CoV's has been investigated in detail, whereas there is limited data on replication of toroviruses and bafiniviruses, this may be of similar replication mechanism followed by these. Virions enter into the target host cell via surface receptors and their spikes (Table 1 ). These virions discharge their genome into host cell by fusion of plasma membrane or endocytic vesicles with viral envelope [19]. The complete replication procedure operates within cytoplasm of host cell. It involves the formation of sub-genome sized and full-length minus-strand RNA intermediates with the viral genome serving as both mRNA for template for minus-strand synthesis and replicase polyproteins [20]. Repilication-transcription complex (poorly characterized) catalyzes the RNA synthesis, comprising of host and viral proteins associated with interconnected modified network of double and intracellular membrane vesicles. These vesicles are ostensibly derived of endoplasmic reticulum [[21], [22], [23]].
Table 1.
Coronavirus hosts with key binding receptors and factors.
| Virus species | Host | Attachment factor | Main receptor |
|---|---|---|---|
| Alphacoronavirus 1 | |||
| Canine coronavirus type I | Dog | Unknown | Unclear |
| Canine coronavirus type II | Dog | Unknown | APN |
| Feline coronavirus type I | Cat | Unknown | Unclear |
| Feline coronavirus type II | Cat | Unknown | APN |
| Transmissible gastroenteritis virus | Pig | Salic acid | APN |
| Betacoronavirus 1 | |||
| Bovine coronavirus | Cow | Unknown | 9-O-Ac Sia? |
| Equine coronavirus | Horse | Unknown | 9-O-Ac Sia? |
| Human coronavirus OC43 | Human | Unknown | 9-O-Ac Sia? |
| Porcine hemagglutinating encephalomyelitis virus | Pig | Unknown | 9-O-Ac Sia? |
| Human coronavirus 229E | Human | Unknown | APN |
| Human coronavirus NL63 | Human | Unknown | ACE2 |
| Murine coronavirus | Mouse | 4-O- or 9-O-Ac Sia | CEACAM1a |
| SARS-related coronavirus | Human | Unknown | ACE2 |
Abbreviations: APN = aminopeptidase N, ACE2 = angiotensin-converting enzyme 2, CEACAM1a = carcinoembryonic antigen adhesion molecule 1.
In addition to this all coronoviridae members share the following characteristics;
-
i.
Genome: positive sense RNA, unimolecular, linear, infectious, capped, polyadenylated, 26–32 kb in length, and polycistronic morphology.
-
ii.
General genomic organization: 5′-UTR-replicase-S-M-N-UTR-3' (genes according to product), with the genome functioning as mRNA for the replicase gene.
-
iii.
Replicase gene: it constitutes of overlapping ORFs (1a and 1b) that code for the production of giant polyproteins (pp1a and pp1ab), these giant proteins are generated autoproteolytically and generation of pp1ab needs programmed 21 ribosomal frame shift.
-
iv.
ORFs downstream of the replicase gene: expression from a 3′ co-terminal nested set of two or more subgenomic mRNAs that are capped and polyadenylated.
-
v
Morphogenesis: virion assembly through budding of preformed nucleocapsids at smooth intracellular membranes of endoplasmic reticulum/early Golgi compartments.
4. Biological role and health disorders caused by coronaviruses
CoV's incorporate pathogens of veterinary, clinical and economic interest, which infect mammals and birds [24]. Viral transmission is species-specific and may occur through fecal-oral or aerogenic and/or fomites. Epithelial cells are the primary targets of CoV's and hence they generally cause respiratory and gastrointestinal infections [25]. These infections may be mild, acute or chronic depends upon the virus species and prolonged infections. In most of the cases, the infections raised by CoV's are generally mild and asymptotic [26]. However, certain CoV's like SARS- CoV-1 and SARS-CoV-2, cause lethal health disorders and even death. The epidemic of 2002–2003 and the ongoing global pandemic both are the outbreaks of coronaviruses, SARS- CoV-1 and SARS-CoV-2, respectively [27]. Murine coronavirus belongs to the genus Beta-CoV's could raise hazardous neurological and hepatic disorders resulting in demyelination and body paralysis [28]. Alpha-CoV's including ferret, canine and feline CoV's, may cause severe immune-mediated systematic infections in hosts. These infections are presumed of infections to monocyte/macrophage linage and generate inflammatory lesions within multiple organs. Farmer discovered human CoV's including Alpha-CoV HCoV-229E and Beta-Cov-1 subspecies HCoV-OC43, were considered least hazardous causing common cold and were considered of meek clinical value. CoV's are now identified with adverse health effects like they cause severe LRTI (lower respiratory tract infections) in elderly and infants [29]. Globally, these CoV's are responsible for hospitalization of 5% of infants from LRTI.
In 2002–2003, SARS-CoV-1 (unknown then) caused a serious epidemic of severe pulmonary disorder in human populations which spread to four continents [30]. 8096 persons got infected and it claimed life of 774 victims, creating a 10% mortality rate [31]. On epidemiological surveillance of the disease it was demonstrated that this novel coronavirus originated from bats, spread to raccoon dogs, Chinese ferret badgers and Himalayan palm civets at Guangdong wet markets, China [32]. Later the virus moved into human population via consumption or handling of these infected species. Although, SARS was believed to be vanished till the latest casualties done by SARS-CoV-2, the episode highlighted the pathogenic potency of CoV's and their probability to emerge as novel coronavirus infections through transmission among cross-species. Various similar episodes were observed thereafter though they imposed miniscule epidemiological outcomes like OC43 human coronavirus (cattle to humans, through single cross transmission of bovine coronavirus), 229E human coronavirus (bats origin?) and canine respiratory coronavirus (bovine coronavirus transmission to dogs) [33]. The SARS epidemic has enhanced the research interest in the field of CoV's. Virus discovery and molecular surveillance have found nearly 60 novel CoV's, out of which two effect human respiratory system including HCoV-NL63 and HCoV-HKU1 [34]. The farmer is key agent of causing bronchiolitis and croup among children. Additionally, these studies showed novel lineage of mostly avian viruses including Munia, Bulbul and Thrush coronavirus with close relatives possibly found in mammals like Chinese ferret badger and Asian leopard cat [35]. These novel avian viruses were placed under a different genius (Delta-CoV's) on the basis of rooted phylogeny (Fig. 1). Bats are exceptionally rich harbors of variety of coronaviruses and hence play a key role in coronavirus evolution and ecology [36]. It has been reported that bats may be the original hosts from which lineages for most of the alpha-CoV's and Beta-CoV's were derived. The habitual features of bats like roosting, migration and population densities favor their virus carrier role [37]. This hypothesis has its merits and in recent studies of virus discovery this hypothesis remained truly Herculean proportions. The actual sampling size of the CoV's is limited though the efforts are primarily concentrated on bats, all of our current insights may be biased.
4.1. Novel coronavirus COVID-19: Epidemiology
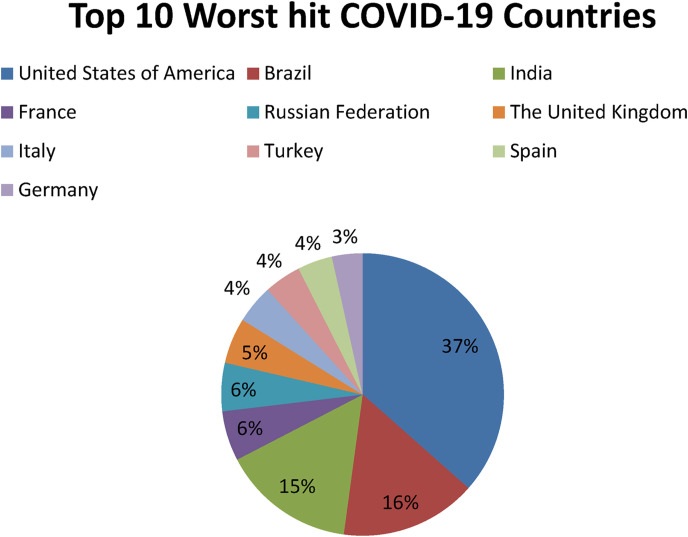
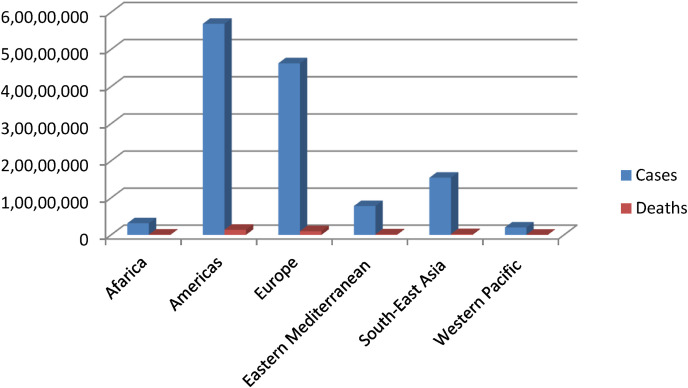
The ongoing lethal pandemic of COVID-19, primarily caused by coronavirus SARSCoV-2, originated from Chinese city of Wuhan, in December 2019 [38]. As the month of December is considered among the months of global festival, this lethal infectious virus got freehand and spread at very high frequency. According to World Health Organization, till April 6, 2021 more than 131,487,572 cases and over 2,857,702 deaths have been reported due SARS-CoV-2 infection globally since the onset of the pandemic (Fig. 3 ). Africa 3,132,143 cases and 78,763 deaths, America 56,880,123 cases, 1,375,139 deaths, Eastern Mediterranean 7,785,717 cases and 161,826 deaths, Europe 46,256,984 cases and 986,342 deaths, South-East Asia 15,438,907 cases and 223,666 deaths and Western Pacific 1,992,953 cases and 31,953 deaths (Fig. 2) [39]. Some of the worst hit countries by SARSCoV-2 have been listed in Table 2 ; America tops the list followed by Brazil and India (Fig. 3). SARSCoV-2 has a minimum incubation period of two weeks or may be longer in some cases. In early stages SARSCoV-2 mostly may remain asymptotic and can cause multiple infections in some patients before becoming apparent [40].
Fig. 3.
Pi-chart representation of top 10 worst hit countries by COVID-19, Situation Report; April 6, 2021.
Fig. 2.
The number of COVID-19 related cases and deaths reported globally, till April 6, 2021, according to WHO data (Coronavirus disease (COVID-19)), Situation Report April 6, 2021.
Table 2.
Some of the worst hit COVID-19 countries in the world, according to WHO data (Coronavirus disease (COVID-19)), Situation Report; April 6, 2021.
| Countries | Number of Cases | Number of Deaths |
|---|---|---|
| United States of America | 30,412,206 | 551,769 |
| Brazil | 12,984,956 | 331,433 |
| India | 12,686,049 | 165,547 |
| France | 4,757,381 | 96,260 |
| Russian Federation | 4,597,868 | 101,106 |
| The United Kingdom | 4,362,154 | 126,862 |
| Italy | 3,678,944 | 111,326 |
| Turkey | 3,529,601 | 32,456 |
| Spain | 3,311,325 | 75,783 |
| Germany | 2,900,768 | 77,103 |
| Poland | 2,456,709 | 55,065 |
| Colombia | 2,446,219 | 64,094 |
| Argentina | 2,393,492 | 56,199 |
| Mexico | 2,250,458 | 204,147 |
| Iran (Islamic Republic of) | 1,945,964 | 63,332 |
| Ukraine | 1,769,164 | 35,017 |
| Peru | 1,582,367 | 52,877 |
| Czechia | 1,555,245 | 27,169 |
| South Africa | 1,552,416 | 52,995 |
| Indonesia | 1, 537,967 | 41,815 |
| Netherlands | 1,307,466 | 16,629 |
The organs adversely affected by SARS-CoV-2 include lungs, kidneys, heart, liver and genitals [41]. Patients with severe COVID-19 infection have been reported in a retrospective investigation to mostly develop ARDS (acute respiratory distress syndrome), abnormal liver functioning, acute cardiac and kidney injury in the proportions of 67.3%, 28.9%, 23.1% and 28.9%, respectively [41]. A mortality rate of 61.5% was observed in patients developing severe COVID-19 infection within four weeks. A great challenge to the medical staff workers not due to the nature of work in ICU's but becoming more vulnerable to the virus and ultimately developing infection. Higher number of medical staff workers has already fallen prey to this deadly virus. SARS-CoV-2 belongs to β-coronaviruses with most likely of animal origin. Recent investigations have demonstrated that SARS-CoV-2 may be a pathogen found in pangolins, snakes and bats. However, on genomic studies it has been revealed that SARS-CoV-2 originated from bats. Genome sequencing has showed that there is a 96% homology between bat coronaviruses and SARS-CoV-2 yet the intermediate host remains unclear [42].
4.2. Transmission
SARS-CoV-2 is transmitted in a number of ways, one of the primary sources is at present hosts and hosts with asymptotic disease [43]. Close contact and respiratory droplets are the key routes of transmission, and it is better to avoid crowding so that asymptotic transmission may be stopped. Studies have marked the presence of SARS-CoV-2 in the air of ICU's and prolonged exposure to closed ICU rooms can enhance vulnerability and aerosolic transmission [44]. Moreover, the SARS-CoV-2 has been detected in tears, saliva, urine, and gastrointestinal tract of patients [45]. The diagnosis of a 72 h child positive with COVID-19 has enhanced the possibility of vertical transmission from mother to fetus. Therefore, much higher precautions and preventive measures should be adopted by the medical staff inside hospitals and ICU's as well.
4.3. Pathogenesis
Till date, the mechanism of pathogenesis of SARS-CoV-2 is uncertain and unknown. It is believed that the ACE2 receptor present in alveolar epithelial cells provides passage to the virus by binding to its spike protein via S protein [46,47]. Once bound to these epithelial cells strong immune responses and direct toxicity gets induced. The resultant inflammation leads to strong cytokine storm generating lethal lung injury which causes lung collapse and ultimately death [48]. Pathological studies have demonstrated that SARS-CoV-2 patients develop hyaline membrane and possess diffused alveolar damage. The rest of the features resemble to that of MERS and SARS [49]. ACE2 is also found in epithelial cells of different organs including intestines, heart, and kidneys. Therefore, once SARS-CoV-2 infection develops it can invade to multi-organs and cause multi-organ failure. Tumor necrosis factor, macrophage inflammatory protein 1α, monocyte chemo-attractant protein-1, interferon gamma-induced protein 10 and levels of IL-2, -6, -7 and -10 granulocyte colony-stimulation factor, all are found highly elevated in case of severe infection and are considered accountable for poor outcomes [50]. In some patients enhancement in pro-inflammatory CCR4+CCR6+Th17 cells and over activation of lymphocytes promote the immune-mediated injury resulting in transformation of mild infection to severe [51]. Predominantly, individuals with reduced immunity and comorbidities are more vulnerable to this infection.
4.4. Diagnosis
Based on recent investigations and data collected from ICU's, COVID-19 patients develop hypoxemia and/or dyspnea within one week of disease onset. Patients with higher viral loads develop even serious complications including acute respiratory distress syndrome, multiple organ dysfunction syndrome, coagulation disorder, metabolic acidosis and septic shock [52]. Patients with SARS-CoV-2 infection may develop cerebral congestion, edema and neuropathy; attention should be paid to neurological symptoms in clinical practice. Some patients have been reported of initial neurological symptoms including myalgia, impaired consciousness, anosmia, headache, and acute cerebrovascular distortion. In future studies, these neurological complexities and their therapeutic modalities should be elucidated. Secondary SARS-CoV-2 infection markers include downregulation of peripheral blood lymphocytes, amplification of blood inflammatory agents like C-reactive protein and IL-6, amplified lactic acid levels and scanning results predicting multilobar or bilateral infiltration, short-term lesions and pleural effusion [53]. Stimulatingly, some researchers found that the neutrophil-to-lymphocyte ratio (NLR) is an impelling factor that can be used for initial identification of the prognosis of patients with severe COVID-19. Lastly, it should be pointed out that chest CT plays particularly a decisive role in COVID-19 diagnosis and the disease severity assessment. Chest CT has high diagnostic value in patients who have negative Reverse Transcription-Polymerase Chain Reaction (RT-PCR) results but whose clinical symptoms, auxiliary test results, and epidemiological history make them highly suspected patients [54].
Polymerase chain reaction (PCR) method is considered as the gold standard for disease diagnostics; however, it requires expensive equipment and knowledgeable manpower.
5. Natural products with promising antiviral potential against coronaviruses
Natural products are a rich and valuable gift of Mother Nature to human and other beings on this planet [55]. Since time immemorial, natural products, their derivate and their synthetic analogs have been used to overcome many human ailments including viral infections, ranging from mild to severe. Herbal medicine possesses an enormous scope in nutraceutical market. Most interestingly, the availability, acceptability and rich biological properties have diverted the interest of researchers from synthetic to nature [56]. Nigella sativa has been reported with substantial suppressive effects against the hepatitis C virus. Several antiviral natural products induce their antiviral effects via targeting the replication mechanism of virus [57]. Moreover, phytochemicals, marine natural products, and biotechnologically synthesized natural products and analogs have been reported to potentially target different infectious viruses. Natural products acts as a pool of chemical compounds explored and developed as medicine against different diseases and pathogens including antiviral, on daily bases [58]. Till date, there are numerous herbal medicines or their constituting chemical agents which have shown promising antiviral properties. However, there is a scarcity of appropriate research for the development of anti-coronavirus drug linked to natural products. Those potential agents may not only help us to overcome coronavirus but also prevents any future viral attack. In humans beings CoV's predominantly result in common cold but complications like SARS and pneumonia can also occur in some cases [59]. The well-known human effecting CoV's include HCoV-HKU1, -NL63, -OC43, -229E and more widely known SARS-CoV-1 and SARS-CoV-2 which caused epidemic in 2002–2003 and ongoing global pandemic, respectively [60]. In addition to this, in 2012 WHO had signified another type of human coronavirus which resulted in MERS (Middle East Respiratory syndrome) and high mortality [61]. There is no 100% vaccine for CoV's infection and efforts are still on to find potential and preventive treatments. Thus there is an immediate need for potential antiviral drugs that can overcome coronavirus infections in human beings.
China is extensively using the Traditional Chinese Medicine (TCM) alongside Western medicine against SARS-CoV-2 in accordance with the COVID-19 treatment guidelines issued by National Health Commission (People's Republic of China) [62]. The applications of plant based treatments against SARS-CoV-2 are a major research pool which led to the screening of 125 Chinese herbal medicines against COVID-19 with a direct potential of inhibition [63].
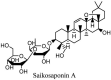
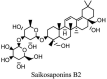
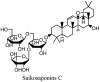
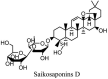
Saikosaponin-A, -B2, C and D have been reported to induce substantial antiviral effects against HCoV-22E9 [64]. These are the naturally occurring triterpene glycosides found and extracted from a number of medicinal plants including Scrophularia scorodonia, Heteromorpha spp. and Bupleurum spp [65]. Siakosaponins effectively targeted the viral attachment and entrance in host thereby preventing primary stage HCoV-22E9 infection. Different extracts of Lindera aggregate, Pyrrosia lingua, Artemisia annua and Lycoris radiata have been reported with substantial antiviral effects against SARS-CoV out of hundreds of Chinese medicinal herbs considered for screening [66]. Different natural inhibitors like scutellarein, myrictin and phenolic compound from Torreya nucifera and Isatis indigotica, of SARS-CoV enzymes such has 3CL protease and nsP13 helicase have shown potential and better results. Other natural medicines for CoV's include the water extract of Houttuynia cordata [10]. This water extract has been reported to induce antiviral effects via variant mechanisms including 3CL protease inhibition and inhibition of RNA polymerase activity. Different plant extracts with their modes of action against different CoV's have been listed in Table 3 .
Table 3.
Plant extracts with anti-CoV potential [10].
| S.no. | Plant extracts | Virus system | Concentration | Targets | IC50/EC50 |
|---|---|---|---|---|---|
| 1. |
Anthemis hyalina, Nigella sativa, and Citrus sinensis extracts |
Coronavirus-infected HeLa-epithelial carcinoembryonic antigen-related cell adhesion molecule 1a cells inoculated with MHV-A59 (mouse hepatitis virus–A59) | 1/50 and 1/100 dilution of ethanolic extract (100 g/200 mL) |
Increased IL-8 level. Significantly changed the expression of TRPA1, TRPC4, TRPM6, TRPM7, TRPM8, and TRPV4 genes. |
Not defined |
| 2. | Herbal extracts (Gentiana scabra, Dioscorea batatas, Cassia tora, Taxillus chinensis, Cibotium barometz) |
SARS-CoV | 25–200 μg/mL | 3CL protease inhibition. | 39 μg/mL and 44 μg/mL (two extracts of Cibotium barometz) |
| 3. | Houttuynia cordata aq. Extract | SARS-CoV | 0–400 μg/mL | 3CL protease and viral Polymerase inhibition. | Not defined |
| 4. | Extract (Rheum officinale Baill., Polygonum multiflorum Thunb.) |
SARS-CoV S spike protein | 0–100 μg/mL | Inhibits the interaction of SARS-CoV S protein and ACE2. |
1–10 μg/mL |
| 5. | Isatis indigotica | SARS-CoV | 1–500 μg/mL | 3CL protease inhibition. | Not defined |
| 6. | Lindera aggregata | SARS-CoV | 10−1–10−4 mg/mL | Unknown | 88.2 ± 7.7 μg/mL |
| 7. | Pyrrosia lingua | SARS-CoV | 10−1–10−4 mg/mL | Unknown | 43.2 ± 14.1 μg/mL |
| 8. | Artemisia annua | SARS-CoV | 10−1–10−4 mg/mL | Unknown | 34.5 ± 2.6 μg/mL |
| 9. | Lycoris radiata | SARS-CoV | 10−1–10−4 mg/mL | Unknown | 2.4 ± 0.2 μg/mL |
6. Compounds of natural origin showing antiviral potency against coronaviruses
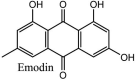
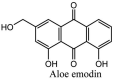
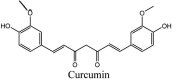
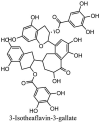
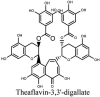
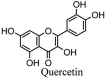
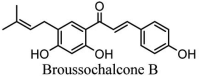
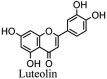
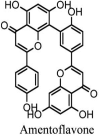
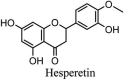
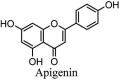
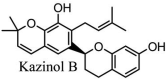
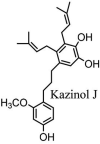
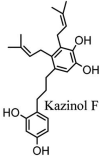
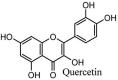
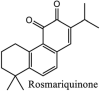
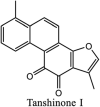
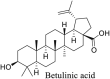
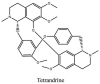
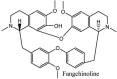
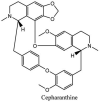
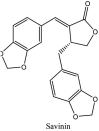
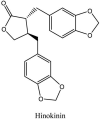
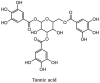
Torreya nucifera-a medicinal plant, is a rich source of flavones and biflavones [67]. These compounds showed potential antiviral activity against SARS-CoV. Out of these compounds, Quercetin, luteolin, apigenin and amentoflavone presented an IC50 value of 23.8, 20.2, 280.8 and 8.3 μM. Three alkaloids including cepharanthine, fangchinoline and tetrandrine, in a recent investigation were shown with inhibitory effects against early stage death stimulated in HCoV-OC43-infected human lung MRC-5 cells with IC50 values of 0.83, 1.01 and 0.33 μM, respectively [68]. The antiviral potency of natural products has inspired many researchers, 221 phytochemicals belonging to different classes were investigated for antiviral propensity against SARS-CoV's [69]. Out of 221, curcumin, five lignans, two triterpenes, two sesquiterpenes and ten diterpenes revealed anti-SARS-CoV activity. Further, savinin lignin and 8b-hydroxyabieta-9(11),-13-dien-12-one diterpenoid were reported with inhibitory effects on 3CLpro activity in SARS-CoV with SI of greated than 667 [69]. Betulinic acid imposed inhibition of 3CLpro activity and was competitive to savinin against SARS-CoV with Ki value 8.2 μM and 9.1 μM, respectively [69]. Tanhinone I and rosmariquinone are the two tanshinones isolated from Salvia miltiorrhiza showed inhibition of replication, PLpro and 3CLpro infections caused by SARS-CoV [70]. Three phenolic compounds; theaflain-3,30-digallate, 3-isotheaflavin-3-gallate and tannic acid, isolated from black tea were reported with inhibition of SARS-CoV 3CLpro. Furthermore, a number of polyphenolic compounds from Isatis indigotica; β-sitosterol, hesperetin, aloe emodin, indigo and singigrin, induced inhibition of SARS-CoV 3CL pro [71]. Research work performed by Yu et al., suggested that scutellarein and myricetin exhibited anti-SARS-CoV 3CLpro activity [72]. Similarly, compounds isolated from Broussonetia papyrifera including kazinol J, kazinol F, broussoflavan A, kanizol B, 30-(3-methylbut-2-enyl)-30,4,7-trihydroxyflavane, papyriflavonol, 4-hydroxyisolonchocarpin, broussochalcone A and broussochalcone B, all displayed inhibitory effects against SARS-CoV PLpro and 3CLpro [73]. Out of these compounds highest inhibition was exerted by papyriflavonol A with an IC50 of 3.7 μM.
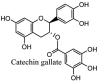
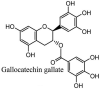
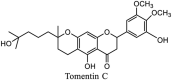
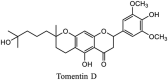
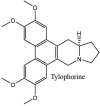
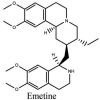
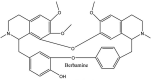
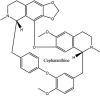
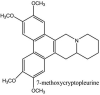
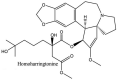
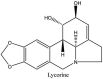
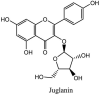
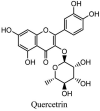
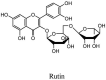
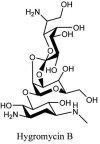
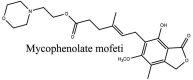
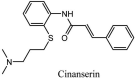
Streptomyces hygroscopicus is rich in hygromycin and showed anti-replicative effects in necrotic liver foci and MHV-A59 (mouse hepatitis virus) in a concentration-reliant fashion [74]. Bacterium Streptomyces parvulus antibiotic, actinomycin D suppressed the penetration and attachment of CoV's into host cell. Likewise, ginsenoside Rb1 one of the active members of ginsenosides isolated from Panax ginseng showed remarkable antiviral activity [75]. NIH clinical collection investigated 727 compounds for antiviral activities against human and murine CoV's, the most active compound showing promising activity was recorded to be macetaxine alkaloid [76]. Tylophora indica is rich in alkaloid content and two alkaloids; 7-methoxycryptopleurine and tylophorine, showed potential inhibition of both S and N protein activity and also inhibited the replication of enteropathogenic coronavirus transmissible gastroenteritis virus [77]. Cepharanthine has been reported to repress the protease enzyme activity in SARS-CoV and recent work performed by Shen et al. showed berbamine substantially inhibited the HCoV-NL63 [78]. The compounds mycophenolate mofetil, emetine and lycorine showed antiviral activities against different human CoV's including MHV-A59, MERS-CoV, HCoV-NL63 and HCoV-OC43. Moreover, emetine and lycorine were observed to arrest cell division, DNA, RNA and protein synthesis whereas mycophenolate mofetil induced immune suppressive effects on mentioned CoV's. Juglanin molecule has been reported with blocking effects on 3a channel in SARS-CoV. In another study, a number of compounds isolated from Paulowinia tomentosa including 6-geranyl-40,- 5,7-trihydroxy-30,50-dimethoxyflavanone, diplacone, mimulone, 40-Omethyldiplacone, 30-O-methyldiplacone, tomentin A, B, C, D, and E, 40-O-methyldiplacol and 30-Omethyldiplacol were shown with antiviral effects against SARS-CoV PLpro. Likewise, (−)-gallocatechin gallate and (−)-catechin gallate repressed nanoparticle based RNA oligonucleotide against SARS-CoV. On the other hand, quercetin, quercetrin, rutin, cinanserin (1 and 2 dpi) isolated from Houttuynia cordata were found to act against murine CoV at 15.63–500 μg/mL [[79], [80], [81]].
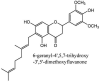
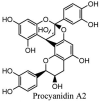
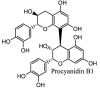
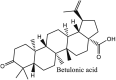
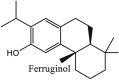
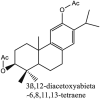
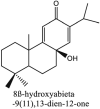
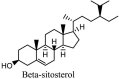
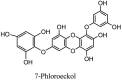
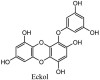
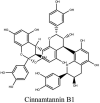
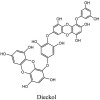
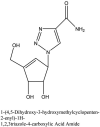
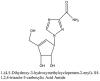
Furthermore, 3β,12-diacetoxyabieta-6,8,11,13-tetraene, 8β-hydroxyabieta-9(11),- 13-dien-12-one, ferruginol, betulinic acid, curcumin, savinin, and hinokinin inhibited the replication of SARS-CoV [82]. On the other hand, viral yields as well as viral titers were diminished by ouabain molecule and also declined the viral RNA copies in number. Similarly, the laboratory derivatives; 1-(4,5-dihydroxy-3-hydroxymethylcyclopenten-2-enyl)-1H-1,2,3-triazole-4 carboxylic acid amide, 1-(4,5-dihydroxy-3-hydroxymethylcyclopenten-2-enyl)-1Himidazole-4-carboxylic acid amide, and 1-(4,5-dihydroxy-3-hydroxymethylcyclopenten-2-enyl)-1H-1,2,4-triazole-3-carboxylic acid amide revealed remarkable anti-SARS-CoV potential [83]. Tylophora indica isolated 7-methoxycryptopleurine and tylophorine showed evident inhibition of replication against CoV-infected swine testicular cells [84]. In this study, 7-methoxycryptopleurine (IC50: 20 μM) was extra operative than the tylophorine (IC50: 58 μM). In another study, tylophorine was also found to target viral RNA replication and cellular JAK2-mediated dominant NF-κB activation in CoV at 0–1000 μM [85]. Structure of some of the important antiviral leads from plants against coronaviruses have been listed in Table 4 .
Table 4.
Molecular structures of some plant based antiviral leads against coronaviruses [10].
| Phenolic compounds | ||
 |
 |
 |
 |
 |
 |
| Flavonoids and Chalcones | ||
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
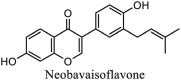 |
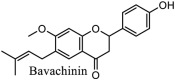 |
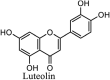 |
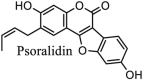 |
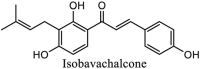 |
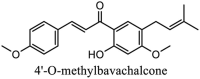 |
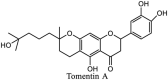 |
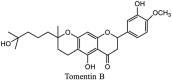 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
| Alkaloids | ||
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
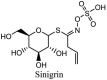
| Glycosides | ||
 |
 |
 |
 |
 |
 |
 |
 |
 |
| Steroids | ||
 |
||
| Lignans | ||
 |
 |
|
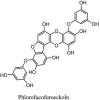
| Tannins | ||
 |
 |
 |
 |
 |
 |
| Miscellaneous | ||
 |
 |
 |
 |
 |
 |
7. Virus-host interactions and mechanism of antiviral action of natural products
Infectious diseases from time to time have caused serious damages to humanity throughout the world. Viruses are one of the serious agents posing infectious diseases in human beings and have attained resistance to prophylaxis or therapy long before any form of life mainly due to their sole dependence upon the host cells they infect and depend upon for survival and replication. This property of theirs has caused serious issues in drug development and chemotherapy. There are only a handful of antiviral agents available to act upon these infectious diseases caused by viruses included natural product based acyclovir. In order to overcome viruses causing devastating and serious health damages to not only humans but insects, animals, plants, bacteria and fungi as well, world is turning towards natural products for antiviral leads sue to diversity in structure and bioactivities. Although the search for naturally occurring products which can interfere with viral infections began with the successful isolation of antibiotics from microorganisms but it has not been as intensive as that of synthetic antiviral agents [86]. This is mainly due to the tendency of most virologists who adopt a rational design of antiviral agents rather than toward empiricism especially with the progress in knowledge of viral replication [87]. Additionally, there are few errors in the screening of crude extracts along with isolations, identifications and purifications of antiviral agents from plant sources. But the recent advancements in the chromatographic and spectroscopic techniques have led down these shortcomings. Some of the antiviral synthatics as well as of natural origin have showed promising antiviral effects in vitro but on in vivo investigations they were found substandard. This was probably thought of hurdles in transportation of these antiviral agents to the sites of infections especially in case were the infected tissue develops inflammation.
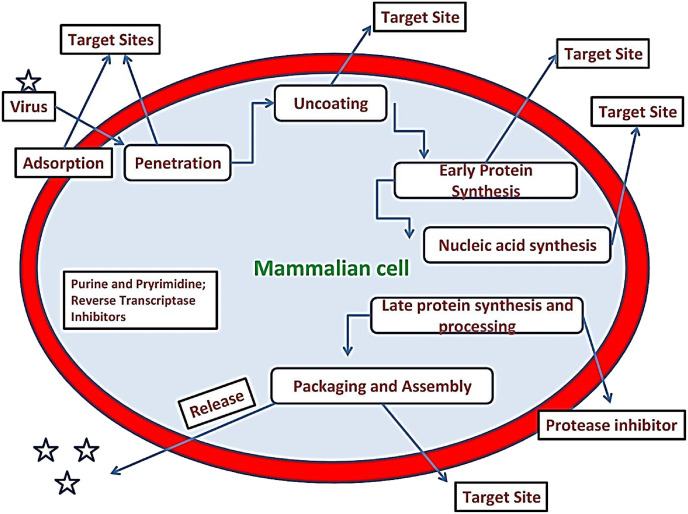
The virus-host interactions begin with the adsorption of virus to the host membrane from where a series of events take place which decide the fate of virus replication as well as drug targets. Viruses mostly replicate by following a temperate or lytic pathway. In a lytic pathway a fast process gets operated in which virus is quickly released as host cell witnesses rupture, lysis and death due to lost content. While in a temperate pathway, virus doesn't kill host cell in fact goes through a period of latency where genetic material of viruses remain inactive inside its host. In some cases of latency the viral genes become integrated into the host's DNA, replicated along with it and passed along to all daughter cells. In time, damage to the DNA or some other event may activate transcription of the viral genes therefore new viral particles can be produced and infected cells are destroyed [6]. Some of the events of virus-host interaction with possible targets are revealed in Fig. 4 . The interaction of virus and host going through a series of events; (a) Attachment (adsorption) (b) Penetration into host (c) Uncoating, release and transport of viral nucleic acid and core proteins (d) Nucleic acid polymerase activation (e) Translation of m-RNA to polypeptides (f) Transcription of m-RNA (g) Replication of nucleic acids (h) Protein synthesis (i) Viral polypeptides cleavage into useful polypeptides (j) Morphogenesis and assemblage of viral capsids and precursors (k) Encapsidation of nucleic acid (l) Envelopment (M) Release.
Fig. 4.
The virus-host relation and possible target sites during pathogenesis.
Natural products show significant potency to act upon different target sites in viral-host association. The different attachment sites for antiviral action of natural products in virus-host association have been shown in Fig. 5 .
Fig. 5.
General mechanism for antiviral activity of different molecular scaffolds from nature.
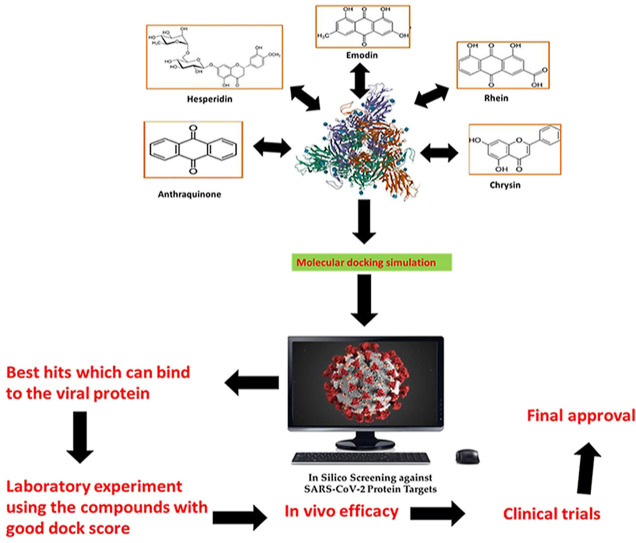
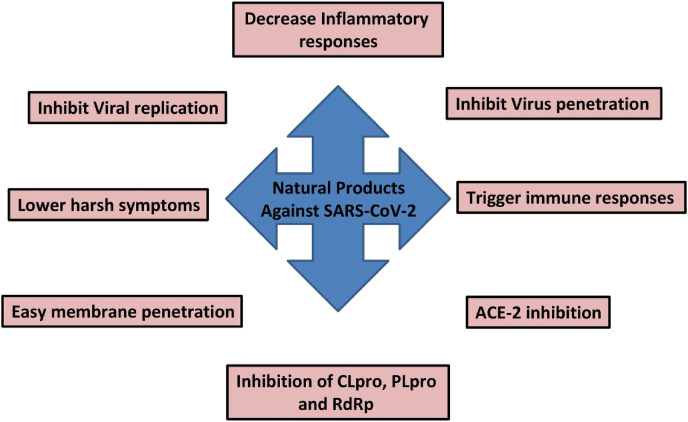
8. Molecular scaffolds from Mother Nature with possible anti-SARS-CoV-2 potential
The development of natural products based drugs against a specific health disorder is more rapid than to develop a potential vaccine. Still, it remains a vertical task due to the enormous structural and chemical varieties of natural metabolites, their chemical extraction and complexity. To save the time involved in natural products research, virtual screening proves to be advantageous for phytochemical screening of natural products and their extracts. This virtual screening is termed as in silico analysis via molecular docking. Since the appearance of COVID-19 as a global pandemic, natural products were subjected to molecular docking to check their role against it. Independent of the class, natural products like steroids, terpenes, quinones, fatty acids, alkaloids, flavonols and flavones revealed similar docking score or binding energy, to that of repurposed drugs like chloroquine and remdesivir, with replicative proteins is SARS-CoV-2 (Fig. 6 ) including TMPRSS2, 3CLpro and ACE2 [88,89]. More emphasis was laid on ACE2 inhibitors as a probable consequence of primary binding concerning COVID-19 replication. Some of the natural products subjected to virtual screening against ACE2 and 3CL Pro docking targets are listed in Table 5, Table 6 .
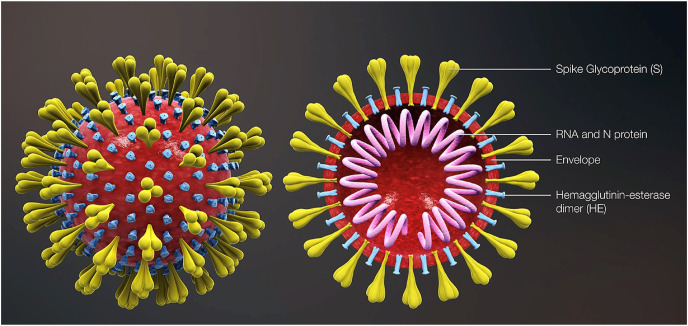
Fig. 6.
Diagramatic representation of the virion structure exhibiting typical spikes of the virus that are reminiscent of solar corona hence the name “corona”. (Source: https://commons.wikimedia.org/wiki/File:3D_medical_animation_corona_virus.jpg).
Table 5.
Phytochemicals screened against ACE2 as docking target and their binding energy. “x” represents the repurposed drugs used for reference.
| S.no. | Phytochemical | Binding energy (Kcal. Mol−1) | Reference |
|---|---|---|---|
| 1. | Zhebeininoside | −6.8 | [90] |
| 2. | Verdine | −6.6 | [90] |
| 3. | Pseudojervine | −6.8 | [90] |
| 4. | Hupehemonside | −7.1 | [90] |
| 5. | Nobiletin | −5.42 | [91] |
| 6. | Neohesperidin | −3.78 | [91] |
| 7. | Hesperetin | −6.09 | [91] |
| 8. | Hesperidin | −4.21 | [91] |
| 9. | Naringenin | −6.05 | [91] |
| 10. | Narigin | −6.85 | [91] |
| 11. | Chloroquinex | −8.01 | [92] |
| 12. | Philligenin | −7.80 | [92] |
| 13. | Hinokinin | −7.11 | [92] |
| 14. | Withaferin A | −9.63 | [92] |
| 15. | Quercetin | −8.66 | [92] |
| 16. | Isoaloresin | −7.83 | [92] |
| 17. | Aloin | −8.38 | [92] |
| 18. | Taiwanhomoflavone A | −7.60 | [93] |
| 19. | Epicatechin-(4β,8)-epicatechin-(4β,6)-catechin | −8.20 | [93] |
| 20. | Epicatechin-4-epigallocatechin | −7.20 | [93] |
| 21. | Quercetin 3-glucosyl-(1,4)-rhamnoside | −6.50 | [93] |
| 22. | Lactucopicrin 15-oxalate | −8.30 | [93] |
| 23. | Lactucopicrin | −8.30 | [93] |
| 24. | Vitetrifolin D | −7.30 | [93] |
| 25. | Myricitrin | −7.10 | [93] |
| 26. | Apigenin | −7.10 | [93] |
| 27. | Kaempferol | −7.20 | [93] |
| 28. | (−)-Asperlicin C | −9.50 | [93] |
| 29. | Cassameridin | −8.10 | [93] |
| 30. | Oriciacridone F | −6.70 | [93] |
| 31. | Remdesivirx | −7.80 | [93] |
| 32. | Afzelin | −7.10 | [93] |
| 33. | Isoquercitrin | −7.80 | [93] |
| 34. | Silybin | −10.5 | [92] |
| 35. | Tetrahydrocurcumin | −8.00 | [92] |
| 36. | Corydine | −6.04 | [92] |
| 37. | Imperialine-3-β-D-glucoside | −7.1 | [90] |
Table 6.
Phytochemicals screened against 3CL Pro as docking target and their binding energy. “x” represents the repurposed drugs used for reference.
| S.no. | Phytochemical | Binding energy (Kcal. Mol−1) | References |
|---|---|---|---|
| 1. | Epicatechin-(4β,8)-epicatechin-(4β,6)-catechin | −10.60 | [93] |
| 2. | Taiwanhomoflavone A | −9.60 | [93] |
| 3. | Quercetin 3-glucosyl-(1,4)-rhamnoside | −9.90 | [93] |
| 4. | Epicatechin-4-epigallocatechin | −10 | [93] |
| 5. | Lactucopicrin | −8.30 | [93] |
| 6. | Lactucopicrin 15-oxalate | −8.20 | [93] |
| 7. | Myricitrin | −8.90 | [93] |
| 8. | Vitetrifolin D | −7.30 | [93] |
| 9. | Kaempferol | −7.80 | [93] |
| 10. | Apigenin | −7.80 | [93] |
| 11. | (−)-Asperlicin C | −9.70 | [93] |
| 12. | Cassameridin | −9.30 | [93] |
| 13. | Oriciacridone F | −9.10 | [93] |
| 14. | Remdesivirx | −8.20 | [93] |
| 15. | Afzelin | −8.80 | [93] |
| 16. | Isoquercitrin | −8.20 | [93] |
| 17. | Amentoflavone | −9.28 | [94] |
| 18. | Glabrolide | −9.16 | [94] |
| 19. | Zeylanone | −9.12 | [94] |
| 20. | 5,7,30,40-tetrahydroxy-2'-(3,3-dimethylallyl) isoflavone | −29.57 | [95] |
| 21. | Mirycitrin | −22.62 | [95] |
| 22. | Methyl rosmarinate | −20.62 | [95] |
| 23. | Amaranthin | −18.14 | [95] |
| 24. | Betulinic acid | −4.23 | [63] |
| 25 | Nelfinavirx | −10.72 | [96] |
| 26. | Apigenine-7-glucoside | −7.83 | [96] |
| 27. | (E)-β-Farnesene | −27.56 | [97] |
| 28. | Epicatechin-gallate | −6.67 | [96] |
| 29. | Gingerol | −5.38 | [96] |
| 30. | Zingerol | −5.40 | [96] |
| 31. | Sugiol | −6.04 | [63] |
| 32. | Cryptotanshinone | −6.23 | [63] |
| 33. | N-cis-feruloyltyramine | −6.25 | [63] |
| 34. | Quercetin | −8.47 | [96] |
| 35. | Kaempferol | −8.58 | [96] |
| 36. | Lupinavir | −9.41 | [96] |
| 37. | α-Copaene | −20.08 | [97] |
Azadirachta indica (Neem), an indigenous Indian medicinal plant used in Traditional Indian Medicine, has been reported of remarkable antiviral potency against avian influenza virus, bursal disease virus, Newcastle disease virus, group B coxsackievirus, dengue virus type-2, duck plague virus, poliovirus type-1 and bovine herpes virus type-1 [98]. Neem has been extensively used to treat asthma, cough, fever and diarrhea in Ayurvedic system of medicine; these are the most prevalent symptoms in SARS-CoV-2 infection [99]. Under viral attack, Neem intake amplifies cell and humoral mediated immune responses. Thus, multidimensional antiviral therapeutic potency of Neem led to hypothesizing its possible implications on COVID-19 along with the modern system of medicine. It is believed that Neem may prove a leading natural agent against COVID-19 but requires translational experimentation and a series of experimental database that can support its activity.
India is putting all its efforts to design and discover natural products against SARS-CoV-2. As a result recently Dr. Shekhar Mande (Director General, Council of Scientific and Industrial Research; CSIR) reported that ACQH (secure drug) a plant based drug has been approved for clinical trials. The medicine has been developed by the Sun Pharmaceutical in association with CSIR Indian Institute of Integrative Medicine (IIIM) Jammu and The International Centre for Genetic Engineering and Biotechnology (ICGEB) New Delhi with the support of Direct Benefit Transfer (DBT) and CSIR. Director General said “We are going to attempt to combat COVID-19 and we hope that it is successful”.
Glycyrrhiza glabra (licorice) may be involved to neutralize the activeness of SARS-CoV-2 due to the presence of active constituents like liquiritin, isoliquiritin, glycyrrhetic acid and glycyrrhizin [100]. An investigation regarding the involvement of natural products against COVID-19 has reported that plant extracted components target –OH (hydroxyl) groups of active components in virus thereby deactivating them through esterification.
In a similar study, the Max Planck Institute and Interfaces (Potsdam, Germany) in collaboration with ArtemiLifer Inc. (US based company) medicinal researchers have shown in laboratory studies that aqueous and ethanolic extracts of specially bred sweet wormwood plants (A. annua) are active against the new coronavirus that has caused the COIVID-19 pandemic [101].
The genomic studies of SARS-CoV-2 have reported that it is more similar to that of SARS-CoV-1, which caused an epidemic in at least 26 countries during 2002 and 2003. Therefore, Huazhong Agricultural University and Guangxi University researchers considered 3D homology model of sequence and screened medicinal plant library of 32,297 capable antiviral TCM compounds and phytochemicals. The outcomes of the investigation specified 9 plant based molecules which may be developed as potential drugs against COVID-19 (Table 7 ) [95].
Table 7.
Molecules and their plant source showing efficacy against SARS-CoV-2.
| Plant source | Name of the plant metabolite | |
|---|---|---|
| 1. | Edible amaranth (Amaranthus tricolor) | Amaranthin |
| 2. | Chinese liquorice (Glycyrrhiza uralensis) | Licoleafol |
| 3. | Myricetin 3-O-beta-d-glucopyranoside | Tea tree (Camellia sinensis) |
| 4. | Chinese flowering ash (Fraxinus sieboldiana) | Calceolarioside B |
| 5. | Indian gooseberry (Phyllanthus emblica) | (2S)-Eriodictyol 7-O-(6″-O-galloyl)-beta-d-glucopyranoside |
| 6. | Common bean (Phaseolus vulgaris) | 3,5,7,3′,4′,5′-hexahydroxy flavanone-3-O-beta-d-glucopyranoside |
| 7. | Marubio oscuro (Hyptis atrorubens Poit) | Methyl rosmarinate |
| 8. | Wax myrtle (Myrica cerifera) | Myricitrin |
| 9. | Mojave indigo bush (Psorothamnus arborescens) | 5,7,3′,4′-Tetrahydroxy-2'-(3,3-dimethylallyl) isoflavone |
Further, Indonesian researchers used main protease (Mpro) as target strategy and tested different inhibitors against Mpro in COVID-19 using molecular docking [101]. They went for bioactive molecules from plant source and came up with lopinavir and nelfinavir as potential treatment for SARS-CoV-2. In addition to this, epicatechin-gallate, catechin, curcumin, oleuropein, apigenin-7-glucoside, demethoxycurcumin and luteolin-7-glucoside, were reported of significant inhibitory potential against COVID-19 MPro [101]. Experimental data are needed to support these virtual molecular docking results.
9. Potential targets of antiviral drugs against coronaviruses
Coronaviruses (CoVs) are a large and diverse family of enveloped and single stranded RNA viruses. This exhibit broad host ranges and infect many mammalian and avian species causing gastrointestinal, upper respiratory, hepatic and central nervous system diseases [102]. Before 2019, only six CoVs were well-known to infect humans and cause health issues like HCoV-229E, HCoV-NL63, HCoV-OC43, HKU1, SARS-CoV and MERS-CoV. With birth of SARS-CoV-2, new member of human coronavirus has been added to this family. Probably the first reported coronavirus-related disease was feline infectious peritonitis in 1912. Nevertheless, until the late 1960s coronaviruses did not receive much attention and were not recognized as lethal for humans. In 2003, human coronaviruses (HCoVs) received global attention with emergence of the severe and acute respiratory syndrome coronavirus (SARS-CoV) which became highly contagious and deadly and infected more than 8000 people in 32 countries, killing about 10%. With SARS-CoV in focus, increased research and studies on coronaviruses soon led to the breakthrough of one more human coronavirus (HCoV-NL63), prevalent in 7% of hospital patients and related with bronchiolitis and, probably, conjunctivitis. HCoVs account for 10%–20% of common colds, and cause respiratory tract infections, gastroenteritis, and rare cases of encephalitis.
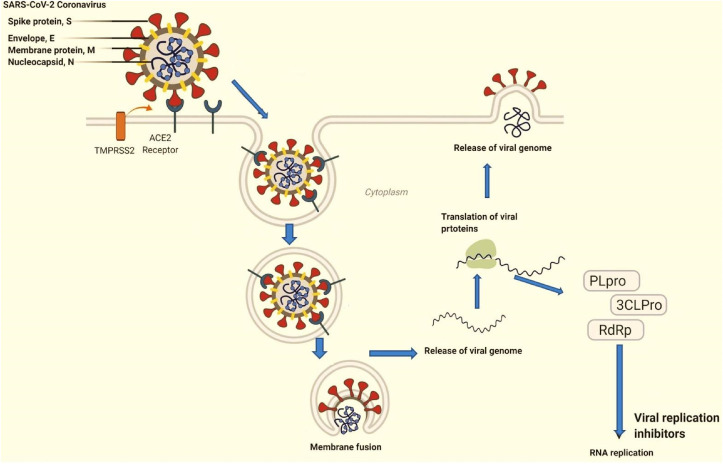
Coronaviruses have the largest genomes among RNA viruses. It is enveloped and packed inside by a helical capsid formed of Nucleocapsid protein (N). Three structural proteins viz. membrane protein (M), envelop protein (E) and spike protein (S) are associated with viral envelop. Membrane protein (M) and envelope protein (E) are involved in viral assembly whereas spike protein (S) has critical role in virus entry into host cells. Besides, some coronaviruses encode hemagglutinin-esterase protein (HE) as well. Moreover, spike protein besides mediating virus entry, determines viral host range, tissue trophism and is a major inducer of immune response in host cells. Spike protein possesses three segments: short intracellular tail, single pass transmembrane anchor and a large ectodomain. Receptor binding subunit S1 and membrane-fusion subunit S2 are part of ectodomain. Studies from electron microscopy have revealed spike as clove-shaped trimer with three S1 heads and trimeric S2 stalk [6,7,[103], [104], [105]]. While entering into the cell, S1 of spike protein binds to cell surface receptor for viral attachment and S2 stalk fuses with host and viral membranes, allocating viral genomes to host cells (Fig. 7 ). These are the initial and critical steps in coronavirus infection cycle and also serve as main targets for human interventions.
Fig. 7.
Representation of virus and host membrane interaction and post association changes inside cytoplasm.
10. Receptor recognition by coronovirus spike proteins
Coronaviruses exhibit a complex pattern for receptor recognition [106]. The alpha coronavirus HCoV-NL63 and beta coronavirus SARS-CoV both identify a zinc peptidase angotensin converting enzyme 2 (ACE2) [107,108]. Other alphacoronavirus such as TGEV, PEDV and PRCV recognize aminopeptidase N (APN0 [[109], [110], [111], [112]]. MERS-CoV and HKU4 recognize a serine peptidase, dipeptidase 4 (DPP4) [113,114]. Similarly, MHV recognizes a cell adhesion molecule, carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) [115,116]. BCoV and OC43 recognize sugar [117]. In addition, alpha coronaviruse TGEV and PDEV and gamma coronavirus IBV also use sugar as receptors or coreceptors [110,[118], [119], [120], [121]]. Besides their role in viral attachment, these receptors have their own physiological functions [[122], [123], [124], [125], [126], [127], [128]]. Receptor diversity is an exceptional feature of coronaviruses. Subunit S1 of coronavirus consists of two domains, N-terminal domain (S1-NTD) and C-terminal domain (S1-CTD), have been identified with one or both of them potentially functioning as receptor binding domains (RBD). S1-NTDs binds sugar [[110], [111], [129], [130]] with beta coronavirus MHV S1-NTD as only exception that recognizes protein receptor CEACAM1 [131]. S1-CTDs recognize protein receptors like ACE2,DPP$ and APN [110,[132], [133], [134], [135], [136], [137]]. Crystal structures of S1 domains along with functional studies have revealed many interesting features about receptor recognition by coronaviruses [6].
SARS-CoV 2 and SARS-CoV both encode large (1253aa; 1273 aa) spike protein [138]. Sequence identity has revealed 76% identity of spike protein between these two origins with large variation at N terminus. For SARS-CoV there is receptor binding domain in the S1 region interacting with ACE2 with high affinity. For SARS-CoV2, it has also come to the fore that it uses S1 RBD to bind ACE2 for entering human host cells. There is 73.5% sequence identity in RBD regions for these two viruses. Nevertheless, many non-conserved mutations have accumulated in two structural regions interacting directly with ACE2 [139]. Crystal and cryo-EM structures of SARS-CoV spike-ACE2 complex have revealed that regions 1 and 2 engage in hydrophobic interactions and hydrogen bonding with ACE2. Surprisingly, some residues in these regions have been replaced in SARS-CoV2, which eventually will lead loss of some of these interactions. Besides, SARS-CoV2 RBD has been predicted to be weekly interacting with ACE2 as compared to SARS-CoV [140]. In case of S2 region, there is no similarity in sequence between these two virus origins. After virus entry, genomic RNA attaches to the host ribosome and translates two large coterminal polyproteins, which are further processed by proteolysis for packaging into new virions [141]. Coronavirus main proteinase (3CLpro) and the papain-like protease (PLpro) are main proteases that participate in proteolysis process [142]. Furthermore, for replication of RNA, CoV encodes RNA dependent RNA polymerase RdRp), a replicase [143]. As these four proteins viz., Spike, CLpro, PLpro and RdRp, are key for pathogenesis of coronaviruses, therefore, therapeutics currently target them for treatment of SARS-CoV2.
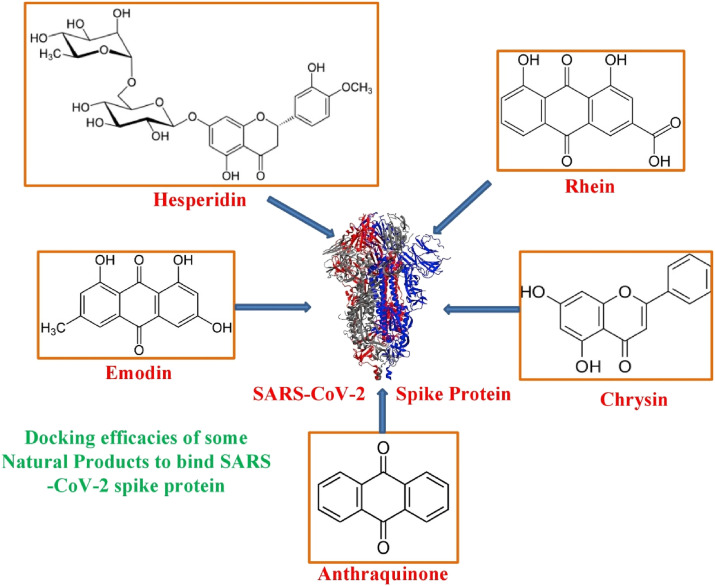
Owing to its role in receptor binding and membrane fusion, spike protein of SARS-CoV is an ideal target for vaccine and antiviral development (Fig. 8 ). Several strategies and approaches like killed SARS-CoV, live-attenuated SARS-CoV, Viral vectored vaccines and DNA vaccines have been successfully experimented against animal SARS-CoVs [[144], [145], [146]]. For SARS-CoV 2, similar approaches could be applied for developing vaccines. Use of SARS-Cov 2 RBD in combination with immunity promoting adjuvants as vaccine to trigger antibodies in human body is an alternative approach to neutralize the virus [147]. Even though, therapeutic antibodies and peptides have been developed to neutralize SARS-CoV spike protein, in case of SARS-CoV 2 these are expected to have little use as interactions between spike RBD for binding ACE2 are quite different in SARS-CoV2 and SARS-CoV. Novel antibodies and therapeutic peptides that potentially interact with RBD of SARS-CoV2 can be exploited to block its interaction with ACE2 [138]. In this regard, several research groups have developed methods for building macrocyclic peptide libraries for applying them to the speedy identification of macrocyclic peptide ligands for drug targets [[148], [149], [150], [151], [152], [153]]. It will potentially lead to quick discovery of anti- SARS-CoV2 macrocyclic peptides. Learning from SARS-CoV studies, possible alternatives include direct use of peptides derived from SARS-CoV2 RBD and ACE2. SARS-CoV peptides derived from its RBD and ACE2 as new therapeutics against SARS-CoV infection by blocking binding of RBD and ACE2. For COVID-19, a possible quick solution to block RBD-ACE2 interaction is to use SARS-CoV 2 based peptides and cocktails thereof [138].
Fig. 8.
Natural products that show significant binding score to spike protein of SARS-CoV-2.
Viruses including human coronaviruses involve host cellular factors for successful replication process during infection [154]. Organized and systematic identification of Virus host protein – protein interactions (PPIs) present an effective way to elucidate mechanisms of viral infection [155]; 88 [138]. Afterwards, targeting cellular antiviral targets like virus host intearactome offer novel strategy for the development of effective treatments against viral infections including, SARS-CoV and MERS-CoV [156].
SARS-CoV 2 and SARS-CoV share about 82% sequence identity at genomic RNA level whereas RdRp proteins have remarkable 96% sequence similarity. 3-chymotrypsin-like protease (3CLpro), RNA-dependent RNA polymerase, papain like protease (PLpro) and spike (S) proteins are major targets of drugs currently employed for SARS-CoV 2 treatment [157]. However, as of now no established antiviral therapies or preventive vaccines are available for its treatment [158]. RdRp with large and deep groove as an active site for RNA polymerization, shows variation at distal site of this active region for SARS-CoV2 as compared to SARS-CoV [159]. High sequence conservation between two enzymes makes it likely targeted with equal efficacy and potency by potent agents developed for SARS-CoV RdRp. Aurintricarboxylic acid (ATA) was one such compound with antiviral activity which targets RdRp. It is an anionic polymer which binds to protein targets and has been demonstrated to prevent replication [[160], [161], [162]]. But, despite computational models authorized against known ATA targets, predicting RdRp as bound target, no experimental evidence has demonstrated this relationship [163]. Several synthetic compounds including hydroxychloroquine and chloroquine [164,165], phosphate have been occasionally used. Antiviral medications such as remdesiver [166,167], lopinavir [168] and arbidol [169] have shown promising results. Nucleoside analogs, peptide EK1 and neuraminidase inhibitors are some other treatments explored for controlling COVID-19 pandemic [170]. Purified natural products and traditional herbal medicines have also been surveyed and explored for developing novel antiviral drugs.
83 compounds were screened in Chinese traditional medicines against RdRp of SARS-CoV 2 [171]. Likewise, Zhang et al. [63] screened 115 compounds for further studies. Most of these were naturally occurring polyphenolic compounds such as kaempferol and quercetin, which have previously received considerable interest for treating other disease [[172], [173], [174]].
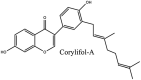
About 64 naturally occurring compounds were screened for activity against SARS-CoV helicase by Ref. [72]. The polyphenolics scutellarein and myricetin were identified as promising candidates. Both compounds did not directly inhibit helicase activity but inhibited SARS-CoV helicase (nsP13) [175]. reported that Chinese herbal medicine Lianhuaqingwen (mixture of plant species) showed antiviral activity against SARS-CoV 2, though EC50 was quite high (−411 μg/mL). Quercetin, a polyphenolic compound found in berries and herbs [176,177] also shows antiviral activity has IC50 value of 8.6 ± 3.2 μM against SARS-CoV PLpro [73]. Ethanolic extracts of Psoralea corylifolia seeds also possess polyphenolic compounds which were found to be responsible for activity against SARS-CoV Plpro [178].
Plant lectins are other naturally occurring compounds that may inhibit SARS –CoV. A total of 33 plant lectins were screened for the activity against SARS-CoV using cytopathicity effect (CPE) assay revealing a EC50 value of 0.45 ± 0.08 μg/mL for Lycoris radiata agglutinin [179]. In clinical trials, some lectins have demonstrated reasonable to good tolerability [180], hence with more testing, they may prove more promising classes of naturally derived compound(s) for the treatment of SARSCoV- 2 and other coronavirus infections.
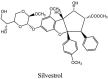
Silvestrol, which is a phytochemical from Aglaia sp., was found potent inhibitor of MERS-CoV replication [181]. It inhibits viral replication by binding RNA helicase eIF4A and preventing formation of replication/transcription complexes [181]. Likewise, Griffithsin, a lectin found in Griffithsia genus, is promising inhibitor of MERS-CoV. Its carbohydrate binding domains allows it to bind specifically to glycans of spike proteins and inhibit attachment of virus to the host cells in in vitro trials against MERS-CoV [182] and several other human corona viruses [183]. Griffithsin also has low systematic toxicity with specificity index against HCoV cells estimated between 30 and 3100 [182], and hence has the potential to be considered for clinical trials against SARS-CoV 2 [184]. Some of the possible targets of natural products are listed in Fig. 9 .
Fig. 9.
Some potential targets of natural products against coronaviruses.
11. Toxicity and adaptability of natural products in antiviral action
A big challenge is to find efficient drug candidates that can overcome infectious diseases with lower or no side-effects. Besides advancements that are being made towards the drug development for many diseases like HIV, AIDS, malaria and cancer, these drugs still cause unwanted side-effects to huge human population globally. Presently, it will be a viable methodology to turn back to “Nature” for answers because of their rich track record in the past. The natural products like taxol, vinblastine, quinine and artemisinin are well established drugs now used in the treatment of cancer and malaria. To address global health issues, the research and drug development concerning natural products plays a significant role in drug discovery. Faced with many stresses and challenges, coupled with being sedentary, plants have developed many molecules to ward off attacks from animals and environmental insults [185]. These phytochemicals show remarkable ability of colours, fragrances and undeniably toxicity. The use of plants by early humans against diseases must have been a trial and error exercise based on necessities. Several plants have been historically used for medicinal purposes including different pathological conditions [[186], [187], [188]]. Natural products are a pool of drugs especially for anticancer and antimicrobial agents. Despite significant potential, traditional medicine systems, which comprises mostly of natural products, is overshadowed by modern medicine. However, an increased interest towards natural products has been seen from previous few decades in both developing and developed countries due to promising health promontory and medicinal effects. Indeed, several plant extracts are now serving as prescribed drugs in some big nations like Germany, UK, China and France [189]. Nearly a quarter of entire EMA and FDA approved drugs are based on phytochemicals, with renowned ones such as morphine and paclitaxel. There are certain drawbacks that make natural products based medicine some times less effective like scarcity of standard procedures, isolation of pure compounds, lesser known biological mechanisms and substandard clinical trials.
The therapeutic activity of plant extracts is usually because of the synergistic and simultaneous action of several chemicals. Given the complex nature of many diseases including cancer and degenerative diseases, it is not surprising that the reliance on single compound-based drug discovery has failed to provide effective cures. Plant-based drug discovery therefore must start with a combinatorial approach when evaluating candidate compounds. The advent of novel technologies including quantum computing, profiling techniques, computational biology techniques, big data, microfluidics and artificial intelligence will enable scientists to use a combinatorial approach to harness the therapeutic properties of plant-based natural products and simultaneously study their molecular effects in physiological conditions [190]. It is however possible that not all components of plant extracts have measurable effects.
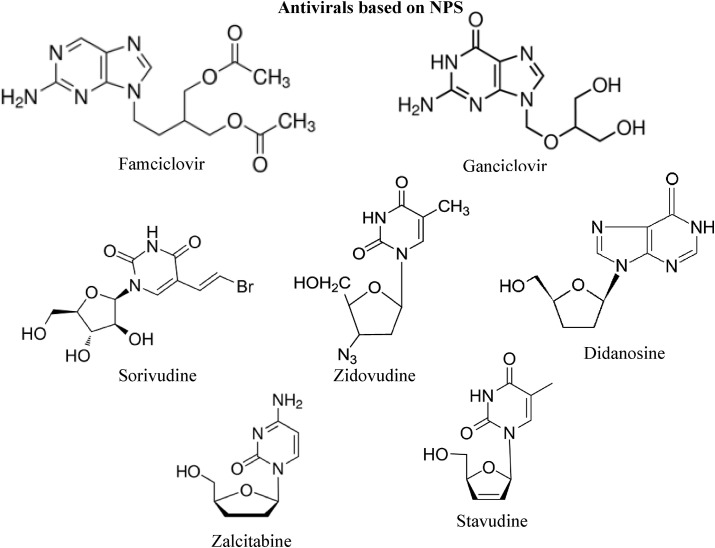
Many antiviral active compounds are too toxic for therapeutic applications. However, natural products remain the best resource for chemically diverse new lead entities that could serve for future development as potent and safe antiviral agents. Recent analysis of the number and sources of antiviral agents reported mainly in the annual reports of medicinal chemistry from 1984 to 1995 indicated that seven out of ten synthetic agents approved by FDA between 1983 and 1994 are modeled on natural product parent. These drugs are: famciclovir, ganciclovir, sorivudine, zidovudine, didanosine, zalcitabine and stavudine (Fig. 10 ).
Fig. 10.
Structure of some approved antiviral drugs based/modeled upon natural products.
The recent COVID-19 outbreak was has effected human race through a number of factors including health and economy. The serious availability of potential drugs and vaccines lacks in almost every corner of the world. Scientists and researchers are working day and night and are leaving no stone unturned in the race for finding efficient and long lasting solutions for COVID-19. Interestingly, more emphasis was laid over exploring natural products against COVID-19 with performing docking and in-vitro investigations. But following the previous track record natural products do not laid us down this time too, they come up with potential agents that can inhibit COVID-19 or prevent its lethal effects. In one such study, a library of about 686 plant based natural products were studied virtually against main protease (Mpro) of SARS-CoV-2 and results showed that 28 had efficient binding energy to Mpro. This study also investigated drug-likeness and toxicity of these natural products. Seven out of 28 drugs were found non-toxic rest showed significant toxicity. Those seven include Mpro-Dehydrtectol, Epsilon-viniferin, Peimisine, Gmelanone, and Isocolumbin were non-toxic. These seven compounds showed significant binding efficacy to Mpro and result in stable complex formation and hence could be used as Mpro inhibitors [191].
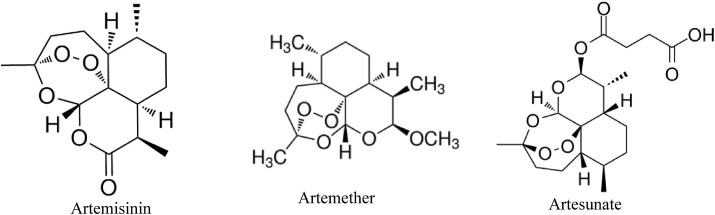
Not only effective but cost effective and affordable treatments are wanted for COVID-19 due its pandemic form effects all sectors of the societies. A study reported by Gilmore et al. showed in vitro activity of Artemisia annua extracts along with some isolates of the plant like artemether, artesunate and artemisinin (Fig. 11 ). Some of these isolates of the plant have already been approved for anti-malarial activity. The results of their study revealed that subsequent dose-response based on immunostaining of SARS-CoV-2 spike glycoprotein, in treatment and pretreatment groups with extracts and these isolates showed preventive effects against SARS-CoV-2 infection in VeroE6 cells. In treatment assays, artesunate (50% effective concentration (EC50): 7 μg/mL) was more potent than the tested plant extracts (128–260 μg/mL) or artemisinin (151 μg/mL) and artemether (>179 μg/mL), while generally EC50 in pretreatment assays were slightly higher [101]. Therefore, Artemisia annua could be a breakthrough against COVID-19 due to easy availibility, cost effective and natural presence.
Fig. 11.
Structure of Artemisinin, Artemether and Artesunate compounds tested effective in vitro against SARS-CoV-2.
12. Conclusion
Since the onset of COVID-19 pandemic natural product researchers have raised their efforts to explore potential drug candidates against this lethal disease. This has increased the number of studies performing on natural products. They have shown promising antiviral effects against several human infectious viruses in clinical studies. Natural products have been reported as lead molecules in different in vitro, in vivo, in silico and clinical studies. Due to immense molecular diversity in nature and structure of these molecules, they show strong antiviral efficacies. Natural products have shown to target virus binding and replication in SARS-CoV-2. Moreover, they suppress the disease symptoms caused by SARS-CoV-2 infection like cough, cold, fever, etc. A lot of effort is still needed in terms of drug design and development in the real sense as most of the current research efforts are based on theoretical calculations and in vitro bioassays. Nonetheless, such research efforts become the foundation stone for any subsequent drug discovery and provide a guide for further in depth in vivo studies.
Declaration of competing interest
The authors declare that there is no conflict of interest to indicate.
References
- 1.Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J. Pediatr. 2020;87(4):281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zubair A.S., McAlpine L.S., Gardin T., Farhadian S., Kuruvilla D.E., Spudich S. Neuropathogenesis and neurologic manifestations of the coronaviruses in the age of coronavirus disease 2019: a review. JAMA Neurol. 2020;77(8):1018–1027. doi: 10.1001/jamaneurol.2020.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brian D.A., Baric R.S. Coronavirus genome structure and replication. Coronav. Replicat. Rever. Genet. 2005:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Y., Peng F., Wang R., Guan K., Jiang T., Xu G., Sun J., Chang C. The deadly coronaviruses: the 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J. Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Estola T. Coronaviruses, a new group of animal RNA viruses. Avian Dis. 1970:330–336. [PubMed] [Google Scholar]
- 6.Li F. Structure, function, and evolution of coronavirus spike proteins. Ann. Rev. Virol. 2016;(3):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531(7592):118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson R.M., Fraser C., Ghani A.C., Donnelly C.A., Riley S., Ferguson N.M., Leung G.M., Lam T.H., Hedley A.J. Epidemiology, transmission dynamics and control of SARS: the 2002–2003 epidemic. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2004;359(1447):1091–1105. doi: 10.1098/rstb.2004.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parsons L.M., Bouwman K.M., Azurmendi H., De Vries R.P., Cipollo J.F., Verheije M.H. Glycosylation of the viral attachment protein of avian coronavirus is essential for host cell and receptor binding. J. Biol. Chem. 2019;294(19):7797–7809. doi: 10.1074/jbc.RA119.007532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Islam M.T., Sarkar C., El‐Kersh D.M., Jamaddar S., Uddin S.J., Shilpi J.A., Mubarak M.S. Natural products and their derivatives against coronavirus: a review of the non‐clinical and pre‐clinical data. Phytother Res. 2020;34(10):2471–2492. doi: 10.1002/ptr.6700. [DOI] [PubMed] [Google Scholar]
- 11.Yan L., Meng B., Xiang J., Wilson I.A., Yang B. Crystal structure of the post-fusion core of the Human coronavirus 229E spike protein at 1.86 Å resolution. Acta Crystallogr. D: Struct. Biol. 2018;74(9):841–851. doi: 10.1107/S2059798318008318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorbalenya A.E., Snijder E.J., Spaan W.J. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 2004;78(15):7863–7866. doi: 10.1128/JVI.78.15.7863-7866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wani A.R., Yadav K., Khursheed A., Rather M.A. An updated and comprehensive review of the antiviral potential of essential oils and their chemical constituents with special focus on their mechanism of action against various influenza and coronaviruses. Microb. Pathog. 2020 doi: 10.1016/j.micpath.2020.104620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jangra M.K., Saxena A. Significance of physiotherapy in “SARS-CoV-2/COVID-19: an epidemic”. Ann. Thorac. Med. 2020;15(3):179. doi: 10.4103/atm.ATM_169_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Groot R.J., Baker S.C., Baric R. The positive sense single stranded RNA viruses. Virus Taxon. –Ninth Rep. Int. Commit. Taxon. Virus. 2012;(Chap. Part II):806–828. [Google Scholar]
- 16.Holmes K.V. Coronaviruses (Coronaviridae) Encyclop. Virol. 1999:291. [Google Scholar]
- 17.Chang C.K., Hou M.H., Chang C.F., Hsiao C.D., Huang T.H. The SARS coronavirus nucleocapsid protein–forms and functions. Antivir. Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sola I., Almazan F., Zuniga S., Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Ann. Rev. Virol. 2015;(2):265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto M., Matsuyama S., Li X., Takeda M., Kawaguchi Y., Inoue J.I., Matsuda Z. Identification of nafamostat as a potent inhibitor of Middle East respiratory syndrome coronavirus S protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob. Agent Chemother. 2016;60(11):6532–6539. doi: 10.1128/AAC.01043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Z., Thackray L.B., Miller B.C., Lynn T.M., Becker M.M., Ward E., Mizushima N., Denison M.R., Virgin I.V.H.W. Coronavirus replication does not require the autophagy gene ATG5. Autophagy. 2007;3(6):581–585. doi: 10.4161/auto.4782. [DOI] [PubMed] [Google Scholar]
- 21.De Wilde A.H., Snijder E.J., Kikkert M., van Hemert M.J. Host factors in coronavirus replication. Roles Host Gene Non-Coding RNA Express. Virus Infect. 2017:1–42. doi: 10.1007/82_2017_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fung T.S., Liu D.X. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014;5:296. doi: 10.3389/fmicb.2014.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snijder E.J., Limpens R.W., De Wilde A.H., De Jong A.W., Zevenhoven-Dobbe J.C., Maier H.J., Faas F.F., Koster A.J., Bárcena M. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biol. 2020;18(6) doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu D.X., Ng Y.L., Fung T.S. Viral Vectors in Veterinary Vaccine Development. Springer; Cham: 2021. Coronaviruses as vaccine vectors for veterinary pathogens; pp. 149–168. [Google Scholar]
- 25.Ziegler C.G., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., Feldman J. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181(5):1016–1035. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J., Pan L., Tang S., Ji J.S., Shi X. Mask use during COVID-19: a risk adjusted strategy. Environ. Pollut. 2020 doi: 10.1016/j.envpol.2020.115099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baay M., Lina B., Fontanet A., Marchant A., Saville M., Sabot P., Duclos P., Vandeputte J., Neels P. SARS-CoV-2: virology, epidemiology, immunology and vaccine development. Biologicals. 2020;66:35–40. doi: 10.1016/j.biologicals.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdel-Moneim A.S. Middle East respiratory syndrome coronavirus (MERS-CoV): evidence and speculations. Arch. Virol. 2014;159(7):1575–1584. doi: 10.1007/s00705-014-1995-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Da Silva E.R., Pitrez M.C., Arruda E., Mattiello R., Sarria E.E., de Paula F.E., Proença-Modena J.L., Delcaro L.S., Cintra O., Jones M.H., Ribeiro J.D. Severe lower respiratory tract infection in infants and toddlers from a non-affluent population: viral etiology and co-detection as risk factors. BMC Infect. Dis. 2013;13(1):1–8. doi: 10.1186/1471-2334-13-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng Z., Monteil V.M., Maurer-Stroh S., Yew C.W., Leong C., Mohd-Ismail N.K., Arularasu S.C., Chow V.T., Lin R.T., Mirazimi A., Hong W. Monoclonal antibodies for the S2 subunit of spike of SARS-CoV-1 cross-react with the newly-emerged SARS-CoV-2. Euro Surveill. 2020;25(28) doi: 10.2807/1560-7917.ES.2020.25.28.2000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar U., Singhal S., Khandia R., Munjal A. Current scenario OF the pandemic COVID-19: an overview. J. Exp. Biol. Agric. Sci. 2020;8(1):158–175. doi: 10.18006/2020.8(spl-1-sars-cov-2).s158.s175. [DOI] [Google Scholar]
- 32.Hassanin A., Grandcolas P., Veron G. Covid-19: natural or anthropic origin? Mammalia. 2020;1 (ahead-of-print) [Google Scholar]
- 33.Ghosh S., Malik Y.S. Drawing comparisons between SARS-CoV-2 and the animal coronaviruses. Microorganisms. 2020;8(11):1840. doi: 10.3390/microorganisms8111840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cui L.J., Zhang C., Zhang T., Lu R.J., Xie Z.D., Zhang L.L., Liu C.Y., Zhou W.M., Ruan L., Ma X.J., Tan W.J. Human coronaviruses HCoV-NL63 and HCoV-HKU1 in hospitalized children with acute respiratory infections in Beijing, China. Adv. Virol. 2011:2011. doi: 10.1155/2011/129134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miłek J., Blicharz-Domańska K. Coronaviruses in avian species–review with focus on epidemiology and diagnosis in wild birds. J. Veterin. Res. 2018;62(3):249–255. doi: 10.2478/jvetres-2018-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L.F., Anderson D.E. Viruses in bats and potential spillover to animals and humans. Curr. Opin. Virol. 2019;34:79–89. doi: 10.1016/j.coviro.2018.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006;19(3):531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Velavan T.P., Meyer C.G. The COVID‐19 epidemic. Trop. Med. Int. Health. 2020;25(3):278. doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.WHO reports . 2021. Coronavirus Disease (COVID-19), Situation Report.https://covid19.who.int/ Data as received by WHO from national authorities by 6th April. Accessed 0n 6 April. [Google Scholar]
- 40.Zahedi Ali, Salehi-Amiri Amirhossein, Smith Neale R., Hajiaghaei-Keshteli Mostafa. Utilizing IoT to design a relief supply chain network for the SARS-COV-2 pandemic. Appl. Soft Comput. 2021 Jun;104:107210. doi: 10.1016/j.asoc.2021.10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie P., Ma W., Tang H., Liu D. Severe COVID-19: a review of recent progress with a look toward the future. Front. Publ. Heal. 2020;8:189. doi: 10.3389/fpubh.2020.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lotfi M., Hamblin M.R., Rezaei N. COVID-19: transmission, prevention, and potential therapeutic opportunities. Clin. Chim. Acta. 2020;508:254–266. doi: 10.1016/j.cca.2020.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phua J., Weng L., Ling L., Egi M., Lim C.M., Divatia J.V., Shrestha B.R., Arabi Y.M., Ng J., Gomersall C.D., Nishimura M. Intensive care management of coronavirus disease 2019 (COVID-19): challenges and recommendations. Lancet Respir. Med. 2020;8(5):506–517. doi: 10.1016/S2213-2600(20)30161-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mohseni A.H., Taghinezhad-S S., Xu Z., Fu X. Body fluids may contribute to human-to-human transmission of severe acute respiratory syndrome coronavirus 2: evidence and practical experience. Chin. Med. 2020;15(1):1–4. doi: 10.1186/s13020-020-00337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 47.Luan J., Lu Y., Jin X., Zhang L. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem. Biophys. Res. Commun. 2020;526(1):165–169. doi: 10.1016/j.bbrc.2020.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu B., Huang S., Yin L. The cytokine storm and COVID‐19. J. Med. Virol. 2021;93(1):250–256. doi: 10.1002/jmv.26232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Costa VG da, ML Moreli, MV Saivish. The emergence of SARS, MERS and novel SARS-2 coronaviruses in the 21st century. Arch. Virol. 2020;165(7):1517–1526. doi: 10.1007/s00705-020-04628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ragab D., Salah Eldin H., Taeimah M., Khattab R., Salem R. The COVID-19 cytokine storm; what we know so far. Front. Immunol. 2020;11:1446. doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Costela-Ruiz V.J., Illescas-Montes R., Puerta-Puerta J.M., Ruiz C., Melguizo-Rodríguez L. SARS-CoV-2 infection: the role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020;54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weissleder R., Lee H., Ko J., Pittet M.J. COVID-19 diagnostics in context. Sci. Transl. Med. 2020;12(546) doi: 10.1126/scitranslmed.abc1931. [DOI] [PubMed] [Google Scholar]
- 53.Ponti G., Maccaferri M., Ruini C., Tomasi A., Ozben T. Biomarkers associated with COVID-19 disease progression. Crit. Rev. Clin. Lab Sci. 2020;57(6):389–399. doi: 10.1080/10408363.2020.1770685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296(2):E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khursheed A., Jain V. Medicinal research progress of natural coumarin and its derivatives. Nat. Prod. J. 2020;10:1. doi: 10.2174/2210315510999201102201552. [DOI] [Google Scholar]
- 56.Khursheed A., Rather M.A., Rashid R. Plant-based natural compounds and herbal extracts as promising apoptotic agents: their implications for cancer prevention and treatment. Adv. Biomed. Pharm. 2016;3(4):245–269. [Google Scholar]
- 57.Kitazato K., Wang Y., Kobayashi N. Viral infectious disease and natural products with antiviral activity. Drug Discov. Ther. 2007;1(1):14–22. [PubMed] [Google Scholar]
- 58.Vougogiannopoulou K., Corona A., Tramontano E., Alexis M.N., Skaltsounis A.L. Natural and nature-derived products targeting human coronaviruses. Molecules. 2021;26(2):448. doi: 10.3390/molecules26020448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu Y.H., Dong J.H., An W.M., Lv X.Y., Yin X.P., Zhang J.Z., Dong L., Ma X., Zhang H.J., Gao B.L. Clinical and computed tomographic imaging features of novel coronavirus pneumonia caused by SARS-CoV-2. J. Infect. 2020;80(4):394–400. doi: 10.1016/j.jinf.2020.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yin Y., Wunderink R.G. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23(2):130–137. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gastanaduy P.A. Update: severe respiratory illness associated with Middle East respiratory syndrome coronavirus (MERS-CoV)—worldwide, 2012–2013. MMWR Morbid. mortal. weekly Rep. 2013;62(23):480. [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang T., He Y., Xu W., Ma A., Yang Y., Xu K.F. Clinical trials for the treatment of Coronavirus disease 2019 (COVID-19): a rapid response to urgent need. Sci. China Life Sci. 2020;63(5):774–776. doi: 10.1007/s11427-020-1660-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang D.H., Wu K.L., Zhang X., Deng S.Q., Peng B. In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. J. Integr. Med. 2020;18(2):152–158. doi: 10.1016/j.joim.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shahzad F., Anderson D., Najafzadeh M. The antiviral, anti-inflammatory effects of natural medicinal herbs and mushrooms and SARS-CoV-2 infection. Nutrients. 2020;12(9):2573. doi: 10.3390/nu12092573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tahir A.H., Javed M.M., Hussain Z. Nutraceuticals and herbal extracts: a ray of hope for COVID-19 and related infections. Int. J. Funct. Nutr. 2020;1(2):1. [Google Scholar]
- 66.Li S.Y., Chen C., Zhang H.Q., Guo H.Y., Wang H., Wang L., Zhang X., Hua S.N., Yu J., Xiao P.G., Li R.S. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antivir. Res. 2005;67(1):18–23. doi: 10.1016/j.antiviral.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Siddiqui A.J., Danciu C., Ashraf S.A., Moin A., Singh R., Alreshidi M., Patel M., Jahan S., Kumar S., Alkhinjar M.I., Badraoui R. Plants-derived biomolecules as potent antiviral phytomedicines: new insights on ethnobotanical evidences against coronaviruses. Plants. 2020;9(9):1244. doi: 10.3390/plants9091244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Refaey M.S., Fayed M.A. Traditional to recent approaches IN herbal medicine therapy OF COVID-19. J. Pharmaceut. Res. 2020;5(5):71–84. [Google Scholar]
- 69.Wen C.C., Kuo Y.H., Jan J.T., Liang P.H., Wang S.Y., Liu H.G., Lee C.K., Chang S.T., Kuo C.J., Lee S.S., Hou C.C. Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J. Med. Chem. 2007;50(17):4087–4095. doi: 10.1021/jm070295s. [DOI] [PubMed] [Google Scholar]
- 70.Park J.Y., Kim J.H., Kim Y.M., Jeong H.J., Kim D.W., Park K.H., Kwon H.J., Park S.J., Lee W.S., Ryu Y.B. Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases. Bioorg. Med. Chem. 2012;20(19):5928–5935. doi: 10.1016/j.bmc.2012.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin C.W., Tsai F.J., Tsai C.H., Lai C.C., Wan L., Ho T.Y., Hsieh C.C., Chao P.D. Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antivir. Res. 2005;68(1):36–42. doi: 10.1016/j.antiviral.2005.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu M.S., Lee J., Lee J.M., Kim Y., Chin Y.W., Jee J.G., Keum Y.S., Jeong Y.J. Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg. Med. Chem. Lett. 2012;22(12):4049–4054. doi: 10.1016/j.bmcl.2012.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Park J.Y., Yuk H.J., Ryu H.W., Lim S.H., Kim K.S., Park K.H., Ryu Y.B., Lee W.S. Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. J. Enzym. Inhib. Med. Chem. 2017;32(1):504–512. doi: 10.1080/14756366.2016.1265519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Macintyre G., Curry B., Wong F., Anderson R. Hygromycin B therapy of a murine coronaviral hepatitis. Antimicrob. Agent Chemother. 1991;35(10):2125–2127. doi: 10.1128/aac.35.10.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song J.H., Choi H.J., Song H.H., Hong E.H., Lee B.R., Oh S.R., Choi K., Yeo S.G., Lee Y.P., Cho S., Ko H.J. Antiviral activity of ginsenosides against coxsackievirus B3, enterovirus 71, and human rhinovirus 3. J. Ginse. Res. 2014;38(3):173–179. doi: 10.1016/j.jgr.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fielding B.C., da Silva Maia Bezerra Filho C., Ismail N.S., Sousa D.P. Alkaloids: therapeutic potential against human coronaviruses. Molecules. 2020;25(23):5496. doi: 10.3390/molecules25235496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang C.W., Lee Y.Z., Kang I.J., Barnard D.L., Jan J.T., Lin D., Huang C.W., Yeh T.K., Chao Y.S., Lee S.J. Identification of phenanthroindolizines and phenanthroquinolizidines as novel potent anti-coronaviral agents for porcine enteropathogenic coronavirus transmissible gastroenteritis virus and human severe acute respiratory syndrome coronavirus. Antivir. Res. 2010;88(2):160–168. doi: 10.1016/j.antiviral.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shen L., Niu J., Wang C., Huang B., Wang W., Zhu N., Deng Y., Wang H., Ye F., Cen S., Tan W. High-throughput screening and identification of potent broad-spectrum inhibitors of coronaviruses. J. Virol. 2019;93(12) doi: 10.1128/JVI.00023-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vrijsen R., Berghe D.V., Vlietinck A.J., Boeye A. Lycorine: a eukaryotic termination inhibitor? J. Biol. Chem. 1986;261(2):505–507. [PubMed] [Google Scholar]
- 80.Sayed A.M., Khattab A.R., AboulMagd A.M., Hassan H.M., Rateb M.E., Zaid H., Abdelmohsen U.R. Nature as a treasure trove of potential anti-SARS-CoV drug leads: a structural/mechanistic rationale. RSC Adv. 2020;10(34):19790–19802. doi: 10.1039/d0ra04199h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chiow K.H., Phoon M.C., Putti T., Tan B.K., Chow V.T. Evaluation of antiviral activities of Houttuynia cordata Thunb. extract, quercetin, quercetrin and cinanserin on murine coronavirus and dengue virus infection. Asian Pacific J. Trop. Med. 2016;9(1):1–7. doi: 10.1016/j.apjtm.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Khare P., Sahu U., Pandey S.C., Samant M. Current approaches for target-specific drug discovery using natural compounds against SARS-CoV-2 infection. Virus Res. 2020 doi: 10.1016/j.virusres.2020.198169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cho J.H., Bernard D.L., Sidwell R.W., Kern E.R., Chu C.K. Synthesis of cyclopentenyl carbocyclic nucleosides as potential antiviral agents against orthopoxviruses and SARS. J. Med. Chem. 2006 Feb 9;49(3):1140–1148. doi: 10.1021/jm0509750. [DOI] [PubMed] [Google Scholar]
- 84.Mohapatra T.K., Nayak R.R., Mondal A., Nanda S.S. A novel and emerging coronavirus infection: repurposing and scale of advances of therapeutics, imunotherapeutics and vaccine development. Drug Discov. 2021;15(35):23–50. [Google Scholar]
- 85.Yang C.W., Lee Y.Z., Hsu H.Y., Shih C., Chao Y.S., Chang H.Y., Lee S.J. Targeting coronaviral replication and cellular JAK2 mediated dominant NF-κB activation for comprehensive and ultimate inhibition of coronaviral activity. Sci. Rep. 2017;7(1):1–3. doi: 10.1038/s41598-017-04203-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vanden Berghe D.A., Vlietinck A.J., Van Hoof L. Plant products as potential antiviral agents. Bull Instit. Pasteur. 1986;84:101–147. [Google Scholar]
- 87.El Sayed K.A. Natural products as antiviral agents. Stud. Nat. Prod. Chem. 2000;24:473–572. (Elsevier) [Google Scholar]
- 88.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280 e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Coban M., Morrison J., Freeman W., S Radisky E., Le Roch K., Caulfield T. Targeting Tmprss2, S-protein: ace2, and 3CLpro for synergetic inhibitory engagement. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12616151.v1. [DOI] [Google Scholar]
- 90.Cheng J., Tang Y., Bao B., Zhang P. Exploring the active compounds of traditional Mongolian medicine agsirga in intervention of novel Coronavirus (2019-nCoV) based on HPLC-Q-Exactive-MS/MS and molecular docking method. ChemRxiv. 2020 doi: 10.26434/chemrxiv.11955273.v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Meneguzzo F., Ciriminna R., Zabini F., Pagliaro M. Review of evidence available on hesperidin-rich products as potential tools against COVID-19 and hydrodynamic cavitation-based extraction as a method of increasing their production. Processes. 2020;8(5):549. [Google Scholar]
- 92.Pandit M., Latha N. 2020. In Silico Studies Reveal Potential Antiviral Activity of Phytochemicals from Medicinal Plants for the Treatment of COVID-19 Infection. PREPRINT (Version 1) available at: Research Square. [DOI] [Google Scholar]
- 93.Joshi R.S., Jagdale S.S., Bansode S.B., Shankar S.S., Tellis M.B., Pandya V.K., Chugh A., Giri A.P., Kulkarni M.J. Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1760137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Olubiyi O.O., Olagunju M., Keutmann M., Loschwitz J., Strodel B. High throughput virtual screening to discover inhibitors of the main protease of the coronavirus SARS-CoV-2. Molecules. 2020;25(14):3193. doi: 10.3390/molecules25143193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.ul Qamar M.T., Alqahtani S.M., Alamri M.A., Chen L.L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharmaceut. Anal. 2020;10(4):313–319. doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Khaerunnisa S., Kurniawan H., Awaluddin R., Suhartati S., Soetjipto S. Potential inhibitor of COVID-19 main protease (Mpro) from several medicinal plant compounds by molecular docking study. Prepr. 2020 doi: 10.20944/preprints202003.0226.v1. [DOI] [Google Scholar]
- 97.da Silva J.K., Figueiredo P.L., Byler K.G., Setzer W.N. Essential oils as antiviral agents, potential of essential oils to treat sars-cov-2 infection: an in-silico investigation. Int. J. Mol. Sci. 2020;21(10):3426. doi: 10.3390/ijms21103426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baildya N., Khan A.A., Ghosh N.N., Dutta T., Chattopadhyay A.P. Screening of potential drug from Azadirachta Indica (Neem) extracts for SARS-CoV-2: an insight from molecular docking and MD-simulation studies. J. Mol. Struct. 2021;1227 doi: 10.1016/j.molstruc.2020.129390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Biswas K., Chattopadhyay I., Banerjee R.K., Bandyopadhyay U. Biological activities and medicinal properties of neem (Azadirachta indica) Curr. Sci. 2002:1336–1345. [Google Scholar]
- 100.Bailly C., Vergoten G. Glycyrrhizin: an alternative drug for the treatment of COVID-19 infection and the associated respiratory syndrome? Pharmacol. Therapeut. 2020 doi: 10.1016/j.pharmthera.2020.107618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.MS Nair, Y Huang, DA Fidock, SJ Polyak, J Wagoner, MJ Towler, PJ Weathers. Artemisia annua L. extracts inhibit the in vitro replication of SARS-CoV-2 and two of its variants. J. Ethnopharmacol. 2020;274:114016. doi: 10.1016/j.jep.2021.114016. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Holmes K.V. In: Fields' Virology. Knipe D., et al., editors. Lippincott Williams & Wilkins; Philadelphia, Pennsylvania, USA: 2001. Coronaviruses; pp. 1187–1203. [Google Scholar]
- 103.Walls A.C., Tortorici M.A., Bosch B.J., Frenz B., Rottier P.J., DiMaio F., Rey F.A., Veesler D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531(7592):114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Beniac D.R., Andonov A., Grudeski E., Booth T.F. Architecture of the SARS coronavirus prefusion spike. Nat. Struct. Mol. Biol. 2006;13(8):751–752. doi: 10.1038/nsmb1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li F., Berardi M., Li W., Farzan M., Dormitzer P.R., Harrison S.C. Conformational states of the severe acute respiratory syndrome coronavirus spike protein ectodomain. J. Virol. 2006;80(14):6794–6800. doi: 10.1128/JVI.02744-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J. Virol. 2015;89(4):1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hofmann H., Pyrc K., Van Der Hoek L., Geier M., Berkhout B., Pöhlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. Unit. States Am. 2005;102(22):7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Delmas B., Gelfi J., L'Haridon R., Sjöström H., Laude H. Aminopeptidase N is a major receptor for the enteropathogenic coronavirus TGEV. Nature. 1992;357(6377):417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu C., Tang J., Ma Y., Liang X., Yang Y., Peng G., Qi Q., Jiang S., Li J., Du L., Li F. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J. Virol. 2015;89(11):6121–6125. doi: 10.1128/JVI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li B.X., Ge J.W., Li Y.J. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virology. 2007;365(1):166–172. doi: 10.1016/j.virol.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Delmas B., Gelfi J., Sjöström H., Noren O., Laude H. Springer; Boston, MA: 1994. Further Characterization of Aminopeptidase-N as a Receptor for Coronaviruses. InCoronaviruses. pp. 293–298. [DOI] [PubMed] [Google Scholar]
- 113.Raj V.S., Mou H., Smits S.L., Dekkers D.H., Müller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A., Thiel V. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang Y., Du L., Liu C., Wang L., Ma C., Tang J., Baric R.S., Jiang S., Li F. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc. Natl. Acad. Sci. Unit. States Am. 2014;111(34):12516–12521. doi: 10.1073/pnas.1405889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dveksler G.S., Pensiero M.N., Cardellichio C.B., Williams R.K., Jiang G.S., Holmes K.V., Dieffenbach C.W. Cloning of the mouse hepatitis virus (MHV) receptor: expression in human and hamster cell lines confers susceptibility to MHV. J. Virol. 1991;65(12):6881–6891. doi: 10.1128/jvi.65.12.6881-6891.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Williams R.K., Jiang G.S., Holmes K.V. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl. Acad. Sci. Unit. States Am. 1991;88(13):5533–5536. doi: 10.1073/pnas.88.13.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schultze B., Gross H.J., Brossmer R., Herrler G. The S protein of bovine coronavirus is a hemagglutinin recognizing 9-O-acetylated sialic acid as a receptor determinant. J. Virol. 1991;65(11):6232–6237. doi: 10.1128/jvi.65.11.6232-6237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schultze B., Cavanagh D., Herrler G. Neuraminidase treatment of avian infectious bronchitis coronavirus reveals a hemagglutinating activity that is dependent on sialic acid-containing receptors on erythrocytes. Virology. 1992;189(2):792–794. doi: 10.1016/0042-6822(92)90608-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cavanagh D., Davis P.J. Coronavirus IBV: removal of spike glycopolypeptide S1 by urea abolishes infectivity and haemagglutination but not attachment to cells. J. Gen. Virol. 1986;67(7):1443–1448. doi: 10.1099/0022-1317-67-7-1443. [DOI] [PubMed] [Google Scholar]
- 120.Schwegmann-Weßels C., Herrler G. Sialic acids as receptor determinants for coronaviruses. Glycoconj. J. 2006;23(1):51–58. doi: 10.1007/s10719-006-5437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Krempl C., Schultze B., Laude H., Herrler G. Point mutations in the S protein connect the sialic acid binding activity with the enteropathogenicity of transmissible gastroenteritis coronavirus. J. Virol. 1997;71(4):3285–3287. doi: 10.1128/jvi.71.4.3285-3287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu C., Yang Y., Chen L., Lin Y.L., Li F. A unified mechanism for aminopeptidase N-based tumor cell motility and tumor-homing therapy. J. Biol. Chem. 2014;289(50):34520–34529. doi: 10.1074/jbc.M114.566802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mina-Osorio P. The moonlighting enzyme CD13: old and new functions to target. Trends Mol. Med. 2008;14(8):361–371. doi: 10.1016/j.molmed.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Boehm M., Nabel E.G. Angiotensin-converting enzyme 2—a new cardiac regulator. N. Engl. J. Med. 2002;347(22):1795–1797. doi: 10.1056/NEJMcibr022472. [DOI] [PubMed] [Google Scholar]
- 125.Kameoka J., Tanaka T., Nojima Y., Schlossman S.F., Morimoto C. Direct association of adenosine deaminase with a T cell activation antigen, CD26. Science. 1993;261(5120):466–469. doi: 10.1126/science.8101391. [DOI] [PubMed] [Google Scholar]
- 126.Wesley U.V., McGroarty M., Homoyouni A. Dipeptidyl peptidase inhibits malignant phenotype of prostate cancer cells by blocking basic fibroblast growth factor signaling pathway. Canc. Res. 2005;65(4):1325–1334. doi: 10.1158/0008-5472.CAN-04-1852. [DOI] [PubMed] [Google Scholar]
- 127.Hammarström S. The carcinoembryonic antigen (CEA) family: structures, suggested functions and expression in normal and malignant tissues. Semin. Canc. Biol. 1999;9(2):67–81. doi: 10.1006/scbi.1998.0119. (Academic Press) [DOI] [PubMed] [Google Scholar]
- 128.Dove A. The bittersweet promise of glycobiology. Nat. Biotechnol. 2001;19(10):913–917. doi: 10.1038/nbt1001-913. [DOI] [PubMed] [Google Scholar]
- 129.Peng G., Xu L., Lin Y.L., Chen L., Pasquarella J.R., Holmes K.V., Li F. Crystal structure of bovine coronavirus spike protein lectin domain. J. Biol. Chem. 2012;287(50):41931–41938. doi: 10.1074/jbc.M112.418210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Promkuntod N., Van Eijndhoven R.E., De Vrieze G., Gröne A., Verheije M.H. Mapping of the receptor-binding domain and amino acids critical for attachment in the spike protein of avian coronavirus infectious bronchitis virus. Virology. 2014;448:26–32. doi: 10.1016/j.virol.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kubo H., Yamada Y.K., Taguchi F. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J. Virol. 1994;68(9):5403–5410. doi: 10.1128/jvi.68.9.5403-5410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279(5):3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lin H.X., Feng Y., Wong G., Wang L., Li B., Zhao X., Li Y., Smaill F., Zhang C. Identification of residues in the receptor-binding domain (RBD) of the spike protein of human coronavirus NL63 that are critical for the RBD–ACE2 receptor interaction. J. Gen. Virol. 2008;89(4):1015–1024. doi: 10.1099/vir.0.83331-0. [DOI] [PubMed] [Google Scholar]
- 134.Hofmann H., Simmons G., Rennekamp A.J., Chaipan C., Gramberg T., Heck E., Geier M., Wegele A., Marzi A., Bates P., Pöhlmann S. Highly conserved regions within the spike proteins of human coronaviruses 229E and NL63 determine recognition of their respective cellular receptors. J. Virol. 2006;80(17):8639–8652. doi: 10.1128/JVI.00560-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Godet M., Grosclaude J., Delmas B., Laude H. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J. Virol. 1994;68(12):8008–8016. doi: 10.1128/jvi.68.12.8008-8016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Du L., Zhao G., Kou Z., Ma C., Sun S., Poon V.K., Lu L., Wang L., Debnath A.K., Zheng B.J., Zhou Y. Identification of a receptor-binding domain in the S protein of the novel human coronavirus Middle East respiratory syndrome coronavirus as an essential target for vaccine development. J. Virol. 2013;87(17):9939–9942. doi: 10.1128/JVI.01048-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mou H., Raj V.S., van Kuppeveld F.J., Rottier P.J., Haagmans B.L., Bosch B.J. The receptor binding domain of the new MERS coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J. Virol. 2013;87(16):9379–9383. doi: 10.1128/JVI.01277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Morse J.S., Lalonde T., Xu S., Liu W.R. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019‐nCoV. Chembiochem. 2020;21(5):730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S., Wong S.K., Huang I.C., Xu K., Vasilieva N., Murakami A. Receptor and viral determinants of SARS‐coronavirus adaptation to human ACE2. EMBO J. 2005;24(8):1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dong N., Yang X., Ye L., Chen K., Chan E.W., Yang M., Chen S. Genomic and protein structure modelling analysis depicts the origin and infectivity of 2019-nCoV, a new coronavirus which caused a pneumonia outbreak in Wuhan, China, 01, 20th ed. bioRxiv. 2020;01:913368. doi: 10.1101/2020.01.20.913368. [DOI] [Google Scholar]
- 141.Baranov P.V., Henderson C.M., Anderson C.B., Gesteland R.F., Atkins J.F., Howard M.T. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology. 2005;332(2):498–510. doi: 10.1016/j.virol.2004.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. Microbiology. 2000;81(4):853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 143.Xu X., Liu Y., Weiss S., Arnold E., Sarafianos S.G., Ding J. Molecular model of SARS coronavirus polymerase: implications for biochemical functions and drug design. Nucleic Acids Res. 2003;31(24):7117–7130. doi: 10.1093/nar/gkg916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cavanagh D. Severe acute respiratory syndrome vaccine development: experiences of vaccination against avian infectious bronchitis coronavirus. Avian Pathol. 2003;32(6):567–582. doi: 10.1080/03079450310001621198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Holmes K.V. SARS coronavirus: a new challenge for prevention and therapy. J. Clin. Invest. 2003;111(11):1605–1609. doi: 10.1172/JCI18819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Navas-Martín S., Weiss S.R. Coronavirus replication and pathogenesis: implications for the recent outbreak of severe acute respiratory syndrome (SARS), and the challenge for vaccine development. J. Neurovirol. 2004;10(2):75–85. doi: 10.1080/13550280490280292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.He R., Adonov A., Traykova-Adonova M., Cao J., Cutts T., Grudesky E., Deschambaul Y., Berry J., Drebot M., Li X. Potent and selective inhibition of SARS coronavirus replication by aurintricarboxylic acid. Biochem. Biophys. Res. Commun. 2004;320(4):1199–1203. doi: 10.1016/j.bbrc.2004.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.O'Neil K.T., Hoess R.H., Jackson S.A., Ramachandran N.S., Mousa S.A., DeGrado W.F. Identification of novel peptide antagonists for GPIIb/IIIa from a conformationally constrained phage peptide library. Proteins: Struct. Funct. Bioinform. 1992;14(4):509–515. doi: 10.1002/prot.340140411. [DOI] [PubMed] [Google Scholar]
- 149.McLafferty M.A., Kent R.B., Ladner R.C., Markland W. M13 bacteriophage displaying disulfide-constrained microproteins. Gene. 1993;128(1):29–36. doi: 10.1016/0378-1119(93)90149-w. [DOI] [PubMed] [Google Scholar]
- 150.Hipolito C.J., Suga H. Ribosomal production and in vitro selection of natural product-like peptidomimetics: the FIT and RaPID systems. Curr. Opin. Chem. Biol. 2012;16(1–2):196–203. doi: 10.1016/j.cbpa.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 151.Palei S., Becher K.S., Nienberg C., Jose J., Mootz H.D. Bacterial cell‐surface display of semisynthetic cyclic peptides. Chembiochem. 2019;20(1):72–77. doi: 10.1002/cbic.201800552. [DOI] [PubMed] [Google Scholar]
- 152.Xiao W., Wang Y., Lau E.Y., Luo J., Yao N., Shi C., Meza L., Tseng H., Maeda Y., Kumaresan P., Liu R. The use of one-bead one-compound combinatorial library technology to discover high-affinity αvβ3 integrin and cancer targeting arginine-glycine-aspartic acid ligands with a built-in handle. Mol. Canc. Therapeut. 2010;9(10):2714–2723. doi: 10.1158/1535-7163.MCT-10-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wang X.S., Chen P.H., Hampton J.T., Tharp J.M., Reed C.A., Das S.K., Wang D.S., Hayatshahi H.S., Shen Y., Liu J., Liu W.R. A genetically encoded, phage‐displayed cyclic‐peptide library. Angew. Chem. Int. Ed. 2019;58(44):15904–15909. doi: 10.1002/anie.201908713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses—drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15(5):327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yang S., Fu C., Lian X., Dong X., Zhang Z. Understanding human-virus protein-protein interactions using a human protein complex-based analysis framework. mSystems. 2019;4(2) doi: 10.1128/mSystems.00303-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dyall J., Coleman C.M., Hart B.J., Venkataraman T., Holbrook M.R., Kindrachuk J., Johnson R.F., Olinger G.G., Jahrling P.B., Laidlaw M., Johansen L.M. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob. Agent Chemother. 2014;58(8):4885–4893. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sohrabi C., Alsafi Z., O'Neill N., Khan M., Kerwan A., Al-Jabir A., Iosifidis C., Agha R. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int. J. Surg. 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10(1):1–9. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.González R.G., Blackburn B.J., Schleich T. Fractionation and structural elucidation of the active components of aurintricarboxylic acid, a potent inhibitor of protein nucleic acid interactions. Biochim. Biophys. Acta Nucleic Acids Protein Synth. 1979;562(3):534–545. doi: 10.1016/0005-2787(79)90116-3. [DOI] [PubMed] [Google Scholar]
- 161.Cushman M., Wang P., Chang S.H., Wild C., De Clercq E., Schols D., Goldman M.E., Bowen J.A. Preparation and anti-HIV activities of aurintricarboxylic acid fractions and analogs: direct correlation of antiviral potency with molecular weight. J. Med. Chem. 1991;34(1):329–337. doi: 10.1021/jm00105a052. [DOI] [PubMed] [Google Scholar]
- 162.He Y., Zhou Y., Liu S., Kou Z., Li W., Farzan M., Jiang S. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem. Biophys. Res. Commun. 2004;324(2):773–781. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yap Y., Zhang X., Andonov A., He R. Structural analysis of inhibition mechanisms of aurintricarboxylic acid on SARS-CoV polymerase and other proteins. Comput. Biol. Chem. 2005;29(3):212–219. doi: 10.1016/j.compbiolchem.2005.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cortegiani A., Ingoglia G., Ippolito M., Giarratano A., Einav S. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J. Crit. Care. 2020;57:279–283. doi: 10.1016/j.jcrc.2020.03.005. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gao J., Tian Z., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;16(14):72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 166.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yao T.T., Qian J.D., Zhu W.Y., Wang Y., Wang G.Q. A systematic review of lopinavir therapy for SARS coronavirus and MERS coronavirus—a possible reference for coronavirus disease‐19 treatment option. J. Med. Virol. 2020;92(6):556–563. doi: 10.1002/jmv.25729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Khamitov R.A., SIa L., Shchukina V.N., Borisevich S.V., Maksimov V.A., Shuster A.M. Antiviral activity of arbidol and its derivatives against the pathogen of severe acute respiratory syndrome in the cell cultures. Vopr. Virusol. 2008;53(4):9–13. [PubMed] [Google Scholar]
- 170.Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV) Biosci. Trends. 2020;14(1):69–71. doi: 10.5582/bst.2020.01020. [DOI] [PubMed] [Google Scholar]
- 171.Lung J., Lin Y.S., Yang Y.H., Chou Y.L., Shu L.H., Cheng Y.C., Liu H.T., Wu C.Y. The potential chemical structure of anti‐SARS‐CoV‐2 RNA‐dependent RNA polymerase. J. Med. Virol. 2020;92(6):693–697. doi: 10.1002/jmv.25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Khan H., Sureda A., Belwal T., Çetinkaya S., Süntar İ., Tejada S., Devkota H.P., Ullah H., Aschner M. Polyphenols in the treatment of autoimmune diseases. Autoimmun. Rev. 2019;18(7):647–657. doi: 10.1016/j.autrev.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tome-Carneiro J., Visioli F. Polyphenol-based nutraceuticals for the prevention and treatment of cardiovascular disease: review of human evidence. Phytomedicine. 2016;23(11):1145–1174. doi: 10.1016/j.phymed.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 174.Cassidy L., Fernandez F., Johnson J.B., Naiker M., Owoola A.G., Broszczak D.A. Oxidative stress in alzheimer's disease: a review on emergent natural polyphenolic therapeutics. Compl. Ther. Med. 2020;49 doi: 10.1016/j.ctim.2019.102294. [DOI] [PubMed] [Google Scholar]
- 175.Runfeng L., Yunlong H., Jicheng H., Weiqi P., Qinhai M., Yongxia S., Chufang L., Jin Z., Zhenhua J., Haiming J., Kui Z. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2) Pharmacol. Res. 2020;156 doi: 10.1016/j.phrs.2020.104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Justesen U., Knuthsen P. Composition of flavonoids in fresh herbs and calculation of flavonoid intake by use of herbs in traditional Danish dishes. Food Chem. 2001;73(2):245–250. [Google Scholar]
- 177.Kaack K., Austed T. Interaction of vitamin C and flavonoids in elderberry (Sambucus nigra L.) during juice processing. Plant Foods Hum. Nutr. 1998;52(3):187–198. doi: 10.1023/a:1008069422202. [DOI] [PubMed] [Google Scholar]
- 178.Kim D.W., Seo K.H., Curtis-Long M.J., Oh K.Y., Oh J.W., Cho J.K., Lee K.H., Park K.H. Phenolic phytochemical displaying SARS-CoV papain-like protease inhibition from the seeds of Psoralea corylifolia. J. Enzym. Inhib. Med. Chem. 2014;29(1):59–63. doi: 10.3109/14756366.2012.753591. [DOI] [PubMed] [Google Scholar]
- 179.Keyaerts E., Vijgen L., Pannecouque C., Van Damme E., Peumans W., Egberink H., Balzarini J., Van Ranst M. Plant lectins are potent inhibitors of coronaviruses by interfering with two targets in the viral replication cycle. Antivir. Res. 2007;75(3):179–187. doi: 10.1016/j.antiviral.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Petersen K.A., Matthiesen F., Agger T., Kongerslev L., Thiel S., Cornelissen K., Axelsen M. Phase I safety, tolerability, and pharmacokinetic study of recombinant human mannan-binding lectin. J. Clin. Immunol. 2006;26(5):465–475. doi: 10.1007/s10875-006-9037-z. [DOI] [PubMed] [Google Scholar]
- 181.Müller C., Schulte F.W., Lange-Grünweller K., Obermann W., Madhugiri R., Pleschka S., Ziebuhr J., Hartmann R.K., Grünweller A. Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona-and picornaviruses. Antivir. Res. 2018;150:123–129. doi: 10.1016/j.antiviral.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.O'Keefe B.R., Giomarelli B., Barnard D.L., Shenoy S.R., Chan P.K., McMahon J.B., Palmer K.E., Barnett B.W., Meyerholz D.K., Wohlford-Lenane C.L., McCray P.B. Broad-spectrum in vitro activity and in vivo efficacy of the antiviral protein griffithsin against emerging viruses of the family Coronaviridae. J. Virol. 2010;84(5):2511–2521. doi: 10.1128/JVI.02322-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Millet J.K., Séron K., Labitt R.N., Danneels A., Palmer K.E., Whittaker G.R., Dubuisson J., Belouzard S. Middle East respiratory syndrome coronavirus infection is inhibited by griffithsin. Antivir. Res. 2016;133:1–8. doi: 10.1016/j.antiviral.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mani J.S., Johnson J.B., Steel J.C., Broszczak D.A., Neilsen P.M., Walsh K.B., Naiker M. Natural product-derived phytochemicals as potential agents against coronaviruses: a review. Virus Res. 2020 doi: 10.1016/j.virusres.2020.197989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Weng J.K., Philippe R.N., Noel J.P. The rise of chemodiversity in plants. Science. 2012;336:1667–1670. doi: 10.1126/science.1217411. [DOI] [PubMed] [Google Scholar]
- 186.Ernst M., Grace O.M., Saslis-Lagoudakis C.H., Nilsson N., Simonsen H.T., Ronsted N. Global medicinal uses of Euphorbia L. (Euphorbiaceae) J. Ethnopharmacol. 2015;176:90–101. doi: 10.1016/j.jep.2015.10.025. [DOI] [PubMed] [Google Scholar]
- 187.Gozubuyuk G.S., Aktas E., Yigit N. An ancient plant Lawsonia inermis (henna): determination of in vitro antifungal activity against dermatophytes species. J. Mycol. Med. 2014;24:313–318. doi: 10.1016/j.mycmed.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 188.Hotwani K., Baliga S., Sharma K. Phytodentistry: use of medicinal plants. J. Compl. Integr. Med. 2014;11:233–251. doi: 10.1515/jcim-2013-0015. [DOI] [PubMed] [Google Scholar]
- 189.Ji S., Fattahi A., Raffel N., Hoffmann I., Beckmann M.W., Dittrich R., Schrauder M. Antioxidant effect of aqueous extract of four plants with therapeutic potential on gynecological diseases; semen persicae, Leonurus cardiaca, Hedyotis diffusa, and Curcuma zedoaria. Eur. J. Med. Res. 2017;22:50. doi: 10.1186/s40001-017-0293-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ozdemir V., Hekim N. Birth of industry 5.0: making sense of big data with artificial intelligence, “the internet of things” and next-generation technology policy. Omics. 2018;22:65–76. doi: 10.1089/omi.2017.0194. [DOI] [PubMed] [Google Scholar]
- 191.Sharma P., Joshi T., Mathpal S., Joshi T., Pundir H., Chandra S., Tamta S. Identification of natural inhibitors against Mpro of SARS-CoV-2 by molecular docking, molecular dynamics simulation, and MM/PBSA methods. J. Biomol. Struct. Dyn. 2020:1–2. doi: 10.1080/07391102.2020.1842806. [DOI] [PMC free article] [PubMed] [Google Scholar]