Abstract
Lung inflammation is a hallmark of coronavirus disease 2019 (COVID-19). In this study, we show that mice develop inflamed lung tissue after being administered exosomes released from the lung epithelial cells exposed to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Nsp12 and Nsp13 (exosomesNsp12Nsp13). Mechanistically, we show that exosomesNsp12Nsp13 are taken up by lung macrophages, leading to activation of nuclear factor κB (NF-κB) and the subsequent induction of an array of inflammatory cytokines. Induction of tumor necrosis factor (TNF)-α, interleukin (IL)-6, and IL-1β from exosomesNsp12Nsp13-activated lung macrophages contributes to inducing apoptosis in lung epithelial cells. Induction of exosomesNsp12Nsp13-mediated lung inflammation was abolished with ginger exosome-like nanoparticle (GELN) microRNA (miRNA aly-miR396a-5p. The role of GELNs in inhibition of the SARS-CoV-2-induced cytopathic effect (CPE) was further demonstrated via GELN aly-miR396a-5p- and rlcv-miR-rL1-28-3p-mediated inhibition of expression of Nsp12 and spike genes, respectively. Taken together, our results reveal exosomesNsp12Nsp13 as potentially important contributors to the development of lung inflammation, and GELNs are a potential therapeutic agent to treat COVID-19.
Keywords: SARS-CoV-2, exosomes, ginger exosome-like nanoparticle, microRNA, lung inflammation, Nsp12, spike, NF-κB, TNF-α, IL-1β, IL-6, ACE2, lung epithelial cells, macrophages
Graphical abstract
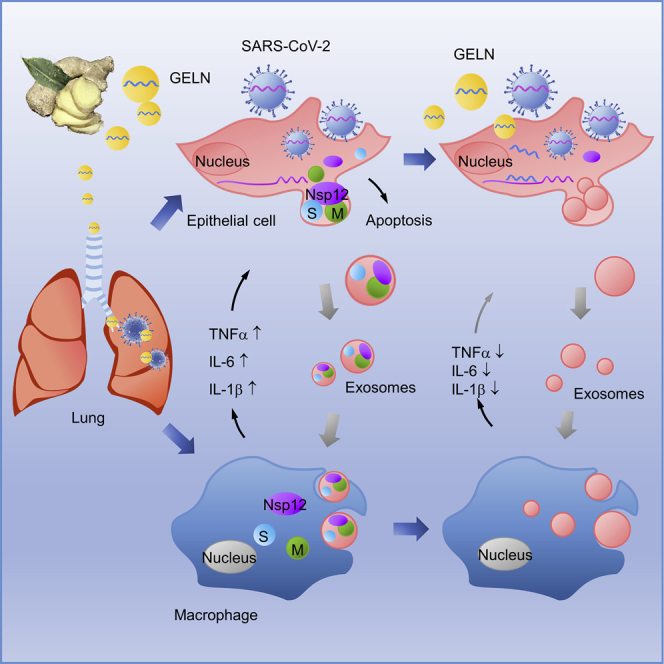
COVID-19 is a disease affecting the pulmonary system with devastating inflammation. Teng et al. have shown that exosomes released from SARS-CoV-2-infected lung epithelia cells are preferentially taken up by lung macrophages that subsequently induce lung inflammation. Ginger exosome-like nanoparticle miRNA inhibits SARS-CoV-2-induced lung inflammation and viral replication.
Introduction
Severe cases of coronavirus disease 2019 (COVID-19) cause a cytokine storm that results in high mortality. Hyperproduction of cytokines ultimately results in tissue damage including apoptosis and necrosis, leading to injury of alveolar epithelial cells and vascular endothelial cells, as well as to lung infiltration sustained by continuously infiltrating immune cells. However, delivering viral-specific therapeutic agents that can inhibit the expression of viral genes that contribute to the cytokine storm is challenging. mRNA encoding severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virulent factors could have homolog sequencing with the host mRNA. Therefore, it is unlikely to target to viral mRNA without an effect on the expression of the host mRNA.
We hypothesize that anti-viral therapeutic agents derived from plants that are not co-evolved within the mammalian kingdom could provide a more potent anti-viral effect with less potential to induce side effects. Recently we1 and others2 have identified exosome-like nanoparticles (NPs) (ELNs) from the tissue of edible plants. The reason we refer to edible plant-derived nanoparticles as plant ELNs is for the following reasons: similar to exosomes, ELNs are nanosized; they consist of proteins, lipids, and RNAs, in particular, small-sized RNA; and they are stable. When we eat food, ELNs are naturally released and they are taken up by host cells and subsequently communicate with ELN recipient cells, which is a biological hallmark of mammalian cell-derived exosomes. Unlike animal exosomes, which are difficult to produce in large quantities, ELNs can easily be isolated and purified in large quantities. Therefore, in this study, we sought to identify the therapeutic factors in ELNs, such as ginger ELN (GELN) microRNAs (miRNAs), that may inhibit the expression of SARS-CoV-2 genes without inducing side effects. Moreover, multiple species of miRNAs can be encapsulated in a single ELN, and each miRNA can potentially bind to multiple sites of the viral genome. Thus, the production of infectious virus is expected to be inhibited via blocking a number of pathways that are critical for generating a viral infection. Exosomes released from virally infected cells contribute to the cytokine storm.3, 4, 5, 6 Whether exosomes released from SARS-CoV-2-infected lung epithelial cells play a role in induction of inflammation cytokines, which further triggers tissue damage, is not known.
Studies in mice and humans have suggested that activation of the nuclear factor κB (NF-κB) pathway contributes to viral factors inducing the lung cytokine storm. The details of whether SARS-CoV-2-derived factors can induce lung inflammation are unknown. The SARS-CoV-2 Nsp12 RNA-dependent RNA polymerase (RdRP) and Nsp13 helicase are non-structural proteins. RdRP is an enzyme that catalyzes the synthesis of the SARS RNA strand complementarily to the SARS-CoV-2 RNA template and is thus essential to the replication of SARS-CoV-2 RNA. Similar to most other RNA viruses, SARS-CoV-2 RdRPs are considered to be highly conserved to maintain viral functions, and for this reason they are targeted in antiviral drug development as well as diagnostic tests.7
In this study, our results support the hypothesis that SARS-CoV-2 Nsp12 induces lung inflammation mediated by exosomes released from lung epithelial cells that can be inhibited by GELN-derived miRNA. Nsp13 has synergistic effects with Nsp12 on lung inflammation. These findings may shed light on therapy development for COVID-19 patients and open a new avenue for studying mechanisms underlying plant kingdom crosstalk with the mammalian kingdom via plant ELNs.
Results
Lung epithelial-derived exosomes containing SARS-COV-2 Nsp12 and Nsp13 have a synergistic effect on the induction of inflammatory cytokines in vitro and in vivo
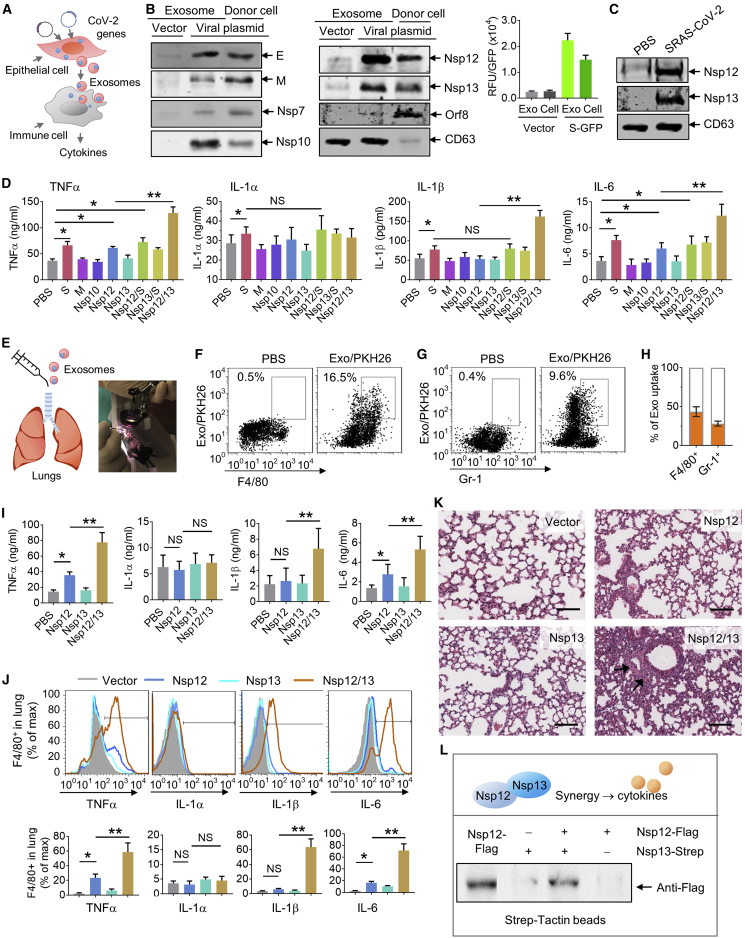
A growing number of reports suggest that infected cells use exosome-mediated intercellular communication to induce inflammation.6,8, 9, 10, 11, 12, 13, 14, 15 We first assessed whether exosomes released from lung epithelial cells expressing SARS-CoV-2 genes are loaded with the viral protein cargo. A mammalian expression vector expressing viral genes encoding for SARS-CoV-2, including spike (S), envelope (E), matrix (M), Nsp7, Nsp10, Nsp12, Nsp13, and orf8, were transfected into human lung epithelial A549 cells (Figure 1A; Table S1). Seventy-two hours after the transfection, exosomes released from the supernatants of cultured human lung epithelial A549 cells were isolated by differential centrifugation and confirmed by the exosome marker CD63 in a western blot assay (Figure 1B, left and middle panels). Using Strep-Tactin-horseradish peroxidase (HRP) conjugate, we found that all of the viral proteins co-expressed with 2xStrep were successfully expressed in A549 cells; the cargo in A549 cell-released exosomes contained viral protein E, Nsp7, Nsp10, Nsp12, Nsp13, and slight protein M, but not orf8 (Figure 1B, left and middle panels). In an independent experiment, we also demonstrated that viral protein S fused with green fluorescent protein (GFP) in the cell-derived exosomes (Figure 1B, right panel; Figures S1A and 1B). To further confirm this result, Vero E2 cells were transfected with plasmid expressing Nsp12 and Nsp13, respectively, for three days, the exosomes in the medium were isolated, and the western blot analyses suggested that the exosomes released from Vero E2 cells contain viral Nsp12 and Nsp13 (Figure 1C). Considering that the lung epithelial cell exosomes can be taken up by lung macrophages,16 we evaluated the impact of the exosomes with viral protein on macrophage activation. The level of cytokines in the medium of human monocyte U937 cells was quantitatively analyzed with a standard enzyme-linked immunosorbent assay (ELISA). Interestingly, as an RNA polymerase, Nsp12 alone slightly induced tumor necrosis factor (TNF)-α and interleukin (IL)-6. Moreover, the synergistic effect of Nsp12 working with Nsp13 was observed where a dramatic induction of TNF-α, IL-1β, and IL-6 occurred when compared to Nsp12 alone, whereas protein M, Nsp13 alone, or Nsp10 did not (Figure 1D). We also found that exosomes containing protein S induce TNF-α, IL-1α, IL-1β, and IL-6, but such exosomes have no synergistic effect with Nsp12 or Nsp13 on the impact of cytokines (Figure 1D). To further confirm that the A549 exosome-mediated induction of inflammation cytokine TNF-α, for example, is viral Nsp12 and Nsp13 specific, U937 cells were exposed to A549-derived exosomes, and ELISA analysis of cytokines suggested no significant influence of protein expression of TNF-α from 1 h up to 12 h post-exposure (Figure S1C).
Figure 1.
Lung epithelial cells release exosomes containing Nsp12 of SARS-CoV-2 that enhances the inflammatory response in lung
(A) Schematic representation of the treatment schedule for the effect of lung epithelial cell-derived exosomes containing SARS-CoV-2 proteins on lung immune cells. (B) SARS-CoV-2 protein expression plasmids transfected into lung epithelial A549 cells. (Left and middle panels) Representative blots of viral proteins in exosomes and cells as well as exosomal marker CD63 by western blot using Strep-Tactin-HRP conjugate and antibody to CD63. (Right panel) Intensity of GFP fused with spike (S) protein expressed in exosomes (Exos) and cells using BioTek’s Synergy microplate reader. (C) Representative western blot of exosomes from Vero E2 cells transfected with Nsp12 and Nsp13 plasmids. (D) Cytokines in the medium assessed by ELISA. (E) Schematic representation of intratracheal injection (left panel) and a mouse undergoing laryngoscopy to expose the vocal cords (right panel). (F and G) Exosomes from mouse lung LLC1 cells transfected with SARS-CoV-2 plasmids administered to C57BL/6 mice (5 × 108/kg, body weight, n = 5) by intratracheal injection. After 24 h, the frequencies of F4/80+ cells (F), Gr-1+ cells (G), and PKH26-labled exosomes in the lung from C57BL/6 mice were assessed using flow cytometry. Numbers in boxes indicate the percentage of exosome/PKH26+ cells. (H) Quantification of percentage of exosome/PKH26+ in F4/80+ cells and Gr-1+ cells. (I) (Top panel) Assessment of cytokines in the lungs using ELISA. (J) Cytokines in the F4/80+ cells assessed by flow cytometry. (Bottom panel) Quantification of data from flow cytometry. (K) Representative hematoxylin and eosin (H&E)-stained sections of formalin-fixed, paraffin-embedded lungs (original magnification, ×400; scale bars, 200 μm) from C57BL/6 mice. (L) A549 cells co-transfected with the plasmids of pAcGFP1-C-Nsp12-FLAG and pLVX-Nsp13-Strep. At 72 h after transfection, Nsp12/13 complex pull-down by Strep-Tactin XT magnetic beads and immunoblot analysis with anti-FLAG antibody are shown. Data are representative of three independent experiments (error bars, SD). ∗p < 0.05, ∗∗p < 0.01 (two-tailed t test).
To further investigate the effect of exosomes with viral protein in vivo, exosomes isolated from mouse lung epithelial LLC1 cells transfected with appropriate viral genes were administered to mice by direct intratracheal injection (Figure 1E). LLC1 cells are immunologically compatible with the murine immune system of C57BL/6 mice. The imaging fluorescence signals indicated that exosomes labeled with fluorescent DiR dye were present in lungs and serum as soon as 1 h after intratracheal administration of C57BL/6 mice (Figure S2A). Twelve hours after intratracheal injection, the fluorescent signals diminished. No significant fluorescence appeared in the brain, heart, liver, kidney, or intestine of mice (Figure S2A).
To further determine whether injected exosomes preferentially targeted immune cells, leukocytes from the lungs of mice treated with the fluorescent dye PKH26-labeled exosomes were isolated. Fluorescence-activated cell sorting (FACS) analysis (Figures 1F and 1G) and immunofluorescence (IF) with confocal microscopy (Figures S2B and S2C) demonstrated that the exosomes were preferentially taken up by F4/80+ cells (Figure 1F; Figure S2B) and moderately taken up by Gr-1+ cells (Figure 1G; Figure S2B), but not by CD11b+ cells (Figure S2C). Among F4/80+ macrophages and Gr-1+ neutrophils, 43.3% ± 6.8% (mean ± standard deviation [SD]) and 28.2% ± 3.7% (mean ± SD) of the cells exhibited exosome and PKH26 positivity, respectively (Figure 1H). To determine whether injected exosomes have an effect on cytokine production, after administering LLC1 exosomes three times via the intratracheal route, murine lung tissues were collected, and cytokines were quantified with a standard ELISA. The results suggested that exosomes containing Nsp12 induced TNF-α and IL-6 in the lung, whereas Nsp13 alone did not, but injecting exosomes containing both Nsp12 and Nsp13 led to a dramatic enhancement in the production of TNF-α, IL-1β, and IL-6 (Figure 1I). These results were also reproducible when mice were treated with exosomes released from Vero E2 cells transfected with Nsp12 and Nsp13 plasmids. The analysis of cytokines in the lung indicated a synergistic effect of Nsp12 and Nsp13 on the induction of TNF-α, IL-1β, and IL-6 (Figure S2D). We further assessed the cytokine expression in lung F4/80+ cells since F4/80+ cells are the predominant exosome recipient cells. Consistent with results from human U937 monocytes, the FACS analysis suggested that Nsp12 induces TNF-α and IL-6 in F4/80+ cells, and Nsp13 enhanced the effect of Nsp12 on the induction of these cytokines (Figure 1J). The inoculation of exosomes containing both Nsp12 and Nsp13 caused lung alveolar wall thickening and lung inflammation (Figure 1K). Lung alveolar wall thickening and lung inflammation did not occur when exosomes containing either Nsp12 or Nsp13 only were used. In addition, the synergistic effect of exosomes Nsp12 and Nsp13 was also evidenced by the fact that increasing levels of TNF-α and IL-6 occur not only in the lung but also in the peripheral blood (Figure S2E) of mice treated with exosomesNsp12Nsp13. The induction of inflammatory cytokines in the C57BL/6 mouse lung was also confirmed by exosomes released from lung primary epithelial cells transfected with Nsp12 and Nsp13 plasmids (Figure S2F).
We then further proposed that Nsp12 might interact with Nsp13 to form the complex that regulates expression of cytokines of exosome recipient cells (Figure 1L). To test our hypothesis, we performed co-immunoprecipitation (coIP) utilizing the Nsp12 with FLAG tag (pGBW-m4046955) and the Nsp13 with Strep tag (pLVX-EF1alpha-SARS-CoV-2-nsp13-2xStrep-IRES-Puro). First, a full-length wild-type Nsp12 gene was constructed from the prokaryotic vector transfected into the eukaryotic vector pAcGFP1-C with a promoter of cytomegalovirus (CMV). After transfection and expression in A549 cells with the plasmids of pAcGFP1-C-Nsp12 and pLVX-Nsp13-Strep, the Nsp13 protein was pulled down with Strep-Tactin XT magnetic beads and probed to determine whether Nsp12 was in the Nsp13 pull-down complex with western blot analysis using anti-FLAG antibody. As expected, the results (Figure 1L) indicated that Nsp12 interacted with Nsp13 in A549 cells. Taken together, our data indicate that Nsp12 is not only capable of mediating RNA synthesis and replication of the viral genome, but it also is cargo in the exosomes and subsequently induces inflammatory cytokines in the exosome recipient cells. Moreover, Nsp12-mediated induction of inflammatory cytokines can be further enhanced by viral Nsp13.
GELN miRNAs inhibit the expression of SARS-CoV-2 S and Nsp12
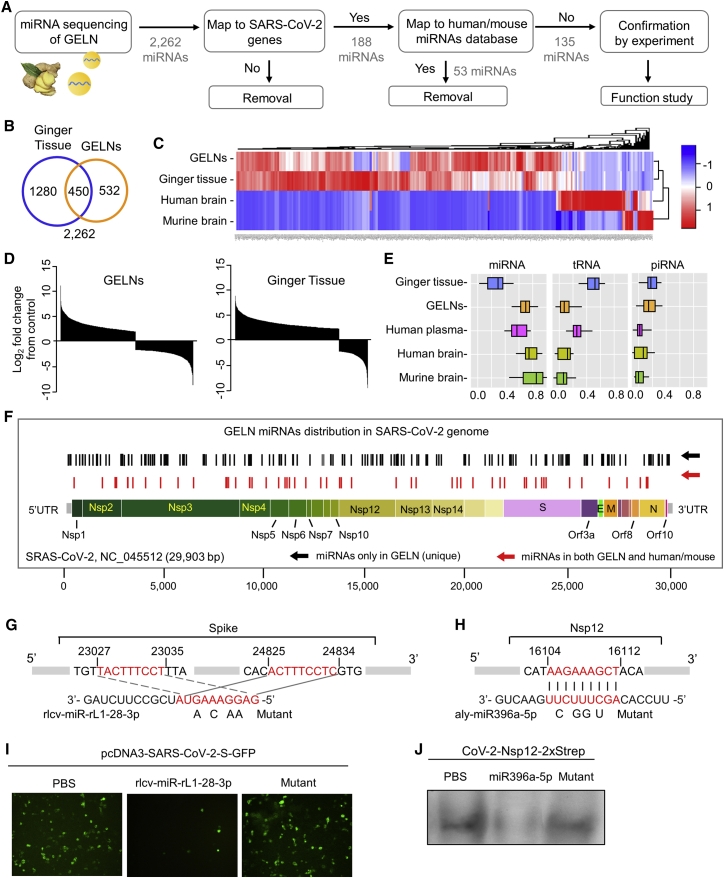
Next, we asked whether a therapeutic strategy could be developed to inhibit the lung cytokine storm and prevent viral Nsp12-induced lung inflammation. We have published results17 that indicate that GELNs can inhibit mouse colitis via GELN miRNA interaction with gut bacterial mRNA. Therefore, in this study, we hypothesize that GELN miRNA may potentially bind to and inhibit SARS-CoV-2 mRNA expression. We first purified the GELNs from ginger root using differential ultracentrifugation and a sucrose gradient technique.1,17, 18, 19 Next-generation sequencing (NGS) analysis of small RNA in the ginger root tissue and in GELNs identified 2.2 million and 3.6 million miRNA readouts of 32 and 42 million total reads, respectively (Figure 2A). Combining our new sequencing data (the sequenced data deposited in NCBI Gene Expression Omnibus [GEO]: GSE153126) and previous sequencing data,17 2,262 of the miRNAs exceeded the minimum confidence thresholds (cutoff of 50 reads) and have been mapped to the entire miRNA database. Of the miRNAs, 532 are higher in GELNs and 1,280 of the miRNAs are higher in ginger tissue (Figure 2B). Further analysis of the cluster (Figure 2C), abundance (Figure 2D), and composition (Figure 2E) of miRNAs revealed significant differences between GELNs and ginger tissue, humans, and mice.20, 21, 22 The miRNA cargo in GELNs is more enriched than in ginger tissue; in contrast, the ginger tissue has more tRNA compared to the GELNs.
Figure 2.
GELN miRNA potentially targets to the RNA of the SARS-CoV-2
(A) Flow diagram depicting the steps taken in identifying unique GELN miRNAs potentially targeting the RNA of SARS-CoV-2. (B) Venn diagram of miRNAs detected in the ginger tissue and GELNs using miRNA sequencing. (C) Heatmap showing miRNAs from ginger tissue, GELNs, and human and mouse brain (n = 3 per group). (D) Waterfall plot showing the differences in the relative abundance of miRNAs between GELNs and ginger tissue normalized by human miRNAs. (E) Distribution of RNA biotype differences. Boxes represent median and interquartile ranges. (F) Schematic diagram and distribution of the putative binding sites of GELN miRNAs in the full-length SARS-CoV-2 genome. UTR, untranslated region. The miRNAs of humans and mice that have the same mapping seed sequences as GELNs are indicated in red and were excluded in further experiments. (G and H) Predicted consequential pairing of target region of spike gene (G, top), Nsp12 gene (H, top), GELN rlcv-miR-rL1-28-3p (G, bottom), and aly-miR396a-5p (H, bottom), respectively. The miRNA seed matches in the target RNAs are mutated at the positions as indicated. (I) A549 cells transfected with CoV-2 S inserted into pcDNA3-GFP and GELN rlcv-miR-rL1-28-3p, mutant RNA. Visualization with confocal fluorescence microscopy. (J) A549 cells transfected with Nsp12 inserted into pLVX-EF1alpha-2xStrep-IRES and GELN aly-miR396a-5p, mutant RNA. Visualization is with Strep-Tactin-HRP conjugate by immunoblot. Data are representative of three independent experiments (error bars, SD). ∗p < 0.05, ∗∗p < 0.01 (two-tailed t test).
To further explore the potential therapeutic effects of miRNA against SARS-CoV-2, a strict seed sequence length of 9 nt in GELN miRNAs was used as a criterion to search for the genome sequence of SARS-CoV-2 (NCBI: NC_045512). We chose 9 nt, although 6 nt is the minimum requirement, and the 6- to 8-nt-long seed sequence of the miRNA is sufficient to bind the target mRNA.23 188 GELN miRNAs (black bars in Figure 2F) that potentially bind to genes of SARS-CoV-2 were identified across the SARS-CoV-2 viral genome except for the genes encoding Nsp7, Nsp11, E, Orf8, and Orf10 (Figure 2F). Considering the world-wide prevalence of SARS-CoV-2 infection and the apparent lack of immunity to prevent SARS-CoV-2 infection, host miRNAs that are sequence homologs to GELN miRNA may not play a critical role in the inhibition of viral gene expression, in particular, Nsp12, which is an essential gene for viral replication. Therefore, we excluded from further study the GELN miRNAs (red bars in Figure 2F) bearing the seed sequences that map to the human or mouse miRNA database. The remaining 135 miRNAs that are unique for GELNs were used as the potential miRNAs to target viral genes. The GELN miRNAs as well as their seed sequences that could potentially bind to the genes of SARS-CoV-2 are listed in Table S2. Interestingly, very few of the human or mouse miRNAs were found to potentially bind to SARS-CoV-2 genes, but more miRNAs with matching sequences were found in GELNs, especially for the SARS-CoV-2 Nsp12 and S genes. Also, some of the GELN miRNAs may bind to multiple sites of a single viral gene (Figure 2F). As a proof of concept in this study, we further tested the effect of GELN rlcv-miR-rL1-28-3p and aly-miR396a-5p on the appropriate viral gene expression. An alignment of sequences of nucleotides using the Basic Local Alignment Search Tool (BLAST) indicated that GELN rlcv-miR-rL1-28-3p potentially binds two sites of the S gene (Figure 2G) and aly-miR396a-5p binds to the Nsp12 gene (Figure 2H). To confirm our prediction, pcDNA3-SARS-CoV-2-S-GFP and CoV-2-Nsp12-2xStrep were co-transfected into A549 cells with GELN rlcv-miR-rL1-28-3p and aly-miR396a-5p, respectively, synthesized and purchased from Eurofins. The expression of S and Nsp12 was significantly downregulated by rlcv-miR-rL1-28-3p and aly-miR396a-5p as visualized with GFP and Strep-Tactin-HRP conjugate, respectively (Figures 2I and 2J).
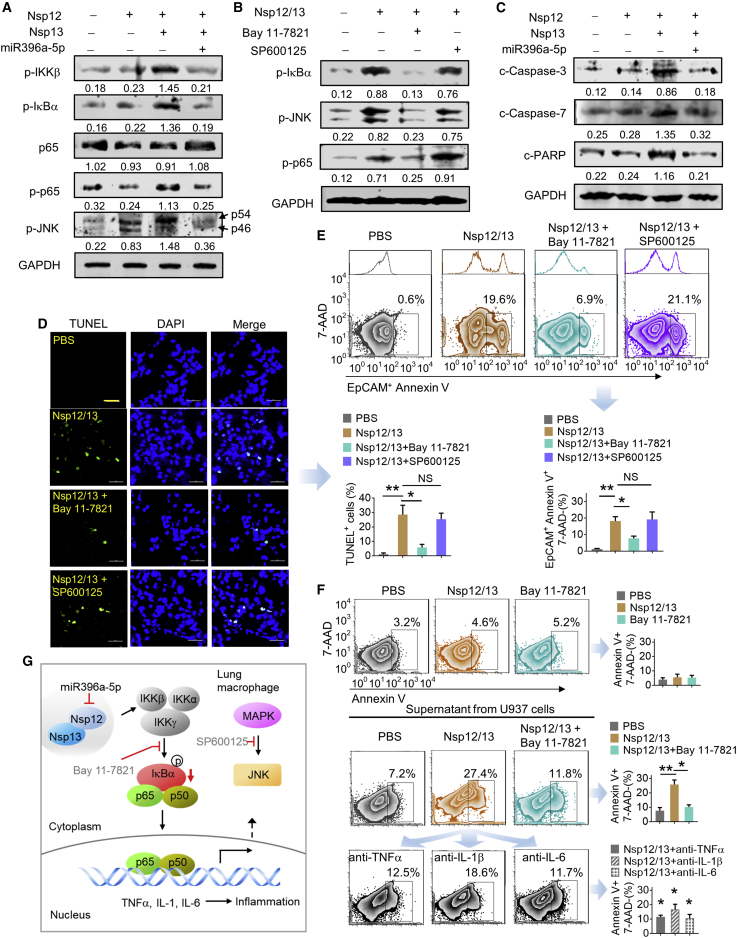
GELN aly-miR396a-5p inhibits NF-κB-mediated inflammation and apoptosis in the lung of mice intratracheally injected with exosomesNsp12Nsp13
Lung inflammation and apoptosis leading to acute respiratory distress syndrome (ARDS) are a hallmark of COVID-19; however, identification of the specific pathways that the viral products induce to elicit lung inflammation and apoptosis are still unknown. Based on the literature, activation of mitogen-activated protein kinase (MAPK, p38), extracellular signal-regulated kinase (ERK)1/2 (p44/42), c-Jun N-terminal kinase (JNK), phosphatidylinositol 3-kinase (PI3K), as well as NF-κB (p65) are all involved in virally induced inflammation and apoptosis. Western blot analysis indicates that mice intratracheally administered exosomesNsp12Nsp13 induce more phosphorylated (p-)p65 (a subunit of NF-κB) and JNK in mouse lung macrophages compared to Nsp12 alone (Figure 3A). There is no evidence suggesting the induction of p-p38, p-ERK1/2, or p-PI3K (Figure S3A). Activation of NF-κB mainly occurs via IκB kinase (IKK)-mediated phosphorylation of IκB-α.24 Our results show that more phosphorylated IKK-β and IκB-α are induced by exosomesNsp12Nsp13 (Figure 3A). Importantly, GELN aly-miR396a-5p treatment prevents the exosomesNsp12Nsp13-mediated activation of NF-κB. Activation of JNK was also detected in the macrophages of mice treated with the exosomesNsp12Nsp13 (Figure 3A), and aly-miR396a-5p treatment prevents the exosomesNsp12Nsp13-mediated activation of JNK as well (Figure 3A). To determine whether the NF-κB activated by exosomesNsp12Nsp13 is initiated from activation of IKK, we treated mice with p-IκB-α inhibitor, Bay 11-7821, or p-JNK inhibitor, SP600125, as a control. The treatments were done daily for 3 days at 1025 or 5 mg/kg/day26 (body weight) before mice were intratracheally injected with exosomesNsp12Nsp13. Lung macrophage NF-κB activity induced by exosomesNsp12Nsp13 was inhibited as a result of the Bay 11-7821 treatment, but not treatment with SP600125, suggesting that IKK activated by exosomesNsp12Nsp13 is essential in NF-κB activation (Figure 3B). Inhibition of NF-κB activation with Bay 11-7821 was also accompanied by attenuation of inflammatory cytokines induced by exosomesNsp12Nsp13 (Figure S3B). Cytokines have been reported to induce lung apoptosis,27,28 which is consistent with our observations that exosomesNsp12Nsp13 treatment enhanced the production of the cleaved caspase-3, caspase-7, and PARP in lung (Figure 3C) and aly-miR396a-5p against the effect of apoptosis induced by Nsp12/13 (Figure 3C). TUNEL (terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick end labeling) staining lung sections further showed that apoptotic cells were induced in the lungs of mice treated with exosomesNsp12Nsp13, and lung apoptosis induced by exosomesNsp12Nsp13 was reduced as a result of treatment with IκB-α inhibitor Bay 11-7821 but not SP600125 (Figure 3D). Moreover, flow cytometry and IF analysis further suggested that higher percentages of apoptotic annexin V+ (Figure 3E) and TUNEL+ cells (Figure S3C) in lung epithelial cells (EpCAM+) were induced as a result of exosomesNsp12Nsp13 treatment, and the induction of lung epithelial cell apoptosis was blocked by Bay 11-7821 but not Sp600125 (Figure 3E). Given that apoptotic cells may generate apoptotic bodies (ABs),29,30 whether ABs may also contribute to induction of inflammatory cytokines was further investigated. We analyzed the effect of ABs released from lung epithelial cells on the immune response of macrophages. Lung epithelial LLC1 cells were transfected with viral genes Nsp12 and Nsp13 as well as aly-miR396a-5p. The exosomesNsp12Nsp13 and exosomesNsp12/13+miR396a-5p from the medium were administrated to mice via intratracheal injection. The lung epithelial cells were isolated and the ABs purified from the cultured medium were quantified with FACS as annexin V+ of 1.0–4.0 μm in size.31 We found that exosomesNsp12Nsp13 significantly induce the ABs and aly-miR396a-5p reduced the exosomesNsp12Nsp-mediated apoptotic effect of lung epithelial cells (Figure S3D). To identify whether the ABs released from the lung epithelial cells have an effect on the activity of lung macrophages, 1 × 108 ABs released from 1 × 106 lung epithelial cells were administrated to mice via intratracheal injection. The analysis of cytokines with ELISA indicated that ABs failed to modulate the cytokine levels in the lung (Figure S3E). Collectively, our data suggest that GELN aly-miR396a-5p treatment prevented exosomesNsp12Nsp13-mediated NF-κB activation and lung epithelial cell apoptosis.
Figure 3.
aly-miR396a-5p reduces NF-κB activated by Nsp12/13 through phosphorylation of IKKβ
(A) Western blot analysis showing the phosphorylation (p) of IKKβ, IκBα, and NF-κB (p65). JNK as well as total NF-κB (p65) are shown in macrophages of the lung in C57BL/6 mice (n = 5) inoculated by intratracheal administration with exosomes (5 × 108/kg, body weight) from LLC1 cells transfected with Nsp12 and/or Nsp13 as well as aly-miR396a-5p. Arrows mark the positions of p54 and p46 subunits of p-JNK. GAPDH served as a loading control. Numbers below western blots represent densitometry values normalized to the loading control. (B) Pretreatment with p-JNK inhibitor (SP600125, 5 mg/kg/day, body weight) and p-IκBα inhibitor (Bay 11-7821, 10 mg/kg/day, body weight) (n = 5) by intraperitoneal injection 3 days following intratracheal administration of exosomes. Western blot analysis shows p-IκBα, p-JNK, and p-p65 in lung macrophages. (C) Western blot analysis of cleaved (c-)caspase-3, c-caspase-7, and c-PARP in the lungs of mice. (D) Analysis of apoptosis by TUNEL staining in lung tissues. The TUNEL assay revealed apoptotic-positive cells in lung marked by GFP staining. The blue DAPI stain marks intact DNA. Original magnification, ×400 (left panel). (Right panel) Quantification of TUNEL-positive cells. The data were collected by counting positive cells from three lung sections of specimens and are shown as mean ± SD versus vehicle. ∗∗p < 0.01. NS, not significant. (E) Analysis of apoptosis by flow cytometry using annexin V-FITC staining in EpCAM+ cells of lungs from mice. (Top panel) Numbers in boxes indicate a representative percentage of EpCAM+ apoptotic cells. The adjunct histograms display the univariate plots that correspond to the EpCAM in the bivariate plot. (Bottom panel) Quantification of percentage of EpCAM+annexin V+7-aminoactinomycin D (7-AAD)− cells. Data are representative of three independent experiments (error bars, SD). ∗p < 0.05, ∗∗p < 0.01 (two-tailed t test). (F) Analysis of apoptosis by flow cytometry in lung epithelial A549 cells presented to Nsp12/13 and Bay 11-7821 (top panel), or culture supernatant from U937 macrophages treated with A549-derived Nsp12/13 exosomes with or without Bay 11-7821 (middle panel), and anti-TNF-α, anti-IL-1β, and anti-IL-6 antibodies (10 ng/mL, bottom panel), respectively. Numbers in boxes indicate a representative percentage of annexin V+7-AAD− apoptotic cells. (Right panel) Quantification of percentage of annexin V+7-AAD− cells. Data are representative of three independent experiments (error bars, SD); versus Nsp12/13 group: ∗p < 0.05, ∗∗p < 0.01 (two-tailed t test). (G) Proposed model for the crosstalk between GELN miR396a-5p that regulates cytokine expression mediated by SARS-CoV-2 Nsp12 in a manner dependent on NF-κB signaling.
We show that exosomesNsp12Nsp13 mediated the induction of inflammatory cytokines via activation of NF-κB in macrophages and lung epithelial cell apoptosis. Whether activated macrophages play a role in the induction of apoptosis of lung epithelial cells was further investigated. FACS analysis indicated that Nsp12/13 and Bay 11-7821 have little influence in lung epithelial A549 cell apoptosis (Figure 3F, top panel). However, the supernatant from human macrophage U937 cells treated with exosomesNsp12Nsp13 significantly induced the apoptosis of lung epithelial cells (Figure 3F, middle panel). The induction of apoptosis was inhibited by Bay 11-7821 (Figure 3F, middle panel), as well as with exosomesNsp12Nsp13-activated macrophage supernatants treated with antibodies against inflammatory cytokines TNF-α, IL-1β, and IL-6 (Figure 3F, bottom panel). Taken together, these data suggest that lung epithelial cells release SARS-CoV-2 exosomes carrying Nsp12/13 as cargo, leading to activation of macrophage NF-κB and subsequent induction of inflammatory cytokines. The cytokine profile induced by exosomes Nsp12 and Nsp13 not only causes lung inflammation but also induces lung epithelial cell apoptosis (Figure 3G). GELN aly-miR396a-5p prevents the inflammatory response and cell apoptosis through specifically targeting the inhibition of expression of the Nsp12 viral gene.
Intratracheal delivery of GELN aly-miR396a-5p inhibits lung inflammation induced by viral Nsp12
To further determine whether GELN miRNA can inhibit the lung inflammation induced by Nsp12, GELN Aly-miR396a-5p was packed into nanoparticles made from GELN-derived total lipids. Along with rare significant adverse effects,17 GELN-derived nanovectors (GNVs) have a number of advantages over nanovectors that are available through commercial markets as demonstrated below.
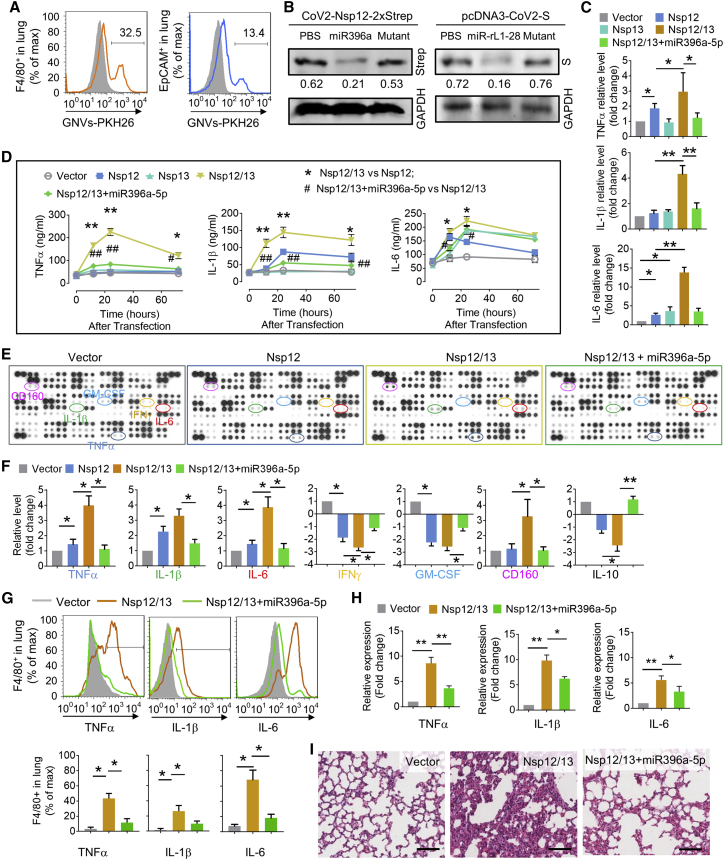
GNVs administrated by intratracheal injection are selectively taken up by lung epithelial cells (host cells for SARS-CoV-2 replication) and macrophages (source for releasing inflammatory cytokines induced by exosomesNsp12Nsp13). First, GELNs were purified with sucrose gradient centrifugation of ginger juice (Figure S4A) using a method as described,17 and GNVs were generated with total lipids extracted from GELNs using an ultrasonication method as described.19 The GNVs were further characterized using NanoSight NS300 for size distribution, concentration (Figure S4B), and yield (Figure S4C) and then electron microscopically examined (Figure S4D). One hour after intratracheal administration of GNVs in mice, the DiR fluorescent dye-labeled GNVs/DiR signal was detectable in the lungs and lasted up to 24 h (Figure S4E, left panel). Imaging of the small intestine excluded misplacement of the esophagus by intratracheal injection (Figure S4E, right panel). When comparing the characteristics of GNVs and GELNs based on size distribution and morphological features from transmission electron microscopy (TEM) analysis, we did not see any visible differences between GNVs and GELNs. FACS analysis indicated that PKH26+ GNVs were taken up by both F4/80+ macrophages and EpCAM+ lung epithelial cells (Figure 4A). This result was further confirmed by fluorescence co-localization analysis using confocal microscopy (Figure S5A). Moreover, the IF revealed that the cells in lung with high expression of ACE2 preferentially take up GNVs (Figure S5B). Given that the lipid of nanoparticles influences target cell uptake,17 we tested the effect of three predominate lipids constituting the nanoparticles, i.e., phosphatidic acid (PA), phosphatidylcholine (PC), phosphatidylethanolamine (PE), on the uptake of GNVs/PKH26 using flow cytometry. We found that additional PE promoted the GNVs uptake by A549 cells, whereas PA and PC inhibited uptake (Figure S5C).
Figure 4.
GELN aly-miR396a-5p suppresses the expression of cytokines mediated by Nsp12/13 synergy
(A) GELN-derived nanovectors (GNVs, 10 mg) administered to C57BL/6 mice (n = 5) by intratracheal injection. Representative flow cytometry plots show GNVs stained with PKH26 in F4/80+ cells (left) and EpCAM+ cells (right) of lungs 12 h after intratracheal injection. (B) Western blot analysis expression of Nsp12-Strep and spike protein in lungs with Strep-Tactin-HRP conjugate and anti-S antibody 48 h after administration of viral plasmid CoV-2-Nsp12-2xStrep and pcDNA3-CoV-2-S, as well as GNVs packing aly-miR396a-5p and rlcv-miR-rL1-28 or appropriate mutant RNA, respectively, by intratracheal injection. GAPDH served as a loading control. Numbers below the western blots represent densitometry values normalized to the loading control. (C) ELISA analysis showing the levels of TNF-α, IL-1β, and IL-6 in human macrophage U937 cells transfected with Nsp12 and/or Nsp13 as well as aly-miR396a-5p. (D) ELISA analysis showing the level of TNF-α, IL-1β, and IL-6 in the lungs inoculated with Nsp12 and/or Nsp13 as well as aly-miR396a-5p through intratracheal administration. Nsp12/13 versus Nsp12 or Nsp13, ∗p < 0.05, ∗∗p < 0.01; Nsp12/13+miR396a-5p versus Nsp12/13, #p < 0.05, ##p < 0.01. (E) Analysis of cytokine levels in lungs from C57BL/6 mice with indicated treatment in the figures through intratracheal administration using a mouse cytokine array (n = 3). (F) Quantification of relative intensity of the selective upregulation and downregulation of cytokines shown in the cytokine array. (G) Cytokines in F4/80+ cells assessed by flow cytometry (top panel). (Bottom panel) Quantification of data from flow cytometry. (H) Cytokines in F4/80+ cells assessed by qPCR. (I) Representative H&E-stained sections of lungs (original magnification, ×400; scale bars, 200 μm). Data are representative of three independent experiments (error bars, SD). ∗p < 0.05, ∗∗p < 0.01 (two-tailed t test).
An anti-inflammation effect as demonstrated in a lipopolysaccharide (LPS)-induced septic mouse model. In contrast to gold nanoparticles that are widely used for chemotherapeutic drug delivery, the GNV anti-inflammatory effect was demonstrated in an LPS-induced lung cytokine storm using the mouse septic model (Figure S5D) without induced side effects observed. To estimate the GNV-related liver toxicity and adverse effects on cells, we measured the level of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) in the serum and cell proliferation. ENVs exhibited neither toxicity in vivo (Figure S5E) nor growth influence on the lung epithelial cells (Figure S5F).
miRNA is effectively packed and delivered to the lung with GNVs
To further test whether GNVs can deliver miRNA to the lung, we first evaluated the packing efficiency of aly-miR396a-5p in GNVs using a quantitative PCR (qPCR) assay. The cargo aly-miR396a-5p in GNVs is 1.22 ± 0.32 μg per 1011 nanoparticles. With 10 μg of aly-miR396a-5p and 200 μmol of GELN-derived total lipids, 4.68 ± 1.03 μg of aly-miR396a-5p per 1011 nanoparticles can be generated (Figure S6A). Moreover, the transfection efficiency of GNVs carrying aly-miR396a-5p was further evaluated. An in vitro PCR analysis indicated that the miRNA level delivered with GNVs was higher than that delivered with poly(ethylenimine) (PEI), which is commonly used to deliver therapeutic mRNAs; the transfection reagent RNAiMAX exhibited the highest transfection efficiency (Figure S6B). In vivo delivery efficiency of aly-miR396a-5p packaged in GNVs was compared with gold nanoparticles. After intratracheal injection, the level of aly-miR396a-5p packed in GNVs was higher than the miRNA packed with gold nanoparticles (Figure S6C). We did not find a difference in the miRNA level between the top and bottom lobes of the lungs (Figure S6C). This result suggested that the GNVs administered by intratracheal injection were distributed throughout the entire lung. Immunoblot and qPCR analyses from in vivo experiments suggested that the expression of viral Nsp12 and S was inhibited by GELN aly-miR396a-5p and rlcv-miR-rL1-28-3p packed in GNVs, respectively (Figure 4B; Figure S6D).
We next sought to determine whether the aly-miR396a-5p delivered by GNVs inhibits the expression of inflammatory cytokines induced by Nsp12 of SARS-CoV-2. Human monocyte U937 cells were transfected with Nsp12 and/or Nsp13, simultaneously, in the presence or absence of aly-miR396a-5p packed in GNVs. ELISA results demonstrated that aly-miR396a-5p remarkably by some means suppressed the induction of TNF-α, IL-1β, and IL-6 (Figure 4C). We then harvested the exosomes from lung epithelial cells transfected with Nsp12, Nsp13, as well as aly-miR396a-5p and exposed mice to these exosomes through intratracheal administration. We found that exosomesNsp12Nsp13-induced TNF-α, IL-1β, and IL-6 in lung peaked 24 h after inoculation with exosomes (Figure 4D). ExosomesNsp12/13+miR396a-5p have no influence on cytokines in the lungs (Figure 4D). Collectively, our data indicated that aly-miR396a-5p had an inhibitory effect of Nsp12/13 on the expression of inflammatory cytokines.
The results generated from cytokine array analysis further demonstrated that besides modulation of TNF-α, IL-1β, and IL-6, exosomesNsp12Nsp13 significantly lowered the levels of interferon (IFN)-γ and IL-10, and aly-miR396a-5p prevented the reduction of IFN-γ (Figures 4E and 4F). Moreover, we found a number of proteins involving cell growth, including granulocyte-macrophage colony-stimulating factor (GM-CSF), G-CSF, M-CSF, and fibroblastic growth factor (FGF)-21, which were downregulated by exosomesNsp12Nsp13 (Figures 4E and 4F; Figures S7A and S7B). The levels of proteins in the chemokine (C-X-C motif) ligand (CXCL) family, including CXCL9, CXCL10, CXCL11, and CXCL16, were decreased as well due to exosomesNsp12Nsp13 treatment, whereas the expression of CD160 that involved cytolytic effector activity on natural killer (NK) cells was increased (Figures 4E and 4F). The effect of exosomesNsp12Nsp13 on cytokine production and the reversal of the effect by aly-miR396a-5p in F4/80+ cells have been confirmed at the protein level by FACS analysis (Figure 4G) as well as in transcription level qPCR analysis (Figure 4H). Histological examination demonstrated that the pulmonary inflammation caused by exosomesNsp12Nsp13 was improved by aly-miR396a-5p (Figure 4I).
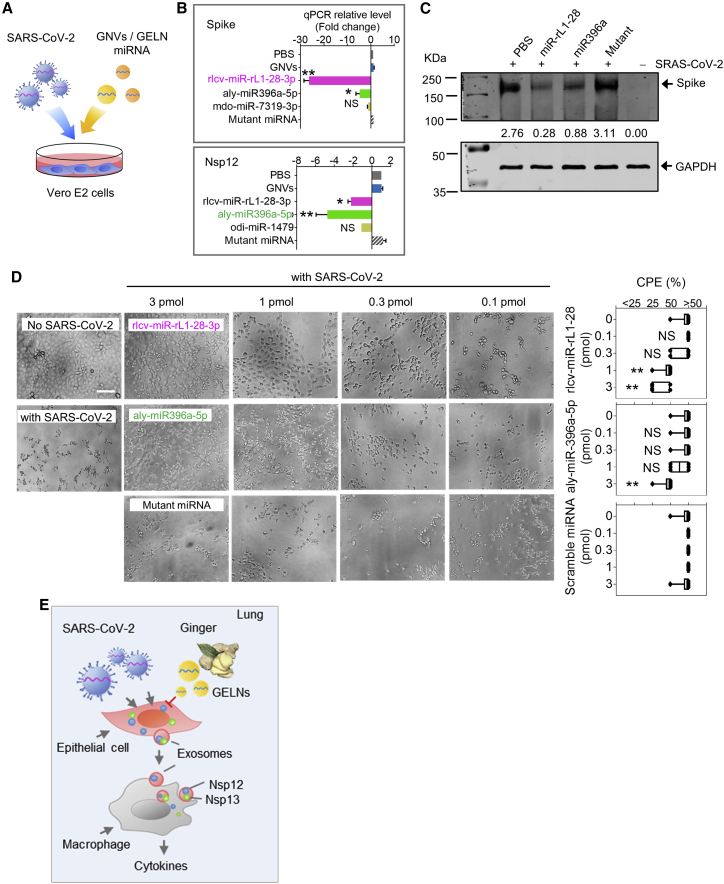
GELN miRNAs inhibit the SRAS-CoV-2 cytopathic effect (CPE) in Vero E6 cells by inhibiting the expression of the viral S and Nsp12
To further investigate whether GELN miRNAs that exhibited potent viral gene inhibitory activity could significantly inhibit viral replication, Vero E6 cells (2 × 104 cells per well) were exposed to SARS-CoV-2 at a concentration of 60 plaque-forming units (PFU) per well for a multiplicity of infection (MOI) of 0.003. GELN rlcv-miR-rL1-28-3p and aly-miR396a-5p, which inhibit the expression of the S gene and Nsp12 genes, respectively, were packed into GNVs and added to virus-infected Vero E6 cells (Figure 5A). On day 3 post-infection, the expression of S protein and Nsp12 in SARS-CoV-2 infected Vero E6 cells with or without GELN miRNAs was estimated by qPCR. The results indicated that both viral S and Nsp12 expression levels were reduced by either rlcv-miR-rL1-28-3p or aly-miR396a-5p (Figure 5B). However, GELN-derived mdo-miR-7319-3p and odi-miR-1479 did not affect the expression of S protein and Nsp12, although they have seed sequencing potential binding sites to S and NSp12, respectively. Western blots further confirmed that both rlcv-miR-rL1-28-3p and aly-miR396a-5p reduced the expression of S protein in transfected Vero E2 cells (Figure 5C). To assess the virus-induced CPE, Vero E2 cells were seeded in 96-well plates at an MOI of 0.003. In contrast to the normal cells, which had no CPE (Figure 5D), the cells infected with SARS-CoV-2 exhibited evident morphological changes, as shown by the rounded cell bodies and their elongated shape (Figure 5D, left panel). These CPE-positive cells detached from the plate and were reduced by rlcv-miR-rL1-28-3p and aly-miR396a-5p in a dose-dependent manner, but the mutant miRNA had no evident influence on SARS-CoV-2-induced CPE (Figure 5D, right panel).
Figure 5.
GELN miRNAs inhibit cytopathic effects (CPEs) of Vero E2 cells infected with SARS-CoV-2
(A) Schematic representation of the treatment schedule for the effect of GELN miRNAs on the CPEs of Vero E2 cells infected with SARS-CoV-2. (B) qPCR analysis of spike gene (top panel) and Nsp12 (bottom panel) expression in Vero E2 cells after a 72-h infection with SARS-CoV-2 at an MOI of 0.003. (C) Western blot analysis of spike protein in transfected Vero E2 cells. Numbers below the western blot represent densitometry values normalized to the loading control. (D) 2 × 104 Vero E2 cells in 96-well plates exposed to 60 PFU of SARS-CoV-2 and GELN miRNAs as well as control indicated in the graph. A representative CPE estimated at 72 h post-infection is shown. Scale bar, 100 μm (left panel). Semiquantitative analysis of CPE at four levels (<25%, 25%, 50%, >50%) from three independent experiments (right panel). (E) Proposed model of SARS-CoV-2 activation of cytokines in lung macrophages mediated by exosome cargo of viral protein from infected epithelial cells. GELN miRNA extinguishes the activation of cytokines in lung by directly targeting the viral gene of SARS-CoV-2. Data are representative of three independent experiments (error bars, SD). ∗p < 0.05, ∗∗p < 0.01 (two-tailed t test). NS, not significant.
Taken together with the evidence that non-human intragenic GELN miRNA can inhibit SARS-CoV-2 replication through specific binding and limiting viral gene expression, including viral S protein and RNA polymerase Nsp12 (Figure 5E), we developed an innovative and safe strategy for inhibition of the SARS-CoV-2 infection.
Discussion
In this study, we discovered a new biological activity of viral Nsp12 by which macrophages are activated through the NK-κB-mediated pathway. Nsp12 is delivered by lung epithelial cell exosomes to macrophages, leading to the activation of the macrophages via NF-κB. Activated macrophages then release a number of inflammatory cytokines that contribute to lung inflammation. We also found that exosomes carrying Nsp13 has a synergistic effect with Nsp12 in terms of activation of NF-κB. The metabolites released from exosomesNsp12Nsp13-activated macrophages causes lung epithelial cell apoptosis.
We discovered that a large number of ginger exosomal miRNAs can potentially bind to multiple sites of the SARS-Cov-2 viral genome and that these ginger miRNAs have no homolog sequences shared with host miRNAs. This finding is significant in that no ginger exosome homolog miRNAs shared sequences with viral host cell-derived miRNAs, which in turn means that it is unlikely that side effects will occur. This finding provides the impetus for future research into therapeutic miRNA-derived applications from the plant kingdom for more broad therapeutic application for the treatment of other diseases. With equal importance is the fact that many edible plants contain ELNs, with more than 500 different species of miRNAs in them, and composition of ELN miRNAs is different among the many ELNs. The finding that GELN miRNA can bind to multiple viral genes and subsequently inhibit the expression of viral genes and viral replication establishes the basis for exploring edible plant ELN miRNA libraries for personalized ELN-based therapy.
The results presented in this study also demonstrate that GNVs can be taken up by lung macrophages and lung epithelial cells with preferential ACE2-positive cells. Targeted delivery of ginger miRNA to lung epithelial cells inhibits the expression of Nsp12 and subsequently prevents exosomesNsp12+-mediated lung inflammation. Targeting to macrophages could lead to inhibiting the activation of macrophages and subsequently altering the composition of the metabolites of exosomesNsp12Nsp13-activated macrophages. Altering the composition of the metabolites leads to a decrease of apoptosis of lung epithelial cells. Interestingly, our data suggest that TNF-α, IL-6, and IL-1β in the context of supernatants from exosomesNsp12Nsp13-activated macrophages play a role in promoting lung epithelial cell apoptosis. Therefore, our data provide a rationale for future studyies as to which molecules in the metabolites can work with these cytokines to induce lung damage. Identifying these factors might be helpful in the treatment of severe COVID-19 patients.
GNV-mediated targeting to ACE2-positive A549 human lung epithelial cells will provide the foundation for further investigations on whether this result generated from a human epithelial cell line can be reproduced in an animal model. Recent studies show that the low rate of infection in A549 cells is the result of low expression of the viral receptor ACE232,33. This low rate of infectivity34 does not induce IFN-α, and INF-β and is beneficial for virus to escape the host immune response upon initial viral infection. The data we present (Figure S5B) show that the GNVs are preferentially taken up by an ACE2-positive sub-population of A549 cells. The result is meaningful since targeting these ACE2-positive subset human epithelial cells with GNVs carrying therapeutic agents will block the SARS-CoV-2 virus replication at the initial stage.
In conclusion, our data prompt further investigations as to whether SARS-Cov-2 can transfer a pathogenic signal from viral host cells to other inflammatory cells such as macrophages infiltrating the lung. In addition, our study highlights the potential to develop a lung-targetable edible plant-derived exosome-like vector to treat lung inflammatory-related disease, which until now has not been attempted or reported. A successful ginger nanoparticle-based therapy will have multiple applications because the activation of NF-κB-mediated pathways plays a crucial role in many inflammation-related diseases, including COVID-19.
Materials and methods
Cell culture
The mouse C57BL/6 lung carcinoma LLC1 and macrophage cell lines, monkey kidney Vero E6 cells, and human alveolar basal epithelial A549 and monocytic U937 cell lines (American Type Culture Collection, Rockville, MD, USA) were grown at 37°C in 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies) and RPMI 1640 medium (Life Technologies), respectively. U937 cells are induced to differentiate into macrophages using phorbol 12-myristate 13-acetate (PMA, Sigma) at 10 ng/mL for 5–7 days prior to use in studies. Media were supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin.
Virus infection
SARS-CoV-2 (strain USA-WA1/2020 isolate; BEI Resources, catalog no. NR-52281) was amplified in Vero E6 cells. Amplified stock virus was stored at −80°C until use. For infection, cells were incubated with virus for 1 h in a CO2 incubator at 37°C, then washed once with PBS. The cells were replenished with media. Virus titration was done by overlaying cells with Avicel overlay media (1% Avicel in DMEM with 10% FBS) and stained at 3 days post-infection with crystal violet staining solution (1% crystal violet, 2% paraformaldehyde, 25% ethanol) for 4 h. Virus titers were determined as 50% tissue culture infectious dose (TCID50)/mL in confluent cells in 96-well microtiter plates.35 Experiments were performed with three replicates per treatment. Experimental procedures with SARS-CoV-2 virus were approved by the Institutional Biosafety Committee of the University of Louisville. All processing of virus was performed in University of Louisville Center for Predictive Medicine, which has state-of-the-art facilities for biosafety level 3 (BSL-3) biocontainment research and in accordance with relevant guidelines and regulations by the Institutional Biosafety Committee of the University of Louisville and US Centers for Disease Control and Prevention (interim guidelines for collecting, handling, and testing clinical specimens for COVID-19).
Mice
8- to 12- week-old male specific pathogen-free (SPF) C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA) and housed under specific pathogen-free conditions. Animal care was performed following the Institute for Laboratory Animal Research (ILAR) guidelines, and all animal experiments were done in accordance with protocols approved by the University of Louisville Institutional Animal Care and Use Committee (Louisville, KY, USA).
Plasmid construction
Nsp12 and S protein genes of SARS2 virus were amplified from plasmids pGBW-m4046955 (no. 145616) and pGBW-m4046887 (no. 145730) (Addgene), respectively, and cloned into plasmid pAcGFP1-C1 (no. 121046) (Addgene) by using the NEBuilder HiFi DNA assembly cloning kit (NEB, catalog no. E5520S) following the manufacturer’s instructions. Plasmid pAcGFP1-C1 was linearized by double digestion of KpnI and BamHI. The primers used to generate construction are listed in Table S3. Positive colonies were confirmed by PCR and Sanger sequencing.
Isolation of lung macrophages in mice
Lung specimen from mice were thoroughly dissected and gently pressed through nylon cell strainers (70 μm in diameter, Fisher Scientific) to obtain single-cell suspensions in RPMI 1640 containing 5% FBS. The cell pellet was resuspended in 40% Percoll and layered onto 70% Percoll in RPMI 1640 with 1× Hanks’ balanced salt solution (HBSS, Thermo Fisher Scientific). After Percoll gradient centrifugation, the layer in the interface between the two Percoll concentrations was collected and washed with PBS. Erythrocytes in the cell suspensions were then removed using ammonium-chloride-potassium (ACK) lysing buffer (0.15 M NH4Cl, 10 mM KHCO3, 0.1 mM EDTA). The cells were allowed to adhere to the tissue culture plate for 24 h at 37°C. Nonadherent cells were removed by gently washing plates three times with warm PBS. At this point the adhering cells were more than 90% macrophages.
Isolation and purification of exosomes from cell culture medium
1 × 107 lung A549 or LLC1 cells were grown in 10 mL of DMEM supplemented with 10% heat-inactivated extracellular vesicle (EV)-depleted FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C in 5% CO2 for 72 h. EV-depleted FBS was prepared by ultracentrifugation overnight. The culture medium was collected and centrifuged at 1,000 × g for 10 min, 2,000 × g for 20 min, 4,000 × g for 30 min, and 10,000 × g for 1 h with the supernatant being retained each time. The exosomes were collected by centrifuging the samples at 100,000 × g for at least 2 h at 4°C and further purified on a sucrose gradient (8%, 30%, 45%, and 60% sucrose in 20 mM HEPES, 20 mM Tris-Cl [pH 7.2]) followed by two PBS washes. Size distribution and concentration of exosomes were analyzed at a flow rate of 0.03 mL per min using a Zetasizer Nano ZS (Malvern Instruments, UK) and NanoSight NS300 (Malvern Panalytical, Westborough, MA), respectively. We have submitted all relevant data of our experiments to the EV-TRACK knowledgebase (EV-TRACK: EV210123).36
Isolation, purification, and electron microscopy of GELNs
Peeled Hawaiian ginger roots (Simply Ginger, PLU no. 4612) were used for isolation and purification of ELNs using a previously described method.17 Briefly, peeled plants were homogenized in a high-speed blender for 1 min. The juice was collected after net filtration. The supernatant was collected after centrifugation at 1,000 × g for 10 min, 2,000 × g for 20 min, 4,000 × g for 30 min, and 10,000 × g for 1 h. The pellets of the plant nanoparticles were spun down at 100,000 × g for 1.5 h at 4°C. The isolated exosomes were further purified in a sucrose gradient (8%, 30%, 45%, and 60% sucrose in 20 mM Tri-Cl [pH 7.2]) followed by centrifugation at 100,000 × g for 1.5 h at 4°C. Purified GELNs were fixed in 2% paraformaldehyde and imaged using a Zeiss EM 900 electron microscope and a previously described method.1 Size distribution and concentration of the GELNs was analyzed at a flow rate of 0.03 mL per min using a Zetasizer Nano ZS (Malvern Instruments, UK).
RNA extraction
Total RNA containing miRNA was isolated from ELNs and murine tissue using a miRNeasy mini kit (QIAGEN) according to the manufacturer’s instructions. In brief, 50 mg of plant-derived ELNs or tissue or culture cells was disrupted in QIAzol lysis reagent. Tissue was homogenized using a tissue grinder before disruption. After mixing the homogenate with 140 μL of chloroform, the homogenate was centrifuged. The upper aqueous phase was mixed with 1.5 vol of ethanol and then loaded into a RNeasy spin column. Flowthrough was discarded and the column was washed with buffers RWT and RPE, respectively. Total RNA was eluted with RNase-free water. Bacteria mRNA was isolated using RiboPure bacteria and MICROBExpress kits (Thermo Fisher Scientific) according to the manufacturer’s instructions. The quality and quantity of the isolated RNA were analyzed using a NanoDrop spectrophotometer and Agilent Bioanalyzer.
Preparation and characterization of GNVs and packaging ELN RNAs in GNVs
The GELN-derived lipids were extracted with chloroform and dried under vacuum. To generate GNVs, 200 nmol of GELN-derived lipid was suspended in 200–400 μL of 155 mM NaCl with or without 10 μg of ELN-derived RNA. After UV irradiation at 500 mJ/cm2 in a Spectrolinker (Spectronic) and a bath sonication (FS60 bath sonicator, Fisher Scientific) for 30 min, the pelleted particles were collected by centrifugation at 100,000 × g for 1 h at 4°C.37,38 The RNA encapsulation efficiency in GNVs was determined using a previously described method.38
Distribution in the mouse respiratory tract with intratracheal intubation
The mice were anesthetized with a 2%–3% isoflurane/oxygen mixture in an anesthesia induction chamber. Mice were secured to the intubation platform. Direct laryngoscopy using an otoscope fitted with a 2.0-mm speculum was used to visualize the glottis following intubation using a 20G intravenous catheter as an endotracheal tube. After confirmation of intubation was established using tubing containing a colored dye, the exosomes or GNVs (5 × 108/kg, body weight, n = 5 per group) were dispensed into the lung in a single fluid motion. The needle/catheter was removed and the mouse allowed to recover from anesthesia.
Exosome and GNV distribution in vivo
The intratracheally administered exosomes or GNVs labeled with DiR dye (5 × 108/kg, body weight, n = 5 per group) were visualized in the lung and other organs of C57BL/6 mice using an Odyssey CLx imaging system (LI-COR Biosciences).
Labeling exosomes and GNVs with PKH26
Exosomes and GNVs were labeled using PKH26 fluorescent cell linker kits (Sigma). GNVs were suspended in 250 μL of diluent C with 5 μL of PKH26 mixed with 250 μL of dye solution for 20 min at room temperature and subsequently incubated with an equal volume of 1% bovine serum albumin (BSA) for 1 min at 22°C. After centrifugation for 1 h at 100,000 × g at 4°C, 20 μL of resuspended labeled GNVs were loaded on a slide for assessment of viability using confocal microscopy (Nikon).
Quantitative real-time PCR analysis of miRNA and mRNA expression
The quantity of mature miRNAs was determined by qPCR using a miScript II RT kit (QIAGEN) and miScript SYBR Green PCR kit (QIAGEN) with QIAGEN 3′ universal primers. The 5′-specific miRNA primers used are listed in Table S3. For analysis of gene mRNA expression, 1 μg of total RNA was reverse transcribed by SuperScript III reverse transcriptase (Invitrogen), and quantitation was performed using SsoAdvanced Universal SYBR Green supermix (Bio-Rad) and the primers listed in Table S3. qPCR was run using the Bio-Rad CFX96 qPCR system with each reaction run in triplicate. Analysis and fold changes were determined using the comparative threshold cycle (Ct) method. After normalizing with an internal control, that is, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression, the change of target gene in miRNA or mRNA expression was calculated as fold change, i.e., relative to PBS or vehicle treated (control).
Flow cytometry
After perfusion via the inferior vena cava with perfusion buffer (Ca2+-Mg2+-free HBSS containing 0.5 mM EGTA, 10 mM HEPES, and 4.2 mM NaHCO3 [pH 7.2]), the lung tissue from mice was incubated in RPMI 1640 supplemented with 15 mM HEPES and 300 U/mL collagenase type VIII (Sigma) for 1 h with gentle shaking. After this, the tissue was gently pressed through nylon cell strainers (70 μm in diameter, Fisher Scientific) to obtain single-cell suspensions in RPMI 1640 containing 5% FBS. Lung leukocytes were isolated from the interface of a 40%/80% colloidal silica particle (Percoll) gradient and washed twice. Erythrocytes in liver and spleen-cell suspensions were then removed using ACK lysing buffer (0.15 M NH4Cl, 10 mM KHCO3, 0.1 mM EDTA). Washed cells were stained for 1 h or overnight at 4°C with the appropriate fluorochrome-conjugated antibodies in PBS with 2% FBS. To detect intracellular antigens, washed cells were incubated in diluted fixation/permeabilization solution (eBioscience, catalog no. 00-5123) at 4°C for 30 min. Characterization and phenotyping of the various lymphocyte subsets from lung were performed by flow cytometry using antibodies against F4/80 (no. 17-4801-82), Gr-1 (no. 11-9668-82), CD-11b (no. 11-0112-41), TNF-α (no. 11-7321-82), IL-1α (no. 50-111-17), IL-1β (no. 50-100-10), IL-6, and EpCAM (no. 11-5791-82) (eBioscience) at a 1:200 dilution with PBS/2% FBS for 1 h on ice. Annexin V-fluorescein isothiocyanate (FITC) was applied to detect non-viable cells, and propidium iodide (PI) staining was used to distinguish apoptotic cells from necrotic and living cells. Data were acquired on a BD FACSCanto (BD Biosciences, San Jose, CA, USA) and were analyzed using FlowJo software (Tree Star, Ashland, OR, USA). Numbers above bracketed lines or boxes in FACS figures indicate the percentage of positive stained cells, and the results of cells stained with an isotype-matched control antibody are shown in gray.
Mouse cytokine array
To investigate the level of the cytokines, the lung tissue extracts were prepared in modified radioimmunoprecipitation assay (RIPA) buffer (Sigma) with the addition of protease and phosphatase inhibitors (Roche). Cytokine proteins were analyzed with a Proteome Profiler mouse XL cytokine array kit (R&D Systems, ARY028). Profiles of mean spot pixel density were created using a transmission-mode scanner, and quantification of the spot intensity in the arrays was conducted with background subtraction using image analysis MasterPlex QT software (MiraiBio Group).
Western blotting
The mice or cells were treated as indicated in individual figure legends and tissues or cells were harvested in RIPA buffer with the addition of a protease inhibitor cocktail (Roche). Proteins of lysates were separated by 10% SDS-PAGE and transferred to Odyssey nitrocellulose membranes (LI-COR Biosciences). Dual-color precision protein molecular weight (MW) markers (Bio-Rad) were separated in parallel. Antibodies were purchased as follows: S of SARS-CoV-2 (catalog no. GTX632604) from GeneTex; CD63 (catalog no. 143902) from BioLegend; p-NF-κB p65 (catalog no. 3031S), p-AKT (catalog no. 9271S), p-JNK (catalog no. 9251S), p-ERK1/2 (catalog no. 4370), and p-p38 MAPK (catalog no. 4511) from Cell Signaling Technology; NF-κB p65 (catalog no. 610869) from BD Biosciences; p-IKKβ (catalog no. ab59195) and p-PI3K (catalog no. ab182651) from Abcam; and p-IκBα (catalog no. sc-8404) and GAPDH (catalog no. sc-47724) from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The secondary antibodies conjugated to Alexa Fluor 790 were purchased from Invitrogen (Eugene, OR, USA). The membranes were incubated with primary antibodies above at a dilution of 1:1,000 with PBST (PBS with 0.1% Tween 20) for 1 h at room temperature. After the secondary antibody incubation at a dilution of 1:10,000 with PBST for 1 h at room temperature, the bands were visualized and analyzed using an Odyssey imager (LI-COR Biosciences, Lincoln, NE, USA).
Transient transfection
A549 or LLC1 cells were grown to 70% confluency in tissue culture plates in antibiotic-free DMEM supplemented with 5% FBS. Cells were transfected with 200 pmol of miRNA or 10 μg of plasmid/well using 30 μL of RNAiMAX or Lipofectamine 3000 (Invitrogen) in antibiotic-free medium and incubated for 48–72 h. As a control, cells were transfected with scramble control miRNA (Ambion) or empty plasmid vector. Expression plasmids used for transfections are listed in Table S1. RNA and protein lysates were prepared for qPCR and western blot analysis.
Immunoblot analysis of viral production from transfected cells
Exosomes and A549 cells transiently transfected with viral plasmids were lysed with RIPA buffer. One μg of lysate was separated by 8%–15% SDS-PAGE and transferred to nitrocellulose membranes (LI-COR Biosciences) and blocked with 5% BSA in PBS at 4°C overnight. Strep fusion protein and CD63 were visualized by Strep-Tactin-HRP conjugate (Bio-Rad) and Alexa Fluor fluorescent-conjugated anti-CD63 antibody, respectively.
Preparation of GELN small RNA libraries and sequencing
Small RNA libraries were generated with 100 ng of total RNA and a QIAseq miRNA library kit (QIAGEN) according to the manufacturer’s instructions. Following reverse transcription, cDNA purification with QIAseq beads, and PCR amplification (16 cycles) with indices, libraries with approximately 180 bp were bead purified and pre-sequencing quality control (QC) was performed with an Agilent Bioanalyzer 2100. Equal amounts of libraries were pooled and sequenced on the Illumina HiSeq 2500 using the Illumina NextSeq sequencing kit (FC-404-2005), followed by demultiplexing and fastq generation with CASAVA v1.8.4. Raw fastq files were preprocessed and miRNA sequences were quantified using QIAGEN’s QIAseq miRNA quantification module (https://geneglobe.qiagen.com/us/). Briefly, 3′ adaptors were trimmed using Cutadapt. Then, all miRNA sequences in the miRBase v21 were used for miRNA sequence quantification. Within the sRNAbench pipeline, mapping was performed with bowtie (v0.12.9), and microRNA folding was predicted with RNAfold from the Vienna package (v2.1.6). To visualize miRNA sequencing (miRNA-seq) results with waterfall plots and heatmaps, low-expressed miRNAs (raw read count <10 in all samples) were removed. Then, miRNA expression levels were normalized using edgeR’s TMM (trimmed mean of M) values.39 To generate a waterfall plot of potential differentially expressed miRNAs, we used a strict fold-change threshold . Heatmaps were generated using the heatmap.2 function from the gplots R package.40 Human and mouse miRNA-seq samples were downloaded from Sequence Read Archive (SRA)41 using accession nos. SRR12338616 and SRR7777390, respectively.
Predicting GELN miRNA potential targeting to SARS-CoV-2 mRNA
After downloading SARS-CoV-2 genomes (NCBI: NC_045512.2) from the NCBI Nucleotide database, viral mRNAs potentially targeted by GELN miRNAs were identified by mapping the reverse complement of the miRNA seed sequence to the SARS-CoV-2 whole genome with the full-length 29,903 bp. Although 6 nt are the minimum requirement, and the 6- to 8-nt-long seed sequence of the miRNA is sufficient to bind the target mRNA,23 the enrichment analysis with 9-nt seed subsequences included an adopted framework that utilizes the first-order Markov model (MM).42 In this framework, the observed k-mer count in the 300-bp region of each bacterial mRNA was compared against the background count derived from the first-order Markov model. A p value was then calculated for each miRNA-mRNA pair to estimate the likelihood of having a functional pair. Once all p values were calculated, the false discovery rate (FDR) was obtained using the Benjamini-Hochberg method43 for multiple comparisons. The plots of miRNA distribution in SARS-CoV-2 genome were generated using a R4.0 programming environment. To determine whether the seed sequences were present in human or mouse miRNA sequences, we downloaded all of the human and mouse microRNA mature sequences (reference sequences) from miRBase database v22.1 Then, we searched for reference sequences that have a perfect match (no mismatches were allowed) of a seed sequence.
Histological analysis
For hematoxylin and eosin (H&E) staining, tissues were fixed with buffered 10% formalin solution (SF93-20; Fisher Scientific, Fair Lawn, NJ, USA) overnight at 4°C. Dehydration was achieved by immersion in a graded ethanol series, i.e., 70%, 80%, 95%, and 100% ethanol for 40 min each. Tissues were embedded in paraffin and subsequently cut into ultra-thin slices (5 μm) using a microtome. The sections were deparaffinized by xylene (Fisher Scientific) and rehydrated by decreasing concentrations of ethanol and PBS. Tissue sections were then stained with H&E and slides were scanned with an Aperio ScanScope. For frozen sections, tissues were fixed with periodate-lysine-paraformaldehyde (PLP) and dehydrated with 30% sucrose in PBS at 4°C overnight. The sections were incubated with anti-F4/80 (Santa Cruz, no. sc-25830), anti-Gr-1 (BioLegend, no. 108404), anti-CD11b (BioLegend, no. 101204), anti-EpCAM (Abcam, ab71916) and anti-angiotensin converting enzyme-2 (ACE-2) (ProSci, no. 3217) at a 1:100 dilution at 4°C overnight. The signal was visualized with the secondary antibodies conjugated to Alexa Fluor 488 or Alexa Fluor 594 (Invitrogen), and nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). The slides were scanned using an Aperio ScanScope or visualized with a confocal laser scanning microscope (Nikon, Melville, NY, USA) as described.44
Apoptosis analysis by TUNEL
Formalin-fixed mouse lung tissues were embedded in paraffin, sectioned, and place on glass slides. TUNEL was used to detect apoptosis in the sections according to the manufacturer’s protocol. Tissue sections were analyzed to detect the localized green fluorescence (GFP) of apoptotic cells using the In Situ Cell Death Detection kit (Roche) and blue fluorescence of cell nuclei using DAPI. The signal was visualized using confocal laser scanning microscopy (Nikon, Melville, NY, USA).
ELISA
The cytokines in cell culture supernatants or mouse lung tissue were quantified using ELISA kits (eBioscience) according to the manufacturer’s instructions. Briefly, a microtiter plate was coated with anti-mouse TNF-α, IL-1α, IL-1β, and IL-6 capture antibody (eBioscience) at 1:200 at 4°C overnight. Excess binding sites were blocked with 200 μL of 1× ELISA/enzyme-linked immunospot (ELISPOT) diluent (eBioscience) for 1 h at 22°C. After washing three times with PBS containing 0.05% Tween 20, the plate was incubated with detective antibody in blocking buffer for 1 h at 22°C. After washing three times, avidin conjugated with HRP and substrate were each added sequentially for 1 h 30 min at 22°C. Absorbance at 405 nm was recorded using a microplate reader (BioTek Synergy HT).
Isolation of ABs
ABs were isolated from culture supernatants as previously described.31 Briefly, cell culture medium was harvested and cells were removed by pelleting at 335 × g for 10min. To remove cell debris, cell-free supernatants were centrifuged at 1,000 × g for 10 min, followed by another centrifugation at 2,000 × g for 30 min to pellet ABs. Pelleted ABs were resuspended and washed with PBS.
CoIP assay
To further confirm the interaction of Nsp12 and Nsp13, A549 cells were co-transfected with the plasmids of pAcGFP1-C-Nsp12-FLAG and pLVX-Nsp13-2xStrep. Seventy-two hours after transfection, Nsp13 was pulled down with Strep-Tactin XT magnetic beads in IP buffer (150 mM NaCl, 10 mM Tris-HCl [pH 7.4], 1 mM EDTA, 1 mM EGTA [pH 8.0], 0.2 mM sodium orthovanadate, 0.2 mM PMSF, 1% Triton X-100, 0.5% Nonidet P-40 [NP-40]). One μg of Nsp12/Nsp13 complex was spotted onto nitrocellulose membranes (LI-COR Biosciences) and blocked with 5% BSA in PBS at 4°C overnight. Nsp12 fusion protein was visualized by rabbit anti-FLAG antibody (Sigma-Aldrich, catalog no. SAB4301135) and Alexa Fluor 790-conjugated anti-rabbit antibody (Invitrogen).
CPEs of SARS-CoV-2
Vero E6 cells (passage 6) were seeded at a density of 2 × 104 cells per well in a 96-well plate in DMEM supplemented with 10% heat-inactivated FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. After attachment of cells to the well bottoms, SARS-CoV-2 was diluted in DMEM to give a final concentration of 60 PFU/well for an MOI of 0.003. GELN miRNA (100 ng) packed in GNVs was added to the wells at dilutions from 1:1 to 1:100. Cells were observed for CPE every day post-infection. On day 3 post-infection, after washing three times with PBS, the cells were collected in RIPA buffer or TRIzol for virus protein and gene expression by western blot and qPCR analysis, respectively.
Statistical analysis
All statistical analyses in this study were performed with SPSS 16.0 software. Data are presented as mean ± SD. The significance of mean values between two groups was analyzed using the Student’s t test. The differences between individual groups were analyzed by a one- or two-way ANOVA test. The differences between percentages of CPEs and signal positive cells in flow cytometer and confocal microscopy were analyzed with a chi-square test. The differences were considered significant when the p value was less than 0.05 or 0.01.
Acknowledgments
We thank Dr. J. Ainsworth for editorial assistance. This work was supported by National Institutes of Health (NIH) grants R01AT008617 and P20GM125504, and by the Robley Rex VA Medical Center Merit Review Grants (to H.-G.Z.). Y.T. is supported by a COBRE Pilot Project (NIH P20GM125504). H.-G.Z. is supported by a Research Career Scientist (RCS) Award. M.L.M., X.Z., and C.J.M. are supported by NIH grants P50AA024337 and P20GM113226. J.W.P. was supported by the National Institute of General Medical Sciences of the National Institutes of Health grant P20GM103436.
Author contributions
Y.T., F.X., J.M., and H.-G.Z. designed the study, analyzed and interpreted the data, and prepared the manuscript; F.X., J.M., X.Z., M.S., X.H., C.L., M.K.S., A.K., K.S., and J.K.W. performed the experiments and interpreted the data; J.K.W., L.Z., J.Y., M.L.M., X.Z., S.Z., S.-y.C., C.J.M., D.C., and K.E.P. interpreted the findings.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2021.05.005.
Contributor Information
Yun Teng, Email: yun.teng@louisville.edu.
Huang-Ge Zhang, Email: h0zhan17@louisville.edu.
Supplemental information
References
- 1.Mu J., Zhuang X., Wang Q., Jiang H., Deng Z.B., Wang B., Zhang L., Kakar S., Jun Y., Miller D., Zhang H.G. Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol. Nutr. Food Res. 2014;58:1561–1573. doi: 10.1002/mnfr.201300729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao J., Feng S., Wang X., Long K., Luo Y., Wang Y., Ma J., Tang Q., Jin L., Li X., Li M. Identification of exosome-like nanoparticle-derived microRNAs from 11 edible fruits and vegetables. PeerJ. 2018;6:e5186. doi: 10.7717/peerj.5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sampey G.C., Saifuddin M., Schwab A., Barclay R., Punya S., Chung M.C., Hakami R.M., Zadeh M.A., Lepene B., Klase Z.A. Exosomes from HIV-1-infected cells stimulate production of pro-inflammatory cytokines through trans-activating response (TAR) RNA. J. Biol. Chem. 2016;291:1251–1266. doi: 10.1074/jbc.M115.662171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miura T.A. Respiratory epithelial cells as master communicators during viral infections. Curr. Clin. Microbiol. Rep. 2019;6:10–17. doi: 10.1007/s40588-019-0111-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukhamedova N., Hoang A., Dragoljevic D., Dubrovsky L., Pushkarsky T., Low H., Ditiatkovski M., Fu Y., Ohkawa R., Meikle P.J. Exosomes containing HIV protein Nef reorganize lipid rafts potentiating inflammatory response in bystander cells. PLoS Pathog. 2019;15:e1007907. doi: 10.1371/journal.ppat.1007907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higuchi H., Yamakawa N., Imadome K.I., Yahata T., Kotaki R., Ogata J., Kakizaki M., Fujita K., Lu J., Yokoyama K. Role of exosomes as a proinflammatory mediator in the development of EBV-associated lymphoma. Blood. 2018;131:2552–2567. doi: 10.1182/blood-2017-07-794529. [DOI] [PubMed] [Google Scholar]
- 7.Aftab S.O., Ghouri M.Z., Masood M.U., Haider Z., Khan Z., Ahmad A., Munawar N. Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach. J. Transl. Med. 2020;18:275. doi: 10.1186/s12967-020-02439-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santiana M., Ghosh S., Ho B.A., Rajasekaran V., Du W.L., Mutsafi Y., De Jésus-Diaz D.A., Sosnovtsev S.V., Levenson E.A., Parra G.I. Vesicle-cloaked virus clusters are optimal units for inter-organismal viral transmission. Cell Host Microbe. 2018;24:208–220.e8. doi: 10.1016/j.chom.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chettimada S., Lorenz D.R., Misra V., Dillon S.T., Reeves R.K., Manickam C., Morgello S., Kirk G.D., Mehta S.H., Gabuzda D. Exosome markers associated with immune activation and oxidative stress in HIV patients on antiretroviral therapy. Sci. Rep. 2018;8:7227. doi: 10.1038/s41598-018-25515-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantoni D., Rossman J.S. Ebolaviruses: New roles for old proteins. PLoS Negl. Trop. Dis. 2018;12:e0006349. doi: 10.1371/journal.pntd.0006349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pastuzyn E.D., Day C.E., Kearns R.B., Kyrke-Smith M., Taibi A.V., McCormick J., Yoder N., Belnap D.M., Erlendsson S., Morado D.R. The neuronal gene Arc encodes a repurposed retrotransposon Gag protein that mediates intercellular RNA transfer. Cell. 2018;172:275–288.e18. doi: 10.1016/j.cell.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raab-Traub N., Dittmer D.P. Viral effects on the content and function of extracellular vesicles. Nat. Rev. Microbiol. 2017;15:559–572. doi: 10.1038/nrmicro.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang H.I., Lin J.Y., Chiang H.C., Huang P.N., Lin Q.D., Shih S.R. Exosomes facilitate transmission of enterovirus A71 from human intestinal epithelial cells. J. Infect. Dis. 2020;222:456–469. doi: 10.1093/infdis/jiaa174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nahand J.S., Mahjoubin-Tehran M., Moghoofei M., Pourhanifeh M.H., Mirzaei H.R., Asemi Z., Khatami A., Bokharaei-Salim F., Mirzaei H., Hamblin M.R. Exosomal miRNAs: Novel players in viral infection. Epigenomics. 2020;12:353–370. doi: 10.2217/epi-2019-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Velandia-Romero M.L., Calderón-Peláez M.A., Balbás-Tepedino A., Márquez-Ortiz R.A., Madroñero L.J., Barreto Prieto A., Castellanos J.E. Extracellular vesicles of U937 macrophage cell line infected with DENV-2 induce activation in endothelial cells EA.hy926. PLoS ONE. 2020;15:e0227030. doi: 10.1371/journal.pone.0227030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee H., Abston E., Zhang D., Rai A., Jin Y. Extracellular vesicle: An emerging mediator of intercellular crosstalk in lung inflammation and injury. Front. Immunol. 2018;9:924. doi: 10.3389/fimmu.2018.00924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teng Y., Ren Y., Sayed M., Hu X., Lei C., Kumar A., Hutchins E., Mu J., Deng Z., Luo C. Plant-derived exosomal microRNAs shape the gut microbiota. Cell Host Microbe. 2018;24:637–652.e8. doi: 10.1016/j.chom.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sundaram K., Miller D.P., Kumar A., Teng Y., Sayed M., Mu J., Lei C., Sriwastva M.K., Zhang L., Yan J. Plant-derived exosomal nanoparticles inhibit pathogenicity of Porphyromonas gingivalis. iScience. 2019;21:308–327. doi: 10.1016/j.isci.2019.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhuang X., Deng Z.B., Mu J., Zhang L., Yan J., Miller D., Feng W., McClain C.J., Zhang H.G. Ginger-derived nanoparticles protect against alcohol-induced liver damage. J. Extracell. Vesicles. 2015;4:28713. doi: 10.3402/jev.v4.28713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berezikov E., Thuemmler F., van Laake L.W., Kondova I., Bontrop R., Cuppen E., Plasterk R.H. Diversity of microRNAs in human and chimpanzee brain. Nat. Genet. 2006;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- 21.Coenen-Stass A.M.L., Magen I., Brooks T., Ben-Dov I.Z., Greensmith L., Hornstein E., Fratta P. Evaluation of methodologies for microRNA biomarker detection by next generation sequencing. RNA Biol. 2018;15:1133–1145. doi: 10.1080/15476286.2018.1514236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Godoy P.M., Bhakta N.R., Barczak A.J., Cakmak H., Fisher S., MacKenzie T.C., Patel T., Price R.W., Smith J.F., Woodruff P.G., Erle D.J. Large differences in small RNA composition between human biofluids. Cell Rep. 2018;25:1346–1358. doi: 10.1016/j.celrep.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellwanger D.C., Büttner F.A., Mewes H.W., Stümpflen V. The sufficient minimal set of miRNA seed types. Bioinformatics. 2011;27:1346–1350. doi: 10.1093/bioinformatics/btr149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Israël A. The IKK complex, a central regulator of NF-κB activation. Cold Spring Harb. Perspect. Biol. 2010;2:a000158. doi: 10.1101/cshperspect.a000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyamoto R., Ito T., Nomura S., Amakawa R., Amuro H., Katashiba Y., Ogata M., Murakami N., Shimamoto K., Yamazaki C. Inhibitor of IκB kinase activity, BAY 11-7082, interferes with interferon regulatory factor 7 nuclear translocation and type I interferon production by plasmacytoid dendritic cells. Arthritis Res. Ther. 2010;12:R87. doi: 10.1186/ar3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim B.J., Silverman S.M., Liu Y., Wordinger R.J., Pang I.H., Clark A.F. In vitro and in vivo neuroprotective effects of cJun N-terminal kinase inhibitors on retinal ganglion cells. Mol. Neurodegener. 2016;11:30. doi: 10.1186/s13024-016-0093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herold S., Steinmueller M., von Wulffen W., Cakarova L., Pinto R., Pleschka S., Mack M., Kuziel W.A., Corazza N., Brunner T. Lung epithelial apoptosis in influenza virus pneumonia: The role of macrophage-expressed TNF-related apoptosis-inducing ligand. J. Exp. Med. 2008;205:3065–3077. doi: 10.1084/jem.20080201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang M., Qian J., Xing X., Kong F.M., Zhao L., Chen M., Lawrence T.S. Inhibition of the tumor necrosis factor-α pathway is radioprotective for the lung. Clin. Cancer Res. 2008;14:1868–1876. doi: 10.1158/1078-0432.CCR-07-1894. [DOI] [PubMed] [Google Scholar]
- 29.Arienti S., Barth N.D., Dorward D.A., Rossi A.G., Dransfield I. Regulation of apoptotic cell clearance during resolution of inflammation. Front. Pharmacol. 2019;10:891. doi: 10.3389/fphar.2019.00891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haanen C., Vermes I. Apoptosis and inflammation. Mediators Inflamm. 1995;4:5–15. doi: 10.1155/S0962935195000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masvekar R., Mizrahi J., Park J., Williamson P.R., Bielekova B. Quantifications of CSF apoptotic bodies do not provide clinical value in multiple sclerosis. Front. Neurol. 2019;10:1241. doi: 10.3389/fneur.2019.01241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harcourt J., Tamin A., Lu X., Kamili S., Sakthivel S.K., Murray J., Queen K., Tao Y., Paden C.R., Zhang J. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg. Infect. Dis. 2020;26:1266–1273. doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361:2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.V-TRACK Consortium. Van Deun J., Mestdagh P., Agostinis P., Akay Ö., Anand S., Anckaert J., Martinez Z.A., Baetens T., Beghein E. EV-TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nat. Methods. 2017;14:228–232. doi: 10.1038/nmeth.4185. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q., Zhuang X., Mu J., Deng Z.B., Jiang H., Zhang L., Xiang X., Wang B., Yan J., Miller D., Zhang H.G. Delivery of therapeutic agents by nanoparticles made of grapefruit-derived lipids. Nat. Commun. 2013;4:1867. doi: 10.1038/ncomms2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teng Y., Mu J., Hu X., Samykutty A., Zhuang X., Deng Z., Zhang L., Cao P., Yan J., Miller D., Zhang H.G. Grapefruit-derived nanovectors deliver miR-18a for treatment of liver metastasis of colon cancer by induction of M1 macrophages. Oncotarget. 2016;7:25683–25697. doi: 10.18632/oncotarget.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warnes G.R., Bolker B., Bonebakker L., Gentleman R., Huber W., Liaw A., Lumley T., Maechler M., Magnusson A., Moeller S. 2009. gplots: Various R programming tools for plotting data. R package version 2.4.1. [Google Scholar]
- 41.Leinonen R., Sugawara H., Shumway M., International Nucleotide Sequence Database Collaboration The sequence read archive. Nucleic Acids Res. 2011;39:D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murphy E., Vanícek J., Robins H., Shenk T., Levine A.J. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: Implications for latency. Proc. Natl. Acad. Sci. USA. 2008;105:5453–5458. doi: 10.1073/pnas.0711910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 44.Teng Y., Ren Y., Hu X., Mu J., Samykutty A., Zhuang X., Deng Z., Kumar A., Zhang L., Merchant M.L. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat. Commun. 2017;8:14448. doi: 10.1038/ncomms14448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.