ABSTRACT
Rickettsia amblyommatis belongs to the spotted fever group of Rickettsia and infects Amblyomma americanum (Lone Star ticks) for transmission to offspring and mammals. Historically, the geographic range of A. americanum was restricted to the southeastern USA. However, recent tick surveys identified the progressive northward invasion of A. americanum, contributing to the increased number of patients with febrile illnesses of unknown etiology after a tick bite in the northeastern USA. While serological evidence strongly suggests that patients are infected with R. amblyommatis, the virulence potential of R. amblyommatis is not well established. Here, we performed a bioinformatic analysis of three genome sequences of R. amblyommatis and identified the presence of multiple putative virulence genes whose products are implicated for spotted fever pathogenesis. Similar to other pathogenic spotted fever rickettsiae, R. amblyommatis replicated intracellularly within the cytoplasm of tissue culture cells. Interestingly, R. amblyommatis displayed defective attachment to microvascular endothelial cells. The attachment defect and slow growth rate of R. amblyommatis required relatively high intravenous infectious doses to produce dose-dependent morbidity and mortality in C3H mice. In summary, our results corroborate clinical evidence that R. amblyommatis can cause mild disease manifestation in some patients.
Keywords: Rickettsia amblyommatis, Amblyomma americanum, spotted fever, pathogenesis, endothelial cell, animal infection model
Rickettsia amblyommatis, an obligate intracellular pathogen transmitted by Lone Star ticks (Amblyomma americanum), infects endothelial cells and causes spotted fever pathogenesis in mice.
INTRODUCTION
Ticks transmit numerous life-threatening infectious diseases that have no preventive vaccines or immune therapeutics and pose ongoing public health threats (Eisen et al. 2017). Several environmental and sociological factors have contributed to the expansion and invasion of multiple tick species, resulting in an increased number of tick-borne infections in the USA (Childs and Paddock 2003; Sonenshine 2018). Recent tick surveillance studies have demonstrated that Amblyomma americanum (Lone Star tick) has rapidly expanded northward and become the dominant tick species, displacing local tick species such as Ixodes scapularis (Deer tick) and Dermacentor variabilis (American Dog tick), in the Northeast and Midwest, where an increased number of patients with tick-borne febrile diseases of unknown etiology has been observed (Dahlgren et al. 2016; Rosenberg et al. 2018; Molaei et al. 2019; Sanchez-Vicente et al. 2019).
Among the several human pathogens transmitted by A. americanum, Rickettsia amblyommatis (formerly known as Candidatus Rickettsia amblyommii), a Gram-negative pathogen that belongs to the spotted fever group (SFG) of Rickettsia, has been identified in a large population of surveyed ticks (Killmaster et al. 2014; Karpathy et al. 2016; De Jesus et al. 2019; Sanchez-Vicente et al. 2019). By contrast, the current prevalence of Rickettsia rickettsii, the causative agent of Rocky Mountain spotted fever (RMSF), in D. variabilis is estimated to be less than 1% (Dahlgren et al. 2016; Hecht et al. 2019). The high prevalence of R. amblyommatis, combined with the aggressive biting behavior of A. americanum, enhances the probability of human infections with R. amblyommatis (Childs and Paddock 2003). Within ticks, R. amblyommatis exhibits obligatory intracellular lifecycle in multiple organ tissues such as ovaries, midgut and salivary glands (Zanetti et al. 2008). Infection in these tick organs facilitates the transmission of R. amblyommatis to offspring and mammalian hosts (Macaluso et al. 2001; Levin, Schumacher and Snellgrove 2018; Suwanbongkot et al. 2019). Unlike strictly endosymbiotic Rickettsia species (e.g. Rickettsia buchneri in I. scapularis), several recently characterized pathogenic Rickettsia species, such as Rickettsia helvetica, Rickettsia slovaca and Rickettsia parkeri, have also been identified in the salivary glands and midgut of ticks, tested for their virulence using in vitro and in vivo infection models, then isolated from humans after tick bites, corroborating that the presence of Rickettsia in the salivary glands in ticks facilitates transmission to mammalian hosts (Raoult et al. 2002; Paddock et al. 2004; Nilsson, Elfving and Pahlson 2010; Zemtsova et al. 2016; Engstrom et al. 2019; Pahlson et al. 2020).
The hallmark of rickettsial diseases is the invasion of Rickettsia into vascular endothelial cells, followed by intracellular replication, vascular pathologies and bacteremia for transmission of Rickettsia into uninfected vectors (Hackstadt 1996; Sahni et al. 2019). Progressive endothelial cell injury leads to the generation of characteristic erythematous rash, vasculitis, cutaneous necrosis and life-threatening symptoms, including sepsis, making Rickettsia one of the most deadly pathogens (Walker and Ismail 2008). Although R. amblyommatis has not been isolated from patients, clinical and serological evidence suggests that this microorganism may be the etiological agent of RMSF-like illness in humans. In 2006, a partially engorged female A. americanum was removed from a patient who developed a macular rash at the tick bite site (Billeter et al. 2007). A PCR test confirmed the presence of R. amblyommatis and the absence of Borrelia, Ehrlichia, Anaplasma, Babesia, Bartonella and other pathogenic Rickettsia species (Billeter et al. 2007). Analysis of paired sera from patients diagnosed with probable RMSF revealed that some patients developed antibodies to R. amblyommatis, but not to R. rickettsii and R. parkeri (the causative agent of R. parkeri rickettsioses), corroborating that R. amblyommatis may cause RMSF-like illnesses in humans (Apperson et al. 2008; Vaughn et al. 2014; Delisle et al. 2016). Those patients with specific reactivity to R. amblyommatis presented typical clinical manifestations of a mild RMSF with fever, headache and myalgia (Delisle et al. 2016). Recent reports found that domestic and wild small mammals (dogs and cats) elicit R. amblyommatis-specific antibodies and may serve as natural hosts for R. amblyommatis transmission (Barrett, Little and Shaw 2014; Costa et al. 2017; Springer et al. 2018; Lopes et al. 2019).
Investigations into the virulence potential of R. amblyommatis generated mixed results. Previous work suggested that R. amblyommatis is non-pathogenic as two strains, WB-8–2 and North Texas, failed to cause clinical diseases in guinea pigs (Burgdorfer, Cooney and Thomas 1974; Blanton et al. 2014). By contrast, another R. amblyommatis strain, 9-CC-3–1, caused vascular inflammation in guinea pigs (Rivas et al. 2015). A recent report also documented that intraperitoneal injection of R. amblyommatis strain Lake Alexander caused mild disease manifestations in guinea pigs (Snellgrove et al. 2021). This is not an unusual finding among Rickettsia, as strains of R. rickettsii displayed drastically different virulence in animal infection models (Clark et al. 2015; Galletti et al. 2021). Interestingly, infections with R. amblyommatis raised broad-spectrum immune responses and provided cross-immune protection in guinea pigs against virulent R. rickettsii infections (Blanton et al. 2014; Rivas et al. 2015). To better understand the biology of R. amblyommatis, we utilized bioinformatic tools, in vitro tissue culture assay and in vivo animal infection models. Our bioinformatic analysis shows that putative virulence genes whose products are characterized for spotted fever pathogenesis in animal infection models are present in R. amblyommatis. Using in vitro tissue culture assays, we demonstrate that R. amblyommatis strain GAT-30V replicates within the cytoplasm of host cells with defective attachment and invasion into microvascular endothelial cells. While assessing its virulence, we determined relatively high infectious doses that produced significant morbidity and mortality in C3H mice. Together, our results support clinical evidence that R. amblyommatis may cause mild rickettsioses in some patients.
MATERIALS AND METHODS
Cell lines and bacterial strains
Vero cells (African green monkey kidney cells, ATCC, Manassas, VA) were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco, Waltham, MA) supplemented with 10% heat-inactivated fetal bovine serum (HI-FBS, HyClone, Marlborough, MA) at 37°C in a 5% CO2 atmosphere. HMEC-1 cells (human dermal microvascular endothelial cells, ATCC) were cultured in MCDB 131 medium (Gibco) supplemented with 10 ng∙ml−1 epidermal growth factor (Gibco), 1 μg∙ml−1 hydrocortisone (Sigma, St. Louis, MO), 10 mM glutamine (Corning, Corning, NY) and 10% HI-FBS at 37°C in a 5% CO2 atmosphere. Stocks of Rickettsia conorii strain Malish 7 (ATCC VR-613) and R. amblyommatis strain GAT-30V (CDC, Dr. Chris Paddock) were generated by growing rickettsiae in Vero cells (5% HI-FBS, DMEM) at 34°C in a 5% CO2 atmosphere. Rickettsiae were purified from Vero cells by differential centrifugation through 33% MD-76R solution (816 mM meglumine diatrizoate, 157 mM sodium diatrizoate hydrate, 1 mM NaH2PO4, pH 7.0; 21 000 × g, 4°C, 20 min) and stored at –80°C in SPG buffer (218 mM sucrose, 3.8 mM KH2PO4, 7.2 mM K2HPO4, 4.9 mM L-glutamate, pH 7.2).
Bioinformatic analysis
Genomic data for Rickettsia were collected from the National Center for Biotechnology Information GenBank database (R. amblyommatis strain GAT-30V, accession no. CP003334.1; R. amblyommatis strain Ac37, accession no. CP012420.1; R. amblyommatis strain An13, accession no. CP015012.1; R. conorii strain Malish 7, accession no. AE006914.1; R. rickettsii strain Sheila Smith, accession no. CP000848.1). Sequences were analyzed with Mauve Multiple Genome Alignment software (ver. snapshot_2015–02–25), Protein BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) to identify conserved and divergent regions in the bacterial genomes, homologs in the designated species and amino acid % identity.
Growth and plaque assay
Growth curves were generated by infecting R. conorii and R. amblyommatis into monolayers of Vero or HMEC-1 cells in six-well plates at a multiplicity of infection (MOI) of 0.01. Infected tissue culture cells were incubated at 34°C in a 5% CO2 atmosphere. At 1-h post-infection and 2-day intervals, host cells in each well were dislodged with 3-mm glass beads and lysed by vortexing with 3-mm glass beads. The infectious titers were determined by infecting fresh monolayers of Vero cells with 10-fold serial dilutions of lysates containing Rickettsia in DMEM supplemented with 5% HI-FBS. Upon infection, Vero cells were incubated at 34°C with 5% CO2 for 60 min to allow attachment and overlaid with DMEM containing 5% HI-FBS and 0.5% agarose. The Rickettsia attachment and invasion levels were determined by dividing the number of Rickettsia infectious titers from the cell lysates by the number of Rickettsia infectious titers in the media.
Microscopy analyses
Cytopathology in Vero and HMEC-1 cell cultures was analyzed by microscopy. On days 2, 4, 6 and 8 post-infection, differential interference contrast (DIC) images of Vero and HMEC-1 cells infected with R. conorii or R. amblyommatis at a MOI of 0.01 in multiple fields of six-well plates were captured with a 12-bit charge-coupled device camera (Eclipse TE300 fluorescent microscope, Laboratory of Comparative Medicine, Stony Brook University) and contrast-adjusted using ImageJ (NIH). Areas of cytopathic Vero cells were assessed by NIH ImageJ software. For confocal laser scanning microscopic studies, HMEC-1 cells were cultured on a eight-well chamber slide (Lab-Tek, Nunc, Rochester, NY), infected with R. amblyommatis at a MOI of 0.01, fixed with 10% formalin for 10 min and post-fixed with 100% methanol for 5 min on Days 4, 6 and 8 of infection. Next, the fixed cells were permeabilized with 0.1% Triton X-100, incubated with mouse anti-β-actin antibody (1:500, Sigma) and rabbit anti-R. conorii LPS (1:500, crossreacts with SFG Rickettsia) overnight (Kim et al. 2019). The secondary antibodies (Alexa Fluor 488 donkey anti-mouse [1:1000, Invitrogen, Waltham, MA] and Alexa Fluor 594 donkey anti-rabbit IgG [1:1000, Invitrogen]) were incubated at room temperature for 1 h. The nuclei of HMEC cells were labeled by Hoechst 33 342 (Prolong Glass Antifade Mountant with NucBlue, Invitrogen). The images were taken by Leica SP8 confocal microscope (Advanced Energy Research & Technology Center, Stony Brook University) and analyzed with ImageJ (NIH).
Mouse model of spotted fever
C3H mice (male and female, 6-week-old, N = 5 per group, Charles River Laboratories, Wilmington, MA) were anesthetized via intraperitoneal injection with 100 mg·ml−1 of ketamine and 20 mg·ml−1 of xylazine per kilogram of body weight. Animals were infected via intravenous retro-orbital injection with Renografin-purified 5 ‒ 50 × 105 PFU R. amblyommatis strain GAT-30V in 0.1 ml SPG buffer or mock-infected with retro-orbital injections of 0.1 ml SPG buffer. Infected mice were monitored twice daily for signs of disease and daily for weight loss. Two days following infection, the heart was removed during necropsy and heart homogenate was analyzed for rickettsial load by plaque assay.
Whole-cell ELISA
Nunc MaxiSorp 96-well plates were coated with 5 × 105 PFU formalin-inactivated R. amblyommatis strain GAT-30V in 0.1 M carbonate buffer (pH 9.5 at 4°C) overnight. The following day, plates were washed three times with 0.05% (v/v) Tween 20 in PBS and blocked with 5% non-fat milk (w/v) in PBS for 1 h at room temperature. For the determination of R. amblyommatis-specific IgG titers, ELISA plates were washed and incubated with dilutions of hyperimmune sera for 1 h at room temperature. Following the wash, HRP-conjugated goat anti-mouse IgG (1:10 000, Rockland, Pottstown, PA) antibody was added and incubation was repeated as above. The wells were developed using an OptEIA kit (BD Lifesciences, Franklin Lakes, NJ) and absorbance at 450 nm was measured to calculate half-maximal titers using Prism software (GraphPad, San Diego, CA).
Biosafety and biosecurity
Animal research was performed in accordance with institutional guidelines following experimental protocol review, approval and supervision by the Institutional Biosafety Committee and the Institutional Animal Care and Use Committee at Stony Brook University. Animals were managed by the Division of Laboratory Animal Resources (Stony Brook University), which is accredited by the American Association for Accreditation of Laboratory Animal Care and the Department of Health and Human Services. Animals were maintained in accordance with the applicable portions of the Animal Welfare Act and the DHHS ‘Guide for the Care and Use of Laboratory Animals’. Veterinary care was under the direction of full-time resident veterinarians boarded by the American College of Laboratory Animal Medicine. Experiments with infectious Rickettsia were performed in biosafety level 3 containment.
Statistical analyses
All statistical analyses were performed using Prism software (GraphPad). Two-tailed Student's t-test was performed to calculate the statistical significance of Rickettsia attachment and invasion into HMEC-1 cells. Two-way ANOVA with Sidak's multiple comparison test was performed to analyze the statistical significance of body weight change and Rickettsia growth data.
Materials and data availability
The data and unique materials that support the findings of this study are available from the corresponding author upon request.
RESULTS
Genetic analysis of R. amblyommatis
Recent analyses of rickettsial genomes and advances in genetic studies of Rickettsia provided insights into the biology of Rickettsia with the identification of conserved and unique genes involved in the rickettsial lifecycle (Andersson et al. 1998; McLeod et al. 2004). The evolution of Rickettsia pathogens seems to follow the strategy of genome reduction, where genes are inactivated or lost via deletions, thereby increasing microbial dependence on host metabolism and nutrients while simultaneously increasing the virulence potential (Diop, Raoult and Fournier 2018, 2019). The genome sequence analyses revealed that SFG genomes (∼1.2–1.6 Mbp) encode ∼1200–1500 proteins, with unique gene rearrangements and single nucleotide polymorphisms in each genome (Ogata et al. 2005; Matsutani et al. 2013; Clark et al. 2015; Londono et al. 2019). SFG rickettsiae present with different clinical characteristics and severity, suggesting that multiple unique rickettsial and host factors engage in clinical pathogenesis. To determine the genomic similarities between R. amblyommatis and other pathogenic SFG rickettsiae, the genetic sequence of R. amblyommatis strain GAT-30V was compared with those of R. rickettsii strain Sheila Smith and R. conorii strain Malish 7. The genome of R. amblyommatis GAT-30V consists of a singular chromosome predicted to encode 1229 proteins with 371 pseudogenes and three circular pRM plasmids containing 31 974, 18 263 and 22 851 bp, respectively (Table 1). While R. rickettsii and R. conorii do not have circular plasmids, their chromosomal DNA sequences are predicted to produce 1234 and 1269 proteins with 150 and 223 pseudogenes (Table 1). Cross-comparison analysis of three spotted fever Rickettsia genome sequences identified high degrees of sequence identity (R. amblyommatis vs. R. rickettsii, 97.03% identity with 88% query coverage; R. amblyommatis vs. R. conorii, 96.09% identity with 89% query coverage), multiple rearranged genetic fragments and regions lost in R. conorii and R. rickettsii (Fig. 1A). Further, numerous putative virulence genes that have been characterized for spotted fever pathogenesis are present with 86–95% amino acid identity in R. amblyommatis (Table 2) (Gillespie et al. 2015). Thus, our bioinformatic analyses suggest that R. amblyommatis has the potential to cause disease in mammalian hosts.
Table 1.
Genomic statistics for R. amblyommatis, R.rickettsii and R. conorii.
| Rickettsia a | Total no. of bases | No. of genes | No. of genes encoding proteins | No. of pseudo genes |
|---|---|---|---|---|
| R. amblyommatis str. GAT-30V | 1 407 796 | 1640 | 1229 | 371 |
| R. rickettsii str. Sheila Smith | 1 257 710 | 1425 | 1234 | 150 |
| R. conorii str. Malish 7 | 1 268 755 | 1532 | 1269 | 223 |
GenBank accession numbers for R. amblyommatis strain GAT-30V, R. rickettsii strain Sheila Smith and R. conorii strain Malish 7 are CP003334.1, CP000848.1 and AE006914.1, respectively.
Figure 1.
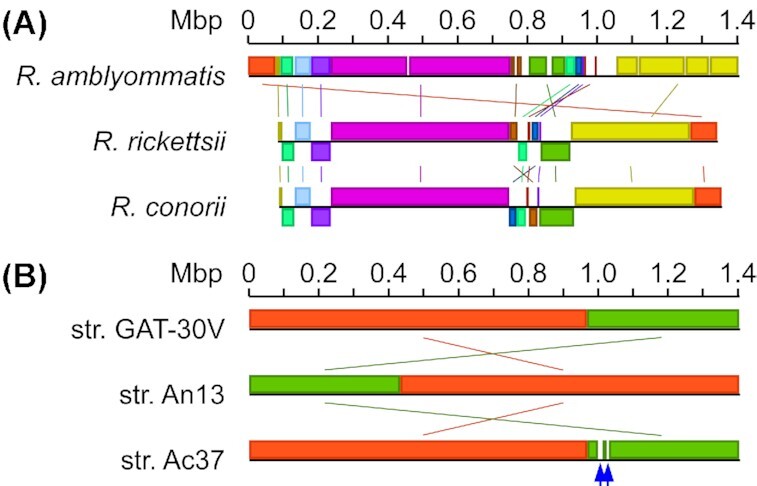
Schematic diagrams of Mauve gene alignments for (A)R. amblyommatis strain GAT-30V (GenBank accession no. CP003334.1), R. rickettsii strain Sheila Smith (GenBank accession no. CP000848.1) and R. conorii strain Malish 7 (GenBank accession no. AE006914.1) and (B)R. amblyommatis strain GAT-30V (GenBank accession no. CP003334.1), R. amblyommatis strain Ac37 (GenBank accession no. CP012420.1) and R. amblyommatis strain An13 (GenBank accession no. CP015012.1). Blue arrows point to genetic regions only present in R. amblyommatis strain Ac37.
Table 2.
Putative virulence genes conserved in R. amblyommatis.
| Putative virulence factorsa | Gene numbers in str. GAT-30Vb | Gene numbers in str. An13c | Gene numbers in str. Ac37d | % identity to R. rickettsiie | Predicted virulence functionsf |
|---|---|---|---|---|---|
| Adr1 | MCE_08 045 | A3305_02 145 | AL573_07 330 | 94.63% | Complement resistance |
| Adr2 | MCE_08 050 | A3305_02 150 | AL573_07 335 | 95.09% | Complement resistance |
| Pat1 | MCE_06 135 | A3305_00 385 | AL573_05 575 | 88.57% | Phagolysosome escape |
| RickA | MCE_06 070 | A3305_00 325 | AL573_05 520 | 91.02% | Actin-based motility |
| RARP2 | MCE_06 205 | A3305_00 455 | AL573_05 645 | 90.13% | Effector protein of T4SS |
| Sca0 | MCE_07 995 | A3305_02 110 | AL573_07 295 | 86.65% | Host cell adhesion |
| Sca1 | MCE_00 840 | A3305_02 870 | AL573_00 730 | 87.67% | Host cell adhesion |
| Sca2 | MCE_01 310 | A3305_03 305 | AL573_01 175 | 86.69% | Actin-based motility |
| Sca4 | MCE_04 250 | A3305_06 025 | AL573_03 910 | 93.89% | Cell-to-cell spread |
| Sca5 | MCE_07 010 | A3305_01 180 | AL573_06 365 | 90.27% | Host cell adhesion |
Annotated virulence factors shown to be involved in spotted fever pathogenesis in animal infection models.
Gene numbers in R. amblyommatis strain GAT-30V (GenBank: CP003334.1).
Gene numbers in R. amblyommatis strain An13 (GenBank: CP015012.1).
Gene numbers in R. amblyommatis strain Ac37 (GenBank: CP012420.1).
Percent identity to amino acids in R. rickettsii strain Sheila Smith (GenBank: CP000848.1).
Predicted virulence functions of each gene products.
Reductive genome evolution of R. amblyommatis
Three genome sequences of R. amblyommatis strains are available in the GenBank database. The GAT-30V strain was isolated from A. americanum in Georgia, a part of historical regions known for A. americanum geographic distribution. The other two R. amblyommatis strains, Ac37 and An13, have been isolated from Amblyomma cajennense in Brazil and Amblyomma neumanni in Argentina. Genetic adaptation to arthropod vectors is predicted to significantly impact vector competence and disease transmission and provide a selective pressure on the genetics of arthropod-borne pathogens (Gooding 1996). Genetic variability among the R. rickettsii strains has been characterized for the basis of differences in the virulence and disease severity in animals and humans (Ellison et al. 2008). Thus, we cross-compared three genetic sequences of R. amblyommatis strains derived from different tick species in three distinct geographical locations to determine the level of genetic stability. While three genomic sequences of R. amblyommatis strains displayed high similarity (>99%), we noted three genetic regions that are present in the strain Ac37 but absent in strains GAT-30V and An13 (Fig. 1B). Genes in these regions in R. amblyommatis Ac37 appear to be undergoing deletions and frameshifts, representing a hot spot for genome degradation in R. amblyommatis (Table 3). Two of the genes in the first region (1 000 184–1 012 987) encode enzymes involved in stringent responses and another encodes IS100 family transposase. Also, there were three non-functional copies of genes predicted to be the remnants of genes encoding transposases and a heat-shock protein. Interestingly, all genes in the second region (1 027 069–1 030 599) are disrupted genes, with a pseudogene homologous to AL573_05 270 that appears in the first region. The last region (1 046 364–1 048 082) only represents one gene, AL573_05 435. Genome reduction has been described for other SFG Rickettsia species, such as R. conorii and R. rickettsii (Ogata et al. 2001; Ellison et al. 2008). Here, we characterized an ongoing process of genome reduction in R. amblyommatis.
Table 3.
Genes present in R. amblyommatis strain AC37.
| Gene numbers in R. amblyommatis strain Ac37a | Coding regionb | Predicted proteinc |
|---|---|---|
| First region (1 000 184–1 012 987) | ||
| AL573_05 270 | 999 477–1 000 205 | Guanosine polyphosphate pyrophosphohydrolase |
| Pseudo gene | 1 000 869–1 000 958 | Transposase |
| AL573_05 280 | 1 001 169–1 002 161 | IS110 family transposase |
| AL573_05 285 | 1 002 491–1 005 442 | Hypothetical protein |
| AL573_05 290 | 1 005 559–1 006 263 | GNAT family N-acetyltransferase |
| AL573_05 295 | 1 006 359–1 006 973 | Bifunctional (p)ppGpp synthetase/guanosine-3',5'-bis(diphosphate) 3'-pyrophosphohydrolase |
| Pseudo gene | 1 007 184–1 007 327 | Heat-shock protein |
| Pseudo gene | 1 007 561–1 008 505 | Transposase |
| AL573_05 310 | 1 008 587–1 008 913 | Hypothetical protein |
| AL573_05 315 | 1 008 960–1 012 943 | Hypothetical protein |
| AL573_05 320 | 1 012 943–1 013 182 | Hypothetical protein |
| Second region (1 027 069–1 030 599) | ||
| Pseudo gene | 1 025 546–1 027 072 | Conjugal transfer protein TraG |
| Pseudo gene | 1 027 108–1 027 997 | Hypothetical protein |
| Pseudo gene | 1 028 082–1 028 883 | DNA methyltransferase |
| Pseudo gene | 1 029 291–1 030 208 | Histidine kinase |
| Pseudo gene | 1 030 362–1 031 021 | Guanosine polyphosphate pyrophosphohydrolase |
| Third region (1 046 364–1 048 082) | ||
| AL573_05 435 | 1 045 757–1 049 095 | tetratricopeptide repeat protein |
Gene numbers in R. amblyommatis strain Ac37 (GenBank accession no. CP012420.1).
Coding regions annotated in GenBank.
Prediction of protein-coding genes in GenBank.
Rickettsia amblyommatis is defective for attachment to endothelial cells and replicates at a slower rate
While Rickettsia targets and invades vascular endothelial cells in humans, multiple in vitro tissue culture systems have supported rickettsial replication and enabled investigators to identify rickettsial and host factors involved in the rickettsial obligate intracellular lifecycle (Haglund et al. 2010; Lehman et al. 2018; Kim et al. 2019). To characterize R. amblyommatis growth phenotypes, we performed pairwise comparisons of bacterial replication at timed intervals in Vero (African green monkey kidney epithelial cells) and HMEC-1 (human dermal microvascular endothelial cells). When inoculated at a multiplicity of infection of 0.01, R. conorii Malish 7 expanded rapidly for the first 2 days of infection (Fig. 2A). By day 6, R. conorii formed large plaques and caused extensive necrosis and destruction of both cell types (Fig. 2C). On day 8 post-infection, the number of infectious R. conorii dropped significantly as no host cells were available to support its obligate intracellular lifecycle on HMEC-1 cells. Importantly, R. conorii replicated at a comparable rate in both cell types (P <>0.05i>). Next, we performed the same growth analysis with R. amblyommatis GAT-30V (Fig. 2B). While both cell types supported R. amblyommatis intracellular lifecycle, R. amblyommatis replicated at a slower rate (R. amblyommatis vs. R. conorii on Vero cells at days 2, 4 and 8, P <<0.05i>; R. amblyommatis vs. R. conorii on HMEC-1 cells at days 0, 2, 4, 6 and 8, P <<0.01i>, Fig. 2D). Furthermore, R. amblyommatis GAT-30V displayed a significant defect in host cell adhesion and invasion on HMEC-1 cells (R.conorii vs. R. amblyommatis, P <0.0001, Fig. 2E). On day 4 of inoculation, R. amblyommatis on Vero cells started to form small plaques, which expanded to 249 ± 13 μm (mean ± SEM, N = 20, Day 6) and 964 ± 30 μm (mean ± SEM, N = 20, Day 8, Fig. 2D). In addition to the attachment defect, R. amblyommatis produced minimal cytopathology on HMEC-1 cell cultures for 8 days of infection (Fig. 2D and E). These data suggest that R. amblyommatis replicates at a slower rate compared with other pathogenic SFG rickettsiae in tissue culture cells, requires specific gene products to survive in endothelial cells and causes minimal cytopathology in endothelial cells.
Figure 2.
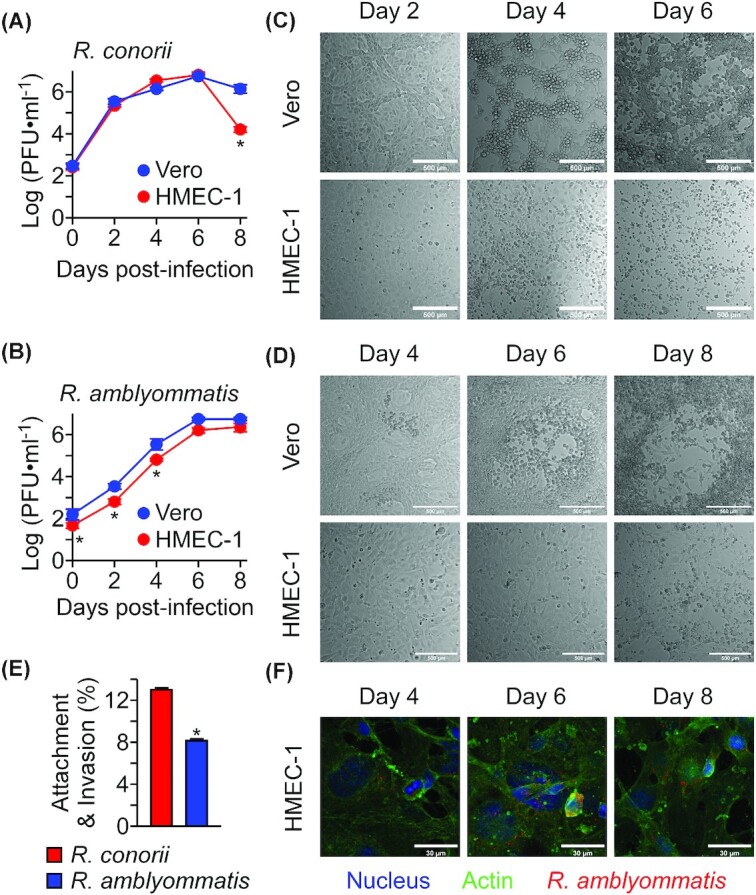
Rickettsia amblyommatis is defective for attachment to host endothelial cells and replicates at a slower rate. (A)Rickettsia conorii strain Malish 7 and (B)R. amblyommatis strain GAT-30V replications in Vero and HMEC-1 cells were quantified with the plaque assay (mean ± SEM, N = 3). Representative differential interference contrast (DIC) microscopic images of Vero and HMEC-1 cells infected with (C)R. conorii or (D)R. amblyommatis on 2, 4, 6 and 8 days post-inoculation (scale bar, 500 µm). (E)Rickettsia conorii strain Malish 7 and R. amblyommatis strain GAT-30V attachment and invasion into HMEC-1 cells were quantified with the plaque assay on Vero cells (mean ± SEM, N = 3). (F) Representative confocal microscopic images of intracellular R. amblyommatis in HMEC-1 cells (scale bar, 30 µm). *, P < 0.05. Data are representative of two independent experiments.
Virulence potential of R. amblyommatis in C3H mice
To investigate whether R. amblyommatis exhibits acute diseases in the mouse model for spotted fever, cohorts of C3H mice were intravenously infected with 5–50 × 105 PFU R. amblyommatis GAT-30V. Animals infected with 5 × 106 PFU R. amblyommatis GAT-30V exhibited severe body weight loss (>20%) and lethal outcome within 3 days of inoculation (Fig. 3A and B). When infected with a sub-lethal dose (5 × 105 PFU), animals exhibited modest body weight losses within 3 days, followed by a fast recovery (Day 3, male, P <<0.05i>; Days 1–3, female, P <<0.05i>; Fig. 3C and D). On the other hand, animals infected with 25 × 105 PFU of R. amblyommatis GAT-30V displayed acute disease with significant body weight loss during the first 2 days of infection followed by a slow recovery for the next 5–10 days (Days 1–14, male, P <<0.01i>; Days 1–6, female, P <<0.01i>; Fig. 3C and D). Following 2 days of infection, we recovered 90 ± 43 PFU∙ml−1 (mean ± SEM, N = 5, male) R. amblyommatis from heart homogenates of animals infected with 25 × 105 PFU of R. amblyommatis. Further, animals developed R. amblyommatis-specific half-maximal IgG titers of 5986 ± 787 (mean ± SEM, N = 5, male) on Day 14 of infection. Taken together, these data suggest that R. amblyommatis is responsible for the observed clinical symptoms in mice. Of note, these infectious doses are ~5–2500-fold higher than the experimental lethal (1 × 106 PFU) and sub-lethal (1 × 103 PFU) doses of R. conorii Malish 7 determined in the mouse infection model for spotted fever (Kim et al. 2019).
Figure 3.
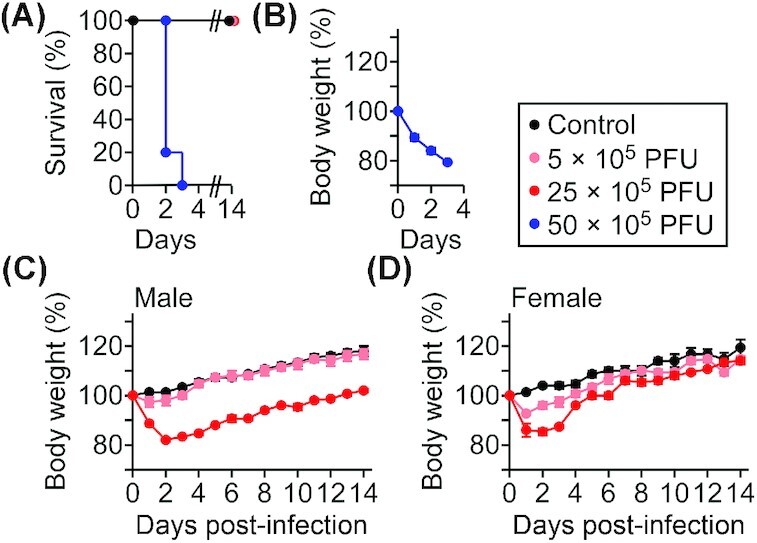
Rickettsia amblyommatis causes spotted fever pathogenesis in C3H mice. (A) Kaplan-Meier analysis for survival and (B) body weight analysis of C3H mice (N = 5) infected with 5–50 × 105 PFU R. amblyommatis strain GAT-30V. (C and D) Body weight analysis of C3H mice (N = 5) infected with 5–25 × 105 PFU of R. amblyommatis. Data are representative of two independent experiments.
DISCUSSION
Our results demonstrate that R. amblyommatis displays defective host cell attachment and invasion phenotype, replicates within host endothelial cells at a slower rate and causes mild spotted fever diseases in mice. Our data corroborate clinical and serological evidence that R. amblyommatis may cause self-limiting mild febrile illnesses in humans and other mammals. Thus far, R. amblyommatis has not been isolated from patients, and the quantitative and qualitative natures of R. amblyommatis transmission during tick blood meal remain unknown (Esteves et al. 2019). Cytotoxic T lymphocyte activities against infected host cells and complement-mediated bacterial killing modulate host-protective immune responses against rickettsial infections (Walker, Olano and Feng 2001; Riley et al. 2018). Recent studies highlight the critical roles of inflammasome and interferons in fighting against infections caused by Rickettsia (Smalley et al. 2016; Burke et al. 2020). Thus, it is possible that patients with immature or compromised immune systems might be at a greater risk of developing clinical symptoms upon infections with R. amblyommatis. While tick saliva proteins are known to reduce local inflammation, pathogenic spotted fever rickettsiae actively modulate host immune responses to establish a replicative niche within the hostile intracellular environment of phagocytes recruited to the tick bite site (Chmelar et al. 2011; Marchal et al. 2011; Curto et al. 2019). Our in vivo data and clinical evidence indicate that infections with R. amblyommatis may invade endothelial cells and cause transient local vasculitis at the tick bite site until professional immune cells, such as macrophages or CD8-positive T cells, clear the invading pathogens. Future R. amblyommatis infection studies with in vitro tissue culture cells or in vivo animal models lacking specific immune components may reveal host cells and immune factors critical for controlling R. amblyommatis infections in humans.
We show here that three strains of R. amblyommatis share highly similar genome sequences and have conserved genes shown to be important for spotted fever pathogenesis in animal infection models. Cross-comparison of spotted fever Rickettsia genome sequences revealed multiple gene rearrangements and gene deletions in R. conorii and R. rickettsii, presumably enhancing their intracellular parasitism to cause spotted fever pathogenesis in mammalian hosts. While all three rickettsial species are predicted to produce comparable numbers of proteins (1229–1269), R. amblyommatis had extra copies of pseudogenes and genetic areas undergoing genome reduction. The precise molecular mechanisms whereby genes and genomes deteriorate in Rickettsia remain mostly unresolved (Blanc et al. 2007). A large proportion of A. americanum ticks is infected with R. amblyommatis in the USA, often exceeding 60% in questing ticks (Jiang et al. 2010; Parola et al. 2013; Karpathy et al. 2016; Sanchez-Vicente et al. 2019). Rickettsia amblyommatis is also closely associated with other species within the genus Amblyomma in South America (Nunes Ede et al. 2015; Mastropaolo et al. 2016). Amblyomma ticks are also capable of transmitting multiple human pathogens, including Ehrlichia chaffeensis (human monocytic ehrlichiosis), Ehrlichia ewingii (human granulocytic ehrlichiosis), Coxiella burnetii (Q fever), Francisella tularensis (tularemia), R. parkeri (spotted fever rickettsiosis) and R. rickettsii (RMSF) (Childs and Paddock 2003). In addition, Amblyomma ticks have been implicated in a life-threatening allergic reaction to α-Gal, also known as meat allergy (Araujo et al. 2016; Hashizume et al. 2018; Mitchell et al. 2020). Recent tick surveys revealed that numerous species in the genus Amblyomma ticks have expanded and invaded into the new geographical regions, creating a potential public health emergency (Childs and Paddock 2003). At the same time, this provides us with a unique opportunity to isolate and characterize multiple R. amblyommatis strains parasitizing Amblyomma ticks in historical areas (regions with warm and humid climates, e.g. South and Central USA) and compare them with those surviving within Amblyomma ticks invading new geographical areas (regions with cold and dry climates, e.g. Northeastern USA). Cross-comparison of genome sequences of R. amblyommatis will provide insights into the molecular and genetic processes of reductive genome evolution in Rickettsia, their interactions with other bacterial pathogens of medical importance, their adaptation to new tick vectors and environment, as well as their impact on vector competence and disease transmission.
ACKNOWLEDGEMENTS
We thank C. Paddock, S. Karpathy and J. Cello for reagents and technical advice; J. Mugavero and L. Sprague for experimental assistance; and members of the Kim Laboratory for comments and discussion. We also thank J. Benach for his encouragement and review of the manuscript
Contributor Information
Wan-Yi Yen, Division of Laboratory Animal Resources, Laboratory of Comparative Medicine, Stony Brook University, Stony Brook, NY 11794, USA.
Kayla Stern, John F. Kennedy High School, Bellmore, NY 11710, USA.
Smruti Mishra, Center for Infectious Diseases, Department of Microbiology and Immunology, Stony Brook University, Stony Brook, NY 11794, USA.
Luke Helminiak, Center for Infectious Diseases, Department of Microbiology and Immunology, Stony Brook University, Stony Brook, NY 11794, USA.
Santiago Sanchez-Vicente, Center for Infectious Diseases, Department of Microbiology and Immunology, Stony Brook University, Stony Brook, NY 11794, USA.
Hwan Keun Kim, Center for Infectious Diseases, Department of Microbiology and Immunology, Stony Brook University, Stony Brook, NY 11794, USA.
AUTHOR CONTRIBUTIONS
H.K.K., S.S.V. conceptualized ideas, H.K.K., W.Y., K.S. designed experiments, W.Y., K.S., S.M., L.H., H.K.K. performed experiments, W.Y., K.S., H.K.K. analyzed data, H.K.K. wrote the manuscript.
FUNDING
This project was supported by funds from Stony Brook University to H.K.K. and a grant AI144136 from the National Institute of Allergy and Infectious Diseases (NIAID), Infectious Diseases Branch to H.K.K.
Conflict of interest
None declared.
REFERENCES
- Andersson SG, Zomorodipour A, Andersson JOet al. . The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–40. [DOI] [PubMed] [Google Scholar]
- Apperson CS, Engber B, Nicholson WLet al. . Tick-borne diseases in North Carolina: is “Rickettsia amblyommii” a possible cause of rickettsiosis reported as Rocky Mountain spotted fever?. Vector-Borne Zoonotic Dis. 2008;8:597–606. [DOI] [PubMed] [Google Scholar]
- Araujo RN, Franco PF, Rodrigues Het al. . Amblyomma sculptum tick saliva: alpha-Gal identification, antibody response and possible association with red meat allergy in Brazil. Int J Parasitol. 2016;46:213–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett A, Little SE, Shaw E. Rickettsia amblyommii” and R. montanensis infection in dogs following natural exposure to ticks. Vector-Borne Zoonotic Dis. 2014;14:20–5. [DOI] [PubMed] [Google Scholar]
- Billeter SA, Blanton HL, Little SEet al. . Detection of Rickettsia amblyommii in association with a tick bite rash. Vector-Borne Zoonotic Dis. 2007;7:607–10. [DOI] [PubMed] [Google Scholar]
- Blanc G, Ogata H, Robert Cet al. . Reductive genome evolution from the mother of Rickettsia. PLos Genet. 2007;3:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton LS, Mendell NL, Walker DHet al. . Rickettsia amblyommii” induces cross protection against lethal Rocky Mountain spotted fever in a guinea pig model. Vector-Borne Zoonotic Dis. 2014;14:557–62. [DOI] [PubMed] [Google Scholar]
- Burgdorfer W, Cooney JC, Thomas LA. Zoonotic potential (Rocky Mountain spotted fever aed tularemia) in the Tennessee Valley region. II. Prevalence of Rickettsia rickettsi and Francisella tularensis in mammals and ticks from Land Between the Lakes. Am J Trop Med Hyg. 1974;23:109–17. [DOI] [PubMed] [Google Scholar]
- Burke TP, Engström P, Chavez RAet al. . Inflammasome-mediated antagonism of type I interferon enhances Rickettsia pathogenesis. Nature Microbiol. 2020;5:688–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs JE, Paddock CD. The ascendancy of Amblyomma americanum as a vector of pathogens affecting humans in the United States. Annu Rev Entomol. 2003;48:307–37. [DOI] [PubMed] [Google Scholar]
- Chmelar J, Oliveira CJ, Rezacova Pet al. . A tick salivary protein targets cathepsin G and chymase and inhibits host inflammation and platelet aggregation. Blood. 2011;117:736–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TR, Noriea NF, Bublitz DeACet al. . Comparative genome sequencing of Rickettsia rickettsii strains that differ in virulence. Infect Immun. 2015;83:1568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa FB, Costa AP, Moraes-Filho Jet al. . Rickettsia amblyommatis infecting ticks and exposure of domestic dogs to Rickettsia spp. in an Amazon-Cerrado transition region of northeastern Brazil. PLoS One. 2017;12:e0179163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curto P, Riley SP, Simões Iet al. . Macrophages infected by a pathogen and a non-pathogen spotted fever group Rickettsia reveal differential reprogramming signatures early in infection. Front Cell Infect Microbiol. 2019;9:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren FS, Paddock CD, Springer YPet al. . Expanding range of Amblyomma Americanum and simultaneous changes in the epidemiology of spotted fever group Rickettsiosis in the United States. Am J Trop Med Hyg. 2016;94:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jesus CE, Ganser C, Kessler WHet al. . A survey of tick-borne bacterial pathogens in Florida. Insects. 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delisle J, Mendell NL, Stull-Lane Aet al. . Human infections by multiple spotted fever group Rickettsiae in Tennessee. Am J Trop Med Hyg. 2016;94:1212–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diop A, Raoult D, Fournier PE. Paradoxical evolution of rickettsial genomes. Ticks Tick-borne Dis. 2019;10:462–9. [DOI] [PubMed] [Google Scholar]
- Diop A, Raoult D, Fournier PE. Rickettsial genomics and the paradigm of genome reduction associated with increased virulence. Microbes Infect. 2018;20:401–9. [DOI] [PubMed] [Google Scholar]
- Eisen RJ, Kugeler KJ, Eisen Let al. . Tick-borne zoonoses in the United States: persistent and emerging threats to human health. ILAR J. 2017;58:319–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison DW, Clark TR, Sturdevant DEet al. . Genomic comparison of virulent Rickettsia rickettsii Sheila Smith and avirulent Rickettsia rickettsii Iowa. Infect Immun. 2008;76:542–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom P, Burke TP, Mitchell Get al. . Evasion of autophagy mediated by Rickettsia surface protein OmpB is critical for virulence. Nature Microbiol. 2019;4:2538–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves E, Bizzarro B, Costa FBet al. . Amblyomma sculptum Salivary PGE2 Modulates the Dendritic Cell-Rickettsia rickettsii Interactions in vitro and in vivo. Front Immunol. 2019;10:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletti M, Paddock CD, Hecht JAet al. . Isolate-dependent differences in clinical, pathological, and transcriptional profiles following in vitro and in vivo infections with Rickettsia rickettsii. Infect Immun. 2021;89:e00626–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JJ, Kaur SJ, Rahman MSet al. . Secretome of obligate intracellular Rickettsia. FEMS Microbiol Lett. 1986;34:47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooding RH. Genetic variation in arthropod vectors of disease-causing organisms: obstacles and opportunities. Clin Microbiol Rev. 1996;9:301–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T. The biology of rickettsiae. Infect Agents Dis. 1996;5:127–43. [PubMed] [Google Scholar]
- Haglund CM, Choe JE, Skau CTet al. +. Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat Cell Biol. 2010;12:1057–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume H, Fujiyama T, Umayahara Tet al. . Repeated Amblyomma testudinarium tick bites are associated with increased galactose-alpha-1,3-galactose carbohydrate IgE antibody levels: a retrospective cohort study in a single institution. J Am Acad Dermatol. 2018;78:1135–41.e3. [DOI] [PubMed] [Google Scholar]
- Hecht JA, Allerdice MEJ, Dykstra EAet al. . Multistate survey of American dog ticks (Dermacentor variabilis) for Rickettsia species. Vector-Borne Zoonotic Dis. 2019;19:652–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Yarina T, Miller MKet al. . Molecular detection of Rickettsia amblyommii in Amblyomma americanum parasitizing humans. Vector-Borne Zoonotic Dis. 2010;10:329–40. [DOI] [PubMed] [Google Scholar]
- Karpathy SE, Slater KS, Goldsmith CSet al. . Rickettsia amblyommatis sp. nov., a spotted fever group Rickettsia associated with multiple species of Amblyomma ticks in North, Central and South America. Int J Syst Evol Microbiol. 2016;66:5236–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killmaster LF, Loftis AD, Zemtsova GEet al. . Detection of bacterial agents in Amblyomma americanum (Acari: ixodidae) from Georgia, USA, and the use of a multiplex assay to differentiate Ehrlichia chaffeensis and Ehrlichia ewingii. J Med Entomol. 2014;51:868–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, Premaratna R, Missiakas DMet al. . Rickettsia conorii O antigen is the target of bactericidal Weil-Felix antibodies. Proc Natl Acad Sci. 2019;116:19659–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman SS, Noriea NF, Aistleitner Ket al. . The Rickettsial Ankyrin repeat protein 2 is a type iv secreted effector that associates with the endoplasmic reticulum. MBio. 2018;9:e00975–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin ML, Schumacher LBM, Snellgrove A. Effects of Rickettsia amblyommatis infection on the vector competence of Amblyomma americanum ticks for Rickettsia rickettsii. Vector-Borne Zoonotic Dis. 2018;18:579–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londono AF, Mendell NL, Valbuena GAet al. . Whole-genome sequence of Rickettsia parkeri strain Atlantic rainforest, isolated from a Colombian tick. Microbiol Resour Announc. 2019;8:e00684–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes MG, Krawczak FS, de Lima JTRet al. . Occurrence of Ehrlichia canis and Hepatozoon canis and probable exposure to Rickettsia amblyommatis in dogs and cats in Natal, RN. Revista Brasileira de Parasitologia Veterinária. 2019;28:151–6. [DOI] [PubMed] [Google Scholar]
- Macaluso KR, Sonenshine DE, Ceraul SMet al. . Infection and transovarial transmission of rickettsiae in Dermacentor variabilis ticks acquired by artificial feeding. Vector-Borne Zoonotic Dis. 2001;1:45–53. [DOI] [PubMed] [Google Scholar]
- Marchal C, Schramm F, Kern Aet al. . Antialarmin effect of tick saliva during the transmission of Lyme disease. Infect Immun. 2011;79:774–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastropaolo M, Tarragona EL, Silaghi Cet al. . High prevalence of “Candidatus Rickettsia amblyommii” in Amblyomma ticks from a spotted fever endemic region in North Argentina. Comp Immunol Microbiol Infect Dis. 2016;46:73–6. [DOI] [PubMed] [Google Scholar]
- Matsutani M, Ogawa M, Takaoka Net al. . Complete genomic DNA sequence of the East Asian spotted fever disease agent Rickettsia japonica. PLoS One. 2013;8:e71861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod MP, Qin X, Karpathy SEet al. . Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J Bacteriol. 2004;186:5842–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell CL, Lin F-C, Vaughn Met al. . Association between lone star tick bites and increased alpha-gal sensitization: evidence from a prospective cohort of outdoor workers. Parasites Vectors. 2020;13:470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molaei G, Little EAH, Williams SCet al. . Bracing for the worst - range expansion of the lone star tick in the northeastern United States. N Engl J Med. 2019;381:2189–92. [DOI] [PubMed] [Google Scholar]
- Nilsson K, Elfving K, Pahlson C. Rickettsia helvetica in patient with meningitis, Sweden, 2006. Emerg Infect Dis. 2010;16;490–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes Ede C, Vizzoni VF, Navarro DLet al. . Rickettsia amblyommii infecting Amblyomma sculptum in endemic spotted fever area from southeastern Brazil. Memórias do Instituto Oswaldo Cruz. 2015;110:1058–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Audic S, Renesto-Audiffren Pet al. . Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science. 2001;293:2093–8. [DOI] [PubMed] [Google Scholar]
- Ogata H, Renesto P, Audic Set al. . The genome sequence of Rickettsia felis identifies the first putative conjugative plasmid in an obligate intracellular parasite. PLoS Biol. 2005;3:e248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddock CD, Sumner JW, Comer JAet al. . Rickettsia parkeri: a newly recognized cause of spotted fever rickettsiosis in the United States. Clin Infect Dis. 2004;38:805–11. [DOI] [PubMed] [Google Scholar]
- Pahlson C, Luc Xi, Ott Met al. . Characteristics of in vitro infection of human monocytes, by Rickettsia helvetica. Microbes Infect. 2020;104776. [DOI] [PubMed] [Google Scholar]
- Parola P, Paddock CD, Socolovschi Cet al. . Update on tick-borne rickettsioses around the world: a geographic approach. Clin Microbiol Rev. 2013;26:657–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoult D, Lakos A, Fenollar Fet al. . Spotless rickettsiosis caused by Rickettsia slovaca and associated with Dermacentor ticks. Clin Infect Dis. 2002;34:1331–6. [DOI] [PubMed] [Google Scholar]
- Riley SP, Fish AI, Del Piero Fet al. . Immunity against the obligate intracellular bacterial pathogen Rickettsia australis requires a functional complement system. Infect Immun. 2018;86:e00139–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas JJ, Moreira-Soto A, Alvarado Get al. . Pathogenic potential of a Costa Rican strain of 'Candidatus Rickettsia amblyommii’ in guinea pigs (Cavia porcellus) and protective immunity against Rickettsia rickettsii. Ticks Tick-borne Dis. 2015;6:805–11. [DOI] [PubMed] [Google Scholar]
- Rosenberg R, Lindsey NP, Fischer Met al. . Vital signs: trends in reported vectorborne disease cases - United States and territories, 2004–2016. MMWR Morb Mortal Wkly Rep. 2018;67:496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahni A, Fang R, Sahni SKet al. . Pathogenesis of Rickettsial diseases: pathogenic and immune mechanisms of an endotheliotropic infection. Ann Rev Pathol: Mech Dis. 2019;14:127–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Vicente S, Tagliafierro T, Coleman JLet al. . Polymicrobial nature of tick-borne diseases. mBio. 2019;10:e02055–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley C, Bechelli J, Rockx-Brouwer Det al. . Rickettsia australis activates inflammasome in human and murine macrophages. PLoS One. 2016;11:e0157231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snellgrove AN, Krapiunaya I, Scott Pet al. . Assessment of the pathogenicity of Rickettsia amblyommatis, Rickettsia bellii, and Rickettsia montanensis in a guinea pig model. Vector-Borne Zoonotic Dis. 2021;21:232–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshine DE. Range expansion of tick disease vectors in North America: implications for spread of tick-borne disease. Int J Environ Res Public Health. 2018;15:478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer A, Montenegro VM, Schicht Set al. . Detection of Rickettsia monacensis and Rickettsia amblyommatis in ticks collected from dogs in Costa Rica and Nicaragua. Ticks Tick-borne Dis. 2018;9:1565–72. [DOI] [PubMed] [Google Scholar]
- Suwanbongkot C, Langohr IM, Harris EKet al. . Spotted fever group Rickettsia infection and transmission dynamics in Amblyomma maculatum. Infect Immun. 2019;87:e00804–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn MF, Delisle J, Johnson Jet al. . Seroepidemiologic study of human infections with spotted fever group Rickettsiae in North Carolina. J Clin Microbiol. 2014;52:3960–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DH, Ismail N. Emerging and re-emerging rickettsioses: endothelial cell infection and early disease events. Nat Rev Microbiol. 2008;6:375–86. [DOI] [PubMed] [Google Scholar]
- Walker DH, Olano JP, Feng HM. Critical role of cytotoxic T lymphocytes in immune clearance of rickettsial infection. Infect Immun. 2001;69:1841–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti AS, Pornwiroon W, Kearney MTet al. . Characterization of rickettsial infection in Amblyomma americanum (Acari: ixodidae) by quantitative real-time polymerase chain reaction. J Med Entomol. 2008;45:267–75. [DOI] [PubMed] [Google Scholar]
- Zemtsova GE, Killmaster LF, Montgomery Met al. . First report of Rickettsia identical to R. slovaca in colony-originated D. variabilis in the United States: detection, laboratory animal model, and vector competence of ticks. Vector-Borne Zoonotic Dis. 2016;16:77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]


