Abstract
Introduction and hypothesis
Growing literature details the critical importance of the microbiome in the modulation of human health and disease including both the gastrointestinal and genitourinary systems. Rectovaginal fistulae (RVF) are notoriously difficult to manage, many requiring multiple attempts at repair before correction is achieved. RVF involves two distinct microbiome communities whose characteristics and potential interplay have not been previously characterized and may influence surgical success.
Methods
In this pilot study, rectal and vaginal samples were collected from 14 patients with RVF. Samples were collected preoperatively, immediately following surgery, 6–8 weeks postoperatively and at the time of any fistula recurrence. Amplification of the 16S rDNA V3-V5 gene region was done to identify microbiota. Data were summarized using both α-diversity to describe species richness and evenness and β-diversity to characterize the shared variation between communities. Differential abundance analysis was performed to identify microbial taxa associated with recurrence.
Results
The rectal and vaginal microbiome in patients undergoing successful fistula repair was different than in those with recurrence (β-diversity, p = 0.005 and 0.018, respectively) and was characterized by higher species diversity (α-diversity, p = 0.07 and p = 0.006, respectively). Thirty-one taxa were enriched in patients undergoing successful repair to include Bacteroidetes, Alistipes and Rikenellaceae as well as Firmicutes, Subdoligranulum, Ruminococcaceae UCG-010 and NK4A214 group.
Conclusions
Microbiome characteristics associated with fistula recurrence have been identified. The association of higher vaginal diversity with a favorable outcome has not been previously described. Expansion of this pilot project is needed to confirm findings. Taxa associated with successful repair could be targeted for subsequent therapeutic intervention.
Keywords: Microbiome, Rectal, Vaginal, Rectovaginal fistula, PROMIS, Bacterial taxa
Introduction
Rectovaginal fistulae (RVF) are characterized by uncontrolled loss of stool and gas from the vagina as a result of an abnormal communication with the rectum. Patients with RVF experience devastating physical and psychosocial effects, including social isolation, sexual dysfunction, vaginitis, cystitis and life-threatening infection. Fistula etiologies include inflammatory bowel disease, infections, radiation therapy, surgical and obstetric trauma [1, 2]. RVFs from obstetric trauma are most common and seen primarily in resource-poor countries as a consequence of unattended obstructed labor; these account for 2 million fistulae worldwide and 50,000–100,000 new cases annually [3].
The microbiome’s influence on human health and disease has been widely investigated through the Human Microbiome Project [4]. The study of the microbiome relies on unique features of the 16S rDNA gene (found only in Archaea and Bacteria) that can be amplified through polymerase chain reaction (PCR) and used to identify individual organisms. Despite increasing evidence for the role of the microbiome in gynecologic and colorectal pathology, characterization of the microbiome in patients with rectovaginal fistula (RVF) has yet to be explored, including any potential effect on RVF treatment success. Correction of RVF may require multiple attempts that cumulatively increase the risk of infectious morbidity, perioperative complications and healthcare costs, and recurrence rates as high as 67% have been reported [5]. It is unknown whether there are pathologic changes in the vaginal and/or rectal microbiome that predispose women to fistula recurrence or prime them for surgical success.
The primary aim of this pilot study is to characterize the vaginal and rectal microbiome in patients with rectovaginal fistula. The secondary aims are to collectively and longitudinally evaluate changes in the microbiome and in quality of life measurements through the perioperative course.
Materials and methods
This single-institution, IRB-approved (17–006551) pilot study involved the longitudinal collection of vaginal and rectal microbial samples from women with RVF planning to undergo surgical treatment from 1 January 2018 to 31 May 2019. Participants with diagnosed rectovaginal fistula were identified and offered enrollment. Women with colovaginal and enterovaginal fistula were not included in the study. Additional exclusion criteria included: current pregnancy, current or recent history (within the past 4 weeks) of antibiotic use, history of chemotherapy within 2 years of fistula surgery, prior pelvic or abdominal radiation therapy, and current or planned intestinal diversion. Patient demographic information was collected and included: age, BMI, menopausal status and tobacco use. Fistula size, etiology, location and history of prior repair were documented as well. Fistula location was designated as low, mid or high [6] at the time of clinical examination. The PROMIS-10 (Patient-Reported Outcomes Measurement Information System) Global Health tool was completed by each patient at enrollment and once again at their scheduled 6–8 week postoperative visit. This is a validated, ten-item measure of mental and physical health-related quality of life that has been used to assess outcomes in several surgical disciplines [7].
Microbial samples were obtained from the rectal and vaginal locations at several time points for each patient. Preoperative (T1) vaginal and rectal samples were collected in clinic within 4 weeks of surgery or in the operating suite prior to initiation of intravenous antibiotics and surgical site preparation. Immediate postoperative (T2) samples were collected by the surgeon at the time of surgical site closure, prior to the patient leaving the operating room. This collection occurred after administration of prophylactic antibiotics, betadine surgical site preparation and intraoperative use of irrigation; mechanical bowel preparation was not utilized. Finally, samples were collected at the scheduled postoperative visit 6–8 weeks following surgery (T3) or earlier if fistula recurrence occurred (T3R). A patient interview was completed 12 weeks following surgery to conclude study participation.
For each collection, two sterile, polyester-tipped swabs were held together and rotated over the site of the fistula within each environment (i.e., vagina or rectum). Swabs were placed in Hologic® Aptima transport tubes containing 2.9 ml of media and hand carried to our Biospecimens Accessioning and Processing (BAP) facility where they were flash frozen to −70°C and stored within 15 min of collection. They were accessioned into the Research and Laboratory Information Management System (RLIMS) at this time for tracking purposes. Once all specimens had been collected, the de-identified samples were sent to the University of Minnesota Genomics Center for DNA extraction and 16S rDNA processing/sequencing. The amount of DNA extracted from each sample was quantified using the Qubit dsDNA HS (High Sensitivity) Assay Kit. The DNA samples were then used to partially (V3-V5) amplify the microbial 16S rDNA genes through a PCR. Controls of both the DNA extraction and microbiome enrichment processes were performed; only one vaginal and one rectal sample had a sequencing depth similar to the negative controls, and they were excluded from analysis. The PCR product was subsequently purified using Agencourt® AMPure® and quantified before sequencing. The 16S rDNA hypervariable tag sequencing was performed using a high-throughput next-generation Illumina MiSeq (San Diego, CA, USA) sequencing platform.
Bioinformatics processing of the 16S rDNA amplicon sequence reads was accomplished with DADA2 [8] (software package that models and corrects amplicon errors); this effectively “de-noises” the sequence reads to identify ASV (amplicon sequence variants) of true biological origin. The SILVA ribosomal RNA gene database project [9] was used to assign taxonomic lineage based on Naïve Bayes Classifier. Finally, FastTree [10] (a bioinformatics method for constructing large phylogenies and for estimating their reliability) was used to construct the phylogenetic tree among ASVs. A total of 5,531,556 sequence reads (2586–137,573 reads per sample) were obtained (mean of 67,458 reads) after quality control.
To analyze the data (ASV table) we used measures of α-diversity, β-diversity and taxa abundance. α-Diversity is used to measure the diversity within a sample; we used two different metrics to calculate α-diversity. The first is the count of the number of different microbes (ASV count) in a sample; this is called species richness. The second, the Shannon Index, measures the richness in a sample as well as the distribution of different microbes within that sample, called evenness. Evenness is a measure of the variation in the relative abundance of different species in the sample. Since the α-diversity of a sample depends on the sequence depth (the total number of reads), we rarefy the sample reads to an equal depth in order to be comparable. We present these indices graphically with rarefaction plots; these plot the number of ASVs versus the number of reads sampled. A linear model or linear mixed effect model was used for testing the association of α-diversity with variables of interest (e.g., recurrence status) while adjusting potential confounders (e.g., BMI).
The second measure, β-diversity, is a term for the comparison of samples to each other. β-Diversity provides a measure of the ecological distance or dissimilarity between bacterial communities. We calculated four different β-diversity measures [weighted UniFrac, unweighted UniFrac, generalized UniFrac (α = 0.5) and Bray-Curtis], each providing a unique view of community structure [11]. Weighted UniFrac measures the abundance of observed ASVs and phylogeny, while the unweighted only compares the presence or absence of ASVs and phylogeny. The generalized UniFrac unifies the weighted and unweighted UniFrac distance into a single framework, while the Bray-Curtis distance is a non-phylogeny-based method that also takes abundance into account. Rarefaction was performed on the ASV table before calculating these distances. We used the PERMANOVA test based on β-diversity measures to test for associations between the overall microbiota composition and variables of interest and PERDISP for testing the difference in dispersion (i.e., between-subject variability) between groups (both 999 permutations) [11]. Within-subject permutation was used for the comparison within the subjects. Principal coordinate analysis plots (PCoA) were used to graphically demonstrate these data. These plots project the distance matrix into a new set of orthogonal axes on which the data are presented. Typically, the first two axes (called PC1, PC2) can be used to depict or explain the maximum amount of variation in the data.
Finally, we performed taxon-level differential abundance analysis at the phylum, class, order, family and genus level to identify differences in the specific microbial genera of each collected cohort. Taxa with prevalence < 10% or with a maximum proportion < 0.2% were excluded to reduce the number of tests. The count data were normalized into relative abundances by the GMPR [12] approach to address variable sequencing depth. To identify differentially abundant taxa associated with variables of interest adjusting for potential confounders, we fit a multiple linear regression based on the square-root transformed normalized abundance. Permutation (999 permutations) based on the F-statistic was used to assess the significance to address potential non-normality of the data. Within-subject permutation was used for the comparison within the subjects. False discovery rate (FDR) control (B-H procedure), which controls the percentage of the false positives in the claimed positives, was used to correct for multiple testing at each taxonomic level, and FDR-adjusted p values or q-values < 0.20 were considered significant (i.e., the expected percentage of the false positives in the result is < 20%). The relatively large FDR cutoff is used so as not to miss differential low-abundance bacteria, which usually have low statistical power.
Results
A total of 14 patients were enrolled into the study over a period of 18 months. Demographic and baseline fistula information is given in Table 1. There were eight fistula recurrences following surgery; all fistulas were of the “low” classification [6]. All surgical repairs included excision of the fistula tract with closure. Concomitant surgeries included the use of a flap in four cases (3 Martius, 1 Gracilis), an endorectal advancement flap in one case and overlapping sphincteroplasty in four cases. Complications included one Martius flap breakdown without fistula recurrence and one sphincteroplasty breakdown that required return to OR for repair and bowel diversion.
Table 1.
Patient demographics and baseline fistula characteristics
| No recurrence (N = 6) | Recurrence (N = 8) | Total (N = 14) | p value | |
|---|---|---|---|---|
| Age | 0.7657 | |||
| Mean (SD) | 43.5 (19.1) | 50.4 (10.4) | 47.4 (14.5) | |
| Range | (23.0–71.0) | (38.0–68.0) | (23.0–71.0) | |
| BMI | 0.0210 | |||
| Mean (SD) | 25.6 (5.1) | 33.1 (5.2) | 29.9 (6.3) | |
| Range | (19.5–34.7) | (24.6–42.8) | (19.5–42.8) | |
| Smoker | 1.0000 | |||
| Current | 1 (16.7%) | 2 (25.0%) | 3 (21.4%) | |
| Never | 2 (33.3%) | 3 (37.5%) | 5 (35.7%) | |
| Prior | 3 (50.0%) | 3 (37.5%) | 6 (42.9%) | |
| Menopausal | 0.6270 | |||
| No | 4 (66.7%) | 4 (50.0%) | 8 (57.1%) | |
| Yes | 2 (33.3%) | 4 (50.0%) | 6 (42.9%) | |
| Fistula size | 0.5105 | |||
| 1 mm | 2 (33.3%) | 5 (62.5%) | 7 (50.0%) | |
| 2 mm | 2 (33.3%) | 1 (12.5%) | 3 (21.4%) | |
| > 6 mm | 2 (33.3%) | 2 (25.0%) | 4 (28.6%) | |
| Fistula etiology | 1.0000 | |||
| Inflammatory | 0 (0.0%) | 1 (12.5%) | 1 (7.1%) | |
| Obstetric | 4 (66.7%) | 4 (50.0%) | 8 (57.1%) | |
| Unknown | 0 (0.0%) | 1 (12.5%) | 1 (7.1%) | |
| Postop/trauma | 2 (33.3%) | 2 (25.0%) | 4 (28.6%) | |
| Fistula location | 1.0000 | |||
| Low | 6 (100%) | 8 (100%) | 14 (100%) | |
| Mid | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | |
| High | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | |
| Prior fistula repair | 0.2774 | |||
| No | 4 (66.7%) | 2 (25.0%) | 6 (42.9%) | |
| Yes (mean no.) | 2 (1) (33.3%) | 6 (3.2) (75.0%) | 8 (57.1%) |
Six of the eight recurrences had an average of three prior repair attempts before being referred to our institution. The only demographic variable that was statistically significant between those experiencing recurrence and those who did not was BMI (mean 33.1 vs. 25.6, respectively, p = 0.02). PROMIS quality of life scores are shown in Table 2; there were significant differences in postoperative physical and mental health scores between patients with and without fistula recurrence (p = 0.006 and p = 0.023, respectively). No differences were appreciated preoperatively between groups.
Table 2.
Perioperative PROMIS* measures
| Total | Recurrent | Non-recurrent | P value | |
|---|---|---|---|---|
| Preop physical health score | 48.3 (9.4) | 45.7 10.4() | 52.5 (6.4) | 0.2222 |
| Preop mental health score | 46.1 (6.7) | 44.5 (7.6) | 48.6 (4.0) | 0.2696 |
| Postop physical health score | 46.2 (9.9) | 39.6 (6.5) | 54.0 (6.5) | 0.0058 |
| Postop mental health score | 45.7 (10.0) | 40.1 (4.1) | 52.3 (11.2) | 0.0233 |
Patient-Reported Outcomes Measurement Information System
T-scores are reported with standard error on T-score metric in parentheses
T-score distributions are standardized such that 50 represents the average (mean) for the US general population, and the standard deviation around that mean is 10 points
Microbiome characterization
The distribution of the samples after quality control is shown in Table S1. The deep sequencing of the V3-V5 16S rDNA region of all 82 samples resulted in the analysis of 16,723 ASVs. The ASVs belong to 15 phyla, 85 families and 339 genera. The dominant taxa of the vaginal microbiome included Lactobacillus (16.2%), Bacteroides (9.4%) and Prevotella (6.4%). The rectal dominant taxa included Bacteroides (14.6%), Parabacteroides (5.8%), Prevotella (5.5%) and Faecalibacterium (5.4%) [Fig. S1].
We identified several unusual relationships not previously described in the literature. First, our PCoA plot based on the weighted UniFrac distance from all time points combined (Fig. 1a) shows a significant degree of overlap and similarity between the vaginal and rectal microbiome across the cohort, with the vaginal microbiome demonstrating greater between-sample diversity (p = 0.004, PERDISP). Some taxa including Lactobacillus remained differential between the vaginal and rectal microbiome [Fig. 1b]. The Bray-Curtis distances, looking at samples collectively at T1 and T2, shows microbiome clustering significantly more by patient than by site, meaning the vaginal microbiome of an individual is more similar to their own rectal microbiome than to the vaginal microbiome of the remaining cohort (and vice versa) (Fig. 2). Typically, in patients without fistula, the rectum has much greater microbial diversity than the vagina, there is little overlap between the two and clustering occurs more by site than by the individual.
Fig. 1.
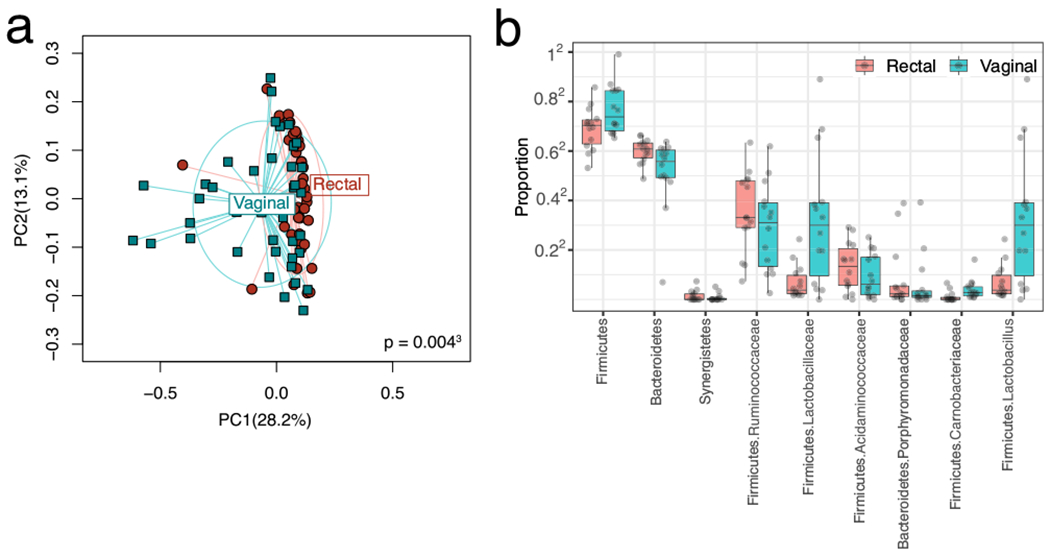
a Rectal and vaginal microbial β-diversity. Principal coordinate analysis depicting the relationship of the rectal and vaginal microbiomes. Weighted UniFrac measure of β-diversity is used. PC1 and PC2: principal coordinate axis 1 and principle coordinate axis 2. Significant overlap between the two microbiomes is seen, yet they are significantly distinct (p = 0.002). Greater between-subject vaginal diversity (p = 0.004). b Differential taxa between the vaginal and rectal microbiomes. Rectal and vaginal diversity by individual patient
Fig. 2.
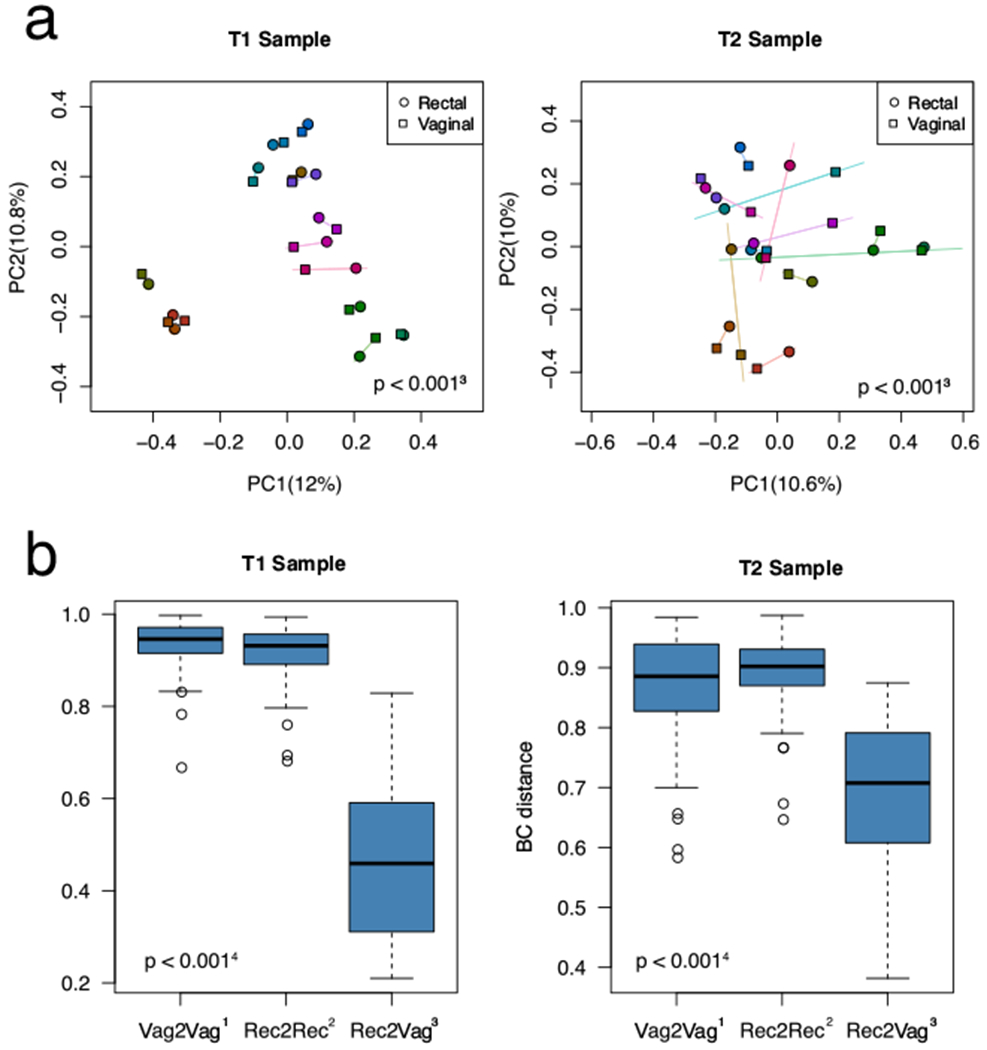
a Principal coordinate analysis demonstrating clustering by individual patient more than microbiome site. Bray-Curtis dissimilarity measure of β-diversity. PC1 and PC2: principal coordinate axis 1 and principle coordinate axis 2. Each subject is represented by a different color. b The vaginal sample resembles the rectal sample from the same subject at T1 and T2. The Y-axis denotes the Bray-Curtis distance; a higher number indicates greater dissimilarity. Vag2Vag: between-subject distance for the vaginal sample; Rec2Rec: between-subject distance for the rectal sample; Rec2Vag: rectal-to-vaginal distance for the same subject. Rec2Vag was significantly more alike or “clustered” than Vag2Vag or Rec2Rec. The microbiome composition of the vaginal T1 sample is associated with surgical outcome
After performing a PERMANOVA analysis again combining all cohort time points, we found some evidence of microbiome association for several clinical variables for both the vaginal and rectal samples. These include menopausal status, BMI and history of vaginal infection in the past 6 months (Fig. S2 and S3). Of these associations, BMI again is the only measure that is significantly different between patients who recur and those who do not (p = 0.029); we therefore adjusted for this potential confounder when comparing the rectal and vaginal microbiota between the two groups.
Vaginal microbiome
We performed a comparative analysis of vaginal samples at each collection time point for those with and without fistula recurrence. Reduced species richness (the number of observed ASVs) approximating significance was seen preoperatively (T1) in patients who would subsequently recur compared to those who would not (p = 0.07, Fig. S4). After adjusting for BMI, the overall microbiota composition is shown to be markedly different between the two groups as revealed by the β-diversity analysis (Bray-Curtis distance, p = 0.005, omnibus p value = 0.013 Fig. 3b). Differential abundance analysis reveals 29 different taxa that are depleted in patients with recurrence (Fig. 3c). The most significant included Firmicutes, Subdoligranulum, Ruminococcaceaea UCG-010 and Ruminococcaceaea NK4A214 group as well as Bacteroidetes, Alistipes and Rikenellaceae (Table S2).
Fig. 3.
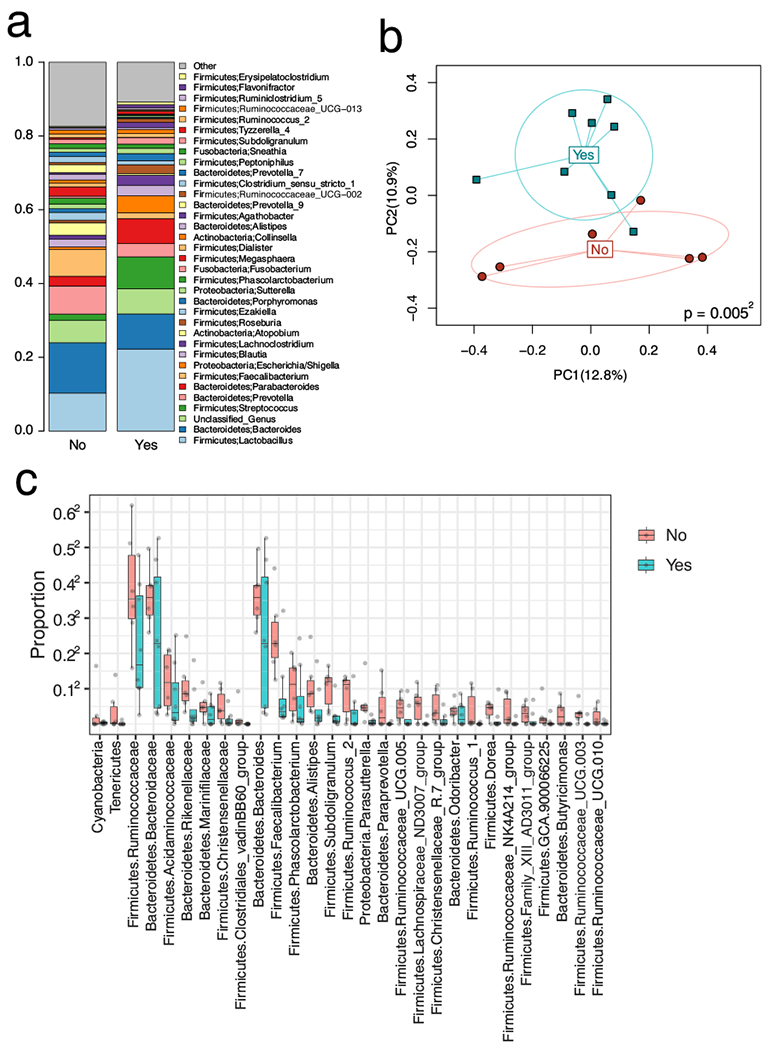
a The colored bars on the left depict the relative proportion of taxa. The taxa are named in the middle portion of the figure and positioned adjacent to their color. b Principal coordinate analysis based on Bray-Curtis dissimilarity. PC1 and PC2 stand for principal coordinate 1 and 2. Yes indicates fistula recurrence; no indicates fistula non-recurrence. p value indicates the distance or difference between the microbiome at T1 of patients who will recur compared to those who will not. c Taxa associated with surgical outcome in the vaginal T1 sample. Plot of the relative proportion of each of the 29 taxa differentially present by surgical outcome. Yes indicates fistula recurrence; no indicates fistula non-recurrence. The microbiome composition of the (or in the) vaginal T1 sample is associated with surgical outcome
When comparing T1 vs. T2 collections based on the β-diversity, we found that surgery changes the overall vaginal microbiome composition (UniFrac, p = 0.001, Fig. S5, S6). The greatest effect was seen with Pseudomonadaceae, Xanthomonadaceae, Pseudomonas, Ralstonia and Serratia, all of which decreased in those with subsequent fistula recurrence. Because these species were present in similar levels in both recurrent and non-recurrent T1 samples, it appears that the surgical effect on T2 may not be related to recurrence. We found no significant differences in recurrent vs. non-recurrent samples collected at T2.
Further analysis of non-recurrent subjects demonstrates a trend toward increased species diversity of T1 relative to T3 (p = 0.07, Fig. S7); no difference in diversity between T2 and T3 is detected at this power (data not shown).
Rectal microbiome
The comparative analysis of the recurrent vs. non-recurrent rectal samples at each collection time point after BMI adjustment also demonstrated a significant reduction in the α-diversity measures of species richness/Shannon index from T1 samples of patients who would subsequently recur compared to those who would not (Fig. 4a). Like the vaginal samples, the overall microbial composition was different between the two groups as revealed by the beta-diversity analysis (Bray-Curtis distance, p = 0.018, Fig. 4c). Differential abundance analysis reveals 13 different taxa depleted in patients with recurrence; this showed significant overlap with those in the vaginal sample with 11 of the 13 taxa in common (Fig. 4d, Table S3).
Fig. 4.
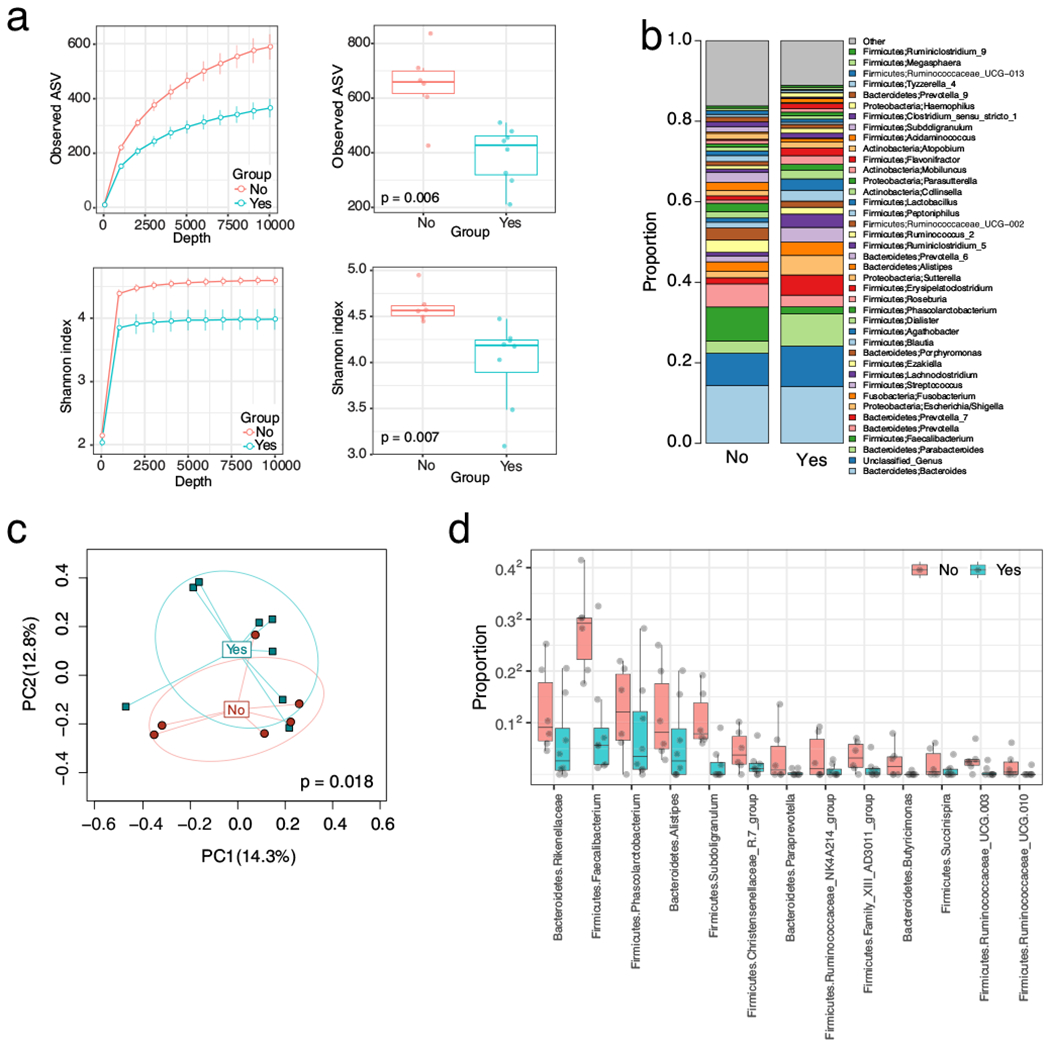
a Species richness and Shannon Index (measures of α-diversity) in rectal visit T1 sample decrease with recurrence. Species richness is depicted in the top two graphs; p value denotes difference in richness between those who will subsequently recur and those who will not. Shannon Index (a measure of both species richness and evenness) is depicted in the bottom two graphs; p value denotes the difference in Shannon Index between those who will recur and those who will not. Yes denotes recurrence; no denotes non-recurrence. The two graphs on the left side of Fig. 4a are rarefaction curves; these plot the number of species as a function of the sequencing depth. The bar on the two right-sided graphs indicates the standard error of the mean. b The colored bars on the left depict the relative proportion of taxa. The taxa are named on the right and positioned adjacent to their color. c Principal coordinate analysis on Bray-Curtis dissimilarity. PC1 and PC2 stand for principal coordinate 1 and 2. Yes indicates fistula recurrence; no indicates fistula non-recurrence. d Taxa associated with surgical outcome in the rectal T1 sample. Plot of the relative proportion of each of the 13 taxa differentially present by surgical outcome. Yes indicates fistula recurrence; no indicates fistula non-recurrence. The microbiome composition in the rectal T1 sample is associated with surgical outcome
We again saw that surgery was associated with reduced species richness at T2 compared to T1, especially for nonrecurrent samples (p = 0.008, Fig. S8). The overall microbiome change affected Actinobacteria and Barnesiellaceae most significantly (Fig. S9). We again see no difference in T2 samples between the two groups.
In the non-recurrent rectal cohort, the increased diversity in T3 relative to T2 (species richness, p = 0.07, Fig. S10) approximated significance but not to T1. This is the opposite of what was seen in the vaginal cohort. Rectal diversity was similarly decreased in all recurrent samples (data not shown).
Discussion
We present a prospective cohort study in which the dominant taxa of the rectal and vaginal microbiome has been characterized for the first time in patients with rectovaginal fistula; 31 taxa were found to be differentially depleted in subjects with recurrence. Though there is significant species overlap, there remain two distinct microbial environments further characterized by greater vaginal than rectal diversity across the cohort with microbiome clustering more by individual than by site. These findings have not been previously described and are the opposite of what is thought to normally occur in a healthy population [13]. Most surprising was seeing increased vaginal diversity associated with a favorable clinical outcome; this (unlike rectal diversity) is not typical.
The dominant taxa of the vaginal microbiome (recurrent or otherwise) was not characteristic of the general population [14, 15]. Studies indicate that it is typically decreased microbial diversity due to a predominance of lactobacillus that confers protection and overall health to the vaginal environment [14–18]. Measures of alpha and beta diversity are greatest in those microbiomes most closely resembling bacterial vaginosis to include those with diagnostic Nugent scores [16, 17]. Though no large epidemiologic studies of vaginal pathology are currently available for verification, several exploratory studies also support this line of thought to include finding a decreased incidence of bacterial STIs, HSV-2, HIV infection and viral shedding as well as high-risk HPV and cervical cancer in populations with a less diverse microbiome and higher levels of Lactobillus [19–24].
The production of lactic acid and bacteriocins by lactobacilli typically maintain a pH of about 4.2 in this particular environment (16); fecal material is basic, with an average pH of 6.6 in otherwise healthy individuals. We suspect that contamination of the vaginal microbiome with fecal material negates the ability to maintain an optimal vaginal pH and thus allows the overgrowth of other microbial species. Vaginal diversity is likely strongly influenced by the neighboring rectal diversity; the degree of diversity in the two microbiomes mirrored each other in our work; where there was greater rectal diversity, there was also greater vaginal diversity. Reasons for this are not clear; perhaps the less mobile, lower pressure environment of the vagina facilitates the establishment and maintenance of less common, protective microbiota that have passed through the fistula from the rectum. This may also explain why higher vaginal diversity was unexpectedly associated with better outcomes in our population. This may have been a reflection of greater rectal diversity, which is known to be associated with health [13, 25–27]. Bacteria typically associated with vaginal dysbiosis or bacterial vaginosis (e.g., Gardnerella, Prevotella, Mycoplasma, Atopobium) were not among the 29 differentially abundant taxa associated with fistula recurrence in our population.
The measurements taken post-surgery (T2) were done to help understand the microbiome progression through the surgical repair process and potentially detect early signs of infection or subclinical infection. It is interesting that at this study power the predictive differences in the microbiome present at T1 were not seen at T2. It appears that the diversity of the non-recurrent group was more greatly impacted by the administration of preoperative antibiotics and vaginal site preparation than the recurrent group. Intuitively, given the study outcomes, a difference between the two groups likely persists at T2 and may be detectable with a better powered study. Perhaps a relatively healthy vaginal/rectal microbiome recovers or maintains its diversity after a single perioperative antibiotic dose better than the microbiome predisposed to recurrence. Finally, it is plausible that the reduced vaginal diversity of T3 seen in the non-recurrent group relative to T1 is due to a relative increase in the contribution of lactobacilli as this microbiome moves toward a more normal postoperative milieu—decreasing its diversity in a healthy manner.
Increased rectal microbial diversity, unlike in the vaginal microbiome, is typically a marker of health and better outcomes [13, 25–27]. Several taxa were associated with surgical success including significantly higher levels of Firmicutes Faecalibacterium (a species with well-described anti-inflammatory properties [28]), Ruminococcus and Christensenellaceae as well as Bacteroides Alistipes and Rikenellaceae. These organisms have been associated with improved outcomes in diverse conditions ranging from inflammatory bowel disease, HIV, non-Hodgkins lymphoma and melanoma [27–31]. All of these, among many others, were differentially abundant in our specimens (Table S2, S3).
In this group we also found that T1 and T3 had similarly increased diversity, while T2 remained distinct from both. This suggests a longitudinal return in diversity to mimic the T1 milieu by the time T3 was collected 6–8 weeks postoperatively. As expected, similarly decreased diversity persisted through all three samples collected in patients that would recur (T1, T2 and T3R).
The trend toward a reduction in diversity at T3 relative to T1 in the vaginal cohort in combination with the trend toward increased diversity at T3 relative to T1 in the rectal cohort appears to describe a postoperative divergence in the two microbiomes to mimic what would likely be seen in the unaffected healthy population. Higher rectal diversity and lower vaginal diversity together are associated with health.
This pilot study is limited by its sample size and its heterogeneity. While it is interesting to see a significant difference in the microbiome based on menopausal status, for example, it does add a degree of confounding that affects the power of an already small study. This will need to be accounted for to ensure subsequent studies are appropriately powered. Even at this power, we were able to demonstrate significant differences between the two cohorts that seem to predict surgical success. It is important to interpret the negative associations with caution, as these may simply be results of underpowering. We are additionally limited by a relatively homogeneous ethnic distribution of patients consistent with our largely mid-western US referral base; this is not representative of the country’s demographics. Hispanic and black ethnicities typically have a more diverse vaginal microbiome and a higher vaginal pH at baseline than Asian and white women [17]; the findings may be different in these groups.
There is a potentially strong microbial influence on the successful repair of rectovaginal fistulas. Organisms with known antimicrobial and anti-inflammatory properties are present in higher proportions in these patients. These would intuitively counter the pro-inflammatory RVF milieu and improve the post-surgical healing process. The composition of the rectal microbiome with its significantly higher microbial burden and higher pressure environment is likely facilitating the establishment and persistence of an atypically diverse vaginal microbiome. The diversity of both areas may be a key modulator of surgical success. The data obtained from this study will be used to guide a power analysis for future studies with the goal of identifying women at risk of recurrence.
There are several potential clinical applications of these findings. Modification of the vaginal or rectal microbiomes through a variety of approaches ranging from oral or vaginal probiotic, prebiotic or synbiotic administration to fecal microbiota transplantation (FMT) have shown promise in a multitude of disease states [32–35]. Using similar approaches pre- and/or postoperatively could significantly improve the likelihood of surgical success in patients with fistula. It is not inconceivable that decreasing inflammation through targeted alteration of the microbiome could facilitate spontaneous closure in some cases. Since many rectovaginal fistulae are the result of obstetric trauma, perhaps modulation of the microbiome after the repair of third- or fourth-degree lacerations would reduce the risk of primary fistula occurrence. Finally, because many probiotic formulations are relatively inexpensive and stable in the absence of refrigeration, they could be readily utilized for humanitarian work in underserved and disproportionately affected populations if benefit is demonstrated. Extending this study to a larger population will allow for verification of our findings and improve the ability to identify and study specifically targeted therapeutics that will increase the likelihood of successful fistula repair on the first attempt.
Supplementary Material
Acknowledgments
Conflict of interest This work was supported by a grant from the International Urogynecological Association.
This project was supported by CTSA grant number KL2 TR002379 from the National Center for Advancing Translational Science (NCATS) and was also supported, in part, by a career enhancement award from NIH grant P50 CA136393. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
MWA is a member of the scientific advisory board of LUCA Biologics, Inc., on research related to urinary tract infections, preterm birth and reproductive medicine. These activities do not overlap with the research presented here.
Dr. Leach is a member of the US Armed Forces; the views and opinions expressed in this article are those of the authors and do not necessarily reflect the official policy or position of this government agency.
Footnotes
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/s00192-020-04580-2.
References
- 1.Pinto RA, Peterson TV, Shawki S, Davila GW, Wexner SD. Are there predictors of outcome following rectovaginal fistula repair? Dis Colon Rectum. 2010;53(9):1240–7. [DOI] [PubMed] [Google Scholar]
- 2.deSouza A, Abcarian H. The Management of Rectovaginal Fistula. In: Cameron JL, Cameron AM, editors. Current surgical therapy. 11th ed.Philadelphia: Saunders; 2014. p. 283–8. [Google Scholar]
- 3.United Nations Population Fund (2015) Obstetric Fistula 2015, from http://www.unfpa.org/obstetric-fistula. Accessed Nov 2017
- 4.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449(7164):804–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wexner SD, Ruiz DE, Genua J, et al. Gracilis muscle interposition for the treatment of rectourethral, rectovaginal, and pouch-vaginal fistulas; results in 53 patients. Ann Surg. 2008;248(1):39–43. [DOI] [PubMed] [Google Scholar]
- 6.Das B, Snyder M. Rectovaginal fistulae. Clin Colon Rectal Surg. 2016;29:50–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones RS, Stukenborg GJ. Patient-reported outcomes measurement information system (PROMIS) use in surgical care: a scoping study. J Am Coll Surg. 2017;224(3):245–54. [DOI] [PubMed] [Google Scholar]
- 8.Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quast C, Pruesse E, Yilmaz P, Gerken J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41(D1):D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Bittinger K, Charlson ES, et al. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics. 2012;28(16):2106–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L, Reeve J, Zhang L, et al. GMPR: a robust normalization method for zero-inflated count data with application to microbiome sequencing data. PeerJ. 2018;6:e4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486: 207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma B, Forney LL, Ravel J. The vaginal microbiome: rethinking health and diseases. Annu Rev Microbiol. 2012;66:371–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyman RW, Fukushima M, Diamond L, Kumm J, Guidice LC, Davis RW. Microbes of the human vaginal epithelium. Proc Natl Acad Sci U S A. 2005;102(22):7952–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim TK, Thomas SM, Ho M, et al. Heterogeneity of vaginal microbial communities within individuals. J Clin Microbiol. 2009;47(4):1181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ravel J, Pawel G, Abdo Z, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108(S1):4680–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White BA, Creedon DJ, Nelson KE, Wilson BA. The vaginal microbiome in health and disease. Trends Endocrinol Metab. 2011;22(10):389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bayigga L, Kateete DP, Anderson DJ, et al. Diversity of vaginal microbiota in sub-Saharan Africa and its effects on HIV transmission and prevention. AJOG. 2019;220(2):155–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bordgorff H, Tsivtsivadze E, Verhelst R, et al. Lactobacillus-dominated cervicovaginal microbiota associated with reduced HIV/STI prevalence and genital HIV viral load in African women. ISME J. 2014;8:1781–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Ma Y, Li R, et al. Associations of Cervicovaginal lactobacilli with high-risk human papillomavirus infection, cervical intraepithelial neoplasia, and cancer: a systematic review and meta-analysis. JID. 2019;220:1243–54. [DOI] [PubMed] [Google Scholar]
- 22.Dols JA, Smit PW, Kort R, et al. PCR-based identification of eight Lactobacillus species and 18 HR-HPV genotypes in fixed cervical samples of south African women at risk of HIV and BV. Diagn Cytopathol. 40:472–7. [DOI] [PubMed] [Google Scholar]
- 23.Spear GT, Sikaroodi M, Zariffard MR, et al. Copmarison of the diversity of the vaginal microbiota in HIV-infected and HIV-uninfected women with or without bacterial vaginosis. J Infect Dis. 198:1131–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell C, Balkus JE, Fredricks D, et al. Interaction between lactobacilli, bacterial vaginosis-associated bacteria, and HIV type 1 RNA and DNA genital shedding in US and Kenyan women. AIDS Res Hum Retrovir. 29:13–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, et al. Decreased diversity of the fecal microbiome in recurrent Clostridium dificile-associated diarrhea. J Infect Dis. 2008;197(3):435–8. [DOI] [PubMed] [Google Scholar]
- 26.Ott SJ, Musfeldt M, Wenderoth DF, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53(5):685–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong D, Gong X, Wang L, Yu X, Dong Q. Involvement of reduced microbial diversity in inflammatory bowel disease. Gastroenterol Res Pract 2016;2016:6951091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verhoog S, Taneri PE Diaz ZM, et al. Dietary factors and modulation of Bacteria strains of Akkermansia muciniphila and Faecalibacterium prausnitzii; a systematic review. Nutrients. 2019;11(7). 10.3390/nu11071565,10.3390/nu11071565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montassier E, Al-Ghalith AG, Ward T, et al. Pretreatment gut microbiome predicts chemotherapy-related bloodstream infection. Genome Med. 2016;8:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dinh DM, Volpe GE, Duffalo C, et al. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis. 2015;211(1):19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dubin K, Callahan MK, Ren B, et al. Intestinal microbioime analysis identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun. 2016;7:10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chase D, Goulder A, Zenhausern F, et al. The vaginal and gastrointestinal microbiomes in gynecologic cancers: a review of applications in etiology, symptoms and treatment. Gynecol Oncol. 2015;138(1):190–200. [DOI] [PubMed] [Google Scholar]
- 33.Lee P, Br Y, Yacyshyn MB. Gut microbiota and obesity; an opportunity to alter obesity through faecal microbiota transplant (FMT). Diabetes Obes Metab. 2019;21(3):479–49. [DOI] [PubMed] [Google Scholar]
- 34.Russel SL Gold MJ, Hartmann M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13(5):440–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nylund L, Nermes M, Isolauri E, et al. Severity of atopic disease inversely correlates with intestinal microbiota diversity and butyrate-producing bacteria. Allergy. 2015;70(2):241–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


