Abstract
Ribonucleic acid (RNA) molecules can be easily attacked by reactive oxygen species (ROS), which are produced during normal cellular metabolism and under various oxidative stress conditions. Numerous findings report that the amount of cellular 8-oxoG, the most abundant RNA damage biomarker, is a promising target for the sensitive measurement of oxidative stress and aging-associated diseases, including neuropsychiatric disorders. Most importantly, available data suggest that RNA oxidation has important implications for various signaling pathways and gene expression regulation in aging-related diseases, highlighting the necessity of using combinations of RNA oxidation adducts in both experimental studies and clinical trials. In this review, we primarily describe evidence for the effect of oxidative stress on RNA integrity modulation and possible quality control systems. Additionally, we discuss the profiles and clinical implications of RNA oxidation products that have been under intensive investigation in several aging-associated medical disorders.
Keywords: Oxidative stress, RNA damage, RNA control systems, Aging, Disorders
Introduction
The components of the cell, especially ribonucleic acid (RNA), are constantly exposed to various kinds of toxic insults from endogenous and exogenous sources. RNA oxidative modifications resulting from these insults must be effectively handled to maintain genome integrity and initiate translational fidelity (Poulsen et al. 2019). However, compared with deoxyribonucleic acid (DNA) oxidation, RNA oxidation and its biological impacts have only recently become apparent. Although this is an area of ongoing investigation, it is speculated that RNA quality control mechanisms have evolved to clear or correct oxidative RNA damage.
These toxic assaults leading to RNA oxidative damage include reactive oxygen species (ROS) and ultraviolet light. Unlike naturally occurring modifications of specific nucleotides in RNA structures, unwanted oxidative modifications generated from chemical reagents typically have deleterious effects on multiple aspects of RNA metabolism and promote RNA pathology (Yan and Zaher 2019). For example, some adducts of RNA oxidation can alter base-pairing properties completely, while others influence RNA-protein interactions. Among the multiple adducts of nucleoside oxidation, the guanosine oxidation products 8-hydroxyguanosine (8-OHG) and 8-oxo-7,8-dihydroguanosine (8-oxoG) are the most abundant and are the best-characterized biomarkers for RNA oxidative lesions (Feyzi et al. 2007). To effectively ensure RNA integrity, cells must have diverse defensive mechanisms to discriminate oxidatively damaged RNA from normal transcripts (Yan et al. 2019a). In fact, there is considerable evidence supporting the involvement of RNA damage in the pathological cascade of human diseases, especially in neuropsychiatric disorders. It has been revealed that RNA oxidation may be a prominent feature in the early stages of neurodegenerative diseases, as illustrated in one recent study of familial Alzheimer’s disease (AD) (Nunomura et al. 2004). Using a mouse model of amyotrophic lateral sclerosis (ALS), Chang et al. observed that mRNA oxidation has been proved as a common event preceding neuron degeneration, which leads to obvious neurodegeneration (Chang et al. 2008). Moreover, the oxidized nucleoside 8-oxoG is routinely used in the clinic as a measure of kidney function in patients. Brown et al. studied brain sections of bipolar disorder (BD) patients and demonstrated increased levels of 8-OHG in all postmortem brain samples (Brown et al. 2014). Employing ultra-performance liquid chromatography and mass spectrometry assays (UPLC-MS/MS), Munkholm et al. (Munkholm et al. 2015) evaluated the 8-oxoG levels in urine from BD patients and found that RNA damage is significantly increased in BD cases, together with an upregulated urinary excretion of 8-oxoG. In addition to these neuropsychiatric disorders, RNA oxidative damage has also been noted in other pathological states, including cancers (Fimognari 2015; Gao et al. 2019). Indeed, these data support the roles of RNA oxidation as potential molecular mechanisms contributing to increased risk of human medical disorders.
It is not surprising that cells have different control mechanisms against chemically altered RNA. Damaged RNA was long thought to be degraded and not repaired. However, although the detailed degradation mechanisms have not been fully illuminated, it appears that these processes are dependent on the ribosome (Hosseini et al. 2018; Ishii and Sekiguchi 2019). Subsequent reports have established that some specific repair mechanisms have evolved to cope with certain oxidative-induced RNA injuries (Nunomura et al. 2017). The findings from Aas’s group (Aas et al. 2003) emphasized the importance of RNA-repair pathways for maintaining cellular homeostasis, suggesting that cells may have a greater instinctive behavior in RNA protection than previously suspected.
In this review, we mainly provide an update on the findings regarding the types of oxidized RNAs and their corresponding quality control mechanisms. We also discuss the well-investigated roles of RNA damage in the pathogenesis and development of several aging-associated human disorders (Fig. 1). Additionally, technologies for the evaluation of damaged RNA in tissues or body fluids for assessing potential clinical implications are also be discussed.
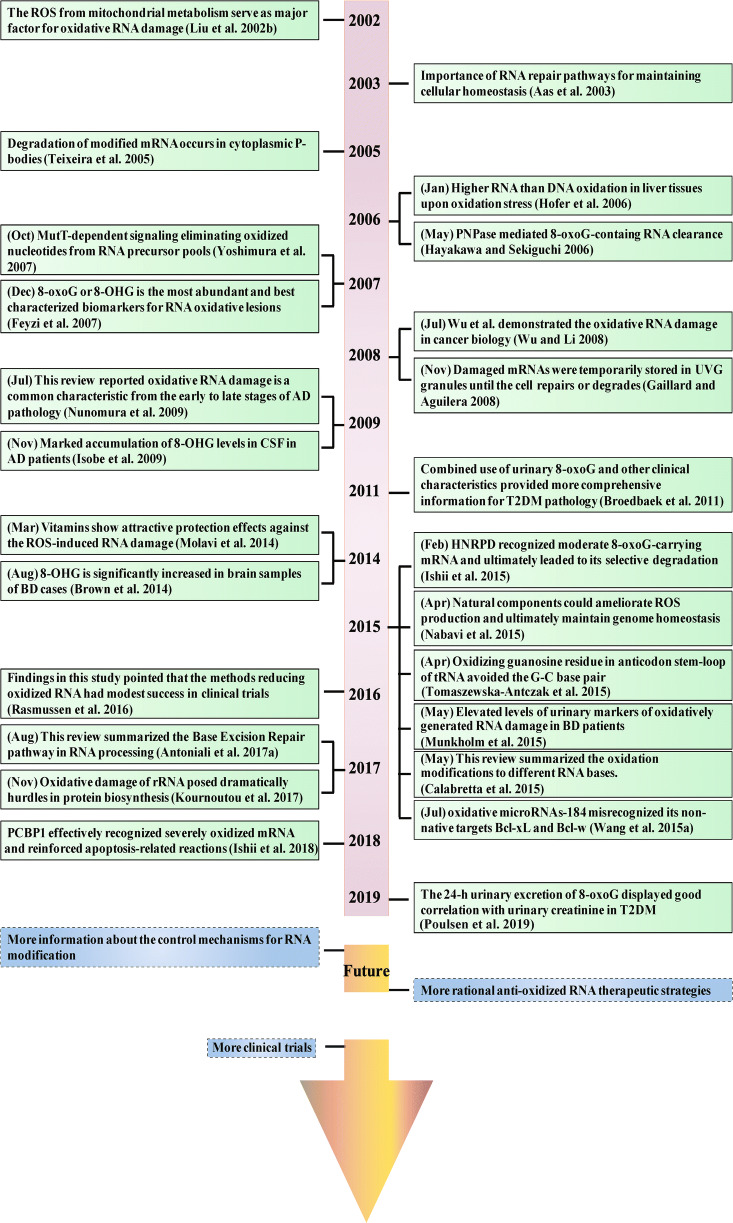
Fig. 1.
Timeline of key events in understanding RNA oxidation modifications. Landmark discoveries and advances in the understanding of RNA oxidation damage, together with its application in human pathophysiological processes
Oxidized RNA damage
Oxidative damage to DNA nucleobases by such sources as ROS and ultraviolet light is recognized as a major threat to genomic stability and plasticity. The reaction of oxygen-free radicals with free nucleobases or oligonucleotides may lead to the generation of numerous distinct chemical modifications in DNA (Cadet and Wagner 2013; Csiszar et al. 2019; Liu et al. 2018). However, base damage by oxidative stress is not restricted to DNA but also occurs in RNA. It is now becoming evident that the base lesions in RNA are very similar to those in DNA. During oxidative stress conditions, cellular metabolic reactions promote ROS formation, resulting in oxidative damage to multiple cellular components (Yang et al. 2018), including RNA. A number of steps in the metabolic reactions involve semiquinone anion radicals that can react with molecular oxygen to generate superoxide anion radicals. In general, through the action of superoxide dismutase (SOD), these superoxide radicals are catalyzed into hydrogen peroxide and molecular oxygen, providing cellular defense against ROS. Subsequently, hydrogen peroxide can be further reduced to water by some specific reactions (Memar et al. 2018). However, in various aging-associated pathologic processes, hydrogen peroxide oxidizes intracellular Fe2+ by the Fenton and Haber-Weiss reactions to produce hydroxyl radicals and Fe3+, thereby causing injurious ROS attack and formation (Liguori et al. 2018). Since Fe2+ has the ability to bind to multiple biomolecules, hydroxyl radicals can be formed in the nucleosides, nucleotides, or nucleobases in the immediate vicinity, thus leading to damaging effects (Wurtmann and Wolin 2009). For example, the oxidized nucleoside 8-oxoG is generated from the reaction of guanine with hydroxyl radicals followed by oxidation (Cejvanovic et al. 2018a). Furthermore, studies of aging skeletal muscle demonstrating abnormal iron homeostasis are consistent with the functions of intracellular metal irons in generating ROS through Fenton and Haber-Weiss chemistry, and in turn leading to RNA oxidative damage (Hofer et al. 2008).
The reaction of ROS with free nucleosides, nucleotides, or nucleobases generates multiple modifications in RNA molecules. Certainly, in past studies, directly oxidized RNA products, such as 8-oxoG, 8-oxo-7,8-dihydroadenosine (8-oxoA), 5-hydroxycytidine (5-HOC), and 5-hydroxyuridine (5-HOU), have been identified in RNA by different detection methods (Calabretta and Kupfer 2015; Simms and Zaher 2016) (Fig. 2). Due to the particular reactivity of guanosine, 8-oxoG has been demonstrated to be the main oxidation product both in disease tissues and in experimental models treated with oxidizing agents (Wurtmann and Wolin 2009). More importantly, upon oxidative stress, oxidized guanosine is able to pair with cytosine (8-oxoG-C) but is more likely to form a mismatched base pair with adenosine (8-oxoG-A) (Simms and Zaher 2016) (Fig. 3).
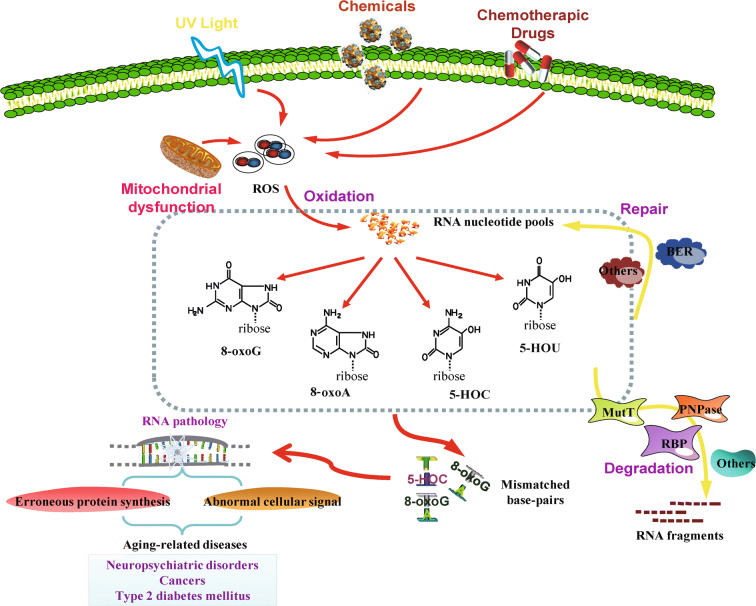
Fig. 2.
Oxidized RNA nucleobases and their potential biological consequences. Under different oxidative stress conditions, oxidized RNA nucleobases result in the formation of mismatched base pairs, which ultimately impair RNA metabolism and promote RNA pathology. Additionally, cells are able to use distinct defense systems to maintain RNA homeostasis, including degradation pathways or repair mechanisms
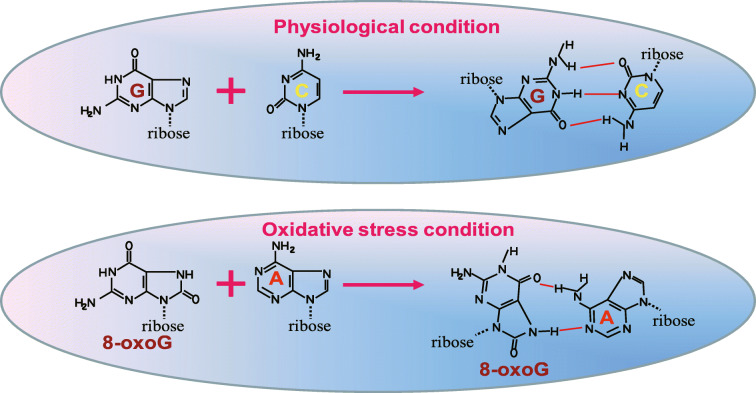
Fig. 3.
Oxidized RNA nucleobases lead to altered base-pairing properties. Due to the particular reactivity of guanosine, 8-oxoG has been demonstrated to be a common oxidation product, both in disease tissue and in experimental models treated with oxidizing agents. It is well known that upon oxidative stress, oxidized guanosine can pair with both cytosine and adenosine during polymerase chain reaction, thus giving rise to 8-oxoG-A mismatches
In fact, compared with DNA, cellular RNA is more frequently vulnerable to oxidative insults in vitro and in vivo. Moreover, it has even been shown that RNA has higher degrees of damage than DNA in multiple studies of oxidative damage. In one recent study, oxidation of RNA and DNA was measured simultaneously using a high-performance liquid chromatography (HPLC)–based assay (Hofer et al. 2006). This study found that in the hepatic tissue of rats, administration of the oxidant generator doxorubicin leads to dramatically increased RNA oxidation but no obvious increase in DNA oxidation. The explanation for the higher degrees of RNA oxidation damage might be that, unlike DNA, RNA is largely a single-stranded structure without protection from specific proteins. Moreover, intracellular RNA molecules are abundantly located close to mitochondria, a major source of free radicals (Kupfer and Leumann 2011; Wang et al. 2019). More importantly, because ROS is mainly generated from the mitochondrial metabolic response, abnormal mitochondrial pathways might supply unavoidable factors for oxidative RNA damage. Indeed, consistent with this hypothesis, feeding old rats with metabolites that improve mitochondrial function has been shown to lead to more predominantly oxidized RNA in the hippocampus comparable to the effect in younger rats (Liu et al. 2002b), suggesting a link between RNA oxidation and mitochondrial dysfunction. Moreover, pretreatment with metabolites inhibits the age-associated increase of oxidative damage to RNA, resulting in delayed stress-induced acceleration of the senescence-like phenotype in human diploid fibroblast cells and neuronal cells (Liu et al. 2002a).
The “oxidative stress theory of aging” has been proposed for a long time, and oxidative stress is closely related to cellular senescence and aging. Several studies demonstrate that various factors and events in oxidative stress, such as RNA oxidative damage, have been implicated in the initiation, regulation, and progression of the aging-associated biological behaviors (Kim et al. 2017). It was shown that oxidative stress induced by ROS mediates cellular senescence phenotypes, including enhanced senescence–associated β-galactosidase (SA-β-gal) activity, a large flat morphology and permanent cell growth arrest (Benameur et al. 2015). Recently, Kuhnel et al. (Kuhnel et al. 2015) found that treatment with the ROS inducer, abnormal savda munziq (ASMq), dramatically induces RNA damage in rat fibroblasts and upregulates the expression of p21, p53, and p16—key players in cellular senescence. Moreover, ASMq has a strong ability to cause cell-cycle arrest and SA-β-gal staining. Inhibition of ROS generation prevents the age-dependent accumulation of 8-oxoG in both nuclear and mitochondrial RNA molecules, along with downregulated SA-β-gal activity (De Luca et al. 2013). Taken together, all these data suggest that under conditions of oxidative stress, the accumulation of RNA oxidative damage plays important role in aging processes.
Defective protein synthesis and cell signaling
It is currently widely accepted that impairment of transcriptional or translational integrity is the main consequence of RNA oxidation damage due to the altered profile of oxidized mRNA or impaired function of some noncoding RNA (ncRNA), such as transfer RNA (tRNA) and ribosomal RNA (rRNA). The accumulation of guanosine damage adducts such as 8-oxoG in bacterial and eukaryotic extracts leads to dramatic effects on mRNA decoding ability, resulting in protein misfolding and short polypeptide formation (Dai et al. 2018; Hudson and Zaher 2015). The oxidant manganese porphyrin/oxone is able to selectively oxidize the guanosine residue in the anticodon stem-loop of tRNA, preventing the formation of a G-C base pair during translation (Tomaszewska-Antczak et al. 2015). Moreover, trace metal-mediated oxidative injuries of 5S and 18S rRNA in the mussel Mytilus galloprovincialis disturbed the structural integrity of large and small ribosomal subunits, respectively, posing dramatic hurdles in protein biosynthesis (Kournoutou et al. 2017). Similar findings were further certified in bacterial 23S rRNA (Willi et al. 2018) and yeast 25S/5.8S rRNAs (Mroczek and Kufel 2008), which resulted in prominently impaired protein homeostasis. Using RNA phosphoramidite chemistry methods, Küpfer et al. (Kupfer and Leumann 2011) incorporated the RNA lesion product 5-HOC into a caged oligoribonucleotide building block. Additionally, further melting curves showed high levels of C-A mismatching when this building block was paired against the complementary sequence under lower temperature conditions. In brief, these base-oxidized lesions might substantially interfere with the homeostasis of protein synthesis.
Additionally, recent progress in genetics has revealed an expanding landscape of RNA beyond its traditional function as indispensable intermediates for the transfer of genetic information from DNA to proteins (Haberle and Stark 2018). Of particular note, some abundant cellular ncRNAs, such as microRNAs, have gained growing recognition for their emerging roles in controlling signaling dynamics (Li and Fu 2019; Ou et al. 2019; Yan et al. 2019b; Yan et al. 2019c). Millan et al. (Millan 2017) have reviewed rapidly accumulating evidence for the roles of oxidative stress on microRNA regulation involved in the pathophysiology of neuropsychiatric disorders. However, several remaining issues need to be addressed: (1) whether microRNAs are oxidatively modified by oxidation stress; and (2) whether oxidative modification actually influences the function of microRNAs in pathophysiological behaviors. To elucidate these questions, a meaningful discovery from Wang’s group (Wang et al. 2015a) has shown that ROS remarkably cause oxidative modification of microRNA and thereby alternate the microRNA-mediated cellular functional signals. These authors found that upon ROS stress oxidative microRNAs-184 misrecognized its non-native targets, Bcl-xL and Bcl-w, thereby initiating abnormal apoptosis in cardiomyocytes.
RNA quality control mechanisms
Given their particular roles in genetic information transfer and cell signaling modulation, a more thorough investigation of the biological significance and control mechanisms of RNA oxidative damage would be highly welcome. Additionally, as the current survey explains, cells are able to use distinct defense systems to maintain RNA integrity (Aas et al. 2003; Burroughs and Aravind 2016), including degradation pathways or directed repair mechanisms for certain forms of damage (Fig. 1).
RNA degradation systems
MutT-type Nudix hydrolase
RNA degradation has been shown to serve as an integral part of cellular RNA homeostasis (Schmid and Jensen 2018). Sanitization of the nucleotide pools is an important defense against the mutagenic consequences of oxidized RNA precursors (Freudenthal et al. 2015). It is well established that the MutT-type Nudix hydrolase (nucleoside diphosphate-linked moiety X motif) superfamily plays major roles in the sanitization of the nucleotide pools in various organisms. In Escherichia coli, the MutT protein specifically hydrolyzes and eliminates 8-oxoG-containing nucleoside triphosphates such as 8-oxo-guanosine-5′-triphosphate (8-oxo-GTP). MutT overexpression has been shown to obviously diminish the misincorporation of 8-oxo-GTP into mRNA by acting on the oxidized ribonucleotide 8-oxoG with high affinity (Sekiguchi et al. 2013). This notion was further demonstrated by increasing 8-oxo-GTP-containing RNA in MutT-deficient cells upon oxidative stress (Taddei et al. 1997). Moreover, disruption of nucleotide pool homeostasis via MutT inhibitors induced a remarkable increase in cellular nucleotide damage (Huber et al. 2014). These findings are of importance for RNA integrity and might be relevant for eukaryotes, as MutT homologs have been verified in several eukaryotic species. MutT homologous proteins, isolated from Arabidopsis (Yoshimura et al. 2007), Zebrafish (Jemth et al. 2018), or Saccharomyces cerevisiae (Nunoshiba et al. 2004), function to eliminate transcriptional errors of genetic information through the sanitization of modified nucleotide pools. MutT homologous enzymes (MTH) in human cells include three types: MTH1 (NUDT1), MTH2 (NUDT15), and MTH3 (NUDT18); and they possess the ability to eliminate oxidized nucleotides from RNA precursor pools (Ishii and Sekiguchi 2019; Takagi et al. 2012). All of these proteins participate in the error-avoiding mechanism mentioned above, and their overproduction significantly suppresses the mutator phenotype in MutT-deficient cells (Lin et al. 2018). Meanwhile, hNUDT1 overexpression notably abolished the senescence phenotype in cultured mouse embryonic fibroblasts (MEFs) and provided a proliferative advantage (De Luca et al. 2013). Thus, different organisms are able to make use of MutT-dependent signaling to protect themselves against RNA oxidative injuries.
PNPase
Another conserved RNA-processing enzyme, polynucleotide phosphorylase (PNPase), has recently aroused widespread research interest due to its central roles in RNA processing and degradation control (Cameron et al. 2019). A previous report showed that the PNPase from Escherichia coli preferentially recognizes and binds a synthetic RNA sequence carrying 8-oxoG (Hayakawa et al. 2001). Surprisingly, the following findings demonstrated that PNPase binds not only synthetic RNA molecules but also the natural RNA sequence that is oxidatively modified upon hydrogen peroxide exposure (Wu et al. 2009). Alternatively, a phylogenetic analysis revealed that PNPase has a high level of evolutionary conservation from bacterial species to other species (Golzarroshan et al. 2018), suggesting its intrinsic and comparable function in biological processes. Similar to its bacterial homolog, human PNPase (hPNPase) has also been identified to specifically bind 8-oxoG RNA with a higher affinity than normal RNA (Hayakawa and Sekiguchi 2006). SiRNA-mediated knockdown of hPNPase significantly increase the 8-oxoG level and decrease cell viability after exposure to hydrogen peroxide in the human cervical cancer HeLa cell lines (Wu and Li 2008), providing direct evidence for cell death induced by RNA damage. Additionally, PNPase is an exoribonuclease from a multienzyme RNA degradosome complex. Studies have found that PNPase mediation of 8-oxoG-containing RNA clearance is dependent on its exoribonuclease activity (Stone et al. 2017) but not its association with other members of the RNA degradosome (Wu et al. 2009). Some factors, such as the Krebs cycle metabolite citrate (Stone et al. 2017), can interact with PNPase and inhibit its exoribonuclease activity, which ultimately interferes with cellular RNA metabolism. Therefore, in the future, it will be interesting to elaborate on whether the critical roles of PNPase in releasing 8-oxoG from RNA are through its own action or are facilitated by other protein co-factors. Moreover, it remains to be elucidated whether the abilities of PNPase to recognize and bind damaged RNA are solely due to its interaction with 8-oxoG specifically or are also involved in other damaged RNA residues.
RNA-binding protein
To date, emerging observations have begun to indicate that cells possess other mechanisms that show a discriminatory activity for destroying 8-oxoG-enriched nucleotides. Human heterogeneous nuclear ribonucleoprotein D (HNRPD), a RNA-binding protein (RBP), is able to specifically bind to 8-oxoG-carrying mRNA, ultimately leading to selective degradation of this oxidized mRNA under mild oxidative stress (Ishii and Sekiguchi 2019). Following hydrogen peroxide exposure, due to the high level of cellular oxidized mRNA, HNRPD-deficient human cancer cells exhibit obvious growth retardation (Ishii et al. 2015). For more severely oxidized mRNA, cells are able to use poly(C)-binding protein PCBP1 to effectively recognize the severely oxidized mRNA and further reinforce apoptosis-related reactions to eliminate unneeded cells, although the KH1 domain of the PCBP1 deletion mutant has totally lost its ability to bind oxidized mRNA and is unable to trigger cell apoptosis-associated reactions (Ishii et al. 2018). However, the detailed mechanisms for RBPs distinguishing oxidized nucleotides remain unknown and need to be clarified in the future.
Other pathways
It is also known that oxidative RNA fragments trigger the formation of cytosolic biomolecular condensates, such as stress granules and processing bodies (P-bodies) (Youn et al. 2019), which benefit cellular fitness by preventing the accumulation of deleterious intracellular products. Indeed, both stress granules and P-bodies are dynamic complexes whose assembly is mainly dependent on the pool of nontranslating mRNA. In yeast, mRNA degradation mainly occurs in defined cytoplasmic P-bodies (Sheth and Parker 2003), controlling translation in early development. The level of oxidized RNAs is markedly increased in yeast cells with the decapping Kllsm4Δ1 mutant, a truncated form of the KlLSM4 subunit from the Lsm1-7 complex in P-bodies (Stirpe et al. 2017). Moreover, P-bodies are present in unstressed cells but are further increased in response to various stresses that lead to mRNA degradation and translation repression. P-bodies have been shown to increase in number and size when mRNA turnover is inhibited under different stress conditions (Teixeira et al. 2005), including oxidative stress (Mazzoni et al. 2007). An important area for future work is to determine how mRNA processing in stress granules or P-bodies is remodeled to affect the fate of mRNA and how this biologic progress affects the translation mechanisms’ response to stress.
RNA-repair systems
RNA rendered dysfunctional by oxidative damage is targeted for quality control in biological evolution by several repair systems (Nandakumar et al. 2008; Yan et al. 2019a), which have only recently become apparent. Indeed, a considerable amount of evidence suggests that cells have developed unique response mechanisms against the detrimental consequences of oxidizing nucleotides.
BER systems
To date, many reports have pointed out a tight linkage between the DNA damage response and RNA quality control (Scott et al. 2017). Some DNA repair factors, especially a large cohort of base-excision repair (BER) members, have been implicated in RNA integrity (Bisht et al. 2017), suggesting that the DNA damage repair system and RNA modification are closely inter-related. Elucidation of these detailed RNA-repair mechanisms would enable interpretation of how cells cope with oxidative-induced RNA damage to effectively prevent cellular dysfunction.
The BER pathway has promising roles in eliminating replicational and translational errors induced by the products of oxidative damage to nucleotides. Several core BER enzymes form an excision repair apparatus capable of repairing damaged bases and abasic sites (Jang et al. 2019). Emerging studies have revealed that the BER enzymes remove lesions from oxidative substrates, promoting the functional resumption of damaged RNA (Antoniali et al. 2017a). This supports a promising role for BER proteins in the RNA damage response. Moreover, oxidative guanine in RNA can be selectively recognized and repaired by specific BER glycosylases, including 8-oxoguanine glycosylase (OGG1) and apurinic/apyrimidinic endodeoxyribonuclease 1 (APE1) (Antoniali et al. 2017a). Preliminary analysis from Manini’s group found that reactive epoxide exposure enhanced the OGG1 level and enzymatic activity, which are closely related to altered concentration of urine 8-oxoG in styrene workers (Manini et al. 2009). Lovell et al. (Lovell and Soman 2011) quantified 8-OHG and OGG1 in neurons in the preclinical stage of AD subjects using immunohistochemistry and demonstrated that OGG1-mediated BER capacities are indispensable for the repair of oxidized guanine 8-OHG. Additionally, APE1 has a pivotal role in the cellular response to oxidative stress, and its mutations are known to be part of the pathological progress (Antoniali et al. 2017b). Upon oxidative stress, siRNA-mediated APE silencing decreases its endonuclease activity, significantly leading to increased 8-OHG-containing rRNA and weakened cell growth rates (Vascotto et al. 2009). Meanwhile, an interatomic study highlighted that APE1 regulates gene expression through its direct control of microRNA processing and stability, participating in tumor development and chemotherapeutic resistance (Antoniali et al. 2017b). Although these data mentioned above have preliminarily outlined a novel and potential role of the BER pathway in RNA metabolism, there are still many open questions regarding the exact composition of the RNA-repair complex. Future investigations on the detailed mechanisms of the BER-mediated RNA-repair system under different oxidative stress conditions will extend our knowledge of RNA homeostasis in both physiological and pathological actions.
Novel identified pathways
Interestingly, other RNA quality control systems have been recently identified in vivo and in vitro. Gaillard et al. (Gaillard and Aguilera 2008) identified a novel class of polyadenylated (poly-A+) mRNA-containing granules in the yeast S. cerevisiae, which were designated as UV-induced mRNA granules (UVGs). UV irradiation leads to a dose-dependent accumulation of potentially damaged poly-A+ mRNA, significantly weakening mRNA stabilization. To safeguard cell viability under UV irradiation conditions, the damaged mRNAs are temporarily stored in UVG granules until the cell repairs or degrades these damaged RNAs at a later time. Fluorescent in situ hybridization identified a small fraction of poly-A+ UVGs colocalized with the P-body marker Dhh1 (Gaillard and Aguilera 2008), revealing that UVGs and P-bodies might interact functionally to some extent. Though these results are meaningful for cellular homeostasis, further work will be required to identify the regulatory signals associated with these granules and whether they represent an extensive mechanism across different species, including humans.
RNA oxidation in pathological disorders
While oxidative damage to RNA is less lethal for cells than mutations in the genome, such moderate insults to cells have been implicated in several aging-associated disease states, especially neuropsychiatric diseases, cancers, and type 2 diabetes mellitus (T2DM) (Fig. 1).
Neuropsychiatric disorders
The mechanisms underlying the progression of nervous system disorders are complex and varied. Recently, both biochemical and immunocytochemical studies of RNA oxidative modifications have provided new insights into these disorders (Nunomura et al. 2009; Nunomura et al. 2017). Indeed, relative to healthy individuals, a significantly increased level of the oxidative RNA damage maker 8-oxoG has been reported in several neuropsychiatric disorders, such as Parkinson’s disease (PD), AD, and schizophrenia. It was shown that these diseases are at least partially due to the occurrence of lesions in RNA (Fimognari 2015; Nunomura et al. 2012a). Of particular interest, it is now becoming evident that oxidative RNA damage is a common characteristic in vulnerable neurons from early (Violet et al. 2015) to late stages (Lovell and Soman 2011) of AD pathology. From the clinical perspective, the involvement of RNA oxidative damage in the etiology and pathogenesis of these disorders would be of great importance for developing valuable diagnostic and therapeutic targets.
Recent in situ and immunohistochemical staining studies showed an obvious increase in cytoplasmic oxidized RNA nucleoside 8-OHG within the hippocampus and temporal neocortex in the earliest stage of AD in patients. Moreover, 8-OHG immunoreactivity was greatly diminished by RNase pretreatment but not by DNase pretreatment (Nunomura et al. 2009; Nunomura et al. 2012b). Other reports also evaluated the concentrations of 8-OHG in cerebrospinal fluid (CSF) and the serum of patients with AD using high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) and found marked accumulation of 8-OHG levels in CSF but not in the serum. Surprisingly, the effect of 8-OHG on the progression of AD pathology showed a negative correlation between 8-OHG concentrations in CSF and the duration of AD illness (Abe et al. 2002; Isobe et al. 2009). Studies have also suggested a higher accumulation of oxidatively generated RNA damage in urine specimens from patients with schizophrenia (Jorgensen et al. 2013a) or depression (Jorgensen et al. 2013b) compared with healthy subjects. These findings collectively support the roles of oxidized RNA within the onset and progression of neuropsychiatric diseases.
However, the detailed scavenging mechanisms of 8-OHG in neuropsychiatric disorders have not been clearly investigated. One common feature might be the inactivation of antioxidant enzymes. In fact, increased oxidative RNA damage in such disorders is often accompanied by dysfunction of cellular antioxidative defense mechanisms in the same subjects. It has previously been shown that autopsied AD brains exhibit reduced enzymatic activity of SOD (Singh et al. 2017), an important antioxidant enzyme for maintaining mitochondrial homeostasis. The increase in oxidized RNA biomarkers during neurodegeneration is significantly impaired by treatment with a SOD mimetic (Sen et al. 2018), further indicating that strong oxidative stress contributes to neuronal dysfunctions. Additionally, other signaling pathways have also been shown to protect nucleic acid from oxidative damage. Lovell et al. demonstrated that neurons in the AD brain exhibit diminished OGG1-mediated BER capacities (Lovell and Soman 2011), which are indispensable for the repair of oxidatively modified guanine. Using mouse models of AD, Violet et al. (Violet et al. 2014) found that Tau, a cellular microtubule-stabilizing nuclear protein, also contributes to the continuous protection of neuronal RNA integrity under oxidative stress. Indeed, investigations aimed at understanding the molecular mechanisms related to RNA oxidation regulation and its consequences would provide crucial insights into the pathogenesis of neurodegenerative disorders and might thereby lead to better therapeutic strategies in the future.
Cancers
Several studies have recently demonstrated that RNA damage rendered by various agents is an important mechanism of their anti-cancer activities (Bellacosa and Moss 2003). The natural compound sulforaphane, a well-characterized isothiocyanate derived from broccoli, induced a significantly high level of RNA damage fragmentation and inhibited cell viability in several human leukemic cells. Meanwhile, coadministration of sulforaphane enhanced the RNA-damaging properties of conventional chemotherapy drugs, such as doxorubicin (Fimognari et al. 2012). A recent study in HeLa cells exposed to hydrogen peroxide revealed that elevated hydrogen peroxide concentration reduces cell viability and increases the 8-oxoG content, indicating RNA oxidative damage induced by oxidative stress (Wu and Li 2008). Although damaged RNA is obviously deleterious for cell survival, whether cell death directly caused by RNA damage remains to be examined by future exploration. To address this problem, Newton et al. (Newton et al. 2001) showed that onconase, a cytotoxic ribonuclease, targets tRNA specifically, and in so doing, induces programmed cell death (apoptosis) through a mitochondria-dependent pathway. Additionally, a xenograft animal model demonstrated that onconase-based therapeutics have few immunotoxic or other side effects. Thus, the RNA damage response is a promising avenue for further investigation to understand the precise mechanisms responsible for its curative effect in cancer patients.
Type 2 diabetes mellitus
The current understanding of the onset and progression of T2DM is complex and varied. It has been reported that RNA oxidative stress provides a novel target (Cejvanovic et al. 2018b; Schottker et al. 2020), although the detailed mechanisms of oxidative damage to RNA and its possible regulatory pathways are still under investigation. To address the effect of the RNA damage marker 8-oxoG in diabetes-associated mortality, Broedbaek et al. (Broedbaek et al. 2011) carried out a study with a population-based cohort of 1381 newly diagnosed T2DM patients and examined their urinary 8-oxoG levels by a UPLC-MS/MS method. The authors found that the 8-oxoG levels in freshly voided morning urine samples predict the hazard radios of long-term all-cause mortality for newly diagnosed T2DM patients. The data also showed that the combined use of urinary 8-oxoG and other known clinical characteristics provides more comprehensive information about disease risk and might be more useful for determining which patients will have a better clinical response from intensified treatment. For diabetic complications, which occur with poor glycemic control (Utumatwishima et al. 2018), another two separate population-based analyses were carried out to characterize the association between RNA oxidative damage and diabetic complications. The findings of these studies unanimously demonstrated significantly higher levels of urinary 8-oxoG in T2DM patients with different complications, especially microangiopathy (Liu et al. 2016) and psychiatric illness (Jorgensen et al. 2018a), compared with the subjects without complications. All of these studies together indicate the clinical application of 8-oxoG as a potential biomarker in patients with T2DM with or without complications. Further elucidation of the exact mechanisms of urinary 8-oxoG and its associated potential functions in diabetes might provide novel clues for diabetic etiology, although its value as a potential biomarker needs to be validated over a longer period.
Compounds that protect against RNA oxidative damage
Oxidative stress provokes severe damage to all kinds of cellular components, including nucleic acids, resulting in abnormal cellular functions and human pathologies. In recent years, examinations of the relationships of exposure to various stimulus conditions with the RNA damage response and human diseases have attracted increasing attention. Highlighted evidence has demonstrated that some noxious chemicals, such as bisphenol A (Yan et al. 2019a), cause ROS upregulation, resulting in significantly increased 8-oxoG in urine. Vitamins, as potential antioxidant reagents in vivo and in vitro, show attractive protection effects against ROS-induced RNA damage. Molavi et al. (Molavi et al. 2014) found that vitamin E administration blocks endogenous peroxidase activity and promotes RNA-repair pathways in ovarian cells, attenuating the cypermethrin-induced impaired structure and function of the ovaries. Similarly, phenylhydrazine exposure induces ROS generation in testicular tissue, leading to poor sperm quality and delayed embryonic development in mammal models. Pretreatment with vitamin C prevents phenylhydrazine-induced biochemical damage and further protects sperm quality (Anbara et al. 2018). Apart from vitamins, natural polymeric compounds have also received increased attention because of their pronounced antioxidant activity. Ginkgolide B, a functional natural component extracted from Ginkgo biloba, presents neuroprotective potential through affecting multiple oxidative stress-associated molecular targets and signaling pathways in the human AD pathological process (Nabavi et al. 2015). Ginkgolide B treatment significantly reactivates antioxidant enzymes (SOD and glutathione reductase, among others) and ameliorates ROS production, ultimately leading to genome homeostasis (Gill et al. 2017). Moreover, data from Fragopoulou’s group (Fragopoulou et al. 2018) provide further evidence that supplementation with a combination of plant substances and vitamins obviously counters the oxidative RNA damage in urine samples, accompanied by increased SOD enzyme activity in serum samples. In brief, lowering intracellular ROS levels by antioxidant compounds, such as the natural products, restores nucleotide pool homeostasis, including reducing the amount of oxidized RNAs and decreasing cellular translational errors (Palermo et al. 2010; Stirpe et al. 2017). All of these reports point toward modulation of antioxidant defense mechanisms by natural extracts and vitamins, e.g., activating antioxidant enzymes (SOD, glutathione reductase) or inhibiting the pro-oxidant enzymes (peroxidase), to significantly lessen oxidative RNA damage.
Biochemical approaches
Modern biomedical research has shed light on the general view that environmental stress has adversely influence human health and accelerates aging-associated diseases. For example, there is recent evidence suggesting that under oxidative stress, elevated levels of the RNA oxidation marker 8-oxoG dramatically increase the risk of T2DM complications, especially diabetic macrovascular complications (Liu et al. 2016). Thus, to elucidate the underlying biological implications of RNA damage, specific and sensitive detection methods need to be developed. Additionally, these analytical techniques for evaluating RNA damage levels might yield information on disease predisposition and development (Table 1).
Table 1.
The methods for RNA damage evaluation in aging-associated disease processes
| Methods | Diseases | Models | Refs. | |
|---|---|---|---|---|
| PCR reaction | ||||
| qRT-PCR | Cervical cancer, Leukemia | Aging-associated cancers | HeLa, Nalm-6 cells | Ishii et al. 2015 |
| Chromatographic methods | ||||
| HPLC | Cervical cancer | Aging-associated cancers | HeLa cells | Wu and Li 2008 |
| HPLC-MS/MS | Alzheimer’s disease | Aging-associated neuropsychiatric disorders | Cerebrospinal fluid | Abe et al. 2002 |
| UPLC-MS/MS | Schizophrenia | Aging-associated neuropsychiatric disorders | Urine | Jorgensen et al. 2013a |
| UPLC-MS/MS | Depression | Aging-associated neuropsychiatric disorders | Urine | Jorgensen et al. 2013b |
| UPLC-MS/MS | Type 2 diabetes mellitus | Aging-associated diabetes mellitus | Urine | Broedbaek et al. 2011 |
| UPLC-MS/MS | Psychiatric illness | Aging-associated neuropsychiatric disorders | Urine | Jorgensen et al. 2018a |
| HPLC-MS/MS | Parkinson’s disease | Aging-associated neuropsychiatric disorders | Cerebrospinal fluid | Abe et al. 2003 |
| UPLC-MS/MS | Bipolar disorder | Aging-associated neuropsychiatric disorders | Urine | Jacoby et al. 2016 |
| Immunological techniques | ||||
| Immunohistochemistry | Alzheimer’s disease | Aging-associated neuropsychiatric disorders | Brain samples | Lovell et al. 2011 |
| Immunofluorescence | Alzheimer’s disease | Aging-associated neuropsychiatric disorders | THY-Tau22 mice | Violet et al. 2015 |
| Immunocytochemistry | Alzheimer’s disease | Aging-associated neuropsychiatric disorders | Brain samples | Nunomura et al. 2012b |
| Immunohistochemistry | Alzheimer’s disease | Aging-associated neuropsychiatric disorders | Brain samples | Sen et al. 2018 |
| Immunohistochemistry | Atherosclerosis | Aging-associated cardiovascular diseases | Atherosclerotic tissue | Martinet et al. 2005 |
| Immunofluorescence | Alzheimer’s disease | Aging-associated neuropsychiatric disorders | Brain samples | Sen and Hongpaisan 2018 |
PCR
In a recent report, Gong et al. developed a method using the quantitative real-time polymerase chain reaction (qRT-PCR) to determine the level of damaged rRNA induced by oxidative stress in the Escherichia coli genome (Gong et al. 2006). Based on the sequence-specific nature of RT-PCR technology, these authors defined significant damage levels of a specific RNA caused by oxidation or other modifications, such as those to 16S rRNA. However, because of a high error rate, qRT-PCR fails to identify all RNA modification products, such as pyrimidine photohydrates produced by photochemical reactions (Qiao and Wigginton 2016). A novel molecular beacon (MB) probe has been developed to detect and quantify many RNA damage sites as a result of various environmental stresses. Based on sequence-specific hybridization features, the MB approach forms unique damaged nucleic acid-MB duplexes by PCR to observe the cellular RNA damage levels in vivo or in vitro with enhanced sensitivity (Yarasi et al. 2005).
Chromatographic methods
To date, chromatographic analysis approaches, such as HPLC-MS/MS (Abe et al. 2003), UPLC-MS/MS (Henriksen et al. 2009; Jacoby et al. 2016), and isotope dilution high-performance liquid chromatography-triple quadruple mass spectrometry (IDLC-MS/MS) (Wang et al. 2015b), provide more accurate means for measuring more than one kind of oxidative nucleic acid in complex fluid samples, with LC-MS/MS having the highest specificity. Using an LC-MS/MS procedure, Joergensen et al. (Joergensen et al. 2011) revealed an age-dependent accumulation of oxidative RNA damage in a population of elderly individuals. However, using the same LC-MS/MS method, Marie’s group (Marie et al. 2009) found no obvious association between environmental stress and urinary 8-oxoG levels. These discrepant findings might be due to the extremely low concentrations of RNA damage makers in the urine from subjects exposed to toxic chemicals, and reanalysis by a more sensitive method such as isotope dilution UPLC-MS/MS might be required. The lower limit of quantitative detection in UPLC-MS/MS is nearly 1 nM for the oxidized nucleoside 8-onoGuo in urine samples (Zhou et al. 2019).
Immunological techniques
Additionally, some immunological techniques, such as ELISA, have been used to quantify oxidized guanosine (8-oxoG) in different body fluids, including urine (Fragopoulou et al. 2018) and cerebrospinal fluid (Jorgensen et al. 2018b). Meanwhile, histological and immunofluorescence technologies have been applied to assess RNA oxidant status in mammalian organic tissues directly. Several groups have successfully performed immunohistochemical staining of 8-oxoG in human carotid endarterectomy specimens (Martinet et al. 2005) and breast cancer tissues (Sova et al. 2010). With 8-oxoG antibody-mediated immunofluorescence staining, Sen et al. demonstrated that the hippocampuses from AD patients are characterized by strong oxidative RNA damage (Sen and Hongpaisan 2018). However, studies have found that the 8-oxoG antibody used for immunological assays sometimes displays probable cross-reactivity with other biomolecules (Nie et al. 2013).
Fluorescence in situ staining
To hamper cross-reactivity, special florescent staining with acridine orange dye was conducted to investigate the levels of damaged nucleic acid in tissues. Under fennel-derived essential oil treatment, the acridine orange test precisely defined cells with damaged RNA, which were marked with a yellowish and/or green fluorescence, in testicular tissue from albino mice (Minas et al. 2018).
Conclusion and future remarks
To date, studies have clearly demonstrated that many species of RNA damage can occur in vitro and in vivo under oxidative stress conditions. Expectedly, ROS species, categorized as toxic byproducts of various oxidation stimuli, have been demonstrated to attack nucleic acids and their precursor nucleotides with oxidizing modifications. Accumulating evidence has begun to indicate that oxidative damage interferes with RNA function, leading to higher error rates in protein synthesis and disturbances in gene regulation. These oxidatively damaged RNA molecules can impair cellular function and ultimately trigger uncontrolled cell proliferation or cell death possibly by error catastrophe. Most importantly, the abnormal proliferation of cells in certain diseases also in turn interferes with the ability to handle damaged RNA, leading to aggravation of deterioration effects (Simms and Zaher 2016).
To maintain genome integrity, RNA stability must be controlled properly in response to a variety of stimuli. As reviewed here, the inability of cells to clear damaged RNAs might contribute to the pathology and etiology of human diseases, such as neurodegenerative disorders and cancers. Recently, emerging studies have focused on the biological significance of oxidative damage to RNA in different physiological and pathological processes and coincidentally have clarified that cells are equipped with several quality control mechanisms for counteracting such RNA modifications. Further research should focus on the detailed functions of RNA quality control mechanisms, in particular by identifying probable macromolecular partners and additional signaling targets of their activities.
Emerging evidence has suggested that oxidative stress is a major causative agent of aging and a number of aging-associated diseases. If oxidative stress exists persistently over time, cellular senescence is a likely consequence and a valuable landmark of aging (Amaya-Montoya et al. 2020; Calcinotto et al. 2019). It is possible that dysfunction of some cells, stemming from the accumulation of oxidative RNA, may be aggravated with increasing age (Hayakawa et al. 2010). One typical model is aging rats, which reveal obvious abnormal mitochondria respiratory functions. In the hippocampus of such rats, the 8-oxoG levels in RNA are higher than those in younger rats (Liu et al. 2002b). Furthermore, the study of aged muscle in rats showing aberrant iron homeostasis are coincident with the function of intracellular metals in producing ROS through Fenton and Haber-Weiss reactions, ultimately leading to oxidative RNA damage (Hofer et al. 2008). Therefore, elevated levels of oxidized RNA evaluated in studies of aging models suggest that at least some fraction of oxidized RNA accumulates in aging-associated diseases. Intriguingly, RNA quality control systems are mechanisms to rectify the purposeful breaks in multiple RNA molecules that occur during cellular oxidative stress and aging (Castellani et al. 2008). For example, the removal of 8-oxoG from the nucleotide pools by the hydrolase MutT precludes the incorporation of these oxidized nucleosides into RNA (Gordon et al. 2014). More importantly, MutT deficiency may be one of the leading causes of accelerated aging (Zheng et al. 2009). Consistent with this hypothesis, a previous study revealed that age-related accumulation of 8-oxoG in RNA is dramatically correlated with the downregulation of MTH1, a MutT-related protein, in the hippocampi of senescence-accelerated (SAMP8) mouse model and AD patients (Song et al. 2011). Overexpression of MTH1 in neurospheres derived from neural progenitor cells prevents the age-dependent accumulation of 8-oxoG, thereby preventing cellular senescence and enhancing proliferative capacity (De Luca et al. 2013). These findings suggest that RNA quality control mechanisms serve as promising antidotes to cytotoxic RNA damage, thereby relieving aging and age-related disorders. Thus, several regulators of RNA oxidative damage involved in aging have been determined so far; however, whether aging is directly regulated by RNA oxidative damage is largely unknown and deserves further investigation. Such important scientific issues may be addressed by pioneering novel work on this topic in the future.
Clearly, the detailed functions of RNA oxidation in clinical settings also need to be explored further in the future. Despite the well-established evidence that oxidative RNA damage is a definite factor in the pathology of multiple human diseases, interventions such as the administration of antioxidants have been only modestly successful in clinical trials (Rasmussen et al. 2016). Given the complexity of ROS metabolism, such treatment methods might be too simplistic and demand more rational strategies not only to carry out the exogenous interventions but also to strengthen the endogenous RNA control systems. Additionally, several other meaningful issues in this emerging field of research also require further clarification: (1) although numerous studies have supported the roles of RNA insults in cell biology, information on potential biomarkers in human health and disease is still limited; (2) with progress in molecular biological techniques, LC-MS/MS has been identified as a widely accepted technology to evaluate the oxidation of RNA products in cells and tissue accurately, but as LC-MS/MS is time-consuming and inconvenient, the use of RNA damage products for diagnosis and prognosis of diseases needs to be tested in much simpler approaches.
Authors’ contributions
Conception and design: ZJ Xu, JZ Huang, M Gao, GJ Guo, SS Zeng, X Chen, X Wang, and ZC Gong. Writing, review, and/or revision of the manuscript: ZJ Xu and YL Yan. Administrative, technical, or material support: X Wang and SS Zeng.
Funding information
This work was supported by the National Natural Science Foundation of China (81703036, 81803035), the China Postdoctoral Science Foundation (2017M610510), and the Natural Science Foundation of Hunan Province, China (2019JJ50932).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aas PA, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- Abe T, Tohgi H, Isobe C, Murata T, Sato C. Remarkable increase in the concentration of 8-hydroxyguanosine in cerebrospinal fluid from patients with Alzheimer’s disease. Journal of Neuroscience Research. 2002;70:447–450. doi: 10.1002/jnr.10349. [DOI] [PubMed] [Google Scholar]
- Abe T, Isobe C, Murata T, Sato C, Tohgi H (2003) Alteration of 8-hydroxyguanosine concentrations in the cerebrospinal fluid and serum from patients with Parkinson’s disease Neuroscience Letters 336:105-108 doi:10.1016/s0304-3940(02)01259-4. [DOI] [PubMed]
- Amaya-Montoya M, Perez-Londono A, Guatibonza-Garcia V, Vargas-Villanueva A, Mendivil CO. Cellular senescence as a therapeutic target for age-related diseases: a review advances in therapy. 2020;37:1407–24. 10.1007/s12325-020-01287-0. [DOI] [PMC free article] [PubMed]
- Anbara H, Shahrooz R, Razi M, Malekinejad H, Najafi G. The effect of vitamin C on mice hemolytic anemia induced by phenylhydrazine: an animal model study using histological changes in testis, pre-implantation embryo development, and biochemical changes. Iranian Journal of Basic Medical Sciences. 2018;21:668–677. doi: 10.22038/IJBMS.2018.25819.6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniali G, Malfatti MC, Tell G. Unveiling the non-repair face of the base excision repair pathway in RNA processing: a missing link between DNA repair and gene expression? DNA Repair. 2017;56:65–74. doi: 10.1016/j.dnarep.2017.06.008. [DOI] [PubMed] [Google Scholar]
- Antoniali G, et al. Mammalian APE1 controls miRNA processing and its interactome is linked to cancer RNA metabolism. Nature Communications. 2017;8:797. doi: 10.1038/s41467-017-00842-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellacosa A, Moss EG. RNA repair: damage control. Current Biology: CB. 2003;13:R482–R484. doi: 10.1016/s0960-9822(03)00408-1. [DOI] [PubMed] [Google Scholar]
- Benameur L, Charif N, Li Y, Stoltz JF, de Isla N. Toward an understanding of mechanism of aging-induced oxidative stress in human mesenchymal stem cells. Bio-medical Materials and Engineering. 2015;25:41–46. doi: 10.3233/BME-141247. [DOI] [PubMed] [Google Scholar]
- Bisht S, Faiq M, Tolahunase M, Dada R. Oxidative stress and male infertility. Nature Reviews Urology. 2017;14:470–485. doi: 10.1038/nrurol.2017.69. [DOI] [PubMed] [Google Scholar]
- Broedbaek K, et al. Urinary markers of nucleic acid oxidation and long-term mortality of newly diagnosed type 2 diabetic patients. Diabetes Care. 2011;34:2594–2596. doi: 10.2337/dc11-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NC, Andreazza AC, Young LT. An updated meta-analysis of oxidative stress markers in bipolar disorder. Psychiatry Research. 2014;218:61–68. doi: 10.1016/j.psychres.2014.04.005. [DOI] [PubMed] [Google Scholar]
- Burroughs AM, Aravind L. RNA damage in biological conflicts and the diversity of responding RNA repair systems. Nucleic Acids Research. 2016;44:8525–8555. doi: 10.1093/nar/gkw722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet J, Wagner JR (2013) DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation Cold Spring Harbor Perspectives in Biology 5 doi:10.1101/cshperspect.a012559 [DOI] [PMC free article] [PubMed]
- Calabretta A, Kupfer PA, Leumann CJ. The effect of RNA base lesions on mRNA translation. Nucleic Acids Research. 2015;43:4713–4720. doi: 10.1093/nar/gkv377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcinotto A, Kohli J, Zagato E, Pellegrini L, Demaria M, Alimonti A. Cellular senescence: aging. Cancer, and Injury Physiological Reviews. 2019;99:1047–1078. doi: 10.1152/physrev.00020.2018. [DOI] [PubMed] [Google Scholar]
- Cameron TA, Matz LM, Sinha D, De Lay NR. Polynucleotide phosphorylase promotes the stability and function of Hfq-binding sRNAs by degrading target mRNA-derived fragments. Nucleic Acids Research. 2019;47:8821–8837. doi: 10.1093/nar/gkz616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellani RJ, Nunomura A, Rolston RK, Moreira PI, Takeda A, Perry G, Smith MA. Sublethal RNA oxidation as a mechanism for neurodegenerative disease. International Journal of Molecular Sciences. 2008;9:789–806. doi: 10.3390/ijms9050789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejvanovic V, et al. Iron induced RNA-oxidation in the general population and in mouse tissue. Free Radical Biology & Medicine. 2018;115:127–135. doi: 10.1016/j.freeradbiomed.2017.11.013. [DOI] [PubMed] [Google Scholar]
- Cejvanovic V, Kjaer LK, Morup Bergholdt HK, Henriksen T, Weimann A, Ellervik C, Poulsen HE. RNA oxidation and iron levels in patients with diabetes. Free Radical Biology & Medicine. 2018;129:532–536. doi: 10.1016/j.freeradbiomed.2018.10.420. [DOI] [PubMed] [Google Scholar]
- Chang Y, Kong Q, Shan X, Tian G, Ilieva H, Cleveland DW, Rothstein JD, Borchelt DR, Wong PC, Lin CLG. Messenger RNA oxidation occurs early in disease pathogenesis and promotes motor neuron degeneration in ALS. PloS one. 2008;3:e2849. doi: 10.1371/journal.pone.0002849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csiszar A, et al. Chemically induced carcinogenesis in rodent models of aging: assessing organismal resilience to genotoxic stressors in geroscience research. GeroScience. 2019;41:209–227. doi: 10.1007/s11357-019-00064-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai DP, et al. Transcriptional mutagenesis mediated by 8-oxoG induces translational errors in mammalian cells, Proceedings of the National Academy of Sciences of the United States of America. 2018;115:4218–22. 10.1073/pnas.1718363115. [DOI] [PMC free article] [PubMed]
- De Luca G, et al. Prolonged lifespan with enhanced exploratory behavior in mice overexpressing the oxidized nucleoside triphosphatase hMTH1. Aging Cell. 2013;12:695–705. doi: 10.1111/acel.12094. [DOI] [PubMed] [Google Scholar]
- Feyzi E, et al. RNA base damage and repair. Current Pharmaceutical Biotechnology. 2007;8:326–331. doi: 10.2174/138920107783018363. [DOI] [PubMed] [Google Scholar]
- Fimognari C. Role of oxidative RNA damage in chronic-degenerative diseases. Oxidative Medicine and Cellular Longevity. 2015;2015:358713. doi: 10.1155/2015/358713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimognari C, Lenzi M, Sestili P, Turrini E, Ferruzzi L, Hrelia P, Cantelli-Forti G. Sulforaphane potentiates RNA damage induced by different xenobiotics. PloS one. 2012;7:e35267. doi: 10.1371/journal.pone.0035267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragopoulou E, et al. Suppression of DNA/RNA and protein oxidation by dietary supplement which contains plant extracts and vitamins: a randomized, double-blind, placebo-controlled trial. Lipids in Health and Disease. 2018;17:187. doi: 10.1186/s12944-018-0836-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenthal BD, Beard WA, Perera L, Shock DD, Kim T, Schlick T, Wilson SH. Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide. Nature. 2015;517:635–639. doi: 10.1038/nature13886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard H, Aguilera A. A novel class of mRNA-containing cytoplasmic granules are produced in response to UV-irradiation. Molecular Biology of the Cell. 2008;19:4980–4992. doi: 10.1091/mbc.E08-02-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, et al. Investigation on potential associations of oxidatively generated DNA/RNA damage with lung, colorectal, breast, prostate and total cancer incidence. Scientific Reports. 2019;9:7109. doi: 10.1038/s41598-019-42596-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill I, Kaur S, Kaur N, Dhiman M, Mantha AK. Phytochemical Ginkgolide B attenuates amyloid-beta1-42 induced oxidative damage and altered cellular responses in human neuroblastoma SH-SY5Y cells. Journal of Alzheimer’s Disease: JAD. 2017;60:S25–S40. doi: 10.3233/JAD-161086. [DOI] [PubMed] [Google Scholar]
- Golzarroshan B, Lin CL, Li CL, Yang WZ, Chu LY, Agrawal S, Yuan HS. Crystal structure of dimeric human PNPase reveals why disease-linked mutants suffer from low RNA import and degradation activities. Nucleic Acids Research. 2018;46:8630–8640. doi: 10.1093/nar/gky642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Tao R, Li Z. Quantification of RNA damage by reverse transcription polymerase chain reactions. Analytical Biochemistry. 2006;357:58–67. doi: 10.1016/j.ab.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Gordon AJ, Satory D, Wang M, Halliday JA, Golding I, Herman C. Removal of 8-oxo-GTP by MutT hydrolase is not a major contributor to transcriptional fidelity. Nucleic Acids Research. 2014;42:12015–12026. doi: 10.1093/nar/gku912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V, Stark A. Eukaryotic core promoters and the functional basis of transcription initiation. Nature Reviews Molecular Cell Biology. 2018;19:621–637. doi: 10.1038/s41580-018-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa H, Sekiguchi M. Human polynucleotide phosphorylase protein in response to oxidative stress. Biochemistry. 2006;45:6749–6755. doi: 10.1021/bi052585l. [DOI] [PubMed] [Google Scholar]
- Hayakawa H, Kuwano M, Sekiguchi M. Specific binding of 8-oxoguanine-containing RNA to polynucleotide phosphorylase protein. Biochemistry. 2001;40:9977–9982. doi: 10.1021/bi010595q. [DOI] [PubMed] [Google Scholar]
- Hayakawa H, Fujikane A, Ito R, Matsumoto M, Nakayama KI, Sekiguchi M. Human proteins that specifically bind to 8-oxoguanine-containing RNA and their responses to oxidative stress. Biochemical and Biophysical Research Communications. 2010;403:220–224. doi: 10.1016/j.bbrc.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Henriksen T, Hillestrom PR, Poulsen HE, Weimann A. Automated method for the direct analysis of 8-oxo-guanosine and 8-oxo-2′-deoxyguanosine in human urine using ultraperformance liquid chromatography and tandem mass spectrometry. Free Radical Biology & Medicine. 2009;47:629–635. doi: 10.1016/j.freeradbiomed.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Hofer T, Seo AY, Prudencio M, Leeuwenburgh C. A method to determine RNA and DNA oxidation simultaneously by HPLC-ECD: greater RNA than DNA oxidation in rat liver after doxorubicin administration. Biological Chemistry. 2006;387:103–111. doi: 10.1515/BC.2006.014. [DOI] [PubMed] [Google Scholar]
- Hofer T, et al. Increased iron content and RNA oxidative damage in skeletal muscle with aging and disuse atrophy. Experimental Gerontology. 2008;43:563–570. doi: 10.1016/j.exger.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini M, Roy P, Sissler M, Zirbel CL, Westhof E, Leontis N. How to fold and protect mitochondrial ribosomal RNA with fewer guanines. Nucleic Acids Research. 2018;46:10946–10968. doi: 10.1093/nar/gky762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KV, et al. Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy. Nature. 2014;508:222–227. doi: 10.1038/nature13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson BH, Zaher HS (2015) O6-Methylguanosine leads to position-dependent effects on ribosome speed and fidelity Rna 21:1648-1659 doi:10.1261/rna.052464.115. [DOI] [PMC free article] [PubMed]
- Ishii T, Sekiguchi M. Two ways of escaping from oxidative RNA damage: Selective degradation and cell death. DNA Repair. 2019;81:102666. doi: 10.1016/j.dnarep.2019.102666. [DOI] [PubMed] [Google Scholar]
- Ishii T, Hayakawa H, Sekiguchi T, Adachi N, Sekiguchi M. Role of Auf1 in elimination of oxidatively damaged messenger RNA in human cells. Free Radical Biology & Medicine. 2015;79:109–116. doi: 10.1016/j.freeradbiomed.2014.11.018. [DOI] [PubMed] [Google Scholar]
- Ishii T, Hayakawa H, Igawa T, Sekiguchi T, Sekiguchi M. Specific binding of PCBP1 to heavily oxidized RNA to induce cell death. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:6715–6720. doi: 10.1073/pnas.1806912115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isobe C, Abe T, Terayama Y. Homocysteine may contribute to pathogenesis of RNA damage in brains with Alzheimer’s disease. Neuro-degenerative Diseases. 2009;6:252–257. doi: 10.1159/000262443. [DOI] [PubMed] [Google Scholar]
- Jacoby AS, Vinberg M, Poulsen HE, Kessing LV, Munkholm K. Increased DNA and RNA damage by oxidation in patients with bipolar I disorder. Translational Psychiatry. 2016;6:e867. doi: 10.1038/tp.2016.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, et al. Damage sensor role of UV-DDB during base excision repair. Nature Structural & Molecular Biology. 2019;26:695–703. doi: 10.1038/s41594-019-0261-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemth AS, et al. MutT homologue 1 (MTH1) catalyzes the hydrolysis of mutagenic O6-methyl-dGTP. Nucleic Acids Research. 2018;46:10888–10904. doi: 10.1093/nar/gky896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joergensen A, Broedbaek K, Weimann A, Semba RD, Ferrucci L, Joergensen MB, Poulsen HE. Association between urinary excretion of cortisol and markers of oxidatively damaged DNA and RNA in humans. PloS one. 2011;6:e20795. doi: 10.1371/journal.pone.0020795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen A, et al. Increased systemic oxidatively generated DNA and RNA damage in schizophrenia. Psychiatry Research. 2013;209:417–423. doi: 10.1016/j.psychres.2013.01.033. [DOI] [PubMed] [Google Scholar]
- Jorgensen A, et al. Systemic oxidatively generated DNA/RNA damage in clinical depression: associations to symptom severity and response to electroconvulsive therapy. Journal of Affective Disorders. 2013;149:355–362. doi: 10.1016/j.jad.2013.02.011. [DOI] [PubMed] [Google Scholar]
- Jorgensen A, Siersma V, Davidsen AS, Weimann A, Henriksen T, Poulsen HE, Olivarius NF. Markers of DNA/RNA damage from oxidation as predictors of a registry-based diagnosis of psychiatric illness in type 2 diabetic patients. Psychiatry Research. 2018;259:370–376. doi: 10.1016/j.psychres.2017.11.017. [DOI] [PubMed] [Google Scholar]
- Jorgensen A, et al. Progressive DNA and RNA damage from oxidation after aneurysmal subarachnoid haemorrhage in humans. Free Radical Research. 2018;52:51–56. doi: 10.1080/10715762.2017.1407413. [DOI] [PubMed] [Google Scholar]
- Kim C, Kang D, Lee EK, Lee JS. Long noncoding RNAs and RNA-binding proteins in oxidative stress, cellular senescence, and age-related diseases. Oxidative Medicine and Cellular Longevity. 2017;2017:2062384–2062321. doi: 10.1155/2017/2062384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kournoutou GG, Giannopoulou PC, Sazakli E, Leotsinidis M, Kalpaxis DL. Oxidative damage of 18S and 5S ribosomal RNA in digestive gland of mussels exposed to trace metals. Aquatic Toxicology. 2017;192:136–147. doi: 10.1016/j.aquatox.2017.09.007. [DOI] [PubMed] [Google Scholar]
- Kuhnel H, et al. Investigations into cytotoxic effects of the herbal preparation abnormal Savda Munziq. Chinese Journal of Integrative Medicine. 2015. 10.1007/s11655-015-2132-3. [DOI] [PubMed]
- Kupfer PA, Leumann CJ. Synthesis, base pairing properties and trans-lesion synthesis by reverse transcriptases of oligoribonucleotides containing the oxidatively damaged base 5-hydroxycytidine. Nucleic Acids Research. 2011;39:9422–9432. doi: 10.1093/nar/gkr673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Fu XD. Chromatin-associated RNAs as facilitators of functional genomic interactions. Nature Reviews Genetics. 2019;20:503–519. doi: 10.1038/s41576-019-0135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liguori I, et al. Oxidative stress, aging, and diseases. Clinical Interventions in Aging. 2018;13:757–772. doi: 10.2147/CIA.S158513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YT, Liu W, He Y, Wu YL, Chen WN, Lin XJ, Lin X. Hepatitis B virus X protein increases 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-Oxodg) level via repressing MTH1/MTH2 expression in hepatocytes. Cellular Physiology and Biochemistry: International Journal of Experimental Cellular Physiology, Biochemistry, and Pharmacology. 2018;51:80–96. doi: 10.1159/000495166. [DOI] [PubMed] [Google Scholar]
- Liu J, Atamna H, Kuratsune H, Ames BN. Delaying brain mitochondrial decay and aging with mitochondrial antioxidants and metabolites. Annals of the New York Academy of Sciences. 2002;959:133–166. doi: 10.1111/j.1749-6632.2002.tb02090.x. [DOI] [PubMed] [Google Scholar]
- Liu J, et al. Memory loss in old rats is associated with brain mitochondrial decay and RNA/DNA oxidation: partial reversal by feeding acetyl-L-carnitine and/or R-alpha -lipoic acid. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:2356–2361. doi: 10.1073/pnas.261709299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X et al. (2016) Elevated levels of urinary markers of oxidative DNA and RNA damage in type 2 diabetes with complications Oxidative Medicine and Cellular Longevity 2016:4323198 doi:10.1155/2016/4323198. [DOI] [PMC free article] [PubMed]
- Liu N, et al. Vitexin compound 1, a novel extraction from a Chinese herb, suppresses melanoma cell growth through DNA damage by increasing ROS levels. Journal of Experimental & Clinical Cancer Research: CR. 2018;37:269. doi: 10.1186/s13046-018-0897-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell MA, Soman S, Bradley MA. Oxidatively modified nucleic acids in preclinical Alzheimer’s disease (PCAD) brain. Mechanisms of Ageing and Development. 2011;132:443–448. doi: 10.1016/j.mad.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manini P, et al. Biomarkers of nucleic acid oxidation, polymorphism in, and expression of, hOGG1 gene in styrene-exposed workers. Toxicology Letters. 2009;190:41–47. doi: 10.1016/j.toxlet.2009.06.862. [DOI] [PubMed] [Google Scholar]
- Marie C, Ravanat JL, Badouard C, Marques M, Balducci F, Maitre A. Urinary levels of oxidative DNA and RNA damage among workers exposed to polycyclic aromatic hydrocarbons in silicon production: comparison with 1-hydroxypyrene. Environmental and Molecular Mutagenesis. 2009;50:88–95. doi: 10.1002/em.20439. [DOI] [PubMed] [Google Scholar]
- Martinet W, De Meyer GR, Herman AG, Kockx MM. RNA damage in human atherosclerosis: pathophysiological significance and implications for gene expression studies. RNA Biology. 2005;2:4–7. doi: 10.4161/rna.2.1.1430. [DOI] [PubMed] [Google Scholar]
- Mazzoni C, D’Addario I, Falcone C. The C-terminus of the yeast Lsm4p is required for the association to P-bodies. FEBS Letters. 2007:581, 4836–4840. 10.1016/j.febslet.2007.09.009. [DOI] [PubMed]
- Memar MY, Ghotaslou R, Samiei M, Adibkia K. Antimicrobial use of reactive oxygen therapy: current insights. Infection and Drug Resistance. 2018;11:567–576. doi: 10.2147/IDR.S142397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan MJ. Linking deregulation of non-coding RNA to the core pathophysiology of Alzheimer’s disease: an integrative review. Progress in Neurobiology. 2017;156:1–68. doi: 10.1016/j.pneurobio.2017.03.004. [DOI] [PubMed] [Google Scholar]
- Minas A, Najafi G, Jalali AS, Razi M. Fennel induces cytotoxic effects against testicular germ cells in mice; evidences for suppressed pre-implantation embryo development. Environmental Toxicology. 2018. 10.1002/tox.22570. [DOI] [PubMed]
- Molavi M, Razi M, Malekinejad H, Amniattalab A, Rezaie H. Vitamin E improved cypermethrin-induced damages in the ovary of rats; evidence for angiogenesis and p53 involvement. Pesticide Biochemistry and Physiology. 2014;110:27–35. doi: 10.1016/j.pestbp.2014.02.004. [DOI] [PubMed] [Google Scholar]
- Mroczek S, Kufel J. Apoptotic signals induce specific degradation of ribosomal RNA in yeast. Nucleic Acids Research. 2008;36:2874–2888. doi: 10.1093/nar/gkm1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkholm K, Poulsen HE, Kessing LV, Vinberg M. Elevated levels of urinary markers of oxidatively generated DNA and RNA damage in bipolar disorder. Bipolar Disorders. 2015;17:257–268. doi: 10.1111/bdi.12245. [DOI] [PubMed] [Google Scholar]
- Nabavi SM, Habtemariam S, Daglia M, Braidy N, Loizzo MR, Tundis R, Nabavi SF. Neuroprotective effects of Ginkgolide B against ischemic stroke: a review of current literature. Current Topics in Medicinal Chemistry. 2015;15:2222–2232. doi: 10.2174/1568026615666150610142647. [DOI] [PubMed] [Google Scholar]
- Nandakumar J, Schwer B, Schaffrath R, Shuman S. RNA repair: an antidote to cytotoxic eukaryal RNA damage. Molecular Cell. 2008;31:278–286. doi: 10.1016/j.molcel.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton DL, et al. Specifically targeting the CD22 receptor of human B-cell lymphomas with RNA damaging agents. Critical Reviews in Oncology/Hematology. 2001;39:79–86. doi: 10.1016/s1040-8428(01)00116-0. [DOI] [PubMed] [Google Scholar]
- Nie B, et al. Age-dependent accumulation of 8-oxoguanine in the DNA and RNA in various rat tissues. Oxidative Medicine and Cellular Longevity. 2013, 2013;303181. 10.1155/2013/303181. [DOI] [PMC free article] [PubMed]
- Nunomura A, et al. Neuronal RNA oxidation is a prominent feature of familial Alzheimer's disease. Neurobiology of Disease. 2004;17:108–113. doi: 10.1016/j.nbd.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Nunomura A, Hofer T, Moreira PI, Castellani RJ, Smith MA, Perry G. RNA oxidation in Alzheimer disease and related neurodegenerative disorders. Acta Neuropathologica. 2009;118:151–166. doi: 10.1007/s00401-009-0508-1. [DOI] [PubMed] [Google Scholar]
- Nunomura A, Moreira PI, Castellani RJ, Lee HG, Zhu X, Smith MA, Perry G. Oxidative damage to RNA in aging and neurodegenerative disorders. Neurotoxicity Research. 2012;22:231–248. doi: 10.1007/s12640-012-9331-x. [DOI] [PubMed] [Google Scholar]
- Nunomura A, et al. The earliest stage of cognitive impairment in transition from normal aging to Alzheimer disease is marked by prominent RNA oxidation in vulnerable neurons. Journal of Neuropathology and experimental Neurology. 2012;71:233–241. doi: 10.1097/NEN.0b013e318248e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunomura A, Lee HG, Zhu X, Perry G. Consequences of RNA oxidation on protein synthesis rate and fidelity: implications for the pathophysiology of neuropsychiatric disorders. Biochemical Society Transactions. 2017;45:1053–1066. doi: 10.1042/BST20160433. [DOI] [PubMed] [Google Scholar]
- Nunoshiba T, Ishida R, Sasaki M, Iwai S, Nakabeppu Y, Yamamoto K. A novel Nudix hydrolase for oxidized purine nucleoside triphosphates encoded by ORFYLR151c (PCD1 gene) in Saccharomyces cerevisiae. Nucleic Acids Research. 2004;32:5339–5348. doi: 10.1093/nar/gkh868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou C, et al. Targeting YAP1/LINC00152/FSCN1 signaling Axis prevents the progression of colorectal cancer. Advanced Science. 2019;1901380. 10.1002/advs.201901380. [DOI] [PMC free article] [PubMed]
- Palermo V, Falcone C, Calvani M, Mazzoni C. Acetyl-L-carnitine protects yeast cells from apoptosis and aging and inhibits mitochondrial fission. Aging Cell. 2010;9:570–579. doi: 10.1111/j.1474-9726.2010.00587.x. [DOI] [PubMed] [Google Scholar]
- Poulsen HE, et al. Oxidatively generated modifications to nucleic acids in vivo: measurement in urine and plasma. Free Radical Biology & Medicine. 2019;145:336–341. doi: 10.1016/j.freeradbiomed.2019.10.001. [DOI] [PubMed] [Google Scholar]
- Qiao Z, Wigginton KR. Direct and Indirect photochemical reactions in viral RNA measured with RT-qPCR and mass spectrometry. Environmental Science & Technology. 2016;50:13371–13379. doi: 10.1021/acs.est.6b04281. [DOI] [PubMed] [Google Scholar]
- Rasmussen ST, et al. Simvastatin and oxidative stress in humans: a randomized, double-blinded, placebo- clinical trial controlled. Redox Biology. 2016;9:32–38. doi: 10.1016/j.redox.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Jensen TH. Controlling nuclear RNA levels. Nature Reviews Genetics. 2018;19:518–529. doi: 10.1038/s41576-018-0013-2. [DOI] [PubMed] [Google Scholar]
- Schottker B, Xuan Y, Gao X, Anusruti A, Brenner H. Oxidatively damaged DNA/RNA and 8-Isoprostane levels are associated with the development of type 2 diabetes at older age: results from a large cohort study. Diabetes Care. 2020;43:130–136. doi: 10.2337/dc19-1379. [DOI] [PubMed] [Google Scholar]
- Scott DD, et al. Nol12 is a multifunctional RNA binding protein at the nexus of RNA and DNA metabolism. Nucleic Acids Research. 2017;45:12509–12528. doi: 10.1093/nar/gkx963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi T, Ito R, Hayakawa H, Sekiguchi M. Elimination and utilization of oxidized guanine nucleotides in the synthesis of RNA and its precursors. The Journal of Biological Chemistry. 2013;288:8128–8135. doi: 10.1074/jbc.M112.418723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, Hongpaisan J. Hippocampal microvasculature changes in association with oxidative stress in Alzheimer’s disease. Free Radical Biology & Medicine. 2018;120:192–203. doi: 10.1016/j.freeradbiomed.2018.03.034. [DOI] [PubMed] [Google Scholar]
- Sen A, Nelson TJ, Alkon DL, Hongpaisan J (2018) Loss in PKC epsilon causes downregulation of MnSOD and BDNF expression in neurons of Alzheimer’s disease hippocampus Journal of Alzheimer’s Disease: JAD 63:1173–1189 doi:10.3233/JAD-171008. [DOI] [PubMed]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms CL, Zaher HS. Quality control of chemically damaged. RNA Cellular and Molecular Life Sciences: CMLS. 2016;73:3639–3653. doi: 10.1007/s00018-016-2261-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Savanur MA, Srivastava S, D'Silva P, Mugesh G. A redox modulatory Mn3 O4 nanozyme with multi-enzyme activity provides efficient cytoprotection to human cells in a Parkinson’s disease model. Angewandte Chemie. 2017;56:14267–14271. doi: 10.1002/anie.201708573. [DOI] [PubMed] [Google Scholar]
- Song XN, et al. Oxidative damage to RNA and expression patterns of MTH1 in the hippocampi of senescence-accelerated SAMP8 mice and Alzheimer’s disease patients. Neurochemical Research. 2011;36:1558–1565. doi: 10.1007/s11064-011-0484-4. [DOI] [PubMed] [Google Scholar]
- Sova H, Jukkola-Vuorinen A, Puistola U, Kauppila S, Karihtala P. 8-Hydroxydeoxyguanosine: a new potential independent prognostic factor in breast cancer. British Journal of Cancer. 2010;102:1018–1023. doi: 10.1038/sj.bjc.6605565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirpe M, Palermo V, Ferrari M, Mroczek S, Kufel J, Falcone C, Mazzoni C. Increased levels of RNA oxidation enhance the reversion frequency in aging pro-apoptotic yeast mutants. Apoptosis: an International Journal on Programmed Cell Death. 2017;22:200–206. doi: 10.1007/s10495-016-1319-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone CM, et al. Inhibition of homologous phosphorolytic ribonucleases by citrate may represent an evolutionarily conserved communicative link between RNA degradation and central metabolism. Nucleic Acids Research. 2017;45:4655–4666. doi: 10.1093/nar/gkx114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei F, Hayakawa H, Bouton M, Cirinesi A, Matic I, Sekiguchi M, Radman M. Counteraction by MutT protein of transcriptional errors caused by oxidative damage. Science. 1997;278:128–130. doi: 10.1126/science.278.5335.128. [DOI] [PubMed] [Google Scholar]
- Takagi Y, Setoyama D, Ito R, Kamiya H, Yamagata Y, Sekiguchi M. Human MTH3 (NUDT18) protein hydrolyzes oxidized forms of guanosine and deoxyguanosine diphosphates: comparison with MTH1 and MTH2. The Journal Of Biological Chemistry. 2012;287:21541–21549. doi: 10.1074/jbc.M112.363010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. Rna. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaszewska-Antczak A, Guga P, Nawrot B, Pratviel G. Guanosine in a single stranded region of anticodon stem-loop tRNA models is prone to oxidatively generated damage resulting in dehydroguanidinohydantoin and spiroiminodihydantoin lesions. Chemistry. 2015;21:6381–6385. doi: 10.1002/chem.201406409. [DOI] [PubMed] [Google Scholar]
- Utumatwishima JN, Chung ST, Bentley AR, Udahogora M, Sumner AE. Reversing the tide - diagnosis and prevention of T2DM in populations of African descent. Nature Reviews Endocrinology. 2018;14:45–56. doi: 10.1038/nrendo.2017.127. [DOI] [PubMed] [Google Scholar]
- Vascotto C, et al. APE1/Ref-1 interacts with NPM1 within nucleoli and plays a role in the rRNA quality control process. Molecular and Cellular Biology. 2009;29:1834–1854. doi: 10.1128/MCB.01337-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Violet M, Delattre L, Tardivel M, Sultan A, Chauderlier A, Caillierez R, Talahari S, Nesslany F, Lefebvre B, Bonnefoy E, Buée L, Galas MC. A major role for Tau in neuronal DNA and RNA protection in vivo under physiological and hyperthermic conditions. Frontiers in Cellular Neuroscience. 2014;8:84. doi: 10.3389/fncel.2014.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Violet M, et al. Prefibrillar tau oligomers alter the nucleic acid protective function of tau in hippocampal neurons in vivo. Neurobiology of Disease. 2015;82:540–551. doi: 10.1016/j.nbd.2015.09.003. [DOI] [PubMed] [Google Scholar]
- Wang JX, et al. Oxidative modification of miR-184 enables it to target Bcl-xL and Bcl-w. Molecular Cell. 2015;59:50–61. doi: 10.1016/j.molcel.2015.05.003. [DOI] [PubMed] [Google Scholar]
- Wang WX, Luo SB, Xia MM, Mao YH, Zhou XY, Jiang P, Jiang HY, Dai DP, Li CB, Hu GX, Cai JP. Analysis of the oxidative damage of DNA, RNA, and their metabolites induced by hyperglycemia and related nephropathy in Sprague Dawley rats. Free Radical Research. 2015;49:1199–1209. doi: 10.3109/10715762.2015.1033416. [DOI] [PubMed] [Google Scholar]
- Wang P, et al. Mapping spatial transcriptome with light-activated proximity-dependent RNA labeling. Nature Chemical Biology. 2019;15:1110–1119. doi: 10.1038/s41589-019-0368-5. [DOI] [PubMed] [Google Scholar]
- Willi J, Kupfer P, Evequoz D, Fernandez G, Katz A, Leumann C, Polacek N. Oxidative stress damages rRNA inside the ribosome and differentially affects the catalytic center. Nucleic Acids Research. 2018;46:1945–1957. doi: 10.1093/nar/gkx1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Li Z. Human polynucleotide phosphorylase reduces oxidative RNA damage and protects HeLa cell against oxidative stress. Biochemical and Biophysical Research Communications. 2008;372:288–292. doi: 10.1016/j.bbrc.2008.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Jiang Z, Liu M, Gong X, Wu S, Burns CM, Li Z. Polynucleotide phosphorylase protects Escherichia coli against oxidative stress. Biochemistry. 2009;48:2012–2020. doi: 10.1021/bi801752p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtmann EJ, Wolin SL. RNA under attack: cellular handling of RNA damage. Critical reviews in Biochemistry and Molecular Biology. 2009;44:34–49. doi: 10.1080/10409230802594043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan LL, Zaher HS. How do cells cope with RNA damage and its consequences? J Biol Chem. 2019;294:15158–15171. doi: 10.1074/jbc.REV119.006513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan LL, Simms CL, McLoughlin F, Vierstra RD, Zaher HS. Oxidation and alkylation stresses activate ribosome-quality control. Nature Communications. 2019;10:5611. doi: 10.1038/s41467-019-13579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Chen X, Wang X, Zhao Z, Hu W, Zeng S, Wei J, Yang X, Qian L, Zhou S, Sun L, Gong Z, Xu Z. The effects and the mechanisms of autophagy on the cancer-associated fibroblasts in cancer. Journal of Experimental & Clinical Cancer Research: CR. 2019;38:171. doi: 10.1186/s13046-019-1172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, et al. Novel function of lncRNA ADAMTS9-AS2 in promoting temozolomide resistance in glioblastoma via Upregulating the FUS/MDM2 ubiquitination axis. Frontiers in Cell and Developmental Biology. 2019;7:217. doi: 10.3389/fcell.2019.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Villani RM, Wang H, Simpson MJ, Roberts MS, Tang M, Liang X. The role of cellular reactive oxygen species in cancer chemotherapy. Journal of Experimental & Clinical Cancer Research: CR. 2018;37:266. doi: 10.1186/s13046-018-0909-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarasi S, McConachie C, Loppnow GR. Molecular beacon probes of photodamage in thymine and uracil oligonucleotides. Photochemistry and Photobiology. 2005;81:467–473. doi: 10.1562/2004-09-02-RA-301. [DOI] [PubMed] [Google Scholar]
- Yoshimura K, Ogawa T, Ueda Y, Shigeoka S. AtNUDX1, an 8-oxo-7,8-dihydro-2′-deoxyguanosine 5′-triphosphate pyrophosphohydrolase, is responsible for eliminating oxidized nucleotides in Arabidopsis. Plant & Cell Physiology. 2007;48:1438–1449. doi: 10.1093/pcp/pcm112. [DOI] [PubMed] [Google Scholar]
- Youn JY, Dyakov BJA, Zhang J, Knight JDR, Vernon RM, Forman-Kay JD, Gingras AC. Properties of Stress granule and P-body proteomes. Molecular Cell. 2019;76:286–294. doi: 10.1016/j.molcel.2019.09.014. [DOI] [PubMed] [Google Scholar]
- Zheng JD, et al. Age-related alterations in the expression of MTH2 in the hippocampus of the SAMP8 mouse with learning and memory deterioration. Journal of the Neurological Sciences. 2009;287:188–196. doi: 10.1016/j.jns.2009.07.027. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Yao Y, Shao Y, Qu W, Chen Y, Jiang Q. Urinary bisphenol analogues concentrations and biomarkers of oxidative DNA and RNA damage in Chinese school children in East China: a repeated measures study. Environmental Pollution. 2019;254:112921. doi: 10.1016/j.envpol.2019.07.089. [DOI] [PubMed] [Google Scholar]