Abstract
The RNA-dependent RNA polymerase of the severe acute respiratory syndrome coronavirus 2 is an important target in current drug development efforts for the treatment of coronavirus disease 2019. Molnupiravir is a broad-spectrum antiviral that is an orally bioavailable prodrug of the nucleoside analogue β-D-N4-hydroxycytidine (NHC). Molnupiravir or NHC can increase G to A and C to U transition mutations in replicating coronaviruses. These increases in mutation frequencies can be linked to increases in antiviral effects; however, biochemical data of molnupiravir-induced mutagenesis have not been reported. Here we studied the effects of the active compound NHC 5’-triphosphate (NHC-TP) against the purified severe acute respiratory syndrome coronavirus 2 RNA-dependent RNA polymerase complex. The efficiency of incorporation of natural nucleotides over the efficiency of incorporation of NHC-TP into model RNA substrates followed the order GTP (12,841) > ATP (424) > UTP (171) > CTP (30), indicating that NHC-TP competes predominantly with CTP for incorporation. No significant inhibition of RNA synthesis was noted as a result of the incorporated monophosphate in the RNA primer strand. When embedded in the template strand, NHC-monophosphate supported the formation of both NHC:G and NHC:A base pairs with similar efficiencies. The extension of the NHC:G product was modestly inhibited, but higher nucleotide concentrations could overcome this blockage. In contrast, the NHC:A base pair led to the observed G to A (G:NHC:A) or C to U (C:G:NHC:A:U) mutations. Together, these biochemical data support a mechanism of action of molnupiravir that is primarily based on RNA mutagenesis mediated via the template strand.
Keywords: coronavirus, SARS-CoV-2, Covid-19, RNA-dependent RNA polymerase, drug development, antiviral agent, mutagen, nucleoside analogue
Abbreviations: NHC, β-D-N4-hydroxycytidine; NHC-MP, NHC monophosphate; NHC-TP, NHC 5’-triphosphate; RdRp, RNA-dependent RNA polymerase; RDV, remdesivir; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2
The discovery and development of potent antiviral drugs for the treatment of infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) remains challenging. The virus can cause severe forms of coronavirus disease 2019 (COVID-19) that require hospitalization. Remdesivir (RDV) targets the viral RNA-dependent RNA polymerase (RdRp) and is currently the only antiviral agent approved by the US Food and Drug Administration (1, 2). Antibody therapies were granted emergency use authorization for the treatment of outpatients who are at high risk for progressing to severe disease and/or hospitalization (3) run up. Both RDV and antibody therapies are intravenously administered, which limits their utility especially for outpatient use. Oral drugs that can be used much earlier in the disease are under investigation, and molnupiravir (MK-4482/EIDD-2801) is perhaps the most advanced candidate compound in this category (4). Molnupiravir is a prodrug of β-D-N4-hydroxycytidine (NHC, EIDD-1931). It is intracellularly metabolized to its triphosphate form (NHC-TP) that can serve as substrate for RNA polymerases (5, 6). NHC shows a broad spectrum of antiviral activities against several positive- and negative-sense RNA viruses (5, 6, 7, 8, 9, 10, 11). More recent studies focused on the development of molnupiravir for the treatment of infection with influenza and coronaviruses, respectively (12, 13, 14, 15, 16).
NHC potently inhibits MERS-CoV, SARS-CoV, and SARS-CoV-2 with EC50 values in the submicromolar range, depending on the specific cell type (13). Molnupiravir was also shown to inhibit SARS-CoV-2 replication in humanized mice (13, 16). Treatment 24 h after exposure to the virus was more efficient than treatment 48 h after virus exposure, and treatment before virus exposure shows the strongest antiviral effects. The large body of preclinical data justified human clinical trials that are presently ongoing, although there have been concerns about its mammalian cell mutagenic potential (5, 11, 17, 18, 19, 20, 21). Molnupiravir was evaluated in a phase 1 clinical study in healthy volunteers, which demonstrated good tolerability and no apparent signs of adverse events after a short duration of treatment and follow-up (22). Knowledge on the mechanism of action is largely derived from cell culture studies. Unlike RDV that inhibits RNA synthesis, molnupiravir seems to act as a mutagen (6, 10, 12, 13). Exposure to NHC increases G to A and C to U transition mutations in MHV, MERS-CoV, and SARS-CoV-2 (10, 13). Increases in mutation frequencies will ultimately yield nonfunctional genomes, which explains the antiviral effect. Other broad-spectrum antiviral agents, such as ribavirin or favipiravir, have also been characterized as mutagenic nucleoside analogues, although the potency of these compounds is generally low with EC50 values in the higher micromolar range (23, 24, 25). The proofreading exonuclease of coronaviruses can excise incorporated nucleotide analogues and diminish the inhibitory effects (26, 27), but recent data have shown that NHC is resistant to this proofreading activity (10). However, it is currently not known how the interaction between the drug and RdRp determines drug potency and the nature of the observed mutations.
Herein, we employed a biochemical approach to study the mechanism of action of molnupiravir. We expressed and purified the RdRp complex with nonstructural proteins nsp7, nsp8, and nsp12 and studied the efficiency of incorporation of NHC-TP in relation to natural NTP pools. NHC-TP is preferentially incorporated as a C-analogue. When the monophosphate (NHC-MP) is embedded in the template, it base pairs with either GTP or ATP. The incorporation of GTP causes subtle inhibition of RNA synthesis, whereas mismatch extension with the incorporated ATP is not inhibited and leads to transition mutations. The collective data presented in this study provide a model that explains the antiviral effects of molnupiravir and NHC.
Results
Selective incorporation of NHC-TP by SARS-CoV-2 RdRp
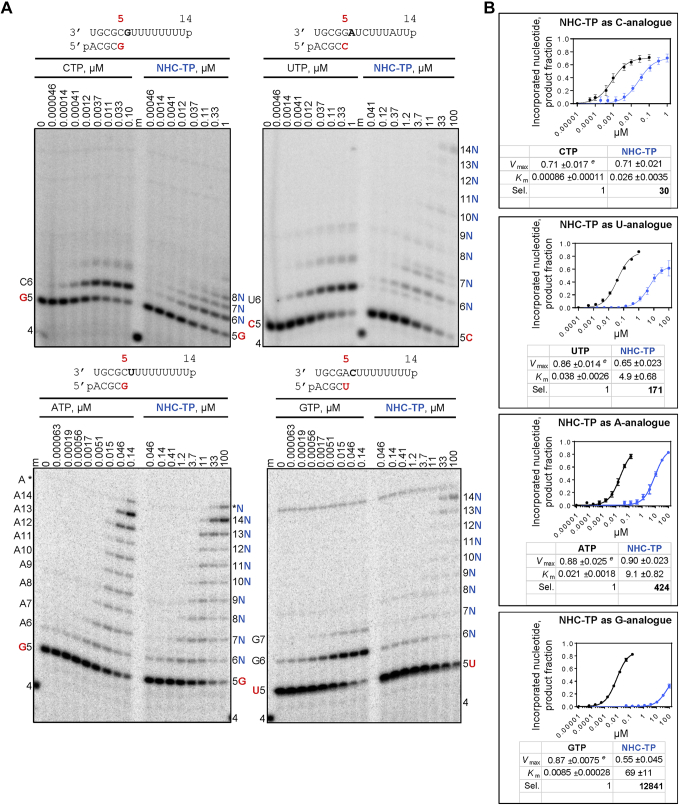
Biochemical data on NHC-TP as a substrate for RNA polymerases are scarce (28). It has been shown that the respiratory syncytial virus RdRp complex accepts NHC-TP as a substrate for incorporation opposite template G. The incorporated NHC-MP does not act as a chain terminator in this case (6). Human mitochondrial DNA-dependent RNA polymerase can use NHC-TP as C- or U-analogue (28). Inhibitory effects following incorporation have not been reported. Here we measured steady-state kinetic parameters for incorporation of NHC-TP by SARS-CoV-2 RdRp (Fig. 1). We have employed the same biochemical approach for RDV-TP, other nucleotides, and other RdRp complexes, which facilitates comparisons (2, 29, 30, 31, 32, 33). RNA synthesis was monitored with a short model primer/template after addition of a single radiolabeled [α-32P]NTP (Fig. 1A). The preference for a natural nucleotide over the analogue is calculated as a ratio of their incorporation efficiencies. The efficiency of incorporation of the natural nucleotide over the efficiency of incorporation of the analogue provides a selectivity value. SARS-CoV-2 RdRp shows a 30-fold preference for CTP over NHC-TP (Fig. 1B). Selectivity values follow the order GTP (12,841) > ATP (424) > UTP (171) > CTP (30), which shows that all NTPs are more efficiently incorporated than the NHC-TP. The data suggest that competition with CTP is most efficient.
Figure 1.
Efficiency of NHC-TP incorporation.A, migration pattern of the products of RNA synthesis catalyzed by SARS-CoV-2 RNA-dependent RNA polymerase along the RNA primer/templates as shown above the panels. The sequences support incorporation of NHC-monophosphate as either a C-, U-, A-, or G-analogue at position 6. G, U, or C indicates incorporation of [α-32P]G-, [α-32P]U-, or [α-32P]CTP at position 5 (red). N indicates incorporation of NHC-monophosphate. Asterisk indicates terminal transferase activity. A 5′-32P-labeled 4-nt primer (4) serves as a size marker (m). B, graphical representation of the data shown in A. Fitting the data points to Michaelis–Menten function and the calculation of the selectivity values (Experimental procedures). Sel., selectivity for a nucleotide substrate analogue is calculated as the ratio of the Vmax/Km values for NTP over NTP analogue. Error bars illustrate standard deviation of the data. ±, standard error of the fit. All reported values have been calculated on the basis of an 8-data point experiment repeated at least three times. NHC-TP, β-D-N4-hydroxycytidine 5’-triphosphate.
Extension of incorporated NHC-MP
We next monitored RNA synthesis following incorporation of NHC-MP opposite template G at position 6 (Fig. 2). The same concentrations of CTP and NHC-TP generate different incorporation patterns at position 6. For CTP, most of the primer is converted to yield the 6-nt product with minimal formation of larger products. Incorporation of CTP opposite subsequent other bases would lead to mismatch formation and is negligible under these conditions. For NHC-TP, both a 6-nt product and a 7-nt product are formed, which points to a certain degree of ambiguous base pairing. Since both products are formed in the absence of ATP and UTP (lane 0), the 7-nt product illustrates NHC-TP misincorporation as a U-analogue opposite AMP in the template. In the presence of increasing concentrations of ATP and UTP that allow full-length product formation, the primer is almost completely extended regardless of whether CMP or NHC-MP was incorporated (Fig. 2, left CTP and NHC-TP panels). This shows that the incorporated NHC-MP is efficiently extended. Higher concentrations of ATP and UTP reduce mismatch formations and generate similar levels of full-template-length products (Fig. 2, right CTP and NHC-TP panels). Moreover, similar levels of terminal transferase activity are also seen at high concentrations of ATP and UTP regardless of whether CMP or NHC-MP are being extended from a 6-nt product (34).
Figure 2.
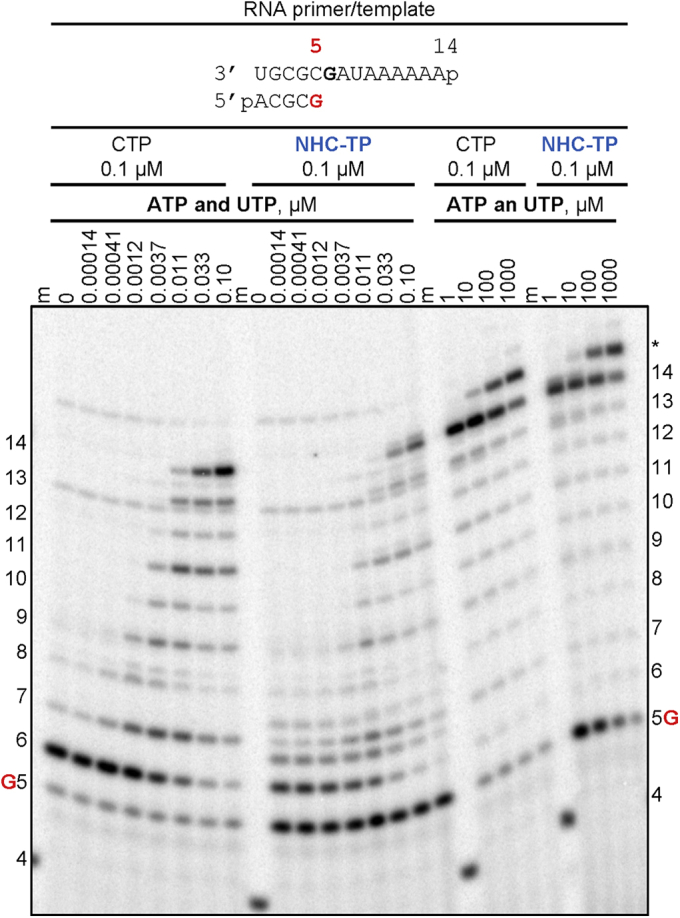
SARS-CoV-2 RNA-dependent RNA polymerase–catalyzed RNA synthesis following incorporation of NHC-monophosphate. Migration pattern of the products of RNA synthesis catalyzed by SARS-CoV-2 RdRp complex along the RNA primer/template as shown at the top of the panel. RNA primer/template supports a single incorporation event of CMP or NHC-monophosphate as a C-analogue at position 6. G indicates incorporation of [α-32P]-GTP at position 5. A 5′-32P-labeled 4-nt primer (4) serves as a size marker (m). Asterisk indicates products of the terminal transferase activity. NHC-TP, β-D-N4-hydroxycytidine 5’-triphosphate.
RNA synthesis using NHC-MP embedded in the template
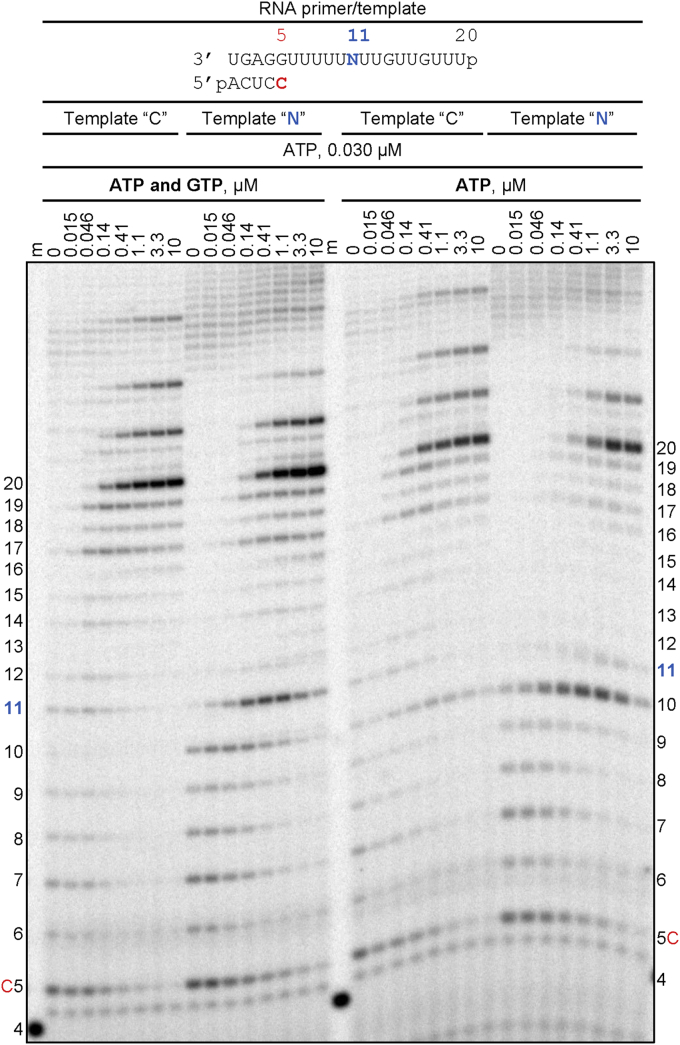
In the absence of significant RNA synthesis inhibition, the copy of the viral genome is likely synthesized in full length and contains embedded NHC-MP residues. Hence, efficiency and fidelity of RNA synthesis may be affected at a later stage when this strand is utilized as a template. To address this question, we have synthesized an RNA template with a single NHC-MP using T7 RNA polymerase (Fig. S1). For comparative purpose, we generated two model RNAs with either a single CMP (Template “C”) or a single NHC-MP (Template “N”) at position 11. To test whether an incorporated NHC-MP still acts as a C-analogue when present in the template we monitored RNA synthesis in the presence of increasing concentrations of GTP (Fig. 3). Incorporation of GTP opposite CMP (Template “C”) is very efficient even at concentrations as low as 0.015 μM. In contrast, the incorporation of GTP opposite NHC-MP is less efficient and concentrations of 0.41 μM (or 27-fold higher) GTP are required to completely convert the 10-nt product to 11-nt product. Note that, even though RNA synthesis past the 11-nt product along template “C” is not very efficient, the full-length products are formed concomitantly with GTP incorporation at position 11. Hence, the full-length product formation depends solely on ATP concentration, which is kept here low at 0.030 μM in order to reduce misincorporations. Another caveat is that concentrations of GTP above 4 μM results in U:G misincorporation on both templates (Fig. 3, asterisks). Full-length product formation along template “N” is less evident than with template “C,” suggesting that overall RNA synthesis might be inhibited. The accumulation of 11-nt product points to a specific site of inhibition.
Figure 3.
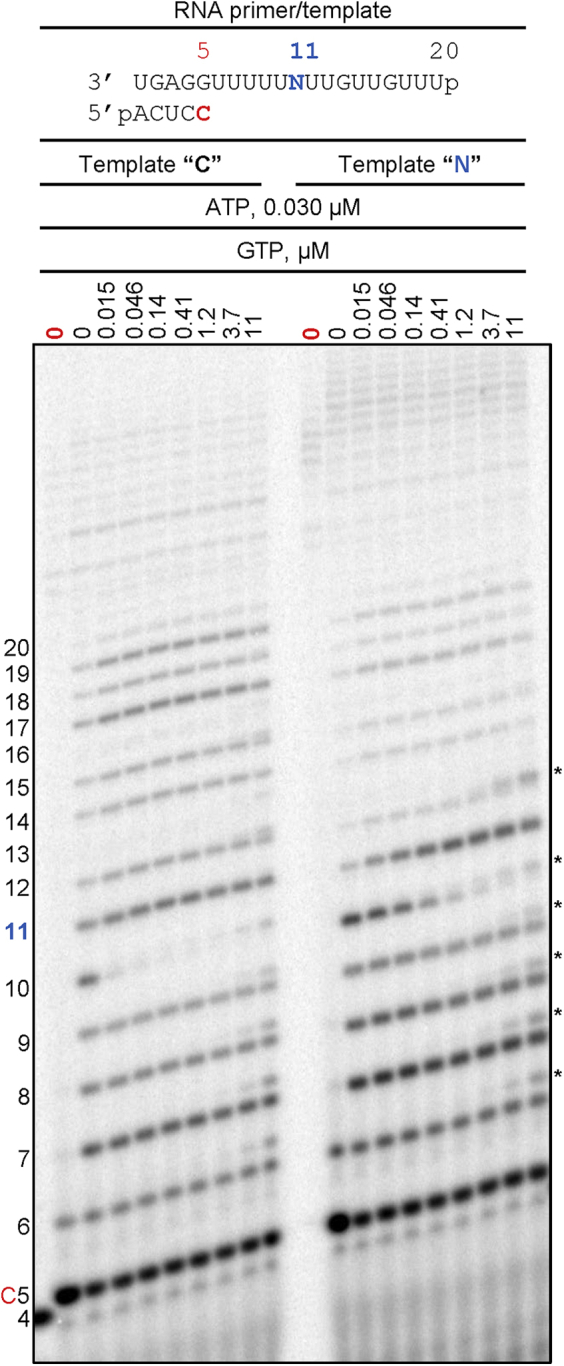
RNA synthesis with NHC-MP in the template strand. Migration pattern of the reaction products catalyzed by SARS-CoV-2 RNA-dependent RNA polymerase. The template contains an embedded NHC-MP at position 11 (Template “N”) or CMP (Template “C”). Reactions with [α-32P]-CTP as the only NTP are indicated by “0” in red. Figure notations are as in Figure 1. Asterisks indicate products of GTP misincorporation opposite U in the template. NHC-MP, β-D-N4-hydroxycytidine 5’-monophosphate.
To avoid the confounding effect of nucleotide misincorporations, we monitored RNA synthesis also in the presence of the other required NTPs. For template N, increasing concomitantly the concentrations of GTP and ATP yields transiently an 11-nt product that is almost completely converted into full-length product at NTP concentrations as low as 10 μM (Fig. 4, panel “ATP and GTP”). The intermediate 11-nt product is not seen with template C, which shows specific inhibition by NHC-MP in the template. The 11-nt product is also not seen when increasing the concentration of ATP in the absence of GTP (Fig. 4, panel “ATP”), which demonstrates that incorporation of GTP opposite NHC-MP is the cause for inhibition. Thus, although both GTP and ATP can be incorporated opposite NHC-MP with similar efficiencies, only GTP causes a subtle inhibitory effect that can be overcome by increasing NTP concentrations. We have also shown that neither CTP nor UTP is incorporated opposite NHC-MP (Fig. S2). Taken together, NHC-MP embedded in the template shows ambiguous base pairing with GTP and ATP. At the level of incorporation, NHC-TP shows a preference for template G.
Figure 4.
Mechanism of template-dependent inhibition of SARS-CoV-2 RNA-dependent RNA polymerase complex. Figure notations are as in Figure 2. Signal accumulation at position 11 illustrates inhibition of nucleotide incorporation right after template-embedded β-D-N4-hydroxycytidine 5’-monophosphate, which can be overcome with increasing concentrations of NTP.
Discussion
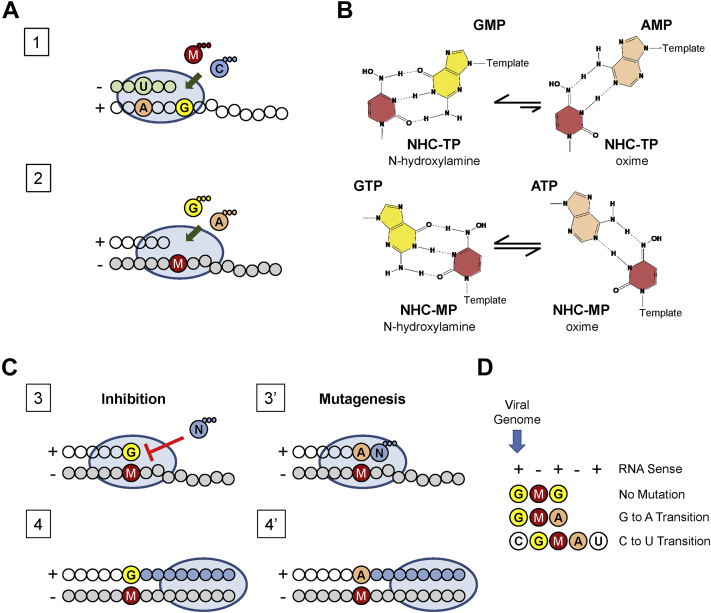
The broad-spectrum antiviral agent molnupiravir, a prodrug of NHC, is currently being evaluated in a clinical phase 3 trial for the treatment of SARS-CoV-2 infections (22). The drug is orally bioavailable and can be given to outpatients early in the disease with the potential to reduce hospitalizations. Preclinical data in cell culture revealed a dose-dependent increase in G to A and C to U transition mutations that correlated with increases in antiviral effects against coronaviruses (10, 13). Molnupiravir is therefore classified as a mutagenic nucleotide analogue. Here we studied the underlying biochemical mechanisms with the purified RdRp complex of SARS-CoV-2. Based on our data, we developed a model that describes effects on both efficiency and fidelity of RNA synthesis (Fig. 5).
Figure 5.
Mutagenesis model of NHC against SARS-CoV-2.A, schematic representation of SARS-CoV-2 RNA-dependent RNA polymerase (oval)-mediated nucleotide incorporation into RNA primer (gray circles)/template (white circles). Plus and minus signs indicate RNA sense. Letters A, C, G, and U refer to natural nucleotide bases. Letter M refers to molnupiravir. Three small circles refer to a triphosphate moiety of the NTP. B, alternative base pairing of NHC base moiety is supported by its tautomerization. The N-hydroxylamine form is dominant when NHC-TP is the substrate, whereas both the N-hydroxylamine and the oxime form are available when NHC-MP is embedded in the template. C, mechanism of viral inhibition and mutagenesis by template-embedded NHC-MP. Blue circles illustrate NTP incorporation past NHC-MP in the template. D, summary of NHC-mediated inhibitory and mutagenic effects on viral replication. MP, monophosphate; NHC, β-D-N4-hydroxycytidine; TP, 5’-triphosphate.
Steady-state kinetic measurements demonstrated that NHC-TP acts predominantly as a C-analogue and is preferentially incorporated opposite template G (Fig. 5A, step 1). Selective incorporation, defined as efficiency of incorporation of CTP over efficiency of incorporation of NHC-TP, is relatively low with a value of 30. By comparison, the selectivity value of RDV-TP is below one, suggesting that RDV-TP is more efficiently incorporated than its natural counterpart ATP (2, 32). However, earlier studies have shown that NHC, in contrast to RDV, is resistant to the coronavirus-associated proofreading exonuclease activity (10). For hepatitis C virus RdRp, the selectivity value of the 5’-triphosphate metabolite of sofosbuvir is also relatively low (45-fold), but in the absence of a proofreading activity, sofosbuvir is an efficient inhibitor of hepatitis C virus replication (35). Thus, the limited opportunities for incorporation of NHC-TP may still have an impact on efficiency and fidelity of viral genome replication.
When NHC-MP is present in the template, base pairing is more ambiguous and both the incoming GTP and ATP are accepted with no significant preference (Fig. 5A, step 1). Like other mutagenic nucleotides, NHC-TP likely exists in different tautomeric forms that affect base pairing (36). The hydroxylamine form acts like C and enables base pairing with G, whereas the oxime form (C=NOH) acts like U and allows base pairing with A (Fig. 5B). Our data suggest that the NHC-TP substrate exists predominantly in its hydroxylamine form and acts like CTP; however, when present as NHC-MP in the template, both tautomeric forms seem to coexist and act like CTP or UTP, favoring incorporation of GTP and ATP, respectively.
Incorporation of GTP opposite NHC-MP inhibits incorporation of the next incoming nucleotide. Increasing NTP concentrations to 10 μM can overcome this obstacle (Fig. 5C, steps 3 and 4). This inhibitory effect on RNA synthesis is not observed with ATP (Fig. 5C, steps 3’ and 4’). Instead, incorporation of ATP yields a G to A transition mutation via G:NHC-TP and NHC-MP:A base pairing or short G:NHC:A (Fig. 5D). C to U transitions are realized if the positive-sense viral RNA contains a C via C:G:NHC:A:U. Thus, the preference for the G:NHC:A pattern is necessary and sufficient to explain the higher frequencies of G to A and C to U transition mutations in the presence of molnupiravir or NHC. The same mechanism may also apply to influenza virus that shows the same transition mutations in the presence of the drug, but not to respiratory syncytial virus that shows a different pattern (6, 12). However, biochemical data are generally lacking, and it will be important to study potential differences among various viral and cellular polymerases that utilize NHC-TP as substrate. In the present study, we have shown that the incorporated NHC-MP can either inhibit RNA synthesis in G:NHC:G or act as a mutagen in G:NHC:A. The mutagenic effect seems to be dominant given that increasing NTP concentrations can overcome G:NHC:G inhibition at relatively low concentrations of GTP. In addition, considering that intracellular concentrations of ATP are several-fold higher than that of GTP (37), the G:NHC:A pairing may also be favored in a cellular environment.
Experimental procedures
Nucleic acids and chemicals
[α-32P]NTP, RNA primers and templates (except RNA templates with embedded NHC-MP) used in this study were 5′-phosphorylated and purchased from PerkinElmer. NHC-TP were from two sources: Dr Schinazi laboratory and from MedChemExpress. NTPs were purchased from GE Healthcare.
Protein expression and purification
The SARS-CoV-2 RdRp complex was produced by expressing nsp-5, -7, -8, and -12 as a polyprotein by employing a baculovirus expression system and purifying the nsp-7-8-12 complex through Ni-NTA affinity chromatography on nsp-8 N-terminal histidine tag as described (2).
NTP incorporation and the effect of primer- or template-embedded NHC-MP on viral RNA synthesis
NTP incorporation by SARS-CoV-2 RdRp and data acquisition and quantification were done as reported by us (2, 32, 38). Enzyme concentration was 100 or 200 nM for single and multiple nucleotide incorporation assays, respectively. RNA synthesis incubation time was 10 min. Data from single nucleotide incorporation assays were used to determine the preference for the natural nucleotide over NHC-TP. The selectivity value is calculated as a ratio of the incorporation efficiencies of the natural nucleotide over the nucleotide analogue. The efficiency of nucleotide incorporation is determined by the ratio of Michaelis–Menten constants Vmax over Km. The substrate for nucleotide incorporation is a 5-nt primer generated by incorporation of [α-32P]NTP into a 4-nt primer. Formation of the 5-nt primer is maximal at a given time point; however, its precise concentration is unknown. Hence, the product generated in the reaction is measured by quantifying the signal corresponding to the 6-nt primer product and dividing it to the total signal in the reaction (5-nt primer and 6-nt primer). This defines the product fraction. The product fraction is commonly multiplied by the total substrate concentration in order to determine the molar units of the Vmax, which is here not possible as explained above. Therefore, the unit of Vmax is reported as product fraction over time. The selectivity value is unitless as it is the ratio of two Vmax/Km measurements with the same units. RNA templates with embedded NHC-MP were produced as described by us (38). NHC-related protocol modifications are explained in Fig. S1.
Data availability
All data are contained within the article.
Supporting information
This article contains supporting information (38).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Emma Woolner and Dr Dana Kocinkova for excellent technical assistance.
Author contributions
M. G., C. J. G., and E. P. T. conceptualization; C. J. G., E. P. T., and R. F. S. methodology; E. P. T. software; M. G., C. J. G., and E. P. T. validation; M. G., C. J. G. and E. P. T. formal analysis; C. J. G. and E. P. T. investigation; M. G. and R. F. S. resources; M. G., C. J. G. and E. P. T. data curation; M. G. and E. P. T. writing-original draft; M. G., C. J. G., E. P. T. and R. F. S. writing-review and editing; M. G., C. J. G., and E. P. T. visualization; M. G. supervision; M. G. project administration; M. G. funding acquisition.
Funding and additional information
This study was supported by grants to M. G. from the Canadian Institutes of Health Research (CIHR, grant number 170343), and from the Alberta Ministry of Economic Development, Trade and Tourism by the Major Innovation Fund Program for the AMR – One Health Consortium. R. F. S. is supported by NIH CFAR grant P30AI050409 and NSF award # 2032273. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Biography

Calvin Gordon is a PhD student in the Department of Medical Microbiology and Immunology at University of Alberta. The focus of Calvin’s research is on broad-spectrum antivirals and their mechanism of action against various RNA viral polymerases. He is currently investigating mutagenic mechanisms of molnupiravir against RNA viruses that possess high epidemic potential.
Edited by Patrick Sung
Supporting information
References
- 1.U.S. Food and Drug Administration . U.S. Food and Drug Administration; Silver Spring, MD: 2020. Fact Sheet for Health Care Providers Emergency Use Authorization (EUA) of Remdesivir (GS-5734TM) [Google Scholar]
- 2.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Gotte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.U.S. Food and Drug Administration . U.S. Food and Drug Administration; Silver Spring, MD: 2020. Fact Sheet for Health Care Providers Emergency Use Authorization (EUA) of Casirivimab and Imdevimab. [Google Scholar]
- 4.Vasudevan N., Ahlqvist G.P., McGeough C.P., Paymode D.J., Cardoso F.S.P., Lucas T., Dietz J.P., Opatz T., Jamison T.F., Gupton F.B., Snead D.R. A concise route to MK-4482 (EIDD-2801) from cytidine. Chem. Commun. (Camb.) 2020;56:13363–13364. doi: 10.1039/d0cc05944g. [DOI] [PubMed] [Google Scholar]
- 5.Stuyver L.J., Whitaker T., McBrayer T.R., Hernandez-Santiago B.I., Lostia S., Tharnish P.M., Ramesh M., Chu C.K., Jordan R., Shi J., Rachakonda S., Watanabe K.A., Otto M.J., Schinazi R.F. Ribonucleoside analogue that blocks replication of bovine viral diarrhea and hepatitis C viruses in culture. Antimicrob. Agents Chemother. 2003;47:244–254. doi: 10.1128/AAC.47.1.244-254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoon J.J., Toots M., Lee S., Lee M.E., Ludeke B., Luczo J.M., Ganti K., Cox R.M., Sticher Z.M., Edpuganti V., Mitchell D.G., Lockwood M.A., Kolykhalov A.A., Greninger A.L., Moore M.L. Orally efficacious broad-spectrum ribonucleoside analog inhibitor of influenza and respiratory syncytial viruses. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.00766-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costantini V.P., Whitaker T., Barclay L., Lee D., McBrayer T.R., Schinazi R.F., Vinje J. Antiviral activity of nucleoside analogues against norovirus. Antivir. Ther. 2012;17:981–991. doi: 10.3851/IMP2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reynard O., Nguyen X.N., Alazard-Dany N., Barateau V., Cimarelli A., Volchkov V.E. Identification of a new ribonucleoside inhibitor of Ebola virus replication. Viruses. 2015;7:6233–6240. doi: 10.3390/v7122934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Urakova N., Kuznetsova V., Crossman D.K., Sokratian A., Guthrie D.B., Kolykhalov A.A., Lockwood M.A., Natchus M.G., Crowley M.R., Painter G.R., Frolova E.I., Frolov I. beta-d-N (4)-Hydroxycytidine is a potent anti-alphavirus compound that induces a high level of mutations in the viral genome. J. Virol. 2018;92 doi: 10.1128/JVI.01965-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agostini M.L., Pruijssers A.J., Chappell J.D., Gribble J., Lu X., Andres E.L., Bluemling G.R., Lockwood M.A., Sheahan T.P., Sims A.C., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G.R., Baric R.S. Small-molecule antiviral beta-d-N (4)-hydroxycytidine inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance. J. Virol. 2019;93 doi: 10.1128/JVI.01348-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehteshami M., Tao S., Zandi K., Hsiao H.M., Jiang Y., Hammond E., Amblard F., Russell O.O., Merits A., Schinazi R.F. Characterization of beta-d-N(4)-hydroxycytidine as a novel inhibitor of Chikungunya virus. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.02395-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toots M., Yoon J.J., Cox R.M., Hart M., Sticher Z.M., Makhsous N., Plesker R., Barrena A.H., Reddy P.G., Mitchell D.G., Shean R.C., Bluemling G.R., Kolykhalov A.A., Greninger A.L., Natchus M.G. Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aax5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schafer A., Dinnon K.H., 3rd, Stevens L.J., Chappell J.D., Lu X., Hughes T.M., George A.S., Hill C.S. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenke K., Hansen F., Schwarz B., Feldmann F., Haddock E., Rosenke R., Barbian K., Meade-White K., Okumura A., Leventhal S., Hawman D.W., Ricotta E., Bosio C.M., Martens C., Saturday G. Orally delivered MK-4482 inhibits SARS-CoV-2 replication in the Syrian hamster model. Nat. Commun. 2021;12:2295. doi: 10.1038/s41467-021-22580-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox R.M., Wolf J.D., Plemper R.K. Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets. Nat. Microbiol. 2021;6:11–18. doi: 10.1038/s41564-020-00835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wahl A., Gralinski L.E., Johnson C.E., Yao W., Kovarova M., Dinnon K.H., 3rd, Liu H., Madden V.J., Krzystek H.M., De C., White K.K., Gully K., Schafer A., Zaman T., Leist S.R. SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801. Nature. 2021;591:451–457. doi: 10.1038/s41586-021-03312-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sledziewska E., Janion C. Mutagenic specificity of N4-hydroxycytidine. Mutat. Res. 1980;70:11–16. doi: 10.1016/0027-5107(80)90053-6. [DOI] [PubMed] [Google Scholar]
- 18.Janion C., Glickman B.W. N-4-Hydroxycytidine - a mutagen specific for at to Gc transitions. Mutat. Res. 1980;72:43–47. doi: 10.1016/0027-5107(80)90218-3. [DOI] [PubMed] [Google Scholar]
- 19.Hernandez-Santiago B.I., Beltran T., Stuyver L., Chu C.K., Schinazi R.F. Metabolism of the anti-hepatitis C virus nucleoside beta-D-N4-hydroxycytidine in different liver cells. Antimicrob. Agents Chemother. 2004;48:4636–4642. doi: 10.1128/AAC.48.12.4636-4642.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janion C. On the different response of Salmonella typhimurium hisG46 and TA1530 to mutagenic action of base analogues. Acta Biochim. Pol. 1979;26:171–177. [PubMed] [Google Scholar]
- 21.Zhou S., Hill C.S., Sarkar S., Tse L.V., Woodburn B.M.D., Schinazi R.F., Sheahan T.P., Baric R.S., Heise M.T., Swanstrom R. beta-D-N 4-hydroxycytidine (NHC) inhibits SARS-CoV-2 through lethal mutagenesis but Is also mutagenic to mammalian cells. J Infect Dis. 2021 doi: 10.1093/infdis/jiab247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Painter W.P., Holman W., Bush J.A., Almazedi F., Malik H., Eraut N., Morin M.J., Szewczyk L.J., Painter G.R. Human safety, tolerability, and pharmacokinetics of molnupiravir, a novel broad-spectrum oral antiviral agent with activity against SARS-CoV-2. Antimicrob. Agents Chemother. 2021 doi: 10.1128/AAC.02428-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie X., Muruato A.E., Zhang X., Lokugamage K.G., Fontes-Garfias C.R., Zou J., Liu J., Ren P., Balakrishnan M., Cihlar T., Tseng C.K., Makino S., Menachery V.D., Bilello J.P., Shi P.Y. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat. Commun. 2020;11:5214. doi: 10.1038/s41467-020-19055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shannon A., Selisko B., Le N.T., Huchting J., Touret F., Piorkowski G., Fattorini V., Ferron F., Decroly E., Meier C., Coutard B., Peersen O., Canard B. Rapid incorporation of Favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis. Nat. Commun. 2020;11:4682. doi: 10.1038/s41467-020-18463-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crotty S., Maag D., Arnold J.J., Zhong W., Lau J.Y., Hong Z., Andino R., Cameron C.E. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 26.Ferron F., Subissi L., Silveira De Morais A.T., Le N.T.T., Sevajol M., Gluais L., Decroly E., Vonrhein C., Bricogne G., Canard B., Imbert I. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E162–E171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith E.C., Blanc H., Surdel M.C., Vignuzzi M., Denison M.R. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: Evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sticher Z.M., Lu G., Mitchell D.G., Marlow J., Moellering L., Bluemling G.R., Guthrie D.B., Natchus M.G., Painter G.R., Kolykhalov A.A. Analysis of the potential for N (4)-hydroxycytidine to inhibit mitochondrial replication and function. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.01719-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tchesnokov E.P., Raeisimakiani P., Ngure M., Marchant D., Gotte M. Recombinant RNA-dependent RNA polymerase complex of Ebola virus. Sci. Rep. 2018;8:3970. doi: 10.1038/s41598-018-22328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tchesnokov E.P., Feng J.Y., Porter D.P., Gotte M. Mechanism of inhibition of Ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11:326. doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tchesnokov E.P., Bailey-Elkin B.A., Mark B.L., Gotte M. Independent inhibition of the polymerase and deubiquitinase activities of the Crimean-Congo Hemorrhagic Fever Virus full-length L-protein. PLoS Negl. Trop. Dis. 2020;14 doi: 10.1371/journal.pntd.0008283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Gotte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020;295:4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deval J., Hong J., Wang G., Taylor J., Smith L.K., Fung A., Stevens S.K., Liu H., Jin Z., Dyatkina N., Prhavc M., Stoycheva A.D., Serebryany V., Liu J., Smith D.B. Molecular basis for the selective inhibition of respiratory syncytial virus RNA polymerase by 2'-fluoro-4'-chloromethyl-cytidine triphosphate. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tvarogova J., Madhugiri R., Bylapudi G., Ferguson L.J., Karl N., Ziebuhr J. Identification and characterization of a human coronavirus 229E nonstructural protein 8-associated RNA 3'-terminal adenylyltransferase activity. J. Virol. 2019;93 doi: 10.1128/JVI.00291-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fung A., Jin Z., Dyatkina N., Wang G., Beigelman L., Deval J. Efficiency of incorporation and chain termination determines the inhibition potency of 2'-modified nucleotide analogs against hepatitis C virus polymerase. Antimicrob. Agents Chemother. 2014;58:3636–3645. doi: 10.1128/AAC.02666-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jena N.R. Role of different tautomers in the base-pairing abilities of some of the vital antiviral drugs used against COVID-19. Phys. Chem. Chem. Phys. 2020;22:28115–28122. doi: 10.1039/d0cp05297c. [DOI] [PubMed] [Google Scholar]
- 37.Traut T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 38.Tchesnokov E.P., Gordon C.J., Woolner E., Kocinkova D., Perry J.K., Feng J.Y., Porter D.P., Gotte M. Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action. J. Biol. Chem. 2020;295:16156–16165. doi: 10.1074/jbc.AC120.015720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.