Abstract
The outbreak of novel coronavirus strain (Covid-19) with a high pandemic threat has predict grave public health and economic concerns. This virus, originating from the Wuhan region in China has spread worldwide affecting millions with no registered persuasive targeted therapy. In this paper, we analyze the three important proteins encoded by the virus, envelope protein 5 × 29, RNA binding nucleocapsid protein 1SSK, and spike glycoprotein 6ACD, for an effective virion accumulation, and remdesivir was the first drug approved by the FDA and EMA for the treatment of COVID-19 cases that require hospitalization, there is still much controversy about its efficacy and also an alternative for novel phytochemicals, deoxynojirimycin, trigoneoside IB, and octanoic acid. The in-silico evaluations were conducted using the PyRx virtual screening tools which lead to the target based on high binding affinity. Trigoneoside IB, derived from Trigonella foenum-graecum (Fenugreek), showed the highest binding affinity and stable interaction with the amino acid residues present in active sites of Covid-19 proteins. Meanwhile, the other two compounds derived from Morus alba (Mulberry) and Morinda citrifolia (Noni), as well as the anti-HIV remdesivir drug exhibited good binding affinity and favorable ADME properties. Thereby offering scope for validation of the new therapeutic components for their in vitro and in vivo efficacy against the Covid-19 proteins.
Keywords: Covid-19, Phytochemicals, Molecular docking, Trigoneoside, Fenugreek
1. Introduction
The recent outbreak of the novel coronavirus (Covid-19) mainly affects the mammalian respiratory system, resulting in pneumonia-like conditions with mild to severe respiratory complications. The SARS‐CoV‐2, responsible for a pandemic now infamously known as the Covid-19 as termed by the World Health Organization in January 2019 (Chhikara et al., 2020; Kumar et al., 2020), belongs to the family of zoonotic viruses that caused the severe acute respiratory syndrome (SARS) and the middle east respiratory syndrome (MERS) in 2003 and 2012, respectively (Chhikara et al., 2020, Yin and Wunderink, 2018). In February 2020, the 2019-nCoV virus was alternatively named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the International Committee on Taxonomy of Viruses (ICTV) (Chan et al., 2020; Lefkowitz et al., 2018). The Covid-19 virus is characterized by clinical symptoms of common cold, cough, breathlessness, dyspnea, and fever in mild cases, with a hint of pneumonia in severe cases which is eventually followed by death (Huang et al., 2020). So far, there is no effective prophylaxis available for the infection and the current global scenario indicating the urgent need for the same.
The agents of the coronavirus family hold up an enveloped RNA-positive genome, categorized into four different α, β, γ, and δ genera (Yudong et al., 2020). The SARS-CoV-2 belongs to the β genus and the major structural proteins encoded by the coronaviral genome are the spike (S) protein, nucleocapsid (N) protein and the envelope (E) protein (Wrapp et al., 2020; Chen et al., 2020, Yang et al., 2020). However, it has been proven that some CoVs do not require the full ensemble of structural proteins to form a complete, infectious virion. Nucleocapsid proteins are phosphoproteins that are capable of binding to the CoV RNA genome and have a vital role in virion structure, replication and transcription of CoVs. The smallest of these key proteins is the E protein which is amply expressed inside the infected cell and is involved in viral assembly, discharge of virions and pathogenesis of the virus (Schoeman et al., 2019). The Spike glycoproteins populate the surface of the virus and determine host tropism by facilitating attachment of the virus to the host cell surface receptors and consequent blend between the viral and host cell membranes to mediate viral entry into the host cell. The coronavirus S protein may exist in two different conformations occurring either prefusion or post-fusion and the mechanism of transmission from the prefusion to post-fusion conformation triggers the formation of trimeric class I fusion protein, a metastable prefusion conformation (Masters et al., 2006; Pradesh et al., 2014) . Treatment of coronavirus infection is broadly classified into two groups based on drug targets (Huang et al., 2020) . While the first group of treatments orients itself towards destabilizing the virus proteins, the second class focusses on stabilizing the human immune system to ward of the virus by enhancing the innate immune response and subsequently blocking the necessary signaling pathways for the viral replication to occur. Such a resilience of the immune system is termed an effective anti-corona virus effect (Siu et al., 2008). The SARS-CoV-2 has been reported to invade the human cells through a viral-host cell surface interaction mediated through glycosylated spike protein (S-protein). The interaction involves the binding of the S-protein to the host cell surface with the aid of the ACE-2 receptor and thereby releasing the RNA-payload into the cell, which eventually occupies the host endoplasmic reticulum (ER) to replicate. Thus, the viral RNA molecules are transported to the host cell surface by the means of vesicles and released via exocytosis (Zhenming et al., 2020).
Since these viruses depend on the adhesion to host cell membranes for their survival, the targeting drugs can be designed to prevent the entry of the lethal SARS-CoV-2 RNA to the inside (Jin et al., 2020). Traditional medicine presents us with a plethora of such plants that are already of therapeutic value to alleviate viral, bacterial, as well as chronic diseases linked with pathogen infection and lifestyle (Dash et al., 2011). Supporting this, evaluation of antiviral capabilities of various phytocompounds has been conducted in the recent past (Zumla et al., 2016) and plants like mulberry, fenugreek, and noni were found to be the rich sources of such antiviral phytoagents of novelty (Naithani et al., 2008; Dash et al., 2011; Bin-Hafeez et al., 2003).
The anti-influenza virus activities of mulberry have been reported by Kim and Chung (2018). The study was conducted using time-of-addition plaque assays and the conclusions indicated the scope for development of a novel plant-derived anti-influenza drug with potent activity against the H1N1-BR59, H1N1-KR01, H3N2-BR10, and FL04 strains (Lee et al., 2014). Mulberry phytochemicals were also reported for their efficacy against the murine norovirus-1 (MNV-1) and feline calicivirus – F9 (FCV-F9) used as the human norovirus surrogates in cytopathic effect inhibition, plaque reduction and RNA expression studies (Jacob et al., 2007). Additionally, the plant was found to be hepatoprotective with capabilities to combat Hepatitis B and C viruses via viral envelope proteins glycosylation inhibition by Deoxynojirimycin. Mohammadi et al. reported that, mulberry leaves used as a traditional Chinese medicine for expectorant, antiphlogistic, antidiabetic etc (Mohammadi et al., 2008). Moreover, the iminosugar derivatives of Morus alba exert antiviral effects which including Japanese encephalitis viruses (RNA virus), HBV (DNA virus), HIV (RNA virus) and Dengue (RNAvirus) (Durantel et al., 2001). In 2015 Dkhil et al., also suggest the anti-herpes simplex virus's activity using morus alba leaves extract (Hamden, et al., 2011; Dkhil et al., 2015). Na et al. reported the inhibition of adenovirus 36 (Ad36) by the anti-inflammatory properties of mulberry extracts (Na et al., 2014). Fenugreek, one of the oldest known plants of significant medicinal values, contains a range of phytochemicals from vitamins and essential volatile oils to alkaloids like Trigonelline and flavonoids like Kaempferol and Luteolin (Sharma et al., 2017). In a study on the efficacy of clove, fenugreek seeds, garlic, onion, ginger, and jalapeno pepper against the foodborne human norovirus, fenugreek treatment resulted in a dose-dependent reduction of the surviving feline calicivirus titers (Ahmad et al., 2016; Aboubakr et al., 2016, Moradi-Kor and Moradi, 2013). The plant belonging to the Fabaceae Family has been extensively studied for its antiviral properties (Oluwaseun et al., 2020). The seeds and leaves of Trigonella foenum-graecum were commonly used a food and they are well known for their traditional medicine across the globe. The biological profiles of Fenugreek include antioxidant, antidiabetic (Oluwaseun et al., 2020), hepatoprotective (Vats et al., 2002; Mathern et al., 2009). The also possesses hypoglycaemic (Bilal et al., 2003), antiulcerogenic and chemoprotectives (Sowmya et al., 1999). It also proven that Trigonella foenum-graecum as a potential immunostimulatory effects (Javan et al., 1997). Likewise, luteolin, a medically important phytochemical found in fenugreek, was also reported to be efficient in alleviating a variety of viral infections such as tick-borne encephalitis (Krylova et al., 2011), Japanese encephalitis (Fan et al., 2016), hand, foot, and mouth disease (HFMD) (Dai, et al., 2019), SARS (Yi et al., 2004) and AIDS (Mehla et al., 2011). In addition, recent molecular docking studies by Khaerunnisa et al. also endorse the antiviral potentials of both kaempferol and luteolin (Khaerunnisa et al., 2020).
Noni has been used for over 2000 years in the folk remedies for diseases ranging from hypotension to cancer. The Polynesian traditional medicinal practices acknowledged the use of the plant for its therapeutic use during viral infections (Wang et al., 2002). Ratnoglik et al. suggest that the noni leaf phytochemicals are active agents against the Hepatitis C virus (Ratnoglik et al., 2014; Suratno et al., 2014). Two novel anthraquinones extracted from the plant were reported to be acting weakly against the proliferation of H1N1, H3N2 as well as S. aureus (Wang et al., 2016). Even a patent filed at the United States of America highlights the antiviral use of noni based formulations, with particular reference to HIV therapeutics (Palu et al., 2010). Selvam et al. concluded that the extract of noni does exhibit a cytopathic effect against both the hepatitis C virus and HIV (Selvam et al., 2009; Baell et al., 2016). Not with standing, more research on the antiviral efficacy of the plant phytochemicals may be deemed imperative.
The SARS-CoV-2 main protease (MPro) was successfully crystallized by Liu et al. opening up the opportunity for the development of target-directed potent drug candidates (Schoeman et al., 2019). In this study, we have considered the RNA-binding domain of SARS-CoV-2 nucleocapsid (N), spike (S), and envelope (E) proteins as potential targets for the development of a drug against the COVID-19. The necessary structural details of the above-mentioned proteins were retrieved from Protein Data Bank (Daniel et al., 2020) . Taking leads from earlier reports, we have considered evaluating the anti-SARS-CoV-2 potential of the anti-viral molecules Deoxynojirimycin, Trigoneoside IB, and Octanoic acid isolates from mulberry leaves, fenugreek, and noni, respectively, alongside the anti-HIV drug remdesivir, in an attempt to evaluate the possibilities of drug repurposing against the COVID-19 disease Fig. 1 .
Fig. 1.
Flowchart representing inhibition of selected 2019-nCoV virus proteins using various phytochemical and anti-viral compounds.
2. Materials and methods
2.1. Ligand's selection and preparation
The two-dimensional structure file of the selected ligand molecules deoxynojirimycin, trigoneoside IB, octanoic acid, and remdesivir were obtained from the PubChem chemistry database The Pubchem Project (Database) 2020 maintained by the National Centre for Biotechnology Information (NCBI), Bethesda, USA (https://pubchem.ncbi.nlm.nih.gov). To do the molecular docking studies the three-dimensional structured .pdb format files of the ligand are a must, hence the 2D structured file .sdf is converted to .pdb format by addition of hydrogen bonds and the coordinates generate using the community-driven, open-source molecular modeling tool Open Babel (http://openbabel.org/). Further geometric augmentation of the ligands was carried out with the free molecular docking program, Argus Lab 4.0.1 (http://www.arguslab.com/).
2.2. Protein preparation
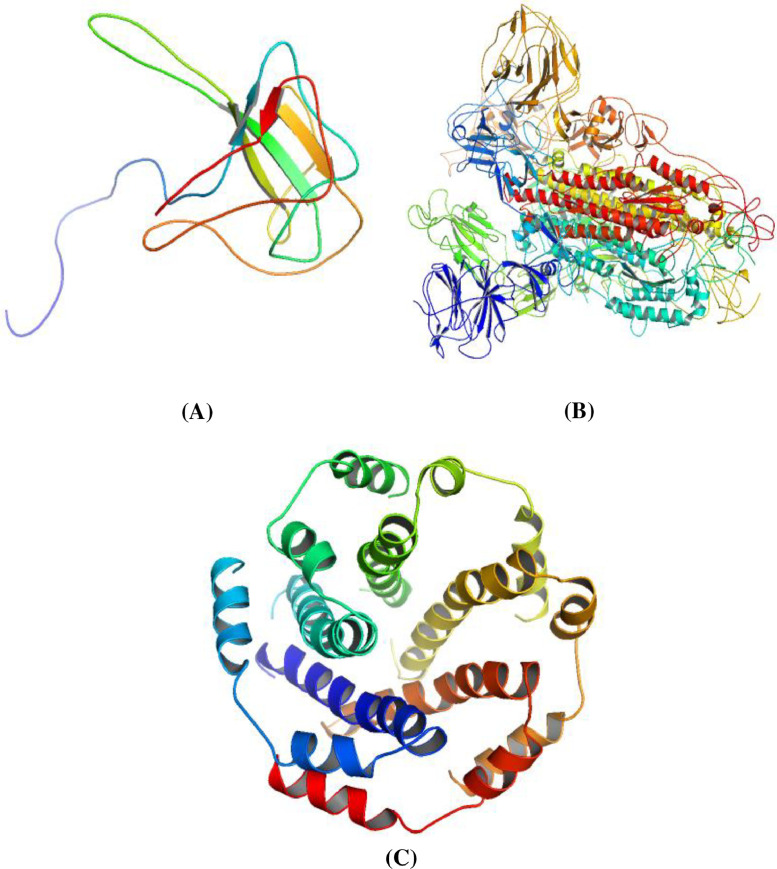
The previously reported 3-dimensional structures of the N-terminal RNA-binding domain of the SARS-CoV nucleocapsid protein (PDB ID: 1SSK), A chain of SARS-CoV spike glycoprotein (PDB ID: 6ACD) and SARS Coronavirus E protein pentameric ion channel (PDB ID: 5 × 29) are retrieved from RCSB Protein data bank (RCSB PDB 2020)(https://www.rcsb.org/). The downloaded protein PDB files were further visualized and edited or modified using chimera software by removing the non-standard amino acid residues present attached to the protein (Pettersen et al., 2000). Before starting the docking analysis, the ligand-binding site pocket residues of the selected proteins were predicted using the In-silico tool Galaxy Site of the Galaxy web server shown in Fig. 2 .
Fig. 2.
Crystal structure of Covid-19, A) 1SSK, B) 6ACD, and C) 5 × 29 were selected from Protein Data Bank and visualized using PyMol software.
2.3. Protein structure validation
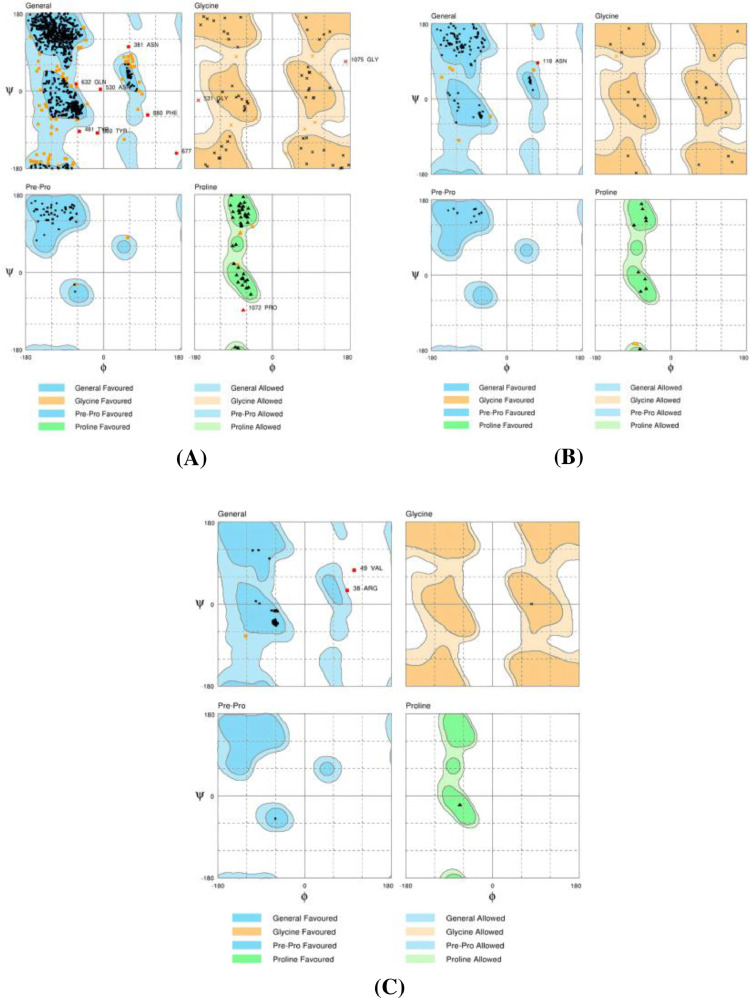
The protein structures were refined using Galaxy protein refinement (http://galaxy.seoklab.org/). The refined protein structures of SARS-CoV-2 were validated and evaluated using the Ramachandran plot, showing energetically allowed regions for backbone dihedral angles ψ against ϕ of amino acid residues in 1SSK, 6ACD, and 5 × 29 protein structures. The plots were generated by RAMPAGE (Bosco et al., 2005; Ho and Brasseur, 2005). The obtained RAMPAGE results of the proteins demonstrated that protein structures are stable (Fig. 3).
Fig. 3.
Ramachandran Plot Generated by RAMPAGE. Ramachandran plot showing energetically allowed regions for backbone dihedral angles ψ against ϕ of amino acid residues in Covid-19 modeled of A)-1SSK, B) −6ACD and C) −5 × 29) proteins structure.
2.4. Molecular docking studies
To understand the interaction between the SARS-CoV-2 proteins against therapeutic compounds, molecule interaction, and docking studies were performed using the PyRx v0.8 open access docking software (Trott et al., 2010; Trott and Olson, 2010). The selected protein and ligand files were uploaded in PDB format and the active binding site residues were shielded with a Grid box; the grid box was set for the selected binding site residues to obtain the best orientation with lowest binding affinity values (kcal/mol) (Table 1 ).
Table 1.
Natural therapeutics compounds with scientific name, molecular formula, compound name, and 2D structures.
| Sl.no | Plant Name | Scientific Name | Molecular Formula | Compound Name | 2D structure of compounds |
|---|---|---|---|---|---|
|
1. |
Mulberry leaves | Morus alba | C6H13NO4 | Deoxynojirimycin | 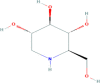 |
| 2. | Fenugreek | Trigonella foenum-graecum | C44H74O19 | Trigoneoside IB | 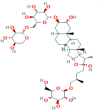 |
| 3. | Noni | Morinda citrifolia | C8H16O2 | Octanoic acid | |
| 4. | A Standard Drug |
Remdesivir | C27H35N6O8P | Remdesivir | 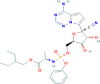 |
2.5. Molecular docking visualization
The obtained docked conformations of the ligand against the specific protein were visualized using PyMol v2.4 (Schrodinger) and LIGPLOT+ v1.4.4. This software was used to visualize the hydrogen bonds, bond length, and hydrophobic interactions between the SARS-CoV-2 proteins and selected compounds (Laskowski et al., 2011). PyMol was used for 3D visualization, whereas the Ligplot+ was used to generate 2D interaction between respective protein and their plant compound.
2.6. Molecular Dynamics simulation
The best protein-ligand docked complex with least binding energy and highest interactions were further analyzed in silico by molecular dynamic simulations using GROMACS software. To determine the conformational stability of protein and complex, simulations were performed for 30 ns. The parameter and topology files for the input ligand files were generated by SwissParam server. GROMOS96 43a1 forcefield was considered with SPC type of water model and triclinic box type was applied during the simulation. The energies and coordinates were saved every 50ps for analysis. Steepest descent minimization process was utilized for energy minimization for 50,000 steps. The plots were generated and visualized by Xmgrace tool (Jain et al., 2021).
3. Results
Since many researchers are struggling for the collaborative and astonishing effort for the development of a potential anti-SARS-CoV-2 drug. At this juncture, we hope that our contribution towards drug repurposing against target SARS-CoV-2 proteins. It can be a breakthrough for spreading this virus (Prajapat et al., 2020). Presently there is no effective vaccine developed yet, thus, in the present studies, we mainly focus on developing potent inhibitors using selected compounds against target SARS-CoV-2 proteins using bioinformatics tools.
3.1. Protein structure validation
The SARS-CoV-2 proteins were evaluated using Rampage. The Ramachandran plot of initial validation of the proteins showed less than 90% of residues in the allowed region but after the refinement of the proteins showed more than 90% of residues in the allowed region and thus making the proteins stable to carry out with the further molecular docking studies. The percentage of residues in favored, allowed, and outlier regions of all the three proteins are shown in Fig. 3 and Table 2 .
Table 2.
Ramachandran plot values showing the number of residues in favored, allowed, and outlier regions through the Rampage evaluation server.
| Sl. No. | COVID-19 Protein structure | Number of residues in the Favoured region (%) | Number of residues in the allowed region (%) | Number of residues in the outliner region (%) |
|---|---|---|---|---|
| 1 | 1SSK | 92.9 | 6.4 | 0.6 |
| 2 | 6ACD | 94.6 | 8.0 | 0.9 |
| 3 | 5 × 29 | 91.1 | 1.8 | 3.6 |
3.2. Molecular docking interactions
The results of this study can be clearly interpreted from Table 3 , the best-docked pose of all the molecular docked compounds was taken into consideration and the lowest corresponding binding affinity was noted down. These docked compounds were visualized and analyzed using PyMol and LIGPLOT+ software which highlights the nearby labeled binding residues. The Covid-19 main protease (1SSK, 6ACD, and 5 × 29) is seen sharing the hydrogen bonds which are < 3 Å in length with semi-circled hydrophobic interactions with selected compounds.
Table 3.
Molecular docking analysis of antiviral compounds against SARS-CoV-2 Major Protease (1SSK, 6ACD and 5 × 29).
| Sl. No. | COVID-19 Main Protease | Compound | Binding affinity (kcal/mol) | Hydrogen bond forming amino acid residues |
|---|---|---|---|---|
| 1 | 1SSK | Deoxynojirimycin | −4.8 | GLY-125, PHE-88, THR-27, TRP-30 |
| Trigoneoside IB | −7.6 | ARG-71, ARG-70, THR-60, SER-29, ALA-28. THR-27, GLy-125, ILE-124, TYR-90 | ||
| Octanoic acid | −4.6 | TYR-90, ILE-124 | ||
| Remdesivir | −7.4 | TYR-87,ARG-70 |
||
| 2 | 6ACD | Deoxynojirimycin | −5.6 | VAL-307,SER-577 |
| Trigoneoside IB | −8.5 | THR-260, ALA-618, SER-289, LYS-946,SER-279, THR-261, THR-616, GLY-288, THR-302, THE-285, CYS-288 | ||
| Octanoic acid | −4.6 | LEU-615 | ||
| Remdesivir | −7.8 | SER-289, THR-302, ASN- 888, THR-285, ASN-863 |
||
| 3 | 5 × 29 | Deoxynojirimycin | −4.5 | TYR-57, LEU-28, THR-35 |
| Trigoneoside IB | −7.5 | ILE-124, TYR-90, ARG-70, THR-60, SER-29, ALA-28, THR-27, GLy-125 | ||
| Octanoic acid | −4.6 | PHE-23 | ||
| Remdesivir | −7.8 | ILE-46, ASN-64, ARG-61 |
3.3. Docking interaction of 1SSK protein with selected ligands
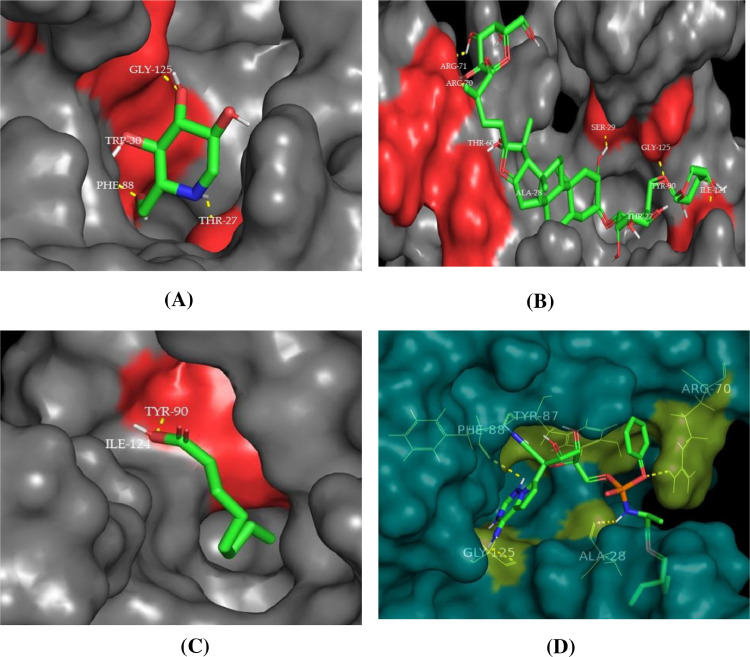
The Molecular Docking analysis and visualization of 1SSK binding protein with all four compounds are shown in Fig. 4 . Out of these compounds, the trigoneoside IB compound exhibited the highest binding affinity (−7.6 Kcal/mol) and ARG-71, ARG-70, THR-60, SER-29, ALA-28. THR-27, GLy-125, ILE-124, TYR-90 are the amino acid residues participating in the interaction at the binding pocket of 1SSK protein. Whereas deoxynojirimycin compound exhibited (−4.8 Kcal/mol), followed by octanoic acid compound exhibited (-4.6 Kcal/mol), finally an anti-HIV remdesivir compound exhibited (−7.4 Kcal/mol) binding affinity with 1SSK proteins and their amino acid residues participating in the interaction at the binding pocket of 1SSK protein were reported in Table 3 and Fig. S1.
Fig. 4.
3D visualization of docking analysis of 1SSK protease binding with A) deoxynojirimycin, B) trigoneoside IB, C) octanoic acid and D) remdesivir.
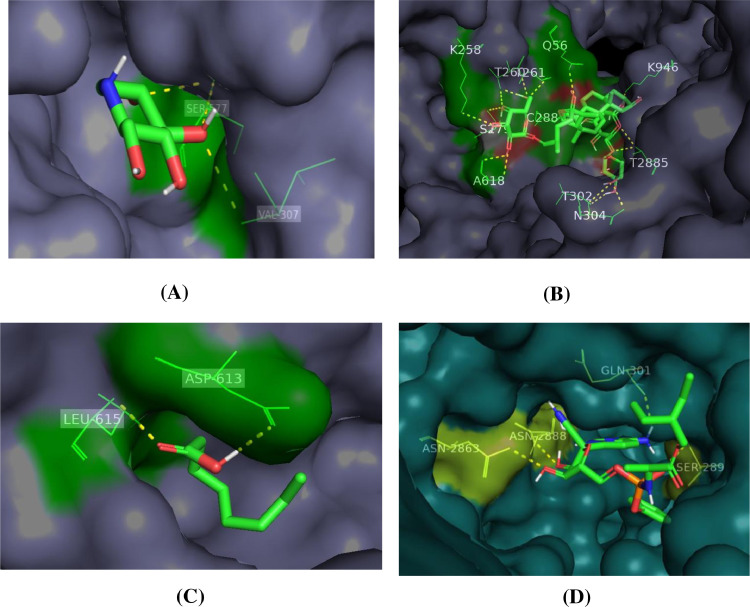
3.4. Docking interaction of 6ACD protein with selected ligands
The Molecular Docking analysis and 3D visualization of 6ACD binding protein with all four compounds are shown in Fig. 5 . Out of these compounds, trigoneoside IB compound exhibited the highest binding affinity (−8.5 Kcal/mol) and THR-260, ALA-618, SER-289, LYS-946, SER-279, THR-261, THR-616, GLY-288, THR-302, THE-285, CYS-288 are the amino acid residues participating in the interaction at the binding pocket of 6ACD protein. Whereas deoxynojirimycin compound exhibited (−5.6 Kcal/mol), followed by octanoic acid compound exhibited (−4.6 Kcal/mol), finally an anti-HIV remdesivir compound exhibited (−7.8 Kcal/mol) binding affinity with 1SSK proteins and their amino acid residues participating in the interaction at the binding pocket of 6ACD protein were reported in Table 3 and Fig. S2.
Fig. 5.
3D visualization of docking analysis of 6ACD protease binding with A) deoxynojirimycin, B) trigoneoside IB, C) octanoic acid and D) remdesivir compounds.
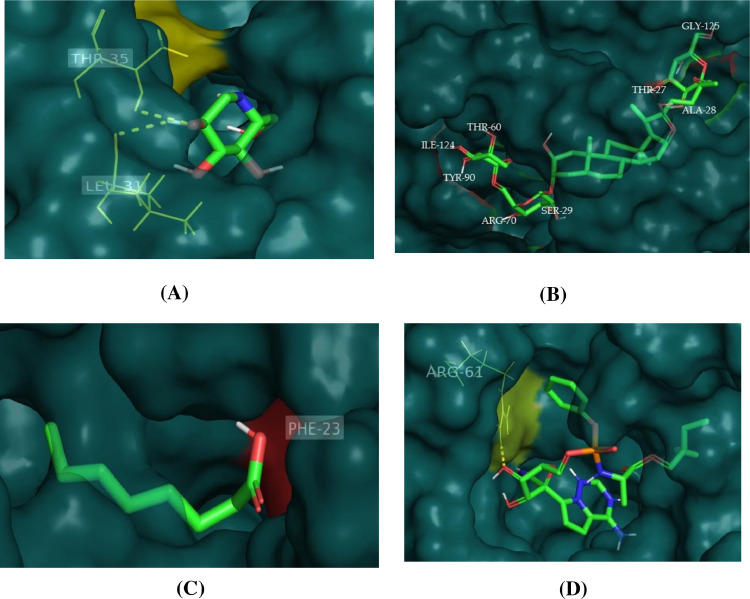
3.5. Docking interaction of 5 × 29 protein with selected ligands
The Molecular Docking analysis and 3D visualization of 5 × 29 binding protein with all four compounds are shown in Fig. 6 . Out of these compounds, the trigoneoside IB compound exhibited the highest binding affinity (−7.5 Kcal/mol) and ARG-71, ILE-124, TYR-90, ARG-70, THR-60, SER-29, ALA-28, THR-27, GLy-125 are the amino acid residues participating in the interaction at the binding pocket of 5 × 29 protein. Whereas deoxynojirimycin compound exhibited (−4.5 Kcal/mol), followed by octanoic acid compound exhibited (−4.6 Kcal/mol), finally an anti-HIV remdesivir compound exhibited (−7.8 Kcal/mol) binding affinity with 5 × 29 proteins and their amino acid residues participating in the interaction at the binding pocket of 5 × 29 protein were reported in Table 3 and Fig.S3.
Fig. 6.
3D visualization of docking analysis of 5 × 29 protease binding with A) deoxynojirimycin, B) trigoneoside IB, C) octanoic acid and D) remdesivir compounds.
3.5. Molecular dynamics simulation analysis
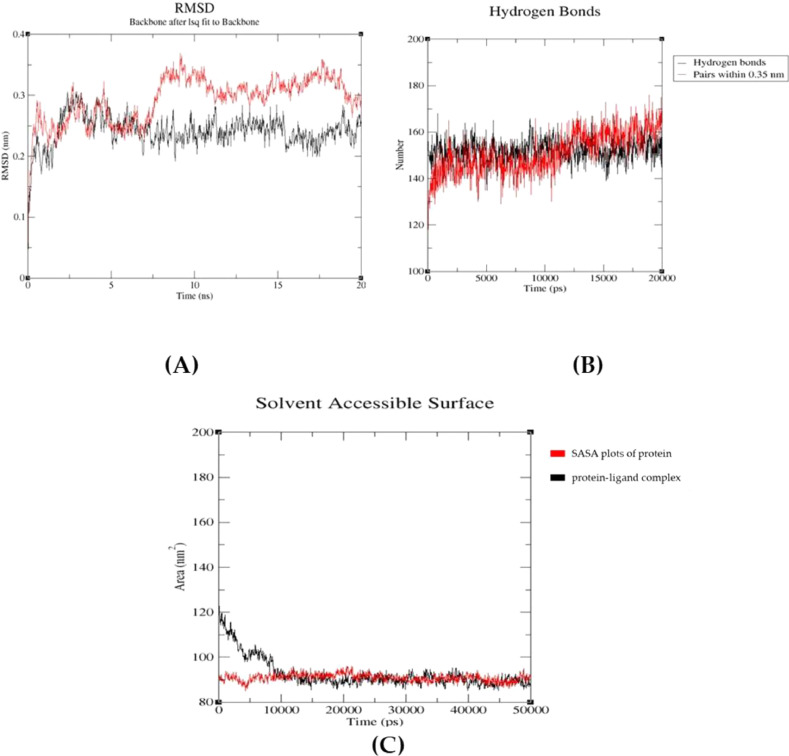
The simulation studies were carried out to ensure the docking results obtained. Based on the docking results, the best docked complex was considered to determine the stability of the complex. Trigoneoside IB was observed to have the least binding energy value of −8.5 Kcal/mol and sharing the highest hydrogen bonds and hydrophobic residues with the A chain of SARS-CoV spike glycoprotein (PDB ID: 6ACD). Hence this complex was further considered for molecular dynamic simulation studies. In the present study, the interaction obtained between protein-ligand complex were analysed inside the solvent (water) system along with NaCl salt type. The results obtained after performing simulation for 30 ns were analysed in Xmgrace software to obtain the RMSD (Root Mean Square Deviation), Hydrogen Bonds and SASA (Solvent Accessible Surface) plots.
RMSD studies were done to analyse the deviation of protein alone and its deviation when it is attached to a ligand. In the RMSD plot, the protein deviated from 0.05 to 0.375 nm wherein the presence of ligand the deviations were seen from 0.1 to 0.3 nm showing the least deviation and attained equilibrium in the end of the simulation Fig. 8A.
Fig. 8.
Representing the A: RMSD, B: Hydrogen Bonds and C: SASA plots of protein alone (Red color) and protein-ligand complex (Black color)
Both Hydrogen bonds and SASA plots when analysed showed least deviations and have attained equilibrium in the end for complex compared to protein Fig. 8B. and Fig. 8C. Thus, the protein-ligand complex was more stable when compared to protein alone.
4. Discussion
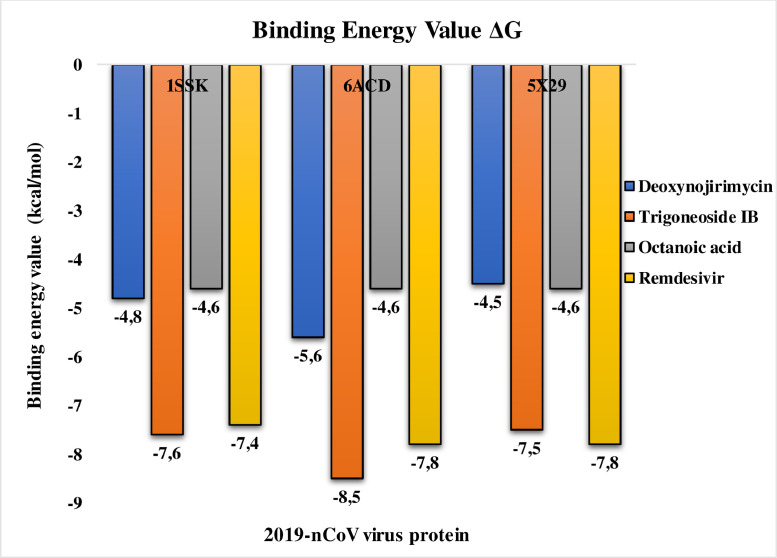
Medicinal plants that produce biologically active compounds have often piqued scientists' interest because they are crucial in the prevention of human diseases (Mohamed et al., 2015). India is known across the world as a place where spices have long been used as a source of medicine. In the current study, molecular docking studies reveal that the selected compounds (deoxynojirimycin, trigoneoside IB and octanoic acid) showed the best binding affinity with the binding pocket residues of proteins (1SSK, 6ACD, and 5 × 29). Lesser the binding affinity, the more strongly the target molecule and the ligand are attached to one another. The obtained results of proteins with selected compound shows a best binding affinity with 4 compounds including the standard drug, ranked by affinity (ΔG); trigoneoside IB > remdesivir > deoxynojirimycin > octanoic acid See Table 3 and Fig. 7 . Trigoneoside IB shows the best binding affinity with selected COVID-19 proteins whereas remdesivir and deoxynojirimycin show a good binding affinity with 1SSK, 6ACD proteins and octanoic acid show the least binding affinity with the selected SARS-CoV-2 proteins. Thereby the obtained results can be based on further studies to use these compounds to treat against SARS-CoV-2 proteases.
Fig. 7.
Histogram showing molecular docking results between 1SSK, 6ACD and 5 × 29 against selected compounds (the binding energy value ΔG is shown in minus kcal/mol).
Trigoneoside has been reported to exhibit in silico antiviral activity against the Zika virus (Byler et al., 2016; Al-Asadi, 2014). Additionally, a patent assigned indicates that the trigoneoside IB has significant biomedical importance as a potent drug candidate (Asadi et al., 2014). Not with standing, it is yet essential to investigate further the antiviral benefits of this phytocompound found in the fenugreek. Meanwhile, remdesivir is currently being tested, at the NIH facility, in combination with Baricitinid, a rheumatoid arthritis prescription drug, to check for their efficacy against the COVID-19. Although the results of this most recent double-blind study are awaited, it may be deduced that the antiviral intensity of the remdesivir drug could be of a greater value owing to the synergistic interactions. Suggesting a novel study design to evaluate the value addition to antiviral efficacies of the compounds in this study when treated in different combinations. Additionally, Wang et al. reported that the remdesivir had inhibitory effects on the zoonotic and anthroponotic coronaviruses (Zumla et al., 2016; Wang et al., 2020). Modified deoxynojirimycin molecules were also reported to be potent anti-influenza virus inhibitors. However, the activity observed was strain-specific due to its dependence on the presence of haemagglutinin (HA) (Hussain et al., 2014). On the other hand, octanoic acid has been found to inactivate lipid enveloped viruses such as the human immunodeficiency virus, pseudorabies virus, bovine viral diarrhea virus, and sindbis virus (Dichtelmüller et al., 2002). The above observations indicate that the test compounds used in the present study do possess significant broad-spectrum antiviral activity which can be investigated further, both experimentally and mechanistically, to repurpose them and mitigate the current COVID-19 pandemic.
5. Conclusions
Enormous human and financial efforts have been carried out, resulting in dozens of COVID-19 vaccines in clinical trials and from those four are already EMA approved like oseltamivir, lopinavir, ritonavir, and remdesivir because these drugs act potently against single-strand RNA viruses. Thus, it is practicable to hypothesize that the anti-HIV drugs also exert a potential activity against SARS-CoV-2. Interestingly, in our bioinformatics studies, an anti-HIV drug remdesivir significantly showed good interaction with two proteins, glycosylated S and Coronavirus E protein, when compared to Oseltamivir, Lopinavir, Ritonavir drugs. Trigoneoside IB shows the best binding interactions when compared to the other two compounds. Based on our in-silico findings, we proposed that the Trigoneoside IB compound of fenugreek needs to be considered as the best potential inhibitor when compared to remdesivir, octanoic acid, deoxynojirimycin compounds. Therefore, our preliminary in-silico analysis could be a base for further in-vitro and in-vivo studies to use these compounds against SARS-CoV-2 proteases.
Declaration of Competing Interest
No potential conflict of interest between authors in publishing this work.
Acknowledgments
Authors thankfully acknowledge the support and infrastructure provided by the JSS Academy of Higher Education and Research (JSSAHER), Mysuru, India. KSP thankfully acknowledge the Director, Amrita Vishwa Vidyapeetham, Mysuru campus for infrastructure support. The authors extend their appreciation to the Researchers Supporting Project (RSP-2020/200), King Saud University, Riyadh, Saudi Arabia.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.envc.2021.100107.
Appendix. Supplementary materials
References
- Aboubakr H.A., Nauertz A., Luong N.T., Agrawal S., El-Sohaimy S.A, Youssef M.M. In Vitro antiviral activity of clove and ginger aqueous extracts against feline calicivirus, a surrogate for human norovirus. J. Food Prot. 2016;79:1001–1012. doi: 10.4315/0362-028X.JFP-15-593. [DOI] [PubMed] [Google Scholar]
- Ahmad A., Alghamdi S.S., Mahmood K., Afzal M. Fenugreek a multipurpose crop: potentialities and improvements. Saudi J. Biol. Sci. 2016;23:300–310. doi: 10.1016/j.sjbs.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Asadi J.N. Therapeutic uses of fenugreek (Trigonella foenum-graecum L.) Am. J. Soc. Issues Hum. 2014;2:21–36. [Google Scholar]
- Baell J.B. Feeling nature's PAINS: natural products, natural product drugs, and pan assay interference compounds (PAINS) J. Nat. Prod. 2016;79:616–628. doi: 10.1021/acs.jnatprod.5b00947. [DOI] [PubMed] [Google Scholar]
- Bin-Hafeez B., Haque R., Parvez S., Pandey S., Sayeed I., Raisuddin S. Immunomodulatory effects of fenugreek (Trigonella foenum graecum L.) extract in mice. Int. Immunopharmacol. 2003;3:257–265. doi: 10.1016/S1567-5769(02)00292-8. [DOI] [PubMed] [Google Scholar]
- Byler K.G., Ogungbe IV., Setzer W.N. In-silico screening for anti-Zika virus phytochemicals. J. Mol. Graph. Model. 2016;69:78–91. doi: 10.1016/j.jmgm.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., Yuan S., Kok K.H., To K.K.W., Chu H., Yang J., Xing F., Liu J., Yip C.C.Y., Poon R.W.S., Tsoi H.W. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736,30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhikara B.S., Rathi B., Singh J., Poonam F.N.U. Corona virus SARS-CoV-2 disease COVID-19: infection, prevention and clinical advances of the prospective chemical drug therapeutics. Chem. Biol. Lett. 2020;7:63–72. [Google Scholar]
- Dai W., Bi J., Li F., Wang S., Huang X., Meng Antiviral efficacy of flavonoids against Enterovirus 71 infection in vitro and in newborn mice. Viruses. 2019;11:625. doi: 10.3390/v11070625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash B.K., Sultana S., Sultana N. Antibacterial activities of methanol and acetone extracts of fenugreek and coriander. Life Sci. Med. Res. 2011;27:1–8. [Google Scholar]
- Dichtelmüller H., Rudnick D., Kloft M. Inactivation of lipid enveloped viruses by octanoic acid treatment of immunoglobulin solution. Biologicals. 2002;30:135–142. doi: 10.1006/biol.2002.0332. [DOI] [PubMed] [Google Scholar]
- Fan W., Qian S., Qian P., Li X. Antiviral activity of luteolin against Japanese encephalitis virus. Virus Res. 2016;220:112–116. doi: 10.1016/j.virusres.2016.04.021. [DOI] [PubMed] [Google Scholar]
- Hamden K., Keskes H., Belhaj S., Mnafgui K., Allouche N. Inhibitory potential of omega-3 fatty and fenugreek essential oil on key enzymes of carbohydrate-digestion and hypertension in diabetes rats. Lipids Health Dis. 2011;10:1–10. doi: 10.1186/1476-511X-10-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho B.K., Brasseur R. The Ramachandran plots of glycine and pre-proline. BMC Struct. Biol. 2005;5:1–11. doi: 10.1186/1472-6807-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y.…Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S., Miller J.L., Harvey D.J., Gu Y., Rosenthal P.B., Zitzmann N., McCauley J.W. Strain-specific antiviral activity of iminosugars against human influenza A viruses. J. Antimicrob. Chemother. 2014;70:136–152. doi: 10.1093/jac/dku349. doi:10.1093/jac/dku349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J.R., Mansfield K., You J.E., Tennant B.C., Kim Y.H. Natural iminosugar derivatives of 1-deoxynojirimycin inhibit glycosylation of hepatitis viral envelope proteins. J. Microbiol. 2007;45:431–440. PMID: 17978803. [PubMed] [Google Scholar]
- Jain A.S., Sushma P., Dharmashekar C., Beelagi M.S., Prasad S.K., Shivamallu C., Prasad A., Syed A., Marraiki N., Prasad K.S. In silico evaluation of flavonoids as effective antiviral agents on the spike glycoprotein of SARS-CoV-2. Saudi J. Biol. Sci. 2021;28:1040–1051. doi: 10.1016/j.sjbs.2020.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javan M., Ahmadiani A., Semnanian S., Kamalinejad M. Antinociceptive effects of Trigonella foenum-graecum leaves extract. J. Ethnopharmacol. 1997;58:125–129. doi: 10.1016/s0378-8741(97)00089-5. [DOI] [PubMed] [Google Scholar]
- Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li X, Zhang L, Peng C, Duan Y, Yu J, Wang L, Yang K, Liu F, Jiang R, Yang X, You T, Liu X, Yang X, Bai F, Liu H, Liu X, Guddat LW, Xu W, Xiao G, Qin C, Shi Z, Jiang H, Rao Z, Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. doi:10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Khaerunnisa S., Kurniawan H., Awaluddin R., Suhartati S., Soetjipto S. Potential inhibitor of COVID-19 main protease (Mpro) from several medicinal plant compounds by molecular docking study. Preprints. 2020 doi: 10.20944/preprints202003.0226.v1. [DOI] [Google Scholar]
- Kim H., Chung M.S. Antiviral activities of mulberry juice and seed against influenza viruses. Evid. Complem. Alternat. Med.: eCAM. 2018 doi: 10.1155/2018/2606583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylova N.V, Am P, Gn L., Aa A, Maistrovskaia O.S. Comparative study of antiviral activity of luteolin and 7, 3’-disulfate luteolin. Antibiot. Khimioter. 2011;56:7–10. PMID: 22856150. [PubMed] [Google Scholar]
- Kumar S., FNU.P. Rathi B. Coronavirus disease COVID-19: a new threat to public health. Curr. Top. Med. Chem. 2020;20:00. doi: 10.2174/1568026620999200305144319. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Swindells M.B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Sy B., MO. Kh, K., Chung M.S. Antiviral effects of mulberry juice and its fractions on foodborne viral surrogates. Foodborne Pathog. Dis. 2014;11:1556–7125. doi: 10.1089/fpd.2013.1633. [DOI] [PubMed] [Google Scholar]
- Lefkowitz E.J., Dempsey D.M., Hendrickson R.C., Orton R.J., Siddell S.G., Smith D.B. Virus taxonomy: the database of the international committee on taxonomy of viruses (ICTV) Nucl. Acids Res. 2018;46:708–717. doi: 10.1093/nar/gkx932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-352766005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathern J.R., Raatz S.K., Thomas W., Slavin J.L. Effect of fenugreek fiber on satiety, blood glucose and insulin response and energy intake in obese subjects. Phytother. Res. 2009;23:1543–1548. doi: 10.1002/ptr.2795. [DOI] [PubMed] [Google Scholar]
- Mehla R., Bivalkar-Mehla S., Chauhan A. A Flavonoid, Luteolin, Cripples HIV-1 by Abrogation of Tat Function. PloS One. 2011;6(11):e27915. doi: 10.1371/journal.pone.0027915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed A., Dkhil., Al-Quraishy. Saleh, Delic. Denis. The antioxidant and anti-herpes simplex viruses activity of morus alba leaves extract. Pakistan J. Zool. 2015;47:1563–1569. [Google Scholar]
- Moradi-Kor N, Moradi K. Physiological and pharmaceutical effects of fenugreek as a multipurpose and valuable medicinal plant. Glob. J. Med. Plant Res. 2013;1:199–206. [Google Scholar]
- Naithani R., Huma L., Holland L., Shukla D., McCormick D., Mehta R. Moriarty, antiviral activity of phytochemicals: a comprehensive review. Mini Rev Med Chem. 2008;8:1106–1133. doi: 10.2174/138955708785909943. [DOI] [PubMed] [Google Scholar]
- Oluwaseun O.T., Rahman S., Alakanse S.O., Akinloye O., Ibrahim S., Makama Y.A., Bankole O. Molecular docking and admet analyses of phytochemicals from nigella sativa (blackseed), trigonella foenum-graecum (fenugreek) and anona muricata (soursop) on sars-cov-2 target. Sci. Open Preprints. 2020 doi.org/10.14293/S2199-1006.1.SOR-.PPKNVFY.v1. [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera? A visualization system for exploratory research and analysis. J. Comput. Chem. 2000;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pradesh U., Upadhayay P.D.D., Vigyan P.C. Coronavirus infection in equines: a review. Asian J. Animal Vet. Adv. 2014;9:164–176. doi: 10.3923/ajava.2014.164.176. [DOI] [Google Scholar]
- Prajapat M., Sarma P., Shekhar N. Drug targets for coronavirus: a systematic review. Indian J. Pharmacol. 2020;52 doi: 10.4103/ijp.IJP_115_20. 56-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnoglik S.L., Aoki C., Sudarmono P., Komoto M., Deng L., Shoji I., Fuchino H., Kawahara N., Hotta H. Antiviral activity of extracts from Morinda citrifolia leaves and chlorophyll catabolites, pheophorbide a and pyropheophorbide a, against hepatitis C virus. Microbiol. Immunol. 2014;58:188–194. doi: 10.1111/1348-0421.12133. &. [DOI] [PubMed] [Google Scholar]
- RCSB PDB: Homepage (Database) cited 2020 Jan 13. Available from: https://www.rcsb.org/.
- Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:1–22. doi: 10.1186/S12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvam P., Murugesh N., Witvrouw M., Keyaerts E., Neyts J. Studies of antiviral activity and cytotoxicity of wrightia tinctoria and morinda citrifolia. Indian J. Pharm. Sci. 2009;71:670–672. doi: 10.4103/0250-474X.59550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Suresh S., Debnath A., Jha S. Trigonella seed extract ameliorates inflammation via regulation of the inflammasome adaptor protein. ASC. Front. Biosci. 2017;9:246–257. doi: 10.2741/799. [DOI] [PubMed] [Google Scholar]
- Siu Y.L., Teoh K.T., Lo J., Chan C.M., Kien F., Escriou N., Tsao S.W., Nicholls J.M., Altmeyer R., Peiris J.S., Bruzzone R., Nal B. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008;82:11318–11330. doi: 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowmya P., Rajyalakshmi P. Hypocholesterolemia effect of germinated fenugreek seeds in human subjects. Plant Foods Hum. Nutr. 1999;53:359–365. doi: 10.1023/a:1008021618733. [DOI] [PubMed] [Google Scholar]
- The Pubchem Project (Database) cited 2020 Jan 13, Available from: https://pubchem.ncbi.nlm.nih.gov/.
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vats V., Grover J.K., Rathi S.S. Evaluation of anti-hyperglycemic and hypoglycemic effect of Trigonella foenum-graecum Linn, Ocimum sanctum Linn and Pterocarpus marsupium Linn in normal and alloxanized diabetic rats. J. Ethnopharmacol. 2002;79:95–100. doi: 10.1016/s0378-8741(01)00374-9. [DOI] [PubMed] [Google Scholar]
- Wang J., Qin X., Chen Z., Ju Z., He W., Tan Y, Liu. Two new anthraquinones with antiviral activities from the barks of Morinda citrifolia (Noni) Phytochem. Lett. 2016;15:13–15. doi: 10.1016/j.phytol.2015.11.006. [DOI] [Google Scholar]
- Wang M.Y., West B.J., Jensen C.J., Nowicki D., Su C., Palu A.K., Anderson G. Morinda citrifolia (Noni): a literature review and recent advances in Noni research. Acta Pharmacol. Sin. 2002;23:1127–1141. [PubMed] [Google Scholar]
- Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicenter trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. doi.org/10.1016/S0140-6736 (20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;82:11318–11330. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Yu Y., Xu J., Shu H., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L., Li Z., Yuan K., Qu X., Chen J., Wang G., Zhang H., Luo H., Zhu L., Jiang P., Chen L., Shen Y., Luo M., Zuo G., Hu J., Duan D., Nie Y., Shi X., Wang W., Han Y.…Xu X. Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. J. Virol. 2004;78:11334–11339. doi: 10.1128/JVI.78.20.11334-11339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Wunderink R.G. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23:130–137. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses - drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15(5):327–347. doi: 10.1038/nrd.2015.37. doi:10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.