Abstract
Stem cell exhaustion plays a major role in the ageing of different tissues. Similarly, in vitro cell ageing during expansion prior to their use in regenerative medicine can severely compromise stem cell quality through progressive declines in differentiation and growth capacity. We utilized non-destructive multispectral assessment of native cell autofluorescence to investigate the metabolic mechanisms of in vitro mesenchymal stem cell (MSC) ageing in human bone marrow MSCs over serial passages (P2–P10). The spectral signals for NAD(P)H, flavins and protein-bound NAD(P)H were successfully isolated using Robust Dependent Component Analysis (RoDECA). NAD(P)H decreased over the course of hMSC ageing in absolute terms as well as relative to flavins (optical redox ratio). Relative changes in other fluorophore levels (flavins, protein-bound NAD(P)H) suggested that this reduction was due to nicotinamide adenine dinucleotide depletion rather than a metabolic shift from glycolysis to oxidative phosphorylation. Using multispectral features, which are determined without cell fixation or fluorescent labelling, we developed and externally validated a reliable, linear model which could accurately categorize the age of culture-expanded hMSCs. The largest shift in spectral characteristics occurs early in hMSC ageing. These findings demonstrate the feasibility of applying multispectral technology for the non-invasive monitoring of MSC health in vitro.
Electronic supplementary material
The online version of this article (10.1007/s11357-020-00250-9) contains supplementary material, which is available to authorized users.
Keywords: Mesenchymal stem cell, Ageing, Autofluorescence, Hyperspectral, Multispectral, Metabolism, NAD
Introduction
Ageing is accompanied by loss of capacity for self-repair, increased chronic disease and ultimately death [1]. Mesenchymal stem cells (MSCs) differentiate to osteoblasts, adipocytes and chondrocytes [2] and have been investigated for the treatment of age-related diseases including arthritis [3], osteoporosis [4] and stroke [5]. Comparatively rare within host tissues [6], MSCs exist with even lower frequency in older organisms [7, 8]. This necessitates increased ex vivo expansion to cultivate therapeutically useful populations. However, expansion further ages MSCs, leading to loss of function [9].
MSCs from old and young individuals have phenotypic differences including morphological changes [10], alterations to transcriptomes [11] and secretomes [10], accelerated senescence [12] and increased levels of reactive oxygen species (ROS) [13]. Older MSCs are predisposed to adipogenesis over osteogenesis [14–16], leading to decreased bone density [17]. A similar shift in differentiation potential is seen for sequentially passaged MSCs, which undergo morphological changes, expanding in size and losing their characteristic spindle-like structures, while losing osteogenic differentiation potential relative to adipogenic [18]. Furthermore, the ex vivo expansion of MSCs leads to loss of function and can result in heterogeneity through spontaneous differentiation [9]. These changes have important implications for the suitability of a hMSC line for transplantation, and a system for the early estimation of the degree to which a line will tolerate expansion would be valuable.
Ageing is linked to cell metabolism and redox state [11]. Key indicators of cells’ redox state are the reduced form of nicotinamide adenine dinucleotide (NADH) and flavin adenine dinucleotide (FAD), which are the principal electron donors and acceptors of oxidative phosphorylation. NADH and FAD are autofluorescent, with NADH having excitation maxima at 290 and 351 nm with emission maxima at 440 and 460 nm, while FAD has an excitation maxima at 450 nm and emission maxima at 535 nm. Other related fluorophores (i.e. NADPH and flavins) have the same excitation/emission profiles and are not spectrally distinct from NADH and FAD. They have lower abundance, however, and make comparatively little contribution to the total signal [19].
In this project, we have applied a multispectral approach to studying the effect of in vitro ageing on the redox state of hMSCs. This technology uses a modified fluorescence microscope with an extended number of spectral channels (20–100) [20, 21] to repeatedly image a single field of view at selected excitation wavelengths, capturing native emissions in specific wavelength ranges. This dataset provides rich cellular details and quantitative information based on cell autofluorescence, which is a direct indicator of intracellular state and activities. As well as giving information on the abundance of specific native fluorophores, the broad-spectrum approach contains rich biomarker information which has been shown to be sensitive to subtle changes in cellular states, including cell cycle stage [20], reactive oxygen species [22] and disease state [23].
Methods
hMSC culture
The hMSC lines ND0094, HE18K00DB, ND0142, ND0338 and ND0106 were derived from healthy adult bone marrow aspirates isolated from the posterior iliac crest of consenting donors aged 18–35 (Royal Adelaide Hospital Human Ethics Committee protocol number 940911a). Purified hMSC were prepared from bone marrow mononuclear cells following immunomagnetic selection for STRO-1 positivity by Mini MACS magnetic separation [24], and characterized as described previously [24]. Approval for use was given by UNSW low-risk HEC (HC180219). Cell lines were maintained in a humidified incubator at 37 °C, 5%CO2 in α-MEM with sodium bicarbonate, without L-glutamine, ribonucleosides and deoxyribonucleosides (M4526, Sigma) supplemented with 10% fetal bovine serum (SH30084, Hyclone), sodium pyruvate (Sigma, S8636), 2 mM l-glutamine (Gibco, 25030081), 100 μM l-ascorbic acid (Sigma, A8960), 50 U/ml penicillin and 50 μg/ml streptomycin (Sigma, P4333). Primary cultures were expanded to passage 1 prior to analysis, making the first observations at passage 2. Media were changed every 3–4 days and cells were subcultured at around 80% confluence by washing cells twice with Dulbecco’s phosphate buffered saline without calcium or magnesium (Gibco, 14190), followed by incubation with TryplE (Gibco, 12604-021)) at 37 °C for up to 5 min with agitation to assist cell detachment. TryplE was then inactivated by the addition of serum containing culture media and removed by centrifugation. Cells were replated at 3.75 × 105 in 75 cm2 culture flasks for continued growth or 0.5 × 105 cells in 35-mm polymer coverslip imaging dishes (Ibidi, 81166). Cells were spectrally imaged 48 h after replating with fresh culture media. Immediately prior to imaging media was replaced with containing spectrally neural Hank’s balanced salt solution (HBSS). Confluency at this timepoint was approximately 50%. Colony-forming unit fibroblast (CFU-F) assays were carried out in culture media as above, but supplemented with 20% FBS. Cells were plated in triplicate in 12 well dishes at 400 or 800 cells (balancing colony overgrowth against the risk of getting no colonies to count) and data are presented as CFUs per 400 cells.
Spectral microscopy
Spectral microscopy was carried out on an Olympus IX83 microscope with a NuVu electron multiplying charge coupling device (EMCCD, hnu1024) camera, a × 40 oil objective lens (UAPON340, Olympus) and LED illumination at 358, 371, 377, 381, 385, 391, 397, 400, 403, 406, 412, 418, 430, 437, 451, 457, 469 and 476 ± 5 nm with emission filters at 414, 451, 575, 594 and 675 ± 20 nm. Methodological details on multispectral microscopy are provided in [25–27] and image preparation in (i.e. the removal of image artifacts including Poisson’s noise, dead and/or saturated pixels, illumination curvature and background fluorescence) [27, 28]. Regions of interest were defined to isolate individual cells within fields of view, and all analyses and assessments have been carried out on a per-cell basis.
Autofluorophore unmixing
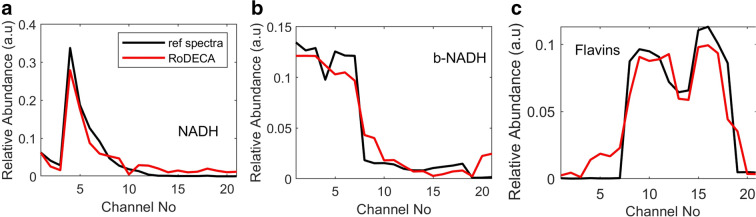
Linear mixed modelling (LMM) was used to compare extracted spectral characteristics to the known characteristics of fluorophores and calculate their abundance. The signal attached to each pixel was assumed to be a linear combination of a small number of endmember component spectra with weights corresponding to the concentration of the fluorophores responsible for these. Channels were chosen to minimize spectral overlap between autofluorophores and avoid interference between complexes (Supp Fig. 1). Relative concentrations were expressed as abundance fractions [29–31], with the unsupervised algorithm Robust Dependent Component Analysis (RoDECA) being used to identify dominant endogenous fluorophores and their abundance. RoDECA has been shown to discriminate individual fluorophores, despite overlapping spectra, in the presence of image noise [23, 28, 32]. Protein bound NAD(P)H was distinguished from free NAD(P)H using the reference spectra of L-Malate Dehydrogenase (Sigma Aldrich #10127248001) bound to NADH (Sigma Aldrich # 10107735001) in Mops (Sigma Aldrich # M1254) [33]. The spectral shift between NADH and protein bound NADH had been observed in the deep UV region, which is carefully covered during the multispectral acquisition [33, 34]. The correlation between reference spectra and the unmixed, RoDECa spectra are shown in Fig. 1.
Fig. 1.
a–c Correlation between reference spectra and unmixed RoDECA spectra. Spectral errors are 0.0175, 0.0114 and 0.0124 for NAD(P)H, b-NAD(P)H and flavins respectively. Channel excitation and emissions matching channel numbers as well as example spectral images are shown in supplementary Figure 1
Multispectral discrimination modelling
Multispectral modelling was performed using a variety of quantitative cellular image features including mean channel intensity, channel intensity ratio [26], colour distribution [35] and textural features [36]. Full definitions of examined features are in [26, 35, 36]. Candidate features (ANOVA p < 0.005) were projected onto an optimal two-dimensional space created by discriminative analysis. This space maximized between-group distance and minimized within-group variance while reducing the dimensions of the feature vectors to two canonical variables that were equal to a linear combination of the selected features [22, 37]. A linear classifier was applied based on a linear predictor function which incorporated a set of weights obtained from the training process [38, 39]. For external validation, this classifier was developed using four hMSC lines and tested on a fifth.
Statistics
All statistical processes were carried out in Matlab (R2017b). Group comparisons were made using the Mann-Whitney U test which ranks data to compare between group differences as data did not meet the assumptions necessary for parametric comparisons (i.e. normal distribution, equal variance). Data were presented as median values and 95% confidence intervals. Groups were accepted as being significantly different at p ≤ 0.05.
Results
Five bone marrow hMSC lines were cultured from passage two to passage ten with multispectral imaging at each subculture. Cells were categorized as young (P2–4), middle (P5–7) and late (P8–10) passage. A main culture was continuously maintained for each line, with samples collected and platted for imaging at each passage. The concentrations of specific fluorophores were identified, and a linear classifier was trained on multispectral data to sort hMSCs into age groupings.
Cell growth dynamics
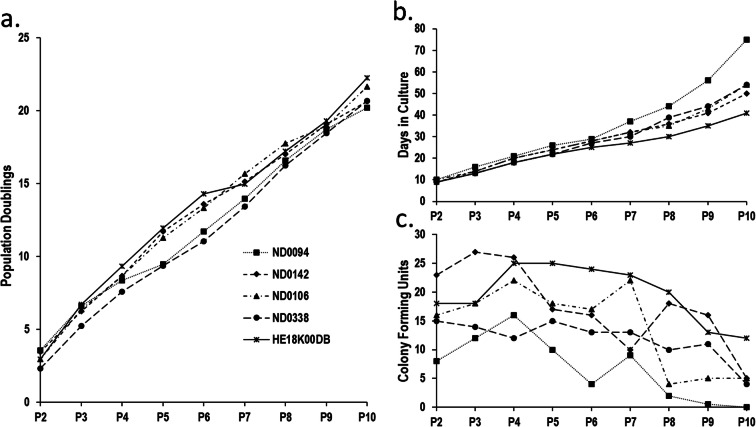
Population doublings were consistent across hMSC lines (Fig. 2a); however, days in culture diverged from passage 6, with ND0094 growing more slowly and HE18K00DB more rapidly (Fig. 2b). All lines produced fewer colony-forming units (CFUs) at their latest passages compared with earlier; however, progression to this point was not smooth (Fig. 2c). Consistent with growth speed (Fig. 2b), ND0094 produced fewer CFUs at most passages and was the only line to produce no CFUs at passage 10, while HE18K00DB produced more CFUs and retained the highest CFU number.
Fig. 2.
Growth characteristics. hMSC lines ND0994, ND0142, ND0106, ND0338 and HE18K00DB included. a Population doublings undergone by hMSC lines during culture at the end of each passage. b Total days in culture at the end of each passage. c Colony-forming unit-fibroblasts per 400 cells plated at the end of each cell passage
Fluorophore unmixing
Free NAD(P)H
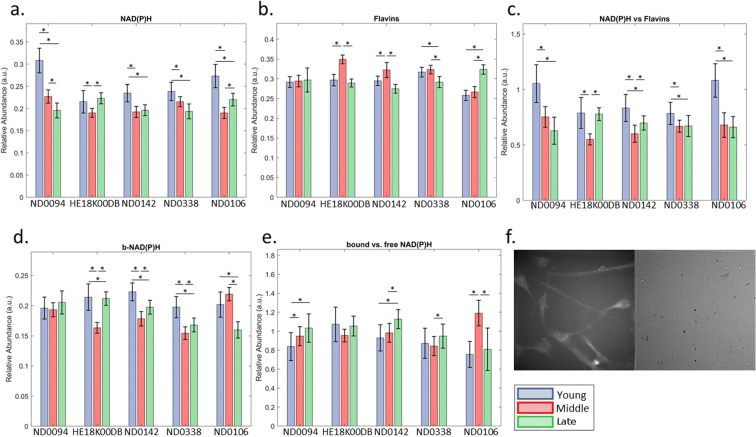
NADH and NADPH are not spectrally distinct and are collectively referred to as NAD(P)H. Middle and late hMSCs had lower NAD(P)H than young (P2–4), apart from HE18K00DB where the difference between young and late cells was not significant (Figs. 3 and 4a). Differences between middle and late hMSCs were inconsistent across lines; ND0094 had a continuation of the decline, HE18K00DB and ND0106 had apparent recoveries, and ND0142 and ND0338 had similar levels (Fig. 4a).
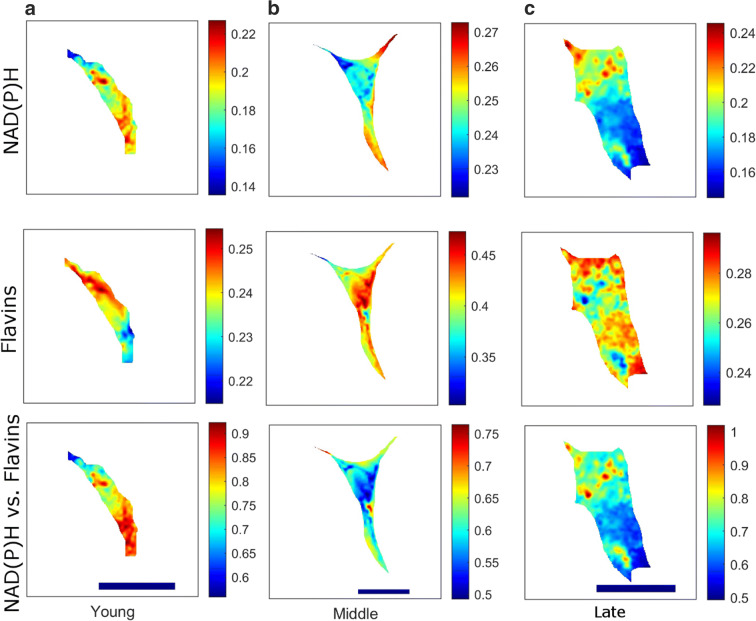
Fig. 3.
Abundance maps of extracted fluorophores (NAD(P)H, flavins and their ratio) from a young, b medium and c late passage hMSCs. Scale bar is 20 μm. Colour bars are different between images to enable legibility. Cells shown are from ND0142
Fig. 4.
Unmixed autofluorophore spectral signals. hMSC lines ND0094, HE18K00DB, ND0142, ND0338 and ND0106 included. Young cells are passages 2–4, middle cells are passages 5–7 and late cells are passages 8–10. Data are median and confidence interval, N ≥ 71 in all groups. Asterisk symbol indicates p < 0.05. (a) Free NAD(P)H spectral signals, (b) flavin spectral signals, (c) the ratio of NAD(P)H spectral signal to flavin, (d) protein-bound NAD(P)H spectral signal, (e) the ratio of protein-bound NAD(P)H spectral signal to free NAD(P)H spectral signal, (f) example spectral image at excitation 358 nm, emission 451 nm from passage 10 ND0142 cells and corresponding DIC image
Flavins
Different flavins are not spectrally distinct, although FAD is the most utilized form [19]. No significant differences existed between age groupings for ND0094 (Figs. 3 and 4b), while HE18K00DB and ND0142 had elevations for middle relative to young or late hMSCs. This pattern was not repeated for ND0338, which had no difference in flavins between young and middle hMSCs, despite a reduction between middle and late hMSCs, or ND0106 where there was no difference between young and middle but an elevation in late hMSCs. Overall, there was no pattern of change with ageing.
Optical redox ratio
The redox ratio (NAD(P)H/Flavins) was generally higher in young hMSCs compared with middle or late hMSCs (Figs. 3 and 4c)—abundance maps are shown in Fig.4. HE18K00DB was an exception with the difference between young and late hMSCs not being significant, although the difference between young and middle persisted. Most lines did not have significant differences between middle and late hMSCs (ND0094, ND0338, ND0106) although HE18K00DB and ND0142 had increases in late hMSCs.
Protein-bound NAD(P)H
The pattern of change in protein-bound NAD(P)H was less consistent than free NAD(P)H but, excluding ND0094, generally showed reduced levels in middle and late hMSCs compared with young (Figs. 3 and 4d). Most hMSC lines (HE18K00DB, ND0142, ND0338) had increased protein-bound NAD(P)H between middle and late hMSCs. For ND0106, there was a significant decrease (Fig.4d). Overall, no consistent trends existed for changes in the ratio of protein-bound NAD(P)H to free NAD(P)H (Fig. 4e).
Multispectral categorization
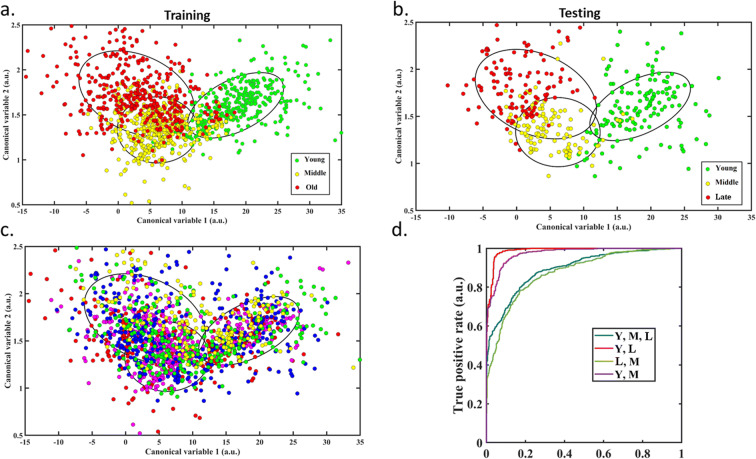
Despite patterns in spectral intensities changing with increasing age, trends were variable with heterogeneity between lines. As such, we used the full multispectral data to train a linear classifier (using data from four cell lines) to classify hMSC in vitro, then tested it on the fifth (ND0142). Strong differentiation was achieved between young and middle as well as young and late hMSCs (Fig. 5a, b, d), although middle and late had more overlap. This effect was quantified by intersection over union (IoU) analysis where the overlap for young and middle hMSCs was 1.9%, young and late was 0.5% and middle and late was 24.7%. The distribution for different hMSC lines (Fig. 5c) showed no bias.
Fig. 5.
Modelling of multispectral data. Categorization of hMSCs into age groups: young (passages 2–4), middle (passages 5–7) and late (passages 8–10). Each point represents a cell. a Results for the training data from hMSC cell lines projected onto a 2-dimensional plane, ellipsis constrain 1 standard deviation around the mean profile. b Results for the testing cell line. c Results for all five cell lines distinguished by different colours demonstrating no clumping or bias for different lines. d ROC curve analysis for the different age categories from the testing data set for the overall accuracy of categorization for each age group as well as the categorization of each age group from each other age group
These finding suggests that the main shift in hMSC characteristics occurred early in their replicative lifespans, with subtler changes later. Correct classification was achieved for 91% of hMSCs (Table 1). Young hMSCs were almost exclusively miscategorized as middle over late, with the inverse being true for late hMSCs (Table 2). Twenty features from twenty-one channels were included in the model (Supp Fig. 2).
Table 1.
Predictive accuracy
| Accuracy (%) | Sensitivity (%) | Specificity (%) | |
|---|---|---|---|
| Young | 93 | 88 | 98 |
| Middle | 87 | 83 | 91 |
| Late | 94 | 97 | 91 |
| Overall | 91 | 89 | 93 |
Table 2.
Age classifications
| Predicted | ||||
|---|---|---|---|---|
| Young | Middle | Old | ||
| True | Young | 88% | 12% | 0% |
| Middle | 5% | 74% | 21% | |
| Old | 0% | 3% | 97% | |
Discussion
As MSCs age, growth and capacity to differentiate become restricted [18]. This has been considered to be a consequence of spontaneous differentiation causing a loss of “stemness”. Prior analysis of directed MSC differentiation found a shift to greater NAD(P)H relative to FAD [40, 41]—the opposite of the pattern observed with increasing age. Meleshina [40] found that changes in FAD made the main contribution to the shift; however, with ageing, we found that the flavin signal was comparatively stable with change driven by decreasing NAD(P)H. A decrease in NAD(P)H relative to FAD during early osteogenic differentiation has been reported [40]; however, Quinn [41] showed that MSCs induced towards adipogenesis had a stable ratio of NAD(P)H to FAD during the initial stages of differentiation, followed by later increases. As MSC ageing is associated with adipogenic differentiation [14–16, 18], spontaneous differentiation should mirror this.
During oxidative phosphorylation, NADH is converted to NAD+ while FAD is generated from non-fluorescent FADH2. The relative concentration of NAD(P)H and flavins therefore tends to be reported as a measure of the cell’s utilization of oxidative phosphorylation relative to glycolysis [40]. Decreasing NAD(P)H relative to flavins (Fig. 3c) could therefore be interpreted as a shift towards oxidative phosphorylation. However, the ratio of bound to free NAD(P)H is also an indicator of cell metabolism [42]. As only free NADH is utilized by the complex I enzymes of the mitochondrial electron transport chain during oxidative phosphorylation, a metabolic shift towards this process is associated with increased bound to free NAD(P)H [43]. In our study, only one cell line, ND0094, showed this pattern (Fig. 3e), suggesting that the decrease in NAD(P)H relative to flavins is not due to increased oxidative phosphorylation. Also opposing this hypothesis is that increased oxidative phosphorylation would generate NAD+, but this metabolite decreases with age [44]. Along with the relatively stable flavin levels (which oxidative phosphorylation would increase), this leads to the conclusion that the reduction in the NAD(P)H spectral signal in late passage hMSCs is most likely the result of depletion of nicotinamide adenine dinucleotide rather than changes in redox balance. An in vivo study of the redox balance of healing epidermal injuries in mice showed an increase in FAD/(NADH+FAD) in aged mice—supporting our observation of a reduced NAD(P)H to FAD in a system related to regeneration [45]. Another study which examined the effect of age on the NAD(P)H to Flavin ratio in ageing mouse muscles was not sufficiently powered to detect significant differences, but observed changes suggestive of declining Flavin and NAD(P)H levels maintaining a stable ratio [46].
Our multispectral model accurately categorized the in vitro age of a hMSC line whose spectral data was not used in its development (Fig. 4b). Misclassifications presented almost exclusively as cells being predicted to belong to adjacent age groupings. As not all cells in culture will divide in step, a system that accurately detected biological age would show some discrepancies with chronologically categorized age. A limitation of the study is that the differences observed in the intensity of the unmixed fluorophores could be the consequence of an emergent subpopulation (e.g. senescent cells) rather than reflective of general changes in ageing cells. A further potential limitation is that artifacts of in vitro culture such as exposure the atmospheric oxygen and repeated trypsinization may have influenced the results observed.
Conclusion
Most hMSC lines isolated for clinical use would be taken from older individuals, particularly those with chronic diseases, and are likely to have high variation in capacity for expansion before their contribution to autologous transplantation is compromised. The ability to prospectively determine the biological age of hMSC lines—and their consequent remaining replicative lifespans—will enable the optimization of expansion on a personalized level and could improve cellular therapies.
Electronic supplementary material
(DOCX 328 kb)
Acknowledgments
The authors would like to thank Dr. Martin Gosnell of Quantitative-biotech Ltd. for his work constructing and maintaining the multispectral microscopy system.
Authors’ contribution
JC contributed to the conception, design of the project, data acquisition and interpretation of the data .They drafted the manuscript, give final approval for publication and agree to be accountable for all aspects of the work. SM contributed to the conception, design of the project, data analysis and interpretation. They revised the manuscript critically, give final approval for publication and agree to be accountable for all aspects of the work. AH contributed to the conception, design of the project, data analysis and interpretation. They revised the manuscript critically, give final approval for publication and agree to be accountable for all aspects of the work. SP contributed to the conception and design of the project. They revised the manuscript critically, give final approval for publication and agree to be accountable for all aspects of the work. SG contributed to the conception and design of the project. They revised the manuscript critically, give final approval for publication and agree to be accountable for all aspects of the work. EG contributed to the conception and design of the project. They revised the manuscript critically, give final approval for publication and agree to be accountable for all aspects of the work.
Funding information
This project was funded by a Discovery Project grant from the Australian Research Council (DP170101863) and the Centre of Excellence scheme (CE140100003).
Data availability
Data will be made available on request.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethics approval statement
Collection of human stem cell lines from consenting patients was approved by the Human Ethics Committee (HEC) of the Royal Adelaide Hospital, 940911a, and their subsequent use in this project by the UNSW low-risk HEC (HC180219).
Consent to participate
Samples were collected from participants with informed consent.
Consent for research
Participants consented for their samples to be used for research purposes, including publication.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Schultz MB, Sinclair DA. When stem cells grow old: phenotypes and mechanisms of stem cell aging. Development. 2016;143(1):3–14. doi: 10.1242/dev.130633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bianco P, Riminucci M, Gronthos S, Robey PG. Bone marrow stromal stem cells: nature, biology, and potential applications. Stem Cells. 2001;19(3):180–192. doi: 10.1634/stemcells.19-3-180. [DOI] [PubMed] [Google Scholar]
- 3.McIntyre JA, Jones IA, Han B, Vangsness CT., Jr Intra-articular mesenchymal stem cell therapy for the human joint: a systematic review. Am J Sports Med. 2018;46(14):3550–3563. doi: 10.1177/0363546517735844. [DOI] [PubMed] [Google Scholar]
- 4.Aghebati-Maleki L, Dolati S, Zandi R, Fotouhi A, Ahmadi M, Aghebati A, Nouri M, Kazem Shakouri S, Yousefi M. Prospect of mesenchymal stem cells in therapy of osteoporosis: a review. J Cell Physiol. 2019;234(6):8570–8578. doi: 10.1002/jcp.27833. [DOI] [PubMed] [Google Scholar]
- 5.Zheng H, Zhang B, Chhatbar PY, Dong Y, Alawieh A, Lowe F, Hu X, Feng W. Mesenchymal stem cell therapy in stroke: a systematic review of literature in pre-clinical and clinical research. Cell Transplant. 2018;27(12):1723–1730. doi: 10.1177/0963689718806846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganguly P, El-Jawhari JJ, Giannoudis PV, Burska AN, Ponchel F, Jones EA. Age-related changes in bone marrow mesenchymal stromal cells: a potential impact on osteoporosis and osteoarthritis development. Cell Transplant. 2017;26(9):1520–1529. doi: 10.1177/0963689717721201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stolzing A, Jones E, McGonagle D, Scutt A. Age-related changes in human bone marrow-derived mesenchymal stem cells: consequences for cell therapies. Mech Ageing Dev. 2008;129(3):163–173. doi: 10.1016/j.mad.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Prat L, Sousa-Victor P, Munoz-Canoves P. Functional dysregulation of stem cells during aging: a focus on skeletal muscle stem cells. FEBS J. 2013;280(17):4051–4062. doi: 10.1111/febs.12221. [DOI] [PubMed] [Google Scholar]
- 9.Li Z, Liu C, Xie Z, Song P, Zhao RC, Guo L, et al. Epigenetic dysregulation in mesenchymal stem cell aging and spontaneous differentiation. PLoS One. 2011;6(6):e20526. doi: 10.1371/journal.pone.0020526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaguchi S, Horie N, Satoh K, Ishikawa T, Mori T, Maeda H, Fukuda Y, Ishizaka S, Hiu T, Morofuji Y, Izumo T, Nishida N, Matsuo T. Age of donor of human mesenchymal stem cells affects structural and functional recovery after cell therapy following ischaemic stroke. J Cereb Blood Flow Metab. 2018;38(7):1199–1212. doi: 10.1177/0271678X17731964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peffers MJ, Collins J, Fang Y, Goljanek-Whysall K, Rushton M, Loughlin J, et al. Age-related changes in mesenchymal stem cells identified using a multi-omics approach. Eur Cell Mater. 2016;31:136–159. doi: 10.22203/eCM.v031a10. [DOI] [PubMed] [Google Scholar]
- 12.Stenderup K, Justesen J, Clausen C, Kassem M. Aging is associated with decreased maximal life span and accelerated senescence of bone marrow stromal cells. Bone. 2003;33(6):919–926. doi: 10.1016/j.bone.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Block TJ, Marinkovic M, Tran ON, Gonzalez AO, Marshall A, Dean DD, Chen XD. Restoring the quantity and quality of elderly human mesenchymal stem cells for autologous cell-based therapies. Stem Cell Res Ther. 2017;8(1):239. doi: 10.1186/s13287-017-0688-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim M, Kim C, Choi YS, Kim M, Park C, Suh Y. Age-related alterations in mesenchymal stem cells related to shift in differentiation from osteogenic to adipogenic potential: implication to age-associated bone diseases and defects. Mech Ageing Dev. 2012;133(5):215–225. doi: 10.1016/j.mad.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 15.Mueller SM, Glowacki J. Age-related decline in the osteogenic potential of human bone marrow cells cultured in three-dimensional collagen sponges. J Cell Biochem. 2001;82(4):583–590. doi: 10.1002/jcb.1174. [DOI] [PubMed] [Google Scholar]
- 16.Moerman EJ, Teng K, Lipschitz DA, Lecka-Czernik B. Aging activates adipogenic and suppresses osteogenic programs in mesenchymal marrow stroma/stem cells: the role of PPAR-gamma2 transcription factor and TGF-beta/BMP signaling pathways. Aging Cell. 2004;3(6):379–389. doi: 10.1111/j.1474-9728.2004.00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veldhuis-Vlug AG, Rosen CJ. Clinical implications of bone marrow adiposity. J Intern Med. 2018;283(2):121–139. doi: 10.1111/joim.12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang YK, Ogando CR, Wang See C, Chang TY, Barabino GA. Changes in phenotype and differentiation potential of human mesenchymal stem cells aging in vitro. Stem Cell Res Ther. 2018;9(1):131. doi: 10.1186/s13287-018-0876-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macheroux P, Kappes B, Ealick SE. Flavogenomics--a genomic and structural view of flavin-dependent proteins. FEBS J. 2011;278(15):2625–2634. doi: 10.1111/j.1742-4658.2011.08202.x. [DOI] [PubMed] [Google Scholar]
- 20.Campbell JM, Habibalahi A, Mahbub S, Gosnell M, Anwer AG, Paton S, Gronthos S, Goldys E. Non-destructive, label free identification of cell cycle phase in cancer cells by multispectral microscopy of autofluorescence. BMC Cancer. 2019;19(1):1242. doi: 10.1186/s12885-019-6463-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell JM, Habibalahi A, Mahbub S, Paton S, Gronthos S, Goldys E. Multispectral characterisation of mesenchymal stem/stromal cells: age, cell cycle, senescence, and pluripotency. LBIS. 2020;11251:112510F. [Google Scholar]
- 22.Habibalahi A, Dashtbani Moghari M, Campbell JM, Anwer AG, Mahbub SB, Gosnell M, et al. Non-invasive real-time imaging of reactive oxygen species (ROS) using multispectral auto-fluorescence imaging technique: a novel tool for redox biology. Redox Biol. 2020;34:101561. doi: 10.1016/j.redox.2020.101561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahbub SB, Guller A, Campbell JM, Anwer AG, Gosnell ME, Vesey G, Goldys EM. Non-invasive monitoring of functional state of articular cartilage tissue with label-free unsupervised hyperspectral imaging. Sci Rep. 2019;9(1):4398. doi: 10.1038/s41598-019-40942-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gronthos S, Zannettino AC, Hay SJ, Shi S, Graves SE, Kortesidis A, et al. Molecular and cellular characterisation of highly purified stromal stem cells derived from human bone marrow. J Cell Sci. 2003;116(Pt 9):1827–1835. doi: 10.1242/jcs.00369. [DOI] [PubMed] [Google Scholar]
- 25.Gosnell ME, Anwer AG, Cassano JC, Sue CM, Goldys EM. Functional hyperspectral imaging captures subtle details of cell metabolism in olfactory neurosphere cells, disease-specific models of neurodegenerative disorders. Biochim Biophys Acta. 2016;1863(1):56–63. doi: 10.1016/j.bbamcr.2015.09.030. [DOI] [PubMed] [Google Scholar]
- 26.Gosnell ME, Anwer AG, Mahbub SB, Menon Perinchery S, Inglis DW, Adhikary PP, Jazayeri JA, Cahill MA, Saad S, Pollock CA, Sutton-McDowall ML, Thompson JG, Goldys EM. Quantitative non-invasive cell characterisation and discrimination based on multispectral autofluorescence features. Sci Rep. 2016;6:23453. doi: 10.1038/srep23453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Habibalahi A, Bala C, Allende A, Anwer AG, Goldys EMJTos. Novel automated non invasive detection of ocular surface squamous neoplasia using multispectral autofluorescence imaging. 2019. [DOI] [PubMed]
- 28.Mahbub SB, Plöschner M, Gosnell ME, Anwer AG, Goldys EM. Statistically strong label-free quantitative identification of native fluorophores in a biological sample. Scientific reports. 2017;7(1):15792. doi: 10.1038/s41598-017-15952-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keshava N, Mustard JF. Spectral unmixing. Signal Processing Magazine, IEEE. 2002;19(1):44–57. doi: 10.1109/79.974727. [DOI] [Google Scholar]
- 30.Keshava N, Kerekes JP, Manolakis DG, Shaw GA, editors. Algorithm taxonomy for hyperspectral unmixing 2000.
- 31.Keshava N. A survey of spectral unmixing algorithms. Lincoln Laboratory Journal. 2003;14(1):55–78. [Google Scholar]
- 32.Mahbub SB. Unsupervised hyperspectral unmixing analysis for label-free quantitative identification of native fluorophores in a biological sample by a Robust Dependent Component Analysis (RoDECA). Macquarie University; 2017.
- 33.Rehman AU, Anwer AG, Gosnell ME, Mahbub SB, Liu G, Goldys EM. Fluorescence quenching of free and bound NADH in HeLa cells determined by hyperspectral imaging and unmixing of cell autofluorescence. Biomedical Optics Express. 2017;8(3):1488–1498. doi: 10.1364/boe.8.001488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lakowicz JR, Szmacinski H, Nowaczyk K, Johnson ML. Fluorescence lifetime imaging of free and protein-bound NADH. Proc Natl Acad Sci U S A. 1992;89(4):1271–1275. doi: 10.1073/pnas.89.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El Aziz MA, Selim IM, Xiong S. Automatic detection of galaxy type from datasets of galaxies image based on image retrieval approach. Scientific Reports. 2017;7(1):4463. doi: 10.1038/s41598-017-04605-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu H, Chu B, Zhang C, Liu F, Jiang L, He Y. Hyperspectral imaging for presymptomatic detection of tobacco disease with successive projections algorithm and machine-learning classifiers. Scientific Reports. 2017;7(1):4125. doi: 10.1038/s41598-017-04501-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Habibalahi A, Allende A, Bala C, Anwer AG, Mukhopadhyay S, Goldys EMJIA. Optimized autofluorescence spectral signature for non-invasive diagnostics of ocular surface squamous neoplasia (OSSN) 2019;7:141343–141351.
- 38.Vapnik V. The nature of statistical learning theory. Springer science & business media; 2013.
- 39.Gosnell ME, Polikarpov DM, Goldys EM, Zvyagin AV, Gillatt DA, editors. Computer-assisted cystoscopy diagnosis of bladder cancer 2018: Elsevier. [DOI] [PubMed]
- 40.Meleshina AV, Dudenkova VV, Bystrova AS, Kuznetsova DS, Shirmanova MV, Zagaynova EV. Two-photon FLIM of NAD(P)H and FAD in mesenchymal stem cells undergoing either osteogenic or chondrogenic differentiation. Stem Cell Res Ther. 2017;8(1):15. doi: 10.1186/s13287-017-0484-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quinn KP, Sridharan GV, Hayden RS, Kaplan DL, Lee K, Georgakoudi I. Quantitative metabolic imaging using endogenous fluorescence to detect stem cell differentiation. Sci Rep. 2013;3:3432. doi: 10.1038/srep03432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Z, Pouli D, Alonzo CA, Varone A, Karaliota S, Quinn KP, et al. Mapping metabolic changes by noninvasive, multiparametric, high-resolution imaging using endogenous contrast. Sci Adv. 2018;4(3):eaap9302. doi: 10.1126/sciadv.aap9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skala MC, Riching KM, Bird DK, Gendron-Fitzpatrick A, Eickhoff J, Eliceiri KW, Keely PJ, Ramanujam N. In vivo multiphoton fluorescence lifetime imaging of protein-bound and free nicotinamide adenine dinucleotide in normal and precancerous epithelia. J Biomed Opt. 2007;12(2):024014. doi: 10.1117/1.2717503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshino J, Baur JA, Imai SI. NAD(+) intermediates: the biology and therapeutic potential of NMN and NR. Cell Metab. 2018;27(3):513–528. doi: 10.1016/j.cmet.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones JD, Ramser HE, Woessner AE, Veves A, Quinn KP. Quantifying age-related changes in skin wound metabolism using in vivo multiphoton microscopy. Adv Wound Care (New Rochelle) 2020;9(3):90–102. doi: 10.1089/wound.2019.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moon L, Frederick DW, Baur JA, Li LZ. Imaging redox state in mouse muscles of different ages. Adv Exp Med Biol. 2017;977:51–57. doi: 10.1007/978-3-319-55231-6_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 328 kb)
Data Availability Statement
Data will be made available on request.