Abstract
The differentiation of neural stem cells (NSCs) into neurons is proposed to be critical in devising potential cell-based therapeutic strategies for central nervous system (CNS) diseases, however, the determination and prediction of differentiation is complex and not yet clearly established, especially at the early stage. We hypothesize that deep learning could extract minutiae from large-scale datasets, and present a deep neural network model for predictable reliable identification of NSCs fate. Remarkably, using only bright field images without artificial labelling, our model is surprisingly effective at identifying the differentiated cell types, even as early as 1 day of culture. Moreover, our approach showcases superior precision and robustness in designed independent test scenarios involving various inducers, including neurotrophins, hormones, small molecule compounds and even nanoparticles, suggesting excellent generalizability and applicability. We anticipate that our accurate and robust deep learning-based platform for NSCs differentiation identification will accelerate the progress of NSCs applications.
Subject terms: Computational neuroscience, Stem cells in the nervous system
The differentiation of neural stem cells (NSCs) into neurons is a critical part in devising potential cell-based therapeutic strategies for central nervous system diseases but NSCs fate determination and prediction is problematic. Here, the authors present a deep neural network model for predictable reliable identification of NSCs fate.
Introduction
Stem cells have the capacity for self-renewal and differentiation, expanding the possible range of cell-based therapy in regenerative medicine; for example, potential applications may include restoring damaged neurons and recomposing tissue1–3. Neural stem cells (NSCs) have the potential to yield several cell types in the anatomy of the brain; therefore, strategies based on NSCs are considered valuable in the treatment of nervous system diseases, such as Alzheimer’s disease, stroke and traumatic brain injury4–7. NSCs are considered the “seed” cells of the central nervous system (CNS), capable of self-renewing and generating neurons and glia during CNS development. NSCs are tripotent cells that generate transit-amplifying progenitors and are later restricted to three lineages: neurons, astrocytes and oligodendrocytes8–10. Neurons are the essential units for information transmission in the nervous system, sending electrical and chemical signals to other cells through axons and receiving such signals via dendrites; neurons cannot reproduce or regenerate once they die11. Transplantation of exogenous NSCs or mobilization of endogenous NSCs, followed by their differentiation into neurons to reconstruct neural circuits damaged by neurological disorders, is a widely explored method for treating neurodegenerative diseases12–16. Astrocytes, the most abundant cell type in the CNS, communicate with and structurally support the growth of neurons, oligodendrocytes and endothelial cells, integrating into tripartite synapses and the neurovascular unit to perform their function. The signalling pathways of astrocytogenesis have attracted great interest as a means of suppressing the generation of astrocytes in many neurological disorders and generating a NSC model as a drug-screening platform17,18. In addition to participating in cellular communication, astrocytes also play an important role in neuroprotection by releasing neurotrophins, such as glial cell line-derived neurotrophic factor (GDNF), and reducing excitotoxicity in neurons19. Properly regulated astrocytes may provide neuroprotection, axonal guidance and vascular integrity after CNS injury20. Oligodendrocytes are myelinating glial cells that are important for neuronal electrical insulation, facilitating saltatory signal conduction21. Oligodendrocytes also provide the axons of neurons with metabolic and trophic support, including lactate, pyruvate and neurotrophic factors such as brain-derived neurotrophic factor (BDNF), via the myelin membrane22. Neurodegeneration can result in immune-mediated myelin sheath damage and subsequent axonal demyelination23. Researchers have made efforts to differentiate NSCs into oligodendrocytes that can contribute to postinjury remyelination, electrically insulating neuronal axons for impulse propagation and providing trophic and metabolic support for neurons24–26.
Therefore, the key mission in therapeutic applications is to efficiently induce NSCs into specific cell types, especially neurons. In this regard, several neurotrophins, small molecule drugs and hormones have proven to be effective, but researchers remain devoted to developing advanced approaches to directionally induce NSC differentiation27–30. Research progress in this area is limited by the fact that common laboratory methods to observe the effectiveness of inducing differentiation can be cumbersome, time consuming and uneconomical; examples include immunofluorescent staining, polymerase chain reaction (PCR) and western blots31,32. Determining the effect of potential inducers on NSC differentiation usually takes several days, and it can be affected by many factors, such as molecular marking techniques, immature laboratory technology, and the operational skill levels of experimental personnel. Sometimes, even limitations on the current understanding of essential biological characteristics of NSCs could become barriers to finding potential regulatory factors, since some mechanisms linked to NSC fate change, such as remodelling or dynamic changes in organelles, are poorly understood33. Current approaches might not be able to correctly identify the factors that act on these unclear mechanisms to affected NSC differentiation. Moreover, the current methods mentioned above are based on specific markers of each cell type, such as NeuN for neurons, GFAP for astrocytes and Olig2 for oligodendrocytes34–38. These markers are applicable only to specific cells, often those that are already at a certain stage of differentiation39. Thus, early identification of NSC differentiation is of great difficulty, limiting the development of related technical means. There is an urgent need for a more efficient, accurate, convenient and less wasting method; furthermore, it is better to have less influence on subjective judgement and less limit to the present understanding of neural development and differentiation.
Major advancements in biomedical applications of deep learning technology have occurred in recent years, including cell biology40,41. Studies have used deep learning to identify cell types, cell states and cell dynamic progress from flow cytometry or microscopy images42–44. Several recent reports have highlighted some noteworthy findings regarding the use of deep learning to observe and predict physiological processes in stem cells. One study showed that differentiation alters the morphology of haematopoietic stem cells. Deep learning is in a position to recognize these changes from microscopy data and predict the development of haematopoietic stem cells in advance by isolating cells earlier than the beginning of the known development progress45. Another study showed that machine learning could distinguish pluripotent stem cells from early differentiating cells46. These studies highlight the possible further application of deep learning in the field of stem cell therapy, and we speculate that machine learning could be used not only to identify the physiological variation among cells but also to judge the biological characteristics and changes caused by differentiation inducers. We hypothesize that NSCs could show similar features in a very early stage of differentiation under inducer treatment, and binding to specific receptors could lead to an initial effect of inducers on the cellular state, which might be extracted from high-throughput data by deep learning. Implementing the idea can help solve the existing experimental restricts of early identification of NSC differentiation and adjust NSCs-based treatment strategies, providing more practical applications of deep learning.
Using flow cytometry to detect cell markers is a common method with notable advantages, namely, high throughput, imageability and relatively high speed47,48. A cell-cycle study used imageable flow cytometry combined with machine learning to distinguish minor changes49, and a label-free imaging flow cytometry technique was developed based on coherent optical implementation of the photonic time stretch concept, achieving the advantages of high accuracy and fast speed42. The aforesaid findings emphasize that these two advanced approaches could be utilized to address the challenge of identifying NSC differentiation; specifically, imageable flow cytometry could provide large datasets to build deep learning networks.
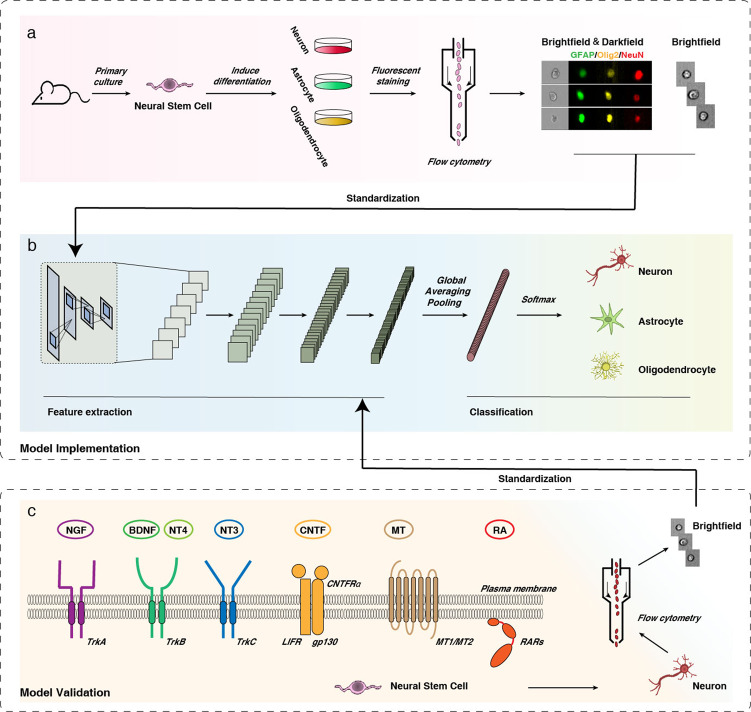
Thus, we developed experiments to exploit the information in brightfield single-cell images for prospective identification of NSC differentiation. Referencing previous studies50–56, through our exploration and experiments, we produced different conditioned culture media to generate specific cell lines from NSCs (Fig. 1a). Several inducers with distinct forms and pathways were selected to test the idea and confirm the value of practical application of our model. Then, the cultured NSCs were marked and collected by imageable flow cytometry; in that way, we obtained 149,428 annotated single-cell images, among which 80% were used as training data to construct deep learning-based darkfield and brightfield models, and the other 20% were used for model testing. We used a convolutional neural network (CNN) to extract local image features, forming a well-architected image classification (Fig. 1b). Recent advances have shown that a CNN can use the convolution operation to establish local connections and to effectively build end-to-end data processing and recognition systems, which have been widely used in image processing43,57,58. CNNs combined with flow cytometry can achieve real-time separation of label-free individual target cells with high accuracy59. In order to further evaluate the general suitability of our system, 59,287 brightfield single-cell images for independent test data were obtained from independent experiments, which were performed using several common types of inducers, including neurotrophic factors, hormones and nanoparticles (Fig. 1c). In this way, we aimed to build a system that allows a CNN to recognize the features of differentiated NSCs via unlabelled brightfield single-cell images, which would provide a quick and simple approach to accurately identify NSC fate at an early phase, screen growth factors/small molecules/nanomedicines with high efficiency, and ultimately promote the medical application of NSCs.
Fig. 1. Overview of workflow.
a NSCs were induced to differentiate into neurons/astrocytes/oligodendrocytes, and the cells were stained with NeuN (red)/GFAP (green)/Olig2 (yellow), collected and subjected to image flow cytometry. b Brightfield and darkfield (labelled) single-cell images were used as training data for the screening system; a schematic of the convolutional neural network (CNN) is presented. c Various inducers in different forms that act on different pathways were used to guide NSCs to differentiate into neurons, and brightfield (unlabelled) single-cell image patches were obtained by flow cytometry. These independent test sets were evaluated with the deep network model to show its generalizability.
Results
A deep learning-based model was established using single-cell images obtained from reliable differentiation experiments
To collect data for model construction, we developed a system that contained differentiating NSCs in different phases, generating neurons, astrocytes and oligodendrocytes and marking them using NeuN, GFAP and Olig2, respectively. To verify the differentiation efficiency of different groups with common laboratory methods, immunofluorescent staining, RT-PCR and western blot were employed. The detection time points were selected according to previously reported studies of differentiation into neurons (1, 3, 5 days), astrocytes (0.5, 1, 2 days), and oligodendrocytes (1, 2, 3 days), separately60–62.
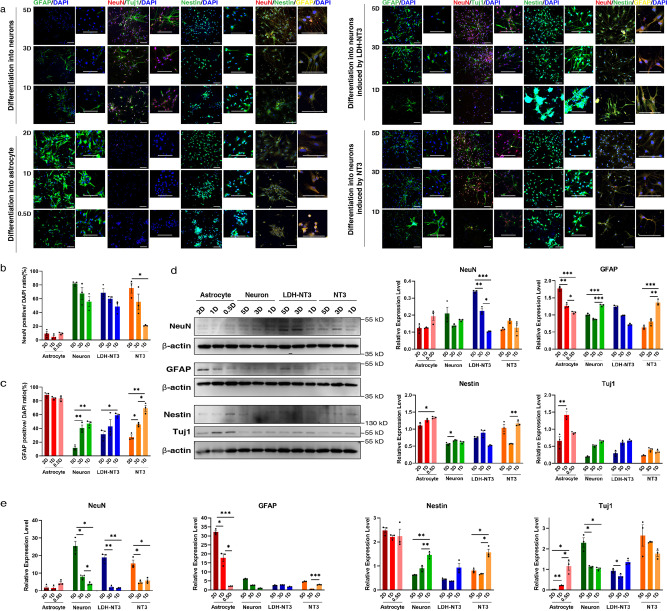
To verify the selected specific markers, we analysed their expression in NSC differentiation process as shown in Fig. 2a and Supplementary Figs. 3 and 4. In agreement with previous studies35,60,63–65, NSCs showed co-expression of GFAP and Nestin, while when treated with neuron differentiation medium, GFAP expression gradually decreased, and the proportion of NeuN+ cells gradually increased. By contrast, with astrocyte differentiation medium, the proportion of GFAP+ cells remained high, while that of NeuN+ cells remained low; over time, GFAP+ cells grew in size and exhibited morphological characteristics of astrocytes. Supplementary Fig. 4e shows that Olig2+ cells increase when treated with oligodendrocyte differentiation medium, while the number of GFAP+ cells is reduced. Results demonstrate that the NeuN, GFAP and Olig2 can clearly label the training data for deep-based model construction.
Fig. 2. Differentiation efficiency of training and testing sets identified by immunofluorescence, western blot and RT-qPCR.
a Images of immunofluorescent staining using NeuN, GFAP, Tuj1 and Nestin as characteristic markers of neuron and astrocyte differentiation induction at 5D (5 days), 3D (3 days), 2D (2 days), 1D (1 day) and 0.5D (0.5 days). Differentiated cells treated with NT3 and LDH-NT3 were stained following the same protocols. Quantification of immunostaining data. The y-axis shows the number of: b NeuN- and c GFAP-positive cells, n = 3 imaging field repeats. d Western blot analysis of NeuN, GFAP, Nestin and Tuj1 protein in NSCs in different states of differentiation, n = 3 biological repeats. e Quantitative real-time PCR detection of NeuN, GFAP, Nestin and Tuj1 gene expression in cells in different states of differentiation, n = 3 biological repeats. Data are shown as mean ± SEM. Statistical significance was determined by two-sided Welch’s ANOVA. *p < 0.05, **p < 0.01, ***p < 0.001. Scale bar = 100 μm. LDH: layered double hydroxide.
As shown in Fig. 2a–c, Supplementary Fig. 4e, f and Supplementary Table 1, under the influence of neuron differentiation medium with retinoic acid (RA) and sonic hedgehog (SHH), 82.073% of NSCs had differentiated into NeuN+ cells after 5 days of induction, 67.427% after 3 days and 55.827% after 1 day. Under the influence of differentiation medium with neurotrophin-3 (NT3), 75.407% of NSCs had differentiated into NeuN+ cells after 5 days of induction, 55.293% after 3 days and 20.997% after 1 day. With layered double hydroxide (LDH) loaded with NT3 nanoparticles (LDH-NT3), 68.823% of NSCs had differentiated into NeuN+ cells after 5 days of induction, 60.06% after 3 days and 48.73% after 1 day. Under the influence of astrocyte differentiation medium, 88.78% of NSCs had differentiated into GFAP+ cells after 2 days of induction, 83.813% after 1 day and 83.357% after 12 h. Under the influence of oligodendrocyte differentiation medium, 63.622% of NSCs had differentiated into Olig2+ cells after 3 days of induction, 55.472% after 2 days and 52.968% after 1 day.
The results of the western blot and RT-qPCR are shown in Fig. 2d, e. The expression of GFAP was highest in the astrocyte induction group and trended upward over time, while the expression of NeuN, Nestin and beta-III-tubulin (Tuj1) decreased over time. The expression of NeuN and Tuj1 was high in the neuron induction group containing RA and SHH, trending upward over time, while the expression of GFAP and Nestin decreased over time. Under the influence of LDH-NT3 and NT3, the expression of these markers had a trend similar to that of the neuron induction group, but weaker.
The above results indicate the reliability of setting differentiation conditions, as well as the clear labelling of the differentiation direction of NSCs. These differentiation methods were used along with flow cytometry, and single-cell images were collected to build and test darkfield and brightfield models. Immunofluorescent staining, RT-qPCR and western blot results demonstrated that the differentiated single-cell data were suitable for deep learning-based model training and testing.
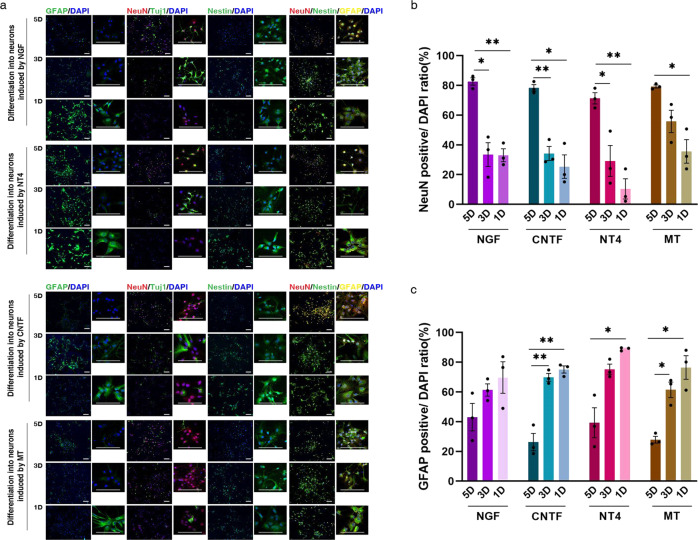
For independent testing of the brightfield model, we obtained images of cells treated with various types of inducers. As shown in Fig. 3 and Supplementary Table 1, the immunofluorescence results show that, under the influence of neuron differentiation medium with neurotrophin-4 (NT4), 71.3333% of NSCs were differentiated into NeuN+ cells after 5 days of induction, 29.2333% after 3 days and 10.4333% after 1 day. Under the influence of neuron differentiation medium with nerve growth factor (NGF), 82.6333% of NSCs were differentiated into NeuN+ cells after 5 days of induction, 33.4667% after 3 days and 33.0333% after 1 day. Under the influence of neuron differentiation medium with melatonin (MT), 79.2667% of NSCs were differentiated into NeuN+ cells after 5 days of induction, 55.7667% after 3 days and 35.6333% after 1 day. Under the influence of neuron differentiation medium with ciliary neurotrophic factor (CNTF), 78.3333% of NSCs were differentiated into NeuN+ cells after 5 days of induction, 34.23% after 3 days and 25.3667% after 1 day. Immunofluorescence results demonstrated the efficiency of the chosen factors as inducers and the suitability of our dataset design.
Fig. 3. Differentiation efficiency of various neuron inducers, identified by immunofluorescence.
a Images of immunofluorescence using NeuN, GFAP, Tuj1 and Nestin as characteristic markers of NGF/CNTF/NT4/MT-treated cells at 5D (5 days), 3D (3 days) and 1D (1 day). Quantification of immunostaining data. The y-axis shows the number of: b NeuN- and c GFAP-positive cells, n = 3 imaging field repeats. Data are shown as mean ± SEM. Statistical significance was determined by two-sided Welch’s ANOVA. *p < 0.05, **p < 0.01, ***p < 0.001. Scale bar = 100 μm.
Deep learning prospectively identifies the differentiation of NSCs at a very early stage
We trained our model on the above-mentioned training set of 119,533 images and tested it on 29,895 images on both darkfield and brightfield channels. To efficiently leverage the dataset, we standardized all images to a size of 45 × 30, and our model achieved high performance on both mixed and independent test sets in terms of accuracy, receiver operating characteristic (ROC)-AUC and precision-recall (PR)-AUC, suggesting its advanced capacity to predict neural differentiation in different scenarios.
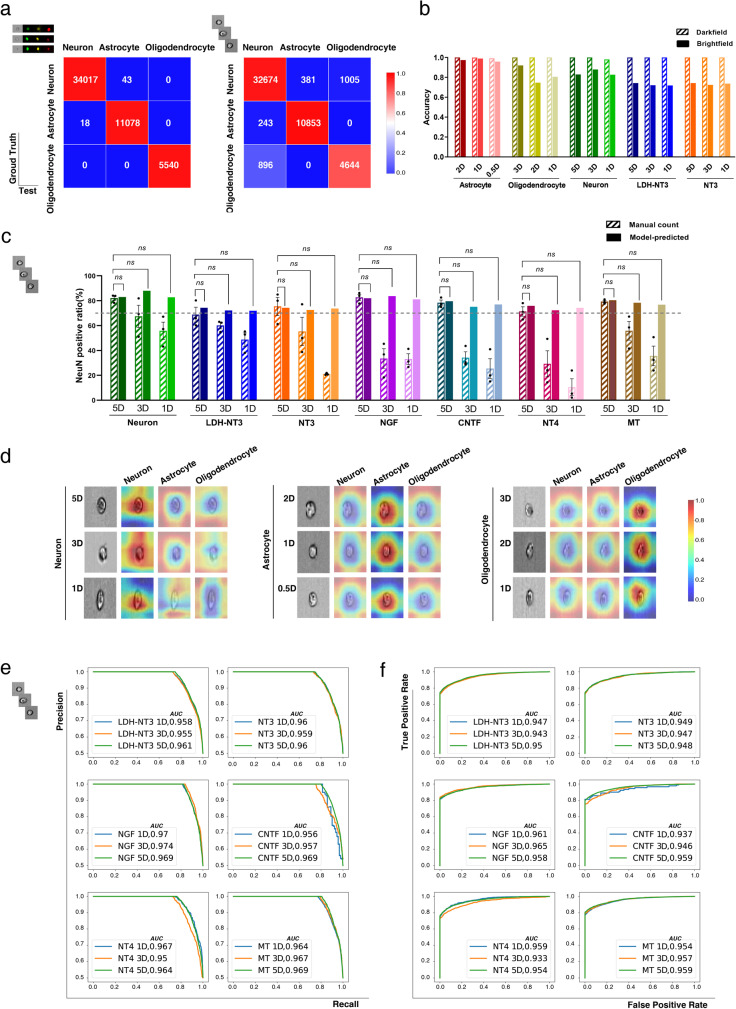
As shown in Fig. 4a, b and Supplementary Table 2, the results suggest no significant difference in total accuracy between the fluorescently marked model (0.998) and the brightfield model (0.923), and the accuracy was high for each differentiation direction at each time point. It is especially interesting that not only the darkfield model but also the brightfield model achieved high accuracy in both mixed test groups (differentiated into neurons, astrocytes and oligodendrocytes; NT3-treated group) and the LDH-NT3-treated group, specifically, illustrating that the brightfield model has strong basic generalizability, which greatly streamlines the application process.
Fig. 4. Deep learning prospectively identifies the differentiation of NSCs at 5D (5 days), 3D (3 days), 2D (2 days), 1D (1 day) and 0.5D (0.5 days).
a Confusion matrices for the darkfield (left) and brightfield (right) models for the classification of each differentiated cell type. b Accuracy of each training set and the independent LDH-NT3 test set in both darkfield and brightfield models, the size of each testing dataset is available in Source Data file. c The proportion of neurons calculated by immunofluorescence and brightfield-based model benchmark, the size of each testing dataset is available in Source Data file. Data are shown as mean ± SEM, statistical significance was determined by two-sided single population t-test. ns: not significant. d The CAM highlights the class-specific discriminative regions of cells, blue represents low attention while red represents high attention. Benchmarking differentiated cell type predictions on independent testing data with: e ROC (receiver operating characteristic) and f PR (precision-recall) curves. LDH: layered double hydroxide.
Surprisingly, as shown in Supplementary Table 1, when NSCs were guided to generate neurons, astrocytes and oligodendrocytes, the brightfield model precisely identified their final fate at 0.5–1 day with very high accuracy (82.72892% for the RA-treated set, 73.67773% for the NT3-treated set, 71.89459% for the LDH-NT3-treated set, 95.86375% for the astrocyte group and 80.59837% for the oligodendrocyte group), consistent with the 5 day experimental observations, whereas common approaches merely demonstrate the instantaneous differentiation status at specific time points by immunostaining. The results indicate that our model can use unlabelled single-cell brightfield images to predict NSC fate within 1 day, which makes it a very valuable approach for NSC research, not only to increase efficiency and reduce cost in the area of molecular screening but also to suppress interference caused by statistical errors, and markers such as GFAP, which expressed in both early differentiated NSCs and astrocytes.
In order to further evaluate the generalization of our system to reliably predict putative NSC differentiation direction in independent experiments that used NGF, NT4, CNTF, MT and LDH-NT3 as inducers for producing neurons, we benchmarked our model on 59,287 datasets with the metrics of accuracy, ROC-AUC and PR-AUC. On all datasets, as shown in Fig. 4c, Supplementary Table 1 and Supplementary Table 2, the brightfield-based model achieves excellent prediction of neuron proportions at 1 day (81.09294% on the NGF set, 74.197% on the NT4 set, 76.92308% on the CNTF set, 76.79027% on the MT set and 71.89459% on the LDH-NT3 set), which is closely consistent with the 5 days immunofluorescence counts. However, common laboratory methods could not identify differentiation at very early stages at 1 day and 3 days, asserting the superiority of our model over existing approaches. Furthermore, the system gained records on both ROC-AUC and PR-AUC, as shown in Fig. 4e, f, suggesting excellent robustness and capability to minimize the imbalance and false positives.
Class activation maps offer insight into the functional mechanisms of medications
To understand how our model interprets the input cell images, we introduced class activation mapping techniques66. The input cell images shown in Fig. 4d are randomly selected from each treated group. Given these images, the activation maps of each layer block were extracted. Of these maps, Supplementary Fig. 5 presents six examples per block due to space limitations, although each layer block has hundreds of activation maps. The layer block-wise activation maps depict the appearance of the input images or the activation maps from the previous layer block processed in units of the current layer block. Moreover, we established the final class activation maps (Fig. 4d). These class activation maps are used to describe the model’s receptive field in the case of given classes, where blue represents low attention while red represents high attention. Results show the attention of the model is concentrated on the cell, including its edges and internal parts, when the given class matches the predicted class, and distracted otherwise, indicating that the model tends to perform its classification based on the details of both edges and internal parts of the cells.
Performance comparison with other neural network architectures
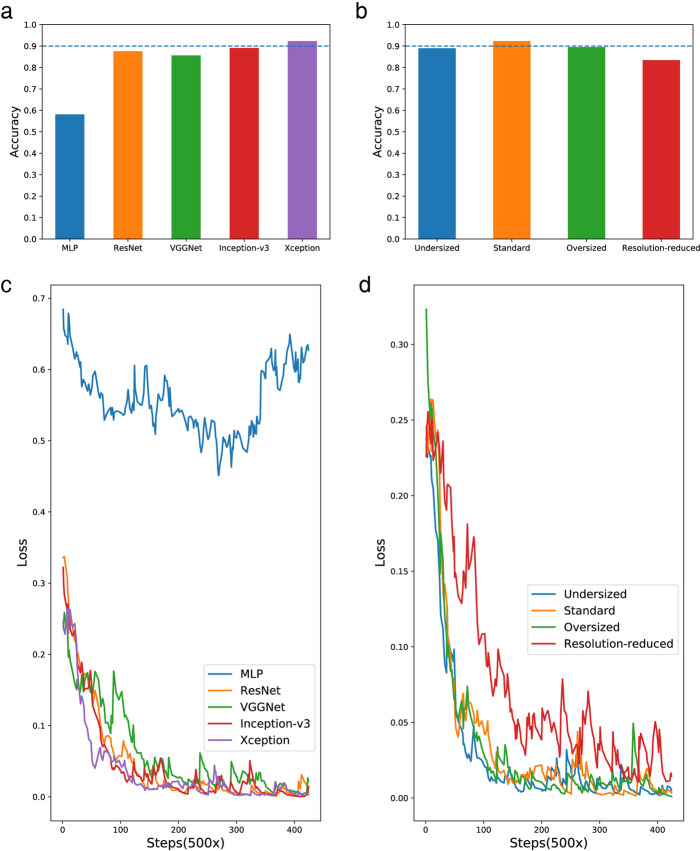
We compared the performance of our Xception-based model67 with models established using other deep learning techniques, including multi-layer perceptron, ResNet68, VGGNet69 and Inception-v370 architectures. These trained models utilize the same training and testing dataset (containing 119,533 images and 29,895 images, respectively) as the Xception-based model above. Figure 5a compares the accuracy of each model on the testing dataset, showing that the Xception-based model is the only one that exceeds 0.9. Additionally, the loss curves of the compared models are depicted in Fig. 5c, showing that the curve of the Xception-based model lowers quickly in the early steps and stabilizes at nearly the smallest loss value among all the network architectures, representing good robustness. These comparisons indicate that the chosen Xception method performs the best among the five deep learning techniques.
Fig. 5. Performance comparison of models established with different structures.
Accuracy comparison of models with: a different architectures and b Xception models with different size and input resolutions. Loss curves comparison of models with: c different architectures and d Xception models with different size and input resolutions. The size of each testing dataset is available in Source Data file. MLP, multi-layer perceptron.
Moreover, we built and evaluated three additional Xception-based models, including an undersized version, an oversized version and a resolution-reduced version, with the same training and testing datasets, where the undersized version was 30% smaller than the standard version while the oversized version was 30% larger. For the resolution-reduced version, the size of the input images was reduced to 15 × 10, in contrast to the standard version, whose input images were 45 × 30. Figure 5b depicts a comparison of accuracy among the four models, showing that the standard version remained the highest. Regarding training robustness, the standard version gave the best performance once again, as shown by the comparison of loss curves in Fig. 5d. The comparison in this part indicates that the undersized version and oversized version, respectively, underfit and overfit the data, in contrast to the standard version of the model; additionally, shrinkage of the input images caused a shortage of effective information, making the corresponding model perform poorly.
Discussion
Transplantation of NSCs offers exciting possibilities for CNS regeneration, but it is challenging to guide the differentiation of NSCs into specific cell types. Biomarkers are commonly used to examine differentiation towards distinct cell fates, relying on the characteristics of cells during the neurogenesis process71. Despite a variety of studies on the factors regulating the fate of NSCs, the neurogenesis process remains to be fully elucidated, especially the cellular changes at the very early stage of differentiation into neurons72,73; thus, it is difficult to identify the direction of differentiation early in the process. An identification process must be generalizable to support the advanced development of efficient agents for neurodegenerative diseases and neurological injuries: it needs to apply to any effective substance, independent of what its pathways may happen to be. Advanced instrumentation facilitates data collection, but one of the most challenging obstacles is that the data are often uninterpretable within the limitations of current devices. Existing approaches are based primarily on human cognition, but differentiating subtle cellular morphologic changes or predicting drug interactions is challenging for human beings74–76. Deep learning enables automatic feature extraction utilizing vast datasets to offer solutions to perplexing problems, including those encountered in the biomedical field77,78. Thus, we decided to take full advantage of abundant single-cell images by using deep learning to prospectively identify the differentiation of NSCs.
First, we innovatively devised and framed a very accurately marked NSC differentiation dataset, ensuring the effectiveness of model training. The NSC fate identification system was built based on the confirmed experimental data, and it is critical to successfully extract the characteristic information of each cell type. An elegant CNN structure produced a highly efficient and strongly generalizable model. We then tested the performance of our system, and the results suggested that our model could estimate the proportion of the final differentiated cell type in the early stages of differentiation, before common laboratory techniques could detect the corresponding alterations.
When gathering the cell images, the repeatability of experiments matters to the further application for the model, thus the cell status for training data is important. The proliferation and differentiation abilities of prolonged cultured NSCs of late passages have differences with early passages, and p0–p2 cells from primary culture may not be able to form enough amount of neurospheres with stable status for collecting high-throughput data, thus researches usually take NSCs from p3 to p979–84. We conducted experiments to evaluate the state of p3–p5 NSCs, results show strong expression of specific markers Nestin and PAX6 in all groups (Supplementary Fig. 2a), indicating certain proliferation capacity. Immunostaining results of measuring differentiation ratio of p3–p5 NSCs demonstrated the consistent differentiation ability (Supplementary Fig. 2b, c), indicating that NSCs at p3–p5 are suitable for model building data collection. Regarding our training data, RA induces NSCs to differentiate into neurons by binding to RA receptors (RARs)85, whereas the combination of thyroid hormone (T3) and platelet-derived growth factor (PDGF) promotes the differentiation of oligodendrocytes through the Wnt/β-catenin signalling pathway86, CNTF, with high concentrations acting via the JAK-STAT signalling pathway to differentiate cells into astrocytes87. Furthermore, we compiled independent testing data on NSC differentiation using a variety of neuronal inducers targeting different receptors (Fig. 1c). First, we used several neurotrophins, including NGF, BDNF, NT4 and NT3, to regulate NSC differentiation into neurons. Neurotrophins are peptide growth factors implicated in neuronal differentiation; they bind to their related receptor tyrosine kinases (Trks), which are located in the plasma membrane of responsive cells. NGF preferentially binds to TrkA, BDNF and NT4 preferentially bind TrkB, and NT3 acts on TrkC88,89. Next, low-concentration CNTF was used as another neuronal inducer. This inducer, which belongs to the haematopoietic cytokine superfamily, acts by binding to CNTF receptor α (CNTFRα) and gp130, ultimately recruiting leukaemia inhibitory factor receptor β (LIFRβ) and activating the mitogen-activated protein kinase pathways87,90. Subsequently, MT was utilized to facilitate neuron differentiation mediated via activation of MT receptors 1 and 2 (MT1 and MT2)91. Moreover, recent studies on the impact of nanoparticles on NSC fate were taken into consideration92,93. LDH has been shown to assist NT3 in regulating stem cell fate94,95; to determine the general validity of our system, we tested it with LDH-NT3 as an unconventional type of inducer with an unclarified mechanism.
Additionally, we performed a study to minimize the duration of inducer treatment in order to improve the efficiency of model training and application; in that study, different time points were carefully selected for cell harvesting. Research indicates that when machine learning determines very early whether embryonic stem cells differentiate or not, its judgements may be based on cell-level variations in biological processes46, suggesting that the physiological process of NSC differentiation is the critical factor that needs to be considered when set the time points. Previously, studies show that it takes different differentiation time when NSCs covert into each lineage, around 7 days for neurons60, 3 days for astrocytes61 and 5 days for oligodendrocytes62, respectively. At these time points, cells start to appear their specific markers expression and functional properties. Our greatest concern is to identify NSC fate in the early stage of differentiation and determine the actions of the inducers, thus shorter time points are set to establish the predictable model. We tested the gene expression levels to observe the changes in NSCs under the action of different inducers to design further data collection procedures for model construction. As the results show in Supplementary Fig. 4, for cells undergoing neuronal differentiation, the expression level of NeuN firstly showed a significant increase at 1 day, between 3 days and 1 day there is an observable difference and at 5 days also there appears a significant increase, while there are no significant changes at earlier time points of 0, 2, 4, 8 and 12 h; thus, 1 day was set as the earliest time point for detection. For astroglial and oligodendroglial differentiation, obvious changes in GFAP and Olig2 appeared at 2 days and 3 days, and significant changes first appeared at 12 h and 1 day, respectively. Therefore, the earliest expected time points for effective prediction by our prediction system were set to 12 h for astrocytes and 1 day for neurons and oligodendrocytes. The results showed that, after these minimum durations, our model could predict the differentiation direction and ratio of cells several days in advance, which may be due to its sensitivity to subtle changes in cell state or organelle morphosis.
In our previous work, we built an Inception 3-based neural network that could analyse the distinction of apoptotic cells; however, we noticed that the complicated structures and numerous manually defined branches increased the difficulty of training, which might affect the efficiency of the optimal model, and the results showed that its performance in brightfield detection had room for improvement96. In the current study, we used a better-constructed neural network based on Xception, which reduced the complexity of the model and made it more concise and clear, and this approach proved to be very effective. The present network builds the optimization model from simple training with the hyperparameters of basic training parameters and default learning rate; in particular, unlabelled brightfield images can fully meet the modelling requirements with excellent accuracy of 0.923. The results of both mixed testing data and multiple independent experiments demonstrated that the mechanism does not influence the effect of the model’s verdict, nor does the form of inducer (Fig. 4a and Supplementary Tables 1 and 2). Our system is able to prospectively identify the direction of NSC differentiation in 1 day using non-fluorescently labelled cell images, and the ROC-AUC/PR-AUC verified its high robustness and precision (Fig. 4e, f and Supplementary Tables 1 and 2). Class activation mapping, shown in Fig. 4d, suggested that our model can identify very small morphological variations in cellular structures; thus, it was able to classify cell fate by discriminating differences between each type, which is the main strength of machine learning. Furthermore, we explored the predictive performance achieved by CNN architectures with various layer structures, branching strategies, network capacities and resolutions of input cell images. The results of the performance comparison on the test set demonstrated the superiority of the architectures that our model adopts.
In addition to the above advantages, our model has the potential for functional expansion to cover a variety of applications; for instance, predicting neuronal subtypes such as motor neurons or dopaminergic neurons would be a very meaningful development. Dopamine neuron transplantation can innervate the striatum and improve motor asymmetry and is a promising therapy for Parkinson’s disease97. Transplantation of motor neurons derived from stem cells can significantly promote motor function recovery in amyotrophic lateral sclerosis98. The development of a more efficient identification system for specific neuron populations will undoubtedly promote cell-based therapy for specific neurodegenerative diseases. At present, however, there are a few hurdles to overcome before this further application. First, the differentiation ratio of NSCs or induced pluripotent stem cells into a specific population is not sufficient to gather a suitable dataset to train the model. According to a previously reported study, under 40% of stem cells treated with several cytokines can differentiate into dopamine neurons or motor neurons64,99–102. In order to build a training set for machine learning, correct classification and labelling are necessary. In addition to isolating the specific subpopulations, flow cytometry/cell sorting should be used to enrich the target cells, but the cells have been secondarily treated before analysis, which introduces uncontrolled variables and reduces training efficiency. Meanwhile, specific markers for neuron subpopulation profiling and selection remain under investigation60, making it difficult to clearly tag data. Another valuable possible extension of the model is to explore the region heterogeneity of NSC differentiation. Heterogeneity describes the regionalization of NSC niches, progenitor cells in different regions express different genes to become different subtypes of cells, appears to be a key feature of NSC development in the CNS, but how it affects NSC fate and function is not fully understood35,103. It is worth considering isolating regional NSC subpopulations, taking advantage of the model to effectively identify subtle cellular variation during the differentiation progress and even explore the impact of different inducers on NSC subtypes development. Some limitations need to be addressed before further exploration, first, to resolve additional layers of heterogeneity, optimized protocols are needed to be established for precise isolation and effective enrichment of regional subtypes.104 Additionally, research shows that existing exploration of regional heterogeneous NSC subtypes is to observe the changes of gene expression using single-cell RNA-sequencing, however, there is still lack of recognized specific markers to identify cells from different regions including ventricular-subventricular zone (V-SVZ) or subgranular zone (SGZ). Besides, lineage-tracing studies indicate that regional specification is established very early in embryonic development, but the progress of its development still needs to be more clearly elucidated in order to set up suitable time points for collecting the regional NSC subtypes105. Though there are difficulties, we believe the extension to the deep-based model application is well worth the efforts due to its strong applied value, suggesting that our system can be used not only in predicting NSC fate and screening neural induction factors but also in research on NSC reactivation and development.
In summary, we provide an advanced tool for the identification of NSC fate; this model has the key features of high precision, a low false positive rate, speed, wide applicability, simple operation and cost-effectiveness. After NSCs are co-cultured for 1 day with the inducer to be studied, our system can offer a predictive differentiation result with a single-cell image. This could be a convenient and well-suited method to investigate the unknown effects of substances on NSCs and rapidly predict potential therapeutic molecules and drugs to promote neural regeneration in CNS diseases. We are also working with further extensions and applications of our system, attempting to develop a multifunctional approach for NSC research.
Methods
Experiments and data collection
Primary NSC processing and culture
Primary NSCs were processed as follows: Embryonic SD rats (E13.5) were chosen for cell extraction. The brain of each embryo was isolated under a stereoscopic microscope, washed with 1× phosphate buffer saline (PBS, Gibco, catalogue no. 10010023) and then incubated with 0.125% trypsin solution (Gibco, catalogue no. 25200056) at 37 °C for 3 min. Next, DMEM containing 10% foetal bovine serum (FBS, Gibco, catalogue no. 040011 A) was applied to neutralize the trypsin activity. The suspension was filtered with 70 μm mesh cell strainers (BD Biosciences, catalogue no. 087712). The filtered cell suspension was centrifuged and resuspended in DMEM/F12 (Gibco, catalogue no. 11320033) containing 2% B27 (Gibco, catalogue no. 17504044), 1% N2 (Gibco, catalogue no. 17502001), 20 ng/ml basic fibroblast growth factor (bFGF, Peprotech, catalogue no. 45033) and 20 ng/ml epidermal growth factor (EGF, Peprotech, catalogue no. 31509). Primary NSCs were cultured in proliferation medium for 2 days until neurosphere formation. The neurospheres were centrifuged and incubated with Accutase (Stem Cell, catalogue no. 7920) for 5 min in a 37 °C incubator to obtain single NSCs for continuous passage culture. NSCs were passaged every 4 days with Accutase after neurospheres formed, and half of the culture solution was refreshed every 2 days. Animals were obtained from Shanghai Slac Laboratory Animal Co., Ltd., and raised in the SPF animal laboratory at Tongji University. All protocols were approved by the Institute of Laboratory Animal Resources of Tongji University and complied with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Ethical and legal approval for this study was obtained from the Institute of Laboratory Animal Resources Animal Care and Use Committee. All efforts were made to minimize animal suffering and sacrifice.
NSC identification and differentiation
NSCs at p3, p4, p5 were identified by immunostaining, using specific markers Nestin and PAX6. For inducing differentiation experiments, cells were seeded onto poly-L-ornithine-coated 6-well plates, and after 12 h of incubation at 37 °C, the proliferation medium was replaced by differentiation medium. The composition of the astrocyte differentiation medium was as follows: DMEM (Gibco, catalogue no. 11965092), 10% FBS, 20 ng/ml CNTF (Peprotech, catalogue no. 45013), 10 ng/ml EGF and 10 ng/ml bFGF. The composition of the oligodendrocyte differentiation medium was as follows: DMEM/F12, 10 ng/ml EGF, 20 ng/ml PDGF (Peprotech, catalogue no. 10014B) and 50 nM T3 (ProSpec-Tany, catalogue no. hor001). The composition of the neuron differentiation medium with RA (Sigma, catalogue no. R2625) and SHH (Peprotech, catalogue no. 31522) was as follows: Neurobasal (Gibco, catalogue no. 10888022), 2% B27, 1% N2, 10 ng/ml BDNF (Peprotech, catalogue no. 45002), 50 ng/ml insulin-like growth factor (IGF, Peprotech, catalogue no. 10011), 0.1 μM c-AMP (Sigma, catalogue no. A9501), 1 μM ascorbic acid (AA, catalogue no. A4403), 1 μM RA and 1 μM SHH. The composition of the neuron differentiation medium with NT4 (Peprotech, catalogue no. 45004) was as follows: Neurobasal, 2% B27, 1% N2, 10 ng/ml BDNF, 50 ng/ml IGF, 0.1 μM c-AMP, 1 μM AA and 20 ng/ml NT4. The composition of the neuron differentiation medium with NGF was as follows: Neurobasal, 2% B27, 1% N2, 10 ng/ml BDNF, 50 ng/ml IGF, 0.1 μM c-AMP, 1 μM AA and 10 ng/ml β-NGF (Peprotech, catalogue no. 45001). The composition of the neuron differentiation medium with CNTF was as follows: Neurobasal, 2% B27, 1% N2, 10 ng/ml BDNF, 50 ng/ml IGF, 0.1 μM c-AMP, 1 μM AA and 100 ng/ml CNTF. The composition of the neuron differentiation medium with MT (Sigma, catalogue no. M5250) was as follows: Neurobasal, 2% B27, 1% N2, 10 ng/ml BDNF, 50 ng/ml IGF, 0.1 μM c-AMP, 1 μM AA and 25 mM MT. The composition of the differentiation medium with NT3 (Peprotech, catalogue no. 45003) was as follows: Neurobasal, 2% B27, 1% N2, 10 ng/ml BDNF, 50 ng/ml IGF, 0.1 μM c-AMP, 1 μM AA and 20 ng/ml NT3. The composition of the differentiation medium with LDH-NT3 was as follows: Neurobasal, 2% B27, 1% N2, 10 ng/ml BDNF, 50 ng/ml IGF, 0.1 μM c-AMP, 1 μM AA, LDH-NT3 with 0.08 ng/ml NT3, and 2 μg/ml LDH.
LDH-NT3 preparation and characterization
LDH was prepared as follows: 0.544 g NaOH was dissolved in 80 ml deionized water (ddH2O) under N2 atmosphere, then a mixture of 1.538 g Mg(NO3)2·6H2O and 0.75 g Al(NO3)3·9H2O was dissolved in 20 ml ddH2O, and added dropwise in the NaOH solution. After 30 min stirring at 500 rpm, the product was washed thrice with ddH2O, and dispersed in ddH2O. After keeping for 16 h hydrothermally at 100 °C, LDH was obtained by centrifugation. NT3 was loaded into LDH using ion exchange intercalation. The morphological features were observed by transmission electron microscopy (TEM). LDH and LDH-NT3 have a size of approximately 100 nm, with a hexagonal lamellar structure (Supplementary Fig. 1).
Flow cytometry
NSCs treated with differentiation medium were collected with Accutase at different stages and washed thrice with PBS before being fixed. The cells were fixed in cold 80% methanol for 5 min and then disrupted using 0.1% Triton X-100 on ice for 20 min. Later, the cells were incubated with anti-GFAP (Bioss, catalogue no. bs-0199R-AF488, 1:200), anti-NeuN (Bioss, catalogue no. bs-10394R-APC, 1:200) and anti-Olig2 (Bioss, catalogue no. bs-11194R-PE, 1:200) antibodies for 2 h on ice. The cells were washed three times with PBS before flow cytometry. Eventually, approximately 20,000 stained cells were collected and photographed. Experiments of the training sets (astrocyte, neuron, oligodendrocyte and NT3-treated groups) were carried out in the Key Laboratory of Spine and Spinal Cord Injury Repair and Regeneration of the Ministry of Education; Experiments of independent test for the darkfield model (LDH-NT3-treated group) were carried out in the laboratory of the School of Life Science and Technology of Tongji University. Images of the training and the darkfield model independent test groups were gathered using a FlowSight apparatus (Merck Millipore) at Tongji University. The brightfield model independent test data (NT4-, NGF-, CNTF-, MT-treated groups) were collected in the laboratory of Tongji Hospital and photographed by ImageStream Mark II apparatus (Merck Millipore) at the Chinese Academy of Sciences Shanghai Institute of Materia Medica. Single-cell images were gained from IDES v6.1. Each laboratory has its own experimenters to conduct the experiments.
Immunofluorescence
Immunofluorescence assays were performed as follows: the cells were washed three times with PBS and fixed with 4% paraformaldehyde for 15 min. Next, the cells were washed and blocked with PBS containing 5% donkey serum and 0.3% Triton X-100 at 37 °C for 1 h, then incubated with primary antibodies at a suitable dilution ratio in blocking solution at 4 °C overnight. Afterward, the cells were washed to remove residual antibody. The secondary antibody was applied at a suitable dilution ratio at 37 °C for 1 h. 4′,6-Diamidino-2-phenylindole (DAPI) was used to stain the nuclei. The antibodies used were as follows: anti-NeuN (Abcam, catalogue no. ab190195, conjugated with Alexa Fluor 488, 1:200), NeuN (Abcam, catalogue no. ab104225, 1:500), anti-NeuN (Millipore, catalogue no. MAB 377, 1:100), anti-GFAP (Abcam, catalogue no. ab53554, 1:1000), anti-Tuj1 (Abcam, catalogue no. ab78078, 1:1000), anti-Nestin (Abcam, catalogue no. ab6320, 1:500), anti-PAX6 (Abcam, catalogue no. ab195045, 1:350), anti-Olig2 (Abcam, catalogue no. ab109186, 1:100), rabbit anti-mouse IgG (YEASEN, catalogue no. 33912ES60, 1:200), donkey anti-mouse IgG (Abcam, catalogue no. ab150105, 1:1000), donkey anti-goat IgG (Abcam, catalogue no. ab175704, 1:1000) and donkey anti-rabbit IgG (Abcam, catalogue no. ab150075, 1:1000). DAPI (Sigma, catalogue no. D9542) was used to stain the nuclei. A Zeiss confocal microscope (LSM 700, Carl Zeiss, Jena, Germany) was applied to observe the fluorescence signal. Image acquisition was done with ZEN 2.3 (blue edition, Carl Zeiss) and micrographs were assembled using Adobe Illustrator CC 2018.
Western blot assay
The total protein content was extracted from cells using nucleoprotein and cytoplasmic protein extraction kit (KeyGen, catalogue no. KGP250) according to the manufacturer’s instructions. Then, a western blot assay was performed as follows: protein was mixed with 5× protein-loading buffer (KeyGen Biotech., Nanjing, China) and boiled at 95 °C for 5 min before being separated on a 12% SDS-PAGE gel. After electrophoresis, proteins were transferred to PVDF membranes (EMD Millipore, Billerica, MA, USA) by applying a current of 300 mA for 100 min in an ice bath. Then, the membranes were blocked with 5% BSA solution containing 0.1% Tween-20 (TBST) for 30–60 min at room temperature. The blocked PVDF membranes were incubated with primary antibodies at 4 °C overnight and washed three times with TBST. The antibodies used were as follows: anti-NeuN (Abcam, catalogue no. ab104225, 1:5000), anti-Nestin (Abcam, catalogue no. ab6320, 1:1000), anti-GFAP (Abcam, catalogue no. ab53554, 1:10,000), anti-Tuj1 (Abcam, catalogue no. ab78078, 1:1000), anti-β-actin (ABWAYS, catalogue no. AB0035, 1:5000), goat anti-mouse HRP (Abcam, catalogue no. ab205719, 1:5000) and goat anti-rabbit HRP (Abcam, catalogue no. ab205718, 1:5000). Blots were visualized with an ImageQuant LAS 4000 mini (GE Healthcare Life Science). Densitometry of protein bands was measured with ImageJ 1.47v and normalized to the respective internal control (β-actin) band. Graphs and statistical analysis were done in GraphPad Prism 8 version 8.4.2.
Quantitative real-time reverse transcription polymerase chain reaction (RT-qPCR)
The total RNA of cells was isolated with RNAiso plus (Takara, catalogue no. 9108). The concentration and purity of RNA samples were measured with a Nanodrop ND-2000 (Thermo Science, MA, USA). cDNA was synthesized with a Primer Script Reverse Transcriptase Kit (Takara, catalogue no. RR037A). Quantitative real-time PCR was performed using a TB GreenTM Premix Ex Taq Kit (Takara, catalogue no. RR820A) on a LightCycler Real-Time PCR System (Roche, 480II). The primer sequences (Sangon Biotech) are listed in Supplementary Table 3. The relative amounts of mRNA were calculated using the ΔΔCt relative quantification method. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as the control gene, and the mRNA levels of specific genes were normalized to GAPDH. Calculations and statistics were done in Microsoft Excel version 16.36; graphs were plotted in GraphPad Prism 8 version 8.4.3.
Image data preprocessing
The cell images were preprocessed before feed into the neural network. The processing details are listed below: first, resize each image to 45 × 30 with OpenCV package; second, divide each resized image pixel value by 255 to squash the value domain down to 0–1.
Deep learning techniques
Convolutional neural network for classification
We implemented a set of CNNs to perform the classification task for each cell image.
These CNNs were constructed with the “Xception” module, which is an upgraded version of the “Google Inception” module. The difference between the “Google Inception” module and a common linearly connected convolutional layer aggregate is that the former adopts a “network-in-network” structure containing branches to extract further useful information from the data flow. The number of branches between two specific layers is usually 2 or 3. Compared with the “Google Inception” module, the “Xception” module uses an extreme policy to perform one spatial convolution for every output channel of the 1 × 1 convoluted input data. With this policy, the “Xception” network surpasses the “Google Inception” network in various benchmarks. In addition, the “Xception” module is much simpler and more compact than the “Google Inception” module, making the whole network simple, clear and highly extendable.
Model training methodology
We used the Adam optimizer combining the momentum and exponentially weighted moving average gradients methods to update the weights of the networks.
The networks were trained with the PyTorch framework on four NVIDIA GTX 1080Ti GPUs. The parameters of the Adam optimizer are as follows: learning rate of 0.001, beta1 of 0.9, beta2 of 0.999 and epsilon of 10−8.
Activation mapping for each layer block
First, we randomly selected a total of 9 cell images, with each treated group providing one image. Given these cell images, the activation maps of each layer block of the model were collected. Due to space limitations, only six activation maps per layer block per image are presented in Supplementary Fig. 5.
Class activation mapping for feature identification
We adopted the class activation mapping technique to generate a comprehensive view with which to understand the recognition paradigm of the networks. For this method, the last three layers were as follows: a feature convolution layer, a global average pooling layer and a 1 × 1 convolution layer without an activation function. In order to produce the class activation map, the weights of the final 1 × 1 feature convolution layer were extracted. Then, the selected input images, which were the same as the input images used by the layer block-wise activation mapping process, were used to perform the forward pass, collecting the feature maps of the feature convolution layer. Finally, the class activation maps were calculated by the weights on the linear combinations of these feature maps, and images were generated through the Python visualization package “Matplotlib” (3.2.2 version).
Other related neural network architectures
We implemented various other types of neural networks to compare with our Xception-based network, including multi-layer perceptron, ResNet, VGGNet and Inception-v3 networks.
The multi-layer perceptron had three layers, namely, the input layer, the hidden layer and the output layer. The input layer took a 45 × 30 image as input and transformed it into a vector of length 512, which was fed into the 512 × 512 hidden layer. Finally, the output layer used the output of the hidden layer to perform the three-way classification.
For the ResNet, VGGNet and Inception-v3 networks, we used the classic layer configuration of each network with customized starting layers to fit the size of the cell images.
Statistics and reproducibility
All values are presented as mean ± SEM calculated by GraphPad Prism 8 version 8.4.3. Two-sided Welch’s ANOVA was used to identify significant differences between different groups in immunofluorescence, RT-qPCR and western blot experiments in Figs. 2 and 3. Two-sided single population t-test was applied to identify significant differences between the proportion of neurons calculated by immunofluorescence and brightfield-based model benchmark in Fig. 4c. p < 0.05 was taken to indicate a statistically significant difference. Statistical significance levels are denoted as follows: *p < 0.05; **p < 0.01; ***p < 0.001. Results of Shapiro-Wilk normality test are shown in Supplementary Fig. 6. p > 0.05 was taken as for passing the normality test. In general, n values refer to the number of biological repeats or imaging fields for a given experiment; details are provided for each experiment in the corresponding figure legend. All data from representative experiments were repeated three times independently with similar results.
We tested the performance of our model on independent datasets using metrics of accuracy, ROC-AUC and PR-AUC, as well as generated the CAM figures given the exemplary cell image without retraining the networks. All attempts at replication were successful with similar results.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This work was financially supported by the National Key Research and Development Program (Grant No. 2016YFA0100800), the National Natural Science Foundation of China (Grant No. 81820108013, 81922039, 81873994, 81901902, 31727801) and Basic Research Project of Shanghai Science and Technology Commission, China (Grant No. 19JC1414700).
Source data
Author contributions
R.Z., L.C. and Y.Z. conceptualized the project. Y.Z. analysed flow cytometry single-cell images and performed Deep Learning techniques. R.H., Z.W. and S.S. optimized cell differentiation. R.H. assisted with confocal imaging and RT-qPCR. Z.W. and S.S. prepared western blot analyses. Y.Z. and R.H. wrote the manuscript. R.Z., Y.Z. and R.H. revised the manuscript. R.Z. and L.C. supervised the project.
Data availability
The main data supporting the findings of this study are available within the article and in the Supplementary Information. The single-cell image data for model building are available through the figshare website with the download link of 10.6084/m9.figshare.13070666.v1. Source data are provided with this paper.
Code availability
The code for model building is available through the link 10.5281/zenodo.4606918
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks Roman Bauer, Maeve Caldwell and Ata Mahjoubfar for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yanjing Zhu, Ruiqi Huang.
Contributor Information
Liming Cheng, Email: limingcheng@tongji.edu.cn.
Rongrong Zhu, Email: rrzhu@tongji.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-021-22758-0.
References
- 1.Lane SW, Williams DA, Watt FM. Modulating the stem cell niche for tissue regeneration. Nat. Biotechnol. 2014;32:795–803. doi: 10.1038/nbt.2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Madl CM, Heilshorn SC, Blau HM. Bioengineering strategies to accelerate stem cell therapeutics. Nature. 2018;557:335–342. doi: 10.1038/s41586-018-0089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizuno H, Tobita M, Uysal AC. Concise review: adipose-derived stem cells as a novel tool for future regenerative medicine. Stem Cells. 2012;30:804–810. doi: 10.1002/stem.1076. [DOI] [PubMed] [Google Scholar]
- 4.Blurton-Jones M, et al. Neural stem cells improve cognition via BDNF in a transgenic model of Alzheimer disease. Proc. Natl Acad. Sci. USA. 2009;106:13594–13599. doi: 10.1073/pnas.0901402106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindvall O, Kokaia Z. Stem cells in human neurodegenerative disorders - time for clinical translation? J. Clin. Invest. 2010;120:29–40. doi: 10.1172/JCI40543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marei HE, et al. Potential of stem cell-based therapy for ischemic stroke. Front. Neurol. 2018;9:34. doi: 10.3389/fneur.2018.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong Y, Mahmood A, Chopp M. Angiogenesis, neurogenesis and brain recovery of function following injury. Curr. Opin. Invest. Drugs. 2010;11:298–308. [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke DL. Neural stem cells. Bone Marrow Transplant. 2003;32:S13–S17. doi: 10.1038/sj.bmt.1703937. [DOI] [PubMed] [Google Scholar]
- 9.Ladran I, Ngoc T, Topol A, Brennand KJ. Neural stem and progenitor cells in health and disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 2013;5:701–715. doi: 10.1002/wsbm.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmer TD, Takahashi J, Gage FH. The adult rat hippocampus contains primordial neural stem cells. Mol. Cell. Neurosci. 1997;8:389–404. doi: 10.1006/mcne.1996.0595. [DOI] [PubMed] [Google Scholar]
- 11.Liu K, Tedeschi A, Park KK, He Z. Neuronal intrinsic mechanisms of axon regeneration. Annu. Rev. Neurosci. 2011;34:131–152. doi: 10.1146/annurev-neuro-061010-113723. [DOI] [PubMed] [Google Scholar]
- 12.Tzeng SF. Neural progenitors isolated from newborn rat spinal cords differentiate into neurons and astroglia. J. Biomed. Sci. 2002;9:10–16. doi: 10.1007/BF02256573. [DOI] [PubMed] [Google Scholar]
- 13.Park SY, et al. Enhanced differentiation of human neural stem cells into neurons on graphene. Adv. Mater. 2011;23:H263–H267. doi: 10.1002/adma.201101503. [DOI] [PubMed] [Google Scholar]
- 14.Deng W, et al. EMSCs build an all-in-one niche via cell-cell lipid raft assembly for promoted neuronal but suppressed astroglial differentiation of neural stem cells. Adv. Mater. 2019;31:1806861. doi: 10.1002/adma.201806861. [DOI] [PubMed] [Google Scholar]
- 15.Li X, et al. A collagen microchannel scaffold carrying paclitaxel-liposomes induces neuronal differentiation of neural stem cells through Wnt/beta-catenin signaling for spinal cord injury repair. Biomaterials. 2018;183:114–127. doi: 10.1016/j.biomaterials.2018.08.037. [DOI] [PubMed] [Google Scholar]
- 16.Lu P, et al. Long-distance growth and connectivity of neural stem cells after severe spinal cord injury. Cell. 2012;150:1264–1273. doi: 10.1016/j.cell.2012.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee H-R, et al. Functional group-dependent induction of astrocytogenesis and neurogenesis by flavone derivatives. Biomolecules. 2019;9:812. doi: 10.3390/biom9120812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee H-R, Lee J, Kim H-J. Differential effects of MEK inhibitors on rat neural stem cell differentiation: Repressive roles of MEK2 in neurogenesis and induction of astrocytogenesis by PD98059. Pharmacol. Res. 2019;149:104466. doi: 10.1016/j.phrs.2019.104466. [DOI] [PubMed] [Google Scholar]
- 19.Vecino E, David Rodriguez F, Ruzafa N, Pereiro X, Sharma SC. Glia-neuron interactions in the mammalian retina. Prog. Retinal Eye Res. 2016;51:1–40. doi: 10.1016/j.preteyeres.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 20.Liu Z, Chopp M. Astrocytes, therapeutic targets for neuroprotection and neurorestoration in ischemic stroke. Prog. Neurobiol. 2016;144:103–120. doi: 10.1016/j.pneurobio.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akkermann R, Jadasz JJ, Azim K, Kuery P. Taking advantage of nature’s gift: can endogenous neural stem cells improve myelin regeneration? Int. J. Mol. Sci. 2016;17:1895. doi: 10.3390/ijms17111895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tognatta R, Miller RH. Contribution of the oligodendrocyte lineage to CNS repair and neurodegenerative pathologies. Neuropharmacology. 2016;110:539–547. doi: 10.1016/j.neuropharm.2016.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song SK, et al. Demyelination increases radial diffusivity in corpus callosum of mouse brain. Neuroimage. 2005;26:132–140. doi: 10.1016/j.neuroimage.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 24.Li S, et al. Rapid and efficient differentiation of rodent neural stem cells into oligodendrocyte progenitor cells. Dev. Neurosci. 2019;41:79–93. doi: 10.1159/000499364. [DOI] [PubMed] [Google Scholar]
- 25.Marton RM, et al. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat. Neurosci. 2019;22:484. doi: 10.1038/s41593-018-0316-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma KD, Pandanaboina SC, Srivatsan M, Xie JY. Predominant differentiation of rat fetal neural stem cells into functional oligodendrocytes in vitro. Neurosci. Lett. 2020;736:135264–135264. doi: 10.1016/j.neulet.2020.135264. [DOI] [PubMed] [Google Scholar]
- 27.Ito H, Nomoto H, Furukawa Y, Furukawa S. Neurotrophins facilitate synthesis of choline acetyltransferase and tyrosine hydroxylase in cultured mouse neural stem cells independently of their neuronal differentiation. Neurosci. Lett. 2003;339:231–234. doi: 10.1016/S0304-3940(03)00015-6. [DOI] [PubMed] [Google Scholar]
- 28.Kapoor R, Desouza LA, Nanavaty IN, Kernie SG, Vaidya VA. Thyroid hormone accelerates the differentiation of adult hippocampal progenitors. J. Neuroendocrinol. 2012;24:1259–1271. doi: 10.1111/j.1365-2826.2012.02329.x. [DOI] [PubMed] [Google Scholar]
- 29.Schneider JW, et al. Small-molecule activation of neuronal cell fate. Nat. Chem. Biol. 2008;4:408–410. doi: 10.1038/nchembio.95. [DOI] [PubMed] [Google Scholar]
- 30.Song Y, Lee S, Jho E-H. Enhancement of neuronal differentiation by using small molecules modulating Nodal/Smad, Wnt/beta-catenin, and FGF signaling. Biochem. Biophys. Res. Commun. 2018;503:352–358. doi: 10.1016/j.bbrc.2018.06.033. [DOI] [PubMed] [Google Scholar]
- 31.Banerjee A, et al. The influence of hydrogel modulus on the proliferation and differentiation of encapsulated neural stem cells. Biomaterials. 2009;30:4695–4699. doi: 10.1016/j.biomaterials.2009.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kashyap V, et al. Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the Nanog, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells Dev. 2009;18:1093–1108. doi: 10.1089/scd.2009.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwata R, Casimir P, Vanderhaeghen P. Mitochondrial dynamics in postmitotic cells regulate neurogenesis. Science. 2020;369:858–862. doi: 10.1126/science.aba9760. [DOI] [PubMed] [Google Scholar]
- 34.Li N, et al. Three-dimensional graphene foam as a biocompatible and conductive scaffold for neural stem cells. Sci. Rep. 2013;3:1604. doi: 10.1038/srep01604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obernier K, Alvarez-Buylla A. Neural stem cells: origin, heterogeneity and regulation in the adult mammalian brain. Development. 2019;146:dev156059. doi: 10.1242/dev.156059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abbaszadeh H-A, Tiraihi T, Delshad AR, Saghedi Zadeh M, Taheri T. Bone marrow stromal cell transdifferentiation into oligodendrocyte-like cells using triiodothyronine as a inducer with expression of platelet-derived growth factor alpha as a maturity marker. Iran. Biomed. J. 2013;17:62–70. doi: 10.6091/ibj.11162.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gusel’nikova VV, Korzhevskiy DE. NeuN as a neuronal nuclear antigen and neuron differentiation marker. Acta Nat. 2015;7:42–47. doi: 10.32607/20758251-2015-7-2-42-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwartz PH, et al. Isolation and characterization of neural progenitor cells from post-mortem human cortex. J. Neurosci. Res. 2003;74:838–851. doi: 10.1002/jnr.10854. [DOI] [PubMed] [Google Scholar]
- 39.Vieira MS, et al. Neural stem cell differentiation into mature neurons: mechanisms of regulation and biotechnological applications. Biotechnol. Adv. 2018;36:1946–1970. doi: 10.1016/j.biotechadv.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 40.Campanella G, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019;25:1301–1309. doi: 10.1038/s41591-019-0508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Falk T, et al. U-Net: deep learning for cell counting, detection, and morphometry. Nat. Methods. 2019;16:67–70. doi: 10.1038/s41592-018-0261-2. [DOI] [PubMed] [Google Scholar]
- 42.Chen CL, et al. Deep learning in label-free cell classification. Sci. Rep. 2016;6:21471. doi: 10.1038/srep21471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moen E, et al. Deep learning for cellular image analysis. Nat. Methods. 2019;16:1233–1246. doi: 10.1038/s41592-019-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H, et al. Deep learning enables cross-modality super-resolution in fluorescence microscopy. Nat. Methods. 2019;16:103–110. doi: 10.1038/s41592-018-0239-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buggenthin F, et al. Prospective identification of hematopoietic lineage choice by deep learning. Nat. Methods. 2017;14:403–406. doi: 10.1038/nmeth.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waisman A, et al. Deep learning neural networks highly predict very early onset of pluripotent stem cell differentiation. Stem Cell Rep. 2019;12:845–859. doi: 10.1016/j.stemcr.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monteiro-Riviere NA, Inman AO, Zhang LW. Limitations and relative utility of screening assays to assess engineered nanoparticle toxicity in a human cell line. Toxicol. Appl. Pharmacol. 2009;234:222–235. doi: 10.1016/j.taap.2008.09.030. [DOI] [PubMed] [Google Scholar]
- 48.Theunissen P, et al. Standardized flow cytometry for highly sensitive MRD measurements in B-cell acute lymphoblastic leukemia. Blood. 2017;129:347–357. doi: 10.1182/blood-2016-07-726307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eulenberg P, et al. Reconstructing cell cycle and disease progression using deep learning. Nat. Commun. 2017;8:463. doi: 10.1038/s41467-017-00623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brunet JF, Allaman I, Magistretti PJ, Pellerin L. Glycogen metabolism as a marker of astrocyte differentiation. J. Cereb. Blood Flow Metab. 2010;30:51–55. doi: 10.1038/jcbfm.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi K-C, et al. Effect of single growth factor and growth factor combinations on differentiation of neural stem cells. J. Korean Neurosurg. Soc. 2008;44:375–381. doi: 10.3340/jkns.2008.44.6.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Darvishi M, Tiraihi T, Mesbah-Namin SA, Delshad A, Taheri T. Motor neuron transdifferentiation of neural stem cell from adipose-derived stem cell characterized by differential gene expression. Cell. Mol. Neurobiol. 2017;37:275–289. doi: 10.1007/s10571-016-0368-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dutton R, Yamada T, Turnley A, Bartlett PF, Murphy M. Sonic hedgehog promotes neuronal differentiation of murine spinal cord precursors and collaborates with neurotrophin 3 to induce islet-1. J. Neurosci. 1999;19:2601–2608. doi: 10.1523/JNEUROSCI.19-07-02601.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mendivil-Perez M, et al. Melatonin enhances neural stem cell differentiation and engraftment by increasing mitochondrial function. J. Pineal Res. 2017;63:e12415. doi: 10.1111/jpi.12415. [DOI] [PubMed] [Google Scholar]
- 55.Park H, Poo M-m. Neurotrophin regulation of neural circuit development and function. Nat. Rev. Neurosci. 2013;14:7–23. doi: 10.1038/nrn3379. [DOI] [PubMed] [Google Scholar]
- 56.Rogister B, Ben-Hur T, Dubois-Dalcq M. From neural stem cells to myelinating oligodendrocytes. Mol. Cell. Neurosci. 1999;14:287–300. doi: 10.1006/mcne.1999.0790. [DOI] [PubMed] [Google Scholar]
- 57.He, K., Gkioxari, G., Dollar, P. & Girshick, R. Mask R-CNN. IEEE Trans Pattern Anal Mach Intell. 42, 386–397 (2020). [DOI] [PubMed]
- 58.Johnson J. W. Automatic nucleus segmentation with mask-RCNN. In Science and Information Conference (eds Arai K. & Kapoor, S.) (Springer, 2020). pp. 399–407.
- 59.Li Y, et al. Deep cytometry: deep learning with real-time inference in cell sorting and flow cytometry. Sci. Rep. 2019;9:11088. doi: 10.1038/s41598-019-47193-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mertens J, Marchetto MC, Bardy C, Gage FH. Evaluating cell reprogramming, differentiation and conversion technologies in neuroscience. Nat. Rev. Neurosci. 2016;17:424–437. doi: 10.1038/nrn.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He F, et al. Tet2-mediated epigenetic drive for astrocyte differentiation from embryonic neural stem cells. Cell Death Discov. 2020;6:30. doi: 10.1038/s41420-020-0264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Imamura O, Arai M, Dateki M, Oishi K, Takishima K. Donepezil-induced oligodendrocyte differentiation is mediated through estrogen receptors. J. Neurochem. 2020;155:494–507. doi: 10.1111/jnc.14927. [DOI] [PubMed] [Google Scholar]
- 63.Xu W, et al. Myelin basic protein regulates primitive and definitive neural stem cell proliferation from the adult spinal cord. Stem Cells. 2017;35:485–496. doi: 10.1002/stem.2488. [DOI] [PubMed] [Google Scholar]
- 64.Shao Z, et al. Induced differentiation of neural stem cells of astrocytic origin to motor neurons in the rat. Stem Cells Dev. 2011;20:1163–1170. doi: 10.1089/scd.2010.0262. [DOI] [PubMed] [Google Scholar]
- 65.Huang W, et al. Origins and proliferative states of human oligodendrocyte precursor cells. Cell. 2020;182:594–608.e11. doi: 10.1016/j.cell.2020.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou, B., Khosla, A., Lapedriza, A., Oliva, A. & Torralba, A. Learning deep features for discriminative localization. In 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2921–2929, 10.1109/CVPR.2016.319 (2016).
- 67.Chollet, F. Xception: deep learning with depthwise separable convolutions. In Proc. 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, 1800–1807 (IEEE, 2017), 10.1109/CVPR.2017.195 (2017).
- 68.Huang Z, Zhu H, Zhou JT, Peng X. Multiple marginal Fisher analysis. IEEE Trans. Ind. Electron. 2019;66:9798–9807. doi: 10.1109/TIE.2018.2870413. [DOI] [Google Scholar]
- 69.Simonyan, K. & Zisserman, A. Very deep convolutional networks for large-scale image recognition. In 3rd Int. Conf. Learning Representations (ICLR 2015) (eds Bengio, Y. & LeCun, Y.) 4 (2015); https://arxiv.org/abs/1409.1556.
- 70.Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J. & Wojna, Z. Rethinking the inception architecture for computer vision. Preprint at https://arxiv.org/abs/1512.00567 (2015).
- 71.Zhang J, Jiao J. Molecular biomarkers for embryonic and adult neural stem cell and neurogenesis. Biomed. Res. Int. 2015;2015:727542. doi: 10.1155/2015/727542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Melo-Braga MN, et al. Comprehensive quantitative comparison of the membrane proteome, phosphoproteome, and sialiome of human embryonic and neural stem cells. Mol. Cell. Proteom. 2014;13:311–328. doi: 10.1074/mcp.M112.026898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang S, et al. Quantitative phosphoproteomic study reveals that protein kinase A regulates neural stem cell differentiation through phosphorylation of catenin beta-1 and glycogen synthase kinase 3 beta. Stem Cells. 2016;34:2090–2101. doi: 10.1002/stem.2387. [DOI] [PubMed] [Google Scholar]
- 74.Baio Y, Zhao X, Wang L, Qian W, Sun J. Morphology-based classification of mycobacteria-infected macrophages with convolutional neural network: reveal EsxA-induced morphologic changes indistinguishable by naked eyes. Transl. Res. 2019;212:1–13. doi: 10.1016/j.trsl.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ragoza M, Hochuli J, Idrobo E, Sunseri J, Koes DR. Protein-ligand scoring with convolutional neural networks. J. Chem. Inf. Model. 2017;57:942–957. doi: 10.1021/acs.jcim.6b00740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sacile R, Montaldo E, Ruggiero C, Nieburgs HE, Nicolo G. A decision support system to detect morphologic changes of chromatin arrangement in normal-appearing cells. IEEE Trans. Nanobiosci. 2003;2:118–123. doi: 10.1109/TNB.2003.813939. [DOI] [PubMed] [Google Scholar]
- 77.Mamoshina P, Vieira A, Putin E, Zhavoronkov A. Applications of deep learning in biomedicine. Mol. Pharm. 2016;13:1445–1454. doi: 10.1021/acs.molpharmaceut.5b00982. [DOI] [PubMed] [Google Scholar]
- 78.Wainberg M, Merico D, Delong A, Frey BJ. Deep learning in biomedicine. Nat. Biotechnol. 2018;36:829–838. doi: 10.1038/nbt.4233. [DOI] [PubMed] [Google Scholar]
- 79.Liu G-H, et al. Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature. 2012;491:603–607. doi: 10.1038/nature11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wachs FP, et al. High efficacy of clonal growth and expansion of adult neural stem cells. Lab. Invest. 2003;83:949–962. doi: 10.1097/01.LAB.0000075556.74231.A5. [DOI] [PubMed] [Google Scholar]
- 81.Pan X, et al. Later passages of neural progenitor cells from neonatal brain are more permissive for human cytomegalovirus infection. J. Virol. 2013;87:10968–10979. doi: 10.1128/JVI.01120-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fu L, et al. Derivation of neural stem cells from mesenchymal stem cells: evidence for a bipotential stem cell population. Stem Cells Dev. 2008;17:1109–1121. doi: 10.1089/scd.2008.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Voelker J, et al. Isolation and characterization of neural stem cells from the rat inferior colliculus. Stem Cells Int. 2019;2019:5831240. doi: 10.1155/2019/5831240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mammolenti M, Gajavelli S, Tsoulfas P, Levy R. Absence of major histocompatibility complex class I on neural stem cells does not permit natural killer cell killing and prevents recognition by alloreactive cytotoxic T lymphocytes in vitro. Stem Cells. 2004;22:1101–1110. doi: 10.1634/stemcells.22-6-1101. [DOI] [PubMed] [Google Scholar]
- 85.Rhinn M, Dolle P. Retinoic acid signalling during development. Development. 2012;139:843–858. doi: 10.1242/dev.065938. [DOI] [PubMed] [Google Scholar]
- 86.Chew L-J, et al. SRY-box containing gene 17 regulates the Wnt/beta-catenin signaling pathway in oligodendrocyte progenitor cells. J. Neurosci. 2011;31:13921–13935. doi: 10.1523/JNEUROSCI.3343-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ding J, et al. Role of ciliary neurotrophic factor in the proliferation and differentiation of neural stem cells. J. Alzheimers Dis. 2013;37:587–592. doi: 10.3233/JAD-130527. [DOI] [PubMed] [Google Scholar]
- 88.Arevalo JC, Wu SH. Neurotrophin signaling: many exciting surprises! Cell. Mol. Life Sci. 2006;63:1523–1537. doi: 10.1007/s00018-006-6010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barnabe-Heider F, Miller FD. Endogenously produced neurotrophins regulate survival and differentiation of cortical progenitors via distinct signaling pathways. J. Neurosci. 2003;23:5149–5160. doi: 10.1523/JNEUROSCI.23-12-05149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Davis S, et al. LIFR-beta and gp-130 as heterodimerizing signal transducers of the tripartite CNTF receptor. Science. 1993;260:1805–1808. doi: 10.1126/science.8390097. [DOI] [PubMed] [Google Scholar]
- 91.Chu J, et al. Effects of melatonin and its analogues on neural stem cells. Mol. Cell. Endocrinol. 2016;420:169–179. doi: 10.1016/j.mce.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 92.Wu Y, et al. Size-dependent effects of layered double hydroxide nanoparticles on cellular functions of mouse embryonic stem cells. Nanomedicine. 2015;10:3469–3482. doi: 10.2217/nnm.15.158. [DOI] [PubMed] [Google Scholar]
- 93.Wu Y, et al. Layered double hydroxide nanoparticles promote self-renewal of mouse embryonic stem cells through the PI3K signaling pathway. Nanoscale. 2015;7:11102–11114. doi: 10.1039/C5NR02339D. [DOI] [PubMed] [Google Scholar]
- 94.Zhang B, et al. Nanomaterials in neural-stem-cell-mediated regenerative medicine: imaging and treatment of neurological diseases. Adv. Mater. 2018;30:e1705694. doi: 10.1002/adma.201705694. [DOI] [PubMed] [Google Scholar]
- 95.Zhu R, et al. pH sensitive nano layered double hydroxides reduce the hematotoxicity and enhance the anticancer efficacy of etoposide on non-small cell lung cancer. Acta Biomater. 2016;29:320–332. doi: 10.1016/j.actbio.2015.10.029. [DOI] [PubMed] [Google Scholar]
- 96.Zhu Y, et al. DeepScreen: an accurate, rapid, and anti-interference screening approach for nanoformulated medication by deep learning. Adv. Sci. 2018;5:1800909. doi: 10.1002/advs.201800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Barker RA, Drouin-Ouellet J, Parmar M. Cell-based therapies for Parkinson disease-past insights and future potential. Nat. Rev. Neurol. 2015;11:492–503. doi: 10.1038/nrneurol.2015.123. [DOI] [PubMed] [Google Scholar]
- 98.Thonhoff JR, Ojeda L, Wu P. Stem cell-derived motor neurons: applications and challenges in amyotrophic lateral sclerosis. Curr. Stem Cell Res. Ther. 2009;4:178–199. doi: 10.2174/157488809789057392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen C, et al. Thyroid hormone-Otx2 signaling is required for embryonic ventral midbrain neural stem cells differentiated into dopamine neurons. Stem Cells Dev. 2015;24:1751–1765. doi: 10.1089/scd.2014.0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ling ZD, Potter ED, Lipton JW, Carvey PM. Differentiation of mesencephalic progenitor cells into dopaminergic neurons by cytokines. Exp. Neurol. 1998;149:411–423. doi: 10.1006/exnr.1998.6715. [DOI] [PubMed] [Google Scholar]
- 101.Kriks S, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shi Y, Kirwan P, Smith J, Robinson HPC, Livesey FJ. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat. Neurosci. 2012;15:477–486. doi: 10.1038/nn.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rushing G, Ihrie RA. Neural stem cell heterogeneity through time and space in the ventricular-subventricular zone. Front. Biol. 2016;11:261–284. doi: 10.1007/s11515-016-1407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mizrak D, et al. Single-cell analysis of regional differences in adult V-SVZ neural stem cell lineages. Cell Rep. 2019;26:394–406.e5. doi: 10.1016/j.celrep.2018.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McCarthy M, Turnbull DH, Walsh CA, Fishell G. Telencephalic neural progenitors appear to be restricted to regional and glial fates before the onset of neurogenesis. J. Neurosci. 2001;21:6772–6781. doi: 10.1523/JNEUROSCI.21-17-06772.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The main data supporting the findings of this study are available within the article and in the Supplementary Information. The single-cell image data for model building are available through the figshare website with the download link of 10.6084/m9.figshare.13070666.v1. Source data are provided with this paper.
The code for model building is available through the link 10.5281/zenodo.4606918