Abstract
Background
Lactate dehydrogenase (LDH) is a critical metabolic enzyme. LDH A (LDHA) overexpression is a hallmark of aggressive malignancies and has been linked to tumour initiation, reprogramming and progression in multiple tumour types. However, successful LDHA inhibition strategies have not materialised in the translational and clinical space. We sought to develop a rational strategy for LDHA suppression in the context of solid tumour treatment.
Methods
We utilised a doxycycline-inducible short hairpin RNA (shRNA) system to generate LDHA suppression. Lactate and LDH activity levels were measured biochemically and kinetically using hyperpolarised 13C-pyruvate nuclear magnetic resonance spectroscopy. We evaluated effects of LDHA suppression on cellular proliferation and clonogenic survival, as well as on tumour growth, in orthotopic models of anaplastic thyroid carcinoma (ATC) and head and neck squamous cell carcinoma (HNSCC), alone or in combination with radiation.
Results
shRNA suppression of LDHA generated a time-dependent decrease in LDH activity with transient shifts in intracellular lactate levels, a decrease in carbon flux from pyruvate into lactate and compensatory shifts in metabolic flux in glycolysis and the Krebs cycle. LDHA suppression decreased cellular proliferation and temporarily stunted tumour growth in ATC and HNSCC xenografts but did not by itself result in tumour cure, owing to the maintenance of residual viable cells. Only when chronic LDHA suppression was combined with radiation was a functional cure achieved.
Conclusions
Successful targeting of LDHA requires exquisite dose and temporal control without significant concomitant off-target toxicity. Combinatorial strategies with conventional radiation are feasible as long as the suppression is targeted, prolonged and non-toxic.
Subject terms: Cancer therapy, Thyroid cancer, Cancer metabolism, Head and neck cancer
Background
The Warburg effect, first described nearly a century ago, consists of an abnormal metabolic phenotype inherent to many solid malignancies in which glycolytic activity persists at a high rate regardless of the presence or absence of oxygen.1 Although energetically inefficient at face value, this altered tumour metabolism has been shown over the intervening decades to be critical for rapid tumour growth, metastasis and even development of treatment resistance.2–4 Therefore, understanding the driving forces and critical metabolic chokepoints that define tumour metabolism could not only impact our understanding of cancer biology but also lead to the development of novel treatment strategies predicated on a therapeutic index generated by differential metabolic activity between tumour and normal tissue. However, efforts to date to translate our knowledge of basic metabolic pathways into effective anti-metabolic strategies have had limited success.5–10 In large part, this lack of translation reflects our incomplete understanding of the variable role that metabolic enzymes, even critical ones, play in tumour cell survival under baseline and treatment-induced stress conditions.
Lactate dehydrogenase (LDH) is an essential enzyme that converts pyruvate into lactate11 and is highly active in tumour cells, which rely on glucose conversion into lactate under both anaerobic and aerobic conditions, in contrast to normal eukaryotic cells.12 Thus, this enzyme is one of the quintessential metabolic drivers of the Warburg effect. The LDH protein is composed of the subunits LDHA and LDHB, generating several isoforms.13 The LDHA gene, encoding the LDH-M protein, has been linked to tumour initiation and growth in multiple tumour types.14–16 High expression of LDHA has been associated with aggressive features in multiple malignancies.12,17,18
We previously showed that the conversion of pyruvate into lactate by LDH represents a critical metabolic nexus that contributes to tumour cell management of oxidative stress.19 Both conventional chemotherapy (e.g. cisplatin, doxorubicin) and external beam radiotherapy (EBRT) generate transient fluctuations in cellular oxidative stress that are balanced through differential metabolic flux through LDH.20 This phenomenon suggests that LDHA plays a critical role in not only tumorigenesis but also treatment response. In this study, we evaluated the impact of LDHA suppression on tumorigenesis and EBRT response in two aggressive malignancies, namely anaplastic thyroid carcinoma (ATC) and head and neck squamous cell carcinoma (HNSCC), which we have previously shown to demonstrate the basic aspects of the classical Warburg effect.19 Rather than using LDH inhibitors, which can be non-specific and generate off-target effects, or constitutive knockdown of LDHA, which can result in aberrant and non-representative clonal expansion, we used an inducible knockdown approach, which provided the most precise window into the effects of differential LDH function in the context of tumour growth and treatment response.
Methods
Note: For ease of reference, in the current manuscript we will maintain the LDHA/LDHB designation for both the gene–mRNA and resulting protein. When referencing the gene–mRNA, the notation will be italicised.
Cell lines
For human papillary thyroid cancer cell lines, K2 cells were maintained in Dulbecco’s modified Eagle’s medium/Nutrient Mixture F-12 (Sigma-Aldrich, St. Louis, MO), and TPC-1 cells were maintained in Roswell Park Memorial Institute-1640 medium (Sigma-Aldrich), along with 10% foetal bovine serum (Sigma-Aldrich), 2 mM l-glutamine, and non-essential amino acids, in a 37 °C incubator supplied with 95% air and 5% CO2. For the ATC cell lines, Hth83 cells were maintained in Roswell Park Memorial Institute-1640 medium, and SW1736 and Hth7 cells were maintained in Minimum Essential Medium (Cambrex BioScience, Walkersville, MD), along with 10% foetal bovine serum, 1 mM sodium pyruvate (Thermo Fisher Scientific, Waltham, MA), 2 mM l-glutamine, and non-essential amino acids (Cambrex BioScience), in a 37 °C incubator supplied with 95% air and 5% CO2. HNSCC cell lines (HN31, HN30, OSC-19, UM-SCC-17A [referred to as 17A], UM-SCC-17B [17B], UM-SCC-11A [11A], FaDu, Detroit 562, UM-SCC-22A [22A]) were maintained in a Dulbecco’s modified Eagle’s medium (Sigma-Aldrich), containing 10% foetal bovine serum, 1 mM sodium pyruvate, 2 mM l-glutamine and non-essential amino acids. Cell line identity was confirmed using short tandem repeat profiling at the Cytogenetics and Cell Authentication Core at The University of Texas MD Anderson Cancer Center (Houston, TX). Cell lines were confirmed to be mycoplasma free using MycoAlert PLUS kit (Lonza, Alpharetta, GA). TPC-1 cells were kindly provided by Dr. Jerome Hershman (VA Greater Los Angeles Healthcare System, Los Angeles, CA). K2 cells were kindly provided by Dr. D. Wynford-Thomas (Cardiff University, Cardiff, UK). Hth83, Hth7, SW1736, HN31, HN30, OSC-19, 17A, 17B, 11A, FaDu, Detroit 562, and 22A cells were kindly provided by Dr. Jeffrey Myers (MD Anderson, Houston, TX). Each cell line was maintained for ten passages at most, following genotyping.
Generation of cell lines with inducible LDHA knockdown
Cells with inducible LDHA knockdown were generated using a lentiviral system (pINDUCER20; Thomas F. Westbrook Laboratory, Baylor College of Medicine, Houston, TX). First, we generated a pCDNA6.2-EmGFP-shLDHA to overexpress shLDHA (5ʹ-GAACTGCAAGTTGCTTATTGT-3ʹ), then transferred the expression cassette into a single inducible lentiviral vector (pINDUCER20) to create a pINDUCER-EmGFP-shLDHA expression vector. Using a previously described protocol,21 we successfully generated doxycycline (DOX)-inducible LDHA knockdown in the ATC cell line Hth83 and the HNSCC cell line 22A.
The Cancer Genome Atlas analysis
We analysed a dataset of 528 patients with a diagnosis of HNSCC from The Cancer Genome Atlas. RSEM-normalised gene expression files, as well as clinical parameters, including tissue histology and survival data, were downloaded directly from the Broad Firehose site (https://gdac.broadinstitute.org/).
Western blot analysis
Western blotting for proteins of interest using anti-LDHA (ab101562, 1:1000, Abcam, Cambridge, UK), anti-LDHB (ab53292, 1:1000, Abcam), anti-β-actin (1:5000, BD Biosciences, San Jose, CA) and β-tubulin (9F3; 1:1000, Cell Signalling Technology, Danvers, MA) was performed as previously described by our group.20
Cell proliferation and viability assays
The MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, VWR, Radnor, PA) assay was used to measure proliferation as previously described by our group under varying experimental conditions.22 Briefly, cells were cultured in 96-well plates for 24 h and then treated with DOX (Thermo Fisher Scientific) for 24–96 h. MTT (5 mg/mL) was added to the wells and incubated for 4 h; the cells were lysed with dimethyl sulfoxide (Sigma-Aldrich), and the release of formazan was quantified with an ELx808 96-well plate reader at 570 nm absorbance (SPECTROstar Nano, Cary, NC).
Clonogenic survival assay
Clonogenic survival assay was performed as previously described.23 Cells were plated at low density, treated with DOX (0.1, 0.5, 1 and 2 µg/mL) for 24 h and incubated for 9–11 days for colony formation. For experiments involving EBRT, cells were treated with 0.1 µg/mL DOX for 24 h prior to irradiation (0, 2, 4 or 6 Gy). Two hours after irradiation, fresh media plus 0.1 µg/mL DOX were placed in each well, and cells were incubated for 9–11 days for clone formation. Cells were then stained with 0.5% crystal violet and colonies that consist of at least 50 cells were counted using a colony counter pen. Plating efficiency (PE) was determined in each experiment for normalisation. PE = number of colonies formed/number of cells seeded × 100%. Surviving fraction (SF) was calculated by the formula [SF = number of colonies formed after treatment/(number of cells seeded × PE)].23 Clonogenic survival curves were constructed from three independent experiments using the GraphPad Prism 8.0 software (GraphPad Software Inc., San Diego, CA).
Biochemical analyses
Cells were plated in 12-well plates for 24 h and then treated with DOX (0, 0.1 and 1 µg/mL) for 24 and 96 h, respectively. LDH activity (Sigma-Aldrich) and lactate (BioVision, Milpitas, CA) levels were measured according to the manufacturer’s instructions.
Hyperpolarised magnetic resonance spectroscopy (HP-MRS)
HP [1-13C]-pyruvate was prepared as previously described by our group.24 Briefly, 8-mg aliquots of [1-13C]-pyruvic acid (Cambridge Isotopes Laboratories, Tewksbury, MA) containing 1.5 mmol/L gadoteridol (Bracco Diagnostics, Monroe Township, NJ) and 15 mmol/L OX63 (GE Healthcare, Chicago, IL) were polarised using a HyperSense dissolution dynamic nuclear polarisation system (Oxford Instruments, Abingdon, UK). Aliquots were cooled to 1.45 K in a 3.35-T magnetic field and irradiated at ~94.13 GHz for 45 min or until solid-state polarisation levels plateaued. The frozen substrate was then dissolved in 4 mL of distilled water at ~180 °C, which also contained 20 mmol/L NaOH, 50 mmol/L NaCl, 0.1 g/L ethylenediaminetetraacetic acid (EDTA) and 40 mmol/L Trizma pre-set crystals (pH 7.6). The final dissolution results in a 37 °C isotonic solution containing 20 mmol/L HP [1-13C]-pyruvate.
Hth83-shLDHA cells were treated with DOX for 48 h to activate the shLDHA construct. Cell suspensions were prepared in parallel, during the wait for polarisation to plateau. Approximately 30 × 106 cells suspended in 900 μL of normal media were supplemented with 100 μL of D2O in a 10-mm Shigemi tube and loaded into a Spectrospin DPX-300 nuclear magnetic resonance (NMR) spectrometer (Bruker, Billerica, MA). Samples were allowed to reach an equilibrium temperature of 310 K over an ~10-min interval, during which automated NMR calibrations, including locking, tuning, and shimming, were carried out. After dissolution, 161 µL of the HP solution was delivered to cell suspensions at the isocentre in the NMR system, using a custom catheter setup, followed by 15 mL of air that was bubbled through to ensure that the entirety of the HP pyruvate bolus was delivered and well mixed to a final HP pyruvate concentration of ~1.8 mmol/L. Dynamic 13C spectra were initiated just prior to the addition of HP pyruvate and acquired using a simple pulse-acquire sequence with an excitation angle of 15° and a repetition time of 2 s. Dynamic signals for HP pyruvate and lactate were calculated using the area of Lorentzian functions fit to their respective peaks, and normalised lactate was calculated as the ratio of the area under the lactate curve to the sum of areas under the pyruvate and lactate curves, divided by the number of cells in each suspension and normalised again to a relative value of 100% in Hth83-shLDHA cells that were not exposed to DOX.
Analysis of tricarboxylic acid and glycolysis pathway metabolites by ion chromatography and high-resolution accurate-mass mass spectrometry (HRAM-MS)
To determine the incorporation of pyruvate carbon (13C3-pyruvate) into intracellular tricarboxylic acid cycle and glycolysis pathways, extracts were prepared and analysed by HRAM-MS. Following exposure to DOX for 48 h, cells were incubated in fresh medium containing 1 mM [U-13C3]-pyruvate (Cambridge Isotopes Laboratories) for 0, 5, 15, 30, 45, 60 and 120 s. Metabolites were extracted using cold 90:10 (v/v) acetonitrile:water and detected on a Thermo Fisher Scientific Dionex ICS-5000+ capillary ion chromatography system containing a Dionex IonPac AS11-HC 250 × 2 mm2 4 μm column. Data were acquired using an Orbitrap Fusion Tribrid Mass Spectrometer (Thermo Fisher Scientific) under electrospray ionisation in negative mode. The raw files were then imported into the Trace Finder software (Thermo Fisher Scientific) for analysis.
Targeted metabolomics
To evaluate the impact of LDHA suppression on tumour metabolism, we generated measurable (~5 × 5 mm2) 22A-shLDHA-5 tumours in the orthotopic of mice as described below. LDHA suppression was induced using DOX for 24 h. DOX-treated tumours (n = 3) and control, untreated tumours (n = 3) were harvested and snap frozen in liquid nitrogen. Metabolites were measured by a targeted approach as described earlier.25,26 Metabolites were extracted from the samples and liver and used as quality controls. The extraction procedure was described earlier.26,27 Briefly, the extraction step involved the addition of 750 µL of ice-cold methanol:water (4:1), containing spiked internal standards, to each tissue sample. Ice-cold chloroform and water were added in a 3:1 ratio for a final proportion of 1:4:3:1 water:methanol:chloroform:water. The extracted samples were then dried and deproteinised prior to mass spectrometry analysis. The data were log 2 transformed and normalised with internal standards on a per-sample, per-method basis. For every metabolite in the normalised dataset, two-sample t tests were conducted to compare expression levels between different groups. Differential metabolites were identified using p values adjusted for multiple testing at a false discovery rate threshold of <0.25.
Mitochondrial respiration
Oxygen consumption rates (OCRs) were assayed under basal conditions (25 mM d-glc, 1 mM pyruvate, 4 mM glutamine, 0% serum) and following administration of various drugs using a Seahorse Bioscience XF96 Extracellular Flux Analyser (Billerica, MA) as previously described by us and others.28,29 Using the 22A-shLDHA #5 clone, we tested 2 conditions: control cells (no treatment) and cells treated with DOX for 48 h (100 ng/mL) prior to OCR measurements. Each experiment was performed in triplicate. ATP production was determined by comparing OCR prior to and following oligomycin treatment. Coupling efficiency was determined by dividing the ATP production rate by the basal respiration rate.
Reactive oxygen species (ROS) measurement
ROS were measured according to the manufacturer’s instructions. Briefly, Total ROS levels were measured using the CellROX Deep Red oxidative stress reagent (Life Technologies, Carlsbad, CA), which are fluorogenic probes designed to reliably measure ROS in live cells. Hth83-shLDHA-4# and 22A-shLDHA-5# cells were pre-treated with DOX (0, 0.1 and 1 µg/mL) for 24 and 48 h to suppress LDHA activity. Fluorescence was measured using flow cytometry according to previously published and validated protocols.30
Orthotopic murine model and treatment
Female immunodeficient athymic nu/nu mice (age 6–8 weeks, weight 20–25 g) were obtained from Envigo (Indianapolis, IN). The mice were fed irradiated mouse chow and housed in laminar flow cabinets under specific pathogen-free conditions. All procedures and care were reviewed and approved by our Institutional Animal Care and Use Committee. The murine orthotopic model of thyroid carcinoma has been described previously by our team.31,32 Hth83-shLDHA-4 and Hth83-shLDHA-31 clones were infected with retrovirus carrying the luciferase gene and injected orthotopically into the thyroid of each mouse (2.5 × 105 cells/mouse). Prior to any surgical procedure, all mice were given intraperitoneal injection of ketamine/xylazine (75 mg/kg; Sigma-Aldrich) as anaesthesia. Mice were randomised to receive control (n = 9) or DOX-containing water (n = 9). Five days after the injection of cells, mice were given DOX-containing water (2 mg/mL) daily to induce LDHA knockdown. The mice were monitored for bioluminescence activity twice a week to assess tumour growth using the IVIS 200 Imaging System (Xenogen Corp., Alameda, CA) with Living Image 3.2 software. For the HNSCC model, 22A-shLDHA-5 and 22A-shLDHA-23 clones were injected into the lateral tongue (5 × 104 cells/mouse) as previously described.33 Mice were randomised to receive control (n = 9) or DOX-containing water (n = 9). Six days after the injection of cells, mice were given DOX-water (2 mg/mL) daily to induce LDHA knockdown. To study the radiosensitisation effect of LDHA suppression, we again injected the 22A-shLDHA-5 clone into the lateral tongue (5 × 104 cells/mouse). Mice were randomised to receive control (n = 8), EBRT (5 Gy × 2) alone (n = 8), DOX-water alone (n = 8) or DOX combined with EBRT (5 Gy × 2) (n = 8). Tumour growth was monitored using a digital Vernier calliper three times a week, and the volume was calculated as π/6 × length × width × height. Mice were sacrificed using the CO2 Euthanex system when loss of body weight reached 20% or when the experiments required termination based on tumour size/burden, consistent with institutional policies. Following mouse euthanasia, xenograft tumours were harvested for subsequent analysis.
Statistical analysis
All in vitro experiments were performed at least triplicate, and every condition was tested at least in triplicate. All statistical analyses for in vitro and in vivo experiments were conducted using the two-tailed Student’s t test, with a p value of 0.05 considered statistically significant. Statistical analysis was conducted using SPSS Statistics 25 (IBM, Armonk, NY). The Kaplan–Meier and Cox regression methods were used to estimate overall survival (OS), while the log-rank test was used to analyse the impact of gene expression on OS and progression-free interval (PFI).
Results
Generation and metabolomic profiling of inducible shLDHA suppression
Using the entire cohort of patients, we performed Cox regression of clinical outcomes as a function of LDHA expression (using log 2-transformed RSEM values) and found a significant inverse correlation between LDHA expression and OS (p = 0.017; Exp(B) 1.234, 95% confidence interval (CI) 1.038–1.476) and PFI (p = 0.045; Exp(B) 1.208, 95% CI 1.003–1.455), but not between LDHB expression and OS or PFI (p = 0.125 and 0.685, respectively). After conversion of RSEM values into Z-scores, patients with a higher Z-score (≥1.0) had decreased OS compared with patients with a lower Z-score (≤−1.0), although this difference did not reach statistical significance (p = 0.051), and patients with a higher Z-score (≥1.0) had significantly decreased PFI compared with patients with a lower Z-score (≤−1.0) (p = 0.021). Using Z-scores of −1.5 and 1.5 to assess the impact on survival, results approached significance for OS (p = 0.051), but were not significant for PFI (p = 0.641) (Supplemental Fig. S1). Consistent with the human data, LDH activity was highly variable in both HNSCC and thyroid cancer cell lines (Supplemental Fig. S2A). However, LDH activity did not correlate with tumorigenesis in the orthotopic murine models of thyroid cancer (Supplemental Fig. S2B) or HNSCC (previously published data).33
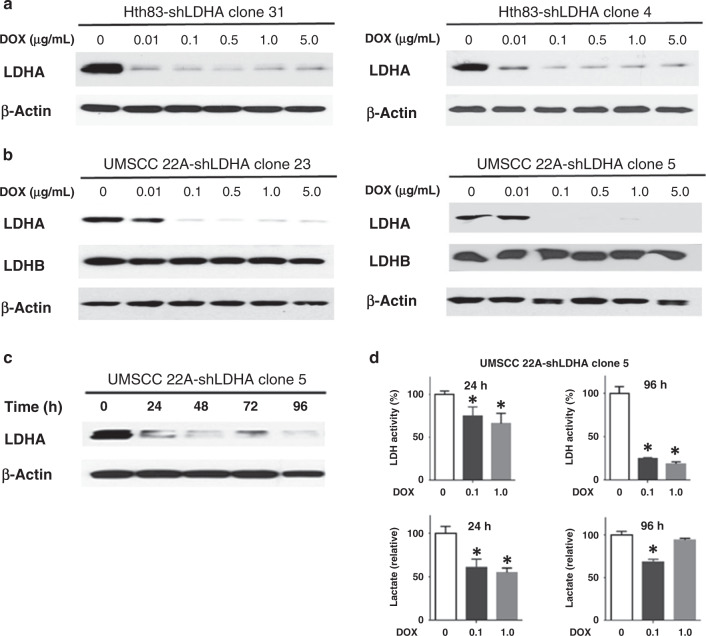
Since LDH is an essential metabolic enzyme present in numerous cell types, constitutive overexpression or knockdown is likely to generate complex and non-physiological metabolic phenotypes. To evaluate the interaction between LDHA, tumorigenesis and treatment response, we applied an inducible targeted knockdown system to two representative cell lines. Hth83 and 22A cells were infected with a lentiviral system (pINDUCER20) containing an overexpressed shLDHA cassette. Single clones were isolated from the parental population and expanded. Following exposure to different DOX doses (0, 0.01, 0.1, 0.5, 1 and 5 µg/mL) for 48 h, protein levels of LDHA, LDHB and β-actin were evaluated using Western blot (Fig. 1a, b). As expected, shLDHA was induced by DOX following doses as low as 0.01 µg/mL in Hth83 cells and 0.1 µg/mL in 22A cells, resulting in decreased LDHA protein levels and no change in LDHB protein levels (Fig. 1b). Following exposure to DOX for 24–96 h of 22A-shLDHA cells, LDHA protein level significantly decreased within 24 until 96 h (Fig. 1c). Reduced LDHA expression led to decreased LDH activity and lactate production within 24 h of activated suppression. However, although LDH activity remained suppressed, lactate levels re-equilibrated at later time points, consistent with the regeneration of metabolic equilibrium (Fig. 1d). We confirmed this phenomenon in the Hth83 cell line (Supplemental Fig. S3).
Fig. 1. Generation and functional validation of LDHA knockdown constructs.
a, b Representative Western blot analysis of clones for LDHA, LDHB and β-actin. Hth83 and 22A cell lines were infected with lentivirus containing shLDHA constructs. Clones were isolated from the parental population and expanded. LDHA was suppressed by DOX for 48 h. c Following exposure to DOX for 24–96 h, LDHA and β-actin levels were evaluated using Western blot. Activation of shLDHA constructs suppressed LDHA protein levels within 24 h of exposure. d LDH activity continued to drop significantly at both 24 and 96 h after activation, but lactate levels equilibrated at 96 h after activation. All experiments were performed at least triplicate, and every condition was tested at least in triplicate. Error bars represent standard deviation. *P < 0.05 compared with control condition.
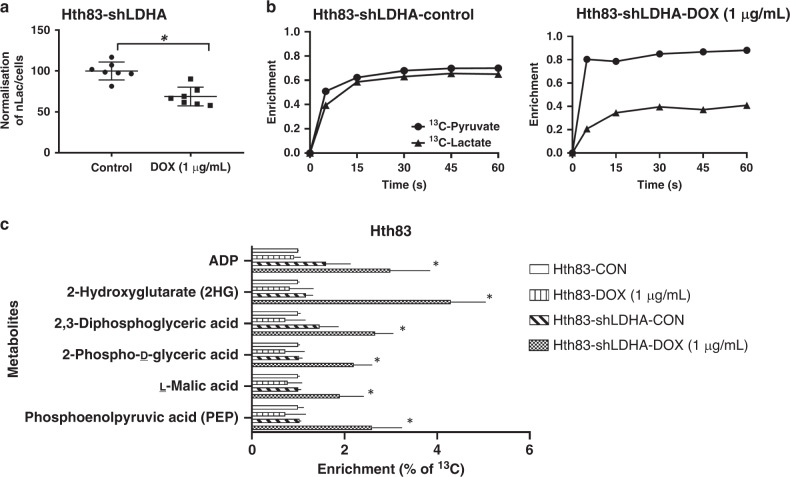
To confirm that LDHA suppression resulted in a direct decrease in the conversion of pyruvate to lactate, we analysed the kinetics of this inducible suppression model using real-time non-invasive measurements including HP-MRS imaging and mass spectrometry-based analysis (Fig. 2). DOX-induced (1 µg/mL) shLDHA activation led to the inhibition of LDH activity and resulted in a 31.9% reduction in the HP lactate signal, compared with lactate levels in control Hth83 cells (Fig. 2a). To confirm our findings, we examined the effect of LDHA knockdown in Hth83 cells using 13C-labelled pyruvate to assess the intracellular pyruvate pool and the release of lactate through mass spectrometry-based analyses. High fractional enrichment after as little as 5 s of exposure indicated that [U-13C3]-pyruvate is rapidly taken in by cells (Fig. 2b). Similar fractional enrichment for both pyruvate and lactate indicated that these pools remain close to isotopic equilibrium in Hth83-shLDHA cells and that inhibition of LDHA by exposure of Hth83-shLDHA cells to DOX (1 µg/mL for 48 h) disrupts that equilibrium (Fig. 2b) and reduces the rate at which the mass label is incorporated into the lactate pool. Taken together, these HP-MRS and mass spectrometry observations confirm inhibition of LDHA in this model system. Inhibition of LDH activity resulted in shunting of labelled carbon into secondary pathways, resulting in the enrichment of labelled malate and 2-hydroxyglutarate (no significant shifts were detected in citrate, isocitrate and fumarate levels; data not shown). Increased labelling of 2,3-diphosphoglyceric acid, 2-phospho-d-glyceric acid and phosphoenolpyruvic acid was also detected, consistent with glycolytic backup and shunting of excess pyruvate into secondary pathways (Fig. 2c).
Fig. 2. Functional validation of LDHA knockdown constructs.
a Activation of shLDHA constructs suppressed LDH activity as measured kinetically using hyperpolarised magnetic resonance spectroscopy for 13C-labelled pyruvate and its labelled by-product lactate in Hth83-shLDHA clone 4. b Mass spectrometry-based analysis of intracellular pool of pyruvate and lactate in Hth83-shLDHA clone 4 using 13C-labelled pyruvate as tracer. c Ion chromatography high-resolution accurate-mass mass spectrometry analysis of intracellular tricarboxylic acid and glycolysis pathways metabolites to determine these pathways’ incorporation of 13C3-pyruvate. All experiments were performed at least in triplicate, and every condition was tested at least in triplicate. Error bars represent standard deviation. *P < 0.05 compared with control condition.
Given the shunting noted following inhibition of LDH activity, we sought to determine whether tumour cells would generate a higher rate of OCR to compensate for decreased LDH activity (Supplemental Fig. S4). Cells with suppressed LDH activity demonstrated a minimal increase in basal respiration and ATP production, but no significant change in maximal respiration and coupling efficiency compared to cells with normal LDH activity levels.
LDHA suppression decreases cellular proliferation and increases EBRT response
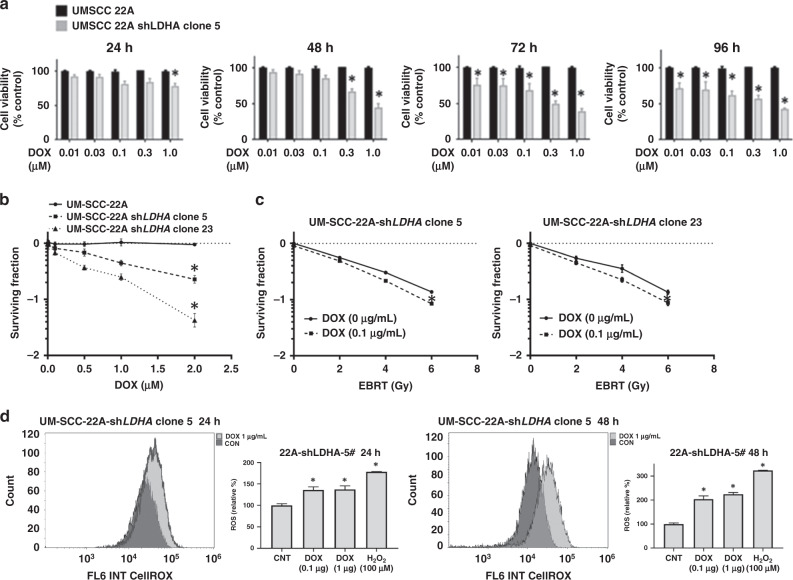
LDHA suppression via short hairpin RNA (shRNA) significantly inhibited the proliferation of 22A (Fig. 3a) and Hth83 (Supplemental Fig. S5A) cells following induction with DOX in a dose-dependent manner. Treatment with DOX resulted in a dose-dependent increase in intracellular ROS at 24- and 48-h post-treatment initiation (Fig. 3d and Supplemental Fig. S5D). Suppression of LDHA activity also significantly decreased the clonogenic surviving fraction of 22A-shLDHA clones 5 and 23 (Fig. 3b), as well as Hth83-shLDHA clones 4 and 31(Supplemental Fig. S5B), suggesting that LDHA is essential for cell survival. To determine whether suppression of LDHA sensitises cells to radiation, we performed clonogenic survival assays with radiation doses of 0, 2, 4 or 6 Gy in combination with 0.1 µg/mL DOX (Fig. 3c and Supplementary Fig. S5C). (DOX treatment alone did not affect radiosensitivity in the parental cell line; data not shown.) Low-dose DOX (0.1 µg/mL) induced suppression of LDHA, which resulted in increased radiation effectiveness in both 22A-shLDHA (p < 0.05) and Hth83-shLDHA clones.
Fig. 3. LDHA is required for viability and survival.
a Suppression of LDHA triggered a time- and dose-dependent decrease in viability. b Clonogenic surviving fraction. c Suppression of LDHA modestly increased radiation effectiveness, measured using clonogenic surviving fraction. d Suppression of LDHA increased intra-cellular ROS level. Activation of shLDHA constructs by DOX increased ROS level in 22A-shLDHA-5 clone at 24 and 48 h. All experiments were performed at least triplicate, and every condition was tested at least in triplicate. Error bars represent standard deviation. *P < 0.05 compared with control condition.
LDHA is required for maximal tumour growth but not essential for tumour persistence
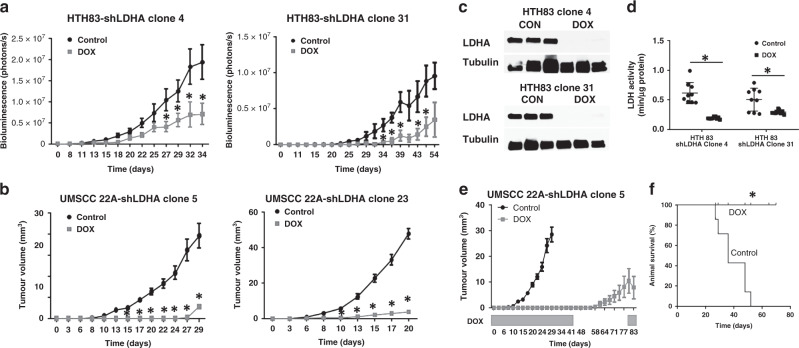
Hth83-shLDHA clones 4 and 31 were used to generate orthotopic xenografts. Tumour growth was monitored by bioluminescence activity (Fig. 4a). Suppression of LDHA significantly decreased bioluminescence activity and suppressed tumour growth in both Hth83-shLDHA clones compared with growth in control mice. Western blot analysis confirmed decreased tumour LDHA protein levels (Fig. 4c), and biochemical analysis confirmed decreased LDH activity in the tumours undergoing LDHA suppression (Fig. 4d). LDHA suppression significantly inhibited tumour growth in tumours from both 22A-shLDHA clones, 5 and 23, compared with growth in control mice. DOX-induced LDHA suppression was associated with increased median survival in mice with 22A tumours; the control group (n = 9) had a median survival of 36 days, while mice treated with DOX-water (n = 9) survived for 80 days after cell injection and were subsequently euthanised to complete this study (Fig. 4f).
Fig. 4. LDHA is required for maximal tumour proliferation and survival.
Hth83 (a) and 22A (b) clones demonstrated significant tumour growth inhibition following LDHA suppression at day 5 post injection, compared with the control, which is consistent with relative LDHA activity levels. c Western blots demonstrating suppression of LDHA protein levels in tumours following treatment with DOX. d LDH activity was significantly decreased following construct activation. e Withdrawal of DOX to restore LDHA expression resulted in tumour growth restoration. f Suppression of LDHA significantly prolonged median survival time. Data for a, b, and e are presented as means with error bars representing standard deviations. *P < 0.05 compared with control condition.
To determine whether LDHA suppression was required for viability in addition to maximal tumour growth, we withdrew DOX, halting LDHA suppression, in 22A-shLDHA clone 5, which demonstrated near-complete abrogation of tumour growth at day 41. As shown in Fig. 4e, removal of suppression allowed for tumour growth resumption, indicating that cell viability was maintained, despite extensive LDHA suppression.
Consistent with the disruption of basic metabolic pathways detected at a cellular level following LDHA suppression (Fig. 2), we detected diffuse metabolic shifts in 22A tumours. Tumours were generated using 22A cells expression shLDHA. Measurable tumours (5 × 5 mm2) were then exposed to DOX for 24 h. Steady-state metabolomics analysis was performed as detailed above, and those individual metabolites that were statistically significantly altered by DOX expose are listed in Supplemental Table S1. Increased levels of pyroglutamic acid, homovanillic acid, n-acetyl-l-aspartic acid are consistent with overall metabolic stress and oxidative stress. Higher levels of spermidine and hydroxyproline were detected, which has putative implications for tumorigenesis (Supplemental Table S1). The remaining metabolites demonstrate fluctuations in amino acid, purine and pyrimidine, as well as fatty acid metabolism, consistent with our conclusion that suppression of LDHA rapidly cascades through cellular and tumour metabolism.
LDHA activity is required for neutralisation of EBRT effects
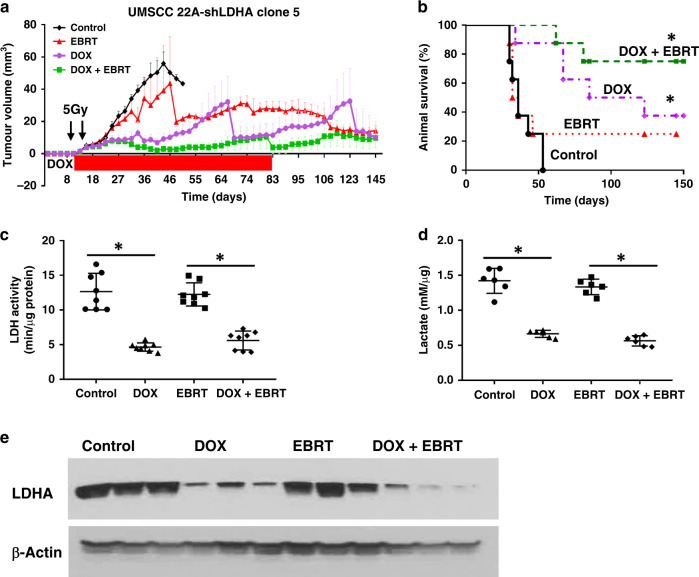
We evaluated the effect of LDHA suppression on relative EBRT sensitivity in vivo. Mice treated with EBRT alone (n = 8) showed no significant inhibition of tumour growth compared with the control group (n = 8), consistent with the relative radiation resistance seen in 22A cells. Suppression of LDHA (n = 8) resulted in significant tumour growth delay. When combined with radiation, suppression of LDHA activity resulted in significant tumour growth inhibition (n = 8). When LDHA suppression was stopped (by withdrawal of DOX treatment) at day 83, tumour volumes remained stable (no tumour growth or shrinkage).
The median survival of mice with 22A tumours was 34 in the control group and 35 days in the radiation-alone group (Fig. 5a). Suppression of LDHA alone significantly prolonged the median survival to 85 days. Combined LDHA suppression and radiation resulted in even longer mouse survival, with 75% of mice remaining alive at day 145 (Fig. 5b). Furthermore, a decrease in tumour volume was observed in mice with LDHA knockdown alone as well as in the combined treatment group at the completion of the experiment at day 145. Specifically, a total of 11 mice remained alive at the end of the study, and six of them had no evidence of disease (two in the LDHA knockdown group and four in the combined treatment group), while the remaining five mice with potential submucosal lesions were histopathologically evaluated and showed no viable residual tumour (Supplemental Fig. S6). LDHA protein levels were confirmed to be suppressed following DOX treatment in the xenograft tumours using Western blot analysis (Fig. 5e). A significant decrease in LDH activity and lactate levels was also measured in the tumour tissue following LDHA suppression in the presence or absence of radiation, but not following radiation treatment alone (Fig. 5c, d).
Fig. 5. LDHA activity is required to neutralise radiation effects.
a Suppression of LDHA activity resulted in a significant increase in tumour growth inhibition when combined with radiation. Cessation of suppression resulted in tumour volume stability. LDHA suppression was induced at day 12 and combined with radiation treatments (5 Gy/day × 2 days) given on days 15 and 18 after cell injection. b Suppression of LDHA significantly prolonged median survival time. c Suppression of LDHA resulted in decreased tumour LDH activity. d Suppression of LDHA led to reduced lactate production in tumour xenografts. e Western blots demonstrating suppression of LDHA protein levels in tumours following treatment with DOX. Data for a are presented as means with error bars representing standard deviations. *P < 0.05 compared with control condition.
Discussion
Effective cancer treatment is predicated on a therapeutic index generated by differential biological activity in tumour cells compared with normal tissue. Targeting of tumour metabolism is similarly grounded in a differential metabolic phenotype that is unique to tumour cells and amenable to manipulation without significant normal tissue toxicity. Since Warburg first described the metabolic activity of tumour cells, a multitude of “biomarkers” of altered tumour metabolism have been described, including both metabolic enzymes (i.e. LDH, isocitrate dehydrogenase, pyruvate kinase) and metabolic regulators (i.e. AMPK, mTOR, LKB1).34,35 In some cases, investigators have correlated differential expression and protein levels with tumorigenesis and treatment response, while in a few select cases, mutations in metabolic enzymes and regulators have been linked to differential tumour activity.17,18,36–38
Drawing from canonical cancer studies in which mutations and differential expression served to generate targets for directed manipulation of cellular signalling (i.e. tyrosine kinase inhibitors), the discovery of metabolic biomarkers led to an initial hope that targeted metabolic inhibition could be used to suppress tumour growth and/or enhance response to conventional therapeutics.39–41 Unfortunately, with the distinct exception of isocitrate dehydrogenase, which remains under active investigation, targeted metabolic inhibition has failed to materialise in the translational and clinical space.42,43 In the current study, we developed a systematic approach to investigate a critical metabolic enzyme that is essential to the Warburg effect in order to identify some potential limitations to our current metabolic targeting approaches.
LDH is a primary metabolic enzyme that catalyses the conversion of pyruvate and lactate, thereby playing an essential role in regulating cellular energy metabolism and nutrient exchange. LDHA expression varies across cancer cell lines and tumours and may be highly variable, as seen in our large cell line panel. Furthermore, LDHA expression has been shown to correlate with differential tumorigenesis, progression and metastasis in multiple tumour types12,15,17,18,44–46 and has also been associated with resistance to chemotherapy47 and radiotherapy48 and with survival, as shown in the current HNSCC The Cancer Genome Atlas dataset. Because of its central role in metabolism, LDHA has been considered a promising potential target for metabolic inhibition and has been investigated in multiple tumour types.8,16,49–52 However, LDHA inhibition has not yet translated into the clinical space.
Data from this study can serve as a useful guide for: (1) how manipulation of critical metabolic enzymes should be undertaken and (2) how manipulation of metabolic enzyme activity must be approached in the translational setting. Since LDHA is an essential enzyme for eukaryotic cells, systematic deletion of LDHA is embryonically lethal.53 Targeted, permanent deletion of LDHA (i.e. via CRISPR/Cas-9 techniques) in tumour cells can give rise to aberrant clonal expansion, which may or may not mimic the effects of transient inhibition; furthermore, deletion cannot generally be titrated.54 Knockdown of LDHA using short interfering RNA has limited application, especially for long-term studies, owing to transient and unpredictable effects.55
To overcome these limitations, we employed an inducible shRNA lentiviral Tet-On system to regulate shRNA expression by DOX.21 Using this system, we successfully established stable HNSCC and ATC cell lines with inducible downregulation of LDHA expression. This novel inducible system provides a number of significant advantages for studies targeting LDHA. The Tet-On-mediated suppression of LDHA is tightly regulated by DOX in a dose- and time-dependent manner. Suppression of LDHA can be fully reversed by DOX withdrawal, which allows us to maintain the transformed phenotype in the LDHA knockdown-stable cell lines. Furthermore, this unique feature also provides a powerful tool to study the function of LDHA in metabolic reprogramming by regulating LDHA expression at different stages for in vivo tumour xenograft models, allowing us to control the temporal component of suppression. The primary limitation of the current model stems from the potential metabolic modulatory effects of DOX tself, which have been previously described in cancer cell lines.56 This limit necessitates extensive utilisation of control conditions for metabolic experiments (Figs. 2 and 3 Supplemental Fig. 4), with the use of DOX in non-infected cell lines and, potentially, scrambled shRNA constructs along with low dosing (0.1–1 mg/mL).
Our results demonstrate the inherent difficulties associated with conventional chemical approaches to the suppression of metabolic enzymes. First, near-complete suppression of LDHA expression does not completely impede LDH activity, as would be expected, since the enzyme is a tetramer of multiple subunits. Second, despite a dramatic drop in LDH activity, lactate levels in tumour cells can re-equilibrate at later time points after suppression, and suppression of the enzyme does not result in comprehensive cell death but, rather, decreased proliferation. This result is explained by the rebalancing of cellular carbon and energetic flux under stress conditions, which would be expected since metabolic flexibility is preserved in tumour cells, as previously demonstrated by our group and others.19 There appear to be inherent limits to this metabolic adaptation as demonstrated by the fairly consistent rates of mitochondrial respiration measured in cells with suppressed LDH activity. This finding highlights two important issues: (1) cells are likely unable to rapidly increase the expression of genes involved in the electron transport chain in order to take advantage of the measured carbon shunting resulting from LDH inhibition and (2) the inducible model can be used to separate metabolic adaptation immediately available to cells upon LDH targeting versus metabolic adaptation, which would require extensive rewiring of cellular metabolism and might be encountered in the context of permanent LDH inhibition/knockout (i.e. stable shLDHA inhibition, CRISPR knockout).
The relatively subtle difference between decreased proliferation and increased cell death generates profound and clinically relevant consequences. As shown in Fig. 4, prolonged LDHA suppression can completely suppress in vivo tumour growth in the HNSCC model. However, removal of the suppression allows for resumption of tumour growth, suggesting persistence of viable tumour cells. Clinically, if LDHA suppression were accomplished chemically, this method would be problematic because of the potential toxicity associated with continued, chronic chemical inhibition and reconstitution of tumours following cessation of treatment. Only when combined with radiation did long-term LDHA suppression result in actual cell death and tumour cures.
Although inhibition of critical metabolic enzymes such as LDHA can generate meaningful antitumour effects and may result in substantial sensitisation to conventional treatments such as radiation, our data demonstrate critical issues related to the efficacy and timing of this inhibition, which can be expected to generate different and translationally relevant results. In addition to relative expression levels and intrinsic dependencies on specific enzymes and pathways, the timing and extent of metabolic targeting, and its integration into translationally relevant algorithms, will be critical to actual clinical efficacy.
Supplementary information
Author contributions
S.Y.L., V.C.S. and Y.C. designed the study. Y.C., A.M., L.T., M.C., X.L., J.S.N., K.A.M., C.J.H., W.L., Y.C.H. and A.S.R.M. performed the experiments and analysed data; S.Y.L., V.C.S., P.L.L., N.P. and J.A.B. analysed data and provided resources. S.Y.L., V.C.S. and Y.C. wrote the initial draft of the paper. A.M., A.S.M., P.L.L., N.P. and J.A.B. reviewed and revised the draft of the paper. J.A.B., V.C.S. and S.Y.L. provided funding support for this study. S.Y.L. supervised the study. All authors reviewed the results and approved the final version of the paper.
Ethics approval and consent to participate
For the animal study, all procedures and care were reviewed and approved by The University of Texas MD Anderson Cancer Center Institutional Animal Care and Use Committee under Institutional Animal Care and Use Committee guidelines.
Consent to publish
Not applicable.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Funding information
The work in this study was funded in part, and V.C.S., S.Y.L., J.A.B., Y.C. and J.S.N. were supported by the Cancer Prevention and Research Institute of Texas (CPRIT) grant RP170366. V.C.S. is supported by the National Institute of Dental and Craniofacial Research through R03DE028858. J.A.B. is supported by R01CA211150. N.P. is supported by the CPRIT Proteomics and Metabolomics Core Facility (RP170005), NIH (P30 CA125123) and Dan L. Duncan Cancer Center. L.T. and P.L.L. were supported by CPRIT grant RP130397 and NIH grants S10OD012304-01, U01CA235510 and P30CA016672. Work performed in the Flow Cytometry and Cellular Imaging Facility is supported in part by the National Institutes of Health through MD Anderson’s Cancer Center Support grant CA016672. Work performed through the Mouse Metabolism and Phenotyping Core (Seahorse) is supported by NIH UM1HG006348 and NIH R01DK114356. The content is solely the responsibility of the authors and does not necessarily represent the official views of their sponsors.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Vlad C. Sandulache, Email: Vlad.Sandulache@bcm.edu
Stephen Y. Lai, Email: sylai@mdanderson.org
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-021-01297-x.
References
- 1.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J. Gen. Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 3.Li XB, Gu JD, Zhou QH. Review of aerobic glycolysis and its key enzymes–new targets for lung cancer therapy. Thorac. Cancer. 2015;6:17–24. doi: 10.1111/1759-7714.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Annibaldi A, Widmann C. Glucose metabolism in cancer cells. Curr. Opin. Clin. Nutr. Metab. Care. 2010;13:466–470. doi: 10.1097/MCO.0b013e32833a5577. [DOI] [PubMed] [Google Scholar]
- 5.Sandulache VC, Skinner HD, Wang Y, Chen Y, Dodge CT, Ow TJ, et al. Glycolytic inhibition alters anaplastic thyroid carcinoma tumor metabolism and improves response to conventional chemotherapy and radiation. Mol. Cancer Ther. 2012;11:1373–1380. doi: 10.1158/1535-7163.MCT-12-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woo SH, Sandulache VC, Yang L, Skinner HD. Evaluating response to metformin/cisplatin combination in cancer cells via metabolic measurement and clonogenic survival. Methods Mol Biol. 2014;1165:11–18. doi: 10.1007/978-1-4939-0856-1_2. [DOI] [PubMed] [Google Scholar]
- 7.Billiard J, Dennison JB, Briand J, Annan RS, Chai D, Colón M, et al. Quinoline 3-sulfonamides inhibit lactate dehydrogenase A and reverse aerobic glycolysis in cancer cells. Cancer Metab. 2013;1:19. doi: 10.1186/2049-3002-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeung C, Gibson AE, Issaq SH, Oshima N, Baumgart JT, Edessa LD, et al. Targeting glycolysis through inhibition of lactate dehydrogenase impairs tumor growth in preclinical models of Ewing sarcoma. Cancer Res. 2019;79:5060–5073. doi: 10.1158/0008-5472.CAN-19-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yecies JL, Manning BD. mTOR links oncogenic signaling to tumor cell metabolism. J. Mol. Med. 2011;89:221–228. doi: 10.1007/s00109-011-0726-6. [DOI] [PubMed] [Google Scholar]
- 10.Shackelford DB, Vasquez DS, Corbeil J, Wu S, Leblanc M, Wu C-L, et al. mTOR and HIF-1α-mediated tumor metabolism in an LKB1 mouse model of Peutz-Jeghers syndrome. Proc. Natl Acad. Sci. USA. 2009;106:11137–11142. doi: 10.1073/pnas.0900465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adeva-Andany M, Lopez-Ojen M, Funcasta-Calderon R, Ameneiros-Rodriguez E, Donapetry-Garcia C, Vila-Altesor M, et al. Comprehensive review on lactate metabolism in human health. Mitochondrion. 2014;17:76–100. doi: 10.1016/j.mito.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Rong Y, Wu W, Ni X, Kuang T, Jin D, Wang D, et al. Lactate dehydrogenase A is overexpressed in pancreatic cancer and promotes the growth of pancreatic cancer cells. Tumor Biol. 2013;34:1523–1530. doi: 10.1007/s13277-013-0679-1. [DOI] [PubMed] [Google Scholar]
- 13.Markert CL, Shaklee JB, Whitt GS. Evolution of a gene. Multiple genes for LDH isozymes provide a model of the evolution of gene structure, function and regulation. Science. 1975;189:102–114. doi: 10.1126/science.1138367. [DOI] [PubMed] [Google Scholar]
- 14.Xie H, Valera VA, Merino MJ, Amato AM, Signoretti S, Linehan WM, et al. LDH-A inhibition, a therapeutic strategy for treatment of hereditary leiomyomatosis and renal cell cancer. Mol. Cancer Ther. 2009;8:626–635. doi: 10.1158/1535-7163.MCT-08-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 16.Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, et al. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc. Natl Acad. Sci. USA. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohajertehran F, Ayatollahi H, Jafarian AH, Khazaeni K, Soukhtanloo M, Shakeri MT, et al. Overexpression of lactate dehydrogenase in the saliva and tissues of patients with head and neck squamous cell carcinoma. Rep. Biochem. Mol. Biol. 2019;7:142–149. [PMC free article] [PubMed] [Google Scholar]
- 18.Thonsri U, Seubwai W, Waraasawapati S, Sawanyawisuth K, Vaeteewoottacharn K, Boonmars T, et al. Overexpression of lactate dehydrogenase A in cholangiocarcinoma is correlated with poor prognosis. Histol. Histopathol. 2017;32:503–510. doi: 10.14670/HH-11-819. [DOI] [PubMed] [Google Scholar]
- 19.Sandulache VC, Chen Y, Skinner HD, Lu T, Feng L, Myers JN, et al. Acute tumor lactate perturbations as a biomarker of genotoxic stress: development of a biochemical model. Mol. Cancer Ther. 2015;14:2901–2908. doi: 10.1158/1535-7163.MCT-15-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu W, Chen Y, Dubrulle J, Stossi F, Putluri V, Sreekumar A, et al. Cisplatin generates oxidative stress which is accompanied by rapid shifts in central carbon metabolism. Sci. Rep. 2018;8:1–12. doi: 10.1038/s41598-018-22640-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le X, Huang AT, Chen Y, Lai SY. Regulation of receptor tyrosine kinases by miRNA overexpression of miRNA using lentiviral inducible expression vectors. Methods Mol Biol. 2015;1233:135–147. doi: 10.1007/978-1-4939-1789-1_13. [DOI] [PubMed] [Google Scholar]
- 22.Wu X, Bhayani MK, Dodge CT, Nicoloso MS, Chen Y, Yan X, et al. Coordinated targeting of the EGFR signaling axis by microRNA-27a. Oncotarget. 2013;4:1388. doi: 10.18632/oncotarget.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 24.Lee J, Ramirez MS, Walker CM, Chen Y, Yi S, Sandulache VC, et al. High-throughput hyperpolarized (13)C metabolic investigations using a multi-channel acquisition system. J. Magn. Reson. 2015;260:20–27. doi: 10.1016/j.jmr.2015.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gohlke, J. H., Lloyd, S. M., Basu, S., Putluri, V., Vareed, S. K., Rasaily, U. et al. Methionine-homocysteine pathway in African-American Prostate Cancer. JNCI Cancer Spectr.3, pkz019 (2019). [DOI] [PMC free article] [PubMed]
- 26.Vantaku, V., Putluri, V., Bader, D. A., Maity, S., Ma, J., Arnold, J. M. et al. Epigenetic loss of AOX1 expression via EZH2 leads to metabolic deregulations and promotes bladder cancer progression. Oncogene10.1038/s41388-019-0902-7 (2019). [DOI] [PMC free article] [PubMed]
- 27.Vantaku V, Dong J, Ambati CR, Perera D, Donepudi SR, Amara CS, et al. Multi-omics integration analysis robustly predicts high-grade patient survival and identifies CPT1B effect on fatty acid metabolism in bladder cancer. Clin. Cancer Res. 2019;25:3689–3701. doi: 10.1158/1078-0432.CCR-18-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu M, Neilson A, Swift AL, Moran R, Tamagnine J, Parslow D, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am. J. Physiol. Cell Physiol. 2007;292:C125–C136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- 29.Sandulache VC, Skinner HD, Ow TJ, Zhang A, Xia X, Luchak JM, et al. Individualizing antimetabolic treatment strategies for head and neck squamous cell carcinoma based on TP53 mutational status. Cancer. 2012;118:711–721. doi: 10.1002/cncr.26321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandulache, V. C., Chen, Y., Lee, J., Rubinstein, A., Ramirez, M. S., Skinner, H. D. et al. Evaluation of hyperpolarized [1-(1)(3)C]-pyruvate by magnetic resonance to detect ionizing radiation effects in real time. PLoS ONE9, e87031 (2014). [DOI] [PMC free article] [PubMed]
- 31.Kim S, Park YW, Schiff BA, Doan DD, Yazici Y, Jasser SA, et al. An orthotopic model of anaplastic thyroid carcinoma in athymic nude mice. Clin. Cancer Res. 2005;11:1713–1721. doi: 10.1158/1078-0432.CCR-04-1908. [DOI] [PubMed] [Google Scholar]
- 32.Ahn SH, Henderson Y, Kang Y, Chattopadhyay C, Holton P, Wang M, et al. An orthotopic model of papillary thyroid carcinoma in athymic nude mice. Arch. Otolaryngol. Head Neck Surg. 2008;134:190–197. doi: 10.1001/archoto.2007.36. [DOI] [PubMed] [Google Scholar]
- 33.Sano D, Xie TX, Ow TJ, Zhao M, Pickering CR, Zhou G, et al. Disruptive TP53 mutation is associated with aggressive disease characteristics in an orthotopic murine model of oral tongue cancer. Clin. Cancer Res. 2011;17:6658–6670. doi: 10.1158/1078-0432.CCR-11-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Green AS, Chapuis N, Lacombe C, Mayeux P, Bouscary D, Tamburini J. LKB1/AMPK/mTOR signaling pathway in hematological malignancies: from metabolism to cancer cell biology. Cell Cycle. 2011;10:2115–2120. doi: 10.4161/cc.10.13.16244. [DOI] [PubMed] [Google Scholar]
- 35.Shackelford DB, Shaw RJ. The LKB1–AMPK pathway: metabolism and growth control in tumour suppression. Nat. Rev. Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li N, Huang D, Lu N, Luo L. Role of the LKB1/AMPK pathway in tumor invasion and metastasis of cancer cells. Oncol. Rep. 2015;34:2821–2826. doi: 10.3892/or.2015.4288. [DOI] [PubMed] [Google Scholar]
- 37.Filipp FV. Cancer metabolism meets systems biology: pyruvate kinase isoform PKM2 is a metabolic master regulator. J. Carcinogen. 2013;12:14. doi: 10.4103/1477-3163.115423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reitman ZJ, Yan H. Isocitrate dehydrogenase 1 and 2 mutations in cancer: alterations at a crossroads of cellular metabolism. J. Natl Cancer Inst. 2010;102:932–941. doi: 10.1093/jnci/djq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 40.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat. Rev. Drug Discov. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 41.Akhenblit PJ, Pagel MD. Recent advances in targeting tumor energy metabolism with tumor acidosis as a biomarker of drug efficacy. J. Cancer Sci. Ther. 2016;8:20. doi: 10.4172/1948-5956.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fiume L, Manerba M, Vettraino M, Di Stefano G. Inhibition of lactate dehydrogenase activity as an approach to cancer therapy. Fut. Med. Chem. 2014;6:429–445. doi: 10.4155/fmc.13.206. [DOI] [PubMed] [Google Scholar]
- 43.Granchi C, Bertini S, Macchia M, Minutolo F. Inhibitors of lactate dehydrogenase isoforms and their therapeutic potentials. Curr. Med. Chem. 2010;17:672–697. doi: 10.2174/092986710790416263. [DOI] [PubMed] [Google Scholar]
- 44.Grimm M, Alexander D, Munz A, Hoffmann J, Reinert S. Increased LDH5 expression is associated with lymph node metastasis and outcome in oral squamous cell carcinoma. Clin. Exp. Metastasis. 2013;30:529–540. doi: 10.1007/s10585-012-9557-2. [DOI] [PubMed] [Google Scholar]
- 45.Kolev Y, Uetake H, Takagi Y, Sugihara K. Lactate dehydrogenase-5 (LDH-5) expression in human gastric cancer: association with hypoxia-inducible factor (HIF-1alpha) pathway, angiogenic factors production and poor prognosis. Ann. Surg. Oncol. 2008;15:2336–2344. doi: 10.1245/s10434-008-9955-5. [DOI] [PubMed] [Google Scholar]
- 46.Semenza GL, Jiang BH, Leung SW, Passantino R, Concordet JP, Maire P, et al. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 1996;271:32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- 47.Koukourakis MI, Giatromanolaki A, Sivridis E, Gatter KC, Trarbach T, Folprecht G, et al. Prognostic and predictive role of lactate dehydrogenase 5 expression in colorectal cancer patients treated with PTK787/ZK 222584 (vatalanib) antiangiogenic therapy. Clin. Cancer Res. 2011;17:4892–4900. doi: 10.1158/1078-0432.CCR-10-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koukourakis MI, Giatromanolaki A, Panteliadou M, Pouliliou SE, Chondrou PS, Mavropoulou S, et al. Lactate dehydrogenase 5 isoenzyme overexpression defines resistance of prostate cancer to radiotherapy. Br. J. Cancer. 2014;110:2217–2223. doi: 10.1038/bjc.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie H, Hanai J, Ren JG, Kats L, Burgess K, Bhargava P, et al. Targeting lactate dehydrogenase-a inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell Metab. 2014;19:795–809. doi: 10.1016/j.cmet.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheng SL, Liu JJ, Dai YH, Sun XG, Xiong XP, Huang G. Knockdown of lactate dehydrogenase A suppresses tumor growth and metastasis of human hepatocellular carcinoma. FEBS J. 2012;279:3898–3910. doi: 10.1111/j.1742-4658.2012.08748.x. [DOI] [PubMed] [Google Scholar]
- 51.Chang CC, Zhang C, Zhang Q, Sahin O, Wang H, Xu J, et al. Upregulation of lactate dehydrogenase a by 14-3-3zeta leads to increased glycolysis critical for breast cancer initiation and progression. Oncotarget. 2016;7:35270–35283. doi: 10.18632/oncotarget.9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lewis BC, Prescott JE, Campbell SE, Shim H, Orlowski RZ, Dang CV. Tumor induction by the c-Myc target genes rcl and lactate dehydrogenase A. Cancer Res. 2000;60:6178–6183. [PubMed] [Google Scholar]
- 53.Merkle S, Favor J, Graw J, Hornhardt S, Pretsch W. Hereditary lactate dehydrogenase A-subunit deficiency as cause of early postimplantation death of homozygotes in Mus musculus. Genetics. 1992;131:413–421. doi: 10.1093/genetics/131.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iriki H, Kawata T, Muramoto T. Generation of deletions and precise point mutations in Dictyostelium discoideum using the CRISPR nickase. PLoS ONE. 2019;14:e0224128. doi: 10.1371/journal.pone.0224128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X, Xu L, Wu Q, Liu M, Tang F, Cai Y, et al. Inhibition of LDHA deliver potential anticancer performance in renal cell carcinoma. Urol. Int. 2017;99:237–244. doi: 10.1159/000445125. [DOI] [PubMed] [Google Scholar]
- 56.Ahler E, Sullivan WJ, Cass A, Braas D, York AG, Bensinger SJ, et al. Doxycycline alters metabolism and proliferation of human cell lines. PLoS ONE. 2013;8:e64561. doi: 10.1371/journal.pone.0064561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.