Summary
Dysregulation of N6-methyladenosine (m6A) modification is associated with cancer development and progression. The m6A modification plays a crucial role in autophagy regulation precipitating anti-cancer drug resistance. In line with this fact, this commentary discusses m6A modification interfering with autophagy machinery as a major contributing factor for drug resistance in cancer.
Subject terms: Cancer epigenetics, Cancer epigenetics
Main
Anti-cancer drug resistance is attributed to multiple mechanisms, including genetic alteration, epigenetic modification, and cell heterogeneity, which appears to hamper cancer treatment and management.1 Autophagy is an evolutionarily conserved catabolic process in which cells self-digest their own damaged organelles, mis-folded proteins, protein aggregates and other macromolecules to maintain cellular homoeostasis.2 Recently, aberrant genetic and epigenetic markers have been identified which critically contribute to the acquisition of anti-cancer drug resistance.1,3 Accumulating evidence indicates that defects in autophagy is associated with tumorigenesis and cancers progression.
Numerous studies have confirmed that autophagy induction promotes anti-cancer drug resistance to facilitate cell survival and that suppression of autophagy could enhance the sensitivity of cancer cells to anti-cancer drugs.4 N6-methyladenosine (m6A) is the most abundant internal modification of eukaryotic RNA. This m6A modification is facilitated by three groups of proteins (m6A regulators) such as m6A “writers” (methyltransferases), “erasers” (demethylases), and “readers” (RNA binding proteins).5–9 m6A regulators play crucial roles in various physiological and pathological processes through the regulation of RNA stability, mRNA splicing, translation or decay, and microRNA processing.5,6 The m6A modification is involved in the initiation and progression of cancers, including head and neck cancer, colorectal cancer, liver cancer, cervical cancer, breast cancer, and glioma.5–7 However, the molecular mechanisms associated with m6A modification involved in autophagy regulation and anti-cancer drug resistance still remain unclear.
The initiation of autophagy and formation of autophagosome is regulated by m6A modifiers. Fat mass and obesity-associated protein (FTO), an m6A demethylase, increased ULK1 expression and autophagy initiation.8 Concurrently, FTO regulates autophagy and adipogenesis by targeting autophagy related gene-5 (ATG5) and ATG7 in an m6A-dependent manner. A concomitant depletion of FTO decreased the expression of ATG5 and ATG7 which led to attenuation of autophagosome formation, thereby inhibiting autophagy.9 The overexpression of FTO in cervical squamous cell carcinoma contributed to resistance to radiotherapy and chemotherapy through upregulation of β-catenin expression.10 Furthermore, it was also demonstrated that FTO depletion increases m6A levels of important oncogenes which promotes their RNA decay through YTHDF2, thereby sensitising melanoma cells to interferon γ (IFNγ) and anti-PD-1 treatment.11
The increased expression of ALKBH5, another m6A demethylase, impaired autophagy, promoted cell proliferation and invasion in epithelial ovarian cancer.12 These findings suggested that m6A regulators play important roles in autophagy regulation in cancer. The suppression of the m6A methyltransferase METTL3 enhanced the sensitivity of pancreatic cancer cells to chemotherapy, especially to cisplatin, gemcitabine, and 5-fluorouracil.13 A recent study indicated that cancer cells with low or no c-MET expression were primarily resistant to chidamide–crizotinib cotreatment and enforced overexpression of c-MET which, in turn, increased the sensitivity of these cells to chidamide–crizotinib cotreatment. Furthermore, chidamide decreased the expression of c-MET by inhibiting mRNA m6A methylation modification through the downregulation of METTL3 and WTAP expression.14 Additionally, the depletion of m6A methyltransferase METTL14 increased autophagy, provoked by cisplatin treatment in pancreatic cancer cells through the mammalian target of rapamycin (mTOR) signalling axis.15 These findings suggested that RNA m6A modification and its regulators may not only affect chemosensitivity of cancer cells, but also serve as novel therapeutic targets for overcoming resistance of cancer cells to anti-cancer therapies.
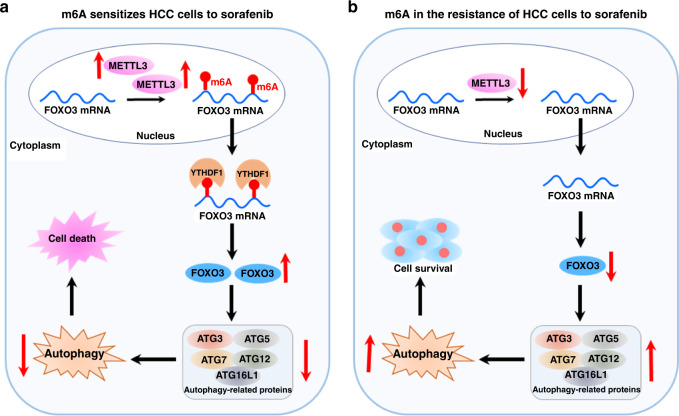
Sorafenib, a multi-kinase inhibitor, exhibits significant inhibitory effects on solid tumours, especially hepatocellular carcinoma (HCC). Interestingly, METTL3 can sensitise HCC cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, ATG5, ATG7, ATG12, and ATG16L1.7 Moreover, depletion of METTL3 in HCC cells promotes sorafenib resistance by the FOXO3-mediated autophagy signalling pathway (Fig. 1).
Fig. 1. Role of m6A modification in sorafenib resistance by modulating autophagy in hepatocellular carcinoma (HCC).
a The m6A “writer” METTL3 sensitises HCC cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner. m6A is deposited on FOXO3 mRNA by METTL3 and translated by YTHDF1, further increased FOXO3 downregulates the transcription of autophagy-related genes, including ATG3, ATG5, ATG7, ATG12, and ATG16L1, thereby, inhibition of autophagy promotes the death of HCC cells. b Depletion of METTL3 and FOXO3 promote autophagy associated sorafenib resistance in HCC.
In conclusion, recent findings indicate that m6A modification and its regulators play vital roles in autophagy-mediated anti-cancer drug resistance. Therefore, targeting m6A modification might provide a novel therapeutic strategy to enhance the treatment response in autophagy-related drug-resistant cancers.
Author contributions
Both the authors wrote the manuscript and performed literature search.
Ethics approval and consent to participate
Not applicable.
Data availability
Not applicable.
Competing interests
The authors declare no competing interests.
Funding information
This study was supported by the Indian Council of Medical Research (Grant No. DHR-GIA, 2020-9530), Government of India.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Quagliano A, Gopalakrishnapillai A, Barwe SP. Understanding the mechanisms by which epigenetic modifiers avert therapy resistance in cancer. Front. Oncol. 2020;10:992. doi: 10.3389/fonc.2020.00992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ouyang, Y., Wu, Q., Li, J., Sun, S. & Sun, S. S‐adenosylmethionine: a metabolite critical to the regulation of autophagy. Cell Prolif. 53, e12891 (2020). [DOI] [PMC free article] [PubMed]
- 3.Xiang M, Liu W, Tian W, You A, Deng D. RNA N-6-methyladenosine enzymes and resistance of cancer cells to chemotherapy and radiotherapy. Epigenomics. 2020;12:801–809. doi: 10.2217/epi-2019-0358. [DOI] [PubMed] [Google Scholar]
- 4.Li X, He S, Ma B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer. 2020;19:12. doi: 10.1186/s12943-020-1138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang H, Weng H, Chen J. m6A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. 2020;37:270–288. doi: 10.1016/j.ccell.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anitha R, Paramasivam A, Vijayashree Priyadharsini J, Chitra S. The m6A readers YTHDF1 and YTHDF3 aberrations associated with metastasis and predict poor prognosis in breast cancer patients. Am. J. Cancer Res. 2020;10:2546–2554. [PMC free article] [PubMed] [Google Scholar]
- 7.Lin Z, Niu Y, Wan A, Chen D, Liang H, Chen X, et al. RNA m6 A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J. 2020;39:e103181. doi: 10.15252/embj.2019103181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin S, Zhang X, Miao Y, Liang P, Zhu K, She Y, et al. m(6)A RNA modification controls autophagy through upregulating ULK1 protein abundance. Cell Res. 2020;28:955–957. doi: 10.1038/s41422-018-0069-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Wu R, Liu Y, Zhao Y, Bi Z, Yao Y, et al. m6A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy. 2020;16:1221–1235. doi: 10.1080/15548627.2019.1659617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou S, Bai ZL, Xia D, Zhao ZJ, Zhao R, Wang YY, et al. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting beta-catenin through mRNA demethylation. Mol. Carcinog. 2018;57:590–597. doi: 10.1002/mc.22782. [DOI] [PubMed] [Google Scholar]
- 11.Yang S, Wei J, Cui YH, Park G, Shah P, Deng Y, et al. M(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 2019;10:2782. doi: 10.1038/s41467-019-10669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu H, Gan X, Jiang X, Diao S, Wu H, Hu J. ALKBH5 in BCL-2hibited autophagy of epithelial ovarian cancer through miR-7 and BCL-2. J. Exp. Clin. Cancer Res. 2019;38:163. doi: 10.1186/s13046-019-1159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taketo K, Konno M, Asai A, Koseki J, Toratani M, Satoh T, et al. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int. J. Oncol. 2018;52:621–629. doi: 10.3892/ijo.2017.4219. [DOI] [PubMed] [Google Scholar]
- 14.Ding N, You A, Tian W, Gu L, Deng D. Chidamide increases the sensitivity of non-small cell lung cancer to crizotinib by decreasing c-MET mRNA methylation. Int. J. Biol. Sci. 2020;16:2595–2611. doi: 10.7150/ijbs.45886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong F, Liu X, Zhou Y, Hou X, He J, Li Q, et al. Downregulation of METTL14 increases apoptosis and autophagy induced by cisplatin in pancreatic cancer cells. Int. J. Biochem. Cell Biol. 2020;122:105731. doi: 10.1016/j.biocel.2020.105731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.