Abstract
Psychological stress has been reported to relate to dysbiosis, imbalance of the intestinal microbiota composition, and contribute to the onset and exacerbation of depression, though, underlying mechanisms of psychological stress-related dysbiosis have been unknown. It has been previously established that α-defensins, which are effector peptides of innate enteric immunity produced by Paneth cells in the small intestine, play an important role in regulation of the intestinal microbiota. However, the relationship between disruption of intestinal ecosystem and α-defensin under psychological stress is yet to be determined. Here we show using chronic social defeat stress (CSDS), a mouse depression model that (1) the exposure to CSDS significantly reduces α-defensin secretion by Paneth cells and (2) induces dysbiosis and significant composition changes in the intestinal metabolites. Furthermore, (3) they are recovered by administration of α-defensin. These results indicate that α-defensin plays an important role in maintaining homeostasis of the intestinal ecosystem under psychological stress, providing novel insights into the onset mechanism of stress-induced depression, and may further contribute to discovery of treatment targets for depression.
Subject terms: Microbiome, Mucosal immunology, Neurological disorders, Experimental models of disease
Introduction
Depression is a serious illness that tends to persist and often relapse, and the number of the patients has been increasing worldwide to cause great social loss1. It has been known that multiple factors are involved in the onset of depression such as the decreased monoamine neurotransmitters including dopamine, the overactivation of hypothalamic–pituitary–adrenal (HPA) axis, and decreased neuroplasticity due to combination of genetic factors and psychological stress2. Among them, psychological stress is considered an important factor contributing to the development of depression3. However, since mechanisms of depression are diverse, the whole picture involving its onset has been still unclear. Relationships between the intestinal microbiota and depression have been reported, recently. The intestinal microbiota has been known to affect immunity of the host via their cell components such as lipopolysaccharide (LPS) or by producing metabolites such as short-chain fatty acids and neurotransmitters including γ-aminobutyric acid (GABA) and serotonin, resulting in changes of brain function to develop depression4. In addition, a cohort study showed that a positive correlation between quality of life of patients with depression and the butyrate-producing bacteria or the synthetic potential of bacterial dopamine metabolites5. Furthermore, it has been reported that transplanting feces of depression patients into mice produces abnormalities in bacterial tryptophan metabolism and production of short-chain fatty acids and further induces behavior characteristic of depression6. Germ-free mice exposed to psychological stress showed the excessive response via HPA axis7. It is known that dysbiosis and changes of the microbial metabolites leading to depression are induced by psychological stress8. However, underlying mechanisms that psychological stress causes the disruption of homeostasis in the bacterial metabolites due to dysbiosis remains to be determined.
α-Defensin, an antimicrobial peptide, produced and secreted by Paneth cells in the crypt of the small intestine is responsible, in part, for innate enteric immunity9–12 and plays a critical role in both elimination and symbiosis in the intestine by killing pathogens while eliciting less bactericidal activities against commensal bacteria13. It has been reported that the absence of active α-defensin alters the composition of small intestinal microbiota in mice14, and oral administration of α-defensin rescues severe dysbiosis of graft-versus-host disease (GVHD) model mice15, indicating that α-defensin plays an important role in maintaining homeostasis of the intestinal microbiota. In addition, it has been known that α-defensin abnormalities cause dysbiosis and disruption of the intestinal metabolism in Crohn’s disease model mice16, 17. Dysbiosis caused by Paneth cell damage with decreased α-defensin has been shown to relate to various diseases18, 19. Thus, it is possible that α-defensin secreted by Paneth cells contributes to the maintenance of systemic homeostasis by regulating the intestinal microbiota and their metabolites. However, relationships between Paneth cell α-defensin and dysbiosis or disruption of homeostasis in the intestinal metabolites in depression have been unknown.
Here, we provide evidence that a decrease of α-defensin due to psychological stress induces dysbiosis and subsequent disruption of homeostasis in microbial metabolites in CSDS model mice, a model of psychological stress-induced depression, and that α-defensin rescue attenuates the observed imbalance of the intestinal microbiota and their metabolites. This study provides new insights into the mechanism of depression and further contributes to the prevention and the discovery of therapeutic targets for depression.
Results
CSDS exposure decreases α-defensin secretion
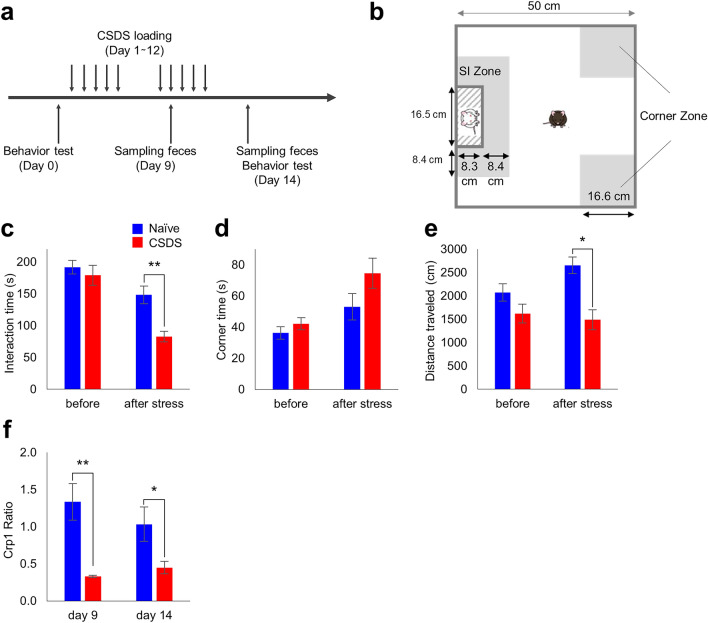
Analysis of behavior changes in the CSDS model (Fig. 1a,b) showed that the interaction time is significantly reduced in CSDS group (82.8 ± 20.3 s.) compared to naïve group (148.5 ± 34.2 s.) (Fig. 1c). In addition, the corner time increased from 53.1 ± 20.7 s. to 74.6 ± 23.8 s. (P = 0.093) (Fig. 1d), and the total action distance significantly decreased from 2654.8 ± 439.6 cm to 1490.7 ± 519.3 cm (Fig. 1e). These results indicated that it was confirmed that a characteristic decrease in sociality occurs in the CSDS model.
Figure 1.
Behavior and fecal cryptdin-1 in CSDS model. (a) Schedule of experiment. (b) The cage for behavior test. (c) Staying time in SI zone. (d) Staying time in corner zone. (e) Total distance traveled. (f) Fecal Crp1 protein ratio vs naïve group in day 1. The data of (c)–(f) were shown as mean ± SE of six independent experiments in each group. Mann–Whitney U tests were used to compare the data. *P < 0.05; **P < 0.01.
Next, to analyze the relationship between the CSDS load and the amount of α-defensin secretion, Crp1 in feces of the CSDS (for 12 days) group was measured. The Crp1 amounts on day 9 and day 14 of the CSDS group were decreased to 33% and 45%, respectively, compared to those on day 1 of the naïve group (Fig. 1f, Table S1).
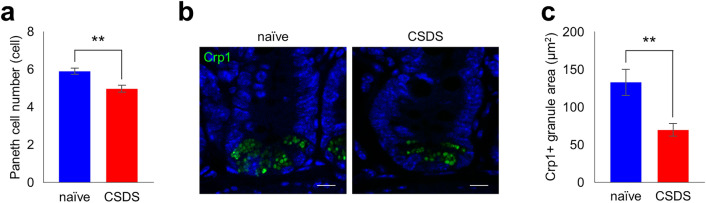
We further conducted immunofluorescent analyses of the small intestinal tissue and found a significant decrease in the number of Paneth cells and Crp1+ granule area in CSDS group (Fig. 2a–c) compared to naïve group. These results were consistent with the decrease in fecal Crp1 in CSDS group (Fig. 1f).
Figure 2.
Decrease of Paneth cell number and cryptdin-1 expression in CSDS model. (a) Paneth cell number per crypt. (b) Representative confocal images of Crp1 (green) with DAPI (blue) of crypt sections. Scale bars indicate 10 µm. (c) Crp1 positive granule area per crypt. The data of a, c were shown as mean ± SE of six independent experiments in each group. Mann–Whitney U tests were used to compare the data. **P < 0.01.
Dysbiosis is induced by α-defensin reduction in CSDS model and recovered by Crp4 administration
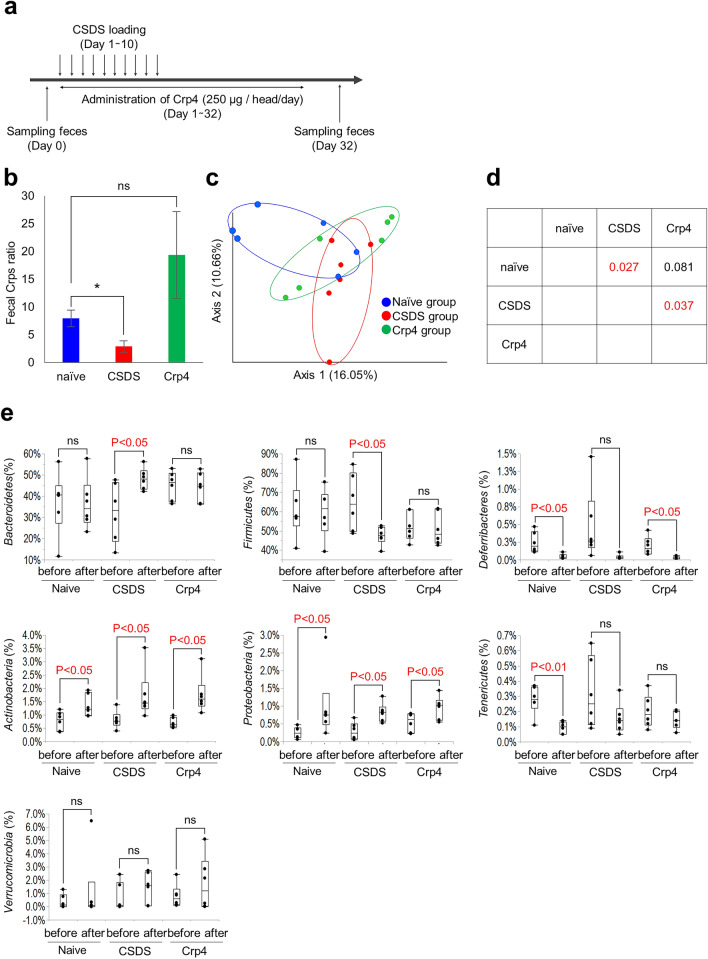
Next, in order to clarify whether the intestinal microbiota changes due to CSDS and the changes depend on decreased α-defensin, Crp4 administration experiment to rescue α-defensin was conducted focusing on the initial phase under the short stress period (Fig. 3a). The three groups; naïve group, CSDS-loaded group (CSDS group), and CSDS-loaded plus Crp4 administration group (Crp4 group) were analyzed. In the CSDS group, fecal α-defensin was significantly decreased compared to the naïve group (Fig. 3b), consistent with data in our previous experiment (Fig. 1f). In contrast, no significant difference was observed between naïve group and Crp4 group, indicating α-defensin (Crps; Crp1 and Crp4) was rescued by oral administration of Crp4 (Fig. 3b). We analyzed the intestinal microbiota of CSDS group and Crp4 group, and β-diversity showed significant differences between naïve group and CSDS group (P = 0.027) and between CSDS group and Crp4 group (P = 0.037). In contrast, no difference was observed between naïve group and Crp4 group (Fig. 3c,d), suggesting that dysbiosis caused by CSDS may be recovered by Crp4 administration. We next analyzed the intestinal microbiota composition at Phylum level in the three groups before and after CSDS (Fig. 3e). Bacteroidetes increased significantly (from 32 to 48%, P = 0.04) and Firmicutes decreased significantly (from 65 to 48%, P = 0.03) in CSDS group. Deferribacteres did not change in CSDS group before and after CSDS loading, whereas significantly decreased in the naïve group (from 0.25 to 0.05%, P = 0.02) and Crp4 group (from 0.19 to 0.03%, P = 0.03). These data indicated that the characteristic changes of Bacteroidetes, Firmicutes, and Deferribacteres by CSDS loading were canceled by Crp4 administration. Actinobacteria and Proteobacteria increased significantly in all groups after CSDS loading. Tenericutes decreased significantly only in naïve group (from 0.3 to 0.1%, P = 0.001). Verrucomicrobia did not change before and after CSDS loading in all groups. These results indicated that CSDS-related dysbiosis is partially rescued by the Cpr4 administration toward the intestinal microbiota in naïve group.
Figure 3.
Change in the composition of gut microbiota in the CSDS model by α-defensin administration. (a) Schedule of experiment. (b) Fecal Crp1 + Crp4 (Crps) protein ratio in day 32 vs before CSDS. Data are shown as the mean ± SE. Steel’s test was used to compare the data (vs naïve group). *P < 0.05. (c) PCoA of β-diversity comparison. (d) The P-values of PERMANOVA test. (e) Changes in the genus compositions before or after CSDS (shown only > 0.1% abundance). Pairs student’s t test was used to compare the data. *P < 0.05. The data of b, c, e were shown of six independent experiments in each group.
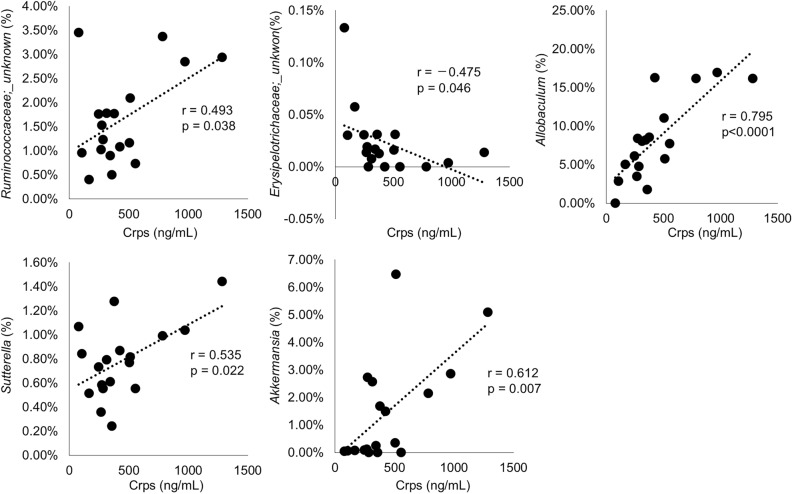
We further determined the intestinal microbiota which change significantly triggered by increase or decrease of α-defensin, by conducting correlation analyses between the amount of α-defensin in feces and the intestinal microbiota composition. Ruminococcaceae (r = 0.493, P = 0.038), Allobaculum (r = 0.795, P < 0.0001), Sutterella (r = 0.535, P = 0.022), and Akkermansia (r = 0.612, P = 0.007) showed positive correlation with α-defensin, whereas Erysipelotrichaceae (r = − 0.475, P = 0.046) showed negative correlation (Table 1, Fig. 4). Taken together, these results including the rescue experiment clarified that α-defensin reduction due to CSDS causes dysbiosis at least partially.
Table 1.
Correlation between fecal Crps and microbiota at genus level.
| Phylum | Class | Order | Family | Genus | r | P value |
|---|---|---|---|---|---|---|
| Actinobacteria | Actinobacteria | Actinomycetales | Actinomycetaceae | Actinomyces | 0.000 | 1.000 |
| Corynebacteriaceae | Corynebacterium | 0.000 | 1.000 | |||
| Microbacteriaceae | Microbacterium | 0.000 | 1.000 | |||
| Nocardiaceae | Rhodococcus | 0.000 | 1.000 | |||
| Bifidobacteriales | Bifidobacteriaceae | Bifidobacterium | − 0.102 | 0.688 | ||
| Coriobacteriia | Coriobacteriales | Coriobacteriaceae | unknown | 0.096 | 0.704 | |
| Coriobacteriaceae | unknown | 0.070 | 0.784 | |||
| Coriobacteriaceae | Adlercreutzia | − 0.017 | 0.947 | |||
| Aquificae | Aquificae | Aquificales | Aquificaceae | Hydrogenobacter | 0.000 | 1.000 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | 0.170 | 0.499 |
| Porphyromonadaceae | Parabacteroides | 0.115 | 0.650 | |||
| Rikenellaceae | unknown | − 0.154 | 0.542 | |||
| S24-7 | unknown | 0.020 | 0.936 | |||
| Paraprevotellaceae | Prevotella | − 0.116 | 0.646 | |||
| Cyanobacteria | Chloroplast | Streptophyta | unknown | unknown | 0.000 | 1.000 |
| Deferribacteres | Deferribacteres | Deferribacterales | Deferribacteraceae | Mucispirillum | − 0.346 | 0.160 |
| Firmicutes | unknown | unknown | unknown | unknown | 0.000 | 1.000 |
| Bacilli | unknown | unknown | unknown | 0.000 | 1.000 | |
| Bacillales | Planococcaceae | Sporosarcina | 0.000 | 1.000 | ||
| Staphylococcaceae | Jeotgalicoccus | 0.000 | 1.000 | |||
| Staphylococcaceae | Staphylococcus | − 0.324 | 0.189 | |||
| Lactobacillales | Enterococcaceae | Enterococcus | 0.000 | 1.000 | ||
| Lactobacillaceae | unknown | − 0.219 | 0.383 | |||
| Lactobacillaceae | Lactobacillus | − 0.350 | 0.154 | |||
| Leuconostocaceae | unknown | 0.000 | 1.000 | |||
| Leuconostocaceae | Weissella | 0.000 | 1.000 | |||
| Streptococcaceae | Streptococcus | − 0.435 | 0.071 | |||
| Turicibacterales | Turicibacteraceae | Turicibacter | − 0.076 | 0.765 | ||
| Clostridia | unknown | unknown | unknown | − 0.237 | 0.343 | |
| Clostridiales | unknown | unknown | − 0.251 | 0.316 | ||
| unknown | unknown | 0.153 | 0.546 | |||
| Christensenellaceae | unknown | − 0.005 | 0.986 | |||
| Clostridiaceae | Candidatus Arthromitus | − 0.168 | 0.504 | |||
| Clostridiaceae | Clostridium | 0.295 | 0.234 | |||
| Dehalobacteriaceae | Dehalobacterium | − 0.216 | 0.390 | |||
| Eubacteriaceae | Anaerofustis | − 0.016 | 0.950 | |||
| Lachnospiraceae | unknown | − 0.328 | 0.184 | |||
| Lachnospiraceae | unknown | − 0.283 | 0.256 | |||
| Lachnospiraceae | Clostridium | − 0.109 | 0.668 | |||
| Lachnospiraceae | Coprococcus | − 0.116 | 0.648 | |||
| Lachnospiraceae | Dorea | − 0.154 | 0.542 | |||
| Lachnospiraceae | Roseburia | − 0.203 | 0.420 | |||
| Lachnospiraceae | Ruminococcus | − 0.122 | 0.631 | |||
| Peptococcaceae | unknown | 0.000 | 1.000 | |||
| Peptococcaceae | unknown | − 0.212 | 0.399 | |||
| Peptococcaceae | rc4-4 | 0.044 | 0.861 | |||
| Ruminococcaceae | unknown | − 0.308 | 0.213 | |||
| Ruminococcaceae | unknown | 0.493 | 0.038* | |||
| Ruminococcaceae | Anaerotruncus | − 0.349 | 0.156 | |||
| Ruminococcaceae | Butyricicoccus | − 0.238 | 0.341 | |||
| Ruminococcaceae | Gemmiger | − 0.343 | 0.164 | |||
| Ruminococcaceae | Oscillospira | − 0.351 | 0.153 | |||
| Ruminococcaceae | Ruminococcus | − 0.037 | 0.884 | |||
| Mogibacteriaceae | unknown | − 0.223 | 0.373 | |||
| Erysipelotrichi | Erysipelotrichales | Erysipelotrichaceae | unknown | − 0.475 | 0.046* | |
| Erysipelotrichaceae | unknown | − 0.151 | 0.551 | |||
| Erysipelotrichaceae | Allobaculum | 0.795 | < .0001** | |||
| Erysipelotrichaceae | Clostridium | − 0.144 | 0.569 | |||
| Erysipelotrichaceae | Coprobacillus | − 0.250 | 0.317 | |||
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Methylobacteriaceae | Methylobacterium | 0.000 | 1.000 |
| Rickettsiales | mitochondria | unknown | 0.000 | 1.000 | ||
| Betaproteobacteria | Burkholderiales | Alcaligenaceae | Sutterella | 0.535 | 0.022* | |
| Oxalobacteraceae | Herbaspirillum | 0.000 | 1.000 | |||
| Deltaproteobacteria | Desulfovibrionales | Desulfovibrionaceae | Desulfovibrio | − 0.137 | 0.588 | |
| Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | unknown | 0.196 | 0.436 | |
| Pseudomonadales | Moraxellaceae | Acinetobacter | 0.000 | 1.000 | ||
| TM7 | TM7-3 | CW040 | F16 | unknown | − 0.154 | 0.542 |
| Tenericutes | Mollicutes | Anaeroplasmatales | Anaeroplasmataceae | Anaeroplasma | − 0.462 | 0.053 |
| RF39 | unknown | unknown | − 0.100 | 0.695 | ||
| Verrucomicrobia | Verrucomicrobiae | Verrucomicrobiales | Verrucomicrobiaceae | Akkermansia | 0.612 | 0.007** |
| unknown | unknown | unknown | unknown | unknown | − 0.246 | 0.325 |
Red; *P < 0.05, **P < 0.01.
Figure 4.
Correlation analysis between Crps and microbiota at genus level. Only pairs with significant correlations are shown in the graph. Horizontal axis shows fecal Crps concentrations after CSDS (day 32) in each mouse. All data were shown of six independent experiments in each group.
The decrease in α-defensin due to CSDS changes fecal metabolites via dysbiosis, and the changes were recovered by Crp4 administration
Next, to determine whether intestinal metabolites were changed due to dysbiosis in the CSDS model and whether α-defensin was involved in the changes, the intestinal metabolites of CSDS group and Crp4 group were analyzed. Metabolites in feces were measured simultaneously using CE-TOFMS to identify 322 candidate compounds (Table S3). In order to clarify fecal metabolites triggered by increase or decrease of α-defensin, correlation analyses between α-defensin and the intestinal metabolites were performed. Thirty- four metabolites were positively correlated and five were negatively correlated with Crps including amino-acids and vitamins (Table 2). Sixteen amino acids or their metabolites and derivatives which showed a positive correlation include glutamic acid (r = 0.49, P = 0.040), lysine (r = 0.58, P = 0.012), and 3-amino butyric acid (r = 0.47, P = 0.049), alanine (r = 0.53, P = 0.025), allo-threonine (r = 0.57, P = 0.014), citrulline (r = 0.49, P = 0.041), isoleucine (r = 0.49, P = 0.038), methionine (r = 0.51, P = 0.030), threonine (r = 0.52, P = 0.028), tyrosine (r = 0.50, P = 0.036), β-alanine (r = 0.61, P = 0.007), N- acetyl glutamic acid (r = 0.56, P = 0.017), carnitine (r = 0.57, P = 0.013), isoglutamic acid (r = 0.50, P = 0.033), N-acetyllysine (r = 0.60, P = 0.008), and N5-ethylglutamine (r = 0.57, P = 0.013). Five vitamins or their derivatives which showed a positive correlation were nicotinic acid (r = 0.47, P = 0.049), pantothenic acid (r = 0.60, P = 0.009), pyridoxamine (r = 0.66, P = 0.003), pyridoxamine 5-phosphate (r = 0.50, P = 0.035), and thiamine phosphate (r = 0.61, P = 0.007). In addition, the other 13 metabolites including uracil (r = 0.51, P = 0.032), 1H-imidazole-4-propionic acid (r = 0.61, P = 0.007), and 2-hydroxypyridine (r = 0.53, P = 0.025), 4-methyl-2-oxovaleric acid or 3-methyl-2-oxovaleric acid (r = 0.47, P = 0.047), 4-methyl-5-thiazoleethanol (r = 0.63, P = 0.005), 5-oxo-2-tetrahydrofurancarboxylic acid (r = 0.53, P = 0.023), ethanolamine (r = 0.54, P = 0.022), fumaric acid (r = 0.56, P = 0.015), hexanoic acid (r = 0.48, P = 0.045), loperamide (r = 0.52, P = 0.026), malic acid (r = 0.58, P = 0.012), orotic acid (r = 0.53, P = 0.023), and succinic acid (r = 0.50, P = 0.035) showed a positive correlation. In contrast, five metabolites including 4-guanidinobutyric acid (r = − 0.54, P = 0.021), cytidine (r = − 0.48, P = 0.044), kynurenic acid (r = − 0.55, P = 0.018), N-methylproline (r = − 0.58, P = 0.011), and sinapic acid (r = − 0.51, P = 0.032) showed negative correlation with α-defensin. These results indicated that the amount of α-defensin in the intestine influences some specific intestinal metabolites.
Table 2.
Significant correlation between fecal Crps and metabolites.
| Category | Metabolites | r | P value |
|---|---|---|---|
| Amino acids and derivatives | Glutamic acid | 0.49 | 0.040* |
| Lysine | 0.58 | 0.012* | |
| 3-Aminobutyric acid | 0.47 | 0.049* | |
| Alanine | 0.53 | 0.025* | |
| allo-Threonine | 0.57 | 0.014* | |
| Citrulline | 0.49 | 0.041* | |
| Isoleucine | 0.49 | 0.038* | |
| Methionine | 0.51 | 0.030* | |
| Threonine | 0.52 | 0.028* | |
| Tyrosine | 0.50 | 0.036* | |
| β-Alanine | 0.61 | 0.007** | |
| N-Acetylglutamic acid | 0.56 | 0.017* | |
| Carnitine | 0.57 | 0.013* | |
| Isoglutamic acid | 0.50 | 0.033* | |
| N-Acetyllysine | 0.60 | 0.008** | |
| N5-Ethylglutamine | 0.57 | 0.013* | |
| Vitamins and derivatives | Nicotinic acid | 0.47 | 0.049* |
| Pantothenic acid | 0.60 | 0.009** | |
| Pyridoxamine | 0.66 | 0.003** | |
| Pyridoxamine 5′-phosphate | 0.50 | 0.035* | |
| Thiamine phosphate | 0.61 | 0.007** | |
| Others | Uracil | 0.51 | 0.032* |
| 1H-Imidazole-4-propionic acid | 0.61 | 0.007** | |
| 2-Hydroxypyridine | 0.53 | 0.025* | |
| 4-Methyl-2-oxovaleric acid or 3-Methyl-2-oxovaleric acid | 0.47 | 0.047* | |
| 4-Methyl-5-thiazoleethanol | 0.63 | 0.005** | |
| 5-Oxo-2-tetrahydrofurancarboxylic acid | 0.53 | 0.023* | |
| Ethanolamine | 0.54 | 0.022* | |
| Fumaric acid | 0.56 | 0.015* | |
| Hexanoic acid | 0.48 | 0.045* | |
| Loperamide | 0.52 | 0.026* | |
| Malic acid | 0.58 | 0.012* | |
| Orotic acid | 0.53 | 0.023* | |
| Succinic acid | 0.50 | 0.035* | |
| 4-Guanidinobutyric acid | -0.54 | 0.021* | |
| Cytidine | -0.48 | 0.044* | |
| Kynurenic acid | -0.55 | 0.018* | |
| N-Methylproline | -0.58 | 0.011* | |
| Sinapic acid | -0.51 | 0.032* |
*P < 0.05, **P < 0.01.
We further conducted correlation analyses between the intestinal microbiota (Fig. 4) and 322 metabolites in order to know the metabolites affected by α-defensin-induced dysbiosis. Seventy-nine metabolites showed positive or negative correlation with at least one of five intestinal bacteria (Table 3). Among them, twenty-two metabolites such as pyridoxamine and β-alanine were identified as metabolites showing positive or negative correlation with α-defensin in Table 2, indicating that dysbiosis induced by α-defensin abnormalities correlates with the specific intestinal metabolites. Finally, to understand the causal relationship between α-defensin reduction and changes of the intestinal metabolites observed in the CSDS model, we analyzed differences in metabolites among each group when α-defensin was administered. Among metabolites shown in Table 2, those showed significant differences between any of the three groups were summarized, then the metabolites listed in Table 3 were shown in Fig. 5a and those not listed in Table 3 were shown in Fig. 5b.
Table 3.
Significant correlation between microbiota which correlate with Crps and metabolites.
| Metabolites | Ruminococcaceae_unknown | Erysipelotrichaceae_unknown | Erysipelotrichaceae_Allobaculum | Alcaligenaceae_Sutterella | Verrucomicrobiaceae_Akkermansia | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| r | P value | r | P value | r | P value | r | P value | r | P value value | |
| 1H-Imidazole-4-propionic acid | 0.250 | 0.317 | − 0.408 | 0.093 | 0.510 | 0.031* | 0.097 | 0.701 | 0.133 | 0.598 |
| 2-Hydroxypyridine | 0.555 | 0.017* | − 0.247 | 0.324 | 0.581 | 0.011* | 0.278 | 0.264 | 0.204 | 0.417 |
| 4-Guanidinobutyric acid | − 0.296 | 0.233 | 0.175 | 0.488 | − 0.512 | 0.030* | − 0.445 | 0.065 | − 0.373 | 0.128 |
| 5-Oxo-2-tetrahydrofurancarboxylic acid | 0.528 | 0.024* | − 0.238 | 0.342 | 0.573 | 0.013* | 0.277 | 0.267 | 0.210 | 0.404 |
| Alanine | 0.214 | 0.395 | − 0.375 | 0.125 | 0.496 | 0.036* | 0.032 | 0.900 | 0.210 | 0.404 |
| Carnitine | 0.074 | 0.772 | − 0.368 | 0.133 | 0.590 | 0.010* | − 0.024 | 0.925 | 0.254 | 0.310 |
| Citrulline | 0.407 | 0.094 | − 0.151 | 0.549 | 0.485 | 0.042* | 0.139 | 0.582 | 0.246 | 0.325 |
| Ethanolamine | 0.112 | 0.658 | − 0.219 | 0.384 | 0.134 | 0.597 | 0.165 | 0.514 | 0.550 | 0.018* |
| Fumaric acid | − 0.032 | 0.900 | − 0.374 | 0.126 | 0.469 | 0.050* | 0.330 | 0.182 | 0.326 | 0.187 |
| Isoleucine | 0.206 | 0.412 | − 0.360 | 0.142 | 0.497 | 0.036* | 0.014 | 0.955 | 0.291 | 0.242 |
| Kynurenic acid | − 0.425 | 0.079 | 0.441 | 0.067 | − 0.531 | 0.023* | − 0.271 | 0.277 | − 0.667 | 0.003** |
| Loperamide | 0.560 | 0.016* | − 0.248 | 0.322 | 0.581 | 0.012* | 0.277 | 0.265 | 0.202 | 0.422 |
| N-Acetyllysine | 0.407 | 0.094 | − 0.263 | 0.292 | 0.541 | 0.020* | 0.218 | 0.385 | 0.306 | 0.217 |
| N-Methylproline | − 0.211 | 0.401 | 0.305 | 0.218 | − 0.630 | 0.005** | − 0.330 | 0.181 | − 0.500 | 0.035* |
| Orotic acid | 0.545 | 0.019* | − 0.244 | 0.330 | 0.579 | 0.012* | 0.278 | 0.264 | 0.207 | 0.410 |
| Pyridoxamine | 0.523 | 0.026* | − 0.058 | 0.818 | 0.311 | 0.210 | 0.271 | 0.277 | 0.353 | 0.151 |
| Pyridoxamine 5′-phosphate | 0.495 | 0.037* | − 0.086 | 0.735 | 0.320 | 0.195 | 0.144 | 0.568 | 0.337 | 0.172 |
| Sinapic acid | − 0.282 | 0.257 | 0.331 | 0.180 | − 0.522 | 0.026* | − 0.304 | 0.220 | − 0.340 | 0.167 |
| Succinic acid | 0.038 | 0.882 | − 0.463 | 0.053 | 0.508 | 0.032* | 0.140 | 0.580 | 0.166 | 0.510 |
| Tyrosine | 0.221 | 0.378 | − 0.369 | 0.132 | 0.531 | 0.023* | 0.050 | 0.845 | 0.234 | 0.350 |
| β-Alanine | 0.485 | 0.041* | − 0.232 | 0.355 | 0.495 | 0.037* | 0.197 | 0.435 | 0.226 | 0.367 |
| 3-Aminobutyric acid | 0.563 | 0.015* | 0.015 | 0.952 | 0.327 | 0.185 | 0.178 | 0.481 | 0.107 | 0.674 |
| 1,3-Diaminopropane | 0.488 | 0.040* | 0.296 | 0.233 | 0.172 | 0.495 | 0.533 | 0.023* | 0.342 | 0.165 |
| 2′ or 5′-Deoxyadenosine | 0.081 | 0.750 | 0.548 | 0.019* | − 0.234 | 0.350 | 0.234 | 0.349 | 0.020 | 0.937 |
| 2′-Deoxycytidine | 0.033 | 0.897 | 0.723 | 0.001** | − 0.318 | 0.199 | 0.078 | 0.758 | − 0.255 | 0.307 |
| 2,4-Diaminobutyric acid | 0.232 | 0.354 | 0.030 | 0.906 | − 0.070 | 0.784 | 0.023 | 0.928 | 0.557 | 0.016* |
| 2,6-Diaminopimelic acid | 0.566 | 0.014* | 0.327 | 0.185 | − 0.019 | 0.939 | 0.374 | 0.126 | − 0.066 | 0.794 |
| 2-Aminoethylphosphonic acid | 0.013 | 0.961 | 0.311 | 0.209 | − 0.478 | 0.045* | − 0.130 | 0.606 | − 0.061 | 0.810 |
| 2-Deoxyribose 1-phosphate | 0.368 | 0.133 | 0.561 | 0.016* | − 0.070 | 0.782 | 0.205 | 0.414 | 0.001 | 0.998 |
| 2-Hydroxy-4-methylvaleric acid | − 0.062 | 0.808 | − 0.575 | 0.013* | 0.295 | 0.234 | − 0.202 | 0.421 | − 0.045 | 0.860 |
| 2-Hydroxyisobutyric acid | 0.118 | 0.640 | 0.062 | 0.807 | − 0.115 | 0.650 | 0.029 | 0.910 | 0.655 | 0.003** |
| 2-Hydroxyvaleric acid | − 0.065 | 0.799 | − 0.503 | 0.033* | 0.271 | 0.277 | − 0.174 | 0.491 | − 0.066 | 0.796 |
| 3′-AMP | 0.120 | 0.635 | 0.276 | 0.267 | − 0.193 | 0.443 | 0.001 | 0.996 | 0.588 | 0.010* |
| 3′ or 2′-CMP | 0.099 | 0.696 | 0.216 | 0.389 | − 0.204 | 0.417 | − 0.058 | 0.820 | 0.517 | 0.028* |
| 3-Phosphoglyceric acid | 0.118 | 0.640 | 0.062 | 0.807 | − 0.115 | 0.650 | 0.029 | 0.910 | 0.655 | 0.003** |
| 5-Hydroxyindoleacetic acid | − 0.470 | 0.049* | − 0.183 | 0.468 | − 0.179 | 0.478 | − 0.240 | 0.337 | − 0.261 | 0.295 |
| AMP | 0.068 | 0.790 | − 0.195 | 0.437 | 0.272 | 0.275 | 0.356 | 0.147 | 0.471 | 0.049* |
| Arginine | 0.131 | 0.605 | − 0.361 | 0.141 | 0.648 | 0.004** | 0.168 | 0.506 | 0.217 | 0.386 |
| Ascorbate 2-glucoside | 0.090 | 0.723 | 0.491 | 0.039* | − 0.429 | 0.076 | 0.009 | 0.972 | − 0.003 | 0.989 |
| Azelaic acid | − 0.288 | 0.247 | − 0.498 | 0.036* | − 0.061 | 0.811 | − 0.377 | 0.123 | − 0.278 | 0.264 |
| CMP | 0.043 | 0.866 | − 0.208 | 0.407 | 0.303 | 0.222 | 0.379 | 0.121 | 0.474 | 0.047* |
| dAMP | 0.153 | 0.544 | − 0.002 | 0.995 | 0.213 | 0.397 | 0.256 | 0.306 | 0.515 | 0.029* |
| Diphenylcarbazide | 0.532 | 0.023* | 0.429 | 0.076 | − 0.128 | 0.614 | 0.400 | 0.100 | 0.056 | 0.827 |
| Ethyl glucuronide | 0.201 | 0.423 | 0.568 | 0.014* | − 0.517 | 0.028* | − 0.091 | 0.719 | − 0.073 | 0.774 |
| Fructose 6-phosphate | 0.223 | 0.374 | − 0.176 | 0.485 | 0.318 | 0.199 | 0.054 | 0.833 | 0.470 | 0.049* |
| Glutamine | 0.242 | 0.334 | − 0.287 | 0.248 | 0.613 | 0.007** | 0.246 | 0.325 | 0.388 | 0.112 |
| Glucose 6-phosphate | 0.215 | 0.392 | − 0.029 | 0.911 | 0.186 | 0.461 | 0.108 | 0.670 | 0.616 | 0.007** |
| Gly-Asp | − 0.179 | 0.478 | − 0.535 | 0.022* | 0.492 | 0.038* | 0.191 | 0.449 | 0.096 | 0.706 |
| Gly-Gly | − 0.161 | 0.525 | − 0.397 | 0.103 | 0.482 | 0.043* | 0.193 | 0.444 | 0.083 | 0.744 |
| Gly-Leu | 0.095 | 0.709 | − 0.364 | 0.138 | 0.472 | 0.048* | 0.068 | 0.788 | 0.099 | 0.696 |
| Glyceric acid | − 0.519 | 0.027* | − 0.237 | 0.344 | − 0.397 | 0.103 | − 0.417 | 0.085 | − 0.155 | 0.540 |
| Homovanillic acid | − 0.032 | 0.901 | − 0.595 | 0.009** | 0.313 | 0.206 | − 0.222 | 0.377 | 0.011 | 0.965 |
| Hydroxyproline | − 0.146 | 0.564 | − 0.629 | 0.005** | 0.231 | 0.357 | − 0.088 | 0.728 | 0.042 | 0.869 |
| IsovalerylalanineN-Acetylleucine | − 0.121 | 0.632 | − 0.655 | 0.003** | 0.394 | 0.106 | − 0.095 | 0.708 | 0.152 | 0.547 |
| Lactic acid | − 0.282 | 0.256 | − 0.611 | 0.007** | 0.150 | 0.553 | − 0.271 | 0.277 | − 0.061 | 0.810 |
| N,N-Dimethylhistidine | 0.366 | 0.135 | − 0.366 | 0.136 | 0.480 | 0.044* | 0.364 | 0.137 | 0.549 | 0.018* |
| N-Acetylasparagine | − 0.509 | 0.031* | 0.048 | 0.849 | − 0.457 | 0.056 | − 0.301 | 0.225 | − 0.362 | 0.141 |
| N-Acetylglucosamine 6-phosphate | 0.474 | 0.047* | 0.877 | < .0001** | − 0.389 | 0.111 | 0.233 | 0.352 | − 0.182 | 0.469 |
| N-Acetylglucosylamine | 0.028 | 0.912 | 0.572 | 0.013* | − 0.138 | 0.585 | 0.042 | 0.869 | − 0.214 | 0.394 |
| N-Acetylmuramic acid | 0.668 | 0.002** | 0.072 | 0.778 | 0.157 | 0.534 | 0.293 | 0.238 | 0.255 | 0.307 |
| p-Hydroxymandelic acid | − 0.411 | 0.090 | − 0.181 | 0.473 | − 0.428 | 0.077 | − 0.540 | 0.021* | − 0.152 | 0.547 |
| Phenylalanine | 0.138 | 0.586 | − 0.407 | 0.094 | 0.530 | 0.024* | 0.028 | 0.913 | 0.207 | 0.410 |
| Picolinic acid | − 0.404 | 0.096 | 0.032 | 0.899 | − 0.479 | 0.044* | − 0.502 | 0.034* | − 0.328 | 0.183 |
| Pimelic acid | − 0.082 | 0.748 | − 0.486 | 0.041* | 0.082 | 0.746 | − 0.247 | 0.324 | − 0.063 | 0.804 |
| Proline | − 0.372 | 0.129 | − 0.525 | 0.025* | 0.297 | 0.232 | − 0.172 | 0.495 | 0.103 | 0.684 |
| Putrescine | − 0.062 | 0.808 | − 0.187 | 0.457 | 0.136 | 0.591 | 0.483 | 0.042* | 0.244 | 0.330 |
| Saccharopine | 0.427 | 0.077 | 0.268 | 0.282 | − 0.017 | 0.946 | 0.255 | 0.308 | 0.473 | 0.047* |
| Sebacic acid | − 0.233 | 0.353 | − 0.472 | 0.048* | − 0.037 | 0.884 | − 0.389 | 0.111 | − 0.049 | 0.846 |
| Sedoheptulose 7-phosphate | 0.237 | 0.345 | − 0.056 | 0.827 | 0.194 | 0.440 | 0.089 | 0.726 | 0.509 | 0.031* |
| Syringic acid | − 0.566 | 0.014* | − 0.294 | 0.237 | − 0.075 | 0.767 | − 0.618 | 0.006** | − 0.485 | 0.041* |
| Thiamine diphosphate | 0.492 | 0.038* | 0.009 | 0.971 | 0.217 | 0.387 | 0.150 | 0.554 | 0.375 | 0.125 |
| Thymidine | 0.372 | 0.128 | 0.649 | 0.004** | − 0.024 | 0.924 | 0.267 | 0.284 | − 0.074 | 0.771 |
| Tryptophan | − 0.110 | 0.665 | − 0.342 | 0.165 | 0.475 | 0.046* | 0.031 | 0.904 | 0.071 | 0.781 |
| Tryptamine | 0.194 | 0.440 | 0.536 | 0.022* | − 0.310 | 0.210 | 0.048 | 0.851 | − 0.269 | 0.280 |
| UMP | 0.214 | 0.394 | − 0.078 | 0.758 | 0.298 | 0.230 | 0.353 | 0.150 | 0.777 | < .0001** |
| Undecanoic acid | 0.476 | 0.046* | 0.031 | 0.903 | 0.115 | 0.650 | 0.107 | 0.672 | 0.229 | 0.360 |
| Urocanic acid | 0.551 | 0.018* | 0.093 | 0.714 | 0.294 | 0.237 | 0.125 | 0.621 | 0.076 | 0.764 |
| Valine | 0.082 | 0.747 | − 0.395 | 0.105 | 0.471 | 0.048* | − 0.042 | 0.869 | 0.175 | 0.489 |
| g-Glu-Val-Gly | 0.070 | 0.782 | 0.736 | 0.001** | − 0.531 | 0.024* | 0.038 | 0.881 | − 0.004 | 0.989 |
Blue: metabolites correlated with Crps, red: *P < 0.05, **P < 0.01.
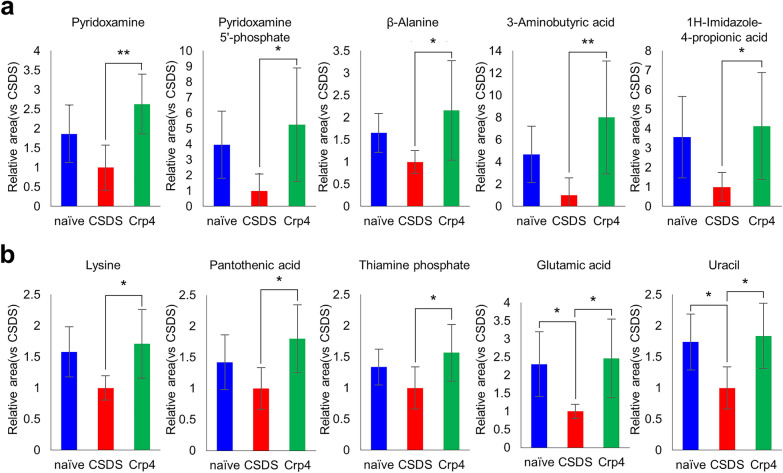
Figure 5.
Changes in fecal metabolites correlated with Crps in each group. (a) Metabolites significantly correlated with Crps and microbiota (blue metabolites in Table 3). (b) Metabolites significantly correlated with Crps (Metabolites in Table 2 other than those shown in (a)). All data were shown as mean ± SE of six independent experiments in each group. Tukey's tests were used to compare the data. *P < 0.05; **P < 0.01.
The metabolites significantly increased in Crp4 group relative to CSDS group were pyridoxamine (P = 0.002), pyridoxamine-5 phosphate (P = 0.027), β-alanine (P = 0.032), 3-aminobutyric acid, (P = 0.008), 1H-imidazole propionic acid (P = 0.044), pantothenic acid (P = 0.019), and thiamine-phosphoric acid (P = 0.044) (Fig. 5a,b). Lysine tended to decrease in CSDS group (P = 0.066), while significantly increased in Crp4 group (P = 0.023). Glutamic acid and uracil were significantly decreased in CSDS group (P = 0.038, P = 0.029), while significantly increased in Crp4 group (P = 0.019, P = 0.014) (Fig. 5b). On the other hand, no significant difference was observed between naïve group and Crp4 group for all those metabolites, indicating that the intestinal metabolites correlated with α-defensin are recovered to the same levels in naïve group by α-defensin administration. In addition, we found metabolites with significant differences in both naïve group and Crp4 group compared to CSDS group, although no correlation was observed with α-defensin (Fig. S1). Among these, three metabolites including N6-acetyllysine, penicillamine, threo-β-methylaspartic acid were significantly decreased by CSDS, and the decrease was significantly attenuated by α-defensin administration. Conversely, seven metabolites including cadaverine, glucaric acid, ferulic acid, mevalonic acid, digalacturonic acid, myo-inositol 2-phosphate, and p-aminophenol or m-aminophenol were significantly increased in CSDS group, and the increase was suppressed by α-defensin administration.
Discussion
In this study, psychological stress reduces α-defensin secreted from Paneth cells, which induces disruption of homeostasis in the intestinal metabolites via imbalance of the intestinal microbiota, dysbiosis in CSDS model. Furthermore, the oral administration of α-defensin rescues dysbiosis and recovers homeostasis in the metabolites, suggesting that psychological stress-induced dysbiosis is associated with Paneth cell dysfunction.
We clarified that the amount of α-defensin secreted into the intestinal lumen decreases due to CSDS loading (Fig. 1f). Since the number of Paneth cells and Crp1+ granule area in the small intestine were decreased in CSDS group (Fig. 2a–c), the decrease in fecal Crp1 may be due to Paneth cell dysfunction. It has been known that psychological stress by mother-infant separation at birth has reduced rat Paneth cells in the small intestine via activation of corticotropin-releasing factor (CRF), and the number of Paneth cells has remained decreased even after weaning20. CRF activation has been also reported in CSDS model21. Our results that α-defensin decreased due to CSDS loading and the decrease lasted for 20 days after the end of CSDS loading are consistent with these previous findings, and importantly, abnormalities of α-defensin, the effector of innate enteric immunity, secreted by Paneth cells were first verified.
α-Defensins regulate the intestinal microbiota composition, and α-defensin deficiency diminishes diversity and affects the composition of the intestinal microbiota such as Firmicutes and Bacteroidetes, resulting in dysbiosis14, 22, 23. Previously, decrease in Firmicutes and increase in Bacteroidetes have been reported in patients with depression24. We revealed that decrease in α-defensin due to CSDS induces decrease of Firmicutes and increase of Bacteroidetes in the intestinal microbiota and further indicated a positive correlation between the amount of α-defensin and the intestinal microbiota including Akkermansia. It has been reported that Akkermansia increases by administration of α-defensin25, 26, and strongly correlates with stress tolerance in CSDS model27, suggesting α-defensin regulates the intestinal microbiota in depression. Although the status of dysbiosis varies depending on certain pathophysiology and models, because β-diversity approached to naïve group by administration of α-defensin, α-defensin may have a function to maintain homeostasis of the intestinal microbiota in response to the changes of the intestinal ecosystem.
The dysbiosis in depression has been known to be diverse between studies due to individual differences in diet, region, race, etc28. On the other hand, metabolic processes are relatively conserved in comparison of high variation of the intestinal microbiota between individuals29, indicating that individuals may have taxonomically different but functionally similar microbiota. Therefore, it is important to analyze intestinal microbial metabolites in this study. Five metabolites including pyridoxamine, pyridoxamine 5′-phosphate, β-alanine, 3-aminobutyric acid, and 1H-imidazole-4-propionic acid are significantly decreased with α-defensin reduction due to CSDS and fully recovered by α-defensin administration. Pyridoxamine, one form of vitamin B6, is important for synthesis of many neurotransmitters such as GABA, serotonin, dopamine, noradrenaline, histamine, glycine, and d-serine, and currently used as therapeutics for autism30. It also has been reported that vitamin B6 administration attenuates depression-like behaviors in dexamethasone-induced depression model mice31. In addition, β-alanine administration has been reported to improve behaviors of depression in post-traumatic stress disorder (PTSD) model mice by increasing carnosine levels in the brain and maintaining hippocampal BDNF expression, an important target of antidepressants32, 33. Our result that those metabolites were recovered to normal by α-defensin administration suggests that α-defensin affects brain function through microbial metabolites by maintaining homeostasis in the intestinal microbiota.
Other metabolites that decreased with α-defensin reduction due to CSDS and recovered to the same levels as naïve group by α-defensin administration include lysine, pantothenic acid, thiamine phosphate, glutamic acid, and uracil. Although these metabolites correlated with α-defensin, no direct correlation with the intestinal microbiota at genus level was observed. We think the reason why no metabolite correlates with microbiota may be because taxonomically similar bacteria often involve in the same function29, and further the metabolism of the intestinal microbiota can be affected by crosstalk between bacteria in addition to phenotypic changes of the bacteria themselves34, 35. In addition, long-term lysine deficiency has been reported to increase anxiety and psychological stress, and lysine-enriched diet interventions improve chronic anxiety36. Pantothenic acid has been known to increase adrenal sensitivities to adrenocorticotropic hormone (ACTH) and stimulate cortisol secretion37. It has been reported that people with higher erythrocyte concentration of thiamine-phosphate have lower symptoms of depression, considering thiamine-phosphate as a potential biomarker for depression38. Glutamic acid has been known as a major excitatory neurotransmitter that regulates higher functions such as memory and learning in the central nervous system of mammals39 and serves as an important material for GABA synthesis in the brain. Furthermore, it has been reported that SNPs localized in uracil-processing genes potentially regulate the onset and development of depression40. Taken together, the decrease of multiple intestinal metabolites which are reported to function in resolving or defending against depression, anxiety, and psychological stress links to the decrease of α-defensin, and these metabolites are recovered to the normal levels by administration of α-defensin in this study, providing the important new findings to improve understanding of depression in relation to the gut-brain axis. There are several metabolites that changed along with α-defensin reduction without correlation of the amount of α-defensin (Fig. S1). Since it has been well known that psychological stress affects many biological events in the host, it remains controversial whether biological disruptions are causes or effects. Especially, psychological stress affects the gastrointestinal function as represented in irritable bowel syndrome41, and the metabolism of the intestinal microbiota and the host closely interacts each other to create a complex metabolic system34. Thus, it is speculated that the change of these metabolites was a secondary effect of improving dysbiosis by α-defensin administration to affect the host intestinal function, and because responses to psychological stress largely vary depending on host, i.e., animal model42, direct correlation with α-defensin could not be observed. However, detailed underlying mechanisms of these findings remain unknown and future study is needed to further understand the gut-brain axis in depression.
In this study, we used Crp4 for administration to CSDS group because Crp4 is known to have the most potent bactericidal activities among Crps13 and the amount of administered Crp4 can be monitored because C57BL/6 mice do not express Crp443. It has been known that Crp1 family consisted of Crp1-3 and 6, is the most abundant α-defensins in mice44–47. Surprisingly, Crp4 administration increased the amount of Crp1 in the CSDS mice (Fig. 3b). Given the results that Crp4 administration rescued the dysbiosis and the impaired intestinal metabolites due to CSDS loading, it is suggested that Crp4 administration improved host microenvironment for Paneth cells and resulted in the increase of Crp1 secretion. This speculation is supported by evidence on the intestinal ecosystem, including that the intestinal commensal microbiota positively affects Paneth cell development and function, i.e., Crp secretions by comparing the germ-free and the conventional mice12, 48, 49.
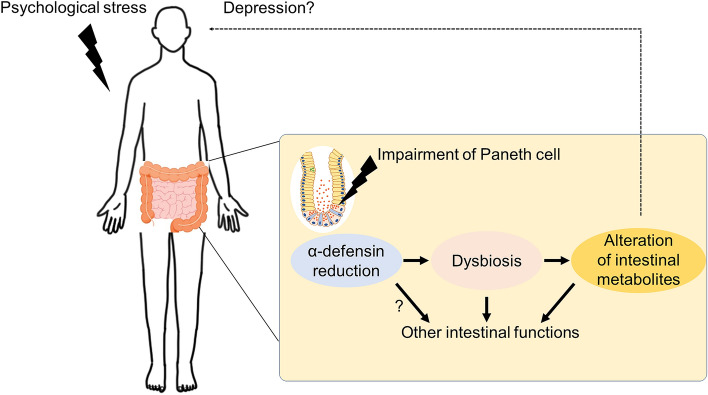
This study provides a novel insight that might likely be relevant to the pathogenesis and pathophysiology of depression. Based on the results in this study, we propose a mechanism leading to depression (Fig. 6). Psychological stress immediately reduces α-defensin secretion from Paneth cells in the small intestine at early stage, resulting in dysbiosis and further disrupting homeostasis of intestinal metabolites. In the gut-brain axis, the disrupted intestinal ecosystem may affect brain through some unclarified pathways to develop or worsen depression. Dysbiosis reported in patients with depression and depression model animals largely varies probably due to individual differences relating such as diet and race28. This study clarified a previously unknown important link between metabolic profiles of the intestinal microbiota and upstream host-derived regulator, α-defensin, and further contribute to understanding mechanisms of depression. Although long-term observation is required to determine systemic effects including behavior in future studies to understand whole picture of the gut-brain axis in depression, the new relationship between depression and α-defensin shown in this study may contribute to development for prevention and therapeutics of depression.
Figure 6.
Hypothesis of a novel mechanism based on α-defensin in the CSDS model. Psychological stress reduces α-defensin produced by Paneth cells in the gut and disrupts the homeostasis of intestinal metabolite composition via dysbiosis. These are normalized by α-defensin administration. Thus, disruption of metabolite homeostasis may affect brain function and lead to the development or exacerbation of depression. However, the direct effects on the brain have not been demonstrated, and are shown as dotted lines.
Methods
Animal
C57BL/6 J (B6J) mice (male) and ICR mice (male) and retirement ICR (Stressor) mice (male, 5 months old and older) were purchased from Charles River Japan (Yokohama, Japan), and B6J mice and ICR mice were subjected at 7 weeks of age to experiments after acclimation and quarantine for more than one week. The bedding was PaperClean (Japan SLC Inc., Hamamatsu, Japan) and B6J mice had received ad libitum drinking water and a diet (CE-2, CLEA Japan, Tokyo, Japan). All animal experiments were approved by the Institutional Animal Care and Use Committee of the National University Corporation at Hokkaido University. All experiments were performed in accordance with Hokkaido University Regulations of Animal Experimentation. All animal experiments were also carried out in compliance with the ARRIVE guidelines.
The experimental design of social defeat stress
CSDS was partially modified from previous reports8. One B6J mouse and one Stressor mouse were placed in one side of a cage divided into two compartments by a clear acrylic plate. Mice were brought into direct contact with each other. The direct contact time was counted from the time the stressor mice made contact, such as biting or covering, and the direct contact time was reduced by 5 min on the first day and by 0.5 min thereafter. In other words, the direct contact time for the 10th time was 0.5 min. Direct contact was performed at PM (13:00–17:00). After direct contact, B6J mice were moved to a neighboring compartment with stressor mice and subjected to visual and olfactory stress until the next direct contact (indirect contact). In the first experiment, five cycles of direct and indirect contact were performed, followed by two days of indirect contact, followed by the remaining five cycles each (Fig. 1a). In the second experiment, 10 cycles of direct and indirect contact were performed (Fig. 3a). Retire ICR mice were selected in order of aggression from 15 aggression tests measuring the number of 3-min bites to B6J mice performed three times a day for five days in advance.
Behavior test
A cage (16.5 cm × 8.3 cm) was placed in a behavioral test box (50 cm × 50 cm), and the perimeter of the cage in the box was defined as the Social Interaction Zone (SI zone, cage perimeter 33.3 cm × 16.7 cm) and as both diagonal corners of the cage (Corner zone 16.6 cm × 16.6 cm). B6J mice were placed in a box with ICR mice present in the cage and allowed to explore freely for 5 min. Behavioral analysis was performed using the image analysis software HOLE BOARD (Muromachi KIKAI, Tokyo, Japan) to calculate the interaction time, corner time and total distance traveled.
Sampling feces
Fresh feces were collected from B6J mice in cages equipped with wire mesh in the floor and stored frozen at − 80℃. Feces during 24 h were collected after cage replacement. We sampled at three time points on day 1, 9, and 14 in the first experiment (Fig. 1a) and at two time points on day 0 and 32 in the second experiment (Fig. 3a).
Immunofluorescent analysis
Mice were euthanized by isoflurane inhalation and the small intestine was harvested at day 14 (Fig. 1a). 10% buffered formalin-fixed the small intestine from naïve and CSDS mice were embedded in paraffin and cut into 4 μm-thick sections. The sections were deparaffinized, rehydrated, and boiled in Dako REAL Target Retrieval Solution (pH 6, Agilent, Santa Clara, CA) for 20 min at 105 °C. After blocking in Block Ace (Dainippon Pharmaceutical, Osaka, Japan) containing 5% goat serum (Sigma-Aldrich, St. Louis, MO) for 30 min at room temperature, the sections were incubated with 1 μg/mL rat monoclonal anti-Crp1 (77-R63) for overnight at 4 °C. Then, the sections were incubated with 5 µg/mL Alexa Fluor 488 goat anti-rat IgG H + L (Thermo Fisher Scientific, Waltham, MA) for 1 h at room temperature. After nucleus staining by 4′, 6-diamidino-2-phenylindole (DAPI, Thermo Fisher Scientific, Waltham, MA) for 5 min, the sections were embedded to the slide grass using RapiClear 1.52 (Sunjin Lab, Hsinchu, Taiwan). Fluorescent images were observed using confocal microscopy (A1, Nikon, Tokyo, Japan). Paneth cell number per crypt and Crp1 positive area of each crypt in binary images based on fluorescent intensity were measured in 5 crypts from the small intestine by using NIS-Elements AR ver.5.11 (Nikon, Tokyo, Japan).
α-Defensin administration
Recombinant Cryptdin-4 (mouse α-defensin) was manufactured and purified according to previous reports50. Samples were dissolved in ultrapure water and administered orally at 250 μg/mouse once daily from day 1 to day 32, and then equal amounts of ultrapure water were administered orally to the naïve group and the CSDS group (Fig. 3a).
ELISA
Fecal α-defensin was measured as previously described23, 51. In brief, samples were air-dried, powdered using a bead beater-type homogenizer(Beads Crusher μT-12;TAITEC) . Fecal extract was collected after blending with PBS using a vortex mixer for 1 h and centrifugation at 20,000 g for 20 min, and levels of Crp1 and Crp4 were measured by sandwich ELISA. ELISA was performed on the feces of C57BL/6 J mice and the Crp1 antibodies detected Crp1-3 and 6, the Crp1 family. The Crp4 antibodies detected administered Crp4 since C57BL/6 J mice do not express Crp4 genetically.
DNA extraction and 16S rRNA sequencing
Genomic DNA was extracted from 100 mg fecal samples using the NucleoSpin Microbial DNA Kit (MACHEREY–NAGEL, Düren, Germany) following the manufacturer’s protocol. Final DNA concentrations were determined at 260 nm using a NanoDrop 2000 spectrometer (Thermo Fischer Scientific). 16S ribosomal RNA genes were amplified by PCR from each fecal DNA sample using universal primer set of Bakt 341F (5′-cctacgggnggcwgcag) and Bakt 805R (5′-gactachvgggtatctaatcc) which covers the V3-V4 variable region16, 52, 53. PCR amplification was performed in 25-μl-volume reaction mixtures containing 12.5 ng of template DNA, 200 nM of each primer, and 1 × KAPA HiFi Hot Start Ready Mix (Kapa Biosystems) under the following conditions: 95 °C for 3 min, 25 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, followed by 72 °C for 5 min. PCR products were purified with AMPure XP beads (Beckman Coulter). After purification, sequencing adapters containing sample-specific 8-bp barcodes were added to the 3′- and 5′- ends by PCR using the Nextera XT Index Kit v2 Set B (Illumina) in 50 μl of reaction mixtures containing 5 μl of PCR amplicon, 5 μl of each indexing primer and 1 × KAPA HiFi Hot Start Ready Mix under the following conditions: 95 °C for 3 min, eight cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, followed by 72 °C for 5 min. Each amplicon was purified, quantified using the Qubit dsDNA HS Assay Kit (Invitrogen), and then adjusted to 4 nM. Amplicons were pooled 4 μl and subjected to quantification using KAPA Library Quantification Kit LightCycler 480 qPCR Mix (Kapa Biosystems) and then diluted to 4 pM. The amplicon library was combined with 5% equimolar PhiX Control v3 (Illumina) and sequenced on a MiSeq instrument using the MiSeq 600-cycle v3 kit (Illumina).
16S rRNA–based taxonomic analysis
Microbiome analysis was performed using the open-source bioinformatics pipeline, QIIME2(version 2019.7). Sequences were quality filtered and denoising into features by DADA2 plugin and remaining contigs were clustered into OTUs with 99% sequence similarity against the SILVA 128 reference database. To acquire taxonomic information for each OTU, representative sequences were aligned to MAFFT and assigned to its database for classification by a naïve-bayes classifier trained on 16S rRNA gene OTUs. β-diversity (unweighted UniFrac distance) was estimated by using QIIME2 workflow. Statistical significance of β-diversity was determined by PERMANOVA test in Qiime2 pipeline.
Measurement of fecal metabolites
Approximately 50 mg of frozen feces were plunged into 1,500 µL of 50% acetonitrile/Milli-Q water containing internal standards (H3304-1002, Human Metabolome Technologies, Inc., Tsuruoka, Japan) at 0 °C in order to inactivate enzymes. The tissue was homogenized thrice at 1,500 rpm for 120 s using a tissue homogenizer (Micro Smash MS100R, Tomy Digital Biology Co., Ltd., Tokyo, Japan) and then the homogenate was centrifuged at 2,300 × g and 4 °C for 5 min. Subsequently, 800 µL of upper aqueous layer was centrifugally filtered through a Millipore 5-kDa cutoff filter at 9,100 × g and 4 °C for 120 min to remove proteins. The filtrate was centrifugally concentrated and re-suspended in 50 µL of Milli-Q water for CE-MS analysis. Metabolome measurements were carried out through a facility service at Human Metabolome Technologies Inc., Tsuruoka, Japan. CE-TOFMS was carried out using an Agilent CE Capillary Electrophoresis System equipped with an Agilent 6210 Time of Flight mass spectrometer, Agilent 1100 isocratic HPLC pump, Agilent G1603A CE-MS adapter kit, and Agilent G1607A CE-ESI–MS sprayer kit (Agilent Technologies, Waldbronn, Germany). The systems were controlled by Agilent G2201AA ChemStation software version B.03.01 for CE (Agilent Technologies, Waldbronn, Germany). The metabolites were analyzed by using a fused silica capillary (50 μm i.d. × 80 cm total length), with commercial electrophoresis buffer (Solution ID: H3301-1001 for cation analysis and H3302-1021 for anion analysis, Human Metabolome Technologies) as the electrolyte. The sample was injected at a pressure of 50 mbar for 10 s (approximately 10 nL) in cation analysis and 25 s (approximately 25 nL) in anion analysis. The spectrometer was scanned from m/z 50 to 1000. Other conditions were as in the described previously54. Peaks were extracted using automatic integration software MasterHands (Keio University, Tsuruoka, Japan) in order to obtain peak information including m/z, migration time for CE-TOFMS measurement (MT) and peak area55. Signal peaks corresponding to adduct ions, and other product ions of known metabolites were excluded, and remaining peaks were annotated with putative metabolites and their isotopic ions from the HMT metabolite database based on their MTs and m/z values determined by TOFMS. The tolerance range for the peak annotation was configured at ± 0.5 min for MT and ± 30 ppm for m/z. In addition, peak areas were normalized against those of the internal standards and then the resultant relative area values were further normalized by sample amount.
Statistical analysis
Statistical analysis was performed using JMP (version 14.0.0) software. Two groups were analyzed by the Mann–Whitney U test and paired t-test and three groups were analyzed by the Steel's test and Turkey's test. Correlation analysis was performed using the Pearson correlation coefficient. For all analyses, differences between groups were considered significant if P-values were < 0.05.
Supplementary Information
Acknowledgements
We thank Ms. Aiko Kuroishi for experimental support. This study was supported by grants from the Japan Society for the Promotion of Science KAKENHI Grant Number 17K11661, 20H04098 (to K.N.) and 18H02788 (to T.A. *), and the Center of Innovation Program from Japan Science and Technology Agency, JST, Grant Number JPMJCE 1301 (to K.N., T.A., and T.A.*).
Author contributions
K.S. developed the conceptual framework of the study, designed the experiments, conducted studies, data analysis and interpretation, and wrote the paper. K.N. designed the experiments, conducted studies, data analysis and interpretation, and reviewed and edited the paper. Y.S. and Y.Y. conducted studies, data analysis and interpretation. S.O. and M.H. conducted data analysis and interpretation. Y.W. and Y.S. manufactured cryptdin-4 peptide. T.A. manufactured cryptdin-4 peptide, conducted data analysis and interpretation. T.A.* developed the conceptual framework of the study, designed the experiments, data analysis and interpretation, and reviewed and edited the paper.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-89308-y.
References
- 1.Smith K. Mental health: a world of depression. Nature. 2014;515:180–181. doi: 10.1038/515180a. [DOI] [PubMed] [Google Scholar]
- 2.Malhi GS, Mann JJ. Depression. Lancet. 2018;392(2299):2312. doi: 10.1016/S0140-6736(18)31948-2. [DOI] [PubMed] [Google Scholar]
- 3.Kendler KS, Karkowski LM, Prescott CA. Causal relationship between stressful life events and the onset of major depression. Am. J. Psychiatr. 1999;156:837–841. doi: 10.1176/ajp.156.6.837. [DOI] [PubMed] [Google Scholar]
- 4.Rieder R, Wisniewski PJ, Alderman BL, Campbell SC. Microbes and mental health: a review. Brain Behav. Immun. 2017;66:9–17. doi: 10.1016/j.bbi.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 5.Colomer MV, et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 2019;4:623–632. doi: 10.1038/s41564-018-0337-x. [DOI] [PubMed] [Google Scholar]
- 6.Kelly JR, et al. Transferring the blues: depression-associated gut microbiota induces neurobehavioural changes in the rat. J. Psychiatr. Res. 2016;82:109–118. doi: 10.1016/j.jpsychires.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 7.Sudo N, et al. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J. Physiol. 2004;558:263–275. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ayako AY, Omics J, et al. Studies of the murine intestinal ecosystem exposed to subchronic and mild social defeat stress. J. Proteome Res. 2016;15:3126–3138. doi: 10.1021/acs.jproteome.6b00262. [DOI] [PubMed] [Google Scholar]
- 9.Ayabe T, et al. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat. Immunol. 2000;1:113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- 10.Yokoi Y, et al. Paneth cell granule dynamics on secretory responses to bacterial stimuli in enteroids. Sci. Rep. 2019;9:2710. doi: 10.1038/s41598-019-39610-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522–526. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura K, et al. Expression and localization of Paneth cells and their a-defensins in the small intestine of adult mouse. Front. Immunol. 2020;11:570296. doi: 10.3389/fimmu.2020.570296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masuda K, Sakai K, Nakamura K, Yoshioka S, Ayabe T. Bactericidal activity of mouse α-defensin cryptdin-4 predominantly affects noncommensal bacteria. J. Innate Immun. 2011;3:315–326. doi: 10.1159/000322037. [DOI] [PubMed] [Google Scholar]
- 14.Salzman NH, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayase E, et al. R-Spondin1 expands Paneth cells and prevents dysbiosis induced by graft-versus-host disease. J. Exp. Med. 2017;214:3507–3518. doi: 10.1084/jem.20170418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimizu Y, et al. Paneth cell α-defensin misfolding correlates with dysbiosis and ileitis in Crohn's disease model mice. Life Sci. Alliance. 2020;3:e201900592. doi: 10.26508/lsa.201900592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komatsu Y, et al. Disease progression-associated alterations in fecal metabolites in SAMP1/YitFc mice, a Crohn's disease model. Metabolomics. 2020;16:48. doi: 10.1007/s11306-020-01671-5. [DOI] [PubMed] [Google Scholar]
- 18.Salzman NH, Bevins CL. Dysbiosis–a consequence of Paneth cell dysfunction. Semin. Immunol. 2013;25:334–341. doi: 10.1016/j.smim.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura K, Sakuragi N, Takakuwa A, Ayabe T. Paneth cell α-defensins and enteric microbiota in health and disease. Biosci. Microbiota Food Health. 2016;35:57–67. doi: 10.12938/bmfh.2015-019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estienne M, et al. Maternal deprivation alters epithelial secretory cell lineages in rat duodenum: role of CRF-related peptides. Gut. 2010;59:744–751. doi: 10.1136/gut.2009.190728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Q, et al. Different effects of chronic social defeat on social behavior and the brain CRF system in adult male C57 mice with different susceptibilities. Behav. Brain Res. 2020;20(384):112553. doi: 10.1016/j.bbr.2020.112553. [DOI] [PubMed] [Google Scholar]
- 22.Eriguchi Y, et al. Graft-versus-host disease disrupts intestinal microbial ecology by inhibiting Paneth cell production of α-defensins. Blood. 2012;120:223–231. doi: 10.1182/blood-2011-12-401166. [DOI] [PubMed] [Google Scholar]
- 23.Eriguchi Y, et al. Decreased secretion of Paneth cell α-defensins in graft-versus-host disease. Transpl. Infect. Dis. 2015;17:702–706. doi: 10.1111/tid.12423. [DOI] [PubMed] [Google Scholar]
- 24.Jiang H, et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun. 2015;48:186–194. doi: 10.1016/j.bbi.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 25.Ehmann D, et al. Paneth cell α-defensins HD-5 and HD-6 display differential degradation into active antimicrobial fragments. Proc. Natl. Acad. Sci. U S A. 2019;116:3746–3751. doi: 10.1073/pnas.1817376116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong W, et al. Paneth cell dysfunction mediates alcohol-related steatohepatitis through promoting bacterial translocation in mice: role of zinc deficiency. Hepatology. 2020;71:1575–1591. doi: 10.1002/hep.30945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGaughey KD, et al. Relative abundance of Akkermansia spp. and other bacterial phylotypes correlates with anxiety- and depressive-like behavior following social defeat in mice. Sci. Rep. 2019;9:3281. doi: 10.1038/s41598-019-40140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheung SG, et al. Systematic review of gut microbiota and major depression. Front. Psychiatry. 2019;10:34. doi: 10.3389/fpsyt.2019.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huttenhower C, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato K. Why is vitamin B6 effective in alleviating the symptoms of autism? Med. Hypotheses. 2018;115:103–106. doi: 10.1016/j.mehy.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 31.Mesripour A, Alhimma F, Hajhashemi V. The effect of vitamin B6 on dexamethasone-induced depression in mice model of despair. Nutr. Neurosci. 2019;22:744–749. doi: 10.1080/1028415X.2018.1442184. [DOI] [PubMed] [Google Scholar]
- 32.Hoffman JR, et al. β-Alanine supplemented diets enhance behavioral resilience to stress exposure in an animal model of PTSD. Amino Acids. 2015;47:1247–1257. doi: 10.1007/s00726-015-1952-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Björkholm C, Monteggia LM. BDNF-a key transducer of antidepressant effects. Neuropharmacology. 2016;102:72–79. doi: 10.1016/j.neuropharm.2015.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ursell LK, et al. The intestinal metabolome: an intersection between microbiota and host. Gastroenterology. 2014;146:1470–1476. doi: 10.1053/j.gastro.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung HH, et al. Evidence of link between quorum sensing and sugar metabolism in Escherichia coli revealed via cocrystal structures of LsrK and HPr. Sci. Adv. 2018;4:7063. doi: 10.1126/sciadv.aar7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smriga M, Ghosh S, Mouneimne Y, Pellett PL, Scrimshaw NS. Lysine fortification reduces anxiety and lessens stress in family members in economically weak communities in Northwest Syria. Proc. Natl. Acad. Sci. U S A. 2004;101:8285–8288. doi: 10.1073/pnas.0402550101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaroenporn S, et al. Effects of pantothenic acid supplementation on adrenal steroid secretion from male rats. Biol. Pharm. Bull. 2008;31:1205–1208. doi: 10.1248/bpb.31.1205. [DOI] [PubMed] [Google Scholar]
- 38.Zhang G, et al. Thiamine nutritional status and depressive symptoms are inversely associated among older chinese adults. J. Nutr. 2013;143:53–58. doi: 10.3945/jn.112.167007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakanishi S, et al. Glutamate receptors: brain function and signal transduction. Brain Res. Brain Res. Rev. 1998;26:230–235. doi: 10.1016/S0165-0173(97)00033-7. [DOI] [PubMed] [Google Scholar]
- 40.Czarny P, et al. Single-nucleotide polymorphisms of uracil-processing genes affect the occurrence and the onset of recurrent depressive disorder. Peer J. 2018;6:e5116. doi: 10.7717/peerj.5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Enck P, et al. Irritable bowel syndrome. Nat. Rev. Dis. Primers. 2016;2:16014. doi: 10.1038/nrdp.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nasca C, et al. Multidimensional predictors of susceptibility and resilience to social defeat stress. Biol. Psychiatr. 2019;86:483–491. doi: 10.1016/j.biopsych.2019.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shanahan MT, Tanabe H, Ouellette AJ. Strain-specific polymorphisms in Paneth cell α-defensins of C57BL/6 mice and evidence of vestigial myeloid α-defensin pseudogenes. Infect. Immun. 2011;79:459–473. doi: 10.1128/IAI.00996-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ouellette AJ, et al. Developmental regulation of cryptdin, a corticostatin/defensin precursor mRNA in mouse small intestinal crypt epithelium. J. Cell Biol. 1989;108:1687–1695. doi: 10.1083/jcb.108.5.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selsted ME, Miller SI, Henschen AH, Ouellette AJ. Enteric defensins: antibiotic peptide components of intestinal host defense. J. Cell Biol. 1992;118:929–936. doi: 10.1083/jcb.118.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huttner KM, Selsted ME, Ouellette AJ. Structure and diversity of the murine cryptdin gene family. Genomics. 1994;19:448–453. doi: 10.1006/geno.1994.1093. [DOI] [PubMed] [Google Scholar]
- 47.Darmoul D, Ouellette AJ. Positional specificity of defensin gene expression reveals Paneth cell heterogeneity in mouse small intestine. Am. J. Physiol. 1996;271:68–74. doi: 10.1152/ajpgi.1996.271.1.G68. [DOI] [PubMed] [Google Scholar]
- 48.Yokoi Y, et al. Simultaneous real-time analysis of Paneth cell and intestinal stem cell response to interferon-γ by a novel stem cell niche tracking method. Biochem. Biophys. Res. Commun. 2021;545:14–19. doi: 10.1016/j.bbrc.2021.01.050. [DOI] [PubMed] [Google Scholar]
- 49.Eriguchi Y, et al. Essential role of IFN-γ in T cell-associated intestinal inflammation. JCI Insight. 2018;3:e121886. doi: 10.1172/jci.insight.121886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomisawa S, et al. Efficient production of a correctly folded mouse α-defensin, cryptdin-4, by refolding during inclusion body solubilization. Protein Expr. Purif. 2015;112:21–28. doi: 10.1016/j.pep.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 51.Nakamura K, Sakuragi N, Ayabe T. A monoclonal antibody-based sandwich enzyme-linked immunosorbent assay for detection of secreted α-defensin. Anal. Biochem. 2013;443:124–131. doi: 10.1016/j.ab.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 52.Nadatan Y, et al. Gastric acid inhibitor aggravates indomethacin-induced small intestinal injury via reducing Lactobacillus johnsonii. Sci. Rep. 2019;9:17490. doi: 10.1038/s41598-019-53559-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gargano ML, et al. Ecology, phylogeny, and potential nutritional and medicinal value of a rare white “Maitake” collected in a mediterranean forest. Diversity. 2020;12:230. doi: 10.3390/d12060230. [DOI] [Google Scholar]
- 54.Soga T, Heiger DN. Amino acid analysis by capillary electrophoresis electrospray ionization mass spectrometry. Anal. Chem. 2000;72:1236–1241. doi: 10.1021/ac990976y. [DOI] [PubMed] [Google Scholar]
- 55.Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer–specific profiles. Metabolomics. 2010;6:78–95. doi: 10.1007/s11306-009-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.