Abstract
The Southwestern Atlantic rocky reef ecosystems are undergoing significant changes due to sun-corals (Tubastraea tagusensis and T. coccinea) invasion. At Búzios Island, on the northern coast of São Paulo State, where the abundance of T. tagusensis is particularly high, some colonies are displaying tissue necrosis, a phenomenon never reported for this invasive nor any other azooxanthellate coral species. Using next-generation sequencing, we sought to understand the relationship between T. tagusensis tissue necrosis and its microbiota. Thus, through amplicon sequencing, we studied both healthy and diseased coral colonies. Results indicate a wide variety of bacteria associated with healthy colonies and an even higher diversity associated with those corals presenting tissue necrosis, which displayed nearly 25% more microorganisms. Also, as the microbial community associated with the seven healthy colonies did not alter composition significantly, it was possible to verify the microbial succession during different stages of tissue necrosis (i.e., initial, intermediate, and advanced). Comparing the microbiome from healthy corals to those in early tissue necrosis suggests 21 potential pathogens, which might act as the promoters of such disease.
Subject terms: Microbial communities, Microbiome
Introduction
Due to the ever-increasing anthropogenic challenges, coral environments are being impacted by pollution, overfishing1, bleaching2–4, and fluctuations and changes associated with the corals’ symbiont community5–11. Such fluctuations and changes are disrupting the long-established species-specific relationships between corals and microorganisms like Bacteria, Archaea, eukaryotes, and viruses, ultimately resulting in several diseases in their coral hosts5,7,9,11–15.
Our understanding of the factors that influence coral's health and, consequently, its symbiont community, has progressed mainly through studies that determined the microbiota associated with healthy16–19 and diseased corals7,11,14,15. In brief, these studies suggested that there are fundamental microorganisms to the host and that the balance of the microbial community is the result of long (co-evolutionary) and short-term (ecological) processes acting simultaneously. Consequently, biotic and abiotic conditions that trigger coral stress unbalance such relationship, which, in turn, may lead to several host diseases20. Nowadays, there are over 20 diseases known to affect scleractinian corals21, but only a few had its pathogens identified20. Other studies have described the changes in the coral microbiota, indicating that the rise in the abundance of microorganisms from the genus Ruegeria leads to several coral diseases10,11,14,22–24.
In Brazilian waters, analyses of the microbial community have been performed on the endemic corals Mussismilia braziliensis7,25 and Mussismilia hispida26,27, as well as in other more widely spread species, such as Madracis decactis26,28, Siderastrea stellata29, and also on the invasive corals Tubastraea coccinea26 and T. tagusensis30. Some of these studies focused on the microbiota associated with healthy corals, but the microbial community associated with bleached and/or diseased colonies of M. braziliensis, S. stellata, and M. decactis were also determined. Nonetheless, besides diseases7 and acute bleaching events31, the Brazilian coral- and rocky-reef environments are being severely impacted by the invasion of sun-corals, T. tagusensis, and T. coccinea32–34. Currently, T. tagusensis is widespread in Brazil, particularly in São Paulo, Rio de Janeiro, Espírito Santo, Bahia, and Ceará states35–40. At Búzios Island, São Paulo State, a place that was known to harbor mainly M. decactis, M. hispida, Palythoa caribaeorum, turf, and sponges41, several rocky shores are saturated (100%) with invasive corals. Although several biological and ecological aspects are known to be key to the invasion success of T. tagusensis, it seems like they may be related to its reproductive characteristics, such as high production of planula42; early reproductive age43; clonality44,45; regeneration capacity46; and quick incubation—all hallmarks of opportunistic species47.
As a result of asexual planulae production, Southwestern Atlantic invasive T. tagusensis displays a high clonal rate44, and therefore a low genetic diversity, a phenomenon previously observed in other invasive species populations48. Such a decrease in diversity is caused by the founding effect—few specimens colonizing a new environment, which can reduce the adaptive potential of the species over time48. This condition of T. tagusensis may be the downside of this invasive species because, in addition to having a high rate of clonality, it also showed the absence of significant differences in the microbial community along a depth gradient30.
In 2014, when several rocky shores of the Búzios Island were already saturated with invasive corals, colonies of T. tagusensis displaying tissue necrosis were observed. Initially, affected colonies were seen in only one small location, but since then, affected colonies have become widespread. Such tissue necrosis is the first report of disease in azooxanthellate scleractinian corals. To better understand this tissue necrosis, here we characterize and compared its bacterial composition during different necrosis stages. Thus, the T. tagusensis microbiome was quali-quantitatively determined during the tissue necrosis progression, but a better understanding of the cause of such lesions requires further studies.
Materials and methods
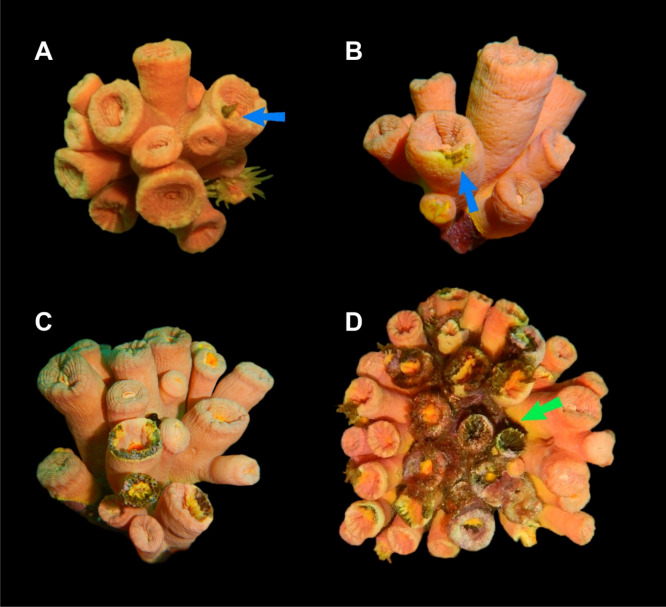
Eighteen specimens of the invasive coral Tubastraea tagusensis were collected in Búzios Island, on the northern coast of the São Paulo State, between 5 and 7 m deep. Sampling was carried out between August 28th and October 09th, 2017. All samples were readily frozen after collection. Part of the samples represented healthy coral colonies (n = 7), and the remaining specimens were from colonies displaying different stages of tissue necrosis: initial (n = 6) defined as colonies with a small necrotic region at the calicular margin (Fig. 1A, B); intermediate (n = 2) defined as polyps with more than 50% of the calicular margin affected (Fig. 1C); and advanced (n = 3). defined as fully necrotic polyps (Fig. 1D).
Figure 1.
Tubastraea tagusensis colonies from Búzios Island—SP, showing signs of tissue necrosis. (A) and (B)—the early stage of tissue necrosis, with a single polyp displaying only a small brown dot (blue arrows). (C)—tissue necrosis progressing throughout the polyp (intermediate stage). (D)—Tissue necrosis at an advanced stage with polyps taken over by the disease (green arrow).
In all analyzed colonies (healthy and diseased) a fragment of approximately 25 mg (~ 5mm2) of the calicular margin containing tissue and skeleton was used for total genomic DNA extraction, for diseased colonies, the extracted portion was exactly the diseased area. DNA was extracted using the DNeasy Blood & Tissue kit (QIAGEN), following the manufacturer's instructions. The quality and purity of extracted DNA were analyzed through agarose gel electrophoresis (1,5%) and spectrophotometry (NanoDrop), respectively.
Using the universal bacterial primers 27F and 519R (V1-V3 region—LANE 1991; Turner et al. 1999), a fragment of the 16S rDNA with approximately 600 bp was amplified using the Advantage HF 2 PCR kit, following concentrations recommended by the manufacturer and two-stage cycling for diseased colonies, as follows: 94 °C for 1 min followed by 38 cycles at 94 °C for 30 s and 68 °C for 1 min. For healthy colonies, PCR reaction was conducted with the following cycling: initial denaturation step 94 °C for 60 s, followed by 30 cycles at 94 °C for 30 s, 56 °C for 40 s, and 68 °C for 33 s, with a final extension step at 68 °C for 33 s.
Amplicons were purified with magnetic beads (Agencourt AMPure XP) following the manufacturer's instructions and eluted in 50 µl of 1X TE buffer (10 mM Tris–HCl, pH 8, 1 mM EDTA). Final sample concentrations were measured using the Qubit dsDNA BR Assay Kit. Libraries were assembled with the NEBNext Ultra II FS DNA Library Prep Kit and their respective concentrations and size distributions were verified using the Qubit dsDNA HS (High Sensitivity) Assay Kit and the Bioanalyzer High Sensitivity DNA Chip, respectively. Sequencing was performed on the MiSeq platform (Illumina) using the MiSeq Nano v2 kit (500 cycles), at the Facility Center for Research from the University of São Paulo (CEFAP-USP). Sequences were deposited in the SRA database (PRJNA637639 and PRJNA675612).
Low-quality strands of each sequence, as well as short reads (< 50pb), were removed using SolexaQA++ software49. Identical sequences were grouped using the Swarm software with d = 150 and then classified on the Mothur platform with a bootstrap cutoff of 8051 using the database 16S Silva—v.132 (SDB)52. Statistical analyses were performed using the STAMP—Statistical Analysis of Metagenomic Profiles software53, after the removal of eukaryotic, chloroplast, mitochondria, and unknown sequences. Statistical tests were performed for multiple groups through variance analyses (ANOVA) and post-hoc Turkey-Kramer tests and Benjamini–Hochberg FDR correction of multiple tests, being 0.01 the adopted p-value filter to identify the relative frequency of OTUs for each necrosis stage and their and their differences. The Whites' two-sided nonparametric t-test, CI method (DP bootstrap, Benjamini–Hochberg FDR multiple test correction, and p-value filter of 0.01) was applied to compare the microbiota between healthy colonies and those at the initial necrosis stage. The diversity parameters were derived from the classification tables (phylum, class, order, family, and genera), such as the wealth estimators Shannon, Neff Shannon, Simpson, and Neff Simpson (Simpson's inverse index). All p-values taken into account were the adjusted p-value after correction.
Results
Tubastraea tagusensis tissue necrosis generally starts at the tissue from the calicular margin like a small brown dot (Fig. 1A, B) that later expands through the polyp (Fig. 1C), sometimes leading to the death of several polyps (Fig. 1D). When colonies with tissue necrosis were manipulated, the skeleton of the diseased region was significantly fragilized and brittle. From the tissue and skeleton of healthy and diseased corals, we obtained 738,462 classified sequences (averaging 220 bp) representing bacteria, chloroplasts, mitochondria, unknown, and eukaryotes. On average, the number of obtained taxonomic classifications from diseased corals (58,564) was four times higher than that from healthy corals (13,465). Sequences from eukaryotes, chloroplasts, mitochondria, and unclassified were removed from statistical analyses. The number of bacterial reads associated with target samples was, on average, 19,268 per coral colony displaying tissue necrosis, and ~ 1538 per healthy coral colony (Table 1).
Table 1.
The number of classified sequences obtained from each analyzed stage of tissue necrosis.
| Status | Sample | Total reads | RI | SHD | Neff Shannon/exp H | D | Neff/D2/Inverse Simpson | |
|---|---|---|---|---|---|---|---|---|
| H | A | 3,635 | 70 | 0.98 | 2.67 | 6.43E−01 | 1.55 | |
| H | B | 1,939 | 70 | 1.07 | 3.05 | 6.04E−01 | 1.65 | |
| H | C | 1,734 | 71 | 1.44 | 4.21 | 3.88E−01 | 2.58 | |
| H | D | 1,110 | 113 | 2.73 | 15.27 | 1.77E−01 | 5.65 | |
| H | E | 865 | 93 | 2.31 | 10.03 | 2.48E−01 | 4.04 | |
| H | F | 793 | 73 | 1.89 | 6.63 | 4.10E−01 | 2.44 | |
| H | G | 676 | 78 | 2.69 | 14.69 | 1.55E−01 | 6.46 | |
| DI | H | 33,752 | 261 | 3.39 | 29.55 | 6.15E−02 | 16.26 | |
| DI | I | 46,855 | 537 | 3.63 | 37.68 | 7.03E−02 | 14.22 | |
| DI | J | 19,994 | 389 | 3.65 | 38.32 | 6.47E−02 | 15.45 | |
| DI | K | 21,844 | 356 | 3.46 | 31.73 | 7.64E−02 | 13.09 | |
| DI | L | 20,039 | 386 | 3.62 | 37.24 | 7.41E−02 | 13.49 | |
| DI | M | 44,301 | 479 | 3.55 | 34.89 | 7.31E−02 | 13.68 | |
| DM | N | 10,705 | 371 | 3.53 | 34.29 | 6.51E−02 | 15.36 | |
| DM | O | 48,436 | 363 | 3.46 | 31.90 | 6.89E−02 | 14.50 | |
| DA | P | 38,101 | 452 | 3.30 | 27.19 | 9.66E−02 | 10.35 | |
| DA | Q | 24,489 | 514 | 3.82 | 45.74 | 6.16E−02 | 16.23 | |
| DA | R | 28,106 | 325 | 3.35 | 28.55 | 8.31E−02 | 12.03 | |
Status: Healthy (H); initial disease (DI); intermediary disease (DM); advanced disease (DA). Richness indices (RI), Shannon H diversity (SHD), Effective number of species (Neff Shannon/exp. H), Simpson index (D), Effective number of species (Neff/D2/inverse Simpson) based on the total reads found in each sample.
The richness index as well as the diversity and the effective number of species in both Shannon and Simpson indexes are higher in those colonies presenting tissue necrosis than that from healthy ones. Richness varies considerably between necrotic colonies (261–537), although there was no observable pattern between different necrosis stages. The greatest richness was found in advanced (514—sample Q) and initial (537—the sample I) stages of the disease. Regarding diversity, there is, on average, a fourfold increase in the number of effective genera from healthy to diseased colonies (Shannon Neff from 8.07 to 34.28 and Simpson Neff Simpson from 3.48 to 14.06).
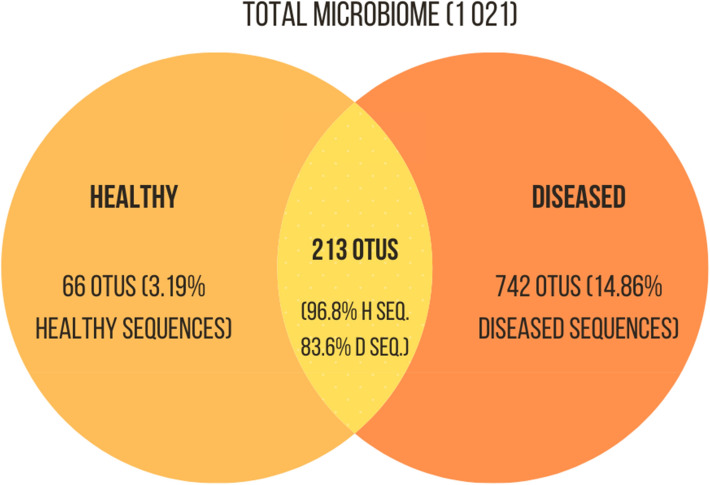
Among Bacteria, 1,021 OTUs were detected, of which 213 occurred in both healthy and diseased colonies. These 213 common OTUs represent 96.8% of the microbiota abundance associated with healthy corals and an average of 83.6% of the abundance of microorganisms associated with diseased colonies. From the 1,021 identified OTUs, only 187 were more abundant in healthy corals, whereas the remaining predominated in diseased ones. In total, 742 genera were exclusively associated with diseased corals, representing 14.86% of their OTUs abundance, while 66 genera were exclusive to healthy colonies and represent 3.19% of their bacterial abundance (Fig. 2). Among the shared OTUs between healthy and diseased coral colonies, 47 displayed significantly different relative frequencies (< 0,01), and, of these, 25 were more frequent in healthy corals.
Figure 2.
Venn Diagram indicating the number of OTUs of Bacteria associated with Tubastraea tagusensis. The microbiota exclusive to healthy colonies is represented in light orange, whereas the microbiota of diseased colonies is represented in dark orange. The intersection, yellow, indicates the microbiota shared by both groups.
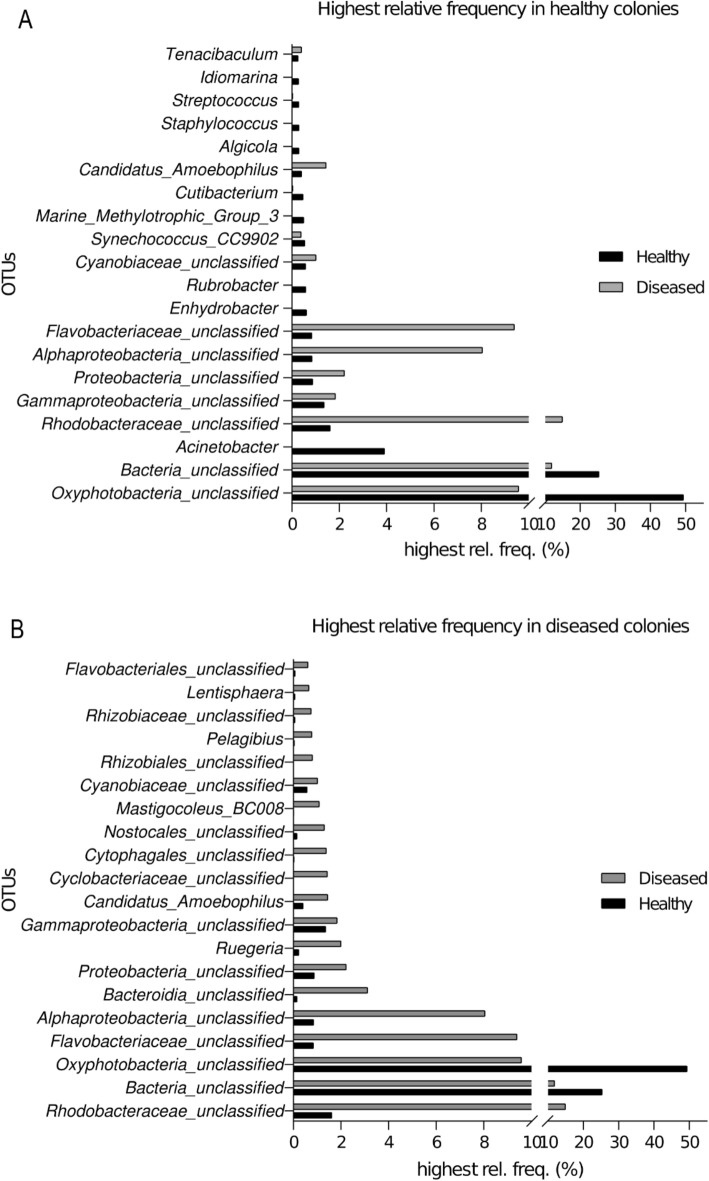
At the genus level, even though most microorganisms have a relatively low frequency, it was possible to infer that some constitute most of the microbiome and markedly differ between coral conditions. In healthy colonies, 20 genera (Fig. 3A) represent 89.2% of the microbiota, with Rubrobacter, Marine_Methylotrophic, and Idiomarina being exclusive to them. The remaining OTUs are significantly abundant in diseased corals, although in a lower proportion (61.1%) of its associated community. Of the 20 most abundant OTUs in diseased colonies (Fig. 3B) (which encompass 74.1% of the detected microbiota), a single one was exclusive to diseased states (Cyclobacteriaceae_unclassified), while the remaining represent 82% of the microbiome associated with healthy colonies.
Figure 3.
The 20 OTUs with the highest relative frequency in (A) healthy corals and their frequencies in diseased corals; and (B) associated with diseased corals and their frequencies in healthy corals. The figure was created using GraphPad Prism 8.0.2 (https://www.graphpad.com/).
Comparisons of the microbiome from healthy and the observed stages of coral tissue necrosis suggest that 15 phyla compose nearly all the microbiota associated with T. tagusensis. Those same 15 phyla represent the majority of taxa, regardless of the disease condition and despite the presence of additional 22 phyla in the diseased colonies. On the other hand, the most abundant ones, Cyanobacteria, Proteobacteria, Bacteroidetes, Actinobacteria, and Firmicutes showed altered frequencies when compared between healthy and diseased colonies (e.g. increased frequency of Proteobacteria and Bacteroidetes and decreased frequency of Cyanobacteria—Table 2).
Table 2.
The relative frequency of the major phyla of Bacteria (calculated with ANOVA) associated with healthy corals (H) and initial (DI), intermediate (DM), and advanced (DA) stages of tissue necrosis, and the p-value.
| Phylum | H (%) | DI (%) | DM (%) | DA (%) | p-value |
|---|---|---|---|---|---|
| Cyanobacteria | 50.54 | 15.81 | 20.45 | 20.4 | 5.11E−02 |
| Proteobacteria | 17.87 | 45.83 | 32.78 | 35.92 | 2.15E−01 |
| Bacteroidetes | 2.81 | 22.37 | 29.66 | 24.98 | 6.53E−03 |
| Actinobacteria | 1.82 | 0.83 | 0.31 | 1.38 | 9.45E−04 |
| Firmicutes | 0.99 | 0.11 | 0.01 | 0.04 | 4.29E−01 |
| Planctomycetes | 0.6 | 2.33 | 2.04 | 2.02 | 1.62E−01 |
| Verrucomicrobia | 0.22 | 0.18 | 0.26 | 0.37 | 4.67E−02 |
| Epsilonbacteraeota | 0.14 | 0.002 | 0.02 | 0.02 | 7.70E−01 |
| Lentisphaerae | 0.09 | 0.62 | 1.46 | 0.91 | 5.82E−01 |
| Chloroflexi | 0.08 | 0.34 | 0.47 | 0.32 | 2.17E−01 |
| Fusobacteria | 0.07 | 0.03 | 0.16 | 0.09 | 2.15E−01 |
| Nitrospinae | 0.04 | 0.01 | 0.03 | 0.04 | 4.41E−01 |
| Acidobacteria | 0.03 | 0.28 | 0.08 | 0.24 | 7.84E−01 |
| Nitrospirae | 0.02 | 0.02 | 0.03 | 0.05 | 2.14E−01 |
| Kiritimatiellaeota | 0.003 | 0.04 | 0.07 | 0.05 | 7.47E−01 |
| Unclassified Bacteria | 24.7 | 10.77 | 11.89 | 12.84 | 2.75E−01 |
| Total | 100 | 99.57 | 99.69 | 99.68 |
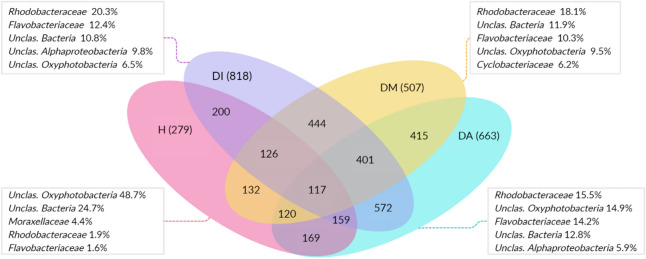
When monitoring the microbial succession in the analyzed stages of tissue necrosis, it is clear that most Bacterial groups present in the microbiome of healthy colonies are also in diseased ones. When the initial signs of tissue necrosis are observed (initial phase), the richness and total reads have a significant increase, but 85.4% (200 OTUs) of microorganisms identified in healthy colonies are also identified in this stage. In the intermediate stage of infection, the number of OTUs increases if compared to healthy colonies (Fig. 4) but decreases 39% concerning the initial stage of the disease. In advanced necrosis stages, a rise in microorganism abundance is observed when compared to the intermediate stage, but 85.4% of the microbiota (169 OTUs) are also found in healthy colonies. When comparing tissue necrosis stages, there is a higher OTUs richness shared between all stages of the disease (Fig. 4), of which 284 OTUs are absent in healthy coral colonies. This group of OTUs represents 12.63%, 18.98%, and 13.41% of abundance from the initial, intermediate, and advanced necrosis stages respectively.
Figure 4.
Venn diagram showing the microbiota (OTUs) of each analyzed stage, of which pink represents healthy colonies (H), purple initial necrosis stage (DI), yellow for colonies in the intermediate necrosis stage (DM), and blue represent colonies in the advanced stage (DA). The numbers of shared OTUs between stages are shown in the intersections. Within the boxes are the five most abundant classifications at the family level for each corresponding group.
Despite the shared microorganisms for each stage, including healthy colonies, a group of bacteria exclusive to each sample is observed: 66 in healthy colonies; 171 in colonies in initial necrosis stage; 43 for the intermediate stage; and 67 in colonies with advanced tissue necrosis.
The microbial core (persistent microorganisms found in all specimens of a particular holobiont species) of T. tagusensis is comprised of 8 genera (Zanotti et. al 2020). Of these, Rubrobacter is absent in all necrotic colonies, Acinetobacter is absent in the intermediate stage, Enhydrobacter is absent in the intermediate and initial stages, and Hydrogenophilus is absent in intermediate and advanced tissue necrosis stages. The microbial core is responsible for only a part of the microbial community of healthy colonies (6.68%), but it is significantly lower in comparison to that from diseased colonies, representing only 2.98%, 2.22%, and 1.39% in colonies at initial, intermediate, and advanced stages respectively. Ruegeria was more frequent in diseased colonies, representing 2.5%, 1.45%, and 1.15% of the microbiota associated with the tissue necrosis initial, intermediate, and advanced stages, respectively (Table 3).
Table 3.
Mean relative frequencies of genera that comprise the microbial core in healthy and at the initial (DI), intermediate (DM), and advanced (DA) stages of the disease in Tubastraea tagusensis.
| Genera | Relative frequencies (%) | |||
|---|---|---|---|---|
| Healthy | DI | DM | DA | |
| Ruegeria | 0.22 | 2.55 | 1.45 | 1.15 |
| Cutibacterium | 0.46 | 0.07 | 0.01 | 0.02 |
| Rubrobacter | 0.57 | 0 | 0 | 0 |
| Acinetobacter | 3.78 | 0.004 | 0 | 0.001 |
| Enhydrobacter | 0.59 | 0 | 0 | 0.001 |
| Hydrogenophilus | 0.22 | 0.008 | 0 | 0 |
| Staphylococcus | 0.29 | 0.005 | 0.002 | 0.003 |
| Synechococcus_CC9902 | 0.52 | 0.34 | 0.76 | 0.21 |
| Total | 6.65 | 2.97 | 2.22 | 1.38 |
To identify potential bacterial triggers of tissue necrosis in T. tagusensis, special attention was given to the community of microorganisms associated exclusively with the initial stage of the disease, and its comparison to those from healthy coral colonies. Among them, 65 significant differences were observed (p value between 0 and 0.0096), but genera that were not linked to disease were excluded, such as those more frequent in healthy colonies (4) and those that were not detected in all diseased colonies in the initial stage (30). Therefore, 31 OTUs fitted these criteria, of which only 13 were classified to the genus level. To improve robustness, genera with less than 10 reads per colony were excluded from further characterization as disease potential triggers. Finally, besides being identified in all colonies during the initial stage of necrosis, these 21 OTUs (Table 4) were also present in all subsequent stages of the disease.
Table 4.
Classification of the 21 OTUs found in all colonies presenting necrosis and with a tendency of increasing relative frequency as the disease progresses in the coral Tubastraea tagusensis.
| Family | Genus | Healthy | DI | p value |
|---|---|---|---|---|
| Alphaproteobacteria_unclassified | 0.847 | 10.08 | 7.24E−04 | |
| Amoebophilaceae | 0.052 | 0.62 | 2.92E−03 | |
| Arenicellaceae | Arenicella | 0.013 | 0.524 | 1.81E−03 |
| Arenicellaceae | 0.004 | 0.112 | 1.21E−03 | |
| Bacteroidia_unclassified | 0.15 | 2.633 | 5.79E−04 | |
| Cyclobacteriaceae | 0 | 0.529 | 1.26E−03 | |
| Deltaproteobacteria_unclassified | 0.004 | 0.183 | 2.66E−03 | |
| Flavobacteriaceae | 0.841 | 9.367 | 6.21E−04 | |
| Flavobacteriaceae | Spongiibacterium | 0 | 0.15 | 2.75E−03 |
| Flavobacteriales_unclassified | 0.07 | 0.53 | 1.75E−03 | |
| Kiloniellaceae | 0 | 0.114 | 1.77E−03 | |
| Myxococcales_unclassified | 0 | 0.124 | 4.11E−03 | |
| Nostocales_unclassified | 0.154 | 1.488 | 6.86E−03 | |
| OM190_fa | 0.017 | 0.219 | 2.02E−03 | |
| Phormidesmiales_unclassified | 0 | 0.526 | 4.04E−03 | |
| Proteobacteria_unclassified | 0.88 | 2.391 | 7.85E−03 | |
| Rhizobiales_unclassified | 0.004 | 0.849 | 7.87E−03 | |
| Rhodobacteraceae | 1.613 | 15.88 | 0 | |
| Rhodobacteraceae | Ruegeria | 0.226 | 2.589 | 1.30E−03 |
| Rhodobacteraceae | Epibacterium | 0.004 | 0.727 | 6.72E−03 |
| Sphingomonadaceae | Sphingorhabdus | 0.023 | 0.298 | 7.67E−04 |
Discussion
Changes in the microorganism community associated with the colonies of the invasive coral Tubastraea tagusensis are significant from the first signs of tissue necrosis. Such changes lead to increased microbial diversity, richness, and the effective number of genera (e.g. bacteria), which is compatible with previous studies on coral diseases54–57. However, most of the microbiota associated with diseased corals (~ 83.6%; 213 OTUs) is present in healthy colonies. Thus, despite the detection of nearly 750 OTUs exclusive in diseased colonies, they did not replace the microbiota associated with healthy hosts but instead reduced it as they grew. Despite the community shift, the relative frequency of bacterial OTUs found exclusively in diseased colonies is low, with the highest disease-specific genus composing 5.1% of the total microbiota in the intermediate stage (unclassified Cyclobacteriaceae). Such low frequency may indicate that the majority of the microbiota in diseased corals is composed of opportunistic species and/or secondary colonizers55,56, which survived in the coral due to the imbalance of the microbial community caused by the primary infection. However, the opposite has also been detected (e.g., 63 OTUs present only in healthy colonies, all with low relative frequencies [an average of ~ 0.097%, representing 3.19 of the total abundance]).
We also observed that the necrotic affected area is not directly related to the diversity of associated microorganisms. In the initial stage, in which it has less than 0.25 cm2, we identified a greater diversity of microorganisms. Also, this is a stage of tissue necrosis that has the most exclusive diversity, suggesting candidate triggers of the disease. In the intermediate stage, which displayed an enlargement of the area affected by the necrosis (Fig. 1C), the number of identified OTUs is lower compared to that from the onset of the disease, possibly indicating a holobiont response. At the advanced stage, the number of identified genera is higher than that in the previous stage, but such increment is not significant compared to the beginning of the infection. Such a variation in the microbiome indicates a rapid destabilization of the symbiont community during the initial stages of T. tagusensis infection, and subsequent colonization by opportunistic microorganisms, as observed in other diseases from zooxanthellate counterparts55,58.
In parallel to the microbiome related to necrosis, attention was given to the microbial core. Among the eight genera considered to be part of the microbial core of T. tagusensis (Zanotti et al. 2020), four were not found in colonies presenting tissue necrosis (Table 3). Such a difference might be a result of the microbiome imbalance caused by the disease. Nonetheless, the relative frequency of four microbial core genera was significantly lower in polyps presenting necrosis. However, Ruegeria had different patterns once it was the most abundant genera in diseased colonies. This genus is known to be associated with several healthy14,59–61 and diseased zooxanthellate corals10,11,14,22–24. Previous studies suggested that Ruegeria inhibits/controls the growth of other bacteria genera through tropodithietic acid62. Also, Ruegeria inhibited a widely known opportunistic coral pathogen, Vibrio coralliilyticus63.
Statistical analyses of the microbiome also indicate that within the bacterial OTUs identified, 21 (Table 4) might represent pathogenic ones. Of these, only five were classified to genus level, one of them being Ruegeria, which as mentioned has no pathogenic profile. Within the remainder, only Arenicella has been associated with coral disease (skeletal growth anomalies in Platygyra carnosa23), although the herein detected Sphingomonadaceae has also been described as a putative pathogen associated with Acropora cervicornis and A. palmata disease64. Besides that, although there is no previous report of Epibacterium in relationship to scleractinian corals (a group described associated with seaweed surfaces65), its detection and increased abundance during T. tagusensis tissue necrosis may be related to the exposure of the coral skeleton to the environment. As the necrosis advances, the exposed skeleton becomes a potential substrate for several organisms, like filamentous algae.
Apart from the aforementioned OTUs, several remained unclassified at the genus level. Among them, the Flavobacteriaceae and Rhodobacteriaceae were found in high abundance in T. tagusensis tissue necrosis and were previously associated with the following scleractinian diseases: White band55; White plague7,66; Yellow-band14; White syndrome67; White-spot syndrome in Porites11; White Plague66; stony coral tissue loss disease68; and Black band10. Additionally, the skeleton fragility in those colonies affected by the tissue necrosis may be related to a higher frequency of Mastigocoleus, a bacterial genus known to have bioerosion capabilities69.
Although concerning as another disease is reported to a scleractinian coral, the described tissue necrosis brings a glimpse of hope in the face of T. tagusensis unprecedented bioinvasion and spreading in the Southwestern Atlantic. Thus, despite its invasion capacity, the founding effect44,70, and the associated microbial community without major significant differences might be a disadvantage for this species, making it highly susceptible to diseases as the tissue necrosis reported herein.
Acknowledgements
The authors are thankful to CEBIMar staff for all their support and the technicians of the GENIAL/CEFAP/USP sequencing facility. MVK thanks the support of the São Paulo Research Foundation (FAPESP, Process #2014/01332-0) and National Research Council (CNPq, Process #301436/2018-5). Support for the work: MSc scholarship to AAZ, Coordination for the Improvement of Higher Education Personnel (CAPES), Finance Code 001. The São Paulo Research Foundation (FAPESP, Process #2014/01332–0). National Research Council (CNPq, Process #301436/2018–5).
Author contributions
This work was done collaboratively by all authors. A.A.Z. in cooperation with M.V.K. defined the research theme and designed the experiment. A.A.Z. obtained sequences, and in cooperation with G.B.G. conducted the analyses. All authors discussed the results, interpretation, presentation, contributed to the writing of the manuscript, and approved the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wilkinson CR. Status of Coral Reefs of the World: 2004. 2004. Australian Institute of Marine Science (AIMS).
- 2.Anthony KRN, Kline DI, Diaz-Pulido G, Dove S, Hoegh-Guldberg O. Ocean acidification causes bleaching and productivity loss in coral reef builders. Proc Natl Acad Sci. 2008;105:17442–17446. doi: 10.1073/pnas.0804478105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben-Haim Y, Zicherman-Keren M, Rosenberg E. Temperature-regulated bleaching and lysis of the coral Pocillopora damicornis by the novel pathogen Vibrio coralliilyticus. Appl Environ Microbiol. 2003;69:4236–4242. doi: 10.1128/AEM.69.7.4236-4242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenberg E, Falkovitz L. The Vibrio shiloi/Oculina patagonica model system of coral bleaching. Annu Rev Microbiol. 2004;58:143–159. doi: 10.1146/annurev.micro.58.030603.123610. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Haim Y, Rosenberg E. A novel Vibrio sp. pathogen of the coral Pocillopora damicornis. Mar Biol 2002; 141: 47–55.
- 6.Denner EBM, Smith GW, Busse HJ, Schumann P, Narzt T, Polson SW, et al. Aurantimonas coralicida gen. nov., sp. nov., the causative agent of white plague type II on Caribbean scleractinian corals. Int J Syst Evol Microbiol 2003; 53: 1115–1122. [DOI] [PubMed]
- 7.Garcia GD, Gregoracci GB, de O. Santos E, Meirelles PM, Silva GGZ, Edwards R, et al. Metagenomic analysis of healthy and white plague-affected Mussismilia braziliensis corals. Microb Ecol 2013; 65: 1076–1086. [DOI] [PubMed]
- 8.Luna GM, Bongiorni L, Gili C, Biavasco F, Danovaro R. Vibrio harveyi as a causative agent of the White Syndrome in tropical stony corals. Environ Microbiol Rep. 2010;2:120–127. doi: 10.1111/j.1758-2229.2009.00114.x. [DOI] [PubMed] [Google Scholar]
- 9.Pantos O, Cooney RP, Le TMDA, Barer MR, Bythell JC, Le Tissier MDA, et al. The bacterial ecology of a plague-like disease affecting the Caribbean coral Montastrea annularis. Environ Microbiol. 2003;5:370–382. doi: 10.1046/j.1462-2920.2003.00427.x. [DOI] [PubMed] [Google Scholar]
- 10.Sekar R, Kaczmarsky LT, Richardson LL. Microbial community composition of black band disease on the coral host Siderastrea siderea from three regions of the wider Caribbean. Mar Ecol Prog Ser. 2008;362:85–98. doi: 10.3354/meps07496. [DOI] [Google Scholar]
- 11.Séré MG, Tortosa P, Chabanet P, Turquet J, Quod J-P, Schleyer MH. Bacterial communities associated with Porites White Patch Syndrome (PWPS) on Three Western Indian Ocean (WIO) Coral Reefs. PLoS ONE. 2013;8:e83746. doi: 10.1371/journal.pone.0083746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patterson KL, Porter JW, Ritchie KB, Polson SW, Mueller E, Peters EC, et al. The etiology of white pox, a lethal disease of the Caribbean elkhorn coral Acropora palmata. PNAS. 2002;99:8725–8730. doi: 10.1073/pnas.092260099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooney RPR, Pantos O, Le TMMDA, Barer MR, O’Donnell AG, Bythell JC, et al. Characterization of the bacterial consortium associated with black band disease in coral using molecular microbiological techniques. Environ Microbiol. 2002;4:401–413. doi: 10.1046/j.1462-2920.2002.00308.x. [DOI] [PubMed] [Google Scholar]
- 14.Apprill A, Hughen K, Mincer T. Major similarities in the bacterial communities associated with lesioned and healthy Fungiidae corals. Environ Microbiol. 2013;15:2063–2072. doi: 10.1111/1462-2920.12107. [DOI] [PubMed] [Google Scholar]
- 15.Meyer JL, Paul VJ, Raymundo LJ, Teplitski M. Comparative metagenomics of the Polymicrobial Black Band Disease of corals. Front Microbiol. 2017;8:618. doi: 10.3389/fmicb.2017.00618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ainsworth TD, Krause L, Bridge T, Torda G, Raina J-B, Zakrzewski M, et al. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME. 2015;9:2261–2274. doi: 10.1038/ismej.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hernandez-Agreda A, Gates RD, Ainsworth TD. Defining the core microbiome in corals’ microbial soup. Trends Microbiol. 2017;25:125–140. doi: 10.1016/j.tim.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Röthig T, Yum LK, Kremb SG, Roik A, Voolstra CR. Microbial community composition of deep-sea corals from the Red Sea provides insight into functional adaption to a unique environment. Sci Rep. 2017;7:1–9. doi: 10.1038/srep44714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosado PM, Leite DCA, Duarte GAS, Chaloub RM, Jospin G, Nunes da Rocha U, et al. Marine probiotics: increasing coral resistance to bleaching through microbiome manipulation. ISME J 2019; 13: 921–936. [DOI] [PMC free article] [PubMed]
- 20.Rosenberg E, Koren O, Reshef L, Efrony R. The role of microorganisms in coral health, disease, and evolution. Nat Rev Microbiol. 2007;5:355–362. doi: 10.1038/nrmicro1635. [DOI] [PubMed] [Google Scholar]
- 21.Raymundo LJ, Couch CS, Bruckner AW, Harvell CD, Work TM, Weil E, et al. Coral Disease Handbook, Guidelines for Assessment, Monitoring & Management, Currie Com. 2008.
- 22.Casey JM, Connolly SR, Ainsworth TD. Coral transplantation triggers shift in microbiome and promotion of coral disease associated potential pathogens. Sci Rep. 2015;5:11903. doi: 10.1038/srep11903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng JCY, Chan Y, Tun HM, Leung FCC, Shin PKS, Chiu JMY. Pyrosequencing of the bacteria associated with Platygyra carnosus corals with skeletal growth anomalies reveals differences in bacterial community composition in apparently healthy and diseased tissues. Front Microbiol. 2015;6:1–14. doi: 10.3389/fmicb.2015.01142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato Y, Ling EYS, Turaev D, Laffy P, Weynberg KD, Rattei T, et al. Unraveling the microbial processes of black band disease in corals through integrated genomics. Sci Rep. 2017;7:40455. doi: 10.1038/srep40455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reis AMM, Araújo SD, Jr, Moura RL, Francini-Filho RB, Pappas G, Jr, Coelho AMA, et al. Bacterial diversity associated with the Brazilian endemic reef coral Mussismilia braziliensis. J Appl Microbiol. 2009;106:1378–1387. doi: 10.1111/j.1365-2672.2008.04106.x. [DOI] [PubMed] [Google Scholar]
- 26.Carlos C, Torres TT, Ottoboni LMM. Bacterial communities and species-specific associations with the mucus of Brazilian coral species. Sci Rep. 2013;3:1624. doi: 10.1038/srep01624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lins-de-Barros MM, Vieira RP, Cardoso AM, Monteiro VA, Turque AS, Silveira CB, et al. Archaea, bacteria, and algal plastids associated with the reef-building corals Siderastrea stellata and Mussismilia hispida from Búzios, South Atlantic Ocean, Brazil. Microb Ecol. 2010;59:523–532. doi: 10.1007/s00248-009-9612-y. [DOI] [PubMed] [Google Scholar]
- 28.Moreira APB, Chimetto Tonon LA, do Valle P. Pereira C, Alves N, Amado-Filho GM, Francini-Filho RB, et al. Culturable heterotrophic Bacteria associated with healthy and bleached scleractinian Madracis decactis and the fireworm Hermodice carunculata from the Remote St. Peter and St. Paul Archipelago, Brazil. Curr Microbiol 2014; 68: 38–46. [DOI] [PubMed]
- 29.Lins-de-Barros MM, Cardoso AM, Silveira CB, Lima JL, Clementino MM, Martins OB, et al. Microbial community compositional shifts in bleached colonies of the brazilian reef-building coral Siderastrea stellata. Microb Ecol. 2013;65:205–213. doi: 10.1007/s00248-012-0095-x. [DOI] [PubMed] [Google Scholar]
- 30.Zanotti AA, Gregoracci GB, Capel KCC, Kitahara MV. Microbiome of the Southwestern Atlantic invasive scleractinian coral, Tubastraea tagusensis. Anim Microbiome. 2020;2:29. doi: 10.1186/s42523-020-00047-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teixeira CD, Leitão RLL, Ribeiro FV, Moraes FC, Neves LM, Bastos AC, et al. Sustained mass coral bleaching (2016–2017) in Brazilian turbid-zone reefs: taxonomic, cross-shelf and habitat-related trends. Coral Reefs. 2019;38:801–813. doi: 10.1007/s00338-019-01789-6. [DOI] [Google Scholar]
- 32.Miranda RJ, Cruz ICS, Barros F. Effects of the alien coral Tubastraea tagusensis on native coral assemblages in a southwestern Atlantic coral reef. Mar Biol. 2016;163:1–12. doi: 10.1007/s00227-016-2819-9. [DOI] [Google Scholar]
- 33.Creed JC. Two invasive alien azooxanthellate corals, Tubastraea coccinea and Tubastraea tagusensis, dominate the native zooxanthellate Mussismilia hispida in Brazil. Coral Reefs. 2006;25:350. doi: 10.1007/s00338-006-0105-x. [DOI] [Google Scholar]
- 34.Silva ICBS, Liparini A, Pereira NS, Braga BLSS, Sial AN, Liu SC, et al. Assessing the growth rate of the South Atlantic coral species Mussismilia hispida (Verrill, 1902) using carbon and oxygen stable isotopes. J South Am Earth Sci. 2019;96:102346. doi: 10.1016/j.jsames.2019.102346. [DOI] [Google Scholar]
- 35.Costa TJF, Pinheiro HT, Teixeira JB, Mazzei EF, Bueno L, Hora MSC, et al. Expansion of an invasive coral species over Abrolhos Bank, Southwestern Atlantic. Mar Pollut Bull. 2014;85:252–253. doi: 10.1016/j.marpolbul.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Creed JC, Eduardo A, Oliveira S, De PAF. Cnidaria, Scleractinia, Tubastraea coccinea Lesson, 1829 and Tubastraea tagusensis Wells, 1982: distribution extension. Check List. 2008;4:297–300. doi: 10.15560/4.3.297. [DOI] [Google Scholar]
- 37.De Paula AF, Creed JC. Two species of the coral Tubastraea (Cnidaria, Scleractinia) in Brazil: a case of accidental introduction. Bull Mar Sci. 2004;74:175–183. [Google Scholar]
- 38.Ferreira CEL, Gonçalves JEA, Coutinho R. Ship hulls and oil platforms as potential vectors to marine species introduction. J Coast Res. 2006;3:1341–1346. [Google Scholar]
- 39.Sampaio CLS, Miranda RJ, Maia-Nogueira R, de Anchieta Nunes JCC. New occurrences of the nonindigenous orange cup corals Tubastraea coccinea and T. tagusensis (Scleractinia: Dendrophylliidae) in southwestern Atlantic. Check List 2012; 8: 528–530.
- 40.Silva AG, De Paula AF, Fleury BG, Creed JC. Eleven years of range expansion of two invasive corals (Tubastraea coccinea and Tubastraea tagusensis) through the Southwest Atlantic (Brazil) Estuar Coast Shelf Sci. 2014;141:9–16. doi: 10.1016/j.ecss.2014.01.013. [DOI] [Google Scholar]
- 41.Mantelatto MC, Creed JC, Mourão GG, Migotto AE, Lindner A. Range expansion of the invasive corals Tubastraea coccinea and Tubastraea tagusensis in the Southwest Atlantic. Coral Reefs. 2011;30:397. doi: 10.1007/s00338-011-0720-z. [DOI] [Google Scholar]
- 42.Luz BLP, Di Domenico M, Migotto AE, Kitahara MV. Life-history traits of Tubastraea coccinea: reproduction, development, and larval competence. Ecol Evol. 2020;10:6223–6238. doi: 10.1002/ece3.6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fenner D, Banks K. Orange cup coral Tubastraea coccinea invades Florida and the Flower Garden Banks, Northwestern Gulf of Mexico. Coral Reefs. 2004;23:505–507. [Google Scholar]
- 44.Capel KCC, Toonen RJ, Rachid CTCC, Creed JC, Kitahara M V., Forsman Z, et al. Clone wars: asexual reproduction dominates in the invasive range of Tubastraea spp. (Anthozoa: Scleractinia) in the South-Atlantic Ocean. PeerJ 2017; 5: 1–21. [DOI] [PMC free article] [PubMed]
- 45.Capel KC, Creed JC, Kitahara MV. Invasive corals trigger seascape changes in the Southwestern Atlantic. Bull Mar Sci. 2020;96:217–218. doi: 10.5343/bms.2019.0075. [DOI] [Google Scholar]
- 46.Luz BLP, Capel KCC, Zilberberg C, Flores AAV, Migotto AE, Kitahara MV. A polyp from nothing: The extreme regeneration capacity of the Atlantic invasive sun corals Tubastraea coccinea and T. tagusensis (Anthozoa, Scleractinia). J Exp Mar Bio Ecol 2018; 503: 60–65.
- 47.De Paula AF. Biologia reprodutiva, crescimento e competição dos corais invasores Tubastraea coccinea e Tubastraea tagusensis (Scleractinia: Dendrophylliidae) com espécies nativas. 2007. Universidade Federal do Rio de Janeiro.
- 48.Dlugosch KM, Parker IM. Founding events in species invasions: Genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol. 2008. John Wiley & Sons, Ltd., 17: 431–449 [DOI] [PubMed]
- 49.Cox MP, Peterson DA, Biggs PJ. SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinform. 2010;11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahé F, Rognes T, Quince C, de Vargas C, Dunthorn M. Swarm: robust and fast clustering method for amplicon-based studies. PeerJ. 2014;2:1–13. doi: 10.7717/peerj.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Closek CJ, Sunagawa S, DeSalvo MK, Piceno YM, DeSantis TZ, Brodie EL, et al. Coral transcriptome and bacterial community profiles reveal distinct Yellow Band Disease states in Orbicella faveolata. ISME J. 2014;8:2411–2422. doi: 10.1038/ismej.2014.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gignoux-Wolfsohn SA, Vollmer SV. Identification of candidate coral pathogens on White Band Disease-Infected Staghorn coral. PLoS ONE. 2015;10:e0134416. doi: 10.1371/journal.pone.0134416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sunagawa S, Desantis TZ, Piceno YM, Brodie EL, DeSalvo MK, Voolstra CR, et al. Bacterial diversity and white Plague disease-associated community changes in the caribbean coral Montastraea faveolata. ISME J. 2009;3:512–521. doi: 10.1038/ismej.2008.131. [DOI] [PubMed] [Google Scholar]
- 57.Pantos O, Bythell JC. Bacterial community structure associated with white band disease in the elkhorn coral Acropora palmata determined using culture-independent 16S rRNA techniques. Dis Aquat Organ. 2006;69:79–88. doi: 10.3354/dao069079. [DOI] [PubMed] [Google Scholar]
- 58.Gignoux-Wolfsohn SA, Aronson FM, Vollmer SV. Complex interactions between potentially pathogenic, opportunistic, and resident bacteria emerge during infection on a reef-building coral. FEMS Microbiol Ecol. 2017;93:1–10. doi: 10.1093/femsec/fix080. [DOI] [PubMed] [Google Scholar]
- 59.Bayer T, Neave MJ, Alsheikh-Hussain A, Aranda M, Yum LK, Mincer T, et al. The microbiome of the Red Sea Coral Stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria. Appl Environ Microbiol. 2013;79:4759–4762. doi: 10.1128/AEM.00695-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harder T, Lau SCK, Dobretsov S, Fang TK, Qian PY. A distinctive epibiotic bacterial community on the soft coral Dendronephthya sp. and antibacterial activity of coral tissue extracts suggest a chemical mechanism against bacterial epibiosis. FEMS Microbiol Ecol 2003; 43: 337–347. [DOI] [PubMed]
- 61.Muller EM, Leporacci NM, Macartney KJ, Shea AG, Crane RE, Hall ER, et al. Low pH reduces the virulence of black band disease on Orbicella faveolata. PLoS ONE. 2017;12:e0178869. doi: 10.1371/journal.pone.0178869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Porsby CH, Nielsen KF, Gram L. Phaeobacter and Ruegeria species of the Roseobacter clade colonize separate niches in a Danish Turbot (Scophthalmus maximus)-rearing farm and antagonize Vibrio anguillarum under different growth conditions. Appl Environ Microbiol. 2008;74:7356–7364. doi: 10.1128/AEM.01738-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miura N, Motone K, Takagi T, Aburaya S, Watanabe S, Aoki W, et al. Ruegeria sp. strains isolated from the Reef-Building Coral Galaxea fascicularis inhibit growth of the temperature-dependent pathogen Vibrio coralliilyticus. Mar Biotechnol 2019; 21: 1–8. [DOI] [PubMed]
- 64.Rosales SM, Miller MW, Williams DE, Traylor-Knowles N, Young B, Serrano XM. Microbiome differences in disease-resistant vs. susceptible Acropora corals subjected to disease challenge assays. Sci Rep 2019; 9: 1–11. [DOI] [PMC free article] [PubMed]
- 65.Penesyan A, Breider S, Schumann P, Tindall BJ, Egan S, Brinkhoff T. Epibacterium ulvae gen. nov., sp. nov., epibiotic bacteria isolated from the surface of a marine alga. Int J Syst Evol Microbiol 2013; 63: 1589–1596. [DOI] [PubMed]
- 66.Roder C, Arif C, Bayer T, Aranda M, Daniels C, Shibl A, et al. Bacterial profiling of White Plague Disease in a comparative coral species framework. ISME J. 2014;8:31–39. doi: 10.1038/ismej.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pollock FJ, Wada N, Torda G, Willis BL, Bourne DG. White Syndrome-Affected corals have a distinct microbiome at disease lesion fronts. Appl Environ Microbiol. 2017;83:e02799–e2816. doi: 10.1128/AEM.02799-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meyer JL, Castellanos-Gell J, Aeby GS, Häse CC, Ushijima B, Paul VJ. Microbial community shifts associated with the ongoing stony coral tissue loss disease outbreak on the Florida Reef Tract. Front Microbiol. 2019;10:2244. doi: 10.3389/fmicb.2019.02244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tribollet A. Dissolution of dead corals by euendolithic microorganisms across the northern Great Barrier Reef (Australia) Microb Ecol. 2008;55:569–580. doi: 10.1007/s00248-007-9302-6. [DOI] [PubMed] [Google Scholar]
- 70.Capel KCC, Creed J, Kitahara MV., Chen CA, Zilberberg C. Multiple introductions and secondary dispersion of Tubastraea spp. in the Southwestern Atlantic. Sci Rep 2019; 9: 1–11. [DOI] [PMC free article] [PubMed]