Abstract
Background
We previously proposed an immune cell score (tumour node metastasis (TNM)-Immune cell score) classifier as an add-on to the existing TNM staging system for non-small cell lung cancer (NSCLC). Herein, we examined how to reliably assess a tertiary lymphoid structure (TLS) score to refine the TNM staging system.
Methods
Using immunohistochemistry (CD8/cytokeratin), we quantified TLS in resected NSCLC whole-tumour tissue sections with three different scoring models on two independent collections (total of 553 patients). In a pilot setting, NanoString gene expression signatures were analysed for associations with TLS.
Results
The number of TLSs significantly decreased in stage III patients as compared to stage II. The TLS score was an independent positive prognostic factor, regardless of the type of (semi)-quantification strategy used (four-scale semi-quantitative; absolute count of total TLS; subpopulation of mature TLS) or the endpoint (disease-specific survival; overall survival; time to recurrence). Subgroup analyses revealed a significant prognostic impact of TLS score within each pathological stage, patient cohort and main histological subtype. Targeted gene expression analysis showed that high TLS levels were associated with the expression of B cell and adaptive immunity genes/metagenes including tumour inflammation signature.
Conclusions
The TLS score increases the prognostic power in each pathological stage and hence has the potential to refine TNM staging in resected NSCLC.
Subject terms: Surgical oncology, Translational immunology, Prognostic markers
Background
The TNM classification system guides the management of non-small cell lung cancer (NSCLC) patients and estimates their survival. Although TNM staging is a well-established system, improvements to guide individualised patient care are sought.1 With increased understanding of the pivotal role of the immune system in cancer, plus the advent of immunotherapy, it is generally accepted that a patient’s in situ immune milieu provides important prognostic and predictive information that has value in clinical management.2,3 Immune classification based on the extent of tumour invasion by the tissue’s immune cells has been found to enhance prognostic accuracy of tumour staging in colorectal cancer (called Immunoscore®) and NSCLC (called TNM-Immune cell score, TNM-I).4,5
In order to refine prognostication for resected NSCLC patients beyond the current TNM staging system, our group previously analysed the presence, density and localisation of various immune cell subsets, such as T- and B-lymphocytes, neutrophils, macrophages (M1 and M2 phenotypes) and immune checkpoint molecules (PD-1, PD-L1, CTLA4, LAG-3 and TIM-3).6–12 Currently, the most promising candidates for supplementing into TNM-I are CD8+ cytotoxic T cells, CD45RO+ memory T cells and tumour-infiltrating lymphocytes (in haematoxylin and eosin (H&E) whole-tissue slides).6–8 However, as recent reports found TLSs to be a predictive biomarker for immunotherapy efficacy in melanoma, sarcoma and renal cell carcinoma,13–15 its potential as a candidate marker for a NSCLC TNM-I model is intriguing.
In human lung tissue, TLSs are also known as inducible bronchus-associated lymphoid tissue, which arise in response to chronic pulmonary inflammation.16,17 Histologically, TLSs highly resemble secondary lymphoid organs, as characterised by immune cell aggregates with central B cell clusters surrounded by T cells. In contrast to secondary lymphoid organs, TLSs appear in ectopic sites of inflammation with highly dynamic structures.18 Structurally, they are present in tissue in two forms: mature and immature. Mature TLSs are well formed and comprise a GC-like structure, including follicular dendritic cells (DCs) and high endothelial venules. Immature TLSs are poorly structured, lack the GC and maybe functionally impaired regarding the humoral response.19,20 TLSs are pivotal sites of adaptive immunity, with the main function of potentiating immune responses through activation and maintenance of local and systemic T and B cell responses, together with (and more often independent from) secondary lymphoid organs.21
To date, several groups have assessed TLSs as prognostic and predictive biomarkers in cancer.13–15,22 Hitherto, published reports have been inconsistent, in part, due to methodology.23 In NSCLC, most studies have identified TLSs by immunohistochemistry (IHC) against TLS-associated immune cell markers such as CD208+ DCs, CD20+ B cells or CD3+ T cells.22 In some studies, TLS were evaluated using gene signatures, including relevant lymphoid chemokines and T-helper 1 phenotypes.24,25 However, the lack of a standardised methodology for assessing their existence, density and maturity limits translation into clinical practice.
In an attempt to resolve such inconsistencies, we aimed to evaluate different histological assessment strategies for TLSs. Further, we wanted to validate TLS score as an approach to refine the TNM staging system in resected NSCLC. As there is limited knowledge about gene signatures representing the TLS status, TLS-associated gene expression profiles in NSCLC were examined in a pilot design.
Methods
Patient cohorts
The study was based on two independent cohorts of NSCLC patients in Norway. The demographic data of both cohorts were described previously.8 Briefly, they were accrued by a serial collection of formalin-fixed paraffin-embedded (FFPE) tissue from the Department of Pathology at the University Hospital of North Norway (cohort #1, n = 295) and the Department of Pathology at Nordland Central Hospital (cohort #2, n = 258). A total of 553 patients were originally included in the study. Of these, 63 patients (11%) were excluded due to inadequate tissue quality on the slides. Hence, 490 patients were available for analyses.
Clinicopathological data and clinical endpoints were retrieved from each patient’s medical journal by an oncologist. Patients were diagnosed between 1990 and 2010. The median follow-up of survivors was 86 months (range 34–267 months). The last follow-up was in October 2013.
The tumour specimens were restaged and reclassified by two pulmonary pathologists according to the most recent updated versions of World Health Organisation (WHO) (2015) and Union for International Cancer Control (2016, 8th edition).26,27 The REMARK guidelines were followed for data reporting of variables, survival and biomarkers.28 The study was ethically approved by the Regional Committee (REK) and Norwegian Data Protection Organization (REK 2016/714).
Immunohistology
FFPE whole-tissue sections were stained using the Discovery Ultra System (Ventana, Tucson, USA). The following prediluted primary antibodies from Ventana were used for co-staining of CD8/cytokeratin (CK) (whole cohort, 553 patients), and multiplexed IHC (CD208/CD8/Ki67/CD20) of selected samples (20 patients): CD8 (clone: SP57, 790-4460), CK (clone: AE1/AE3/PCK26, 760-2595), CD20 (clone: L26, 760-2531) and Ki67 (clone: 30-9, 790-4286). DCs were labelled with mouse monoclonal primary antibody CD208/DC-LAMP (clone:104G4, DDX0190P-100; Nordic BioSite).
Quantification of TLS
Two observers (M.R., S.J.), blinded to each other’s scores and patient outcomes, evaluated TLSs in digitalised whole-tissue slides, immunostained with CD8/CK, using three different scoring models. TLS was regarded as both lymphoid aggregates and follicles. A lymphoid aggregate was defined as the accumulation of lymphocytes and plasma cells without a germinal centre (GC, immature TLS). Lymphoid follicles were defined as aggregates of lymphocytes with a GC (mature TLS).22,24 Other lymphoid patterns were ignored (see scoring criteria).
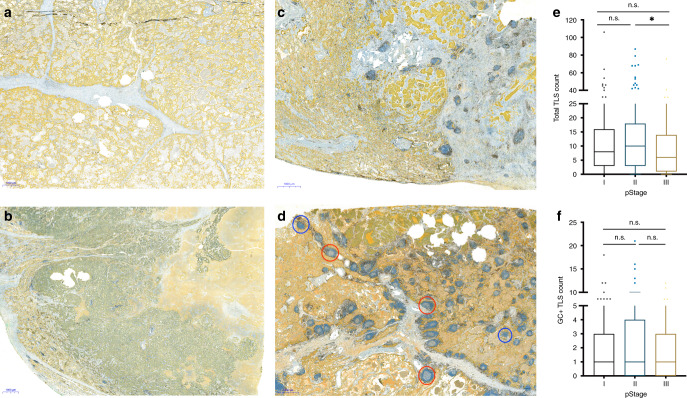
Model 1: In this semi-quantitative approach, the amount of TLS overall in the tissue, regardless of the compartment, was scored on a four-tiered scale: 0, none or one TLS; 1, sparse TLSs; 2, moderate presence of TLSs; 3, strong and heavy presence of TLSs, equivalent to that seen in a lymph node (Fig. 1a–d).
Fig. 1. Distribution of tertiary lymphoid structures (TLS) in tissue and across TNM stages of NSCLC.
a–d Examples of different levels of TLS in resected primary tumours using CD8 (brown)/cytokeratin (CK, yellow) antibodies. Cytotoxic T cells labelled with CD8 and epithelial cells with CK. a No, b low, c moderate and d high TLS based on semi-quantitative model of scoring. TLSs with germinal centres (GC+ TLS, red circle) and without germinal centres (blue circle) are indicated. e, f Box-plots showing the distribution of total TLS (e) and GC+ TLS (f) count across TNM stages (stage I, n = 213; stage II, n = 159; stage III, n = 118) in NSCLC. Statistical analysis was performed with Kruskal–Wallis and Dunn’s multiple comparison tests. n.s. not significant, * adjusted P < 0.05.
Model 2: This quantitative method for evaluating TLSs was based on the manual absolute count in both the tumour periphery and tumour core. TLSs in the ‘tumour periphery’ were defined as lymphoid structures localised in the stroma surrounding the tumour nest (from tumour nest border to normal tissue). The ‘tumour core’ TLSs were defined as lymphoid structures localised deep in the tumour and in direct central contact with the tumour epithelial cells. The total lymphoid follicles and aggregates were counted separately in each tumour compartment (core and periphery).
Model 3: In the second quantitative model, lymphoid follicles (with GC) were counted as a separate variable in each tumour compartment. TLSs were regarded as GC+ when the lymphoid structure exhibited reactive proliferating centrocytes and centroblasts.
Scoring criteria for all models:
TLSs within normal alveolar epithelial areas were excluded from the evaluation (Supplementary Fig. S1), since it is believed that TLSs reside in the alveolar epithelium as a result of chronic inhalation of alum or cigarette smoke.29,30
The areas consisting of non-contiguous lymph nodes were excluded from the evaluation, particularly in cases where the tumour was anatomically located on paratracheal or subcarinal zones (Supplementary Figure S1).
The areas with highly diffused lymphocytic infiltration, without any recognisable lymphoid follicles/aggregates architecture, were excluded from the analysis (Supplementary Figure S2).
The nonnodular-shaped lymphoid aggregates including irregularly shaped (unstructured and intermediately organised31) as well as long and narrow structures were ignored.
The lymphoid aggregates with <150 μm in diameter (or major axis in oval-shaped TLSs) were not included in the count. TLS’s T and B cell zones identification was challenging for aggregates under this size threshold.
Models 2 and 3 were based on an ‘absolute count’ of aggerates and/or follicles without considering the ratio of tumour area occupied by those lymphoid structures.
For each patient, a single slide with the largest tumour–stromal interface was assessed for TLSs.
To avoid bias, semi-quantitative and quantitative scoring were performed by each investigator after a 1-month interval. Discordant cases were reviewed by a pulmonary pathologist and followed by a conclusive judgement.
Gene expression profiling
For RNA extraction and analysis, 14 samples were selected consecutively from the latest inclusion period (2010) based on high vs. low TLS scores. Two serial 10-μm-thick sections were cut from the same FFPE block that was used for IHC. RNA was extracted using High Pure FFPET RNA Isolation Kit (Roche, Basel, Switzerland) according to the manufacturer’s instructions. The concentration and quality of the RNA were evaluated by NanoDrop and the Agilent 2100 bioanalyzer. Based on the DV200 values, all samples passed through quality control. NanoString gene expression analysis was performed using the nCounter FLEX with Dx Enablement platform and the PanCancer Immuno-Oncology (IO) 360 panel.
Data analysis
Normalisation was in two steps. The first step depended on whether or not the genes are in the tumour inflammation signature (TIS). Genes were normalised by using a ratio of the expression value to the geometric mean of all 20 housekeeping genes on the panel. Genes in the TIS signature were normalised by using a ratio of the expression value to the geometric mean of the ten housekeeper genes used for the TIS signature alone. Genes were further normalised by using a ratio of the housekeeper-normalised data and a panel standard run on the same cartridge as the observed data. The housekeeper- and panel standard-normalised data were log(2) transformed.
Comparisons of differential gene expression data were made between patients with high vs. low TLS counts. Differential expression was fitted on a per-gene or per-signature basis using a linear model for the analyses without a blocking factor. The P values were adjusted within each analysis, gene or signature, using the method of Benjamini and Yekutieli to control the false discovery rate. All models were fitted using the limma package in R.32
Survival analysis
The clinical endpoints in this study were based on the following definitions: disease-specific survival (DSS, time from surgical resection to the date of death from lung cancer); overall survival (OS, time from resection to date of death from any cause); and time to recurrence (TTR, time from resection to date of locoregional or distant relapse). Survival differences were compared using the log-rank test and plotted by the Kaplan–Meier method. To model the associations between survival and confounders, multivariable Cox regression models were calculated, using a stepwise backward conditional method, with 0.10 and 0.05 as entry and exit points, for variable selection.
Statistics
The correlation between TLS count and clinicopathological variables was evaluated using either Fisher’s exact or χ2 tests. Spearman’s correlation coefficients were calculated to examine the associations between two continuous variables. Inter-observer agreements were assessed using a two-way random-effects model with an absolute agreement definition and Cohen’s kappa coefficient with equal weights. Passing–Bablok regression and Bland–Altman analyses were performed for inter-observer variability on continuous scale variables. A Kruskal–Wallis test was applied for statistical comparisons across pathological stages for a given continuous variable, followed by Dunn–Bonferroni post hoc test. Both the survival and statistical analyses were performed using SPSS and R packages. P values of <0.05 were considered statistically significant.
Results
Variation in distribution of total and mature TLS subsets in tissue and across pathological stages
Clinical and histopathological variables for both cohorts #1 (N = 295) and #2 (N = 258) are presented in Supplementary Table S1. In the overall cohort, including all scoring models, there were no statistically significant correlations between clinicopathological characteristics and patients TLS (or GC + TLS) stratification, except for T stage with semi-quantitative scoring model (Supplementary Table S2).
The majority of TLSs were located in the tumour periphery. A remarkable variation in size and form of the TLSs was noted (Fig. 1a–d). The highest TLS score for the tumour periphery and core regions were 87 (interquartile range, IQR, Q1 = 0, Q2 = 7, Q3 = 14) and 22 (IQR, Q1 = 0, Q2 = 0, Q3 = 2), respectively. A similar finding regarding localisation was observed for GC + TLSs with highest score of 21 (IQR, Q1 = 0, Q2 = 1, Q3 = 3) for tumour periphery and 9 (IQR, Q1 = 0, Q2 = 0, Q3 = 0) for tumour core. However, only 53% of patients had GC + TLSs, while 74% had TLSs—they were considered positive if more than two TLSs were observed in their whole-tissue section slide, based on quantitative scoring model 2.
Interestingly, we observed some cases with an abundance of TLSs and very low CD8+ T cells, and vice versa; that is, high CD8 infiltration with low to no TLSs (Supplementary Figure S2). However, this finding requires further investigation to assess the strength of the association between CD8 infiltration and TLS count.
TLS distribution was evaluated across the TNM stages. Interestingly, the total TLS count decreased from stages II to III, while the frequency of the GC+ TLS phenotype was not significantly changed across TNM stages (Fig. 1e, f). No significant changes were observed with total TLS count or GC+ TLS phenotype comparing main histological subgroups, lung squamous cell carcinoma (LUSC) and adenocarcinoma (LUAD; Supplementary Figure S3).
In order to find out the impact of heterogeneity in size of the tissue with TLS count (scoring models 2 and 3), we measured the area of tissue per slide using image analysis tools (QuPath, v0.2.0-m8, Queen’s University, Belfast, Northern Ireland). In the whole cohort, the median tissue area was 317 mm2 (range: 151–566 mm2). We observed a very weak correlation between TLS absolute count and tissue area (r = 0.13; P = 0.002). Not surprisingly, in regression analysis, very low relationship was detected between tissue size and TLS quantity (R2 = 0.02, Supplementary Figure S4), which may represent that TLS count is independent of tissue area in the slide. Similarly, there was a very low association between GC+ TLS subset and tissue size (R2 = 0.01, data not shown).
B cell and adaptive immunity genes/signatures are significantly associated with high TLS levels
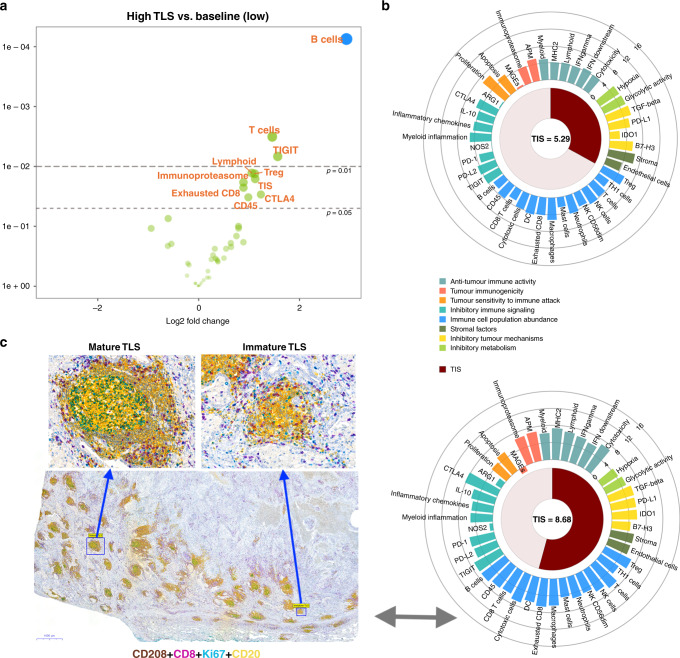
To further characterise the immune cellular composition and signatures, based on low (seven samples, scoring range 0–3) and high (seven samples, scoring range 25–68) TLS IHC stratification, we performed a NanoString assay consisting of 770 immune-specific genes. The assay consists of 46 signatures based on a single gene or metagene setting assessment and have been used in previous publications.33,34 The normalised expression levels of genes and signatures are shown in Supplementary Figure S5. At the metagene level, the predominant immune cell profile in the high TLS group was a B cell signature. In addition to B cells, inhibitory immune signalling (TIGIT and CTLA4) and adaptive immunity signatures (T cells, T regulatory, TIS, lymphoid, CD45, immunoproteasome and exhausted CD8) were upregulated in the high TLS group compared with baseline values (Fig. 2a).
Fig. 2. Relationship between immune gene signatures and tertiary lymphoid structures (TLS).
a Volcano plot of ‘All Signatures’ displays the significance of signature difference between high TLS group vs. baseline, low TLS group. Each dot represents a signature, the x-axis shows the log 2 fold change and the y-axis shows log 2 fold change P values. Adjusted and unadjusted P values are coloured blue and green, respectively. Signatures that have greater statistical significance produced points that are both larger and darker in hue, in addition to appearing higher on the plot. b Examples of low (upper) vs. high (lower) TIS scores corresponding to TLS status. The wheel plot depicts the relative expression of each signature for two samples selected from high and low TLS groups. Signatures are grouped based on the biological process in which they belong, and the TIS score is shown as a radial arc around its numeric score. c The panel represents: first, the tissue landscape with regards to TLS frequency from the same individual case with high TIS score; second, an example of multiplex IHC thath was used for differentiation of morphologically mature (GC+) from immature (GC−) TLSs on 20 samples—in order to validate the scoring model 3. Well-formed TLSs showing CD208+ mature dendritic cells (brown), CD8+ T cells (purple), Ki67+ proliferating cells (teal) and CD20+ B cells (yellow). Germinal centre positivity account for areas with B cells clusters with proliferation activity, as appeared with tertiary green colour. GC+ TLSs were easily differentiated from immature TLSs with very low magnification (×1).
In order to identify gene expression patterns associated with the TIS score across the high vs. low TLS patients, a weighted linear combination of the 18 genes algorithm was computed.35 Figure 2b shows the lowest and highest TIS scores in this subset of patients. Interestingly, the median TIS score was significantly different in the low vs. high TLS groups (6.4 ± 2.4 vs. 7.6 ± 1.8; t test P = 0.02, Supplementary Figure S6). No significant difference was detected for TIS score across main histological subtypes (Supplementary Figure S6).
Accuracy of identifying GC+ TLS phenotypes and inter-observer and inter-method concordance with scoring models
To rule out the subjectivity with respect to the differentiation of GC+ TLS from immature TLS by CD8/CK, a broader panel of TLS-specific antigens were immunostained. The multiplexed IHC consisted of Ki67 (proliferation), DC-LAMP/CD208 (mature DC), CD8 (cytotoxic T cell) and CD20 (B cell) (Fig. 2c). This approach was tested in 20 tissue samples, randomly selected from the high-scoring GC+ TLS population. Significant concordance was observed between the paired section scores for GC+ TLS (Spearman’s rho, r = 0.77, P < 0.001). Likewise, there was a high correlation between the paired section scores (4-plex vs. 2-plex IHC) for total TLS counts (r = 0.82, P < 0.001). Further, we observed very low numbers of CD208+ DCs in lymphoid aggregates, even in mature TLS subpopulations. This was confirmed by DC-LAMP/CD208 single staining.
The semi-quantitative scoring model (1) revealed an excellent inter-observer agreement (intraclass correlation coefficient = 0.97, kappa = 0.84). For the scoring models (2 and 3) with TLS as a continuous scale, there was also an excellent agreement between the two scorers, with a greater consensus on the tumour periphery than the tumour core (Supplementary Table S3). Bland–Altman analysis confirmed this correlation for scoring models 2 and 3, and pointed to some degree of bias toward under-scoring the number of GC+ TLS (Supplementary Figure S7). However, Passing–Bablok regression analysis confirmed a good agreement, with no major intercept or slope shift between observers for the GC+ TLS scores (data not shown). In addition, estimated inter-method correlation coefficients confirmed a strong correlation between scoring models 1 and 2 (r = 0.92; 95% confidence interval (CI) 0.86–0.94, P < 0.001).
Total TLS and GC+ TLS subset scores are robust predictors of survival, regardless of the cohort, scoring strategy, clinical endpoint and histological subtype
Univariable analyses
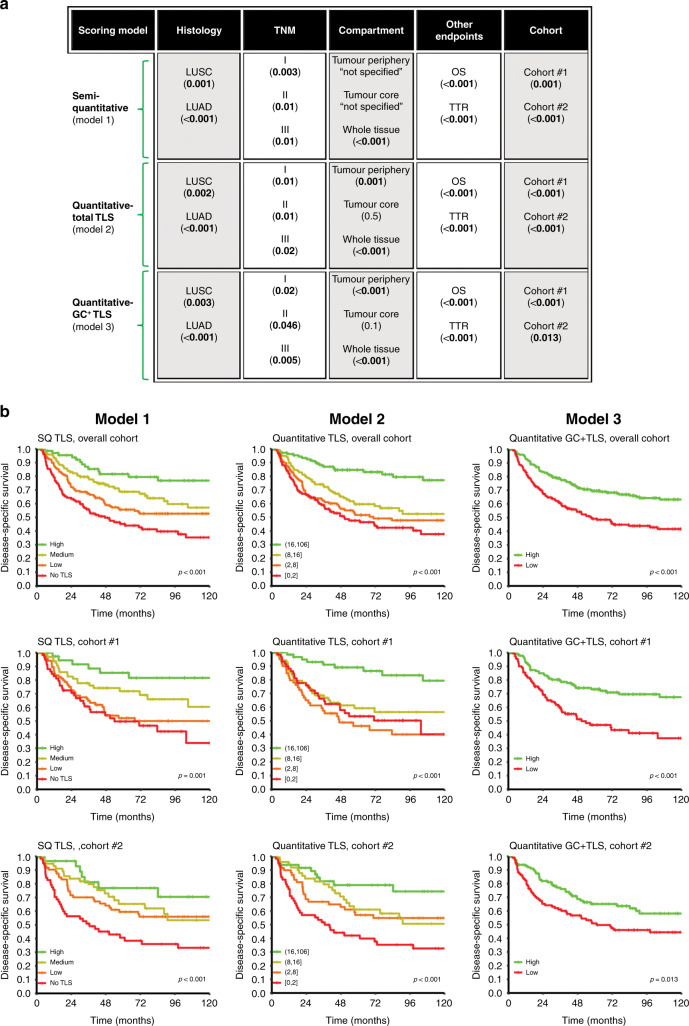
Generally, all three models showed TLS to be a significant positive prognostic factor. The prognostic impact of TLS score was evaluated in both separate and merged analyses of cohort #1 and #2 (Supplementary Table S4). Figure 3a presents a glance view of the positive prognostic impact of TLS in the overall cohort using different scoring models, including other fundamental variables. In scoring models 2 and 3, no advantage on results was observed when TLSs were scored separately based on regional localisation (periphery vs. core). Therefore, in models 2 and 3, the combined tumour periphery and core scores were used.
Fig. 3. Tertiary lymphoid structures (TLS) score and clinical outcome.
Glance overview on the positive prognostic impact of TLS in the overall cohort of NSCLC by different (semi)-quantification methods and stratification based on cohorts, other endpoints (OS overall survival, TTR time to recurrence), compartment, histology (LUSC lung squamous cell carcinoma, LUAD lung adenocarcinoma) and TNM subgroups. Log-rank P value of 5-years disease-specific survival (DSS) in parentheses. b DSS curves of the entire cohort and sub-cohorts (cohort #1 and #2) according to different scoring model of TLS. Left column, semi-quantitative (SQ); middle column, quantitative; right column, for germinal centre-positive subset (GC+ TLS) scoring models. The TLS scores were stratified using quartiles (models 1 and 2) and median cut-off (model 3) as indicated in the figure insets.
In model 1 (using the TLS score as a stepwise discrete variable), increasing survival was seen with elevated TLS levels. In model 2 (quantitative), quartiles were used as a cut-off for survival analysis. Consistent with model 1, high counts of TLS were significantly associated with better DSS in both cohorts. In model 3 (with the median cut-off of 1), we found a similar prognostic association with GC+ TLS phenotypes (Fig. 3).
In the subgroup analysis based on histology, high TLS levels were associated with longer DSS in both LUSC and LUAD histotypes (Supplementary Figure S8). Moreover, all potential cut-points for TLS absolute counts in the overall cohort, and in the LUSC and LUAD subgroups, are visualised in Supplementary Figure S9. The data show that TLS count is significant for all possible cut-points for both histologies. Interestingly, the most discriminating prognostic information was found for LUAD patients.
In addition, when other clinical endpoints were explored, TLS scores were associated with OS and TTR (Supplementary Figure S10).
Multivariable analysis
In the overall cohort, TLS score was found to be an independent positive prognostic factor regardless of the scoring model and endpoints including DSS (Table 1), OS and TTR (data not shown). Consistent with this, the sub-cohort analysis further revealed a significant association between survival and TLS score, by any of the models, independent of any known prognostic factors (Supplementary Table S5).
Table 1.
Cox regression analysis summarising significant independent prognostic factors for DSS in the overall cohort using different scoring models.
| Model 1 (SQ-TLS) | HR (95% CI) | P | Model 2 quantitative TLS) | HR (95% CI) | P | Model 3 (quantitative GC+ TLS) | HR (95% CI) | P |
|---|---|---|---|---|---|---|---|---|
| TLS | <0.001 | <0.001 | ||||||
| No TLS | 1 | 0–2 | 1 | Low | 1 | |||
| Low | 0.7 (0.5–1.05) | 0.09 | 3–8 | 0.9 (0.6–1.3) | 0.8 | High | 0.5 (0.4–0.7) | <0.001 |
| Medium | 0.5 (0.36–0.8) | 0.003 | 9–16 | 0.7 (0.5–1.1) | 0.1 | |||
| High | 0.3 (0.1–0.5) | <0.001 | 17–106 | 0.3 (0.1–0.5) | 0.001 | |||
| TNM | <0.001 | <0.001 | <0.001 | |||||
| I | 1 | 1 | 1 | |||||
| II | 1.6 (1.1–2.3) | 0.01 | 1.6 (1.1–2.3) | 0.009 | 1.5 (1.1–2.2) | 0.03 | ||
| III | 4.02 (2.8–5.7) | <0.001 | 4.2 (2.8–5.7) | <0.001 | 4.1 (2.8–5.7) | <0.001 | ||
| Sex | ||||||||
| Male vs. female | 1.5 (1.05–2.05) | 0.02 | 1.5 (1.1–2.05) | 0.02 | 1.4 (1.03–1.9) | 0.03 | ||
| ECOG | 0.01 | 0.006 | 0.007 | |||||
| 0 | 1 | 1 | 1 | |||||
| 1 | 1.5 (1.1–2.1) | 0.004 | 1.6 (1.2–2.2) | 0.002 | 1.6 (1.2–2.1) | 0.003 | ||
| 2 | 1.6 (0.8–3.03) | 0.1 | 1.7 (0.9–3.2) | 0.1 | 1.6 (0.8–2.9) | 0.1 | ||
| Histology | 0.06 | 0.08 | 0.1 | |||||
| LUSC | 1 | 1 | 1 | |||||
| LUAD | 1.4 (1.06–1.9) | 0.02 | 1.4 (1.03–1.8) | 0.03 | 1.3 (1.02–1.8) | 0.03 | ||
| Others | 1.1 (0.3–4.8) | 0.8 | 1.2 (0.3–4.9) | 0.8 | 0.8 (0.2–3.4) | 0.7 | ||
| Vascular invasion | ||||||||
| Yes vs. no | 1.9 (1.3–2.7) | 0.001 | 1.8 (1.3–2.6) | 0.001 | 1.9 (1.4–2.7) | <0.001 |
SQ semi-quantitative, HR hazard ratio, Cl confidence interval, ECOG Eastern Cooperative Oncology Group, LUAD lung adenocarcinoma, LUSC lung squamous cell carcinoma. Significant p-values in bold (threshold p < 0.05).
TLS score as a potential candidate marker for inclusion in the NSCLC TNM-I classifier
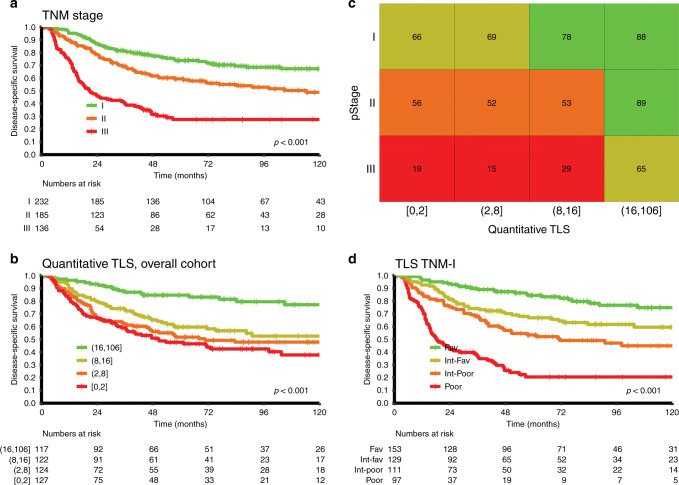
To test the eligibility of TLS as a potential refinement of the TNM system, the overall cohort was stratified according to pStage. With the scoring method in model 2, high TLS scores had a significant prognostic impact in all pStages. Figure 4 demonstrates how an integrated TLS adds greater prognostic accuracy than pStage alone. The 5-year DSS of the four TNM TLS score subgroups were: 84% (favourable, n = 153), 67% (intermediate-favourable, n = 129), 53% (intermediate-poor, n = 111), and 21% (poor, n = 97).
Fig. 4. Inclusion of TLS in NSCLC TNM-Immune cell score (TNM-I) classifier.
a–d Disease-specific survival (DSS) curves of the entire cohort, according to TNM stages (a) and TLS score (from scoring model 2) (b). The combination of A and B results in a TNM-I survival table (c), in which classifying subgroup of patients based on 5-year DSS differences within the same TNM stages. The colour of the squares in the table represent patient subgroups with similar survival (favourable with green, intermediate-favourable with moss green, intermediate-poor with orange and poor prognosis with red). d Kaplan–Meier survival curves of TNM-I classifier illustrating the significant prognostic value of TLS-derived Immune score across each pathological stage.
Discussion
This study incorporates several factors in the histological evaluation of TLSs in NSCLC whole-tumour tissue sections. First, three different scoring models for manual TLS (semi)-quantification were devised and applied. Second, TLS score became an independent prognostic factor regardless of scoring model, endpoint, cohort and histology (LUSC and LUAD). Third, the TLS score increases prognostic power in each pathological stage and therefore has the potential to refine TNM staging in resected NSCLC.
The clinical relevance of TLSs in a variety of solid cancers has been extensively documented.36 Supplementary Table S6 summarises previous studies that assessed the prognostic impact of TLS in NSCLC. Despite the heterogeneity of the methods applied for TLS evaluation, most of the studies concluded that a high TLS count is associated with a favourable prognosis, in line with our present study. The methodologies varied in factors such as the choice of IHC marker (and on gene signatures), the use of semi-quantitative (rather than fully quantitative) analysis of TLS, and the lack of descriptions of histological criteria for TLS evaluation. For example, Silina et al.24 showed that superior outcomes in NSCLC were associated with high TLS counts, using a full-quantification approach in H&E-stained slides, while Buisseret et al.37 presented TLS to be more accurately and consistently scored (with high inter-observer reliability), in breast cancer, using IHC but not with H&E staining.37 The classical method for TLS detection is IHC staining specifically for mature DCs (CD208). Although a widely used marker, a relatively low number of CD208+ DCs reside in TLSs compared to other relevant immune cell subsets,23,38 also in line with our observations. Thus, it is challenging to identify TLSs precisely. In this study, we labelled CD8 for easier morphological TLS identification as used in a previous report.39 An advantage with this method is that, to date, in situ CD8+ cytotoxic T cells have been proved to have the strongest prognostic impact of the immune cell subpopulations in various malignancies. There is an ongoing worldwide effort (Immunoscore®) to implement this knowledge clinically.2,40 As well as its use for quantifying CD8 within the Immunoscore® or TNM-I framework, TLSs can also be easily identified and integrated into this model without requiring extra staining protocols.
We quantified TLS with three different, easily reproducible scoring models. All three correlated well with inter-observer agreements. As expected, the quantitative models (2 and 3) showed consistent results between observers, even though a good correlation was found between the two quantitative (model 2) and semi-quantitative (model 1) methods. In addition, digital pathology has recently shown real promise in translational medicine.41 Obviously, using an estimate of TLS as a continuous variable is more suitable for machine-learning models, overcoming the limitations associated with manual scoring. Further, the results we obtained from scoring model 3 were in line with a previous report on survival benefit with mature TLS phenotypes.24 We suggest that different quantification strategies like those proposed and elaborated on in this study may help to reduce confusion, improve the inter-observer reproducibility and reliability, and provide a better prognostic accuracy for TLSs in NSCLC.
One striking finding was that the frequency of total TLSs significantly drops in pathological stage III. This supports the notion that during cancer progression, tumour cells evade the immune response particularly via the dysregulation of specific chemokine pathways that promote the formation of TLS.42 On this basis, it would be interesting to further elucidate which genomic aberrations or transcriptomic profile losses impact on TLS scarcity in the more advanced stages of cancer.
Although application of IHC, chromogen or immunofluorescence is specific and sensitive for reliable identification of TLSs, some reports propose various TLS-specific gene expression profiles. These mainly relate to adaptive immune cell lineages and chemokines that link to TLS formation and maturation.22 However, in this context, there is a need to determine which transcriptomic signature(s) are the most reliable and technically reproducible for detecting TLSs. We detected a significant upregulation of 21 genes corresponding to B and T cell and co-inhibitory receptor signatures (CTLA4, TIGIT) in the high TLS patient group. Consistent with this, B cell signatures were previously used for TLS detection.13,15,43 A plausible explanation underlying the expression of immune checkpoint-related genes is that TLS-rich tumours are more infiltrated by CD8+ T cells. These T cells can become exhausted, explaining the correlation of the expression of immune checkpoints (such as CTLA4 and TIGIT) with TLSs.15 Further, Ayers et al.35 proposed a specific signature called TIS, which is well-validated in FFPE tissue as a predictive marker in patients undergoing anti-PD-1/PD-L1 therapy. This signature contains genes related to interferon signalling, antigen-presenting cells, natural killer and T cell abundance and inhibitory pathways.35,44 Our study found a significant association between TIS score and the presence of TLSs, suggesting that TLS scoring could be a potential alternative marker for differentiating hot (T cell-inflamed) from cold (non-T cell-inflamed) tumour phenotypes.44 Taken together, from the result of our RNA data on the rather small size sample, B cell signature or immunogenic hot/cold models, derived from TIS score, may reflect the high/low TLS status at the tissue level, and could therefore be used as a surrogate biomarker for TLS. Indeed, further investigations in larger cohorts are warranted on this basis.
In NSCLC and colorectal cancer, different immune-based tumour classifications have shown the potential to be an important supplement to TNM staging.4,5 In this study, the subgroup analysis based on TNM stage revealed that high TLS counts were a significant prognostic factor within all pathological stages, regardless of the scoring model. The TLS score revealed considerable and significant differences in outcome between the four score groups (none, low, moderate and high) and was in the multivariate analysis a robust indicator for improved prognosis—using scoring model 2. In addition, the results were consistent when scoring models 1 and 3 tested for TNM-I stratification (data not shown). In scoring model 2, the most robust form of cut-off was applied, in which patients grouped based on four quantiles derived from a continuous measure, rather than arbitrary dichotomisation. Advantage of using the quartile cut-point is preservation from risk of type I and II errors and is more valid and reliable for potential application into the routine. Considering scoring model 2 was performed independent of tissue size, and notably almost no association was found between TLS density and tissue area in our resected NSCLC cohort (Supplementary Figure S4). Clearly, larger studies need to be undertaken for validation of this scoring method. One major weakness of our study is the retrospective nature of collected samples, since the cohort predates immune checkpoint blockade therapy that is now available. Likely, the prognostic impact of tumour microenvironment factors such as TLSs may interact with those of immune-modulating therapy, so the results require validation in a contemporary cohort. But notably, our research team is currently conducting a prospective multi-institutional clinical study (NCT03299478) that aims to implement the NSCLC TNM-I classifier into routine practice. Our results suggest that TLS has as much potential as CD8 for further validation and most likely integration into a NSCLC TNM-I model.
In conclusion, this study provides a detailed description of the histological assessment of TLS in a sizeable number of NSCLC patients from two independent cohorts. We found the TLS score to be a robust and independent positive prognosticator, irrespective of scoring model, and hence being a promising candidate to refine the TNM staging in NSCLC.
Supplementary information
Acknowledgements
We thank Dr. Wenjie Xu at NanoString Data Analysis Service (DAS) and Anette Skogstad at Molecular Pathology Lab (UNN) for NanoString analysis assistance.
Author contributions
Conception and design: M.R. and T.D. Methodology (tissue processing, mIHC and NanoString): M.R., T.B. and M.B. Methodology (histopathology and development of immune scoring): M.R., L.-T.R.B., S.A.-S., F.P., E.R. and S.J. Clinical annotations including follow-up and tumour restaging: S.A., T.D., M.P., R.M.B. and E.-E.P. Analysis and interpretation of data: M.R. and T.K.K. Data visualisation: M.R., D.J.K. and T.K.K. Writing, review and editing: all authors. Project administration, ethical approval and funding acquisition: T.D., L.-T.R.B. and R.M.B.
Ethics approval and consent to participate
This study was performed in accordance with the Declaration of Helsinki. The study was ethically approved by the Regional Committee (REK) and Norwegian Data Protection Organization (REK 2016/714). The ethics committee waived the need for patient’s consent in this retrospective study.
Consent to publish
This manuscript does not contain any individual person’s data.
Data availability
The datasets used and analysed during this study are available from the corresponding author on reasonable request, and most of the original data are included within the article and its Supplementary Materials.
Competing interests
The authors declare no competing interests.
Funding information
This work was financially supported by Norwegian Cancer Society and the Northern Norway Health Region Authority. The funders had no role in study design, data collection/analysis, decision to publish and preparation of the manuscript.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-021-01307-y.
References
- 1.Goldstraw P, Crowley J, Chansky K, Giroux DJ, Groome PA, Rami-Porta R, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM classification of malignant tumours. J. Thorac. Oncol. 2007;2:706–714. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- 2.Fridman WH, Zitvogel L, Sautès–Fridman C, Kroemer G, Sautès-Fridman C, Kroemer G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 2017;25:717–734. doi: 10.1038/nrclinonc.2017.101. [DOI] [PubMed] [Google Scholar]
- 3.Bremnes RM, Busund L-T, Kilvær TL, Andersen S, Richardsen E, Paulsen EE, et al. The role of tumor-infiltrating lymphocytes in development, progression, and prognosis of non-small cell lung cancer. J. Thorac. Oncol. 2016;11:789–800. doi: 10.1016/j.jtho.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Pagès F, Mlecnik B, Marliot F, Bindea G, Ou FS, Bifulco C, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391:2128–2139. doi: 10.1016/S0140-6736(18)30789-X. [DOI] [PubMed] [Google Scholar]
- 5.Donnem T, Kilvaer TK, Andersen S, Richardsen E, Paulsen EE, Hald SM, et al. Strategies for clinical implementation of TNM-immunoscore in resected nonsmall-cell lung cancer. Ann. Oncol. 2016;27:225–232. doi: 10.1093/annonc/mdv560. [DOI] [PubMed] [Google Scholar]
- 6.Donnem T, Hald SM, Paulsen E-E, Richardsen E, Al-Saad S, Kilvaer TK, et al. Stromal CD8+ T-cell density—a promising supplement to TNM staging in non-small cell lung cancer. Clin. Cancer Res. 2015;21:2635–2643. doi: 10.1158/1078-0432.CCR-14-1905. [DOI] [PubMed] [Google Scholar]
- 7.Paulsen E-E, Kilvaer T, Khanehkenari MR, Maurseth RJ, Al-Saad S, Hald SM, et al. CD45RO(+) memory T lymphocytes-a candidate marker for TNM-immunoscore in squamous non-small cell lung cancer. Neoplasia. 2015;17:839–848. doi: 10.1016/j.neo.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rakaee M, Kilvaer TK, Dalen SM, Richardsen E, Paulsen E-E, Hald SM, et al. Evaluation of tumor-infiltrating lymphocytes using routine H&E slides predicts patient survival in resected non-small cell lung cancer. Hum. Pathol. 2018;79:188–198. doi: 10.1016/j.humpath.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 9.Rakaee M, Busund LTR, Jamaly S, Paulsen EE, Richardsen E, Andersen S, et al. Prognostic value of macrophage phenotypes in resectable non–small cell lung cancer assessed by multiplex immunohistochemistry. Neoplasia. 2019;21:282–293. doi: 10.1016/j.neo.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paulsen E-E, Kilvaer TK, Rakaee M, Richardsen E, Hald SM, Andersen S, et al. CTLA-4 expression in the non-small cell lung cancer patient tumor microenvironment: diverging prognostic impact in primary tumors and lymph node metastases. Cancer Immunol. Immunother. 2017;66:1449–1461. doi: 10.1007/s00262-017-2039-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paulsen E-E, Kilvaer TK, Khanehkenari MR, Al-Saad S, Hald SM, Andersen S, et al. Assessing PDL-1 and PD-1 in non-small cell lung cancer: a novel immunoscore approach. Clin. Lung Cancer. 2017;18:220–233.e8. doi: 10.1016/j.cllc.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Rakaee M, Busund L-T, Paulsen E-E, Richardsen E, Al-Saad S, Andersen S, et al. Prognostic effect of intratumoral neutrophils across histological subtypes of non-small cell lung cancer. Oncotarget. 2016;7:72184–72196. doi: 10.18632/oncotarget.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helmink BA, Reddy SM, Gao J, Zhang S, Basar R, Thakur R, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020;577:549–555. doi: 10.1038/s41586-019-1922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cabrita R, Lauss M, Sanna A, Donia M, Skaarup Larsen M, Mitra S, et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature. 2020;577:561–565. doi: 10.1038/s41586-019-1914-8. [DOI] [PubMed] [Google Scholar]
- 15.Petitprez F, de Reyniès A, Keung EZ, Chen TWW, Sun CM, Calderaro J, et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature. 2020;577:556–560. doi: 10.1038/s41586-019-1906-8. [DOI] [PubMed] [Google Scholar]
- 16.Moyron-Quiroz JE, Rangel-Moreno J, Kusser K, Hartson L, Sprague F, Goodrich S, et al. Role of inducible bronchus associated lymphoid tissue (iBALT) in respiratory immunity. Nat. Med. 2004;10:927–934. doi: 10.1038/nm1091. [DOI] [PubMed] [Google Scholar]
- 17.Randall, T. D. Bronchus-associated lymphoid tissue (BALT). Structure and function. Adv. Immunol.107, 187–241 (2010). [DOI] [PMC free article] [PubMed]
- 18.Pitzalis, C., Jones, G. W., Bombardieri, M. & Jones S. A. Ectopic lymphoid-like structures in infection, cancer and autoimmunity. Nat. Rev. Immunol. 14, 447–462 (2014). [DOI] [PubMed]
- 19.Victora GD, Nussenzweig MC. Germinal centers. Annu. Rev. Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 20.Bruno, T. C. New predictors for immunotherapy responses sharpen our view of the tumour microenvironment. Nat. Res.577, 474–476 (2020). [DOI] [PMC free article] [PubMed]
- 21.Teillaud, J. L. & Dieu-Nosjean, M. C. Tertiary lymphoid structures: an anti-tumor school for adaptive immune cells and an antibody factory to fight cancer? Front. Immunol.8, 830 (2017). [DOI] [PMC free article] [PubMed]
- 22.Sautès-Fridman, C., Petitprez, F., Calderaro, J. & Fridman, W. H. Tertiary lymphoid structures in the era of cancer immunotherapy. Nat. Rev. Cancer19, 307–325 (2019). [DOI] [PubMed]
- 23.Sautès-Fridman, C., Lawand, M., Giraldo, N. A., Kaplon, H., Germain, C., Fridman, W. H. et al. Tertiary lymphoid structures in cancers: prognostic value, regulation, and manipulation for therapeutic intervention. Front. Immunol. 7, 407 (2016). [DOI] [PMC free article] [PubMed]
- 24.Silina K, Soltermann A, Attar FM, Casanova R, Uckeley ZM, Thut H, et al. Germinal centers determine the prognostic relevance of tertiary lymphoid structures and are impaired by corticosteroids in lung squamous cell carcinoma. Cancer Res. 2018;78:1308–1320. doi: 10.1158/0008-5472.CAN-17-1987. [DOI] [PubMed] [Google Scholar]
- 25.Goc J, Germain C, Vo-Bourgais TKD, Lupo A, Klein C, Knockaert S, et al. Dendritic cells in tumor-associated tertiary lymphoid structures signal a th1 cytotoxic immune contexture and license the positive prognostic value of infiltrating CD8+ t cells. Cancer Res. 2014;74:705–715. doi: 10.1158/0008-5472.CAN-13-1342. [DOI] [PubMed] [Google Scholar]
- 26.Goldstraw P, Chansky K, Crowley J, Rami-Porta R, Asamura H, Eberhardt WEE, et al. The IASLC lung cancer staging project: proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM Classification for lung cancer. J. Thorac. Oncol. 2016;11:39–51. doi: 10.1016/j.jtho.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 27.Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, et al. The 2015 World Health Organization Classification of lung tumors. J. Thorac. Oncol. 2015;10:1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 28.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumor MARKer prognostic studies (REMARK) Breast Cancer Res. Treat. 2006;100:229–235. doi: 10.1007/s10549-006-9242-8. [DOI] [PubMed] [Google Scholar]
- 29.Eustace A, Smyth LJC, Mitchell L, Williamson K, Plumb J, Singh D. Identification of cells expressing IL-17A and IL-17F in the lungs of patients with COPD. Chest. 2011;139:1089–1100. doi: 10.1378/chest.10-0779. [DOI] [PubMed] [Google Scholar]
- 30.Kuroda E, Ozasa K, Temizoz B, Ohata K, Koo CX, Kanuma T, et al. Inhaled fine particles induce alveolar macrophage death and interleukin-1α release to promote inducible bronchus-associated lymphoid tissue formation. Immunity. 2016;45:1299–1310. doi: 10.1016/j.immuni.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 31.Schaadt, N. S., Schönmeyer, R., Forestier, G., Brieu, N., Braubach, P., Nekolla, K. et al. Graph-based description of tertiary lymphoid organs at single-cell level. PLoS Comput. Biol.16, e1007385 (2020). [DOI] [PMC free article] [PubMed]
- 32.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Damotte D, Warren S, Arrondeau J, Boudou-Rouquette P, Mansuet-Lupo A, Biton J, et al. The tumor inflammation signature (TIS) is associated with anti-PD-1 treatment benefit in the CERTIM pan-cancer cohort. J. Transl. Med. 2019;17:1–10. doi: 10.1186/s12967-019-2100-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asleh K, Brauer HA, Sullivan A, Lauttia S, Lindman H, Nielsen TO, et al. Predictive biomarkers for adjuvant capecitabine benefit in early-stage triple-negative breast cancer in the FinXX Clinical Trial. Clin. Cancer Res. 2020;26:2603–2614. doi: 10.1158/1078-0432.CCR-19-1945. [DOI] [PubMed] [Google Scholar]
- 35.Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dieu-Nosjean M-C, Giraldo NA, Kaplon H, Germain C, Fridman WH, Sautès-Fridman C. Tertiary lymphoid structures, drivers of the anti-tumor responses in human cancers. Immunol. Rev. 2016;271:260–275. doi: 10.1111/imr.12405. [DOI] [PubMed] [Google Scholar]
- 37.Buisseret L, Desmedt C, Garaud S, Fornili M, Wang X, Van Den Eyden G, et al. Reliability of tumor-infiltrating lymphocyte and tertiary lymphoid structure assessment in human breast cancer. Mod. Pathol. 2017;30:1204–1212. doi: 10.1038/modpathol.2017.43. [DOI] [PubMed] [Google Scholar]
- 38.Dieu-Nosjean, M. C., Goc, J., Giraldo, N. A., Sautès-Fridman, C. & Fridman W. H. Tertiary lymphoid structures in cancer and beyond. Trends Immunol.35, 571–580 (2014). [DOI] [PubMed]
- 39.Remark R, Lupo A, Alifano M, Biton J, Ouakrim H, Stefani A, et al. Immune contexture and histological response after neoadjuvant chemotherapy predict clinical outcome of lung cancer patients. Oncoimmunology. 2016;5:e1255394. doi: 10.1080/2162402X.2016.1255394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galon, J., Mlecnik, B., Bindea, G., Angell, H. K., Berger, A., Lagorce, C. et al. Towards the introduction of the “immunoscore” in the classification of malignant tumours. J. Pathol.232, 199–209 (2014). [DOI] [PMC free article] [PubMed]
- 41.Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019;16:703–715. doi: 10.1038/s41571-019-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pimenta E, Barnes B. Role of tertiary lymphoid structures (TLS) in anti-tumor immunity: potential tumor-induced cytokines/chemokines that regulate TLS formation in epithelial-derived cancers. Cancers. 2014;6:969–997. doi: 10.3390/cancers6020969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hennequin A, Derangère V, Boidot R, Apetoh L, Vincent J, Orry D, et al. Tumor infiltration by Tbet+ effector T cells and CD20+ B cells is associated with survival in gastric cancer patients. Oncoimmunology. 2016;5:e1054598. doi: 10.1080/2162402X.2015.1054598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Danaher P, Warren S, Lu R, Samayoa J, Sullivan A, Pekker I, et al. Pan-cancer adaptive immune resistance as defined by the tumor inflammation signature (TIS): results from The Cancer Genome Atlas (TCGA) J. Immunother. Cancer. 2018;6:63. doi: 10.1186/s40425-018-0367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analysed during this study are available from the corresponding author on reasonable request, and most of the original data are included within the article and its Supplementary Materials.