Abstract
In recent years, many studies have shown that the intestinal microflora has various effects that are linked to the critical physiological functions and pathological systems of the host. The intestinal microbial community is widely involved in the metabolism of food components such as protein, which is one of the essential nutrients in diets. Additionally, dietary protein/amino acids have been shown to have had a profound impact on profile and operation of gut microbiota. This review summarizes the current literature on the mutual interaction between intestinal microbiota and protein/amino acid metabolism for host mucosal immunity and health.
Keywords: Gut microbiota, Amino acid, Metabolism, Mucosal immunity, Host
1. Introduction
The gastrointestinal tract (GIT) harbors large amounts of microorganisms, which play an essential role in many critical physiological host functions (Lin et al., 2017), including metabolic and nutritional homeostasis, maturation and stimulation of the immune system, and brain activity (Agus et al., 2018). The gut microbiota performs an important role in facilitating the course of protein/amino acid digestion and absorption, by decomposing complex subunits and changing the metabolic mechanisms of the host cell. This renders the nutrients easily absorbable by the host and subsequently changes the metabolic mechanisms of the host cell (Armstrong et al., 2019; Ibrahim and Anishetty, 2012). Additionally, bacteria colonizing the intestine has the capacity to facilitate the de novo synthesis of essential amino acids, which are implicated in amino acid homeostasis in the host (Collins et al., 2012).
The profile of the gut microbiota is influenced by numerous factors, including host-based factors and environmental factors. One of fundamental factors is the host diet (Rist et al., 2013). Previous studies have shown that moderating dietary protein/amino acids is a strategic approach for the control of amino acid-fermenting bacterial species and their metabolic pathways, which in turn could have an impact on the metabolism of the host (Shen et al., 2010; Rist et al., 2013; Vital et al., 2014).
The purpose of the present review is to outline the complex interaction between gut microbiota and dietary protein/amino acids for host mucosal immunity and health.
2. Regulation of intestinal microorganisms on protein/amino acid metabolism and synthesis
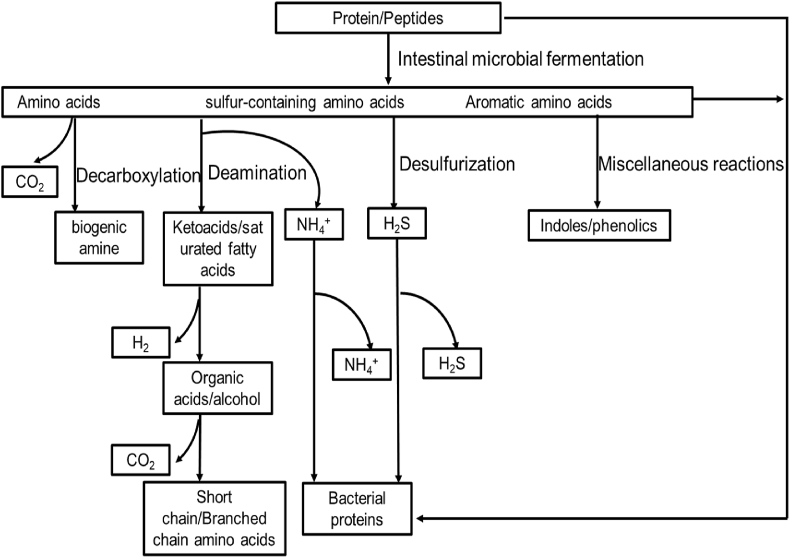
The intestinal flora regulates various metabolic pathways in the host, including those related to glucose, lipid, and lipid metabolism. Bacterial metabolites in the GIT are extremely complicated. Many metabolites produced by the intestinal microflora use amino acids as substrates (Wikoff et al., 2009). The amino acids present in food or those synthesized by the host can provide nutrition for the intestinal flora for protein synthesis. Amino acids can be directly incorporated into bacterial cells as amino acid residues in proteins and enter the catabolic pathway. Proteins are hydrolyzed into amino acids and peptides by extracellular proteases and peptidases secreted by intestinal microorganisms. When specific transporters are present, amino acids and peptides can enter microbial cells (Gottschalk, 1979). However, these biomolecules can meet different fates according to the physiological conditions of the body that they encounter (Fig. 1; Davila et al., 2013). The first step in amino acid catabolism is transamination or deamination; amino acids undergo oxidation, reduction, or both (redox). Metabolic ammonia can be used either as a nitrogen source by microorganisms or excreted (Blachier et al., 2007). Deaminases and decarboxylases are enzymes involved in amino acid metabolism. Amino acids are metabolized into biogenic amines by a decarboxylation reaction. Davila et al. (2013) describe many complex amino acids, such as aromatic amino acids that are resistant to the enzymes stated above, which can be metabolized through a series of reactions such as fission, deamination, decarboxylation, oxidation, and reduction to produce a variety of structurally related indoles and phenols.
Fig. 1.
Protein catabolism pathway of colonic microorganisms. Amino acids produced by microbial metabolism can be directly used by microorganisms to synthesize bacterial protein, and can also enter the catabolic pathway, which is the primary metabolic mode of amino acids in the colon. In microbial metabolism, most amino acids produce corresponding ketoacids or saturated fatty acids through transamination or deamination such as short-chain fatty acids, organic acids, ethanol, H2, and CO2. Some amino acids are metabolized into biogenic amines by decarboxylation. Many complex amino acids, such as aromatic amino acids, do not occur only in the above general reactions. They can be metabolized by a series of reactions such as fission, deamination, decarboxylation, oxidation, and reduction to produce various indoles and phenols.
Mafra et al. (2013) reported that Bacteroides and Propionibacterium were the main proteolytic bacteria in fecal samples; Clostridium, Streptococcus, Staphylococcus, and Bacillus are also common proteolytic bacteria. Colonic microorganisms decompose proteins and peptides to produce a variety of end products, including short-chain fatty acids (SCFA), ammonia, amines, phenols, indoles, mercaptans, carbon dioxide, hydrogen, and hydrogen sulfide; many of which are toxic (Mafra et al., 2013). The production of these metabolites is generally considered a marker for evaluating the level of protein fermentation in the colon. In addition, colonic flora has been reported to accelerate the metabolism of undigested proteins and the content of bacterial metabolites, including amino acids, phenols, indoles, hydrogen sulfide, and branched-chain fatty acids (isobutyrate, isovalerate, and 2-methylbutyrate), which are increased in the colon epithelium (Thom and Lean, 2017).
The amino acids produced or metabolized by bacteria can be used by the host, and these may compensate for the lack of essential amino acid in a low-quality protein diet. A genome-wide analysis has shown that Clostridium perfringens lacks a number of genes for the biosynthesis of amino acids like glutamic acid, arginine, histidine, lysine, methionine, serine, and threonine as well as aromatic and branched-chain amino acids (Portune et al., 2016), whereas other Clostridium species such as Clostridium acetobutylicum have a complete set of genes for amino acid biosynthesis (Nölling et al., 2001). Therefore, it is necessary to understand the pathways for the biosynthesis of amino acids in microorganisms in order to elucidate the functions of different amino acids according to bacterial species. The metabolic cost of amino acid synthesis is as high as the carbon skeleton of all amino acids that come from common metabolic intermediates, which are involved in the tricarboxylic acid cycle, pentose phosphate pathway, and glycolysis (Mu et al., 2015). However, it has also been reported that the residual dietary proteins that reach the large intestine can provide nitrogen and amino acids for the growth and fermentation of glycolytic bacteria (Mafra et al., 2013). Gamma aminobutyric acid is a neuroactive substance produced by several Lactobacillus and Bifidobacterium strains (Barrett et al., 2012). Lactobacillus reuteri can produce histamine (Hemarajata et al., 2014), and both bacteria and fungi produce acetylcholine. In addition, tryptophan decarboxylase was also detected in the human intestinal flora, indicating that tryptophan may be produced by the intestinal flora (Marcel et al., 2017).
3. Effects of dietary protein/amino acid metabolism on intestinal microorganisms
Many factors regulate the composition and physiological functions of microbiota; one of the main factors that leads to an imbalance in the composition of the intestinal microbiota is diet (Moszak et al., 2020). Some suggest that variations in dietary protein cause changes in the composition and function of intestinal flora (Wang et al., 2020). A high-protein diet (HPD) has an important effect on the composition of intestinal microflora. Butyric acid-producing bacteria, which are estimated to account for 2% to 15% of the total bacteria, are found in Lactobacillaceae (Clostridium XIVa). In one study, the abundance of these bacteria decreased significantly in human volunteers who were given an HPD, and their abundance was significantly correlated with fecal butyric acid levels (Duncan et al., 2007). An increased intake of dietary protein has been reported to decrease fecal microbial diversity in most mice but to increase the microbial diversity of milk microbial and pup cecum, and also increase the number of potentially health-promoting bacteria, such as Lactobacillus spp. (Warren et al., 2019). A high-protein diet changed the composition of intestinal microbiota as the abundance of beneficial bacteria (such as Bifidobacterium or Rothia) decreased (Salonen et al., 2014). In a study by Mu et al. (2017), adult rats were fed a regular protein diet or HPD for six weeks, and their feces were collected at weekly intervals and analyzed for microbiological composition. Compared with the fecal samples of the rats which were fed a regular protein diet, the fecal samples of those fed an HPD had an increased number of Escherichia coli but decreased number of Akermania mucilaginosa, Bifidobacterium, Prevotella, and Brucella. Moreover, those fed an HPD had reduced concentrations of acetate, propionate, and butyrate in their feces. In previous studies, an HPD was reported to increase the number of Clostridium species in the colon of piglets and the number of E. coli, Shigella, and Streptococcus, and reduce the number of Rumen cocci, Ackermann's bacteria, and Faecalbacterium prausnitzii in the colon of rats, but reduce the number of Rose sac/Eubacterium in human feces (Pi et al., 2020; Wang et al., 2020).
In addition, protein or amino acid restriction and amino acid supplementation also changed the composition of intestinal microbiota. Wang et al. (2019) reported that low-protein diets with casein hydrolysate reduced the concentrations of putrescine, phenol, and indole compounds in the colonic digesta of pigs and the relative abundance of L. reuteri in the colon of pigs at the species level, when compared with diets added with free amino acids. Lysine restriction enhanced the relative abundances of Actinobacteria, Saccharibacteria, and Synergistetes abundances in the ileum of piglets at the phylum level and Moraxellaceae, Halomonadaceae, Shewanellaceae, Corynebacteriaceae, Bacillaceae, Comamonadaceae Microbacteriaceae, Caulobacteraceae, and Synergistaceae in the ileum at the family (Yin et al., 2017). Yin et al. (2018) found that dietary lysine restriction enhanced the relative abundances of Escherichia-Shigella, Aquabacterium, and Candidatus Methylomirabilis in the ileum of piglets at the genus level, while reducing the abundance of Streptococcus, Bacteroides, Bacillus, Pasteurella, Clostridium sensu stricto, Faecalibacterium, Paucisalibacillus, and Lachnoclostridium. A low-protein diet decreased the relative richness of Firmicutes in the colon contents of weaned piglets at the phylum level, yet increased the relative richness of Proteobacteria (Wan et al., 2020). In a study by Raza et al. (2020), when antibiotic-treated flies were exposed to 10 °C conditions, the mitochondria in their intestinal cells were severely damaged; the administration of l-arginine and l-proline through microinjection significantly prolonged the survival time of antibiotic-treated flies. These results indicate that intestinal microflora plays a vital role in promoting resistance in the host against stress induced by low temperatures via the stimulation of the arginine and proline metabolic pathways.
4. Immunoregulation of protein/amino acid metabolites on the intestinal mucosa
The function of the gut is complicated in that it can selectively allow nutrients to pass through and enter into the circulatory system and internal environment of the body; on the other hand, it can prevent the invasion of toxins, pathogens, and inflammatory factors (Song et al., 2010). The latter function is also known as the intestinal mucosal barrier function (König et al., 2016). The intestinal mucosal barrier is the body's first barrier against the external environment. A healthy intestinal mucosal barrier is important to ensure adequate nutrition and prevent related intestinal diseases (Yang et al., 2017).
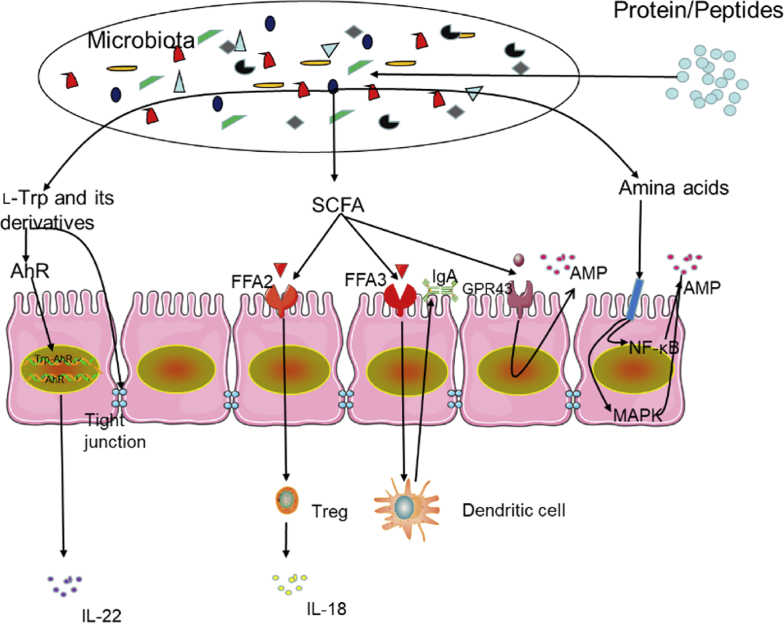
Previous studies have also revealed that bacterial metabolites such as SCFA and aryl hydrocarbon receptor (AHR) ligands benefit the intestinal immune system (Fig. 2). Butyric acid regulates immunity and tissue inflammation by controlling intestinal epithelial cells (IEC), promoting the development of regulatory T cells, enhancing the secretion of mucus by goblet cells, and promoting the flow of IL-18 in T cells (Goldsmith and Sartor, 2014; Koh and Bäckhed, 2020). Free fatty acid receptor-2 (FFA2) and FFA3, which are G protein-coupled receptors, partly mediate the extracellular effects of SCFA. Free fatty acid receptor-2 and FFA3 are essential regulators of intestinal inflammation and the epithelial barrier function. Free fatty acid receptor-2 regulates the development of regulatory T cells; moreover, the impact of FFA2 on dendritic cells can promote the production of intestinal IgA, which provides additional protection for IEC against pathogenic microorganisms (Holota et al., 2019). Different from FFA2, FFA3 (which is expressed in intestinal non-immune cells) enhances the expression of transforming growth factor-α, IL-6, and other cytokines and chemokines mainly through the mitogen-activated protein kinase/extracellular signal-regulated kinase pathway (Kim et al., 2013). Indole derivatives such as indole-3-aldehyde, indole-3-acid-acetic acid, indole-3-propanoic acid, indole-3-acetaldehyde, and indole acrylic acid play an essential role in intestinal homeostasis by participating in IEC renewal, barrier integrity, and many different types of immune cell differentiation (Agus et al., 2018).
Fig. 2.
Immunomodulatory effect of protein/amino acid metabolites on the colon. Tryptophan promotes the expression of the tight junction protein to achieve the intestinal barrier function. Aryl hydrocarbon receptor (AHR), which is a crucial ligand-dependent transcription regulator in the cytoplasm, can bind to tryptophan and its downstream metabolites. The resulting complex is transported to the nucleus, where the AHR is activated. This regulates the proliferation and differentiation of intestinal inflammatory cells and the transcription and expression of inflammatory-related factors (such as interleukin-22, IL-22). Short-chained fatty acids regulate protective immunity and tissue inflammation by controlling intestinal epithelial cells (IEC), promoting the development of regulatory T cells (Treg), enhancing the secretion of mucus by goblet cells, and promoting IL-18 in T cells. Short-chain fatty acid receptor 2 and 3 (FFA2 and FFA3) are essential regulators of intestinal inflammation and epithelial barrier function. Short-chain fatty acid receptor 2 regulates the Treg development. Short-chain fatty acid receptor 2 on dendritic cells can also promote the production of intestinal IgA. Different from FFA2, FFA3 (which is expressed in the intestinal nonimmune cell population) enhances the expression of transforming growth factor-α, IL-6, and other cytokines and chemokines mainly through mitogen-activated protein (MAPK)/extracellular signal-regulated kinases pathway; short-chained fatty acids can promote IEC to produce antimicrobial peptides (AMP) through FFA2. Some amino acids significantly stimulate the expression of β-defensin in epithelial cells by blocking nuclear factor kappa-B (NF-κB) and MAPK inflammatory signaling pathways and activating the mammalian target of rapamycin signaling pathway.
l-Tryptophan, which is used in the nutritional fortification of foods, plays a vital role in maintaining the balance between intestinal immune tolerance and intestinal microbiota maintenance. Recent findings have highlighted the changes in the microbiome that regulate the host's immune system through the regulation of the tryptophan metabolism. Additionally, tryptophan, endogenous tryptophan metabolites, and bacterial tryptophan metabolites have been reported to exhibit profound effects on the host's immune system-intestinal microbiota interaction (Gao et al., 2018). One study showed that l-tryptophan enhanced tight junction protein expression in pig IEC in vitro (Wang et al., 2015). Moreover, the indole derived from l-tryptophan has been reported to increase the resistance of epithelial cells to tight junctions. Indole propionate, which is another bacterial metabolite of tryptophan, can effectively reduce intestinal permeability in rodents (Dodd et al., 2017). Recent studies have found that tryptophan may enhance the intestinal barrier function by promoting the expression of zonula occludens 1 (Dodd et al., 2017). The derivatives of tryptophan such as indoleacetic acid and indole-3-propionic acid are produced by Streptococcus and Clostridium sporogenes, respectively, and regulate the intestinal barrier function via the exogenous pregnane X receptor (Elias et al., 2014). Studies have shown that tryptophan and its downstream metabolites can bind to AHR; the resulting complex is transported into the nucleus, where AHR is activated, and this regulates intestinal inflammation, cell proliferation and differentiation, and the transcription and expression of inflammation-related factors (Lanis et al., 2017).
Antimicrobial peptides (AMP) produced by IEC play a crucial role in regulating intestinal homeostasis by controlling microbial populations. Dietary amino acid supplementation could improve intestinal AMP expression, maintain the homeostasis of intestinal microorganisms, and reduce susceptibility to intestinal inflammation (Hashimoto et al., 2012). As a traditional essential amino acid in mature mammals, arginine is utilized in the synthesis of proteins and many bioactive molecules. l-Arginine alleviates inflammatory response and intestinal injury by blocking the activation of nuclear factor kappa-B and mitogen-activated protein kinase inflammatory signaling pathways, significantly stimulating the expression of β-defensin in epithelial cells by activating the mTOR signaling pathway, and participates in the innate immune response (Lan et al., 2020). Butyrate can promote the production of AMP, such as regenerative islet derived protein III γ and defensin, in the IEC of mice through the SCFA receptor (G protein-coupled receptor 43) (Chen et al., 2020). Butyrate also significantly increases the expression of porcine β-defensin-2 and β-defensin-3 (Zeng et al., 2013).
5. Protein/amino acid metabolites in the progression of gastrointestinal diseases
The host's innate response to the microbiota plays an essential role in the homeostasis of the intestinal immunity and inflammatory bowel disease (IBD) pathogenesis. However, in most model systems, defects in the intrinsic pathway do not cause homeostasis destruction and the development of IBD (Elson and Cong, 2012). The interaction between the host and its rich intestinal microflora is complex; some symbionts, such as myxomycetes and Helicobacter spp., are usually called pathological organisms as they can cause diseases under certain conditions. The composition of the intestinal microbiota has a wide range of inter- and intra-individual variability (Huttenhower et al., 2012), which is considered a key determinant of the host's susceptibility to various diseases, including IBD (Caruso et al., 2020).
Excessive protein fermentation produces metabolites such as amines, hydrogen sulfide, p-cresol, and ammonia, which can damage the colon epithelium (Arumugam et al., 2011). Some of these products may induce DNA damage, intestinal leakage, colon cancer, IBD, and other diseases (Marchesi et al., 2016). Recent advances in high-throughput sequencing have linked the imbalance of the microbial community caused by HPD with many gastrointestinal diseases. Liu et al. (2014) found that although HPD changed the composition of intestinal microflora and increased the utilization rate of substrates, the increase in protein volume content in the large intestine did not affect butyrate concentration, thus maintaining the metabolic homeostasis of the colon epithelium. However, HPD showed a reduction in fecal butyrate concentration in a repetitive manner. The intake of high-fat and high-protein diets can lead to a dysfunction in the intestinal microbiota; its metabolite biosynthesis may lead to an imbalance, which subsequently causes intestinal inflammation that may further lead to IBD (Shi et al., 2020). Caspase recruitment domain-containing protein 9 is one among many IBD-susceptible genes, promoting the recovery of colitis by activating the interleukin-22 pathway (Lamas et al., 2016). Recently, it was found that deficiencies of the microbiota of caspase recruitment domain-containing protein 9 in mice caused reduced levels of bacteria with tryptophan catabolism, which regulated the production of local interleukin-22 and affected the balance between the microbiota and host cells (Behnsen et al., 2014; Zenewicz et al., 2013).
The gastrointestinal brain axis is a 2-way communication network between the GIT and the central nervous system. Studies over the past 30 years have described the role of brain–gut interaction in functional gastrointestinal diseases such as irritable bowel syndrome (IBS) (Mayer et al., 2006). In recent years, intestinal microflora has been considered a critical mediator of the brain–gut axis signaling (Cryan and Dinan, 2012), which plays an important role in many aspects of brain function and behavior (Sampson and Mazmanian, 2015). Tryptophan and other amino acids are neurotransmitters in the central nervous system. Tryptophan can also be metabolized into kynurenine and indole, resulting in the regulation of neuroendocrine and intestinal immune responses (Gao et al., 2020). After tryptophan is metabolized to kynurenine, it is further metabolized via two different pathways: kynurenic acid and quinolinic acid pathways. The metabolites produced via the kynurenic acid pathway are called “kynurenine” (Lukić et al., 2019), which may have anti-inflammatory properties in the GIT and participate in immune regulation (Kennedy et al., 2017). It is also a neuroactive metabolite that inhibits N-methyl-d-aspartate and α7 nicotinic acetylcholine receptors, which can cross the blood–brain barrier and reach the central nervous system. The dysregulation of 5-hydroxytryptamine has been observed in gastrointestinal diseases such as IBD (Gracie et al., 2019) and IBS, which are functional gastrointestinal diseases accompanied by severe psychiatric complications and autism spectrum disorder. A recent study of BTBR T+ tf/J (BTBR) mice, using an atrial septal defect-like behavior model, showed microbiota-related damage caused by the production of intestinal 5-hydroxytryptamine (Golubeva et al., 2017). Atrial septal defect is usually associated with gastrointestinal symptoms and thus may be related to disorders of tryptophan metabolism in the gut (Gheorghe et al., 2019). Therefore, they are considered neuromodulators in various physiological and pathological processes of the brain and gastrointestinal dysfunction (Gao et al., 2020).
6. Conclusions
Intestinal microbes can achieve a mutual relationship with the host by facilitating the course of protein/amino acid digestion and absorption. The amino acids, either from the diet or produced by the host, can provide nutrition for the intestinal flora and support protein synthesis. This mutual interaction results in the maintenance of the composition and physiological functions of microbial communities and the homeostasis of the host's gut immune response. An imbalance in the composition of the intestinal microflora is one of the causes of various gastrointestinal diseases. An imbalance in protein/amino acid intake leads to an imbalance in the composition of the intestinal flora, which may lead to IBD, IBS, or other gastrointestinal diseases. In conclusion, a more comprehensive understanding of the crosstalk between gut microbiota and host through the elucidation of protein/amino acid metabolism, is essential for the development of new therapeutic interventions.
Author contributions
Liuting Wu and Zhihong Sun drafted the manuscript. Huiyuan Chen, Zhongxiang Ren, Qi Ding, and Kaiyang Liang reviewed the manuscript. Zhiru Tang conceptualized the study and proofread the manuscript.
Conflict of interest
The authors declare that there is no conflict of interest.
Acknowledgements
This study was funded by grants from the National Natural Science Foundation, China (31872370); Fundamental Research Funds for the Central Universities, China (XDJK2019B014); Natural Science Foundation Project of CQ CSTC (cstc2018jcyjAX0025).
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Contributor Information
Zhiru Tang, Email: tangzhiru2326@sina.com.
Zhihong Sun, Email: sunzh2002cn@aliyun.com.
References
- Agus A., Planchais J., Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. 2018;23:716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- Armstrong L.E., Casa D.J., Belval L.N. Metabolism, bioenergetics and thermal physiology: influences of the human intestinal microbiota. Nutr Res Rev. 2019;32:205–217. doi: 10.1017/S0954422419000076. [DOI] [PubMed] [Google Scholar]
- Arumugam M., Raes J., Pelletier E., Paslier D.L., Yamada T., Mende D.R. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett E., Ross R.P., O'Toole P.W., Fitzgerald G.F., Stanton C. γ-Aminobutyric acid production by culturable bacteria from the human intestine. J Appl Microbiol. 2012;113:411–417. doi: 10.1111/j.1365-2672.2012.05344.x. [DOI] [PubMed] [Google Scholar]
- Behnsen J., Jellbauer S., Wong C.P., Edwards R.A., George M.D., Ouyang W. The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity. 2014;40:262–273. doi: 10.1016/j.immuni.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachier F., Mariotti F., Huneau J.F., Tomé D. Effects of amino acid-derived luminal metabolites on the colonic epithelium and physiopathological consequences. Amino Acids. 2007;33:547–562. doi: 10.1007/s00726-006-0477-9. [DOI] [PubMed] [Google Scholar]
- Caruso R., Lo B.C., Núñez G. Host-microbiota interactions in inflammatory bowel disease. Nat Rev Immunol. 2020;20:411–426. doi: 10.1038/s41577-019-0268-7. [DOI] [PubMed] [Google Scholar]
- Chen J., Zhai Z., Long H., Yang G., Deng B., Deng J. Inducible expression of defensins and cathelicidins by nutrients and associated regulatory mechanisms. Peptides. 2020;123:170–177. doi: 10.1016/j.peptides.2019.170177. [DOI] [PubMed] [Google Scholar]
- Collins S.M., Surette M., Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol. 2012;10:735–742. doi: 10.1038/nrmicro2876. [DOI] [PubMed] [Google Scholar]
- Cryan J.F., Dinan T.G. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- Davila A.M., Blachier F., Gotteland M., Andriamihaja M., Benetti P.H., Sanz Y. Intestinal luminal nitrogen metabolism: role of the gut microbiota and consequences for the host. Pharmacol Res. 2013;68:95–107. doi: 10.1016/j.phrs.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Dodd D., Spitzer M.H., Van Treuren W., Merril BDl, Hryckowian A.J., Higginbottom S.K. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature. 2017;551:648–652. doi: 10.1038/nature24661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan S.H., Belenguer A., Holtrop G., Johnstone A.M., Flint H.J., Lobley G.E. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol. 2007;73:1073–1078. doi: 10.1128/AEM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias A., High A.A., Mishra A., Ong S.S., Wu J., Peng J. Identification and characterization of phosphorylation sites within the pregnane X receptor protein. Biochem Pharmacol. 2014;87:360–370. doi: 10.1016/j.bcp.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson C.O., Cong Y. Host-microbiota interactions in inflammatory bowel disease. Gut Microb. 2012;3:332–344. doi: 10.4161/gmic.20228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Xu K., Liu H., Liu G., Bai M., Peng C. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front Cell Infect Microbiol. 2018;8:13. doi: 10.3389/fcimb.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao K., Mu C.L., Farzi A., Zhu W.Y. Tryptophan metabolism: a link between the gut microbiota and brain. Adv Nutr. 2020;11:709–723. doi: 10.1093/advances/nmz127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheorghe C.E., Martin J.A., Manriquez F.V., Dinan T.G., Cryan J.F., Clarke G. Focus on the essentials: tryptophan metabolism and the microbiome-gut-brain axis. Curr Opin Pharmacol. 2019;48:137–145. doi: 10.1016/j.coph.2019.08.004. [DOI] [PubMed] [Google Scholar]
- Goldsmith J.R., Sartor R.B. The role of diet on intestinal microbiota metabolism: downstream impacts on host immune function and health, and therapeutic implications. J Gastroenterol. 2014;49:785–798. doi: 10.1007/s00535-014-0953-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubeva A.V., Joyce S.A., Moloney G., Burokas A., Arboleya S., Sherwin E. Microbiota-related changes in bile acid & tryptophan metabolism are associated with gastrointestinal dysfunction in a mouse model of autism. EBioMedicine. 2017;24:166–178. doi: 10.1016/j.ebiom.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk G. Springer series in microbiology; 1979. Bacterial metabolism. [Google Scholar]
- Gracie D.J., Hamlin P.J., Ford A.C. The influence of the brain-gut axis in inflammatory bowel disease and possible implications for treatment. Lancet Gastroenterol Hepatol. 2019;4:632–642. doi: 10.1016/S2468-1253(19)30089-5. [DOI] [PubMed] [Google Scholar]
- Hashimoto T., Perlot T., Rehman A., Trichereau J., Ishiguro H., Paolino M. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemarajata P., Spinler J.K., Balderas M.A., Versalovic J. Identification of a proton-chloride antiporter (EriC) by Himar1 transposon mutagenesis in Lactobacillus reuteri and its role in histamine production. Antonie Leeuwenhoek. 2014;105:579–592. doi: 10.1007/s10482-014-0113-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holota Y., Dovbynchuk T., Kaji I., Vareniuk I., Dzyubenko N., Chervinska T. The long-term consequences of antibiotic therapy: role of colonic short-chain fatty acids (SCFA) system and intestinal barrier integrity. PloS One. 2019;14 doi: 10.1371/journal.pone.0220642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C., Gevers D., Knight R.J., Badger, Creasy H.H., Fitzgerald M.G. The human microbiome project (HMP) consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim M., Anishetty S. A meta-metabolome network of carbohydrate metabolism: interactions between gut microbiota and host. Biochem Biophys Res Commun. 2012;428:278–284. doi: 10.1016/j.bbrc.2012.10.045. [DOI] [PubMed] [Google Scholar]
- Kennedy P.J., Cryan J.F., Dinan T.G., Clarke G. Kynurenine pathway metabolism and the microbiota-gut-brain axis. Neuropharmacology. 2017;112:399–412. doi: 10.1016/j.neuropharm.2016.07.002. [DOI] [PubMed] [Google Scholar]
- Kim M.H., Kang S.G., Park J.H., Yanagisawa M., Kim C.H. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology. 2013;145:396–406. doi: 10.1053/j.gastro.2013.04.056. [DOI] [PubMed] [Google Scholar]
- Koh A., Bäckhed F. From association to causality: the role of the gut microbiota and its functional products on host metabolism. Mol Cell. 2020;78:584–596. doi: 10.1016/j.molcel.2020.03.005. [DOI] [PubMed] [Google Scholar]
- König J., Wells J., Cani P.D., García-Ródenas C.L., MacDonal T., Mercenier A. Human intestinal barrier function in health and disease. Clin Transl Gastroenterol. 2016;7:196–210. doi: 10.1038/ctg.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamas B., Richard M.L., Leducq V., Pham H., Michel M., Costa G.D. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Dou X., Li J., Yang Y., Xue C., Wang C. l-arginine ameliorates lipopolysaccharide-induced intestinal inflammation through inhibiting the TLR4/NF-κB and MAPK pathways and stimulating β-defensin expression in vivo and in vitro. J Agric Food Chem. 2020;68:2648–2663. doi: 10.1021/acs.jafc.9b07611. [DOI] [PubMed] [Google Scholar]
- Lanis J.M., Alexeev E.E., Curtis V.F., Kitzenberg D., Kao D., Battista K. Tryptophan metabolite activation of the aryl hydrocarbon receptor regulates IL-10 receptor expression on intestinal epithelia. Mucosal Immunol. 2017;10:1133–1144. doi: 10.1038/mi.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Liu W., Piao M., Zhu H. A review of the relationship between the gut microbiota and amino acid metabolism. Amino Acids. 2017;49:2083–2090. doi: 10.1007/s00726-017-2493-3. [DOI] [PubMed] [Google Scholar]
- Liu X., Blouin J.M., Santacruz A., Lan A., Andriamihaja M., Wilkanowicz S. High-protein diet modifies colonic microbiota and luminal environment but not colonocyte metabolism in the rat model: the increased luminal bulk connection. Am J Physiol Gastrointest Liver Physiol. 2014;307:459–470. doi: 10.1152/ajpgi.00400.2013. [DOI] [PubMed] [Google Scholar]
- Lukić I., Getselter D., Koren O., Elliott E. Role of tryptophan in microbiota-induced depressive-like behavior: evidence from tryptophan depletion study. Front Behav Neurosci. 2019;13:123. doi: 10.3389/fnbeh.2019.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mafra D., Barros A.F., Fouque D. Dietary protein metabolism by gut microbiota and its consequences for chronic kidney disease patients. Future Microbiol. 2013;8:1317–1323. doi: 10.2217/fmb.13.103. [DOI] [PubMed] [Google Scholar]
- Marcel V.D.W., Schellekens H., Dinan T.G., Cryan J.F. Microbiota-gut-brain axis: modulator of host metabolism and appetite. J Nutr. 2017;147:727–745. doi: 10.3945/jn.116.240481. [DOI] [PubMed] [Google Scholar]
- Marchesi J.R., Adams D.H., Fava F., Hermes G., Hirschfield G., Hold G. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65:330–339. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer E.A., Tillisch K., Bradesi S. Review article: modulation of the brain-gut axis as a therapeutic approach in gastrointestinal disease. Aliment Pharmacol Ther. 2006;24:919–933. doi: 10.1111/j.1365-2036.2006.03078.x. [DOI] [PubMed] [Google Scholar]
- Moszak M., Szulińska M., Bogdański P. You are what you eat-the relationship between diet, microbiota, and metabolic disorders-a review. Nutrients. 2020;12:1096. doi: 10.3390/nu12041096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu C., Yang Y., Luo Z., Zhu W. Metabolomic analysis reveals distinct profiles in the plasma and urine of rats fed a high-protein diet. Amino Acids. 2015;47:1225–1238. doi: 10.1007/s00726-015-1949-6. [DOI] [PubMed] [Google Scholar]
- Mu C., Yang Y., Luo Z., Zhu W. Temporal microbiota changes of high-protein diet intake in a rat model. Anaerobe. 2017;47:218–225. doi: 10.1016/j.anaerobe.2017.06.003. [DOI] [PubMed] [Google Scholar]
- Nölling J., Breton G., Omelchenko M.V., Makarova K.S., Zeng Q., Gibson R. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J Bacteriol. 2001;183:4823–4838. doi: 10.1128/JB.183.16.4823-4838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi Y., Mu C., Gao K., Liu Z., Peng Y., Zhu W. Increasing the hindgut carbohydrate/protein ratio by cecal infusion of corn starch or casein hydrolysate drives gut microbiota-related bile acid metabolism to stimulate colonic barrier function. mSystems. 2020;5 doi: 10.1128/mSystems.00176-20. 176-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portune K.J., Beaumont M.D.A.M., Tomé D., Blachier F., Sanz Y. Gut microbiota role in dietary protein metabolism and health-related outcomes: the two sides of the coin. Trends Food Sci Technol. 2016;57:213–232. [Google Scholar]
- Raza M.F., Wang Y., Cai Z., Bai S., Yao Z., Awan U.A. Gut microbiota promotes host resistance to low-temperature stress by stimulating its arginine and proline metabolism pathway in adult Bactrocera dorsalis. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rist V.T., Weiss E., Eklund M., Mosenthin R. Impact of dietary protein on microbiota composition and activity in the gastrointestinal tract of piglets in relation to gut health: a review. Anim Int J Anim Biosci. 2013;7:1067–1078. doi: 10.1017/S1751731113000062. [DOI] [PubMed] [Google Scholar]
- Salonen A., Lahti L., Salojärvi J., Grietje H., Katri K., Sylvia H. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J. 2014;8:2218–2230. doi: 10.1038/ismej.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson T.R., Mazmanian S.K. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe. 2015;17:565–576. doi: 10.1016/j.chom.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q., Chen Y.A., Tuohy K.M. A comparative in vitro investigation into the effects of cooked meats on the human faecal microbiota. Anaerobe. 2010;16:572–577. doi: 10.1016/j.anaerobe.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Shi J., Zhao D., Song S., Zhang M., Li C. High-meat-protein high-fat diet induced dysbiosis of gut microbiota and tryptophan metabolism in wistar rats. J Agric Food Chem. 2020;68:6333–6346. doi: 10.1021/acs.jafc.0c00245. [DOI] [PubMed] [Google Scholar]
- Song W.B., Wang Y.Y., Meng F.S., Meng F.S., Zhang Q.H., Zeng J.Y. Curcumin protects intestinal mucosal barrier function of rat enteritis via activation of MKP-1 and attenuation of p38 and NF-κB activation. PloS One. 2010;5 doi: 10.1371/journal.pone.0012969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thom G., Lean M. Is there an optimal diet for weight management and metabolic health? Gastroenterology. 2017;152:1739–1751. doi: 10.1053/j.gastro.2017.01.056. [DOI] [PubMed] [Google Scholar]
- Vital M., Howe A.C., Tiedje J.M. Revealing the bacterial butyrate synthesis pathways by analyzing (meta) genomic data. mBio. 2014;5 doi: 10.1128/mBio.00889-14. e00889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan K., Li Y., Sun W.Z., An R., Tang Z.R., Wu L.T. Effects of dietary calcium pyruvate on gastrointestinal tract development, intestinal health and growth performance of newly weaned piglets fed low-protein diets. J Appl Microbiol. 2020;128:355–365. doi: 10.1111/jam.14494. [DOI] [PubMed] [Google Scholar]
- Wang H., Ji Y., Wu G., Kaiji S., Sun Y., Wang B. l-tryptophan activates mammalian target of rapamycin and enhances expression of tight junction proteins in intestinal porcine epithelial cells. J Nutr. 2015;145:1156–1162. doi: 10.3945/jn.114.209817. [DOI] [PubMed] [Google Scholar]
- Wang H., Shen J., Pi Y., Gao K., Zhu W. Low-protein diets supplemented with casein hydrolysate favor the microbiota and enhance the mucosal humoral immunity in the colon of pigs. J Anim Sci Biotechnol. 2019;10:79. doi: 10.1186/s40104-019-0387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.Z., Yu Y.J., Adeli K. Role of gut microbiota in neuroendocrine regulation of carbohydrate and lipid metabolism via the microbiota-gut-brain-liver axis. Microorganisms. 2020;8:527. doi: 10.3390/microorganisms8040527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren M.F., Hallowell H.A., Higgins K.V., Liles M.R., Hood W.R. Maternal dietary protein intake influences milk and offspring gut microbial diversity in a rat (rattus norvegicus) model. Nutrients. 2019;11:2257. doi: 10.3390/nu11092257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff W.R., Anfora A.T., Liu J., Schultz P.G., Lesley S.A., Peters E.C. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Bibi S., Du M., Suzuki T., Zhu M.J. Regulation of the intestinal tight junction by natural polyphenols: a mechanistic perspective. Crit Rev Food Sci Nutr. 2017;57:3830–3839. doi: 10.1080/10408398.2016.1152230. [DOI] [PubMed] [Google Scholar]
- Yin J., Han H., Li Y., Liu Z., Zhao Y., Fang R. Lysine restriction affects feed intake and amino acid metabolism via gut microbiome in piglets. Cell Physiol Biochem. 2017;44:1749–1761. doi: 10.1159/000485782. [DOI] [PubMed] [Google Scholar]
- Yin J., Li Y.Y., Han H., Liu Z.J., Zeng X.F., Li T.J. Long-term effects of lysine concentration on growth performance, intestinal microbiome, and metabolic profiles in a pig model. Food & Function. 2018;9:4153–4163. doi: 10.1039/c8fo00973b. [DOI] [PubMed] [Google Scholar]
- Zenewicz L.A., Yin X., Wang G., Elinav E., Hao L., Zhao L. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J Immunol. 2013;190:5306–5312. doi: 10.4049/jimmunol.1300016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Sunkara L.T., Jiang W., Bible M., Carter S., Ma X. Induction of porcine host defense peptide gene expression by short-chain fatty acids and their analogs. PloS One. 2013;8 doi: 10.1371/journal.pone.0072922. [DOI] [PMC free article] [PubMed] [Google Scholar]