Abstract
Necrotic enteritis (NE) is an important enteric disease in poultry and has become a major concern in poultry production in the post-antibiotic era. The infection with NE can damage the intestinal mucosa of the birds leading to impaired health and, thus, productivity. To gain a better understanding of how NE impacts the gut function of infected broilers, global mRNA sequencing (RNA-seq) was performed in the jejunum tissue of NE challenged and non-challenged broilers to identify the pathways and genes affected by this disease. Briefly, to induce NE, birds in the challenge group were inoculated with 1 mL of Eimeria species on day 9 followed by 1 mL of approximately 108 CFU/mL of a NetB producing Clostridium perfringens on days 14 and 15. On day 16, 2 birds in each treatment were randomly selected and euthanized and the whole intestinal tract was evaluated for lesion scores. Duodenum tissue samples from one of the euthanized birds of each replicate (n = 4) was used for histology, and the jejunum tissue for RNA extraction. RNA-seq analysis was performed with an Illumina RNA HiSeq 2000 sequencer. The differentially expressed genes (DEG) were identified and functional analysis was performed in DAVID to find protein–protein interactions (PPI). At a false discovery rate threshold <0.05, a total of 377 DEG (207 upregulated and 170 downregulated) DEG were identified. Pathway enrichment analysis revealed that DEG were considerably enriched in peroxisome proliferator-activated receptors (PPAR) signaling (P < 0.01) and β-oxidation pathways (P < 0.05). The DEG were mostly related to fatty acid metabolism and degradation (cluster of differentiation 36 [CD36], acyl-CoA synthetase bubblegum family member-1 [ACSBG1], fatty acid-binding protein-1 and -2 [FABP1] and [FABP2]; and acyl-coenzyme A synthetase-1 [ACSL1]), bile acid production and transportation (acyl-CoA oxidase-2 [ACOX2], apical sodium–bile acid transporter [ASBT]) and essential genes in the immune system (interferon-, [IFN-γ], LCK proto-oncogene, Src family tyrosine kinase [LCK], zeta chain of T cell receptor associated protein kinase 70 kDa [ZAP70], and aconitate decarboxylase 1 [ACOD1]). Our data revealed that pathways related to fatty acid digestion were significantly compromised which thereby could have affected metabolic and immune responses in NE infected birds.
Keywords: Necrotic enteritis, Clostridium perfringens, Transcriptome, Fatty acid metabolism, Broiler, Challenge
1. Introduction
Necrotic enteritis (NE), is an enteric disease in poultry caused by NetB producing Clostridium perfringens strains (Keyburn et al., 2008). The global economic loss of the subclinical and/or clinical form of the disease has been estimated to be 6 billion dollars annually due to weight loss, impaired feed conversion, mortality, and management costs (Wade and Keyburn 2015). For decades, in-feed antimicrobials have been used to control the outbreaks of NE in chickens. However, due to the occurrence of antibiotic-resistant bacteria, regulatory and voluntary restrictions on the use of in-feed antibiotics have been set in place, which have negatively impacted poultry production due to the increase of NE outbreaks in flocks (McDevitt et al., 2006). The bacterium C. perfringens is naturally present in the intestine of healthy chickens; however, C. perfringens strains expressing NetB toxin, along with the action of predisposing factors such as Eimeria infection, changes in immunity, mycotoxins, diet composition, and animal protein levels can provide a suitable environment for the proliferation of C. perfringens that leads to gastrointestinal epithelium damage and eventually the occurrence of NE in chickens (M'Sadeq et al., 2015; Moore 2016; Van Immerseel et al., 2004).
Deeper understanding of the responses to NE infection in the host cells is essential for developing preventive strategies such as averting bacteria binding with specific cells, producing specific antibodies for disease control, or developing vaccines. Many studies have observed the effects of NE on intestinal microbiota, pH, epithelium structure and immune responses in broilers (Gharib-Naseri et al., 2019a; Golder et al., 2011; Liu et al., 2012). However, global investigation of the gene expression at transcriptomic level in response to the NE challenge has been scarce. Information on differentially expressed genes (DEG) provides a better understanding of the pathogenesis of diseases and identification of genes affected by the infection (Campbell et al., 2010). A powerful tool for the detection of DEG is the sequencing of RNA (RNA-Seq) in a whole-transcriptome, which provides a holistic view of the metabolic pathways and relevant genes in response to these factors (Su et al., 2011). Using such analyses, Kim et al. (2014) and Truong et al. (2017) have reported the activation of many pathways related to immunity responsive to NE infections in broiler chicken intestine.
Intestinal tissue samples used in the present study were obtained from a previous NE challenge experiment conducted by Rodgers et al. (2015) which showed that the NE challenge negatively affected weight gain (WG) and feed conversion ratio (FCR) of the broilers. Stanley et al. (2014) further evaluated the cecal microbiota changes in the challenged birds of the same experiment and reported that the NE challenge significantly altered the gut microbiota and short-chain fatty acids profile compared to the non-challenged birds. Furthermore, Kitessa et al. (2014) investigated the mucosal mRNA gene expression from same birds used in the Rodgers et al. (2015) study with the candidate gene approach and reported an upregulation in the expression of tumor necrosis factor alpha (TNF-α) and downregulated mucin2 (MUC2) and cluster of differentiation 36 (CD36) gene in the NE infected birds. To provide further insight into NE pathogenesis, the current study aimed to characterize the effects of NE infection on the transcriptomic profile in the intestine of challenged and non-challenged chickens sampled from the Rodgers et al. (2015) study. We hypothesize that NE infections can alter the expression of intestinal genes involved in the pathways related to digestion and immunity of the chickens. Such gene expression changes may underlie, at least partially, the negative effect of NE on the performance of broiler chickens.
2. Materials and methods
2.1. Animal management and NE challenge
The live phase of the study was approved by and conducted according to the conditions set by the Animal Ethics Committee of the University of New England, Armidale, Australia. Management was conducted in compliance with the Model code of practice for the welfare of animals: domestic poultry (Agriculture and Resource Management Council of Australia and New Zealand, 2002) and the Australian code of practice for the care and use of animals for scientific purposes (Naujoks et al., 2016).
Tissue samples used in this study were obtained from a previous study conducted by Rodgers et al. (2015) which investigated the roles of C. perfringens, Eimeria and fishmeal in broilers in introducing NE challenge. However, only samples from the challenged and non-challenged birds fed the same basal diet were used from this study. In brief, Rodgers et al. (2015) used 300-day-old Ross 308 male broilers, obtained from Baiada Country Road Hatchery, Tamworth, New South Wales, Australia. The chicks were assigned to 12 pens with 25 birds in each pen (6 pens for each treatment). Birds were raised for 35 days in a temperature-controlled room at Kirby Research Station, the University of New England, Australia. Lighting, humidity and temperature program for the experimental period followed the Ross 308 recommendations (Aviagen 2011). On day 9 birds in the challenged groups were inoculated with 1 mL of phosphate-buffered saline (PBS) suspension containing Eimeria (suspension of 5,000 sporulated oocyst strains of E. acervulina, and Eimeria maxima, and 2,500 sporulated oocysts of E. brunetti from Bioproperties Pty. Ltd., Werribee, VIC, Australia). The non-challenged groups were gavaged with 1 mL sterile PBS. On days 14 and 15 the birds from the challenged group were inoculated per os with 1 mL of C. perfringens suspension (approximately 108 to 109 CFU/mL), strain EHE-NE18 (Commonwealth Scientific and Industrial Research Organization, Geelong, Australia). Birds had ad libitum access to the same starter (day 0 to 21) and grower (day 22 to 35) diets during the experimental period and the diet compositions and nutrients were described previously (Rodgers et al., 2015).
2.2. Lesion score evaluation
On day 16, 2 birds per pen were randomly chosen and euthanized by cervical dislocation and dissected to determine intestinal NE lesion scores in the whole intestinal tract. The NE lesions were assessed according to Prescott et al. (1978), and the severity of the lesions was scored from 0 to 4. Lesions in the intestinal tract were scored as follows: 0, no lesions; 1, thin-walled or friable small intestine; 2, focal necrosis or ulceration; 3, larger patches of necrosis; 4, severe, extensive necrosis.
2.3. Histology
Approximately 2 cm of duodenum tissue was fixed in 10% formalin in 30-mL containers. Fixed samples were then dehydrated, cleared, and embedded in paraffin for sectioning and subsequent histological analysis. Consecutive longitudinal sections (8 μm) were placed individually onto Superfrost slides (Thermo Scientific, Rockville, MD) and stained with hematoxylin and eosin. Villus height and crypt depth were measured by the VideoPro 32 program (Leading Edge Pty Ltd, Adelaide, Australia). The height of 20 villi and the depth of 20 crypts were measured from each replicate. The villus height-to-crypt depth (VH:CD) ratio was determined accordingly.
2.4. Intestinal tissue collection and RNA extraction
On day 16, 2 cm of jejunum tissue was separated from one of the euthanized birds, flushed with chilled sterile PBS and collected in 2-mL Eppendorf cap-lock tubes. The samples were immediately snap-frozen in liquid nitrogen and were preserved at −80 °C until RNA extraction. For RNA-seq analysis, the samples from one bird of all 4 replicates of each experimental group (NE challenged and non-challenged) were used. For total RNA extraction, each sample was homogenized with 1 mL of TRIsure (Bioline, Sydney, Australia) using an IKA T10 basic Homogenizer (Wilmington, NC, USA) in accordance with manufacturer's instructions. An RNAeasy mini Kit (Qiagen, GmbH, Hilden, Germany) was used to further purify the TRIsure extracted RNA. The total RNA purity and quantity were determined using a NanoDrop ND-8000 spectrophotometer (Thermo Fisher Scientific, Waltham, USA), and integrity measured by RNA integrity number (RIN) assayed by using an RNA 6000 Nano Kit (Agilent Technologies, Inc., Waldbronn, Germany) with Bioanalyzer (Agilent Technologies, Waldbronn, Germany) following the procedure provided by the manufacturer. The samples were deemed as high-quality if the value of A260/A230 (the ratios of the readings at the 260 nm and 230 nm of the Nanodrop spectrophotometer) was >1.8, A260/A280 value was between 2.0 and 2.2, and RIN > 7.5. Samples with RIN higher than 7.5 were considered high quality. In the present study, RIN values obtained from each sample were above 8.2.
2.5. cDNA libraries preparation
RNA samples were submitted to the Australian Genome Research Facility (AGRF) for complementary DNA (cDNA) library preparation and sequencing. To process the RNA samples, Illumina's TruSeq Stranded mRNA Prep Kit was used to purify and fragment the mRNA, and cDNA was synthesized following the manufacturer instructions. For each sample, one library was created (total of 8 libraries) for sequencing. Sequencing of the libraries was performed on an Illumina HiSeq 2000 sequencing system (Illumina, San Diego, CA) using 100 bp paired reads.
2.6. Sequencing quality control and read mapping
The quality control (QC) of reads was assessed using the software FASTQC v0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), whereas trimmomatic v0.36 (Bolger et al., 2014) was used to remove the low-quality reads and adaptors. The cleaned reads were mapped to the chicken reference genome (Gallus gallus) using TopHat v2.1.1 (Trapnell et al., 2009). The gene counts were calculated using the HTSeq v0.6.1 (Anders et al., 2015). Genes with low expression were removed, and the trimmed mean of M-values (TMM) normalization was performed with the edgeR package in R software. A principal component analysis (PCA) was carried out using prcomp R function on the logarithmic copies per million (log2 CPM) to explore the transcriptomic profile between the control and challenged birds. Differential expression analysis was performed using voom function from the limma R package v 3.32.2 (Ritchie et al., 2015). The DEG between the control and challenge groups were identified with a significant threshold of adjusted P-value ≤ 0.05 and log2 fold change ≥1).
2.7. Pathway enrichment analysis and protein–protein interaction network and heatmap
The chicken Ensembl gene ID were converted to official gene symbols using the DAVID (Database for Annotation, Visualization and Integrated Discovery, version 6.8 https://david.ncifcrf.gov/). The official gene symbols were then used for functional clustering and enrichment relative to the whole genome. Pathway analysis of DEG was carried out using the Kyoto encyclopedia of genes and genomes (KEGG) database (Kanehisa et al., 2006). In addition, the list of DEG was used to identify the interaction network in the jejunum tissue. The PPI interaction network using the STRING v10.5 database (Search Tool for the Retrieval of Interacting Genes/Proteins) was employed to obtain the protein interaction scores where a network of interactions from a variety of sources was generated, including different interaction databases, text mining, genetic interactions, and shared pathway interactions (Franceschini et al., 2012). The software Cytoscape was used to visualize the network. Average gene counts were considered for the top 100 significant down and up-regulated genes when performing the hierarchical clustering. The clustering was performed in gplots (version 3.0.1.1) of R packages (version 3.5.3), and the results are presented as a heat map.
2.8. Quantitative PCR validation, primer design, and cDNA preparation
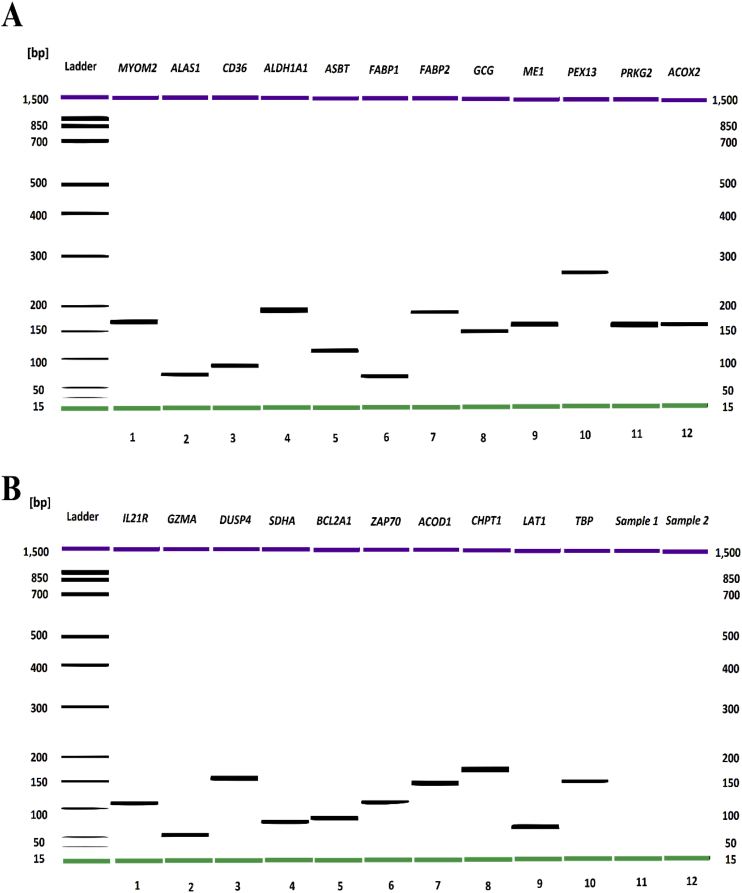
Quantitative PCR (qPCR) analysis was performed to validate the expression of a subgroup of DEG. Twenty genes (Table 1) were selected according to their roles in specific pathways and the fold-change values determined in the RNA-seq analysis. The primers for l-type amino acid transporter-1 (LAT1) (Gilbert et al., 2007), TATA-box binding protein (TBP) were sourced from literature. The primers for all other DEG were designed in this study using the NCBI Primer-BLAST tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). The specificity of primers was then determined using melting curve analysis on a Rotrogene 6000 real-time PCR machine (Corbett, Sydney, Australia) and by the analysis of their amplicons with Agilent 2100 Bioanalyzer DNA 1000 Kit (Agilent Technologies, Inc., Germany) (Fig. 1A and B). Prior to the gene expression evaluation, cDNA was synthesized from RNA through reverse-transcription using the QuantiTect Reverse Transcription Kit (Qiagen, GmbH, Hilden, Germany) according to the manufacturer's instructions, with gDNA eliminated by the addition of DNA. The cDNA was diluted 10 times with nuclease-free water and stored at −20 °C until required.
Table 1.
Primers for candidate target and reference genes in expression studies by qPCR in the jejunum.
| Genes | Gene name | Size, pb | Annealing temperature, °C | Accession No. | Sequence (5′ to 3′) | Reference |
|---|---|---|---|---|---|---|
| ALAS1 | 5’-aminolevulinate synthase-1 | 70 | 60 | NM_001018012.1 | F: CAGGTGGACAGGAAAGGTAAAGA R: CAGCCTCAGTGTTCCTCACCAC |
This study |
| MYOM2 | Myomesin-2 | 167 | 60 | XM_015284819.2 | F: CACTTGCCACTGTCGTATGATG R: TCTCTATACCACTCAGCACGAGG |
This study |
| CD36 | CD36 molecule | 85 | 60 | XM_025147449.1 | F: CTGACATTTGCAGATCCATCTATGG R: CGAGGAACTGTGAAACGATACAGT |
This study |
| ALDH1A1 | Aldehyde dehydrogenase 1 family, member A1 | 191 | 60 | XM_015280233.2 | F: GCAGATGCAGACTTGGATGAAG R: CCTTGCTGTACACCAGGTAACAG |
This study |
| FABP1 | Fatty acid binding protein 1 | 86 | 60 | NM_204192.3 | F: GAAGGTAACAACAAACTGGTTGCT R: GTCATTGTATGGGTGATGGTGTC |
This study |
| FABP2 | Fatty acid binding protein 2 | 184 | 60 | NM_001007923.1 | F: GCAATGGGCGTGAATGTGA R: AGCCTGAAAGTTCAGTCCCGT |
This study |
| GCG | Glucagon | 144 | 60 | NM_205260.5 | F: CGGATCAGCTCTCAAGTAATGC R: CTCGTCCATTCACTAACCAAGC |
This study |
| ME1 | Malic enzyme 1 | 164 | 60 | NM_204303.1 | F: GCTGTCAAAAGGATATGGATGGT R: GCAGCGACTCCTATTAGCACA |
This study |
| PEX13 | Peroxisomal biogenesis factor 13 | 264 | 60 | NM_001199421.1 | F: TGGGAGAACCGGCGATTAGT R: CAAGCCACCGTATCCATAACTG |
This study |
| MSRB1 | Methionine sulfoxide reductase B1 | 164 | 60 | NM_001346746.2 | F: TCGAGCCGGGTGTGTATGTA R: CTTGCCACAGGACACCTTTAAG |
This study |
| PRKG2 | Protein kinase cGMP-dependent 2 | 161 | 60 | XM_025149790.1 | F: CGACATGCCAAAAGAAGATCC R: GTTCAACCCTTCCGAACCCA |
This study |
| ACOX2 | Acyl-CoA oxidase 2 | 163 | 60 | XM_004944647.3 | F: CACTGTGCCCAGGTATAACTGC R: GACCCACGCCTTACATAGGTG |
This study |
| ASBT | Solute carrier family 10 member 2 | 119 | 60 | NM_001319027.1 | F: GTGGGTTATCACACCTAAGTTATG R: CACTGTACGACATCTGCTCCAAG |
This study |
| ACOD1 | Aconitate decarboxylase 1 | 146 | 60 | XM_015275372.2 | F: AAGGTTTCACCCACCAACAGTG R: GAGTTGCTGCGTTAGCCAGTG |
This study |
| IL21R | Interleukin 21 receptor | 111 | 60 | XM_025155100.1 | F: CTGGGAGACTCAGAAGATCAAATC R: GGTCTGGCTCTCACTTGGAATTC |
This study |
| GZMA | Granzyme A | 56 | 60 | NM_204457.1 | F: CTTCCTGGAGATTTGTGCGTG R: GAGTGTGGTGCTACTTCATGTCCT |
This study |
| CHPT1 | Choline phosphotransferase 1 | 183 | 60 | XM_015285488.2 | F: CGAGCAGGCACCTTTTTGG R: GCTATGCAGGATCCAAGGACA |
This study |
| DUSP4 | Dual specificity phosphatase 4 | 155 | 60 | NM_204838.1 | F: ATCACAGCCCTGCTGAACGT R: CAGCACTCTTTCACTGAGTCGATG |
This study |
| BCL2A1 | BCL2 related protein A1 | 78 | 60 | NM_204866.1 | F: GGTGGCTGGGAAAACGGT R: TGGTACTCTCTGAACAAGGAAAGAAC |
This study |
| LAT1 | Neutral amino acid transporter | 70 | 60 | KT876067.1 | F: GATTGCAACGGGTGATGTGA R: CCCCACACCCACTTTTGTTT |
Gilbert et al. (2007) |
| ZAP70 | Zeta chain of T cell receptor associated protein kinase 70 kDa | 110 | 60 | NM_001321556.1 | F: GCTGGACCTACAGTTGGGAAGA R: CAATGCTGTAGTAGTAGGTGCGGA |
This study |
| TBP | TATA-box binding protein | 147 | 62 | NM_20510383127 | F-TAGCCCGATGATGCCGTAT R- GTTCCCTGTGTCGCTTGC |
Li et al. (2005) |
| SDHA | Succinate dehydrogenase complex flavoprotein subunit A | 74 | 60 | NM_00127739 | F-ATACGGGAAGGAAGGGGTTG R-TGCTGGGGTGGTAAATGGTG |
Barzegar Nafari (2019) |
Fig. 1.
TDNA gen electrophoresis of the qPCR products showing that the primers were specific in amplification. (A) Ladder: DNA ladder; MYOM2, 167 bp; ALAS1, 70 bp; CD36, 85 bp; ALDH1A1, 191 bp; ASBT, 119 bp; FABP1, 86 bp; FABP2, 184 bp; GCG, 144 bp; ME1, 164 bp; PEX13, 264 bp; PRKG2, 161 bp; ACOX2, 163 bp. (B) Ladder: DNA Ladder; IL21R, 111; GZMA, 56 bp; DUSP4, 155 bp; SDHA, 74 bp; BCL2A1, 78 bp; ZAP70, 110 bp; ACOD1, 146 bp; CHPT1, 183 bp; LAT1, 70 bp; TBP, 147 bp. MYOM2 = myomesin-2; ALAS1 = 5'-aminolevulinate synthase-1; CD36 = cluster of differentiation 36; ALDH1A1 = aldehyde dehydrogenase-1 family member A1; ASBT = apical sodium–bile acid transporter; FABP1 and FABP2 = fatty acid-binding protein-1 and -2; GCG = glucagon; ME1 = malic enzyme; PEX13 = peroxisomal biogenesis factor-13; PRKG2 = protein kinase cGMP-dependent 2; ACOX2 = acyl-CoA oxidase-2; IL21R = interleukin-21 receptor; GZMA = granzyme A; DUSP4 = dual-specificity phosphatase-4; SDHA = succinate dehydrogenase complex flavoprotein subunit A; BCL2A1 = BCL2 related protein A1; ZAP70 = zeta chain of T cell receptor associated protein kinase 70 kDa; ACOD1 = aconitate decarboxylase 1; CHPT1 = choline phosphotransferase-1; LAT1 = L-type amino acid transporter-1; TBP = TATA-box binding protein.
Quantitative PCR was performed in duplicates using an SYBR Green kit SensiFAST SYBR No-ROX (Bioline, Sydney, Australia) with a Rotorgene 6000 real-time PCR machine (Corbett Research, Sydney, Australia). The PCR reaction was performed in a volume of 10 μL containing 5 μL of 2 × SensiFAST, 400 mmol/L of each primer and 2 μL of 10 × diluted cDNA template. The relative quantity of mRNA of the target genes was calculated by qBase + version 3.0 (Biogazelle, Zwijnbeke, Belgium) software with SDH and TBP as reference genes that were optimized from 10 widely used house-keeping genes prior to the analysis of target genes. The qBase + applied arithmetic mean method to transform the logarithmic cycle threshold (Cq) value to a relative linear quantity using the exponential function for relative quantification of target genes (Hellemans et al., 2007, Vandesompele et al., 2002). The qPCR results were then presented as relative expression levels of the genes in respective treatment groups as means of normalized relative quantities (NRQ). The fold changes for each particular gene analyzed with qPCR were calculated using the mean NRQ values of the NE challenged and non-challenged groups.
2.9. Statistical analysis
Statistical analysis was performed using the SPSS statistics version 24 (IBM SPSS, UK). The student's t-test was applied to determine differences between means of the two groups using SPSS 24 statistics software on the performance parameters and villus height. Intestinal lesion scoring data were analyzed by the non-parametric Kruskal–Wallis test (Kruskal and Wallis, 1952) as the data was not normally distributed.
3. Results
3.1. Effectiveness of the necrotic enteritis challenge
Table 2 shows the performance (14 to 35 days), jejunal lesion score, and duodenal villus height (16 days) of birds with and without NE challenge. The chickens with NE challenge had lower WG and higher FCR (P < 0.01), higher lesion score (P < 0.05), lower duodenal villus height (P < 0.05) and VH:CD ratio (P < 0.01) compared to the non-challenged birds. However, feed intake was not affected by the NE challenge during 14 and 35 days (P > 0.05).
Table 2.
Effect of NE challenged on WG, FI, FCR of birds (day 14 to 35), and duodenum morphology and jejunum lesion scores on day 16.
| Treatments | Performance |
Histology, μm |
Lesion score1 | ||||
|---|---|---|---|---|---|---|---|
| WG, g/bird | FI, g/bird | FCR | VH | CD | VH-to-CD ratio | ||
| Non-challenged | 1,708a | 2,911 | 1.710b | 1,420a | 179 | 7.96a | 0.00b |
| NE challenged | 1,437b | 2,762 | 1.899a | 1,011b | 193 | 5.27b | 0.50a |
| P-value | 0.005 | 0.239 | 0.045 | 0.021 | 0.457 | 0.005 | 0.011 |
NE = necrotic enteritis, WG = weight gain; FI = feed intake; FCR = feed conversion ratio, VH = villus height, CD = crypt depth.
a, b Means sharing the same superscripts are not significantly different from each other at P < 0.05.
Jejunum lesion score day 16.
3.2. Quality of RNA-seq reads and deferentially expressed genes
On average, 21, 543, 431 reads with a length of 100 bp were generated from the 8 cDNA libraries. Table 3 presents a summary for read features, including the alignment rates for both groups, which all exceed 80% and were mapped successfully to the chicken reference genome. The average number of sequence reads was 22.3 for non-challenged, and 20.7 million for the NE challenged birds. On average, more than 97% of the reads passed the quality control and cleaning process (Table 3).
Table 3.
Number of reads obtained from the RNA sequencing and the mapped reads to the reference genome (Gallus gallus) in the non-challenged and necrotic enteritis (NE) challenged broiler groups.
| No. | Group | Raw reads | Cleaned reads | Mapped | Mapping rate, % |
|---|---|---|---|---|---|
| 1 | Non-challenged | 23,735,190 | 22,326,785 | 18,833,668 | 84.9 |
| 2 | Non-challenged | 23,102,585 | 21,879,408 | 18,468,203 | 84.4 |
| 3 | Non-challenged | 20,558,626 | 19,502,432 | 16,538,279 | 84.8 |
| 4 | Non-challenged | 21,960,653 | 20,844,944 | 17,795,298 | 85.4 |
| 5 | NE challenged | 20,853,776 | 19,737,566 | 16,031,560 | 81.2 |
| 6 | NE challenged | 20,710,878 | 19,665,303 | 16,028,299 | 81.5 |
| 7 | NE challenged | 20,396,891 | 19,046,762 | 15,581,264 | 81.8 |
| 8 | NE challenged | 21,028,852 | 19,662,408 | 15,893,653 | 80.8 |
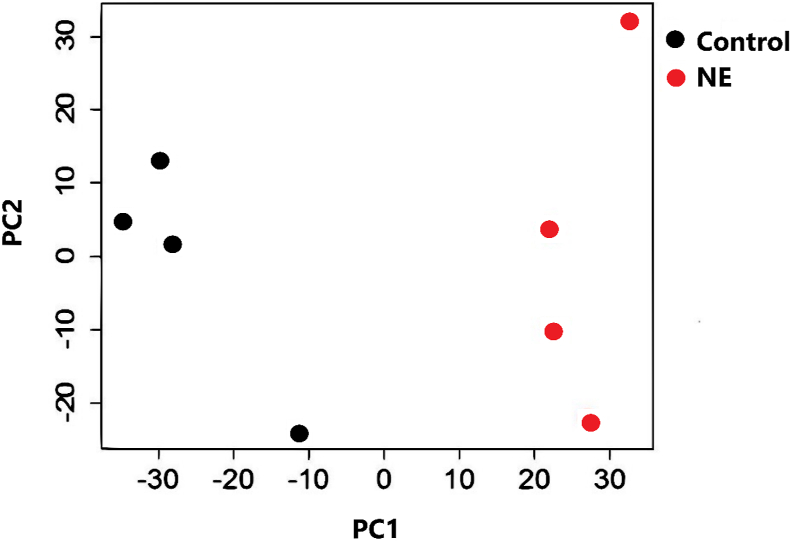
The principal component analysis (PCA) plot showed the transcriptomic profile in the jejunum tissue with a clear separation of samples by the principal component 1 (PC1) which explained over 36% of the variance and principal component 2 (PC2) which accounted for 16% of the total variance between the samples (Fig. 2).
Fig. 2.
Principal component analysis plot of differentially abundant transcripts. The plot shows log2 counts per million mapped reads (CPM) for both groups. The horizontal and vertical axes show 2 principal components that respectively explain variation between different groups. Control, non-challenged birds (black); NE, necrotic enteritis challenged birds (red); PC1, the axis showing the first principal direction along which the samples show the largest variation; PC2, the axis showing the second most important direction which is orthogonal to the PC1 axis.
Differential gene expression (DEG) analysis showed 377 DEG (log2 fold change > 1; false discovery rate < 0.05) differentially expressed genes between the two groups. Among the DEG, 207 were significantly up-regulated, and 170 genes were significantly down-regulated by NE challenge (Appendix Table 1). The top three up and down-regulated genes in the challenged birds were myosin binding protein C-1 (MYBPC1), apical sodium–bile acid transporter (ASBT) and aconitate decarboxylase-1 (ACOD1), and phosphoenolpyruvate carboxykinase-1 (PCK1), myomesin-2 (MYOM2) and protein kinase cGMP-dependent 2 (PRKG2), respectively. A full list of the top 35 significantly up- and down-regulated genes are shown in Table 4, Table 5, respectively. Table 6 illustrates the effect of NE challenge on fold change of genes related to immunity compared to the non-challenged birds.
Table 4.
Top 35 upregulated DEG in NE challenged birds relative to non-challenged birds.
| Gene symbol | Gene name | Fold change | P-value | FDR |
|---|---|---|---|---|
| MYBPC1 | Myosin binding protein C, slow type | +3.616 | 0.0006 | 0.0128 |
| SLC10A2, ASBT | Solute carrier family 10 member 2 | +3.254 | 0.0005 | 0.0115 |
| ACOD1 | Aconitate decarboxylase 1 | +2.652 | 0.0002 | 0.0064 |
| ANKF1 | Ankyrin repeat and fibronectin type III domain containing 1 | +2.600 | 0.0000 | 0.0026 |
| IFN-γ | Interferon gamma | +2.590 | 0.0043 | 0.0352 |
| WASF1/wave 1 | WAS protein family member 1 | +2.199 | 0.0000 | 0.0027 |
| BEND4/ccdc4 | BEN domain containing 4 | +2.197 | 0.0003 | 0.0086 |
| TM4SF19 | Transmembrane 4 L six family member 19 | +2.110 | 0.0001 | 0.0056 |
| RNF208 | Ring finger protein 208 | +1.998 | 0.0000 | 0.0012 |
| KRT19 | Keratin 19 | +1.989 | 0.0053 | 0.0400 |
| LNX1 | Ligand of numb-protein X 1 | +1.932 | 0.0064 | 0.0449 |
| WNT5B | Wnt family member 5B | +1.914 | 0.0068 | 0.0463 |
| SLC7A5 | Solute carrier family 7 member 5 | +1.893 | 0.0000 | 0.0040 |
| SCN3B | Sodium voltage-gated channel beta subunit 3 | +1.888 | 0.0028 | 0.0276 |
| FRMD3 | FERM domain containing 3 | +1.874 | 0.0001 | 0.0054 |
| NEURL1B | Neuralized E3 ubiquitin protein ligase 1B | +1.804 | 0.0003 | 0.0084 |
| PLCD4 | Phospholipase C delta 4 | +1.717 | 0.0021 | 0.0235 |
| DUSP4 | Dual specificity phosphatase 4 | +1.659 | 0.0000 | 0.0039 |
| GZMA | Granzyme A | +1.655 | 0.0003 | 0.0082 |
| CEBPB | CCAAT enhancer binding protein beta | +1.634 | 0.0009 | 0.0152 |
| ASS1 | Argininosuccinate synthase 1 | +1.631 | 0.0001 | 0.0051 |
| HYAL3 | Hyaluronoglucosaminidase 3 | +1.551 | 0.0018 | 0.0221 |
| BCL2A1 | BCL2 related protein A1 | +1.545 | 0.0039 | 0.0334 |
| KCNJ5 | Potassium voltage-gated channel subfamily J member 5 | +1.516 | 0.0024 | 0.0254 |
| EOMES | Eomesodermin | +1.506 | 0.0020 | 0.0230 |
| THEMIS2 | Thymocyte selection associated family member 2 | +1.486 | 0.0000 | 0.0038 |
| SLC11A1 | Solute carrier family 11 member 1 | +1.486 | 0.0009 | 0.0154 |
| DCT | Dopachrome tautomerase | +1.479 | 0.0024 | 0.0253 |
| NRG1 | Neuregulin 1 | +1.474 | 0.0002 | 0.0076 |
| PROKR2 | Prokineticin receptor 2 | +1.473 | 0.0019 | 0.0223 |
| SCL6A7/LAT1 | Solute carrier family 6 member 7 | +1.458 | 0.0032 | 0.0300 |
| INSRR | Insulin receptor related receptor | +1.453 | 0.0000 | 0.0026 |
| ART1 | ADP-ribosyltransferase 1 | +1.429 | 0.0011 | 0.0168 |
| IL21R | Interleukin 21 receptor | +1.414 | 0.0018 | 0.0217 |
| NEXMIF | Neurite extension and migration factor | +1.389 | 0.0034 | 0.0305 |
DEG = differentially expressed genes; NE = necrotic enteritis; FDR = false discovery rate.
Table 5.
Top 35 downregulated DEG in NE challenged birds relative to non-challenged birds.
| Gene symbol | Gene name | Fold change | P-value | FDR |
|---|---|---|---|---|
| PCK1 | Phosphoenolpyruvate carboxykinase 1 | −3.052 | 5.89E-05 | 0.0043 |
| MYOM2 | Myomesin 2 | −2.935 | 0.000558 | 0.0120 |
| PRKG2 | Protein kinase cGMP-dependent 2 | −2.929 | 0.000144 | 0.0063 |
| FAXDC2 | Fatty acid hydroxylase domain containing 2 | −2.676 | 0.001366 | 0.0189 |
| AADAC | Arylacetamide deacetylase | −2.633 | 4.06E-07 | 0.0011 |
| CYP2AC1 | Cytochrome P450, family 2, subfamily AC, polypeptide 1 | −2.582 | 1.85E-05 | 0.0029 |
| AGMO | Alkylglycerol monooxygenase | −2.520 | 0.000651 | 0.0129 |
| DDO | d-aspartate oxidase | −2.448 | 0.006803 | 0.0463 |
| PDK4 | Pyruvate dehydrogenase kinase 4 | −2.040 | 0.000659 | 0.0130 |
| ABCB11 | ATP binding cassette subfamily B member 11 | −1.979 | 0.005847 | 0.0425 |
| ENPP7 | Ectonucleotide pyrophosphatase/phosphodiesterase 7 | −1.952 | 0.000643 | 0.0128 |
| CYP4V2 | Cytochrome P450 family 4 subfamily V member 2 | −1.904 | 0.000259 | 0.0082 |
| ALDH1A1 | Aldehyde dehydrogenase 1 family member A1 | −1.898 | 5.93E-05 | 0.0043 |
| CD36 | CD36 molecule | −1.851 | 0.003539 | 0.0312 |
| ME1 | Malic enzyme 1 | −1.779 | 1.26E-05 | 0.0027 |
| FETUB | Fetuin B | −1.760 | 4.63E-05 | 0.0040 |
| DHRS7 | Dehydrogenase/reductase 7 | −1.738 | 0.00028 | 0.0085 |
| DNM1 | Dynamin 1 | −1.737 | 8.97E-05 | 0.0051 |
| GSTT1 | Glutathione S-transferase theta 1 | −1.713 | 0.006629 | 0.0454 |
| BBOX1 | Gamma-butyrobetaine hydroxylase 1 | −1.708 | 0.00021 | 0.0074 |
| FTCD | Formimidoyltransferase cyclodeaminase | −1.690 | 3.1E-05 | 0.0037 |
| GPX4 | Glutathione peroxidase 4 | −1.624 | 5.68E-05 | 0.0043 |
| ACO12 | Acyl-CoA thioesterase 12 | −1.619 | 0.000127 | 0.0058 |
| FABP2 | Fatty acid binding protein 2 | −1.606 | 5.01E-07 | 0.0011 |
| PDXDC1 | PX domain containing 1 | −1.579 | 1.6E-05 | 0.0028 |
| SUSD2 | Sushi domain containing 2 | −1.572 | 4.67E-05 | 0.0040 |
| ACMSD | Aminocarboxymuconate semialdehyde decarboxylase | −1.567 | 0.00392 | 0.0333 |
| CDA | Cytidine deaminase | −1.561 | 2.03E-06 | 0.0016 |
| ANO5 | Anoctamin 5 | −1.551 | 0.000163 | 0.0066 |
| HESX1 | HESX homeobox 1 | −1.541 | 0.001476 | 0.0197 |
| PDE6C | Phosphodiesterase 6C | −1.509 | 0.005998 | 0.0431 |
| ACSL1 | Acyl-CoA synthetase long chain family member 1 | −1.499 | 1.04E-06 | 0.0012 |
| FAM83B | Family with sequence similarity 83 member B | −1.476 | 0.002718 | 0.0272 |
| FABP1 | Fatty acid binding protein 1 | −1.473 | 9.25E-05 | 0.0052 |
| HSD17B4 | Hydroxysteroid 17-beta dehydrogenase 4 | −1.469 | 0.00012 | 0.0056 |
| MLKL | Mixed lineage kinase domain like pseudokinase | −1.456 | 0.002283 | 0.0247 |
| KL | Klotho | −1.438 | 0.000212 | 0.0074 |
DEG = differentially expressed genes; NE = necrotic enteritis; FDR = false discovery rate.
Table 6.
Several immune-related genes significantly affected by the necrotic enteritis challenge.
| Gene | Gene name | Gene function | log2FC |
|---|---|---|---|
| MSRB1 | Methionine sulfoxide reductase B1 | Innate immune response | −0.718 |
| TLR3 | Toll-like receptor 3 | Immune system | −0.578 |
| PEX13 | Peroxisomal biogenesis factor 13 | Fatty acid alpha-oxidation | −0.480 |
| PDK4 | Pyruvate dehydrogenase kinase 4 | ATP binding, kinase activity | −1.894 |
| INF-γ | Interferon-gamma | Macrophage activation | +2.600 |
| LCK | LCK proto-oncogene, Src family tyrosine kinase | Regulation of T-cell activation | +1.115 |
| TMEM173 | Transmembrane protein 173 | Innate immune response | +1.217 |
| IL21R | Interleukin 21 receptor | Protein binding | +1.415 |
| ZAP70 | Zeta-chain T cell receptor associated protein kinase 70 kDa | Regulation of T-cell differentiation | +1.179 |
| PTPN22 | Protein tyrosine phosphatase non-receptor type 22 | T-cell differentiation | +1.039 |
FC = fold change.
3.3. Pathway enrichment, protein–protein interaction
In total, 2 KEGG pathways were significantly enriched, including the peroxisome proliferator-activated receptors (PPAR) signaling pathways and fatty acid degradation pathway. Table 7 shows the significant KEGG pathways related to DEG. It was shown that all genes enriched in both pathways were down-regulated by the NE challenge.
Table 7.
Metabolic pathways affected by the necrotic enteritis challenge.
| Pathway | P-value | Benjamini | DEG |
|---|---|---|---|
| PPAR signaling | 0.00015 | 0.008 | CD36, ACOX2, ACSBG1, ACSL1, EHHADH, FABP1, FABP2, ACAA1, PCK1 |
| Fatty acid degradation | 0.0017 | 0.035 |
ACAA1, ACSL1, EHHADH ACSBG1, ECI2, ALDH2 |
DEG = differentially expressed genes; PPAR = peroxisome proliferator-activated receptors; CD36 = cluster of differentiation 36; ACOX2 = acyl-CoA oxidase-2; ACSBG1 = acyl-CoA synthetase bubblegum family member-1; ACSL1 = acyl-coenzyme A synthetase-1; EHHADH = enoyl-CoA hydratase and 3-hydroxyacyl CoA dehydrogenase; FABP1 and FABP2 = fatty acid-binding protein-1 and -2; ACAA1 = acetyl-CoA acyltransferase 1, PCK1 = phosphoenolpyruvate carboxykinase 1; ECI2 = enoyl-CoA delta isomerase-2, ALDH2 = aldehyde dehydrogenase-2.
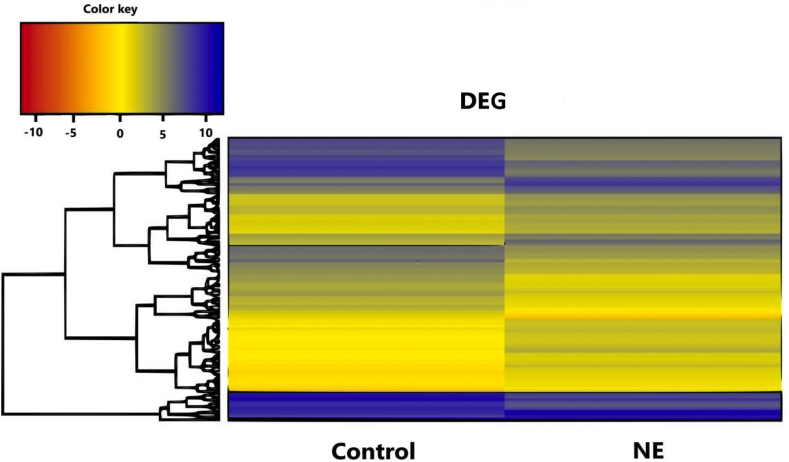
Hierarchical clustering analysis was performed using the top 100 most extreme DEG in the 2 groups of birds. The pattern of expression for the 100 genes is presented in the heat map as shown in Fig. 3. The list of genes of the heat map is provided in the supplementary file (Appendix Table 2).
Fig. 3.
Heat map showing the top 100 differentially expressed genes (DEG). Each horizontal row represents a single gene, and each column represents 1 of the 2 experimental groups. Colors indicate a positive (blue) or negative (red) fold-change. The log2 counts per million mapped reads (CPM) values were used to cluster all the presented differentially expressed genes in Java TreeView by hierarchical clustering using Euclidean distance and pairwise average linkage methods.
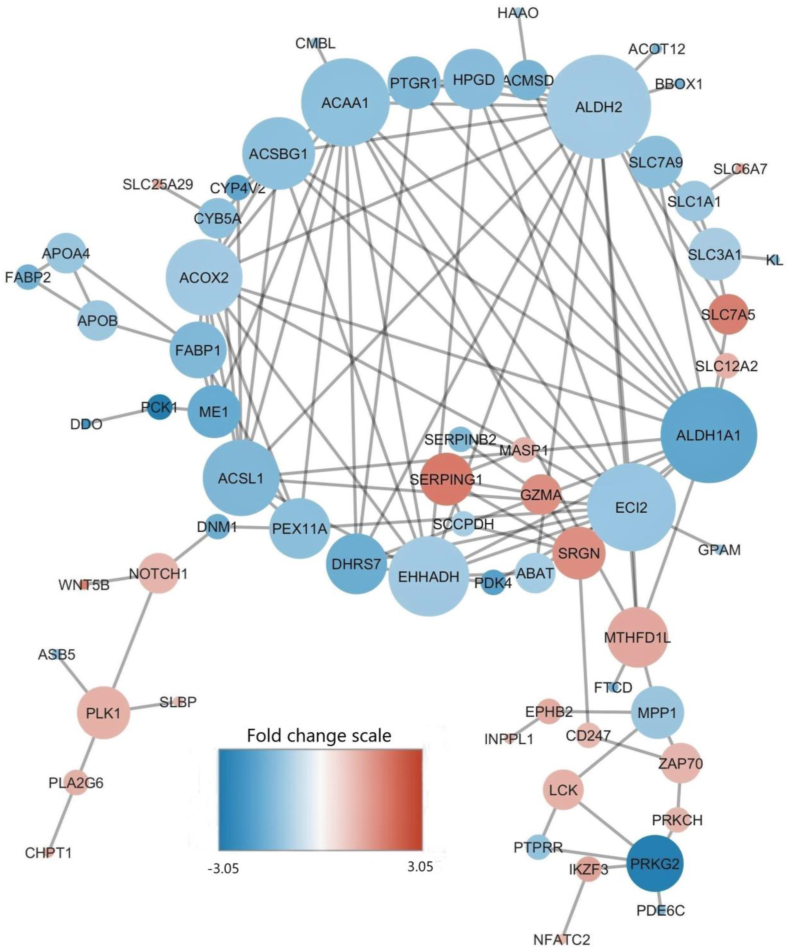
To further explore the interactions between the DEG identified using the edgeR/cuffdiff, these DEG were subjected them to STRING analysis for PPI network analysis. The nodes in these networks are genes, and the links between them (edges) represent co-regulation (Fig. 4). The size of the nodes indicates the number of interactions of each gene with other DEG genes, therefore, any changes in gene expression of proteins with high interactions in a network may result in deep dysfunction of the interactome system (Zamanian-Azodi et al., 2016). Genes such as aldehyde dehydrogenase-2 (ALDH2), aldehyde dehydrogenase-1 family member A1 (ALDH1A1), enoyl-CoA delta isomerase-2 (ECI2), acyl-CoA synthetase-1 (ACSL1), enoyl-CoA hydratase and 3-hydroxyacyl CoA dehydrogenase (EHHADH), acyl-CoA synthetase bubblegum family member-1 (ACSBG1), and acyl-CoA oxidase-2 (ACOX2) show the highest interaction with other DEG, and could indicate their importance related to the negative responses observed in the infected chickens. Aldehyde dehydrogenase-2 and ALDH1A1 are among the genes with the highest number of interactions.
Fig. 4.
Visualization for the predicted results of protein–protein interactions of DEG in the jejunum of NE challenged and non-challenged broilers. Each node represents a protein, and each line refers to an interaction. Line thickness reflects the strength of PPI. Node colors represent fold change, and node font sizes and circle sizes of the corresponding genes indicate the number of PPI. The largest cluster mostly represents genes related to the PPAR and β-oxidation pathways. The network is generated using the Cytoscape tool. DEG = differentially expressed genes; PPI = protein–protein interactions; PPAR = peroxisome proliferator-activated receptors.
3.4. Validation of differential gene expression data by qPCR
To validate the DEG identified by sequencing analysis, a qPCR analysis was performed for 20 genes including 9 down-regulated genes, i.e., ACOD1, ASBT, BCL2 related protein A1 (BCL2A1), choline phosphotransferase-1 (CHPT1), dual-specificity phosphatase-4 (DUSP4), granzyme A (GZMA), interleukin-21 receptor (IL21R), LAT1, zeta-chain (TCR) associated protein kinase 70 kDa (ZAP70) and 11 upregulated genes: ACOX2, 5'-aminolevulinate synthase-1 (ALAS1), ALDH1A1, CD36, glucagon (GCG), fatty acid-binding protein-1 (FABP1), fatty acid-binding protein-2 (FABP2), malic enzyme 1 (ME1), MYOM2, peroxisomal biogenesis factor-13 (PEX13) and PRKG2. All the 20 selected genes for validation showed consistent up or down-regulation in response to the NE challenge with a strong positive correlation (R2 = 0.813; P < 0.001) in the fold change between qPCR and the RNA-seq data (Table 8). The qPCR results confirm the reliability of the differential expression analysis using RNA-Seq data.
Table 8.
Comparison of the gene expression data between RNA-seq and qPCR1.
| Gene | Fold change |
|
|---|---|---|
| qPCR | RNA-seq | |
| ACOD1 | +13.5 | +2.65 |
| ASBT | +58.6 | +3.25 |
| BCL2A1 | +5.02 | +1.54 |
| CHPT1 | +10.9 | +1.32 |
| DUSP4 | +7.14 | +1.66 |
| GZMA | +11.3 | +1.65 |
| IL21R | +7.92 | +1.41 |
| LAT1 | +6.18 | +1.46 |
| ZAP70 | +8.02 | +1.06 |
| ACOX2 | −5.95 | −1.04 |
| ALAS1 | −2.75 | −1.10 |
| ALDH1A1 | −4.35 | −1.90 |
| CD36 | −14.0 | −1.85 |
| GCG | −2.86 | −1.12 |
| FABP1 | −2.52 | −1.48 |
| FABP2 | −5.16 | −1.61 |
| ME1 | −3.80 | −1.78 |
| MYOM2 | −11.2 | −2.93 |
| PEX13 | −4.20 | −1.32 |
| PRKG2 | −11.2 | −2.93 |
qPCR = quantitative PCR;ACOD1 = aconitate decarboxylase 1; ASBT = apical sodium–bile acid transporter; BCL2A1 = BCL2 related protein A1; CHPT1 = choline phosphotransferase-1; DUSP4 = dual-specificity phosphatase-4; GZMA = granzyme A; IL21R = interleukin-21 receptor; LAT1 = L-type amino acid transporter-1; ZAP70 = zeta chain of T cell receptor associated protein kinase 70 kDa; ACOX2 = acyl-CoA oxidase-2; ALAS1 = 5’-aminolevulinate synthase-1; ALDH1A1 = aldehyde dehydrogenase-1 family member A1; CD36 = cluster of differentiation 36; GCG = glucagon; FABP1 and FABP2 = fatty acid-binding protein-1 and -2; ME1 = malic enzyme; PEX13 = peroxisomal biogenesis factor-13; PRKG2 = protein kinase cGMP-dependent 2.
For the qPCR gene expression normalization, SDH and TBP genes were used as reference genes. Positive signs show upregulation, and negative signs show downregulation of genes in necrotic enteritis challenged birds.
4. Discussion
The current study identified pathways and corresponding genes in the jejunum of broilers that are responsive to NE infection. Two main metabolic pathways that were responsive to the challenge were involved in fatty acid production and utilization. Most down-regulated genes, affected by the NE infection, were related to enzymes active in oxidation and fatty acid transportation. The NE challenge also up-regulated genes related to immunity. The results confirmed our hypothesis that NE infections can alter the expression of intestinal genes involved in the pathways related to digestion and immunity of the chickens, possibly in a negative way, which leads to negative changes in the performance of the challenged birds.
4.1. Fatty acid related genes
It is well known that the small intestine is an important organ for digestion, absorption, and transportation of dietary nutrients including lipid. Lipids play a key role in many biological processes such as energy sourcing and storage and inflammatory responses, and they participate in overall homeostasis as signaling molecules (Grygiel-Górniak 2014; Robinson and Mazurak 2013). Nine genes from the PPAR pathway and six genes in the β-oxidation pathway were suppressed in the NE challenged birds, indicating reduced lipid metabolism and fatty acid utilization in the challenged birds. The PPAR pathway (categorized as α, β/σ, and γ types) is closely associated with lipid and carbohydrate metabolism, energy balance, intestinal inflammation and hemostasis and ameliorating insulin sensitivity (Dupont et al., 2012; Ricote et al., 2004; Varga et al., 2011; Zhou et al., 2016). The NE infected broilers have previously shown to have dysregulated lipid metabolism in the liver through the reduced expression of PPAR-γ and PPAR-α genes (Qing et al., 2017).
Additionally, the β-oxidation pathway, the main metabolic pathway for energy supply from fatty acids in cells (Bartlett and Eaton 2004), was also suppressed by the NE challenge. Among the DEG in the PPAR pathways, FABP1, FABP2, FAT/CD36, ACOX2, and PCK1 were significantly down-regulated in our study. The FABP1 and FABP2 genes are involved in the uptake of long-chain fatty acids (Pelsers et al., 2005) and can also modulate critical lipid–sensitive pathways in adipocytes and macrophages in humans that are important upon the occurrence of intestinal inflammation (Levy et al., 2001). Downregulation of FABP1 and FABP2 in NE infected birds is in agreement with previous reports where same observation was described (Guo et al., 2013). In the current study, the intestinal histology results confirmed significant villus damage in the challenged birds. It is suggested that down-regulation of FABP1 and FABP2 in the jejunum of challenged birds could be attributed to structural damage and intestinal epithelium loss (Chen et al., 2015). The gene CD36 is also involved in the regulation of fatty acid uptake, and is a necessary compound for transporting cholesterol and triglyceride from the intestine to the liver (Abumrad and Davidson 2012). Down-regulation of CD36 has resulted in subclinical inflammation and impaired barrier integrity in mice (Cifarelli et al., 2017). Altogether, the down-regulation of genes such as FABP1, FABP2 and CD36 can reduce fatty acid utilization and suppress immunity in the intestinal tissue.
In the current study, three DEG were common in both PPAR and β-oxidation pathways which are ACSBG1, ACSL1 and EHHADH. The ACSBG1 and ACSL1 genes are a strong activator for long fatty acid substrates (Pei et al., 2003). It has been reported that fruit flies with an inoperative ACSBG1 gene showed an accumulation of saturated very-long-chain fatty acids in their body (Pei et al., 2006), and the down-regulation of this gene has been previously reported in NE infected chickens (Qing et al., 2018) indicating the possible role of the gene in the fatty acid metabolic pathway. Lower expression of ACSL1 is known to reduce triglyceride synthesis and fatty acids in the cells and lead to inflammation in the tissue (Yan et al., 2015). Furthermore, EHHADH is mostly known for its important role in the peroxisomal β-oxidation pathway (Houten et al., 2012). Therefore, the down-regulations of these genes may indicate reduced fatty acid metabolism and energy expenditure and activated immunity reactions. The PPI analysis showed that ALDH2 and ALDH1A1 are among the genes with the highest number of interactions. These two genes are part of the aldehyde dehydrogenases (ALDH) family which are involved in a variety of biological processes such as inflammation, mitochondrial respiration, and xenobiotic metabolism (Chen et al. 2009, 2016). Down-regulation of the gene can induce inflammation, apoptosis and necrosis by the activation of ROS pathway (Fang et al., 2018; Liang et al., 2017).
4.2. Glycogenesis related genes
The strongest down-regulated gene found in the challenged group compared to non-challenged birds is PCK1. The PCK1 protein plays a key role in glutamine uptake, the main precursor of gluconeogenesis in the intestine, and thus the production of glucose (Croset et al., 2001; Rajas et al., 2000). Tissue inflammation is an energy-consuming process that increases the energy metabolic rate (Martin et al., 2003), and the fuel needed for such increased demand is glutamine (Wilmore 2001). It is also important to note that the primary source of energy for T-cells is glucose and glutamine (Bental and Deutsch 1993); thus, the down-regulation of PCK1 may lead to lower T-cell activity in the body (Ho et al., 2015) resulting in compromised immunity. Another strongly down-regulated gene in the current results is Pyruvate dehydrogenase lipoamide kinase isozyme 4 (PDK4). Inactivation of PDK4 plays an essential role in cellular energy metabolism (Sugden and Holness 2006) and can also regulate the response of macrophages in immunity activated situations (Min et al., 2019). Park et al. (2018) reported that deficiency of PDK4 in vitro and in vivo can reduce the expression of gluconeogenic genes which can subsequently diminish glucose production. These reports support the finding in the present study that the reduction of PDK4 was associated with the reduced expression of glucagon (GCG), a key hormone stimulating gluconeogenesis and the uptake of glucose. Down-regulation of GCG is also observed in inflammatory bowel disease and the secretion of inflammatory cytokines (Gu et al., 2018). Thus, GCG down-regulation observed in the current study may be an indication of inflammation and a disease condition in general.
4.3. Bile acid production and uptake related genes
Among the highest up-regulated genes in the NE challenged birds, was the ASBT. This protein is a primary intestinal bile salt transporter, and is a critical component of the enterohepatic circulation of bile acids (Hagenbuch and Dawson 2004). C. perfringens is known to have bile acid hydrolyze activity, which can hydrolyze the amide bond of conjugated bile salts and produce free bile salts. Free bile salts that have lower detergent properties for emulsifying fat (Cole and Fuller 1984; Hill 1995), thus, lead to lower fat absorption. On the other hand, the down-regulation of two genes essential in cholesterol and bile acid production, ACOX2 and ME1, suggest a lower production of cholesterol (Vilarinho et al., 2016). Furthermore, the ACOX2 protein, is believed to mediate the first step in β-oxidation and act as a bile acid intermediate (Baumgart et al., 1996). It should be noted that a portion of the bile salts secreted by hepatocytes are not newly synthesized and have already undergone enterohepatic recycling. Due to the hydrolyses effects of C. perfringens on the produced bile acids and the reduced synthesized amount of these substances in the liver, the increased expression of ASBT could be a response to enable the body to maximize the reabsorption of bile acids and enable the body to compensate for the possible deficiency of this product.
4.4. Immunity related genes
The NE infection significantly up-regulated several immunity-related genes such as ACOD1, INF-γ, Src family tyrosine kinase (LCK) and ZAP70. Among the highly up-regulated genes, ACOD1 is identified as a liposaccharide-inducible gene within macrophages that suggests an important role of this gene in the immune system (Lee et al., 1995). Macrophages form a heterogeneous population of immune cells that can respond efficiently to environmental and microbial signals. Up-regulation of interferon-γ (IFN-γ) has been previously observed under NE infection (Park et al., 2008; Wu et al., 2018) which is in agreement with the observation in the current study. The INF-γ protein capacitates macrophages to release a microbicidal product hydrogen peroxide (H2O2) (Nathan et al., 1983) which is an effective tool against bacteria, fungi and parasites (Liew et al., 1990). The ACOD1 gene has recently been shown to produce enzymes that act as an antimicrobial peptide and have bacteriostatic effects on some harmful bacteria (Michelucci et al., 2013). It is believed that the expression of this gene is largely dependent on the functional IFN signaling (Naujoks et al., 2016). Furthermore, LCK-null mice have shown blocked T-cell development and it seems that the expression of this gene is the initial event in the activation of T-cell antigen receptor complex (van Oers et al., 1996). The expression of LAT1 has shown to be increased in NE challenged broilers (Gharib-Naseri et al., 2019b), which is also observed in the current study. Hayashi et al. (2013) reported that LAT1 is a major transporter for essential amino acid uptake in activated T-cells. Altogether, it seems that the NE infection has up-regulated many genes related to T-cells activation. The up-regulation of some nutrient transporter genes could be due to the increasing demand for the essential amino acids from the T-cell function.
4.5. Skeletal muscle related genes
Negative effects of NE infections are primarily observed in the lower body weight which is related to reduced meat yield compared to the non-infected birds (Xue et al., 2017). Current results have shown that a number of genes related to skeletal muscle were differentially expressed between challenged and non-challenged birds. The genes PRKG2 and MYOM2 were among the strongly down-regulated genes, while MYBPC1 was strongly up-regulated in the NE challenged birds. The PRKG2 encodes a protein connected to organized columnar growth plates, and the knockout of this gene can leading to dwarfism in both mice and cattle (Koltes et al., 2009; Pfeifer et al., 1996). MYOM2 is a myosin-binding protein expressed in muscle sarcomeres (Van Der Ven et al., 1996), and has a critical role in muscle development in broilers (Liu et al., 2016). However, MYOM2 is also known to be associated with the immune system because it is an intracellular member of the immunoglobulin superfamily (Van Der Ven et al., 1996). Liu et al. reported that blocking TNF-α activity up-regulated MYOM2 expression. Furthermore, MYBPC1 belongs to the myosin-binding protein C family of proteins that are expressed in straight muscles (Ackermann et al., 2013) and plays an important structural and regulatory role in muscle function (James and Robbins 2011). It has been shown that the expression of MYBPC1 is counteracted by TNF-α in skeletal muscles (Meyer et al., 2015). The altered expression of these genes could have also affected the muscle growth and could be a reason for poor growth in the challenged birds. However, because RNA samples were obtained from the intestinal tissue, these changes are only the reflection of intestinal muscle development, and in some genes related to muscles, the changes can be due to the tissue damage caused by the infection. Further investigation is needed to identify the exact role of these genes and detect if expression changes of these genes are also observed in the skeletal muscles of challenged birds or not.
5. Conclusion
In conclusion, the transcriptomic analysis on the intestinal tissue of NE challenged birds showed significant changes in the expression of genes involved in their respective metabolic pathways that cause distinct responses to the disease. The analysis also revealed that fatty acid metabolism and absorption are widely affected by the infection. Immunity genes, especially those related to macrophage activity, were activated in infected birds. This study has taken a further step in identifying the responses of NE infected broilers at transcriptomic level, which can help develop therapeutic treatments by targeting the relevant pathways in the host and thereby aiding poultry producers to control this disease in the post-antibiotic era. There are many remaining questions concerning the regulation of lipid metabolism and the immune system, and thus further investigations are warranted for a better understanding of how the predisposing factors and NetB-producing C. perfringens modulate the expression of the genes and respective pathways and their functions.
Author contributions
Kosar Gharib-Naseri: Data curation, Formal analysis, Investigation, Writing- Original draft preparation; Sara de Las Heras-Saldana: Data curation, Formal analysis, Methodology, Investigation, Reviewing and Editing; Sarbast Kheravii: Methodology, Investigation, Data curation, Reviewing and Editing. Lihong Qin: Investigation, Data curation, Reviewing and Editing. Jingxue Wang: Investigation, Data curation, Reviewing and Editing. Shu-Biao Wu: Conceptualization, Data curation, Investigation, Project administration. Reviewing and Editing.
Conflict of interest
We declare that we have no financial or personal relationships with other people or organizations that might inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgements
The authors would like to thank Dr. Nickolas Rodgers for performing the animal trial, Dr Robert Moore of CSIRO, for providing C. perfringens strain for the challenge experiment, and Dr Rima Youil, Clodualdo Villaflor, and Wayne Woods of Bioproperties Pty. Ltd., Glenelg, for providing Eimeria spp. This research was conducted within the Poultry CRC, established and supported under the Australian Government’s Cooperative Research Centres Program (Project 2.1.5).
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aninu.2020.08.003.
Appendix. Supplementary data
The following is the supplementary data to this article:
References
- Abumrad N.A., Davidson N.O. Role of the gut in lipid homeostasis. Physiol Rev. 2012;92:1061–1085. doi: 10.1152/physrev.00019.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann M.A., Patel P.D., Valenti J., Takagi Y., Homsher E., Sellers J.R., Kontrogianni-Konstantopoulos A. Loss of actomyosin regulation in distal arthrogryposis myopathy due to mutant myosin binding protein-C slow. FASEB J. 2013;27:3217–3228. doi: 10.1096/fj.13-228882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agriculture and Resource Management Council of Australia and New Zealand . Model code of practice for the welfare of animals. CSIRO; Collingwood, Australia: 2002. [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviagen Ross broiler management manual. Manual. 2011:10–35. http://ptaviagencom/assets/Tech Center/Ross Broiler/Ross Broiler [Google Scholar]
- Bartlett K., Eaton S. Mitochondrial β-oxidation. Eur J Biochem. 2004;271:462–469. doi: 10.1046/j.1432-1033.2003.03947.x. [DOI] [PubMed] [Google Scholar]
- Barzegar Nafari S. University of New England; Armidale , Australia: 2019. Metabolism of energy and implementation of net energy system in laying hens. PhD thesis. [Google Scholar]
- Baumgart E., Vanhooren J.C., Fransen M., Marynen P., Puype M., Vandekerckhove J. Molecular characterization of the human peroxisomal branchedchain acyl-CoA oxidase: cDNA cloning, chromosomal assignment, tissue distribution, and evidence for the absence of the protein in Zellweger syndrome. Proc Natl Acad Sci. 1996;93:13748–13753. doi: 10.1073/pnas.93.24.13748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bental M., Deutsch C. Metabolic changes in activated T cells: an NMR study of human peripheral blood lymphocytes. Magn Reson Med. 1993;29:317–326. doi: 10.1002/mrm.1910290307. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S.J., Gaulton A., Marshall J., Bichko D., Martin S., Brouwer C., Harland L. Visualizing the drug target landscape. Drug Discov Today. 2010;15:3–15. doi: 10.1016/j.drudis.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Chen C.H., Joshi A.U., Mochly-Rosen D. The role of mitochondrial aldehyde dehydrogenase 2 (ALDH2) in neuropathology and neurodegeneration. Acta Neurol Taiwan. 2016;25:111–123. [PMC free article] [PubMed] [Google Scholar]
- Chen J., Tellez G., Richards J.D., Escobar J. Identification of potential biomarkers for gut barrier failure in broiler chickens. Front Vet Sci. 2015;2:14. doi: 10.3389/fvets.2015.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Mehta G., Vasiliou V. Antioxidant defenses in the ocular surface. Ocul Surf. 2009;7:176–185. doi: 10.1016/s1542-0124(12)70185-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifarelli V., Ivanov S., Xie Y., Son N.-H., Saunders B.T., Pietka T.A. CD36 deficiency impairs the small intestinal barrier and induces subclinical inflammation in mice. Cell Mol Gastroenterol Hepatol. 2017;3:82–98. doi: 10.1016/j.jcmgh.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C., Fuller R. Bile acid deconjugation and attachment of chicken gut bacteria: their possible role in growth depression. Br Poult Sci. 1984;25:227–231. doi: 10.1080/00071668408454861. [DOI] [PubMed] [Google Scholar]
- Croset M., Rajas F., Zitoun C., Hurot J.-M., Montano S., Mithieux G. Rat small intestine is an insulin-sensitive gluconeogenic organ. Diabetes. 2001;50:740–746. doi: 10.2337/diabetes.50.4.740. [DOI] [PubMed] [Google Scholar]
- Dupont J., Reverchon M., Cloix L., Froment P., Ramé C. Involvement of adipokines, AMPK, PI3K and the PPAR signaling pathways in ovarian follicle development and cancer. Int J Dev Biol. 2012;56:959–967. doi: 10.1387/ijdb.120134jd. [DOI] [PubMed] [Google Scholar]
- Fang T., Cao R., Wang W., Ye H., Shen L., Li Z. Alterations in necroptosis during ALDH2-mediated protection against high glucose-induced H9c2 cardiac cell injury. Mol Med Rep. 2018;18:2807–2815. doi: 10.3892/mmr.2018.9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A. STRING v9. 1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2012;41:808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharib-Naseri K., Kheravii S.K., Keerqin C., N. M, Swick R., Wu S. Two different Clostridium perfringens strains produce different levels of necrotic enteritis in broiler chickens. Poult Sci. 2019;98:6422–6432. doi: 10.3382/ps/pez480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharib-Naseri K., Kheravii S.K., Swick R., Choct M., Morgan N., Wu S. 22nd European symposium on poultry nutrition. Gdańsk; Poland: 2019. Different strains of Clostridium perfringens cause different effects on expression of genes encoding nutrient transporters in intestine of broilers under necrotic enteritis infection. [Google Scholar]
- Gilbert E., Li H., Emmerson D., Webb K., Wong E. Developmental regulation of nutrient transporter and enzyme mRNA abundance in the small intestine of broilers. Poult Sci. 2007;86:1739–1753. doi: 10.1093/ps/86.8.1739. [DOI] [PubMed] [Google Scholar]
- Golder H.M., Geier M.S., Forder R.E., Hynd P.I., Hughes R.J. Effects of necrotic enteritis challenge on intestinal micro-architecture and mucin profile. Br Poult Sci. 2011;52:500–506. doi: 10.1080/00071668.2011.587183. [DOI] [PubMed] [Google Scholar]
- Grygiel-Górniak B. Peroxisome proliferator-activated receptors and their ligands: nutritional and clinical implications-a review. J Nutr. 2014;13:1–10. doi: 10.1186/1475-2891-13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Liu J., Huang T., Zhang W., Jia B., Mu N. The protective and anti-inflammatory effects of a modified glucagon-like peptide-2 dimer in inflammatory bowel disease. Biochem Pharmacol. 2018;155:425–433. doi: 10.1016/j.bcp.2018.07.027. [DOI] [PubMed] [Google Scholar]
- Guo J., Shu G., Zhou L., Zhu X., Liao W., Wang S. Selective transport of long-chain fatty acids by FAT/CD36 in skeletal muscle of broilers. Animal. 2013;7:422–429. doi: 10.1017/S1751731112001619. [DOI] [PubMed] [Google Scholar]
- Hagenbuch B., Dawson P. The sodium bile salt cotransport family SLC10. Pflügers Archiv. 2004;447:566–570. doi: 10.1007/s00424-003-1130-z. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Endou H., Anzai N. Induction of expression of LAT1, an essential amino acid transporter, in activated T cells. J Immunol. 2013;190:4080–4085. doi: 10.4049/jimmunol.1300923. [DOI] [PubMed] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Vandesompele J. Qbase relative quantification framework and software for management and automated analysis of real-time quantitative pcr data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill M. Taylor and Francis; London: 1995. Role of gut bacteria in human toxicology and pharmacology. [Google Scholar]
- Ho P.C., Bihuniak J.D., Macintyre A.N., Staron M., Liu X., Amezquita R. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell. 2015;162:1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houten S.M., Denis S., Argmann C.A., Jia Y., Ferdinandusse S., Reddy J.K., Wanders R.J. Peroxisomal L-bifunctional enzyme (EHHADH) is essential for the production of medium-chain dicarboxylic acids. J Lipid Res. 2012;53:1296–1303. doi: 10.1194/jlr.M024463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James J., Robbins J. Signaling and myosin-binding protein C. J Biol Chem. 2011;286:9913–9919. doi: 10.1074/jbc.R110.171801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Hattori M., Aoki-Kinoshita K.F., Itoh M., Kawashima S. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyburn A.L., Boyce J.D., Vaz P., Bannam T.L., Ford M.E., Parker D. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog. 2008;4:1–11. doi: 10.1371/journal.ppat.0040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.K., Lillehoj H.S., Jang S.I., Lee S.H., Hong Y.H., Cheng H.H. Transcriptional profiles of host-pathogen responses to necrotic enteritis and differential regulation of immune genes in two inbreed chicken lines showing disparate disease susceptibility. PloS One. 2014;9 doi: 10.1371/journal.pone.0114960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitessa S.M., Nattrass G.S., Forder R.E., McGrice H.A., Wu S.-B., Hughes R.J. Mucin gene mRNA levels in broilers challenged with Eimeria and/or Clostridium perfringens. Avian Dis. 2014;58:408–414. doi: 10.1637/10757-122313-Reg.1. [DOI] [PubMed] [Google Scholar]
- Koltes J.E., Mishra B.P., Kumar D., Kataria R.S., Totir L.R., Fernando R.L. A nonsense mutation in cGMP-dependent type II protein kinase (PRKG2) causes dwarfism in American Angus cattle. Proc Natl Acad Sci. 2009;106:19250–19255. doi: 10.1073/pnas.0904513106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruskal W.H., Wallis A. Use of ranks in one-criterion variance analysis. J Amer Stat Assoc. 1952;47:583–621. [Google Scholar]
- Lee C.G., Jenkins N.A., Gilbert D.J., Copeland N.G., O'Brien W.E. Cloning and analysis of gene regulation of a novel LPS-inducible cDNA. Immunogenetics. 1995;41:263–270. doi: 10.1007/BF00172150. [DOI] [PubMed] [Google Scholar]
- Levy E., Ménard D., Delvin E., Stan S., Mitchell G., Lambert M. The polymorphism at codon 54 of the FABP2 gene increases fat absorption in human intestinal explants. J Biol Chem. 2001;276:39679–39684. doi: 10.1074/jbc.M105713200. [DOI] [PubMed] [Google Scholar]
- Li Y.P., Bang D.D., Handberg K.J., Jorgensen P.H., Zhang M.F. Evaluation of the suitability of six host genes as internal control in real-time RT-PCR assays in chicken embryo cell cultures infected with infectious bursal disease virus. Vet Microbiol. 2005;110:155–165. doi: 10.1016/j.vetmic.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Liang W., Chen M., Zheng D., Li J., Song M., Feng J. The opening of ATP-sensitive K+ channels protects H9c2 cardiac cells against the high glucose-induced injury and inflammation by inhibiting the ROS-TLR4-necroptosis pathway. Cell Physiol Biochem. 2017;41:1020–1034. doi: 10.1159/000461391. [DOI] [PubMed] [Google Scholar]
- Liew F., Millott S., Parkinson C., Palmer R., Moncada S. Macrophage killing of Leishmania parasite in vivo is mediated by nitric oxide from L-arginine. J Immunol. 1990;144:4794–4797. [PubMed] [Google Scholar]
- Liu D., Guo S., Guo Y. Xylanase supplementation to a wheat-based diet alleviated the intestinal mucosal barrier impairment of broiler chickens challenged by Clostridium perfringens. Avian Pathol. 2012;41:291–298. doi: 10.1080/03079457.2012.684089. [DOI] [PubMed] [Google Scholar]
- Liu J., Fu R., Liu R., Zhao G., Zheng M., Cui H. Protein profiles for muscle development and intramuscular fat accumulation at different post-hatching ages in chickens. PloS One. 2016;11 doi: 10.1371/journal.pone.0159722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M'Sadeq S.A., Wu S., Swick R.A., Choct M. Towards the control of necrotic enteritis in broiler chickens with in-feed antibiotics phasing-out worldwide. Anim Nut. 2015;1:1–11. doi: 10.1016/j.aninu.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L.B., Scheuerlein A., Wikelski M. Immune activity elevates energy expenditure of house sparrows: a link between direct and indirect costs? Proc R Soc Lond B Biol Sci. 2003;270:153–158. doi: 10.1098/rspb.2002.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt R., Brooker J., Acamovic T., Sparks N. Necrotic enteritis; a continuing challenge for the poultry industry. World’s Poult Sci J. 2006;62:221–247. [Google Scholar]
- Meyer S.U., Krebs S., Thirion C., Blum H., Krause S., Pfaffl M.W. Tumor necrosis factor alpha and insulin-like growth factor 1 induced modifications of the gene expression kinetics of differentiating skeletal muscle cells. PloS One. 2015;10 doi: 10.1371/journal.pone.0139520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelucci A., Cordes T., Ghelfi J., Pailot A., Reiling N., Goldmann O. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc Natl Acad Sci. 2013;110:7820–7825. doi: 10.1073/pnas.1218599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min B.-K., Park S., Kang H.J., Kim D.W., Ham H.J., Ha C.M. Pyruvate dehydrogenase kinase is a metabolic checkpoint for polarization of macrophages to the M1 phenotype. Front Immunol. 2019;10:944. doi: 10.3389/fimmu.2019.00944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R.J. Necrotic enteritis predisposing factors in broiler chickens. Avian Pathol. 2016;45:275–281. doi: 10.1080/03079457.2016.1150587. [DOI] [PubMed] [Google Scholar]
- Nathan C.F., Murray H.W., Wiebe M.E., Rubin B.Y. Identification of interferon-gamma as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J Exp Med. 1983;158:670–689. doi: 10.1084/jem.158.3.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naujoks J., Tabeling C., Dill B.D., Hoffmann C., Brown A.S., Kunze M. IFNs modify the proteome of Legionella-containing vacuoles and restrict infection via IRG1-derived itaconic acid. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B.Y., Jeon J.H., Go Y., Ham H.J., Kim J.E., Yoo E.K. PDK4 deficiency suppresses hepatic glucagon signaling by decreasing cAMP levels. Diabetes. 2018;67:2054–2068. doi: 10.2337/db17-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.S., Lillehoj H.S., Allen P.C., Park D.W., FitzCoy S., Bautista D.A., Lillehoj E.P. Immunopathology and cytokine responses in broiler chickens coinfected with Eimeria maxima and Clostridium perfringens with the use of an animal model of necrotic enteritis. Avian Dis. 2008;52:14–22. doi: 10.1637/7997-041707-Reg. [DOI] [PubMed] [Google Scholar]
- Pei Z., Jia Z., Watkins P.A. The second member of the human and murine “bubblegum” family is a testis-and brainstem-specific acyl-CoA synthetase. J Biol Chem. 2006;281:6632–6641. doi: 10.1074/jbc.M511558200. [DOI] [PubMed] [Google Scholar]
- Pei Z., Oey N.A., Zuidervaart M.M., Jia Z., Li Y., Steinberg S.J. The Acyl-CoA Synthetase “Bubblegum”(Lipidosin). Further characterization and role in neuronal fatty acid β-oxidation. J Biol Chem. 2003;278:47070–47078. doi: 10.1074/jbc.M310075200. [DOI] [PubMed] [Google Scholar]
- Pelsers M.M., Hermens W.T., Glatz J.F. Fatty acid-binding proteins as plasma markers of tissue injury. Clin Chim Acta. 2005;352:15–35. doi: 10.1016/j.cccn.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Pfeifer A., Aszódi A., Seidler U., Ruth P., Hofmann F., Fässler R. Intestinal secretory defects and dwarfism in mice lacking cGMP-dependent protein kinase II. Science. 1996;274:2082–2086. doi: 10.1126/science.274.5295.2082. [DOI] [PubMed] [Google Scholar]
- Prescott J., Sivendra R., Barnum D. The use of bacitracin in the prevention and treatment of experimentally-induced necrotic enteritis in the chicken. Can Vet J. 1978;19:181–183. [PMC free article] [PubMed] [Google Scholar]
- Qing X., Zeng D., Wang H., Ni X., Lai J., Liu L. Analysis of hepatic transcriptome demonstrates altered lipid metabolism following Lactobacillus johnsonii BS15 prevention in chickens with subclinical necrotic enteritis. Lipids Health Dis. 2018;17:93. doi: 10.1186/s12944-018-0741-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing X., Zeng D., Wang H., Ni X., Liu L., Lai J. Preventing subclinical necrotic enteritis through Lactobacillus johnsonii BS15 by ameliorating lipid metabolism and intestinal microflora in broiler chickens. AMB Express. 2017;7:139. doi: 10.1186/s13568-017-0439-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajas F., Croset M., Zitoun C., Montano S., Mithieux G. Induction of PEPCK gene expression in insulinopenia in rat small intestine. Diabetes. 2000;49:1165–1168. doi: 10.2337/diabetes.49.7.1165. [DOI] [PubMed] [Google Scholar]
- Ricote M., Valledor A.F., Glass C.K. Decoding transcriptional programs regulated by PPARs and LXRs in the macrophage: effects on lipid homeostasis, inflammation, and atherosclerosis. Arterioscler Thromb Vasc Biol. 2004;24:230–239. doi: 10.1161/01.ATV.0000103951.67680.B1. [DOI] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson L.E., Mazurak V.C. N-3 polyunsaturated fatty acids: relationship to inflammation in healthy adults and adults exhibiting features of metabolic syndrome. Lipids. 2013;48:319–332. doi: 10.1007/s11745-013-3774-6. [DOI] [PubMed] [Google Scholar]
- Rodgers N.J., Swick R.A., Geier M.S., Moore R.J., Choct M., Wu S.-B. A Multifactorial Analysis of the extent to which Eimeria and fishmeal predispose broiler chickens to necrotic enteritis. Avian Dis. 2015;59:38–45. doi: 10.1637/10774-011614-reg.1. [DOI] [PubMed] [Google Scholar]
- Stanley D., Wu S.B., Rodgers N., Swick R.A., Moore R.J. Differential responses of cecal microbiota to fishmeal, Eimeria and Clostridium perfringens in a necrotic enteritis challenge model in chickens. PloS One. 2014;9 doi: 10.1371/journal.pone.0104739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z., Li Z., Chen T., Li Q.-Z., Fang H., Ding D. Comparing next-generation sequencing and microarray technologies in a toxicological study of the effects of aristolochic acid on rat kidneys. Chem Res Toxic. 2011;24:1486–1493. doi: 10.1021/tx200103b. [DOI] [PubMed] [Google Scholar]
- Sugden M.C., Holness M.J. Mechanisms underlying regulation of the expression and activities of the mammalian pyruvate dehydrogenase kinases. Arch Physiol Biochem. 2006;112:139–219. doi: 10.1080/13813450600935263. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong A.D., Hong Y., Ban J., Park B., Hoang T.C., Hong Y.H., Lillehoj H.S. Analysis of differentially expressed genes in necrotic enteritis-infected fayoumi chickens using RNA sequencing. J Poult Sci. 2017;54:121–133. doi: 10.2141/jpsa.0160053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Ven P.F., Obermann W.M., Weber K., Fürst D.O. Myomesin, M-protein and the structure of the sarcomeric M-band. Adv Biophys. 1996;33:91–99. doi: 10.1016/0065-227x(96)81666-2. [DOI] [PubMed] [Google Scholar]
- Van Immerseel F., De Buck J., Pasmans F., Huyghebaert G., Haesebrouck F., Ducatelle R. Clostridium perfringens in poultry: an emerging threat for animal and public health. Avian Pathol. 2004;33:537–549. doi: 10.1080/03079450400013162. [DOI] [PubMed] [Google Scholar]
- van Oers N.S., Killeen N., Weiss A. Lck regulates the tyrosine phosphorylation of the T cell receptor subunits and ZAP-70 in murine thymocytes. J Exp Med. 1996;183:1053–1062. doi: 10.1084/jem.183.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A. Accurate normalization of real-time quantitative rt-pcr data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. research0034.1–research0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga T., Czimmerer Z., Nagy L. PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim Biophys Acta. 2011;1812:1007–1022. doi: 10.1016/j.bbadis.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilarinho S., Sari S., Mazzacuva F., Bilgüvar K., Esendagli-Yilmaz G., Jain D. ACOX2 deficiency: a disorder of bile acid synthesis with transaminase elevation, liver fibrosis, ataxia, and cognitive impairment. Proc Natl Acad Sci. 2016;113:11289–11293. doi: 10.1073/pnas.1613228113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade B., Keyburn A. The true cost of necrotic enteritis. World Poult. 2015;31:16–17. [Google Scholar]
- Wilmore D.W. The effect of glutamine supplementation in patients following elective surgery and accidental injury. J Nutr. 2001;131:2543–2549. doi: 10.1093/jn/131.9.2543S. [DOI] [PubMed] [Google Scholar]
- Wu Y., Shao Y., Song B., Zhen W., Wang Z., Guo Y. Effects of Bacillus coagulans supplementation on the growth performance and gut health of broiler chickens with Clostridium perfringens-induced necrotic enteritis. Rev Bras Cienc Avic. 2018;9:1–14. doi: 10.1186/s40104-017-0220-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue G.D., Wu S.B., Choct M., Swick R.A. Effects of yeast cell wall on growth performance, immune responses and intestinal short chain fatty acid concentrations of broiler in an experimental necrotic enteritis model. Anim Nut. 2017;3:399–405. doi: 10.1016/j.aninu.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S., Yang X.F., Liu H.L., Fu N., Ouyang Y., Qing K. Long-chain acyl-CoA synthetase in fatty acid metabolism involved in liver and other diseases: an update. World J Gastroenterol. 2015;21:3492–3498. doi: 10.3748/wjg.v21.i12.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian-Azodi M., Mortazavi-Tabatabaei S.A., Mansouri V., Vafaee R. Metabolite-protein interaction (MPI) network analysis of obsessive-compulsive disorder (OCD) from reported metabolites. Arvand J Health Med Sci. 2016;1:112–120. [Google Scholar]
- Zhou M.C., Yu P., Sun Q., Li Y.X. Expression profiling analysis: uncoupling protein 2 deficiency improves hepatic glucose, lipid profiles and insulin sensitivity in high-fat diet-fed mice by modulating expression of genes in peroxisome proliferator-activated receptor signaling pathway. J Diabetes Investig. 2016;7:179–189. doi: 10.1111/jdi.12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.