Abstract
Considering the importance of the poultry industry and the increasing interest in alternative growth promoters, probiotics are considered as a potential candidate for use in the poultry industry. In this study, Lactobacillus species were isolated from 21 rectal swabs of 11 healthy 6-day-old and 10 healthy 21-day-old chickens and their fecal and feed samples. The isolates were characterized and their probiotic characteristics, including resistance to gastric acid and bile salts, biofilm formation and adherence to epithelium or mucus, amylase and protease activity and production of inhibitory compounds, were assessed. From 31 acid and bile resistant lactobacilli, only 2 Lactobacillus brevis and 1 Lactobacillus reuteri strains showed significant probiotic properties. These isolates indicated detectable attachment to Caco-2 cells and significant antibacterial activities against Gram-positive and Gram-negative pathogens. Additionally, phenotypic and genotypic diversity of lactobacilli isolates were studied by Phene Plate (PhP) system (PhP-LB) and random amplified polymorphic DNA (RAPD)-PCR, respectively. PhP-LB results of 24 L. brevis isolates showed a high phenotypic variation among the isolates. In comparison, results of RAPD-PCR highlighted a low diversity. Therefore, it seems that combination of the 2 techniques (PhP and RAPD-PCR) could result in a significant discriminatory power than each of them used alone.
Keywords: Probiotic, Lactobacilli, Poultry, Feed, Fecal
1. Introduction
The Iranian poultry industry is the largest in the Middle East with nearly 1.2 million tons of output (meat and eggs). Therefore, this industry has a special status in the Iranian industry (Shariatmadari, 2000). The significant concerns related to this industry are health issues that threaten not only animal production, but also the people using their products (Griggs and Jacob, 2005; Nava et al., 2005; Zhang et al., 2018). Gastrointestinal diseases are one of the most important threats, as they lead to lost productivity, increased mortality, and contamination of poultry products for human consumption (Patterson and Burkholder, 2003).
The balance among the gastrointestinal microbiota plays a significant role in maintaining the normal physiology of host animals. Gastrointestinal microbiota help direct the normal formation or development of gut structure and morphology, support immune responses, offer protection from intestinal pathogens, and play an active role in the digestion of nutrients (Slizewska et al., 2020; Rodrigues et al., 2020). In the past, using antibiotics to promote the growth of animals and manage gut microbiota was a norm. Feeding of antibiotics to food animals has been recognized as one leading cause of the spread of antimicrobial resistance in human populations. The gradual emergence of populations of antibiotic resistant bacteria has become a major public health problem of global proportions. Due to this concern, since 2006 the European Union banned the use of antibiotics as growth promoters in animal feed (de Souza et al., 2018). Therefore, several alternative strategies have been proposed with some success that mimic the functions of antibiotics. Probiotics have been widely studied because of their ability to modulate gut microbiota and immunological systems in both humans and livestock. They have been used to increase milk production and to reduce diarrhoea both in cattle and pigs, and to control the colonisation of the intestinal tract by pathogenic bacteria (Alayande et al., 2020).
The microbes which are suitable for probiotic purposes in human and animals are mainly members of a metabolically defined group of Gram-positive bacteria, known as lactic acid bacteria (LAB) (Naidu et al., 1999). These microbes are widely distributed in the environment and play a significant role in the gastrointestinal tract (GIT) of a diverse array of animals (Bermudez-Brito et al., 2012; Butel, 2014). A main part of the candidate strains, which have been introduced for probiotic purposes, fall into the genus Lactobacillus which is a major genus of LAB and harbor more than 200 species (Goktepe et al., 2005). Lactobacillus species, with a record of safe use as probiotics in humans and animals, are among the common inhabitants of the broiler GIT (Lu et al., 2003). In poultry, administration of the probiotic Lactobacillus strains improves not only the feed digestion, but also the nutrient uptake. In addition, probiotics increase the growth performance, neutralizing various enterotoxins and enhancing immune responses (Ghadban, 2002; Al-Khalaifa et al., 2019). Additionally, probiotics reduce the risk of gastrointestinal colonization by foodborne pathogens, such as Campylobacter (Ghareeb et al., 2012; Khan et al., 2019; Neal-McKinney et al., 2012), Clostridium (Li et al., 2017) and Salmonella (Kizerwetter-Swida and Binek, 2009; Tellez et al., 2012), and increase the safety of poultry-based foods (Gaggìa et al., 2010). Such antagonistic activities against the pathogens is highly linked to lactic acid produced by Lactobacillus strains, which can be toxic for many bacteria, can compete for nutrients, and affect cell attachment capabilities of the beneficial microbes to the intestinal epithelium (Patterson and Burkholder, 2003).
Despite the importance of the poultry industry in Iran, little attention has been given to the isolation of probiotic bacteria and determination of their biological activities in this country. In this study, we aimed to isolate and characterize Lactobacillus species from indigenous poultry farms with a special focus on their probiotic properties.
2. Materials and methods
2.1. Isolation of lactic acid bacteria
A total of 21 rectal swabs of 11 healthy 6-day-old and 10 healthy 21-day-old chickens, their fecal and their feed samples, were collected from 2 poultry farms near Tehran, Iran. The chicken breed was Ross and diets were standard according to the breed requirements, containing maize and soybean balanced with minerals, vitamins, and amino acids without growth promoting antibiotics. To isolate LAB from fecal samples, 6 chicken feces from 2 poultry farms (each sample was 1 g) were collected randomly in Man Rogosa and Sharpe (MRS) broth (Merck, Germany). The samples were serially diluted in phosphate-buffered saline (PBS) pH 7.4 and aliquots of them were plated on MRS agar medium in 8-cm plates. The plates were incubated for 48 h at 37 °C in microaerobic conditions. From each sample, different morpho-type colonies were selected for further purification and then the LAB isolates were cryopreserved at −80 °C in MRS broth containing 20% glycerol under defined designations.
2.2. Resistance to low pH and bile salts
Tolerance of isolates to low acidity and bile salts were determined in triplicate experiments as described by Cano Roca et al. (2014). Briefly, exponentially growing cells in MRS broth were washed by centrifugation (4,000 × g at 25 °C for 10 min) and re-suspended in PBS. After serial dilutions, an initial dilution of the bacterial suspension was prepared for plating on MRS agar. To investigate the reaction of the isolates to low pH values, 100 μL of the cell suspension (108 CFU/mL) was added to 900 μL of sterile PBS (pH = 3) in a 1.5-mL microtube. The endurable cell counts were measured after 3 h of incubation at 37 °C. A similar procedure was performed using PBS (pH = 7) as a control. To determine the tolerance of isolates to bile salts, 50 μL of bacterial suspension was added into tubes containing 4,950 μL of MRS broth (Merck, Germany) with 0.4% (wt/vol) of bile salts (Merck, Germany) and incubate at 37 °C for 6 h.
The harvested cells from the acid in both bile salt stress experiments were washed in PBS (pH = 7.4) and cultured on MRS agar and finally counting was performed. Based on 2, 2–4, 4–6 and >6 log reduction in comparison to the initial suspension after 3 and 6 h of incubation in acid and bile salts, isolates were grouped as strongly resistant, resistant, intermediate and susceptible, respectively.
2.3. Phenotypic classification
All of the acid-bile resistant isolates were subjected to a biochemical fingerprinting with the PhPlate system according to the manufacturer's instructions (PhPlate Micro-plate Techniques AB, Stockholm, Sweden) which was modified for isolates typing (PhP-LB). The microplates contained 4 sets of dehydrated reagents (23 different sugars including arabinose, xylose, galactose, maltose, cellobiose, trehalose, palatinose, sucrose, lactose, melibiose, manose, melezitose, inosin, mannitol, arbutin, sorbitool, gallac, sorbose, rhamnose, taghatose, amigdalin, gluconate, salicin), which have been specifically selected for phenotypic typing of Lactobacillus species. After the incubation of PhP-LB plates at 37 °C, the utilization of the substrates in each well was measured by scanning the images after 24, 48, and 72 h. Scanned images were analyzed by software package PhPWIN (PhPlate micro-plate techniques AB, Sweden). The mean similarity between duplicate assays of all strains ±2 SD was calculated as the identification level (ID), which was 0.975 and strains with >0.975 similarities were grouped into the same Phene Plate (PhP) type.
2.4. Molecular identification
Total DNA of the acid-bile resistant isolates was extracted using a peqGOLD Bacterial DNA Kit (peQlab, Germany) according to the manufacturer's instruction. Preliminary characterization of lactobacilli was performed based on the phenotype. Then, molecular identification of the Lactobacillus spp. was performed using primers which were specific for amplification of a 247 bp region of the 16S rRNA gene in the genus Lactobacillus (McOrist et al., 2002). The PCR amplification program was as follows: a single initial denaturation cycle (5 min at 94 °C) followed by 30 cycles (30 s at 94 °C [denaturation], 30 s at 57 °C [annealing], and 30 s at 72 °C [elongation]), with a final extension of 7 min at 72 °C. As the next step, multiplex PCR amplifications were used for Lactobacillus species identification. Hence, the previously designed species specific primer pairs which were already confirmed for detection of Lactobacillus acidophilus, Lactobacillus casei, Lactobacillus delbrueckii, Lactobacillus gasseri, Lactobacillus rhamnosus, Lactobacillus plantarum, and Lactobacillus reuteri were used to identify the Lactobacillus isolates (Kwon et al., 2004). Multiplex PCR reactions for amplifications entailed a cycle of 94 °C for 5 min, followed by 40 cycles (94 °C for 30 s, 51 °C for 40 s and 72 °C for 30 s), plus one additional cycle with a final 7 min chain elongation at 72 °C. The amplified genomic regions of Lactobacillus brevis strains were sequenced. Additionally, species specific primers were used for identification of L. brevis strains as described by Guarneri et al. (2001).The thermo-cycling conditions for the L. brevis specific PCR reaction was as follows: after one cycle at 94 °C for 2 min, 25 cycles of 94 °C for 1 min, 40 °C for 1 min and 72 °C for 1 min, followed by an additional 10 min cycle of extension (Table 1).
Table 1.
Sequence of primers used to identify different species of Lactobacillus bacteria.
| Target bacteria | Sequence | References |
|---|---|---|
| All Lactobacillus | 5′ TGGAAACAGGTGCTAATACCG 3′ 5′ CCATTGTGGAAGATTCCC 3′ |
McOrist et al. (2002) |
|
L. casei-group L. acidophilus L. delbrueckii L. gasseri L. reuteri L. plantarum L. rhamnosus |
5′ CCACCTTCCTCCGGTTTGTCA 3′ 5′ AGGGTGAAGTCGTAACAAGTAGCC 3′ 5′ TGGTCGGCAGAGTAACTGTTGTCG 3′ 5′ AACTATCGCTTACGCTACCACTTTGC 3′ 5′ CTGTGCTACACCTAGAGATAGGTGG 3′ 5′ ATTTCAAGTTGAGTCTCTCTCTC 3′ 5′ ACCTGATTGACGATGGATCACCAGT 3′ 5′ CTAGTGGTAACAGTTGATTAAAACTGC 3′ 5′ GCCAACAAGCTATGTGTTCGCTTGC 3′ |
Kwon et al. (2004) |
| L. brevis | 5′ CTTGCACTGATTTTAACA 3′ 5′ GGGCGGTGTGTACAAGGC 3′ |
Guarneri et al. (2001) |
2.5. RAPD-PCR genotypic classification
Acid-bile resistant isolates, which were identified as lactobacilli, were subjected to Random amplified polymorphic DNA (RAPD)-PCR using a previously designed oligonucleotide by Tilsala et al. (1998) with some modifications. The PCR amplification conditions were as follows: 2 min at 94 °C for initial denaturation followed by 40 cycles of 30 s at 94 °C for denaturation, 30 s at 37 °C for annealing, and 2 min at 72 °C for elongation. The final extension at 72 °C was prolonged to 10 min. PCR reaction was performed for each primer in a separate tube and run in the same well in 1.5% agarose gel to increase the discrimination. The UPGMA method using the software Gel compare II version 4.0 was used to compare banding patterns (Applied Maths, Sint-Martens-Latem, Belgium).
2.6. Biofilm assay
Biofilm formation of lactobacilli was studied as previously described by Lebeer et al. (2007) with minor modifications. For each strain, 200 μL aliquots of a modified tryptic soy broth (TSB) medium (15 g/L TSB enriched with 20 g/L Bacto proteose peptone), which was already inoculated by approximately 3 × 107 CFU of a Lactobacillus isolate, were added into 96-well plates (8 wells for each strain) and incubated at 37 °C. After 72 h of incubation, the wells were washed with PBS and stained for 30 min with 200 μL crystal violet (0.1%) in an isopropanol-methanol-PBS solution (1:1:18). After washing with double distilled water, the wells were air-dried for 30 min at room temperature. Extraction of the dye bound to the adherent cells was done with 200 μL ethanol-acetone (80:20) solution. The optical density of 135 μL of each well was measured at 570 nm. Data were normalized to the indicated positive control, which was taken as 100% to compare different experiments. The results are presented as means ± SD. Additionally, the sterile medium and Pseudomonas aeruginosa were used as negative and positive controls, respectively.
2.7. Attachment to Caco-2 cells
Detection of adhesion ability in lactobacilli isolates was performed according to Jacobsen et al. (1999). A monolayer of Caco-2 cells was cultured in Roswell Park Memorial Institute (RPMI) medium (Gibco, Carlsbad, CA, USA), supplemented with 20% (vol/vol) fetal calf serum (Gibco, Life Technology, USA), penicillin (100 U/mL) and streptomycin (100 mg/mL), and incubated at 37 °C in 5% CO2 atmosphere. At first, 3 mL of Caco-2 cells containing 1.5 × 105 cells/mL were seeded on a 6-well cell culture plate and after confluency, the cells were washed twice with 3 mL PBS. After adding 2 mL of RPMI (without antibiotics) to each well, the plates were incubated for 3 h at 37 °C. Overnight cultures of the isolates (cell concentration of approximately 109 CFU/mL) were suspended in 1 mL RPMI1640 medium (without antibiotics) and added to different wells and incubated for 1 h at 37 °C. The wells were washed 4 times with PBS to remove the unbound bacteria. Then, the cells were fixed with 3 mL of methanol and incubated for 5 to 10 min at room temperature for the removal of methanol. Staining was made with 3 mL of Giemsa stain solution (1:20) (Sigma–Aldrich Co., Mo, USA) and incubated for 30 min at room temperature. After washing the plates with distilled water, the air-dried plates were examined microscopically under oil immersion. Adherent isolates were counted in 20 random microscopic fields. Cells showing <40, between 40 and 100, and >100 attached bacteria were regarded as non-adhesive, adhesive, and strongly adhesive, respectively.
2.8. Detection of amylase and protease activities
Enzymatic activity of 31 acid-bile resistant lactobacilli was determined according to the method described by Taheri et al. (2009) with minor modifications. For assessment of the amylase activity, the selected Lactobacillus strains were cultured on modified MRS broth described by Taheri et al. (2009) (0.25% starch instead of glucose), and inoculated on a medium containing starch (2%), meat peptone (0.5%), yeast extract (0.7%), NaCl (0.2%), and agar (1.5%). After 48 h of incubation at 37 °C, lugol's solution (5 g iodine [Merck, Germany] and 10 g potassium iodide [KI] [Merck, Germany] in 100 mL distilled water) was poured over the agar for detection of any clear zones as indicative of amylolytic activities.
For detection of proteolytic activity, Lactobacillus strains were inoculated into MRS broth and were incubated at 37 °C for 24 h. Bacterial suspension (30 μL) was moved onto a disc placed over a MRS agar containing 1% skim milk. Finally, the halo zone surrounding each disc was measured.
2.9. Antimicrobial activity
The antimicrobial activity of the lactobacilli, which showed a detectable attachment to Caco-2 cells, was studied against Shigella soneii (ATCC 12022), Pesudomonas aeruginosa (ATCC 27853), Salmonella typhi (ATCC, 19430), Proteus mirabilis (ATCC 25933), Yersinia enterocolitica (ATCC 23715), Streptococcus agalactiea (ATCC 12386), Listeria monocytogenes (ATCC, 19113), wild types of Escherichia coli strains belonging to 3 pathotypes i.e. enteropathogenic E. coli (EPEC) (ATCC 43887), enterotoxigenic E. coli (ETEC) and enteroaggregative E. coli (EAEC). These strains were provided by the Microbial Collection of Iran (Davoodabadi et al., 2015; Shahrokhi et al., 2011). The antimicrobial activity was observed based on the well diffusion method as described by Fernández et al. (2003). Suspensions containing approximately 108 CFU/mL of the abovementioned pathogens were poured on Muller Hinton agar medium in 8-cm plates, except for L. monocytogenes where brain heart infusion (BHI) agar medium was used. Then, 100 μL of an overnight culture of the selected Lactobacillus strains in MRS broth was poured into 6 mm agar wells created by punching in 8-cm plates. After 24 h of incubation at 37 °C, the antimicrobial activity was measured as the zone of growth inhibition around the wells. The inhibition zones of 1, 2, 2 to 5 mm, and more than 5 mm were classified as strains of no (−), mild (+), strong (++), and very strong (+++) inhibition, respectively.
2.10. Antibiotic susceptibility testing
The susceptibility of isolates to different antibiotics including penicillin G (10 μg), gentamicin (120 μg), erythromycin (15 μg), tetracycline (30 μg), amoxicillin (25 μg), ciprofloxacin (5 μg), chloramphenicol (30 μg), oxacillin (1 μg), and streptomycin (10 μg) (MAST Diagnostics, U.K.) was determined by the agar disc diffusion method on MRS agar plates instead of the Muller Hinton agar. Lactic acid bacteria require special growth conditions and conventional media, as Mueller Hinton agar are not uniformly suitable for to susceptibility test them (Klare et al., 2005). After incubation of plates at 37 °C for 48 h, the diameter of the inhibition zones was measured and the results were expressed as sensitive or resistant according to CLSI standard (Institute, 2009).
2.11. Plasmid profiles
The isolation of plasmid DNA from the selected bacterial strains was performed by GF-1 plasmid DNA extraction kit (Vivantis, Malaysia). E. coli V517 was used as a positive control. Electrophoresis of the extracted plasmids was performed in a 1% agarose gel and the plasmids were visualized with UV trans-illumination in a Gel Doc apparatus.
3. Results
3.1. Isolation and identification
A total of 168 LAB were isolated from rectal swabs, fecal and feed samples, among which 89 (53%) and 79 (47%) isolates were from 21 to 6 days old chickens, respectively. Furthermore, out of 168 LAB isolates, 51 (30.3%) isolates were resistant to low pH (pH 3.00) and bile salts (0.4%), among which 20 and 31 isolates were identified as members of the genera Pediococcus and Lactobacillus, respectively.
The results of the molecular identification showed that 31 isolates belonged to the genus Lactobacillus. Out of these, 24 (77.4%), 3 (9.6%), 2 (6.4%), and 2 (6.4%) were identified as L. brevis, L. plantarum, L. reuteri and L. vaginalis, respectively (Table 2). Interestingly, a considerable part of the isolates which originated from the rectal-swabs were identified as L. brevis. The same results were observed in fecal samples.
Table 2.
Determination of acid and bile resistant Lactobacillus species isolated from different samples.
| Item |
L. brevis |
L. plantarum |
L. reuteri |
L. vaginalis |
||||
|---|---|---|---|---|---|---|---|---|
| 21-day-old | 6-day-old | 21-day-old | 6-day old | 21-day-old | 6-day-old | 21-day-old | 6-day-old | |
| Feed sample | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 0 |
| Rectal swab | 14 | 6 | 0 | 0 | 1 | 1 | 0 | 2 |
| Fecal sample | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
3.2. Phenotypic and genotypic classification
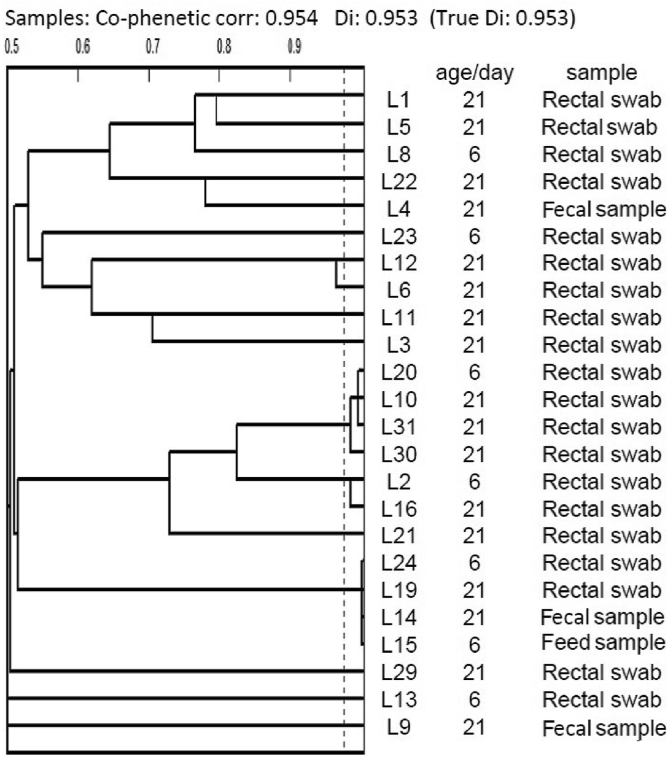
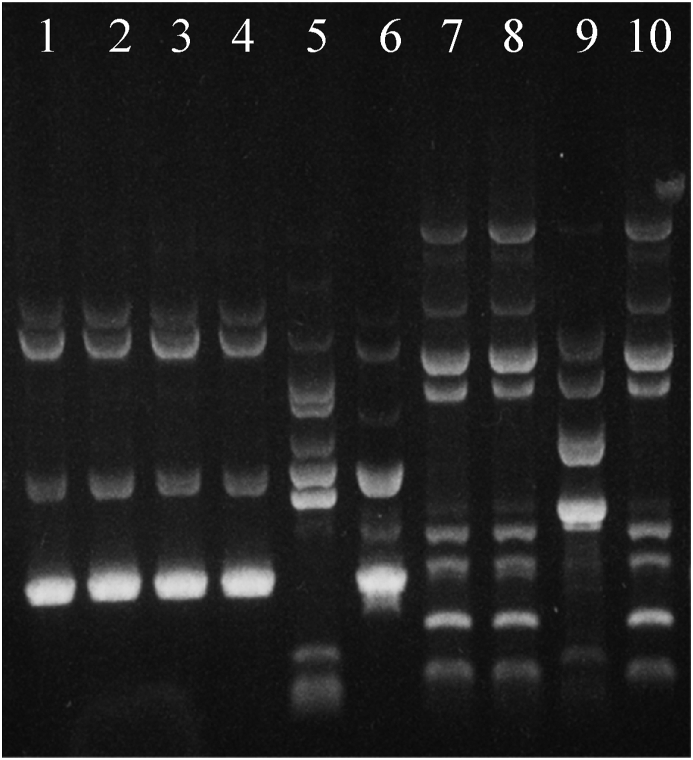
Acid-bile resistant lactobacilli were typed using Phene Plate system (PhP-LB) followed by genotype-based identification using RAPD-PCR. PhP-LB results of 24 L. brevis isolates showed a high phenotypic diversity and most of the isolates gave unique phenotypes, denoted as single types (3 common types with 10 isolates and 14 single types) (Fig. 1). On the other hand, the results of RAPD-PCR showed a low genetic diversity in L. brevis species (3 common types with 20 isolates and 4 single types) (Fig. 2).
Fig. 1.
Cluster analysis of Phene plate (PhP) assay of 24 bile and acid resistant Lactobacillus brevis.
Fig. 2.
Agarose gel electrophoreses of RAPD-PCR products of Lactobacillus brevis strains. RAPD = random amplified polymorphic DNA.
3.3. Biofilm and attachment to Caco-2 cells assay
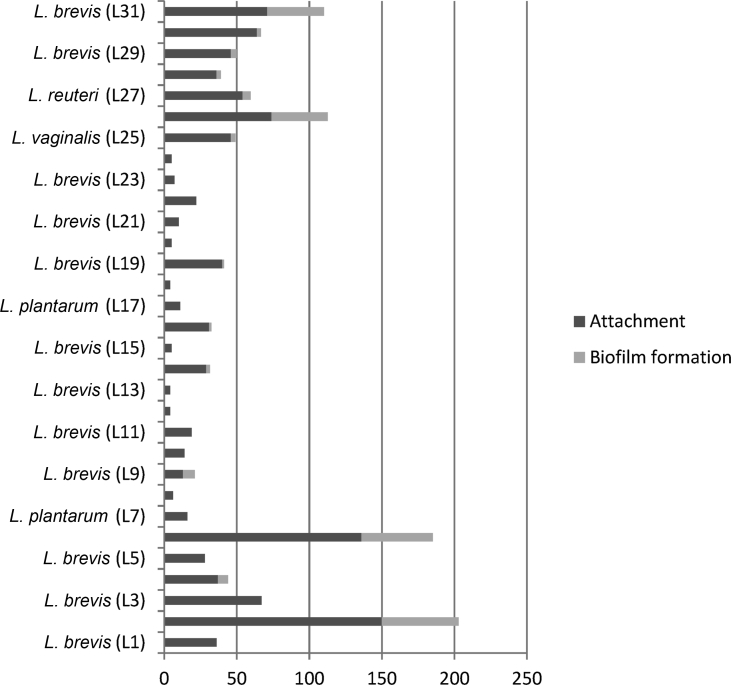
Out of the 31 acid-bile resistant lactobacilli strains examined in this study, L. brevis L2, L6, L31, and L. reuteri L26 had the biofilm formation capability. Adhesion to Caco-2 cells showed that 22 (71%) isolates were non-adhesive (with less than 40 bacteria attached in 20 microscopic fields), 7 (22.5%) isolates (L. brevis L3, L29, L30, L31, L. reuteri L26, L27, and L. vaginalis L25) were adhesive (with 41 to 100 bacteria) and 2 (6.4%) isolates (L. brevis L2 and L6) were strongly adhesive (>101 bacteria) (Fig. 3).
Fig. 3.
Biofilm formation and attachment in 31 bile and acid resistant lactobacilli. Dark gray bars show the number of attached lactobacilli in 20 microscopic fields to Caco-2 cell line and light gray bars show the percentage of biofilm formation.
3.4. Antibacterial activity
The assay of antagonistic activity of lactobacilli with an acceptable ability of attachment to Caco-2 cells showed that L. brevis strains L2, L6, L31 and L. reuteri strains L26 and L27 have a strong inhibitory effect against different serotypes of E. coli (EAEC, EPEC, and ETEC) (Table 3). L. brevis strains L2 and L6 had antibacterial activities against all the tested pathogenic bacteria with the exception of L. monocytogenes and Staphylococcus aureus.
Table 3.
Spectrum of antimicrobial activity exhibited by Lactobacillus strains L2, L3, L6, L25, L26, L 27, L 29, L 30 and L 31.
| Item | L2 (L. brevis) | L3 (L. brevis) | L6 (L. brevis) | L25 (L. vaginalis) | L26 (L. reuteri) | L27 (L. reuteri) | L29 (L. brevis) | L30 (L. brevis) | L31 (L. brevis) |
|---|---|---|---|---|---|---|---|---|---|
| ETEC | + | – | + | + | + | + | – | – | + |
| EPEC | ++ | – | ++ | – | ++ | ++ | – | – | ++ |
| EAEC | ++ | – | ++ | – | + | + | – | – | ++ |
| Salmonella entritidis | ++ | – | ++ | + | + | + | – | – | + |
| Salmonella typhi | + | – | + | – | – | – | – | – | – |
| Pseudomonas aeruginosa | + | – | + | – | – | – | – | + | – |
| Shigella flexneri | + | – | + | – | + | + | – | – | – |
| Klebsiella pneumonia | + | – | + | – | – | – | – | – | – |
| Citrobacter freundii | + | – | + | – | – | – | + | + | – |
| Proteus mirabilis | + | – | + | – | – | – | + | + | – |
| Yersinia. enterocolitica | + | – | + | – | – | – | – | – | – |
| Listeria monocytogenes | – | – | – | – | – | – | – | – | – |
| Staphylococcus aureus | – | – | – | – | – | – | – | – | – |
| Staphylococcus saprophyticus | + | – | + | – | – | – | – | – | – |
| Streptococcus group A | + | – | + | – | – | – | – | – | – |
ETEC = enterotoxigenic E. coli; EPEC = enteropathogenic E. coli; EAEC = enteroaggregative E. coli.
Note: no (−), mild (+), and strong (++) inhibition.
3.5. Detection of amylase and protease activities
Measuring the size of the halo zones surrounding the colonies as an indicative of extracellular enzyme level showed that all 31 lactobacilli isolates investigated here were protease positive but extracellular-amylase negative.
3.6. Plasmid profiles and antibiotic susceptibility testing
No plasmid was found in L. brevis strain L6 but L. brevis L2 and L. reuteri L26 harbored a single plasmid. Antibiotic susceptibility tests showed that the Lactobacillus isolates were sensitive to augmentin, amoxicillin, erythromycin, penicillin G, chloramphenicol, and rifampin and were resistant to ciprofloxacin, amikacin, tobramycin, oxacillin and streptomycin. L. reuteri L26 was sensitive to tetracycline but L. brevis L2 and L6 were resistant to this antibiotic.
4. Discussion
Probiotics have been emerging as a safe alternative to antibiotics for increasing performance in livestock. Administration of probiotic strains may have a significant effect on absorption and utilization of feed, and increase the body weight of various animals, including chicken, piglets, sheep and cattle (Markowiak et al., 2018). Lactobacillus species with a record of safe use as a probiotic in humans and animals is regarded as a significant part of chicken-GIT microbiota (Wei et al., 2013; Yadav, 2019). Among such a diverse array of Lactobacillus species, some defined species have been frequently reported as chicken-GIT inhabitants. L. reuteri, L. salivarius and L. johnsonii are among the most detected lactobacilli in chicken-GIT samples (Adhikari and Kwon, 2017; Dec et al., 2016; Pan and Yu, 2014). Interestingly, L. reuteri, L. salivarius and L. johnsonii have been repeatedly isolated from the GIT samples of a wide range of hosts and this harsh environment is among their preferred ecological niches (Lebeer et al., 2008; Walter, 2008; Pokorná, 2019). However, L. brevis have also been found in GIT samples in a diverse range of warm-blooded animals (Feyereisen et al., 2019, Fraunhofer, 2019). Considering the results of this study, 3 lactobacilli strains were isolated, which showed promising probiotic characteristics. Those strains fall into L. brevis (L2 and L6) and L. reuteri (L26) species. L. brevis shows a significant prevalence in the GIT of chickens although the GIT is not the preferred ecological niche for this species. The species L. brevis falls into a Lactobacillus phylogenetic group which harbors mostly foodborne species (Papizadeh et al., 2017). Strains of this species have been isolated from a diverse array of samples, including water, feces of various animals, and various food-associated samples (Feyereisen et al., 2019). Hence, the findings of this study shed more light on the ecological distribution of L. brevis.
Considering the results, L. brevis species were isolated from the rectal swabs of both 21- and 6-day-old chickens and also from their feed and fecal samples. Hence, it can be inferred that this species has the ability to survive on a wide range of substrates (Ramos et al., 2013).
Phenotypic characterization of the isolates indicated a high intra-species diversity among L. brevis isolates. In comparison, results of RAPD-PCR highlighted a low diversity. Therefore, it seems that combination of 2 techniques (PhPlate and RAPD-PCR) could result in a more significant discriminatory power than each of them used alone. In this study, biochemical fingerprinting of lactobacilli was used primarily for the screening of biodiversity in lactobacilli strains to reduce their number for the next tests; a significant number of single types indicated that PhP system alone cannot serve as a method for determining relationships between Lactobacillus strains (Skelin et al., 2012).
Adhesion to mucosal surfaces has been used as a criterion for the selection of probiotic bacteria because this character has a major role in the colonization of the GIT by these bacteria (Broderick and Duong, 2016; Kosin and Rakshit, 2006). Additionally, mucosal adhesion is important for pathogenic antagonism, modulation of the immune system and healing of damaged gastric mucosa (Oelschlaeger, 2010; Ohland and MacNaughton, 2010; Monteagudo-Mera et al., 2019). In this study, 2 strongly adhesive strains (L. brevis strains L2 and L6) showed the highest biofilm formation capacity. Furthermore, we observed specific correlation between adhesion to Caco-2 cells and biofilm formation by lactobacilli isolates.
Antimicrobial resistance poses a serious global threat of growing concern to human, animal and environmental health. This is due to the consumption of antibiotics in animals (raised for food or kept as pets) and humans (Aslam et al., 2018). Therefore, probiotics with antibacterial activity against pathogens are a promising alternative to antibiotics (Baldwin et al., 2018). The strong antibacterial activity of L. brevis strains L2, L6, L31 and L. reuteri strains L26 and L27 against various serotypes of E. coli (EAEC, EPEC and ETEC), which is shown in Table 3, have highlighted the probiotic capabilities of these strains. Interestingly, such a capability among the isolates of this study was highly detectable in cases of L. brevis strains L2 and L6, which showed significant antibacterial activities against all the tested pathogenic bacteria (except for L. monocytogenes and S. aureus). Such an antibacterial activity of Lactobacillus isolates is essentially associated with the production of bacteriocins, H2O2, lactic acid and other metabolites which inhibit the growth of pathogens (Vieco-Saiz et al., 2019). Considering the fact that various serotypes of E. coli (EAEC, EPEC, and ETEC) are considered as the most important cause of enteric bacterial infections in poultry, the use of such isolates with functional probiotic competence can significantly reduce the infection rate. Another finding was that all 31 lactobacilli isolates investigated in this study, were protease positive with no extracellular amylase activity and this has shed light on the importance of these isolates since amylase, lipase, and protease enzymes play very important roles in the digestion of nutrient materials.
The probable existence of transferable resistant genes in the 3 probiotic Lactobacillus strains was observed by the determination of antibiotic resistance patterns and plasmid profiling, but no plasmid was detected in L. brevis strains L2 and L6 and only a single plasmid was detected in L. reuteri strain L26. Furthermore, L. reuteri L26 was sensitive to tetracycline, but L. brevis L2 and L6 were resistant to this antibiotic. The properties of antibiotic resistance in various Lactobacillus species seem to be associated with drug resistant genes which are mainly located on the chromosome.
According to the criteria, the potential probiotic strains, which are assumed for animal or human applications, have to be non-pathogenic and from the same origin (host). Additionally, such strains should resist intestinal tract, gastric and bile acids, adhere to the epithelium or mucus, and produce inhibitory compounds. Among the lactobacilli isolated in this study, we found 3 lactobacilli strains with probiotic characteristics, L. brevis (L2 and L6) and L. reuteri (L26), which could be considered probiotic strains for use in the poultry industry.
5. Conclusion
In this study, the most common acid and bile resistant lactobacilli strains isolated from chickens belonged to the L. brevis species, with a high intra-species phenotypic diversity. In vitro evaluation in this study showed that 3 Lactobacillus strains (2 L. brevis strains and 1 L. reuteri strain) could be considered as probiotic. Further in vivo evaluation for determination of the beneficial effects of our isolates in natural conditions could be highly advantageous to the Iranian poultry industry.
Author contributions
Nasrin Noohi: writing - original draft, visualization, investigation, formal analysis; Moslem Papizadeh: writing - review & editing; Mahdi Rohani: investigation; Malihe Talebi: project administration, methodology; Mohammad R. Pourshafie: conceptualiza, supervision.
Conflict of interest
We declare that we have no financial or personal relationships with other people or organizations that might inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgements
This study was support by a grant (No. 90007454) from Iran National Science Foundation.
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Contributor Information
Malihe Talebi, Email: talebi.m@iums.ac.ir.
Mohammad R. Pourshafie, Email: pour@pasteur.ac.ir.
References
- Adhikari B., Kwon Y.M. Characterization of the culturable subpopulations of Lactobacillus in the chicken intestinal tract as a resource for probiotic development. Front Microbiol. 2017;8:1389. doi: 10.3389/fmicb.2017.01389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khalaifa H., Al-Nasser A., Al-Surayee T., Al-Kandari S., Al-Enzi N., Al-Sharrah T., Ragheb G., Al-Qalaf S., Mohammed A. Effect of dietary probiotics and prebiotics on the performance of broiler chickens. Poultry Sci. 2019;98:4465–4479. doi: 10.3382/ps/pez282. [DOI] [PubMed] [Google Scholar]
- Alayande K.A., Aiyegoro O.A., Ateba C.N. Probiotics in animal husbandry: applicability and associated risk factors. Sustainability. 2020;12:1087. [Google Scholar]
- Aslam B., Wang W., Arshad M.I., Khurshid M., Muzammil S., Rasool M.H., Nisar M.A., Alvi R.F., Aslam M.A., Qamar M.U., Farooq Salamat M.K., Baloch Z. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist. 2018;11:1645–1658. doi: 10.2147/IDR.S173867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin S., Hughes R.J., Hao Van T., Moore R.J., Stanley D. At-hatch administration of probiotic to chickens can introduce beneficial changes in gut microbiota. PloS One. 2018;13(3) doi: 10.1371/journal.pone.0194825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez-Brito M., Plaza-Díaz J., Muñoz-Quezada S., Gómez-Llorente C., Gil A. Probiotic mechanisms of action. Ann Nutr Metabol. 2012;61:160–174. doi: 10.1159/000342079. [DOI] [PubMed] [Google Scholar]
- Broderick T., Duong T. Mechanisms OF lactobacillus persistence and colonization IN the gastrointestinal tract OF poultry, a review. Int J Probiotics Prebiotics. 2016;11:15–28. [Google Scholar]
- Butel M.J. Probiotics, gut microbiota and health. Med Maladies Infect. 2014;44:1–8. doi: 10.1016/j.medmal.2013.10.002. [DOI] [PubMed] [Google Scholar]
- Cano Roca C.L. University of Nebraska - Lincoln; 2014. Characterization of commercial probiotics: antibiotic resistance, acid and bile resistance, and prebiotic utilization. [Google Scholar]
- Davoodabadi A., Abbaszadeh M., Oloomi M., Bouzari S. Phenotypic and genotypic characterization of enteroaggregative Escherichia coli strains isolated from diarrheic children in Iran. Jundishapur J Microbiol. 2015;8 doi: 10.5812/jjm.22295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza L.F.A., Araujob D.N., Stefania L.M., Giomettia I.C., Cruz-Polycarpoc V.C., Polycarpoc G., Burbarellid M.F. Probiotics on performance, intestinal morphology and carcass characteristics of broiler chickens raised with lower or higher environmental challenge. Aus J Vet Sci. 2018;50:35–41. [Google Scholar]
- Dec M., Puchalski A., Nowaczek A., Wernicki A. Antimicrobial activity of Lactobacillus strains of chicken origin against bacterial pathogens. Int Microbiol. 2016;19:57–67. doi: 10.2436/20.1501.01.264. [DOI] [PubMed] [Google Scholar]
- Fernández M.F., Boris S., Barbes C. Probiotic properties of human lactobacilli strains to be used in the gastrointestinal tract. J Appl Microbiol. 2003;94:449–455. doi: 10.1046/j.1365-2672.2003.01850.x. [DOI] [PubMed] [Google Scholar]
- Feyereisen M., Mahony J., Kelleher P., Roberts R., O'Sullivan T., Geertman J.A., Sinderen D. Comparative genome analysis of the Lactobacillus brevis species. BMC Genom. 2019;20:416. doi: 10.1186/s12864-019-5783-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraunhofer M.E., Geißler A.J., Behr J., Vogel R.F. Comparative genomics of Lactobacillus brevis reveals a significant plasmidome overlap of brewery and insect isolates. Curr Microbiol. 2019;76:37–47. doi: 10.1007/s00284-018-1581-2. [DOI] [PubMed] [Google Scholar]
- Gaggìa F., Mattarelli P., Biavati B. Probiotics and prebiotics in animal feeding for safe food production. Int J Food Microbiol. 2010;141:15–28. doi: 10.1016/j.ijfoodmicro.2010.02.031. [DOI] [PubMed] [Google Scholar]
- Ghadban G. Probiotics in broiler production-a review. Archiv fur Geflugelkunde. 2002;66:49–58. [Google Scholar]
- Ghareeb K., Awad W., Mohnl M., Porta R., Biarnes M., Böhm J., Schatzmayr G. Evaluating the efficacy of an avian-specific probiotic to reduce the colonization of Campylobacter jejuni in broiler chickens. Poultry Sci. 2012;91:1825–1832. doi: 10.3382/ps.2012-02168. [DOI] [PubMed] [Google Scholar]
- Goktepe I., Juneja V.K., Ahmedna M. CRC Press; 2005. Probiotics in food safety and human health. [Google Scholar]
- Griggs J., Jacob J.P. Alternatives to antibiotics for organic poultry production. J Appl Poultry Res. 2005;14:750–756. [Google Scholar]
- Guarneri T., Rossetti L., Giraffa G. Rapid identification of Lactobacillus brevis using the polymerase chain reaction. Lett Appl Microbiol. 2001;33:377–381. doi: 10.1046/j.1472-765x.2001.01014.x. [DOI] [PubMed] [Google Scholar]
- Institute C.L.S. 2009. Performance standards for antimicrobial susceptibility testing; 19th informational supplement. CLSI document M100-S18. [Google Scholar]
- Jacobsen C.N., Nielsen V.R., Hayford A., Moller P.L., Michaelsen K., Paerregaard A., Sandström B., Tvede M., Jakobsen M. Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl Environ Microbiol. 1999;65:4949–4956. doi: 10.1128/aem.65.11.4949-4956.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Anjum A.A., Nawaz M., Awan A.R., Ali M.A. Effect of newly characterized probiotic lactobacilli on weight gain, immunomodulation and gut microbiota of Campylobacter jejuni challenged broiler chicken. Pak Vet J. 2019;39:473–478. [Google Scholar]
- Kizerwetter-Swida M., Binek M. Protective effect of potentially probiotic Lactobacillus strain on infection with pathogenic bacteria in chickens. Pol J Vet Sci. 2009;12:15–20. [PubMed] [Google Scholar]
- Klare I., Konstabel C., Muller-Bertling S., Reissbrodt R., Huys G., Vancanneyt M., Swings J., Goossens H., Witte W. Evaluation of new broth media for microdilution antibiotic susceptibility testing of lactobacilli, pediococci, lactococci, and bifidobacteria. Appl Environ Microbiol. 2005;17:8982–8986. doi: 10.1128/AEM.71.12.8982-8986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosin B., Rakshit S.K. Microbial and processing criteria for production of probiotics: a review. Food Technol Biotechnol. 2006;44:371–379. [Google Scholar]
- Kwon H.-S., Yang E.-H., Yeon S.-W., Kang B.-H., Kim T.-Y. Rapid identification of probiotic Lactobacillus species by multiplex PCR using species-specific primers based on the region extending from 16S rRNA through 23S rRNA. FEMS Microbiol Lett. 2004;239:267–275. doi: 10.1016/j.femsle.2004.08.049. [DOI] [PubMed] [Google Scholar]
- Lebeer S., Vanderleyden J., De Keersmaecker S.C. Genes and molecules of lactobacilli supporting probiotic action. Microbiol Mol Biol Rev. 2008;72:728–764. doi: 10.1128/MMBR.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeer S., Verhoeven T.L., Vélez M.P., Vanderleyden J., De Keersmaecker S.C. Impact of environmental and genetic factors on biofilm formation by the probiotic strain Lactobacillus rhamnosus GG. Appl Environ Microbiol. 2007;73:6768–6775. doi: 10.1128/AEM.01393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wang W., Liu D., Guo Y. Effects of Lactobacillus acidophilus on gut microbiota composition in broilers challenged with Clostridium perfringens. PloS One. 2017;12 doi: 10.1371/journal.pone.0188634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Idris U., Harmon B., Hofacre C., Maurer J.J., Lee M.D. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl Environ Microbiol. 2003;69:6816–6824. doi: 10.1128/AEM.69.11.6816-6824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowiak P., Slizewska K. The role of probiotics, prebiotics and synbiotics in animal nutrition. Gut Pathog. 2018;10:21. doi: 10.1186/s13099-018-0250-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McOrist A.L., Jackson M., Bird A.R. A comparison of five methods for extraction of bacterial DNA from human faecal samples. J Microbiol Methods. 2002;50:131–139. doi: 10.1016/s0167-7012(02)00018-0. [DOI] [PubMed] [Google Scholar]
- Monteagudo-Mera A., Rastall R.A., Gibson G.R., Charalampopoulos D., Chatzifragkou A. Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health. Appl Microbiol Biotechnol. 2019;103:6463–6472. doi: 10.1007/s00253-019-09978-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidu A., Bidlack W., Clemens R. Probiotic spectra of lactic acid bacteria (LAB) Crit Rev Food Sci Nutr. 1999;39:13–126. doi: 10.1080/10408699991279187. [DOI] [PubMed] [Google Scholar]
- Nava G., Bielke L., Callaway T., Castaneda M. Probiotic alternatives to reduce gastrointestinal infections: the poultry experience. Anim Health Res Rev. 2005;6:105–118. doi: 10.1079/ahr2005103. [DOI] [PubMed] [Google Scholar]
- Neal-McKinney J.M., Lu X., Duong T., Larson C.L., Call D.R., Shah D.H., Konkel M.E. Production of organic acids by probiotic lactobacilli can be used to reduce pathogen load in poultry. PloS One. 2012;7 doi: 10.1371/journal.pone.0043928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelschlaeger T.A. Mechanisms of probiotic actions–a review. Int J Med Microbio. 2010;300:57–62. doi: 10.1016/j.ijmm.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Ohland C.L., MacNaughton W.K. Probiotic bacteria and intestinal epithelial barrier function. Am J Physiol Gastrointest Liver Physiol. 2010;298:G807–G819. doi: 10.1152/ajpgi.00243.2009. [DOI] [PubMed] [Google Scholar]
- Pan D., Yu Z. Intestinal microbiome of poultry and its interaction with host and diet. Gut Microb. 2014;5:108–119. doi: 10.4161/gmic.26945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papizadeh M., Rohani M., Nahrevanian H., Javadi A., Pourshafie M.R. Probiotic characters of Bifidobacterium and Lactobacillus are a result of the ongoing gene acquisition and genome minimization evolutionary trends. Microb Pathog. 2017;111:118–131. doi: 10.1016/j.micpath.2017.08.021. [DOI] [PubMed] [Google Scholar]
- Patterson J.A., Burkholder K.M. Application of prebiotics and probiotics in poultry production. Poultry Sci. 2003;82:627–631. doi: 10.1093/ps/82.4.627. [DOI] [PubMed] [Google Scholar]
- Pokorna A., Manakova T., Cizek A. Properties of potentially probiotic Lactobacillus isolates from poultry intestines. Acta Vet. 2019;88:73–84. [Google Scholar]
- Ramos C.L., Thorsen L., Schwan R.F., Jespersen L. Strain-specific probiotics properties of Lactobacillus fermentum, Lactobacillus plantarum and Lactobacillus brevis isolates from Brazilian food products. Food Microbiol. 2013;36:22–29. doi: 10.1016/j.fm.2013.03.010. [DOI] [PubMed] [Google Scholar]
- Rodrigues D.R., Briggs W., Duff A., Chasser K., Murugesan R., Pender C., Ramirez S., Petri D., Bielke Comparative effectiveness of probiotic-based formulations on cecal 3 microbiota modulation in broilers. PloS One. 2020;15 doi: 10.1371/journal.pone.0225871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahrokhi N., Bouzari S., Jafari A. Comparison of virulence markers and antibiotic resistance in enterotoxigenic Escherichia coli isolated ten years apart in Tehran. J Infec Dev Countries. 2011;5:248–254. doi: 10.3855/jidc.1206. [DOI] [PubMed] [Google Scholar]
- Shariatmadari F. Poultry production and the industry in Iran. World Poultry Sci J. 2000;56:55–65. [Google Scholar]
- Slizewska K., Markowiak-Kopec P., Zbikowski A., Szeleszczuk P. The effect of synbiotic preparations on the intestinal microbiota and her metabolism in broiler chickens. Sci Rep. 2020;10:4281. doi: 10.1038/s41598-020-61256-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelin A., Fuka M.M., Majhenič A.Č., Redžepović S., Samaržija D., Matijašic B.B. Phenotypic and genotypic characterization of indigenous Lactobacillus community from traditional Istrian Ewe's cheese. Food Technol Biotechnol. 2012;50:362–370. [Google Scholar]
- Taheri H., Moravej H., Tabandeh F., Zaghari M., Shivazad M. Screening of lactic acid bacteria toward their selection as a source of chicken probiotic. Poultry Sci. 2009;88:1586–1593. doi: 10.3382/ps.2009-00041. [DOI] [PubMed] [Google Scholar]
- Tellez G., Pixley C., Wolfenden R., Layton S., Hargis B. Probiotics/direct fed microbials for Salmonella control in poultry. Food Res Int. 2012;45:628–633. [Google Scholar]
- Tilsala-Timisjarvi A., Alatossava T. Strain-specific identification of ProbioticLactobacillus rhamnosus with randomly amplified polymorphic DNA-derived PCR primers. Appl Environ Microbiol. 1998;64:4816–4819. doi: 10.1128/aem.64.12.4816-4819.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieco-Saiz N., Belguesmia Y., Raspoet R., Auclair E., Gancel F., Kempf I., Drider D. Benefits and inputs from lactic acid bacteria and their bacteriocins as alternatives to antibiotic growth promoters during food-animal production. Front Microbiol. 2019;10:57. doi: 10.3389/fmicb.2019.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J. Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl Environ Microbiol. 2008;74:4985–4996. doi: 10.1128/AEM.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S., Morrison M., Yu Z. Bacterial census of poultry intestinal microbiome. Poultry Sci. 2013;92:671–683. doi: 10.3382/ps.2012-02822. [DOI] [PubMed] [Google Scholar]
- Yadav S., Jha R. Strategies to modulate the intestinal microbiota and their effects on nutrient utilization, performance, and health of poultry. J Anim Sci Biotechnol. 2019;10:2. doi: 10.1186/s40104-018-0310-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Lv J., Pan L., Zhang Y. Roles and applications of probiotic Lactobacillus strains. Appl Microbiol Biotechnol. 2018;102:8135–8143. doi: 10.1007/s00253-018-9217-9. [DOI] [PubMed] [Google Scholar]