Abstract
Background
Activation of mTORC1 plays a significant role in cancer development and progression. However, the metabolic mechanisms to sustain mTORC1 activation of cancer cells within stressed environments are still under-appreciated. We recently revealed high autophagy activity in tumour cells with mTORC1 hyper-activation. Nevertheless, the functions and mechanisms of autophagy in regulating mTORC1 in glioma are not studied.
Methods
Using glioma patient database and human glioma cells, we assessed the mechanisms and function of selective autophagy to sustain mTORC1 hyper-activation in glioma.
Results
We revealed a strong association of altered mRNA levels in mTORC1 upstream and downstream genes with prognosis of glioma patients. Our results indicated that autophagy-mediated lipid catabolism was essential to sustain mTORC1 activity in glioma cells under energy stresses. We found that autophagy inhibitors or fatty acid oxidation (FAO) inhibitors in combination with 2-Deoxy-D-glucose (2DG) decreased energy production and survival of glioma cells in vitro. Consistently, inhibition of autophagy or FAO inhibitors with 2DG effectively suppressed the progression of xenografted glioma with hyper-activated mTORC1.
Conclusions
This study established an autophagy/lipid degradation/FAO/ATP generation pathway, which might be used in brain cancer cells under energy stresses to maintain high mTORC1 signalling for tumour progression.
Subject terms: CNS cancer, Autophagy, Molecular biology
Background
Glioblastoma is the most common and deadly type of brain cancer. The 5-year survival rate of the most aggressive grade IV glioma, glioblastoma multiforme (GBM) has remained at 4–5% for the last 30 years.1 The therapeutic outcome of glioblastoma is still dismal, despite advances in surgical and pharmacological treatments with the best modalities available.2–4 The quick development of therapy resistance leads to inevitable tumour recurrence and treatment failure in glioblastoma therapy.5–7 Therefore, finding alternative therapeutic methods can only be achieved with further understanding of the genetic, molecular and metabolic mechanisms of glioblastoma. Modulation of cancer cell metabolism, one of the therapeutic targets proposed in recent years, has the potential to produce novel approaches for glioblastoma therapy.
Many glioblastomas with mutations in p53, Pten, Nf1, Pik3ca and/or EGFR exhibit high mechanistic target of rapamycin complex 1 (mTORC1) activity.8,9 mTORC1 is a master regulator in cell growth, proliferation, differentiation and metabolism.10 mTORC1 has important functions in tumorigenesis and progression of glioblastoma.9,11 mTOR inhibitors have shown anti-cancer effects in cell lines and in animal models.12–15 However, the efficacy of mTORC1 inhibitors, such as Rapamycin, is limited in glioblastoma therapy.16,17 One possible reason for Rapamycin resistance is that it activates autophagy in cancer cells.18
Hyper-activation of mTORC1 increases glucose uptake and glycolysis,19,20 amplifies nucleotide production,21 promotes lipid synthesis, reduces lipolysis22–24 and accelerates anaplerotic entry of glutamine to the TCA cycle.25 It is established that metabolic adaptations are important for cancer cell survival and proliferation,26 dramatically increasing the malignancy of glioblastoma.27 Lipid metabolic abnormalities in tumour cells have become recognised in the past few years.28 Redirecting lipid metabolism in tumour cells and its role in tumour progression and metastasis has received widespread attention.29 Lipids are stored in form of droplets (LDs) and high content of LDs is associated with poor prognosis and disease progression for multiple cancers,30–32 including glioblastoma.33 Considering that mTORC1 hyper-activation increases lipid synthesis and reduces lipid catabolism,34 these general mechanisms favour increased lipid storage in glioblastoma cells. Utilisation of LDs can occur to meet the bioenergy needs of the tumour cells and this catabolic reprogramming could help cells survive stringent conditions.35 Previously, lipolysis of lipids stored in LDs was considered to be solely carried out by cytosolic lipases.36 Only recently, studies demonstrates that autophagic degradation of lipids by acidic lipases (lipophagy) serves for LDs degradation.37 However, whether the functions of lipophagy exist in GBM to facilitate tumour cell survival is not yet examined.
Autophagy, a catabolic process for degradation of cytoplasmic content, is responsible for the metabolic plasticity of cancer cells.38 Autophagy plays an essential role in survival of cancer cells in oxygen, pH and nutrient stresses.39 Autophagy is negatively regulated by mTORC1 and hyper-activation of mTORC1 suppresses autophagy through phosphorylation of Ulk1 at Serine residue 757.40,41 Interestingly, our recent studies indicated that under energy stress conditions, autophagy activity is higher in Tsc1-deficient breast cancer cells than that in wild-type breast cancer cells.42 These studies seem to contradict to the prevailing model; however, they raised previously unrecognised questions: what are the functions of autophagy in mTORC1 hyper-activated cancer cells and could we still target autophagy for treatment of these cancers?
Studies from our group and others revealed tumour promoting functions of autophagy-mediated metabolisms for different cancer in various mouse and xenograft models.43–46 Our most recent work also indicated a role of lipophagy in the tumorigenesis of low grade gliomas in a Tsc1-deficient mouse model.47 However, whether there is a function of autophagy in lipid metabolism in mTORC1 hyper-activated glioblastoma still needs investigation. In this study, we investigated the autophagy-mediated lipid catabolism in glioblastoma cells under energy stresses, and the potential therapeutic effects targeting lipophagy and its downstream FAO in GBM treatment. Our results provide insights into the significance of hitherto under-appreciated role of lipophagy in glioblastoma cell metabolism and tumour progression.
Methods
Animals
Eight-weeks-old nude mice (Envigo, Indianapolis) were housed and handled according to local, state and federal regulations. All experimental procedures were carried out according to the guidelines of Institutional Animal Care and Use Committee at University of Cincinnati.
Cell culture
Human glioblastoma cell lines are gifted from Dr. David R Plas in the Department of Cancer Biology, University of Cincinnati College of Medicine. These cells were cultured in DMEM with 10% FBS under growth conditions. Energy stresses were carried out by using glucose-free media (Gibco) and DMEM containing 25 mM 2DG (Cayman, MI) without FBS as in our recent publication.47
shRNA and Crispr lentivirus production and infection
Human scramble shRNA and TSC1 shRNAs (#1 5′‐ ACT CTT TCA TCG CCT TTA TCT CGA GAT AAA GGC GAT GAA AGA GTG‐3′; #2 5′‐ CAA GAA AGA CCA CCT TCT TCT CGA GAA GAA GGT GGT CTT TCT TGG‐3′) were encoded in pGIPZ lentiviral vector (Open Biosystem). Package vectors of pSPAX2 and pMD2.G were mixed with shRNA vectors to transfect HEK293 cells for virus production.47 Recombinant lentiviral particles encoding six nucleotide mismatches shRNA-resistant human TSC1 (ACT CTT TCA TCG CCT TTA TCG CGC GAT AAA GGC GAT GAA AGA GTG) or control plasmid were generated according to our previous publication.48 sgRNA targeting human Fip200 (5′-CAGGTGCTGGTGGTCAATGG-3′)49 was subcloned into lentiCRISPR v2 (Addgene, 52961). The CRISPR plasmids were packaged into lentiviruses to infect TSC1 KD-1 LN229 cells with puromycin for selection. The selected clones were cultured in DMEM with 10% FBS.
Measurements of cell viability and ATP content
Cell viability was measured by Vybrant® MTT Cell Proliferation Assay kit (Thermo Fischer Scientific). Cells were seeded in 12-well tissue culture plates (5.0 × 104 cells per well) 24 h prior to treatment. Five days after treatment, the medium was removed, and fresh medium containing 0.5 mg/mL MTT was added. The experimental procedure was performed according to the manufacturer’s instructions.
ATP content was measured by using a CellTiter-Glo Luminescent Cell Viability Assay Kit (Promega). The experimental procedure was performed according to the manufacturer’s instructions. The measurement was carried out on a microplate reader (Synergy 2 Multi-Mode, BioTEK).
Measurements of TG and FFA
LN229 and GaMg (4 × 106) cells were lysed in 0.2 ml lysis buffer (5% NP-40 in H2O) and assayed for TG using a Triglyceride Quantification Colorimetric Kit (Biovision) according to the manufacturer’s instructions. LN229 (2 × 106) cells were extracted by homogenisation with 200 μl of chloroform-Triton X-100 (1% Triton X-100 in pure chloroform) and assayed for FFA using a Free Fatty Acid Quantification Colorimetric/Fluorometric Kit (Biovision). For each assay, 50 μl of the extracted sample was used. The measuring procedures were performed according to the instructions provided.
Patient data analysis
Patient and gene expression data were downloaded from the R2: genomics analysis and visualisation platform (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi); Repository for Molecular Brain Neoplasia Data (http://www.betastasis.com/glioma/rembrandt/, GSE108476). The Cancer Genome Atlas (TCGA) IlluminaHiSeq RNA-seq cohort data of GBMs were downloaded from https://tcga-data.nci.nih.gov/ via Xena Browser developed by UCSC. Kaplan–Meier survival analysis was conducted based on the median gene(s) expression cut-off. p values (log-rank test) were obtained from the website.
Subcutaneous and orthotopic xenograft models
2 × 105 cells were subcutaneously injected into the right and left sides of lateral backside of nude mice. Callipers were used to measure the tumour volume, which was calculated using the formula Volume (π/6) × length × width2.
1 × 105 LN229 TSC1 KD-1 and 2KD cells were implanted into the right frontal lobe of brains in nude mice. Thirty-six nude mice were grouped randomly for six different treatments. The body conditions and tumour behaviours were monitored. The time for sample collection is based on the criteria for humane euthanise, such as body weight loss of 20% and body condition score of 2 or less. We set 65 days after treatment as an endpoint to euthanise all the live mice. For the quantification of intracranial tumour volume, we sliced the brain with mouse brain matrices (model 69-2165-1, Agnthos, Sweden) at 1-mm interval to cover the whole allograft. Brain sections were prepared at 100-μm intervals and tumour regions were identified on H&E-stained slides. Tumour areas were measured from the beginning to the end of each fragment. The average area was multiplied by 1 mm in each fragment and the tumour volume was added up from all fragments containing tumour.
Antibodies and reagents
The details for primary antibodies and secondary used in this study were listed in Supplementary Table 1. BODIPY 493/503 was purchased from Invitrogen. DAPI was purchased from Millipore. DMEM, glucose-free DMEM and HBSS were from Thermo Fisher Scientific. ATP, BSA, palmitate, glucose, glutamine, sodium pyruvate, bafilomycin A1, chloroquine and Spautin-1 were purchased from Sigma. 2DG, etomoxir, ranolazine and trimetazidine were purchased from Cayman.
Histology, immunofluorescence, and BODIPY (493/503) staining
Mice were euthanised using CO2 and a complete tissue set was harvested during necropsy. Fixation was carried out for 16 h at 4 °C using 4% (wt/vol) freshly made, pre-chilled PBS-buffered paraformaldehyde (PFA). The tissues were embedded in paraffin and sectioned into 5-μm sections. H&E-stained sections were examined under a BX41 light microscope (Olympus America, Center Valley), and images were captured with an Olympus digital camera (model DP70) using DP Controller software (Version 1.2.1.10 8). For IF and IHC, the procedure was carried out as described previously.44 The quantification of data was carried out by counting all the positive cells in the field and divided by the total cell number. For BODIPY staining, samples were fixed by 4% PFA followed by three washes with PBS. Samples were incubated in 1 μg/ml BODIPY 493/503 for 20 min at room temperature.
2DG, CQ, Eto, Rano and TRMZ administration
PBS, 2DG (500 mg/kg), CQ (50 mg/kg), 2DG + CQ, Eto (50 mg/kg), Rano (25 mg/kg), TRMZ (5 mg/kg), 2DG + Eto, 2DG + Rano, 2DG + TRMZ were dissolved in PBS and administrated intra-peritoneal (IP) into mice every other day.
Protein extraction, SDS-PAGE and western blotting
Cells were homogenised in modified radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.4, 1% Triton X-100, 0.2% sodium deoxycholate, 0.2% SDS, 1 mm sodium EDTA) supplemented with protease inhibitors (Sigma). Cell debris were removed by centrifugation at 13,000 rpm for 10 min at 4 °C. The lysates were boiled for 5 min in 1× SDS sample buffer (50 mm Tris-HCl, pH 6.8, 12.5% glycerol, 1% SDS, 0.01% bromophenol blue) containing 5% β-mercaptoethanol. They were then analysed by SDS-PAGE followed by western blotting as described previously.47
Seahorse extracellular assay for oxygen consumption rate (OCR)
Measurement of OCR from intact cells was made using an XF24 Extracellular Flux Analyzer (Agilent, CA). Twenty thousands cells per well were seeded for mitochondria stress tests. The compounds of Eto (200 μM), Rano (400 μM) or TRMZ (1 mM) prepared in assay medium (75 µl) were preloaded into reagent delivery chambers and were injected after FCCP. Data acquisition, normalisation and analysis were carried out as described before.47
Statistical analysis
Statistical significance was evaluated by student’s t-test, one-way or two-way ANOVA, Kaplan–Meier analysis with p < 0.05 as indicative of statistical significance using Graph Pad Prism (Version 5.0). The number of experiment repeats and animals was indicated in the figure legends.
Results
Analysis of TSC/mTORC1 pathway in glioma patients
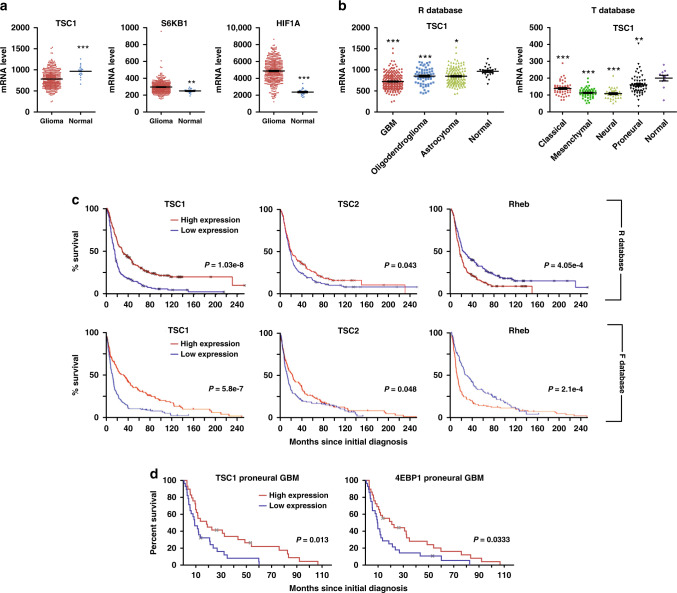
The expression levels of mTORC1 upstream regulators and downstream effectors in glioma and their correlation with glioma progression are not well studied. We conducted a search using databases of REMBRANDT (designated as R database),50 TCGA (designated as T database)8 and Tumour Glioma–French-284 (designated as F database)51 to examine the upstream regulators and downstream effectors of mTORC1 in human brain tumours. First, we analysed gliomas in R database irrespective of their WHO grades. We found the TSC1 mRNA expression was lower while expressions of mTORC1 target genes of S6K1 and HIF1α were higher in glioma samples relative to normal tissues (Fig. 1a). Next, we analysed TSC1 mRNA expression in astrocytoma, oligodendrocytoma and GBM. We found decreased TSC1 mRNA level in gliomas, most prominent among glioblastomas (Fig. 1b). Furthermore, TSC1 mRNA expression was significantly lower in all GBM subtypes (classic, pro-neural, mesenchymal and neural) (Fig. 1b). The mRNA level of TSC2 did not show obvious changes in GBM samples in R database (Fig. S1A) but it decreased in all GBM subtypes of T database (Fig. S1A). This discrepancy between the databases might be caused by different methods to detect gene expression (i.e. microarray used in R database but RNA-seq used in T database). The mRNA level of Rheb was higher in GBM (Fig. S1A). These results suggested a frequent mechanism to increase mTORC1 activation through decreasing TSC function in human gliomas.
Fig. 1. Analysis of mTORC1 upstream regulators and downstream effectors in glioma patients.
a mRNA expression levels of Tsc1, S6KB1 and HIF1α in glioma versus normal brain tissues are extracted from dataset of REMBRANDT. b mRNA expression levels of Tsc1 in GBM (left panel) and subtypes of GBM (right panel) versus normal brain tissues are extracted from datasets of REMBRANDT and TCGA–GBM, respectively. c Kaplan–Meier analysis of glioma patients’ survival with different mRNA expression levels of Tsc1, Tsc2 and Rheb. Data were extracted from datasets of REMBRANDT (R database, upper panels) and Tumour Glioma–French 284 (F database, lower panels). d Kaplan–Meier analysis of pro-neural subtype GBM patients’ survival with different mRNA expression levels of Tsc1 and 4EBP1. Data were extracted from datasets of TCGA- GBM. Data were analysed by two-tailed student’s t-test (a), one-way analysis of variance (ANOVA) with Tukey’s post-hoc test (b) or Kaplan–Meier estimator (c, d). *p < 0.05, **p < 0.01, ***p < 0.001.
Our analysis showed that lower mRNA level of TSC1 and TSC2, and higher Rheb mRNA expression, correlated with poor progression-free survival in glioma patients (Fig. 1c). Higher expression level of mTORC1 substrates RPS6KB1, RPS6KB2, Grb10 and VEGFA all correlated with poor patient’s survival (Fig. S1B). Focusing on GBM, TSC1 (Fig. S1C) and 4EBP1 mRNA levels (data not shown) did not correlate with survival in unstratified tumours. Nevertheless, we found that lower levels of TSC1 and 4EBP1 mRNA correlated with poor survival of pro-neural GBM (Fig. 1d). Moreover, higher mRNA expression of Grb10 correlated with worse prognosis of pro-neural GBM (Fig. S1C). Together, these results confirmed the positive correlation of mTORC1 activation status with glioma progression.
Generation of ATP in energy-stressed GBM cells through autophagic degradation of LDs
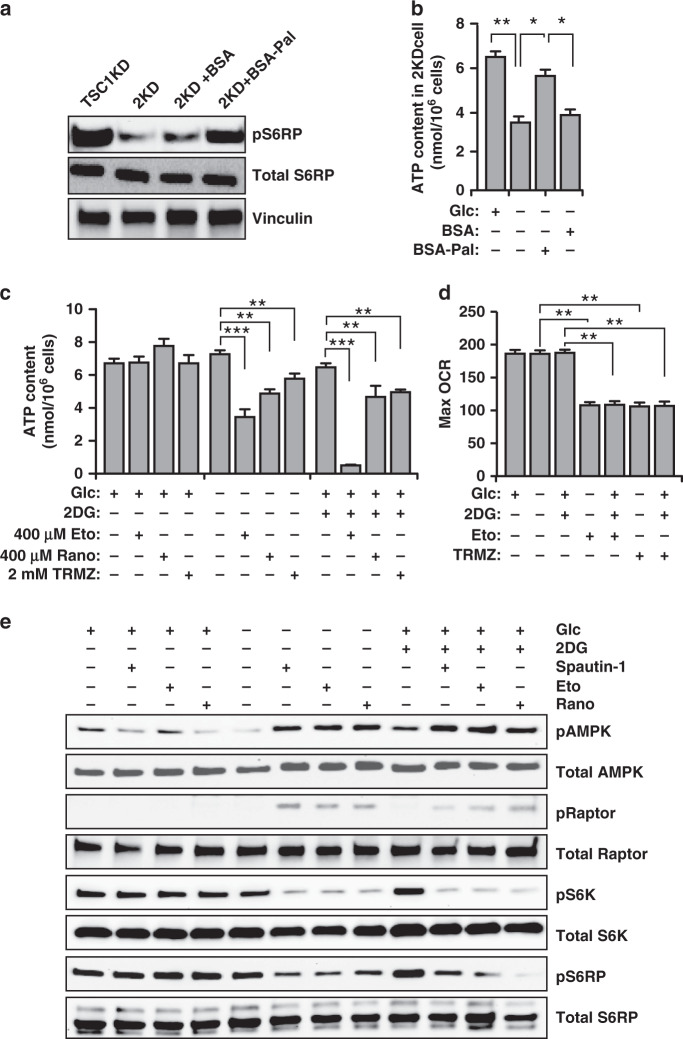
We used six human GBM cell lines (8MGBA, LN18, U87MG, A172, LN229 and GaMg) and we found mTORC1 activity varied in GBM cells under growth conditions (Fig. S2A). GBM could maintain mTORC1 activation upon removal of glucose (glc-free) from or under incubation of glycolysis inhibitor 2-Deoxy-D-glucose (2DG) in media (Fig. S2B). In contrast, primary astroglia reduced mTORC1 activity under 1-h energy stresses (Fig. S2B). Next, we examined ATP content in LN229 and GaMg cells and we found energy stresses had no effect on ATP level in these cells (Fig. 2a). Then, we analysed their mitochondrial oxygen consumption rate (OCR) and we found glc-free and 2DG did not change ATP-related OCR or Max OCR in GBM cells (Fig. 2b, c). Since energy stresses could activate autophagy, we incubated GBM cells with autophagy inhibitors Spautin-1 to inhibit LC3-II generation52 and chloroquine (CQ) to block LC3-II degradation (Fig. S2C). Spautin-1 and CQ had no effect on ATP content under normal conditions; however, they decreased ATP content and Spautin-1 also reduced OCR in energy-stressed GBM cells (Fig. 2a–c). These results suggested that GBM cells could maintain OxPHOS to generate ATP through autophagy under energy stresses.
Fig. 2. Maintenance of mTORC1 hyper-activity in GBM cells upon energy stresses through autophagy-mediated lipid degradation.
a Mean ± SE of the ATP content of 20 μM CQ and 10 μM Spautin-1-treated LN229 and GaMg cells in normal media, glc-free media and 25 mM 2DG media for 2 h were shown. The autophagy inhibitors were pre-incubated for 12 h before experiments. n = 6 independent experiments. b, c Mean ± SE of ATP-related OCR (b) and maximum OCR (c) of Spautin-1-treated LN229 and GaMg cells under normal and glc-free conditions were shown. n = 6 independent experiments. d, e Mean ± SE of triglycerides (d) and free fatty acids (e) of Ctrl LN229 cell, Spautin-1-treated TSC1 KD LN229 cells, and 2KD cells under normal and glc-free conditions were shown. n = 5 independent experiments. f BODIPY staining of lipid droplets in Ctrl LN229 cell, Spautin-1-treated TSC1 KD LN229 cells and 2KD cells under normal and glc-free conditions. g Mean ± SE of lipid droplets number of Ctrl LN229 cell, Spautin-1 treated TSC1 KD LN229 cells and 2KD cells under normal and glc-free conditions were shown. n = 100 cells from 5 independent experiments. Data were analysed by one-way analysis of variance (ANOVA) with Tukey’s post-hoc test. ns no significance, *p < 0.05, **p < 0.01, ***p < 0.001. Bar = 20 μm.
Next, we constructed recombinant lentiviruses encoding Ctrl shRNA and two human TSC1 shRNA sequences to infect LN229 cells, obtaining Ctrl LN229 cells and two TSC1 knockdown LN229 cells (designated as TSC1 KD-1 and TSC1 KD-2) with hyper-activated mTORC1 (Fig. S2D). We also used Crispr-Cas9 technology to knockout Fip200 in TSC1 KD-1 LN229 cells (designated as 2KD cells) to repress their autophagy as shown by decreased LC3-II conversion and increased p62 level in 2KD cells (Fig. S2E). Lack of FIP200 did not affect mTORC1 in 2KD cells under normal condition but these cells exhibited lower mTORC1 activity and diminished autophagy under 24 h glc-free conditions (Fig. S2E). Consistent with previous report,22 we found higher TG content in TSC1 KD-1 cells (Fig. 2d) and glc-free decreased TG content to upregulate free fatty acids (FFAs). Blocking autophagy by Spautin-1 or KO Fip200 counteracted these changes (Fig. 2d, e). We also found more LDs in TSC1 KD-1 cells and glc-free reduced LDs number in TSC1 KD-1 cells in an autophagy-dependent manner (Fig. 2f, g). Together, these results indicate that autophagy-facilitated FFAs production from LDs to generate ATP in energy-stressed GBM cells.
Maintenance of mTORC1 activity in energy-stressed GBM cells through FAO
Our recent studies in postnatal neural stem cells indicated that FFAs generated through autophagy are essential for ATP production to maintain mTORC1 hyper-activation during energy crisis.47 To test this, we added BSA or BSA-conjugated palmitate to energy-stressed 2KD cells. In glc-free media, mTORC1 activity and ATP content were decreased in 2KD cells, while supplementation with BSA–palmitate but not BSA alone restored their mTORC1 activation and ATP levels (Fig. 3a, b). These results supported that autophagy-mediated FAO played an important role to maintain mTORC1 activation. Next, we treated LN229 cells with FAO inhibitors of Etomoxir (Eto), Trimetazidine (TRMZ) and Ranolazine (Rano) to measure their ATP content. FAO inhibitors significantly decreased ATP content and Max OCR under energy stresses but not under normal conditions (Fig. 3c, d). FAO inhibitors increased LC3 puncta in LN229 cells under glc-free conditions (Fig. S2F), which suggested generation of FFAs from LDs by autophagy. These results indicated that autophagy-mediated FAO might serve as an ATP-generating mechanism in energy-stressed GBM cells.
Fig. 3. Maintenance of mTORC1 hyper-activity in GBM cells upon energy stresses through autophagy-mediated fatty acid oxidation.
a Lysates were extracted from TSC1 KD LN229 and 2KD LN229 cells treated with BSA or BSA-conjugated palmitate under glc-free conditions for 2 h. The protein levels were examined by western blot using phosphorylated S6RP, total S6RP and vinculin antibodies. Three independent experiments yielded similar results. b Mean ± SE of the ATP content of 2KD LN229 cells treated with BSA or BSA-conjugated palmitate under normal and glc-free conditions for 2 h. n = 5 independent experiments. c Mean ± SE of the ATP content of 400 μM Eto, 400 μM Rano and 2 mM TRMZ-treated LN229 cells in normal media, glc-free media, and 25 mM 2DG media for 2 h were shown. The FAO inhibitors were pre-incubated for 12 h before experiments. n = 5 independent experiments. d Mean ± SE of Max OCR of Eto and TRMZ-treated LN229 cells under normal media, glc-free media, and 25 mM 2DG media treatment conditions were shown. n = 5 independent experiments. e Lysates were extracted from LN229 treated with vehicle, Spautin-1, Eto, and Rano under glc-free and 25 mM 2DG conditions for 2 h. The protein levels were examined by Western blot using antibodies against phosphorylated AMPK, total AMPK, phosphorylated Raptor, total Raptor, phosphorylated S6K, total S6K, phosphorylated S6RP and total S6RP. Three independent experiments yielded similar results. Data were analysed by one-way analysis of variance (ANOVA) with Tukey’s post-hoc test. *p < 0.05, **p < 0.01, ***p < 0.001.
Consistent with decreased ATP production, Eto and Rano increased AMPK phosphorylation level at Threonine 172 (Fig. 3e). Activated AMPK phosphorylates Raptor (regulatory-associated protein of mTOR) at Serine 792 to suppress mTORC1 pathway.53 FAO inhibitors and Spautin-1 increased the Serine 792 phosphorylation of Raptor and they suppressed mTORC1 activation in glc-free or 2DG treated LN229 cells (Fig. 3e). Together, these results revealed the essential functions of autophagy to supply FAO for ATP production to suppress AMPK for mTORC1 hyper-activation in GBM cells under energy crisis.
Targeting autophagy-mediated FAO with 2DG inhibited GBM cells growth in vitro
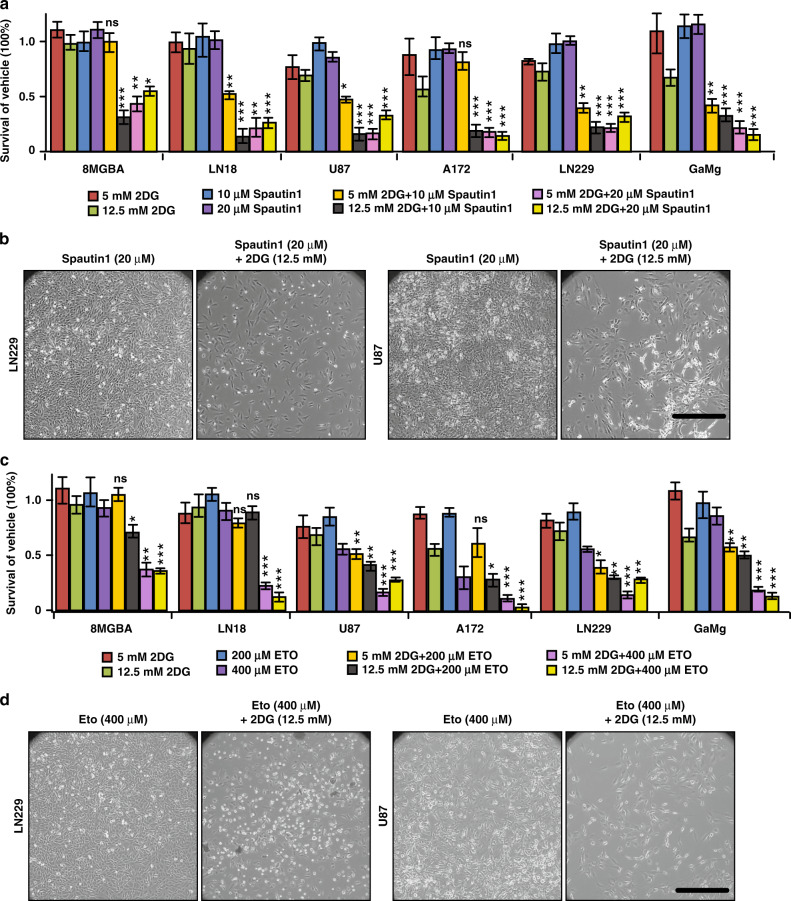
Our studies suggested that targeting autophagy-mediated FAO with glycolysis might suppress GBM progression because of deficient ATP and decreased mTORC1 activity. Next, we treated six GBM cell lines with glycolysis inhibitor 2DG either alone or in combination with autophagy inhibitors (Spautin-1 and CQ) or with FAO inhibitors (Eto, Rano and TRMZ). 5 mM and 12.5 mM 2DG had minimal effect on GBM cell growth and all GBM cells reached to confluency (Fig. 4a, c, S3A). Similarly, autophagy inhibitors and FAO inhibitors used as single agents had little effect on the growth of GBM cells, except CQ at a higher dose of 20 μM for 8MGBA and LN18 cells (Fig. 4a–d, S3C, S3E, S3G). Interestingly, we found 10 μM Spautin-1 with 12.5 mM 2DG (Fig. 4a) and 15 μM CQ with 5 mM 2DG (Fig. S3B) significantly decreased GBM cell survival. The combinations of FAO inhibitors with 2DG also decreased GBM cells survival (Fig. 4c, d, S3D–S3G). We found only a few live cells after combinational treatments (Fig. 4b, d, S3C, S3E, S3G). These results suggested a possible new strategy targeting autophagy-mediated lipid catabolism with glycolysis for GBM treatment.
Fig. 4. Therapeutic target of autophagy-mediated lipid catabolism in glioblastoma cells.
a Survival of human GBM cell lines 8MGBA, LN18, U87MG, A172, LN229 and GaMg in 5 days treatment of vehicle, Spautin-1, 2DG, 2DG + Spautin-1. The dose of each compound was listed below the bar graph. n = 4 independent experiments. Data were normalised with vehicle treated group for each GBM. b Phase contrast images of LN229 and U87 cells after 5 days treatment of Spautin-1 and 2DG + Spautin-1. c Survival of human GBM cell lines 8MGBA, LN18, U87MG, A172, LN229 and GaMg in 5 days treatment of vehicle, Eto, 2DG, 2DG + Eto. The dose of each compound was listed below the bar graph. n = 4 independent experiments. Data were normalised with vehicle treated group for each GBM. d Phase contrast images of LN229 and U87 cells after 5 days treatment of Eto and 2DG + Eto. Data were analysed by two-way analysis of variance (ANOVA). ns no significance, *p < 0.05, **p < 0.01, ***p < 0.001. Bar = 100 μm.
Targeting autophagy-mediated FAO and glycolysis inhibited GBM progression in vivo
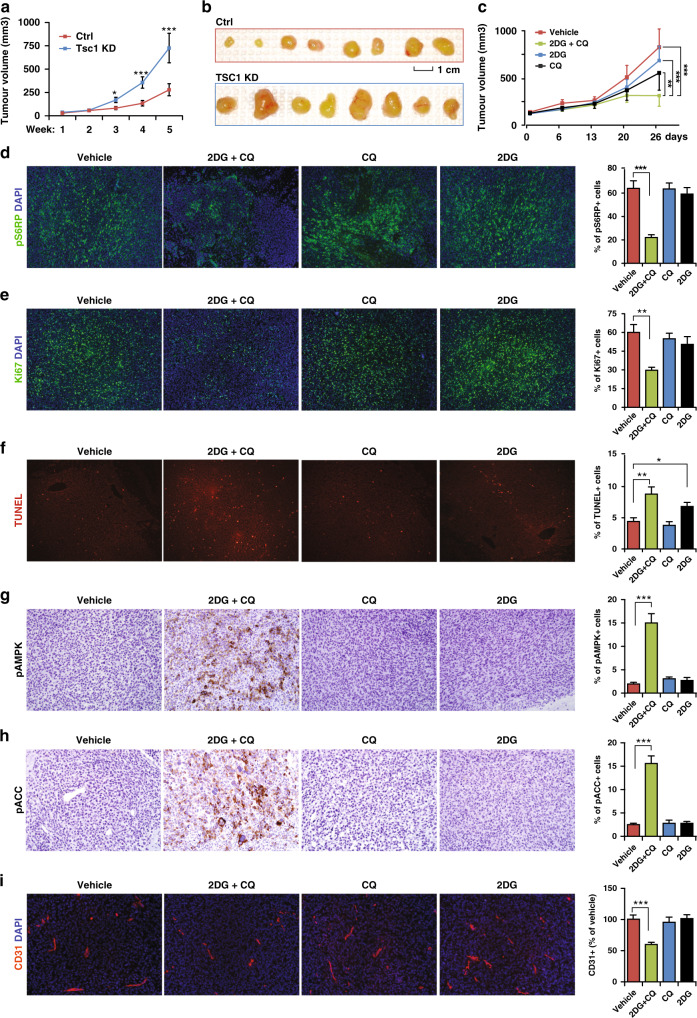
Next, we performed subcutaneous transplantation of Ctrl, #1 and #2 TSC1 KD LN229 cells into nude mice. The time to develop palpable tumours was similar between these cells. Two weeks after transplantation, tumours from TSC1 KD-1 cells began to grow faster than tumours from Ctrl cells (Fig. 5a). Both #1 and #2 TSC1 KD tumours were larger than Ctrl tumours at 5 weeks after transplantation (Fig. 5b, S4A–S4C). To minimise shRNA off-target effects, we re-expressed shRNA-resistant human TSC1 (hTSC1) cDNA in TSC1 KD-1 LN229 cells. Re-expression of hTSC1 in TSC1 KD-1 cells restored TSC1 protein level and mTORC1 activation to comparable levels as in Ctrl cells (Fig. S4D) and hTSC1 re-expression generated smaller tumours (Fig. S4E–S4G). TSC1 KD-1 tumours sustained higher mTORC1 activity, more Ki67+ proliferation, reduced TUNEL+ apoptosis, and more CD31+ vascular structures than Ctrl tumours, which were comparable as in hTSC1 re-expressing tumours (Fig. S5A–S5H). We did not find differences in the percentages of pAMPK+ cells and pACC+ (an AMPK substrate) cells from Ctrl, TSC1 KD-1 and hTSC1 re-expressing tumours (Fig. S5I–S5L). These findings suggested that TSC1-deficient glioblastomas had a growth advantage in vivo.
Fig. 5. Targeting autophagy and glycolysis in a subcutaneously transplanted GBM model.
a Growth curve of subcutaneously transplanted tumours from Ctrl LN229 cells and Tsc1 KD-1 LN229 cells for 5 weeks. b Images of tumours from Ctrl LN229 cells and Tsc1 KD-1 LN229 cells taken out 5 weeks after transplantation. c Growth curve of subcutaneously transplanted tumours from Tsc1 KD-1 LN229 cells treated with vehicle, 2DG, CQ and 2DG + CQ for 26 days. d–i Immunofluorescence (d–f, i) or immunohistochemistry images (g, h) and mean ± SE of the percentage (right bar graphs) of pS6RP+ cells (d), Ki67+ cells (e), TUNEL+ cells (f), pAMPK+ cells (g), pACC+ cells (h) of total cells in a section and relative CD31+ area (i) in TSC1 KD-1 LN229 tumours treated with vehicle, 2DG, CQ and 2DG + CQ for 26 days were shown. Data were analysed by two-way ANOVA (a, c) or one-way ANOVA with Tukey’s post-hoc test (d–i). n = 5 animals. *p < 0.05; **p < 0.01; ***p < 0.001. Bar = 1 cm for B and 100 μm for d–i.
We tested the efficacy of coordinated inhibition of glycolysis and autophagy to oppose progression in TSC1 KD-1 tumours when they were ~150 mm3. Mice were intra-peritoneally (I.P.) injected with vehicle (PBS), 2DG (500 mg/kg), CQ (50 mg/kg) or 2DG + CQ every other day for 3 weeks. The combination of 2DG with CQ, but not either compound alone, significantly inhibited tumour progression (Fig. 5c). 2DG + CQ reduced the percentages of pS6RP+ cells and Ki67+ cells and increased apoptotic rate in the tumours (Fig. 5d–f). 2DG alone only increased TUNEL+ cells but to a much lesser extent than the combinational treatment (Fig. 5f). The combination significantly elevated the number of pAMPK+ cells and pACC+ cells (Fig. 5g, h), suggesting a severe energy crisis. 2DG + CQ also decreased CD31+ blood vessels structure in TSC1 KD-1 tumours (Fig. 5i). Neither CQ nor 2DG affected mTORC1 activity, cell proliferation, AMPK activation or blood vessel (Fig. 5d, e, g–i). Together, these results suggested that targeting both autophagy and glycolysis was effective to treat GBM in vivo.
Next, we treated subcutaneous tumours with (I.P.) PBS, Eto (50 mg/kg), Rano (25 mg/kg), TRMZ (5 mg/kg), 2DG (500 mg/kg), 2DG + Eto, 2DG + Rano or 2DG + TRMZ. The combination of FAO inhibitors with 2DG reduced the growth of TSC1 KD-1 tumours while single agents of FAO inhibitors or 2DG had little effect in reducing tumour progression (Fig. S6A). 2DG + FAO inhibitors significantly reduced the number of pS6RP+ and Ki67+ cells and increased apoptotic rate in TSC1 KD-1 tumours (Fig. S6B–S6D). This combination strategy also activated AMPK pathways and decreased blood vessel structures (Fig. S6E–S6G). In contrast, single agent had no effects on the parameters for tumour growth, mTORC1, AMPK and blood vessel formation (Fig. S6A–S6G). Together, these results suggested a novel and efficient therapy for GBM treatment, especially those with high mTORC1 activity.
Targeting autophagy in orthotopic xenograft GBM model
Nutrient availability in the brain is regulated by the bioenergetic demands of the neurons and glia, and the transport of nutrients across the blood brain barrier.54 To test the effects of autophagy inhibition on cerebral glioblastomas, we injected 1 ×105 LN229 TSC1 KD-1 and 2KD cells into mouse brain and treated with PBS or 2DG 4 days after surgery. As indicated by the KM curve, mice bearing 2KD tumour survived longer than mice bearing TSC1 KD-1 tumours and 2DG treatment further increased the lifespan of mice receiving 2KD tumours (Fig. S7A). H&E staining revealed smaller 2KD tumours than TSC1 KD-1 tumours in the brain and 2DG further decreased 2KD tumour volume (Fig. S7B). We found fewer Ki67+ cells, more TUNEL+ cells, more pAMPK+ cells, decreased pS6RP+ signalling and CD31+ blood vessel, as well as fewer nestin+ progenitors in 2KD tumours (Fig. S7C–S7H). 2DG treatment further decreased Ki67+ cells and pS6RP+ signalling in 2KD tumours without affecting other markers (Fig. S7C–S7H). Taken together, these findings suggested that targeting autophagy might be an effective strategy to treat GBMs.
Targeting autophagy-mediated lipid catabolism in brain GBM
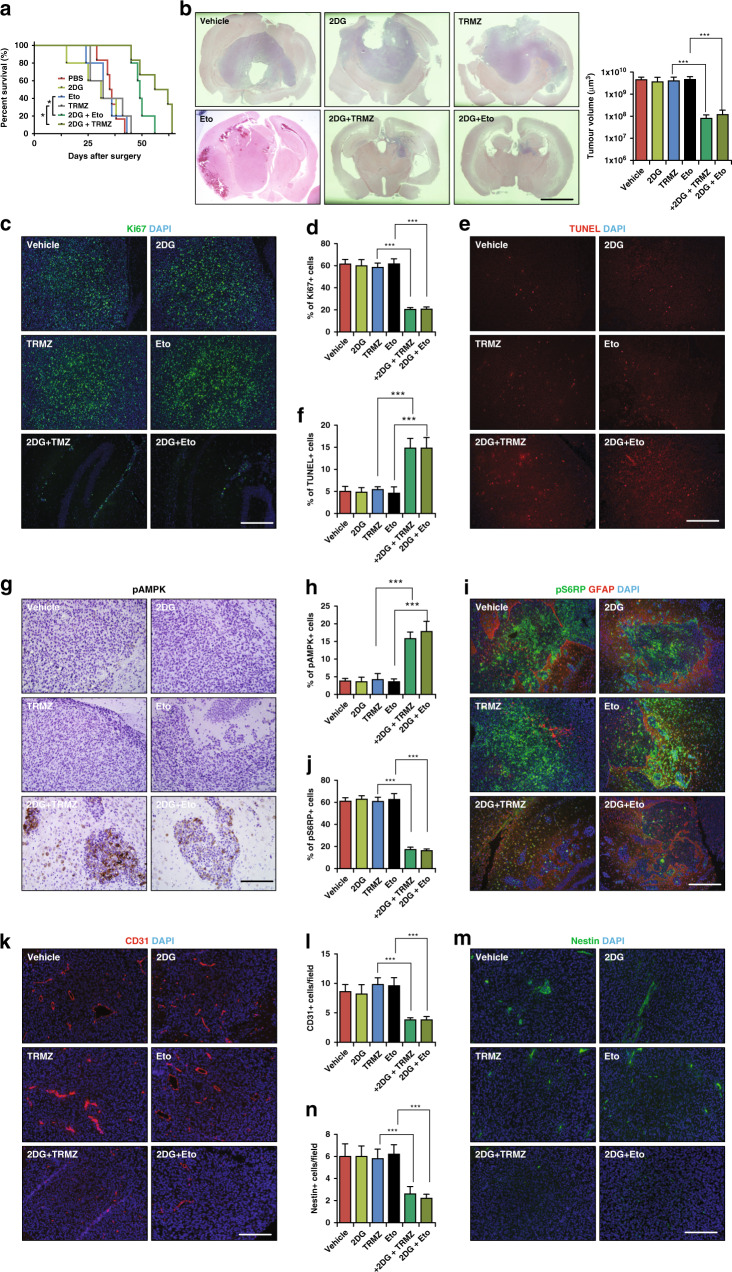
Next, we treated mice bearing TSC1 KD-1 tumour with 2DG, Eto, TRMZ, 2DG + Eto and 2DG + TRMZ 4 days after brain implantation. As indicated by the KM curve, 2DG + Eto and 2DG + TRMZ significantly delayed the death of tumour bearing mice compared to those receiving single treatments of 2DG, Eto, TRMZ or PBS (Fig. 6a). We found 2DG + Eto and 2DG + TRMZ significantly decreased tumour volume and some tumours only contained an estimated 1000 cells in a section (Fig. 6b). In contrast, single agent ETO and TRMZ had little effect on orthotopic tumour growth (Fig. 6b). We examined TSC1 expression and we found that all tumours had comparable level of TSC1 despite drug treatments (Fig. S8A). We found fewer Ki67+ cells, more TUNEL+ cells, fewer pS6RP+ cells, fewer CD31+ blood vessel structure, and reduced number of nestin+, more pAMPK+ cells and more pACC+ cells (Figs. 6c–n, S8B and S8C) in TSC1 KD-1 tumours after combinational treatments. Single agent treatment with 2DG, Eto, or TRMZ was comparable to vehicle control in measurements of parameters for tumour growth, mTORC1 and AMPK activity (Fig. 6d–o). Taken together, these findings indicated that targeting glycolysis and FAO was effective in orthotopic GBM therapy.
Fig. 6. Targeting autophagy-mediated fatty acid β-oxidation in an intracranial transplanted GBM model.
a Kaplan–Meier analysis of survival of mice intracranial transplanted Tsc1 KD-1 tumours treated with vehicle, 2DG, Eto, TRMZ, 2DG + Eto and 2DG + TRMZ. b H&E staining of coronal sectioned LN229 Tsc1 KD-1 in intracranial transplanted tumours treated with vehicle, 2DG, Eto, TRMZ, 2DG + Eto and 2DG + TRMZ. Mean ± SE of the tumour volumes were shown on the right. c–f Immunostaining of Ki67 (c) and TUNEL (e) with DAPI in intracranial transplanted Tsc1 KD-1 LN229 tumour treated with vehicle, 2DG, Eto, TRMZ, 2DG + Eto and 2DG + TRMZ. Mean ± SE of the percentage of Ki67+ cells (d) and TUNEL+ cells (f) of total cells in a section in tumours were shown. g Immunohistochemistry of pAMPK in in intracranial transplanted Tsc1 KD-1 LN229 tumours treated with vehicle, 2DG, Eto, TRMZ, 2DG + Eto and 2DG + TRMZ. h Mean ± SE of the percentage of pAMPK+ cells of total cells in a section in tumours were shown. i Immunostaining of pS6RP with DAPI in intracranial transplanted Tsc1 KD-1 LN229 tumours treated with vehicle, 2DG, Eto, TRMZ, 2DG + Eto and 2DG + TRMZ. j Mean ± SE of the percentage of pS6RP+ cells of total cells in a section in tumours were shown. k–n Immunostaining of Cd31 (k) and Nestin (m) with DAPI in intracranial transplanted tumours treated with vehicle, 2DG, Eto, TRMZ, 2DG + Eto and 2DG + TRMZ. l, n Mean ± SE of the percentage of Cd31+ cells (l) and Nestin+ cells (n) of total cells in a section in tumours treated with vehicle, 2DG, Eto, TRMZ, 2DG + Eto and 2DG + TRMZ were shown. Data were analysed by Kaplan–Meier test (a), one-way analysis of variance (ANOVA) with Tukey’s post-hoc test (b–n). n = 5 animals. ns no significance, *p < 0.05, **p < 0.01, ***p < 0.001. Bar = 5 mm in b and 100 μm in c–m.
Discussion
The hyper-activation of mTORC1 indicates a poor prognosis for brain tumours (ref. 55 and Fig. 1) and is a compelling target in molecular therapy.11,45 mTORC1 functions as an integrator of intracellular and extracellular signals in eukaryotic cells to coordinate anabolism and catabolism.56,57 It is not surprising that glioma cells utilise the mitochondrial TCA cycle/OxPHOS as major energy production pathway in addition to aerobic glycolysis. Nevertheless, the function of the TCA cycle in supporting glioma viability and stress resistance to maintain mTORC1 hyper-activation is not well studied. To overcome mTORC1-mediated tumour progression, we identified that autophagy-facilitated lipid catabolism provides metabolic plasticity and bioenergy production in glioblastoma under energy stress. Our studies provided evidence that co-targeting autophagy-mediated FAO and glycolysis could be an efficacious treatment for malignant GBM.
Autophagy activation in cancer cells allows metabolic rewiring as a compensatory response to stress58 and resistance to cancer therapies.59 Although mTORC1 suppresses autophagy through phosphorylating Ulk1,40,41 several recent studies suggested that autophagy could still occur in mTORC1 hyper-activated fibroblasts, cancer cells,42 neurons and neural stem cells.47,60 Our recent studies indicate that mTORC1 hyper-activated cells (with Tsc1 or Tsc2 deletion in neural stem cell and fibroblast as models) induce lipophagy to degrade LDs, which helps to sustain mTORC1 activity under bioenergetic stresses.47 Results from the current study further showed that GBM could resist energy crisis through lipophagy to maintain ATP production and mTORC1 hyper-activation.
The fact that high mRNA levels of FAO genes correlates with poor prognosis in GBM patients suggests FAO as a potential target for therapy.61 However, in our study, all GBM cells and tumours tolerated well FAO inhibitors as monotherapy (Figs. 3–6). Lin et al. also reported that Etomoxir only moderately reduced progression of xenografted malignant gliomas.61 Although different GBM cells were used in the previous study, it is possible that FAO is an alternative ATP production pathway in cancer cells, and the dependence on FAO is only heightened in nutrient- and oxygen-depleted environments.28,62 FAO inhibitors are also known to promote glucose utilisation in heart muscle cells in treating angina patients.63 Consistent with autophagy-mediated FAO, the autophagy inhibitor CQ promotes ECAR while suppressing OxPHOS in macrophages.64 Our results indicated that CQ and BafA1 had no effect on mTORC1 activation and LAMP2 expression on lysosome when used alone (Fig. S8D and S8E), indicating that the effect of autophagy inhibition is not mediated through disruption of mTORC1 signalling.
Previous studies indicate that the inhibition of either glycolysis or mitochondrial OxPHOS only achieves modest outcome to treat cancers, including glioblastomas.65 For effective therapeutic approach, simultaneous inhibition of glycolysis and OxPHOS in a GBM model achieves better outcome.66 Moreover, the selection of a specified energy production mechanism, e.g. lipophagy, instead of suppressing whole OxPHOS process will likely reduce off-target effects. As a result of mTORC1 hyper-activation, ~60% of carbon skeletons from glucose are used for lipid synthesis in GBM cells.67 Besides lipogenesis, we found that increased mRNA level of fatty acids binding proteins or transport proteins in GBM (data not shown). These lipid metabolic pathways have been identified as part of the ‘lipogenic phenotype’ in cancer, which provides proliferation and survival advantages.68,69 Regardless of the lipid sources, the ability of GBM cells to store LDs and to activate lipophagy suggests that metabolic reprogramming serves to sustain growth in GBM.
Our study provides translational insight to develop new GBM therapy. 2DG and its derivatives were tested in combination with radiotherapy70 and chemotherapy71 in neoplastic diseases. The dosage of CQ administrated in our experiment is at a level well-tolerated and previously used in patients. Several clinical trials using CQ or HCQ as mono- or combination-therapy for cancers are under way (NCT04397679, NCT02333890). There are at least two Phase-3 clinical trials using TRMZ (commercial name Vastarel®) to treat intermediate or advanced hepatocellular carcinoma (NCT03274427 and NCT03278444). Rano (commercial name RANEXA®) was approved by the FDA for the treatment of patients with chronic angina. Our preclinical evidence strongly suggests that combining FAO inhibitors with 2DG in GBM patients for clinical trials. Taken together, our studies laid a mechanistic basis for an essential role of lipophagy to meet high energy demand in GBM and reveal a valuable strategy for potential GBM therapy.
Supplementary information
Acknowledgements
We thank Dr. Xin Tang and Mr. Harsh Patel in Dr. Wang’s lab and other people from Dr. Guan’s lab for careful reading of the manuscript and helpful suggestions. We thank Glenn Doerman for his help in the preparation of figures. We thank the support from Brain Tumor Center of University of Cincinnati College of Medicine.
Author contributions
C.W.: developed idea, designed and executed experiments, analysed data and wrote manuscript; M.H.: assisted in the mice transplantation and edited the manuscript; S.Y.: contributed in the development of molecular mechanism, the manuscript review and correction; R.P.: assisted in experiments and edited the manuscript; F.Y.: assisted in experiments and edited the manuscript; S.V.: assisted in the mice transplantation and edited the manuscript; X.Q.: designed experiments, analysed data and edited the manuscript; D.P.: contributed in the development of molecular mechanism, analysed data and edited the manuscript; J.-L.G.: developed idea, analysed data and edited the manuscript.
Ethics approval and consent to participate
Mice were housed and handled according to local, state and federal regulations. All animal studies were carried out according to the protocols approved by the Institutional Animal Care and Use Committee at University of Cincinnati (Cincinnati, OH, USA). Human GBM cell lines were gifts from Dr. David R. Plas (University of Cincinnati College of Medicine) as described in the Methods section.
Data availability
All original data and materials generated during the current study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Funding information
This study was supported by NIH grants (NS103981-01) to C.R.W. and (NS094144-02, CA211066) to J.L.G.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chenran Wang, Michael A. Haas
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-021-01294-0.
References
- 1.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310:1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 2.Clarke J, Butowski N, Chang S. Recent advances in therapy for glioblastoma. Arch. Neurol. 2010;67:279–283. doi: 10.1001/archneurol.2010.5. [DOI] [PubMed] [Google Scholar]
- 3.Thomas AA, Brennan CW, DeAngelis LM, Omuro AM. Emerging therapies for glioblastoma. JAMA Neurol. 2014;71:1437–1444. doi: 10.1001/jamaneurol.2014.1701. [DOI] [PubMed] [Google Scholar]
- 4.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 5.Liang BC, Thornton AF, Jr., Sandler HM, Greenberg HS. Malignant astrocytomas: focal tumor recurrence after focal external beam radiation therapy. J. Neurosurg. 1991;75:559–563. doi: 10.3171/jns.1991.75.4.0559. [DOI] [PubMed] [Google Scholar]
- 6.Petrecca K, Guiot MC, Panet-Raymond V, Souhami L. Failure pattern following complete resection plus radiotherapy and temozolomide is at the resection margin in patients with glioblastoma. J. Neuro Oncol. 2013;111:19–23. doi: 10.1007/s11060-012-0983-4. [DOI] [PubMed] [Google Scholar]
- 7.Sneed PK, Gutin PH, Larson DA, Malec MK, Phillips TL, Prados MD, et al. Patterns of recurrence of glioblastoma multiforme after external irradiation followed by implant boost. Int. J. Radiat. Oncol. Biol. Phys. 1994;29:719–727. doi: 10.1016/0360-3016(94)90559-2. [DOI] [PubMed] [Google Scholar]
- 8.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akhavan D, Cloughesy TF, Mischel PS. mTOR signaling in glioblastoma: lessons learned from bench to bedside. Neuro Oncol. 2010;12:882–889. doi: 10.1093/neuonc/noq052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laplante M, Sabatini DM. mTOR signaling at a glance. J. Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pachow D, Wick W, Gutmann DH, Mawrin C. The mTOR signaling pathway as a treatment target for intracranial neoplasms. Neuro Oncol. 2015;17:189–199. doi: 10.1093/neuonc/nou164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang S, Houghton PJ. Targeting mTOR signaling for cancer therapy. Curr. Opin. Pharmacol. 2003;3:371–377. doi: 10.1016/S1471-4892(03)00071-7. [DOI] [PubMed] [Google Scholar]
- 13.Dowling RJ, Topisirovic I, Fonseca BD, Sonenberg N. Dissecting the role of mTOR: lessons from mTOR inhibitors. Biochim. Biophys. Acta. 2010;1804:433–439. doi: 10.1016/j.bbapap.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Guertin DA, Sabatini DM. An expanding role for mTOR in cancer. Trends Mol. Med. 2005;11:353–361. doi: 10.1016/j.molmed.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 15.Fan Q, Aksoy O, Wong RA, Ilkhanizadeh S, Novotny CJ, Gustafson WC, et al. A kinase inhibitor targeted to mTORC1 drives regression in glioblastoma. Cancer Cell. 2017;31:424–435. doi: 10.1016/j.ccell.2017.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cloughesy TF, Yoshimoto K, Nghiemphu P, Brown K, Dang J, Zhu S, et al. Antitumor activity of rapamycin in a Phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med. 2008;5:e8. doi: 10.1371/journal.pmed.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwanami A, Gini B, Zanca C, Matsutani T, Assuncao A, Nael A, et al. PML mediates glioblastoma resistance to mammalian target of rapamycin (mTOR)-targeted therapies. Proc. Natl Acad. Sci. USA. 2013;110:4339–4344. doi: 10.1073/pnas.1217602110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takeuchi H, Kondo Y, Fujiwara K, Kanzawa T, Aoki H, Mills GB, et al. Synergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidylinositol 3-kinase/protein kinase B inhibitors. Cancer Res. 2005;65:3336–3346. doi: 10.1158/0008-5472.CAN-04-3640. [DOI] [PubMed] [Google Scholar]
- 19.Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat. Cell Biol. 2013;15:555–564. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Q, Chen X, Ma J, Peng H, Wang F, Zha X, et al. Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proc. Natl Acad. Sci. USA. 2011;108:4129–4134. doi: 10.1073/pnas.1014769108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valvezan AJ, Turner M, Belaid A, Lam HC, Miller SK, McNamara MC, et al. mTORC1 couples nucleotide synthesis to nucleotide demand resulting in a targetable metabolic vulnerability. Cancer Cell. 2017;32:624–638 e5. doi: 10.1016/j.ccell.2017.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chakrabarti P, English T, Shi J, Smas CM, Kandror KV. Mammalian target of rapamycin complex 1 suppresses lipolysis, stimulates lipogenesis, and promotes fat storage. Diabetes. 2010;59:775–781. doi: 10.2337/db09-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee G, Zheng Y, Cho S, Jang C, England C, Dempsey JM, et al. Post-transcriptional regulation of de novo lipogenesis by mTORC1-S6K1-SRPK2 signaling. Cell. 2017;171:1545–58 e18. doi: 10.1016/j.cell.2017.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Csibi A, Lee G, Yoon SO, Tong H, Ilter D, Elia I, et al. The mTORC1/S6K1 pathway regulates glutamine metabolism through the eIF4B-dependent control of c-Myc translation. Curr. Biol. 2014;24:2274–2280. doi: 10.1016/j.cub.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat. Rev. Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 27.Agnihotri S, Zadeh G. Metabolic reprogramming in glioblastoma: the influence of cancer metabolism on epigenetics and unanswered questions. Neuro Oncol. 2016;18:160–172. doi: 10.1093/neuonc/nov125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beloribi-Djefaflia S, Vasseur S, Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 2016;5:e189. doi: 10.1038/oncsis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo X, Cheng C, Tan Z, Li N, Tang M, Yang L, et al. Emerging roles of lipid metabolism in cancer metastasis. Mol. Cancer. 2017;16:76. doi: 10.1186/s12943-017-0646-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Accioly MT, Pacheco P, Maya-Monteiro CM, Carrossini N, Robbs BK, Oliveira SS, et al. Lipid bodies are reservoirs of cyclooxygenase-2 and sites of prostaglandin-E2 synthesis in colon cancer cells. Cancer Res. 2008;68:1732–1740. doi: 10.1158/0008-5472.CAN-07-1999. [DOI] [PubMed] [Google Scholar]
- 31.Qiu B, Ackerman D, Sanchez DJ, Li B, Ochocki JD, Grazioli A, et al. HIF2alpha-dependent lipid storage promotes endoplasmic reticulum homeostasis in clear-cell renal cell carcinoma. Cancer Discov. 2015;5:652–667. doi: 10.1158/2159-8290.CD-14-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yue S, Li J, Lee SY, Lee HJ, Shao T, Song B, et al. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab. 2014;19:393–406. doi: 10.1016/j.cmet.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geng F, Cheng X, Wu X, Yoo JY, Cheng C, Guo JY, et al. Inhibition of SOAT1 suppresses glioblastoma growth via blocking SREBP-1-mediated lipogenesis. Clinical cancer research: an official journal of the American Association for. Cancer Res. 2016;22:5337–5348. doi: 10.1158/1078-0432.CCR-15-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laplante M, Sabatini DM. An emerging role of mTOR in lipid biosynthesis. Curr. Biol. 2009;19:R1046–R1052. doi: 10.1016/j.cub.2009.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walther TC, Farese RV., Jr. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zechner R, Zimmermann R, Eichmann TO, Kohlwein SD, Haemmerle G, Lass A, et al. FAT SIGNALS-lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012;15:279–291. doi: 10.1016/j.cmet.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimmelman AC, White E. Autophagy and tumor metabolism. Cell Metab. 2017;25:1037–1043. doi: 10.1016/j.cmet.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y, Wei H, Liu F, Guan JL. Hyperactivation of mammalian target of rapamycin complex 1 (mTORC1) promotes breast cancer progression through enhancing glucose starvation-induced autophagy and Akt signaling. J. Biol. Chem. 2014;289:1164–1173. doi: 10.1074/jbc.M113.526335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen S, Wang C, Yeo S, Liang CC, Okamoto T, Sun S, et al. Distinct roles of autophagy-dependent and -independent functions of FIP200 revealed by generation and analysis of a mutant knock-in mouse model. Genes Dev. 2016;30:856–869. doi: 10.1101/gad.276428.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 2011;25:1510–1527. doi: 10.1101/gad.2051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson CM, Macleod KF. Autophagy and cancer cell metabolism. Int. Rev. Cell Mol. Biol. 2019;347:145–190. doi: 10.1016/bs.ircmb.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poillet-Perez L, White E. Role of tumor and host autophagy in cancer metabolism. Genes Dev. 2019;33:610–619. doi: 10.1101/gad.325514.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang C, Haas MA, Yang F, Yeo S, Okamoto T, Chen S, et al. Autophagic lipid metabolism sustains mTORC1 activity in TSC-deficient neural stem cells. Nature. Metabolism. 2019;1:1127–1140. doi: 10.1038/s42255-019-0137-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoo Y, Wu X, Guan JL. A novel role of the actin-nucleating Arp2/3 complex in the regulation of RNA polymerase II-dependent transcription. J. Biol. Chem. 2007;282:7616–7623. doi: 10.1074/jbc.M607596200. [DOI] [PubMed] [Google Scholar]
- 49.Chen Q, Shao X, Hao M, Fang H, Guan R, Tian Z, et al. Quantitative analysis of interactive behavior of mitochondria and lysosomes using structured illumination microscopy. Biomaterials. 2020;250:120059. doi: 10.1016/j.biomaterials.2020.120059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol. Cancer Res. 2009;7:157–167. doi: 10.1158/1541-7786.MCR-08-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gravendeel LA, Kouwenhoven MC, Gevaert O, de Rooi JJ, Stubbs AP, Duijm JE, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69:9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 52.Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147:223–234. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wagle N, Kesari S. Breaking down the blood brain barrier. Neuro Oncol. 2021;23:6. doi: 10.1093/neuonc/noaa274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li X, Wu C, Chen N, Gu H, Yen A, Cao L, et al. PI3K/Akt/mTOR signaling pathway and targeted therapy for glioblastoma. Oncotarget. 2016;7:33440–33450. doi: 10.18632/oncotarget.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo JY, Xia B, White E. Autophagy-mediated tumor promotion. Cell. 2013;155:1216–1219. doi: 10.1016/j.cell.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sui X, Chen R, Wang Z, Huang Z, Kong N, Zhang M, et al. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis. 2013;4:e838. doi: 10.1038/cddis.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Di Nardo A, Wertz MH, Kwiatkowski E, Tsai PT, Leech JD, Greene-Colozzi E, et al. Neuronal Tsc1/2 complex controls autophagy through AMPK-dependent regulation of ULK1. Hum. Mol. Genet. 2014;23:3865–3874. doi: 10.1093/hmg/ddu101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin H, Patel S, Affleck VS, Wilson I, Turnbull DM, Joshi AR, et al. Fatty acid oxidation is required for the respiration and proliferation of malignant glioma cells. Neuro Oncol. 2017;19:43–54. doi: 10.1093/neuonc/now128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nat. Rev. Cancer. 2013;13:227–232. doi: 10.1038/nrc3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kolwicz SC, Jr., Purohit S, Tian R. Cardiac metabolism and its interactions with contraction, growth, and survival of cardiomyocytes. Circ. Res. 2013;113:603–616. doi: 10.1161/CIRCRESAHA.113.302095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen D, Xie J, Fiskesund R, Dong W, Liang X, Lv J, et al. Chloroquine modulates antitumor immune response by resetting tumor-associated macrophages toward M1 phenotype. Nat. Commun. 2018;9:873. doi: 10.1038/s41467-018-03225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kalender A, Selvaraj A, Kim SY, Gulati P, Brule S, Viollet B, et al. Metformin, independent of AMPK, inhibits mTORC1 in a rag GTPase-dependent manner. Cell Metab. 2010;11:390–401. doi: 10.1016/j.cmet.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim EH, Lee JH, Oh Y, Koh I, Shim JK, Park J, et al. Inhibition of glioblastoma tumorspheres by combined treatment with 2-deoxyglucose and metformin. Neuro Oncol. 2017;19:197–207. doi: 10.1093/neuonc/now174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lunt SY, Vander, Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 68.Currie E, Schulze A, Zechner R, Walther TC, Farese RV., Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 70.Singh D, Banerji AK, Dwarakanath BS, Tripathi RP, Gupta JP, Mathew TL, et al. Optimizing cancer radiotherapy with 2-deoxy-d-glucose dose escalation studies in patients with glioblastoma multiforme. Strahlenther. Onkol. 2005;181:507–514. doi: 10.1007/s00066-005-1320-z. [DOI] [PubMed] [Google Scholar]
- 71.Raez LE, Papadopoulos K, Ricart AD, Chiorean EG, Dipaola RS, Stein MN, et al. A phase I dose-escalation trial of 2-deoxy-D-glucose alone or combined with docetaxel in patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2013;71:523–530. doi: 10.1007/s00280-012-2045-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All original data and materials generated during the current study are available from the corresponding author upon reasonable request.