Identification of Candida auris is challenging and requires molecular or protein profiling-based approaches, availability of which is limited in many routine diagnostic laboratories, necessitating the development of a cost-effective, rapid, and reliable method of identification. The objective of this study was to develop a selective medium for C. auris identification.
KEYWORDS: Candida auris, invasive candidiasis, rapid identification, selective media
ABSTRACT
Identification of Candida auris is challenging and requires molecular or protein profiling-based approaches, availability of which is limited in many routine diagnostic laboratories, necessitating the development of a cost-effective, rapid, and reliable method of identification. The objective of this study was to develop a selective medium for C. auris identification. Eighteen C. auris and 30 non-C. auris yeasts were used for the standardization of the selective medium. Sodium chloride (10% to 13% concentration) and ferrous sulfate (8 mM to 15 mM) were added to yeast extract-peptone-dextrose (YPD) agar in various combinations followed by incubation at 37°C, 40°C, or 42°C for 2 to 3 days. For validation, 579 yeast isolates and 40 signal-positive Bactec blood culture (BC) broths were used. YPD agar comprising 12.5% NaCl and 9 mM ferrous sulfate incubated at 42°C for 48 h, named Selective Auris Medium (SAM), allowed selective growth of C. auris. A total of 95% (127/133) of C. auris isolates tested grew on the standardized media within 48 h, and the remaining 6 isolates grew after 72 h, whereas the growth of 446 non-C. auris yeast isolates was completely inhibited. The specificity and sensitivity of the test medium were both 100% after 72 h of incubation. The positive and negative predictive values were also noted to be 100% after 72 h of incubation. The formulated selective medium can be used for the detection and identification of C. auris. The SAM is inexpensive, can easily be prepared, and can be used as an alternative to molecular diagnostic tools in the clinical microbiology laboratory.
INTRODUCTION
Following the global emergence of Candida auris, a clinical alert was issued by the Centers for Disease Control and Prevention (CDC), Atlanta, GA, USA, highlighting it as a serious threat to public health (1). Since the first report in 2009 from Japan, C. auris infection has been reported from six continents (2, 3). Based on whole-genome sequencing, C. auris has been classified into four major geographical clades, i.e., clade I (South Asia), clade II (East Asia), clade III (South Africa), and clade IV (South America), and a possible fifth clade in Iran (4, 5). C. auris infection represents a formidable challenge in health care due to its propensity for rapid nosocomial spread, unknown environmental niche, and ability to form adherent biofilms on clinically important substrates (6–10). Other major issues include its resistance to various classes of antifungals and high mortality due to the infection, ranging from 30% to 70% (2, 11–13).
Despite the alarming rise in reports of C. auris infections over the past decade, the reported numbers represent perhaps an underestimation of the true incidence since the fungus is difficult to identify by conventional phenotypic and biochemical tests in routine clinical laboratory practice (14). The commercial microbial identification systems, such as Vitek-2 YST, API AUX 20C, BD Phoenix, and MicroScan, may misidentify C. auris (12, 15).
Accurate identification of Candida species is important to initiate appropriate antifungal treatment and prevent its spread within the hospital. Various tests developed for rapid identification of C. auris use either DNA-based or matrix-assisted laser desorption ionization–time of flight (MALDI-TOF)-based protein profiling (2, 16–18). The majority of the clinical laboratories performing routine assays are not equipped with such advanced facilities. Further, sending the unidentified yeast to reference laboratories adds to the turnaround time. While major attention and efforts have focused on rapid detection using advanced molecular techniques, the development of an easy-to-prepare, low-cost, and yet reproducible and sensitive method for resource-limited settings is largely ignored. In this study, we attempted to develop a selective medium which could significantly reduce the time and cost associated with the identification of C. auris even in low-resource health care settings. The first phase of the study was physiological stress induction, and the result from this served as the concept behind formulating the selective medium.
MATERIALS AND METHODS
Isolates, culture, and antifungal susceptibility testing.
For standardization, a total of 18 C. auris isolates, including 6 fluconazole-resistant, 6 fluconazole-susceptible, 2 caspofungin-resistant, 2 caspofungin-susceptible, and 2 reference isolates, were used (Table 1). To confirm resistance or susceptibility, we perform microbroth dilution antifungal susceptibility testing following Clinical and Laboratory Standards Institute (CLSI) M-60 guidelines (19). A fluconazole MIC of ≥32 μg/ml and a caspofungin MIC of ≥2 μg/ml were considered to represent resistance per the breakpoints proposed by the Centers for Disease Control and Prevention (CDC), Atlanta, GA (20).
TABLE 1.
List of 18 C. auris isolates included in the standardization of the mediuma
| Sl no. | Isolate category | Isolation site | No. of isolates | MIC (μg/ml) |
|---|---|---|---|---|
| 1 | Fluconazole resistant | Blood | 2 | 512 (both isolates) |
| Hospital environment (ECG leads) | 2 | 256 (both isolates) | ||
| Patient’s skin and groin | 2 | 256 (both isolates) | ||
| 2 | Fluconazole susceptible | Blood | 2 | 4 (both isolates) |
| Hospital environment (temp probes) | 2 | 0.5 (both isolates) | ||
| Patient’s skin and axilla | 2 | 0.5 (both isolates) | ||
| 3 | Caspofungin resistant | Blood | 1 | 16 |
| Blood | 1 | 16 | ||
| 4 | Caspofungin susceptible | Blood | 1 | 0.125 |
| Blood | 1 | 0.125 | ||
| 5 | Reference strain (JCM 15448, Japan); fluconazole susceptible, caspofungin susceptible |
Ear | 1 | 1 0.5 |
| 6 | Reference strain (CBS 12372, South Korea); fluconazole susceptible, caspofungin susceptible |
Blood | 1 | 2 0.5 |
CBS, Centraalbureau voor Schimmelcultures, Utrecht, Netherlands; ECG, echocardiogram; JCM, Japan Collection of Microorganisms; Sl, serial.
Additionally, 30 clinical non-C. auris yeast isolates (1 isolate each of Candida albicans, Candida tropicalis, Candida krusei [syn. Pichia kudriavzevii], Candida glabrata [syn. Nakaseomyces glabrata], Candida parapsilosis, Candida metapsilosis, Candida orthopsilosis, Candida guilliermondii [syn. Meyerozyma guilliermondii], Candida utilis [syn. Cyberlindnera jadinii], Candida lusitaniae [syn. Clavispora lusitaniae], Candida viswanathii, Candida rugosa [syn. Diutina rugosa], Candida pararugosa [syn. Wickerhamiella pararugosa], Candida kefyr [syn. Kluyveromyces marxianus], Candida ciferrii [syn. Trichomonascus ciferrii], Candida famata [syn. Debaryomyces hansenii], Kodamaea ohmeri, Wickerhamomyces anomalus, Saccharomyces cerevisiae, Cryptococcus neoformans var. grubii, Cryptococcus neoformans var. gatti, Cryptococcus neoformans var. laurentii [syn. Papiliotrema laurentii], Geotrichum silvicola, Lodderomyces elongisporus, Cyberlindnera fabianii, Trichosporon asahii, Trichosporon dohaense, Candida haemulonii, Candida pseudohaemulonii, and Candida duobushaemulonii) from the National Culture Collection for Pathogenic Fungi (NCCPF), Chandigarh, India, were included in the study to evaluate the specificity of test medium. C. albicans SC5314 was used as a control for the phenotypic stress studies. All strains were cultured fresh on yeast extract-peptone-dextrose (YPD) agar plates (HiMedia, India), and a single pure colony was inoculated into 20 ml YPD broth medium (HiMedia, India) and incubated at 30°C overnight (16 to 24 h) by shaking at a speed of 200 rpm. Cells from overnight cultures were washed twice with sterile normal saline solution, and the optical density at 600 nm (OD600) was measured. The cells were diluted in YPD broth medium to obtain a cell count of ∼1 × 106 cells/ml, corresponding to an OD600 of 0.3. Those aliquots (with ∼1 × 106 cells/ml) were then exponentially diluted from 10-fold to 1,000-fold, and 10 μl of each diluted cell suspension was spotted on YPD agar with different modifications depending on the stress being studied.
Stress induction.
Initially, as a part of a pilot study, 53 C. auris isolates were spotted (∼106 cells/ml) onto YPD agar plates containing sodium chloride at concentrations ranging from 8% to 18% (with 0.5% increments). After 48 to 72 h of incubation, the majority of the isolates showed moderate to heavy growth until a 16% to 17% concentration of sodium chloride was reached (see Fig. S1 in the supplemental material). Similarly, metal stress induction was performed for 15 C. auris isolates by addition of ferrous sulfate to YPD agar at final concentrations ranging from 1 mM to 17 mM (with 0.5 mM increments). The C. auris isolates could tolerate 13 mM to 15 mM ferrous sulfate.
Dual stress at the various combinations of 10% to 13% sodium chloride (with 0.5% increments) and 8 mM to 15 mM ferrous sulfate (with 0.5 mM increments) was evaluated in the present study. All stock solutions except the sodium chloride solutions were freshly prepared immediately before use and were filtered by the use of 0.2-μm-pore-size sterile syringe filters (Acrodisc syringe filters with Supor membrane; Pall Corporation) before being added to the media. Sodium chloride was directly added to the YPD agar media before autoclaving. All plates were incubated at 37°C, 40°C, and 42°C for 2 to 3 days.
Salt, metal, and temperature tolerance.
As the 18 C. auris isolates used during standardization were all clade I isolates, 2 C. auris clade II isolates (JCM 15448, Japan; CBS 12372, South Korea), 1 C. auris clade III isolate (CDC AR 0383), and 1 C. auris clade IV isolate (CDC AR 0385) were evaluated for salt, metal, and temperature tolerance. The levels of metal and salt tolerance were checked on YPD agar plates with the stress of 9 mM ferrous sulfate at 37°C or of sodium chloride (8% to 18% concentration) at 37°C and 42°C.
Growth kinetics.
For growth curve analysis, a fresh overnight (16-to-24-h) culture consisting of ∼1 × 106 cells/ml cell suspension in YPD broth was used. Subsequently, 200 μl of that suspension was added to 96-well flat-bottomed plates (Nunclon surface; Nunc, Denmark) and incubated in a kinetic enzyme-linked immunosorbent assay (ELISA) microplate reader (Epoch 2 microplate reader; BioTek Instruments, Inc., USA) for 30 h at 35°C and 42°C separately with continuous orbital shaking. The reader was programmed to read the results at 600 nm at 30-min intervals. All experiments were performed in triplicate.
Preparation of standardized selective medium.
On the basis of the results of the dual-stress induction, the best combination of stress factors yielding isolation of C. auris was optimized for the preparation of selective medium. Briefly, YPD broth was prepared per the manufacturer’s directions, after which agar powder was added (Agar Ultra-pure; HiMedia, India) at a concentration of 2.5%. Sodium chloride was added to obtain a final concentration of 12.5%. The medium was autoclaved for 15 min at 121°C. A ferrous sulfate solution (1 M) was freshly prepared (no more than 10 to 15 min before addition to the autoclaved medium to avoid aerial oxidation) and sterilized by syringe filtration. Once the agar-containing medium cooled to 80°C, the ferrous sulfate solution (9 ml per liter) was added to obtain a final concentration of 9 mM. The medium was then mixed thoroughly (to avoid the necessity for vortex mixing) and manually poured into a sterile petri plate (90-mm diameter) to obtain a thickness of 5 to 6 mm. The plates were dried at room temperature followed by sterility checking using two plates from each batch incubated at 25°C and 37°C for 2 days.
Validation of the standardized selective medium.
For validation of the selective medium, yeast isolates recovered from the clinical laboratory at the Department of Medical Microbiology, Postgraduate Institute of Medical Education and Research (PGIMER), Chandigarh, India, and the isolates received from various centers across the country through June to November 2019 as a part of National Antimicrobial Resistance Surveillance (AMR) were included. An isolated yeast colony was touched with a sterile stainless-steel inoculation loop and streaked directly on the standardized selective medium. The minimum inoculum was picked and streaked to ensure the lack of visibility of that inoculum at the primary streaking area.
For signal-positive blood culture (BC) samples (maintained using BD Bactec blood culture vials [Bactec Plus Aerobic/F and Bactec Peds Plus/F, BD Diagnostics, Heidelberg, Germany]), one loopful of the broth was used as the inoculum. The isolates were identified by matrix-assisted laser desorption ionization–time of flight mass spectroscopy (Microflex LT Biotyper system [Bruker Daltonics, Bremen, Germany]) per a previously described protocol (18).
Genotypic characterization.
Fluorescent amplified fragment length polymorphism (FAFLP) analysis was performed to determine the genetic similarity of strains used in the study. A total of 22 C. auris isolates, including 17 isolates from the present study (9 blood isolates, 6 colonization isolates [3 aggregative and 3 nonaggregative], and 2 environmental isolates [1 aggregative and 1 nonaggregative]) and 5 from other sources (1 C. auris clade I isolate [NCCPF 470146]; 2 clade II isolates; 1 isolate each from clades III and IV [as mentioned above]) were used. The FAFLP analysis was performed per the previously standardized protocol (21). Briefly, EcoRI and HindIII restriction enzymes (New England Biolabs, Ipswich, MA, USA) and corresponding adapters were used. Amplification was performed using preselective primers of EcoRI (5’GACTGCGTACCAGCTT-3′) and HindIII (5’GACTGCGTACCAATTC-3′). Both a 6-carboxyfluorescein (FAM)-labeled EcoRI primer with one selective residue (5′-GACTGCGTACCAGCTTT-3′) and a HindIII primer with two selective residues (5′-GACTGCGTACCAATTCAC-3′) were used. Capillary electrophoresis of the amplified products along with standard marker LIZ600 was performed in an ABI automated DNA sequencer (model 3500 Dx genetic analyzer; Applied Biosystems, USA). Typing data were imported to BioNumerics v 6.6 software (Applied Maths, Ghent, Belgium). The similarity coefficient was determined by Pearson correlation, with negative similarities clipped to zero. Cluster analysis was performed by the use of the unweighted pair group method using average linkages and BioNumerics software.
RESULTS
Single-stress and dual-stress induction.
Only C. auris and five other Candida species (C. albicans, C. tropicalis, C. krusei, C. glabrata, and C. guilliermondii) could grow on YPD medium at 42°C without salt and metal stress. Salt (12.5% sodium chloride) tolerance at 37°C after 24 h of incubation was noted in C. albicans, C. tropicalis, C. krusei, C. glabrata, C. guilliermondii, C. utilis, C. lusitaniae, C. viswanathii, C. pararugosa, C. kefyr, C. ciferrii, C. famata, S. cerevisiae, C. neoformans, C. gattii, C. laurentii, G. silvicola, L. elongisporus, C. fabianii, T. asahii, T. dohaense, and 18 C. auris isolates. Metal (9 mM ferrous sulfate) tolerance at 37°C after 24 h of incubation was noted in C. auris and all non-C. auris yeast species (except C. parapsilosis). The members of clades I, II, III, and IV tolerated salt concentrations of 16%, 12%, 16%, and 16%, respectively. No differences in temperature and metal stress tolerance were observed among the clades. Tolerance at the stress levels represented by 12.5% sodium chloride and 9 mM ferrous sulfate as shown by growth at 42°C was seen only in C. auris (all four clades) except in two clade II isolates (Japanese C. auris JCM 15448 and South Korean C. auris 12372) and in no non-auris yeast isolates. Finally, YPD medium containing 12.5% sodium chloride and 9 mM ferrous sulfate, used with incubation at 42°C, was chosen as the selective medium (Selective Auris Medium [SAM]) for C. auris in the validation study. The control plates with YPD media and its modifications, maintained at different temperatures, are shown in Fig. S2 in the supplemental material. Representative images depicting the results of combined stress inductions in 13 common non-C. auris yeasts are presented in Fig. 1.
FIG 1.
Results of dual-stress induction in C. auris and 13 common non-C. auris yeast isolates.
Growth kinetics.
Growth curve analysis of one reference C. auris isolate each from clades I, II, III, and IV showed that the four clades had different growth patterns at 42°C. The doubling times for clades I, II, III, and IV at 42°C were 6.769 ± 0.388, 12.670 ± 0.337, 5.136 ± 0.868, and 3.426 ± 0.940 h, respectively (means ± standard deviations [SD]) (Fig. S3). The doubling time of the clade II isolate at 42°C was significantly higher than that seen with each of other three clade isolates (compared with clade I, P = 0.0032; compared with clade III, P = 0.0019; compared with clade IV, P = 0.0026).
Validation of selective medium.
During validation, a total of 579 yeast isolates were evaluated on the SAM (Fig. 2).
FIG 2.

(A) Validation of results of the use of selective medium with C. auris and non-C. auris yeast species by growth at 48 h of incubation. (B) Directly streaked C. auris clinical isolates at positions 1 to 12 and positions 14 to 16, showing heavy confluent growth on the selective medium after 3 days of incubation. Position 13 represents non-C. auris yeast (C. parapsilosis) showing no growth.
Of 133 C. auris isolates (clinical, n = 117; environmental, n = 8; patient colonizer, n = 8) tested, 127 C. auris isolates grew on the selective medium plates, showing moderate to heavy growth after 48 h of incubation, whereas the remaining 6 isolates exhibited growth only after 72 h of incubation (Fig. 3). None of the other yeast isolates tested grew on this selective medium at 42°C even after incubation for 72 h. The specificity of the selective isolation of C. auris by the test medium was 100% (95% confidence interval [CI], 99.18% to 100%) whereas the sensitivity was 95.5% (95% CI, 90.44% to 98.33%) at 48 h, and specificity and sensitivity were 100% each at 72 h of incubation. The positive predictive value (PPV) for correct identification was 100%, with a negative predictive value of 98.67% (95% CI, 97.14% to 99.4%) at 48 h, and the values were 100% each at 72 h of incubation.
FIG 3.

(A) Validation of results of the use of selective medium for isolation of C. auris directly from positive automated blood culture vials. (B) Confluent growth of C. auris obtained from direct inoculation from one C. auris-positive blood culture vial (position 1) after 48 h of incubation.
A total of 40 yeast-positive BD Bactec blood culture broths were inoculated onto the selective medium. Only one showed confluent growth on the selective medium, which was later confirmed as C. auris by MALDI-TOF mass spectrometry (MS). The results of selective medium validation for direct isolation of C. auris from yeast-positive blood culture vials are shown in Fig. 3.
FAFLP.
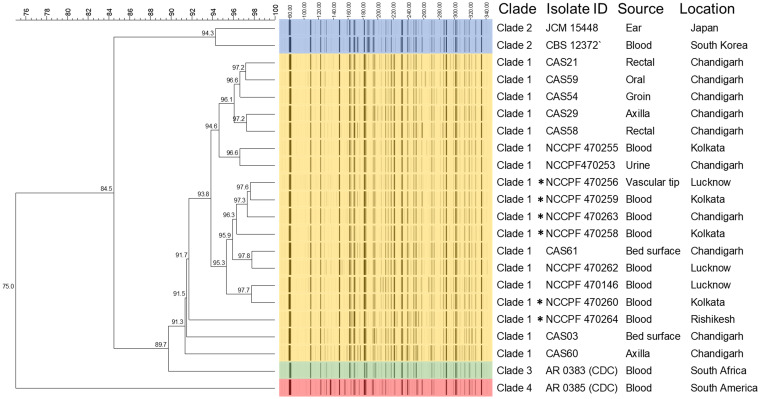
FAFLP analyses revealed four distinct clusters (A, B, C, and D) (Fig. 4) with percent similarity of >74% and intercluster difference of <8%. The C. auris isolates in cluster B were clustered with the standard clade I isolate. The isolates were from different locations of India and from clinical samples (blood, urine, vascular tip), colonizing sites (oral, rectal, axilla), and environmental (bed surface) sources. The reference strains of clade II (JCM 15448 and CBS 12372), clade III (CDC-AR 0383), and clade IV (CDC-AR 0385) separated distinctly from each other, forming clusters A, C, and D, respectively.
FIG 4.
Fluorescent amplified fragment length polymorphism (FAFLP) analysis of 22 isolates of C. auris (9 clinical isolates [NCCPF], 6 colonizing isolates [Chemical Abstracts Service {CAS} no. 21, 29, and 54, aggregative; CAS no. 58, 59, and 60, nonaggregative], 2 environmental isolates [CAS 03, aggregative; CAS 61, nonaggregative], 1 C. auris clade I isolate [NCCPF 470146], 2 C. auris clade II isolates [JCM 15448, Japan; CBS 12372, South Korea], 1 C. auris clade III isolate [AR 0383, CDC], and 1 C. auris clade IV isolate [AR 0385, CDC]). *, six isolates which grew after prolonged incubation (72 h) on standardized medium.
DISCUSSION
In the present study, a simple, cost-effective, and sensitive selective medium was developed for the isolation and identification of C. auris. This medium was successfully validated in a large number of clinical yeast isolates, including C. auris from the four clades described above. Additionally, direct isolation of C. auris was performed from positive automated blood culture vials, with encouraging results.
The worldwide emergence of C. auris has emphasized the need for identifying Candida to the species level. This is essential for prompt initiation of appropriate antifungal therapy in the context of predictable species-specific susceptibility profiling of Candida species. Moreover, the CDC (Atlanta, GA) advises species-level identification of Candida species isolated from clinical sample, especially when C. auris infection or colonization has been detected in any health care facility or when a patient had had an overnight hospital stay in a country with C. auris transmission in the previous 6 months (2).
Identification of C. auris by conventional phenotypic methods is challenging, and the results are often inconclusive (22). On commercial chromogenic media such as CHROMagar Candida, BBL CHROMagar Candida, CAN2 chromogenic, Candida ID, and Brilliance Candida agar, the colony color of C. auris ranges from beige and pink to dark purple, making it difficult to differentiate from other yeasts, including C. parapsilosis, whose colonies appear similar in color (23, 24). Commercial identification systems also misidentify or fail to identify this pathogen (23). C. auris isolates are mistakenly identified by commercial systems as C. famata, Candida sake, Rhodotorula glutinis, and S. cerevisiae by API 20C AUX (bioMérieux); as C. haemulonii, C. duobushaemulonii, C. famata, and C. glabrata by the older version of Vitek 2 (bioMérieux); as C. albicans, C. parapsilosis, C. tropicalis, C. famata, C. lusitaniae, C. guilliermondii, and Candida catenulate by MicroScan (Beckman Coulter); and as C. haemulonii, C. parapsilosis, and C. catenulata by BD Phoenix (Becton, Dickinson) (12, 23–28).
Identification by ribosomal DNA (rDNA) sequencing, C. auris-specific PCR/quantitative PCR (qPCR), or MALDI-TOF MS is accurate. However, such advanced molecular methods are not available at the majority of routine clinical laboratories, especially in resource-limited settings (29). This highlights the need for adopting a more practical solution for C. auris identification in those developing countries (30).
Supplemented or modified media have been used previously for the screening of C. auris with limited success (23, 31, 32). A previous study claimed that C. auris could be distinguished from the C. haemulonii species complex on CHROMagar Candida medium supplemented with Pal’s agar (24). While C. auris forms creamy white colonies on this medium when incubated at 37°C, C. haemulonii complex forms light pink colonies with growth inhibition at 42°C. However, variations between different clades of C. auris have been seen with this medium, limiting its applicability.
In another study, Welsh et al. developed an enrichment broth procedure based on the ability of C. auris to survive under high-salt and high-temperature conditions, facilitating its isolation (31). They observed that among all Candida isolates tested, only C. auris grew at a temperature of 40°C and salinity of 10% (wt/vol) in Sabouraud’s or yeast nitrogen base broth in the presence of dulcitol or mannitol (31). In the present study, a 12.5% sodium chloride concentration and a high (42°C) incubation temperature were not enough to inhibit a few non-C. auris yeast species when used individually. However, a combination of 12.5% sodium chloride with 9 mM ferrous sulfate added to YPD and incubation at 42°C inhibited all common non-C. auris yeast isolates. Tolerance of high salt stress and metal stress in C. auris might be due to higher expression of the Hog1 gene than has been seen with other species (33). We did not find selective inhibition of C. glabrata and C. guilliermondii in the presence of high salinity, metal stress, and high temperature. However, on raising the temperature to 42°C, we were able to overcome this hindrance, resulting in a highly specific medium for the selective isolation of C. auris. Similarly to our findings, Welsh et al. reported observable growth of C. glabrata in 10% salt-containing Sabouraud’s broth whereas growth of all other Candida species, including C. haemulonii complex, was inhibited.
We also isolated C. auris on this medium directly from a signal-positive Bactec culture BC broth in one confirmed case of infection. This could enable rapid identification from the blood culture broth, but future studies with a large number of samples are needed. This selective medium may cost ∼22 to 25 Indian rupees (INR) (approximately 0.5 U.S. dollars) per plate, which is less expensive than any of the other chromogenic or supplemented media described earlier. Simple inexpensive chemicals, an easy preparation method, and direct inoculation (streaking) of isolated culture would make this medium user-friendly.
In the present study, all of the C. auris isolates except the Japanese isolate (C. auris JCM 15448) and the South Korean isolate (C. auris 12372) showed growth on SAM. The differences in the characteristics of the strains in terms of origin or clade might represent the possible cause of this growth inhibition. All the Indian isolates of C. auris and reference isolates of clade III and clade IV grew well on this medium. To assess the reasons for the inability of the two clade II C. auris isolates to grow on the standardized medium, we employed growth kinetics, temperature tolerance (42°C), and metal stress (9 mM FeSO4) assays. C. auris clade II isolates showed significantly higher doubling times at 42°C than the other 3 clades, which could possibly be one of the reasons for its absence of growth on this selective medium when additional stresses were added. Those two clade II isolates were able to grow at a maximum level of salt tolerance of 12%. Hence, in the regions where clade II is circulating more extensively, we suggest reducing the salt concentration and prolonging the period of incubation at 42°C or lower. Though, in view of clade-wise variations in the expression levels of stress response pathway genes, the lower level of expression of the Hog1 gene in clade II isolates may be one of the reasons for the lack of growth described above, the hypothesis needs further confirmation. Nonetheless, the applicability of this medium in the global scenario needs to be validated using a greater number of C. auris isolates from different geographic locations. Another minor limitation of this medium is its short shelf life (2 to 4 days, at 4°C) due to gradual oxidation and precipitation of iron, necessitating fresh preparation before use.
Conclusion.
We successfully developed a selective medium, Selective Auris Medium (SAM), for the rapid detection and reliable identification of C. auris. This could be especially useful in diagnostic settings where costly molecular tests are not available. Due to its high sensitivity and specificity, this medium can be applied for the routine screening of suspected C. auris isolates. It could be particularly useful in regions with a preponderance of clades I, III, and IV of C. auris. For those regions with a predominance of clade II, the same medium can be used with a lower salt concentration and longer period of incubation at 42°C or lower. A study on the utility of this medium for direct isolation of C. auris from a signal-positive commercial isolation system is warranted.
Supplementary Material
ACKNOWLEDGMENTS
We thank Neeraj Chauhan, Rutgers, NJ, USA, for providing clade II, III, and IV reference C. auris strains. We are also thankful to the mycology staff members of the AMR project for their cooperation and assistance in collection of routine yeast isolates and of signal-positive automated blood culture vials.
We declare no conflict of interest.
This work was supported by the Indian Council of Medical Research (grant AMR/160/2018-ECD-II).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Centers for Disease Control and Prevention (CDC). 2019. Global emergence of invasive infections caused by the multidrug-resistant yeast Candida auris. https://www.cdc.gov/fungal/diseases/candidiasis/candida-auris-alert.html. Accessed 20 December 2019.
- 2.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CDC. Tracking Candida auris. 2019. https://www.cdc.gov/fungal/candida-auris/tracking-c-auris.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Ffungal%2Fdiseases%2Fcandidiasis%2Ftracking-c-auris.html#world. Accessed 7 December 2019.
- 4.Chow NA, De Groot T, Badali H, Abastabar M, Chiller TM, Meis JF. 2019. Potential fifth clade of Candida auris, Iran, 2018. Emerg Infect Dis 25:1780–1781. doi: 10.3201/eid2509.190686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abastabar M, Haghani I, Ahangarkani F, Rezai MS, Taghizadeh Armaki M, Roodgari S, Kiakojuri K, Al-Hatmi AMS, Meis JF, Badali H. 2019. Candida auris otomycosis in Iran and review of recent literature. Mycoses 62:101–105. doi: 10.1111/myc.12886. [DOI] [PubMed] [Google Scholar]
- 6.Chowdhary A, Sharma C, Meis JF. 2017. Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog 13:e1006290. doi: 10.1371/journal.ppat.1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lone SA, Ahmad A. 2019. Candida auris-the growing menace to global health. Mycoses 62:620–637. doi: 10.1111/myc.12904. [DOI] [PubMed] [Google Scholar]
- 8.Sherry L, Ramage G, Kean R, Borman A, Johnson EM, Richardson MD, Rautemaa-Richardson R. 2017. Biofilm-forming capability of highly virulent, multidrug-resistant Candida auris. Emerg Infect Dis 23:328–331. doi: 10.3201/eid2302.161320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biswal M, Rudramurthy SM, Jain N, Shamanth AS, Sharma D, Jain K, Yaddanapudi LN, Chakrabarti A. 2017. Controlling a possible outbreak of Candida auris infection: lessons learnt from multiple interventions. J Hosp Infect 97:363–370. doi: 10.1016/j.jhin.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Singh R, Kaur M, Chakrabarti A, Shankarnarayan SA, Rudramurthy SM. 2019. Biofilm formation by Candida auris isolated from colonising sites and candidemia cases. Mycoses 62:706–709. doi: 10.1111/myc.12947. [DOI] [PubMed] [Google Scholar]
- 11.Osei Sekyere J. 2018. Candida auris: a systematic review and meta-analysis of current updates on an emerging multidrug-resistant pathogen. Microbiologyopen 7:e00578. doi: 10.1002/mbo3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeffery-Smith A, Taori SK, Schelenz S, Jeffery K, Johnson EM, Borman A, Candida auris Incident Management Team, Manuel R, Brown CS. 2018. Candida auris: a review of the literature. Clin Microbiol Rev 31:e00029-17. doi: 10.1128/CMR.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rudramurthy SM, Chakrabarti A, Paul RA, Sood P, Kaur H, Capoor MR, Kindo AJ, Marak RSK, Arora A, Sardana R, Das S, Chhina D, Patel A, Xess I, Tarai B, Singh P, Ghosh A. 2017. Candida auris candidaemia in Indian ICUs: analysis of risk factors. J Antimicrob Chemother 72:1794–1801. doi: 10.1093/jac/dkx034. [DOI] [PubMed] [Google Scholar]
- 14.Cortegiani A, Misseri G, Giarratano A, Bassetti M, Eyre D. 2019. The global challenge of Candida auris in the intensive care unit. Crit Care 23:150. doi: 10.1186/s13054-019-2449-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lockhart SR, Jackson BR, Vallabhaneni S, Ostrosky-Zeichner L, Pappas PG, Chiller T. 2017. Thinking beyond the common Candida species: need for species-level identification of Candida due to the emergence of multidrug-resistant Candida auris. J Clin Microbiol 55:3324–3327. doi: 10.1128/JCM.01355-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma C, Kumar N, Pandey R, Meis JF, Chowdhary A. 2016. Whole genome sequencing of emerging multidrug resistant Candida auris isolates in India demonstrates low genetic variation. New Microbes New Infect 13:77–82. doi: 10.1016/j.nmni.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lockhart SR, Berkow EL, Chow N, Welsh RM. 2017. Candida auris for the clinical microbiology laboratory: not your grandfather’s Candida species. Clin Microbiol Newsl 39:99–103. doi: 10.1016/j.clinmicnews.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh AK, Paul S, Sood P, Rudramurthy SM, Rajbanshi A, Jillwin TJ, Chakrabarti A. 2015. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for the rapid identification of yeasts causing bloodstream infections. Clin Microbiol Infect 21:372–378. doi: 10.1016/j.cmi.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute. 2019. M60 reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard, 2nd ed. https://clsi.org/standards/products/microbiology/documents/m60/. [PubMed]
- 20.CDC. 2019. Candida auris: antifungal susceptibility testing and interpretation. https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html.
- 21.Chakrabarti A, Rudramurthy SM, Kale P, Hariprasath P, Dhaliwal M, Singhi S, Rao KLN. 2014. Epidemiological study of a large cluster of fungaemia cases due to Kodamaea ohmeri in an Indian tertiary care centre. Clin Microbiol Infect 20:O83–O89. doi: 10.1111/1469-0691.12337. [DOI] [PubMed] [Google Scholar]
- 22.Caceres DH, Forsberg K, Welsh RM, Sexton DJ, Lockhart SR, Jackson BR, Chiller T. 2019. Candida auris: a review of recommendations for detection and control in healthcare settings. J Fungi (Basel) 5:111. doi: 10.3390/jof5040111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizusawa M, Miller H, Green R, Lee R, Durante M, Perkins R, Hewitt C, Simner PJ, Carroll KC, Hayden RT, Zhang SX. 2017. Can multidrug-resistant Candida auris be reliably identified in clinical microbiology laboratories? J Clin Microbiol 55:638–640. doi: 10.1128/JCM.02202-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar A, Sachu A, Mohan K, Vinod V, Dinesh K, Karim S. 2017. Diferenciación sencilla y de bajo coste de Candida auris del complejo Candida haemulonii con el empleo del medio CHROMagar Candida enriquecido con medio de Pal. Rev Iberoam Micol 34:109–111. doi: 10.1016/j.riam.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Spivak ES, Hanson KE. 2017. Candida auris: an emerging fungal pathogen. J Clin Microbiol 56:e01588-17. doi: 10.1128/JCM.01588-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pekard-Amenitsch S, Schriebl A, Posawetz W, Willinger B, Kölli B, Buzina W. 2018. Isolation of Candida auris from ear of otherwise healthy patient. Emerg Infect Dis 24:1596–1597. doi: 10.3201/eid2408.180495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bidaud AL, Chowdhary A, Dannaoui E. 2018. Candida auris: an emerging drug resistant yeast – a mini-review. J Mycol Med 28:568–573. doi: 10.1016/j.mycmed.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 28.Girard V, Mailler S, Chetry M, Vidal C, Durand G, van Belkum A, Colombo AL, Hagen F, Meis JF, Chowdhary A. 2016. Identification and typing of the emerging pathogen Candida auris by matrix-assisted laser desorption ionisation time of flight mass spectrometry. Mycoses 59:535–538. doi: 10.1111/myc.12519. [DOI] [PubMed] [Google Scholar]
- 29.Chindamporn A, Chakrabarti A, Li R, Sun P-L, Tan B-H, Chua M, Wahyuningsih R, Patel A, Liu Z, Chen Y-C, Chayakulkeeree M. 2018. Survey of laboratory practices for diagnosis of fungal infection in seven Asian countries: an Asia Fungal Working Group (AFWG) initiative. Med Mycol 56:416–425. doi: 10.1093/mmy/myx066. [DOI] [PubMed] [Google Scholar]
- 30.Durante AJ, Maloney MH, Leung VH, Razeq JH, Banach DB. 2018. Challenges in identifying Candida auris in hospital clinical laboratories: a need for hospital and public health laboratory collaboration in rapid identification of an emerging pathogen. Infect Control Hosp Epidemiol 39:1015–1016. doi: 10.1017/ice.2018.133. [DOI] [PubMed] [Google Scholar]
- 31.Welsh RM, Bentz ML, Shams A, Houston H, Lyons A, Rose LJ, Litvintseva AP. 2017. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J Clin Microbiol 55:2996–3005. doi: 10.1128/JCM.00921-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.S2 Media. 2018. 5067: salt Sabouraud dulcitol broth (SSDB) package insert. S2 Media, Spokane, WA. https://www.inciensa.sa.cr/inciensa/centros_referencia/cursos_cnrb/taller%20regional/diagnostico%20y%20psa%20de%20candida/lab%20protocols/Medio%20selectivo%20C.%20auris.pdf. [Google Scholar]
- 33.Day AM, McNiff MM, da Silva Dantas A, Gow NAR, Quinn J. 2018. Hog1 regulates stress tolerance and virulence in the emerging fungal pathogen Candida auris. mSphere 3:e00506-18. doi: 10.1128/mSphere.00506-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.