Rapid point-of-care tests (POCTs) for detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-specific antibodies vary in performance. A critical need exists to perform head-to-head comparisons of these assays.
KEYWORDS: SARS-CoV-2 serology, point-of-care test, performance, cross-reactivity
ABSTRACT
Rapid point-of-care tests (POCTs) for detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-specific antibodies vary in performance. A critical need exists to perform head-to-head comparisons of these assays. The performances of 15 different lateral flow POCTs for the detection of SARS-CoV-2-specific antibodies were compared on a well-characterized set of 100 samples. Of these, 40 samples from known SARS-CoV-2-infected, convalescent individuals (collected an average of 45 days after symptom onset) were used to assess sensitivity. Sixty samples from the prepandemic era (negative control) that were known to represent infections with other respiratory viruses (rhinoviruses A, B, and C and/or coronavirus 229E, HKU1, and NL63 OC43) were used to assess specificity. The timing of seroconversion was assessed using five lateral flow assays (LFAs) and a panel of 272 longitudinal samples from 47 patients for whom the time since symptom onset was known. Among the assays that were evaluated, the sensitivity and specificity for any reactive band ranged from 55% to 97% and from 78% to 100%, respectively. Assessing the performance of the IgM and the IgG bands alone, sensitivity and specificity ranged from 0% to 88% and 80% to 100% for IgM and from 25% to 95% and 90% to 100% for IgG, respectively. Longitudinal testing revealed that the median times after symptom onset to a positive result were 7 days (interquartile range [IQR], 5.4 to 9.8) for IgM and 8.2 days (IQR, 6.3 to 11.3) for IgG. The testing performances differed widely among LFAs, with greatest amount of variation related to the sensitivity of the assays. The IgM band was the band most likely to misclassify prepandemic samples. The appearances of IgM and IgG bands occurred almost simultaneously.
INTRODUCTION
The respiratory illness coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection (1). The COVID-19 pandemic has challenged the diagnostic testing capacity of the global health care industry. Though the initial burden of disease was most pronounced in high-income countries, the pandemic has since spread to middle- and low-income countries that lack substantial laboratory infrastructure. Despite major efforts to contain and slow the viral spread, the limited testing capability of hospitals, public health laboratories, and government agencies remains a major challenge. Accurate serological tests for SARS-CoV-2 infection are used to estimate the numbers of individuals who have been infected and have developed a humoral immune response (seroconverted). Understanding seroprevalence is important to determine the spread of the disease and to identify populations with a high burden of infection (2). Furthermore, if previous infection provides immunity to the disease, these assays could be used to identify those who would be vulnerable to or protected from infection.
Broadly, there are two types of assay formats used to detect antibodies against SARS-CoV-2 infection: enzyme-linked immunosorbent assays (ELISAs) and serologic lateral flow assays (LFA). ELISAs, with or without a chemiluminescent signal, offer high-throughput testing but require substantial laboratory infrastructure and trained personnel for operation (3). LFAs that detect antibodies against SARS-CoV-2 are easy to use, rapid, and portable and often qualify as point-of-care tests (POCTs) that can be used outside a centralized laboratory facility (4). POCTs can be used at home or in a doctor’s office and take minutes to complete. Unfortunately, there is a great deal of variation in the performance of these POCT assays for the accurate detection of antibodies to SARS-CoV-2 infection (5). Serologic LFAs can have wide-ranging performances based on the viral antigens used, on how they were elaborated, and on the construction of the cassette.
Comparison of these different LFAs for SARS-CoV-2 antibody detection has been initiated (6). Initial reports are mixed: some report LFAs as being unsuitable for use, while others profess their potential for rapid screening of patients (7–9). Many of these studies were constrained by small sample sizes, failure to evaluate for cross-reactivity, and failure to assess sensitivity of the assays by duration of infection, all of which could influence the findings. The U.S. Food and Drug Administration has developed criteria to allow emergency use authorization (10). These include evaluation of cross-reactivity (specificity of >95% with respect to other circulating coronaviruses), sensitivity approaching 100%, and high (≥90%) positive/negative predictive agreement.
SARS-CoV-2 antibody reactivity or presence varies after onset of symptoms (11). While consensus on the optimal time to perform the POCT for SARS-CoV-2 antibody detection is lacking, the majority of reports suggest that the tests are best undertaken >14 days after symptom onset (12–17). Furthermore, studies on samples from convalescent plasma donors who had a documented positive real-time PCR (RT-PCR) test result demonstrate that some individuals have undetectable antibody responses (18). In terms of specificity, false-positive results may occur for a variety of reasons, particularly due to cross-reactivity to other coronaviruses (229E, HKU1, NL63, and OC43) (9, 12–21).
Despite increasing reports on the performance of individual POCTs to detect SARS-CoV-2 antibodies, the overall performance of all the commercially available POCTs is still unclear. To further expand POCT evaluation, we compared the performances of multiple LFAs for SARS-CoV-2 antibody detection. To this end, we used the same set of samples from known infected and uninfected individuals to perform a head-to-head analysis of 15 LFAs. We further evaluated 7 of these assays to assess the time window between the onset of symptoms and detection of antibodies to SARS-CoV-2 infection. Overall, the goal of our work is highlighting the performance characteristics of a series of 15 LFAs to further expand general understanding of LFA utility and serve as an informative reference for potential deployment efforts.
MATERIALS AND METHODS
Characteristics of individuals studied. (i) Ethics statement.
The parent studies were approved by The Johns Hopkins University School of Medicine Institutional Review Board (IRB00247886, IRB00250798, and IRB00091667). All samples were deidentified prior to testing. The parent studies were conducted according to the ethical standards of the Helsinki Declaration of the World Medical Association. This report includes an analysis of stored samples and data from those studies. No additional samples were collected for the current study.
(ii) Convalescent SARS-CoV-2 samples.
Determinations of the sensitivity of the 15 LFAs were performed on 40 samples from convalescent plasma donors (Table 1; see also Table S1 in the supplemental material) (20). These individuals had to have been RT-PCR positive for SARS-CoV-2 and asymptomatic for at least 28 days. The time interval between date of symptom onset and the sample drawn for this study was 45 days (standard deviation [SD], ±7.5 days). All subjects were human immunodeficiency virus (HIV) and hepatitis C virus (HCV) negative (Table S1).
TABLE 1.
Description of assays evaluateda
| Lateral flow assay |
Test format | Antigen(s) | No. of convalescent samples |
No. of prepandemic samples |
Manufacturer(s) | Regulation |
|---|---|---|---|---|---|---|
| AllTest | Separate IgM, IgG, and control bands | N, S | 40 | 60 | Hangzhou AllTest Biotech Co., Ltd. | CE |
| AYTU | Separate IgM, IgG, and control bands | N, S | 40 | 60 | AYTU Biosciences | CE |
| Clarity | Separate IgM, IgG, and control bands | N, S | 40 | 60 | Alfa Scientific Designs Inc. | IVD |
| CoronaChek | Separate IgM, IgG, and control bands | RBD | 40 | 60 | Hangzhou Biotest Biotech Co., Ltd. | EUA, CE-IVD |
| Covisure | Separate IgM, IgG, and control bands | ? | 38 | 59 | W.H.P.M., Inc. | IVD |
| DNA Link | Separate IgM, IgG, and control bands | ? | 40 | 60 | ||
| Nirmidas | Separate IgM, IgG, and control bands | S | 40 | 60 | Nirmidas Biotech, Inc., and Lows Health | |
| Premier Biotech | Separate IgM, IgG, and control bands | N, S | 40 | 60 | Hangzhou Biotest Biotech Co., Ltd. | CE |
| Ready Result | Separate IgM, IgG, and control bands | ? | 40 | 60 | CE | |
| SafeCare | Separate IgM, IgG, and control bands | ? | 40 | 60 | Safecare Biotech (Hangszhou) Co., Ltd. | IVD |
| Sensing Self | Separate IgM, IgG, and control bands | N, S | 40 | 60 | Sensing Self, PTE. Ltd. | CE |
| Smart Screen | Separate IgM, IgG, and control bands | ? | 40 | 60 | Intelligent Endoscopy | |
| TBG | Separate IgM, IgG, and control bands | ? | 40 | 60 | TBG Biotechnology Corp. | CE-IVD |
| Wondfo | Combined IgM/IgG bands; 1 control band | ? | 40 | 60 | Wondfo Biotechnology | CE-IVD |
| Zeus | Separate IgM, IgG, and control bands | N, S | 40 | 60 | Zeus Scientific, Inc. |
Abbreviations: S, spike protein; N, nucleocapsid; RBD, receptor binding domain; ?, information not provided by manufacturer; CE, certification mark; IVD, in vitro diagnostic; EUA, emergency use authorization.
(iii) Prepandemic challenge samples.
Specificity of LFAs was assessed with 60 samples from prepandemic time points of individuals known to be uninfected by SARS-CoV-2 (Table 1; see also Table S1). These samples came from a study of patients presenting to the Johns Hopkins Hospital Emergency Department with symptoms of an acute respiratory tract infection between January 2016 and June of 2019 as part of the Johns Hopkins Center for Influenza Research and Surveillance study (22). At the time of illness, nasopharyngeal swabs and sera were obtained at the same time. Nasopharyngeal swabs were tested for influenza A/B viruses utilizing the Cepheid GeneXpert Xpress Flu A/B/RSV assay (Cepheid, Sunnyvale, CA) and were subsequently tested for respiratory viral and bacterial coinfections as well as noninfluenza respiratory viruses and bacterial pathogens utilizing Genmark ePlex RP RUO cartridges (Genmark, Carlsbad, CA). The sera from these time points were tested with the VirScan assay, as previously described (23, 24), to identify samples with IgG reactivity to other coronaviruses. For the analysis performed in this study, only data related to coronaviruses 229E, HKU1, NL63, and OC43 were analyzed. Any sample that was reactive to any peptide for these viruses was considered to have antibodies present against these viruses (Table 1; see also Table S1 and Fig. S1 in the supplemental material).
(iv) Longitudinal study samples.
To determine the sensitivity of antibody testing by duration of infection, plasma specimens obtained from individuals with known date of symptom onset who had serial specimens were tested. Samples (n = 272) came from 47 hospitalized SARS-CoV-2 RT-PCR-confirmed patients and were used to determine the sensitivity by duration of infection for a subset of SARS-CoV-2 point-of-care antibody test kits evaluated. Patient characteristics for the longitudinal study are included in Table S2.
Serology testing for antibodies to SARS-CoV-2 infection.
A list of the 15 LFAs evaluated along with information regarding antibody detection, target antigen, and manufacturer is provided in Table 1. All LFAs were performed according to the manufacturers’ protocols. Any detectable band (IgM and/or IgG) was considered a positive result. All LFAs, except Wondfo, had separate bands for IgM and IgG detection. Results were considered invalid when the control band was not visible. Samples were also tested using the Euroimmun Anti-SARS-CoV-2 IgG ELISA (Mountain Lakes, NJ) and the Epitope Diagnostic IgM ELISA (San Diego, CA) per the manufacturers’ protocols. The ELISAs were used for semiquantitative assessment of IgM and IgG levels of the samples evaluated with the LFAs.
Analysis.
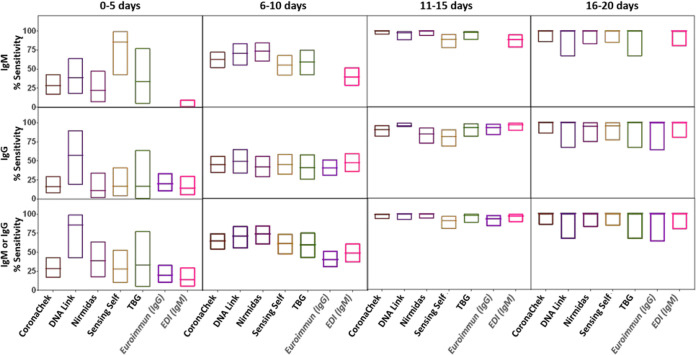
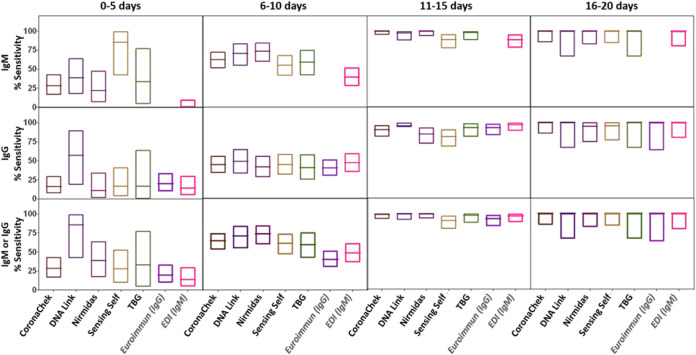
The McNemar test was used to calculate test performance difference (two-tailed P values) between the lateral flow assays with the RT-PCR-confirmed results (Fig. 1; see also Data Set S1 and Tables S3 to S5). Each sensitivity and specificity value determined includes a 95% confidence interval (CI), which was determined using a 95% binomial exact CI. Sensitivity of the assays was calculated on the basis of samples from convalescent individuals (n = 40) and from the longitudinally followed individuals. For the longitudinal samples, sensitivity was calculated at four different time intervals, 0 to 5, 6 to 10, 11 to 15, and 16 to 20 days after symptom onset. Sensitivity was calculated for IgM and IgG separately or for IgM or IgG individually as indicated in Table 1.
FIG 1.
Analytical sensitivity and specificity toward IgM and IgG for the evaluated SARS-CoV-2 antibody-based assays. The boxes represent the lower and upper 95% confidence intervals (95% binomial exact CI), and the lines inside the boxes indicate the values determined for each assay.
Specificity was assessed for the prepandemic sample set (n = 60). The impact of seroreactivity to other coronaviruses was determined. Specificity was calculated for IgM and IgG separately or for IgM or IgG individually as indicated in Table 1. Assay agreement was calculated with the samples from both the convalescent-phase sample group and the prepandemic-era sample group. The Kappa statistic was determined and interpreted according to the method described previously by Landis and Koch (25) to assess agreement between the different lateral flow assays and ELISAs. All analysis was performed using GraphPad Prism (26).
RESULTS
Sensitivity and specificity of IgM and IgG vary by SARS-CoV-2 antibody-based LFA.
Performance results varied across the different assays (Fig. 1; see also Tables S3 to S5). Considering the detection of IgM or of IgG or of either to be representative of a positive result, the sensitivity of the assays ranged from 55% (95% CI, 38% to 71%) to 98% (95% CI, 87% to 100%) and the specificity from 80% (95% CI, 67% to 89%) to 100% (95% CI, 94% to 100%). Of the assays tested, DNA Link and Clarity exhibited the highest sensitivity (98%; 95% CI, 87% to 100%), while for specificity, CoronaChek, Nirmidas, Premier Biotech, and Sensing Self were the best performers (100%; 95% CI, 94% to 100%). The lowest sensitivity was obtained from Wondfo (55%; 95% CI, 38% to 71%), followed by Zeus (58%; 95% CI, 41% to 73%), while the lowest specificity was obtained from DNA Link (80%; 95% CI, 67% to 89%). In general, the IgM band results showed lower sensitivity among the samples from convalescent individuals (0% to 88%) than the IgG band results (25% to 95%). Only for the Clarity and Smart Screen assay was sensitivity reversed. Overall, the specificity was much lower for the IgM band than the IgG band (P < 0.05).
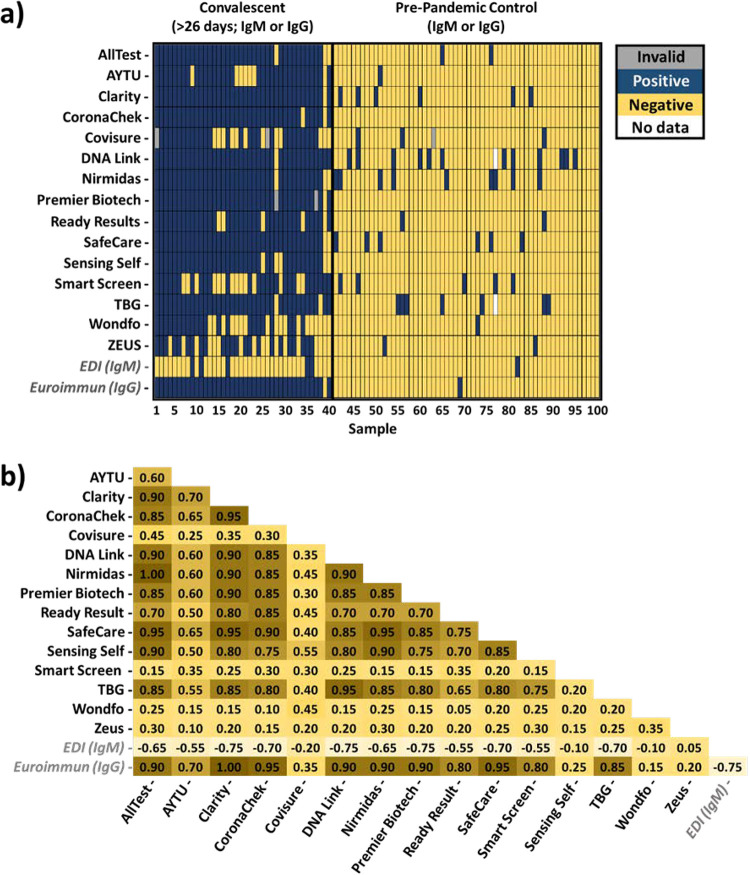
Of the 15 assays evaluated, CoronaChek, Nirmadas, Premier Biotech, and Sensing Self were the only tests without false-positive results in testing the designated negative SARS-CoV-2 samples (Fig. 2a). Fourteen of 60 samples generated false-positive results on two or more LFAs. Among the specimens that generated a false-positive result, most did so in four different tests. Similarly, false-negative results were obtained in testing samples from patients known to be RT-PCR positive. Twenty-eight of 40 samples generated a false-negative result on two or more LFAs. Of note, one specimen generated a false-negative result for all but two of the tests.
FIG 2.
Comparison of 15 evaluated LFA and two ELISA-based assay results obtained by testing the designated negative or positive plasma sample. (a) Results obtained from evaluating prepandemic (negative) and convalescent (positive) plasma. Any detection of IgM or IgG or both is shown as a positive result (blue), whereas lack of detection is shown as a negative result (yellow).Those marked in gray indicate an invalid result, while those marked in white represent missing data for comparison. (b) Agreement (IgM or IgG) between each LFA and ELISAs (in italics). The values shown represent the kappa agreement values, which are interpreted as representing no agreement (<0), slight agreement (0.00 to 0.20), fair agreement (0.021 to 0.40), moderate agreement (0.41 to 0.060), substantial agreement (0.61 to 0.80), almost perfect agreement (0.81 to 1.00), or perfect agreement (1.00).
Cross-reactivity with other viral infections.
To further evaluate the specificity of the different assays, a set of challenge specimens (n = 60) were tested. These specimens comprised prepandemic samples obtained between January 2016 and June 2019 from patients known to be infected with other non-SARS-CoV-2 viruses. False-positive results were obtained with all the LFAs tested (see Fig. S1 in the supplemental material). Cross-reactivity was more pronounced (>55% of samples) with sera from patients infected with different strains of coronaviruses (229E, HKU1, NL63, and OC43); little cross-reactivity was observed with sera from patients known to have influenza A, B, or C and parainfluenza (5% and 2% of samples, respectively).
Convalescent-phase samples (n = 40) from patients confirmed to be positive for SARS-CoV-2 via PCR-based testing were also evaluated for potential viral coinfection (Fig. S2). None of the convalescent individuals were infected with HIV, HBV, or HCV; however, all but three patients were seropositive for at least one of the four coronavirus strains evaluated (229E, HKU1, NL63, and OC43). Four patients were infected with all four of the coronavirus strains evaluated. Therefore, cross-reactivity cannot be ruled out for the convalescent-phase samples.
Agreement between assays.
Agreement among the evaluated LFAs on matched patient samples ranged from slight to perfect (kappa 0.05 to 1.00), according to Landis and Koch interpretation (25).The majority of the assays’ results ranged from moderate to perfect agreement (kappa 0.50 to 1.00), but four assays (Covisure, Smart Screen, Wondfo, and Zeus) had slight to fair agreement (kappa 0.05 to 0.45). The lowest level of agreement was obtained between Wondfo and Ready Result (kappa 0.05), while a higher level of agreement was obtained between Nirmidas and AllTest (“perfect”; kappa 1.00), AllTest and Safe Care (“almost perfect”; kappa 0.95), Safe Care and Clarity (“almost perfect”; kappa 0.95), Safe Care and Nirmidas (“almost perfect”; kappa 0.95), CoronaChek and Clarity (“almost perfect”; kappa 0.95), and TBG and DNA Link (“almost perfect”; kappa 0.95). The IgM results showed lower levels of agreement than the IgG results (Fig. S3a and b).
Two ELISA-based tests, EDI IgM and the Euroimmun IgG, were used for semiquantitative assessment of IgM and IgG levels, respectively, of the samples evaluated with the LFAs (Fig. 2a). The EDI IgM ELISA had 35 false-negative results among the 40 convalescent plasma samples tested. Euroimmun IgG ELISA testing resulted in one false-negative result (sample 39), which was IgG negative by all but three of the LFAs evaluated. Both ELISAs generated one false-positive result in testing the prepandemic samples (negative control), and both of these were obtained for different samples. Evaluating the agreement between the LFAs and ELISAs (Fig. 2b) yielded results that ranged from no agreement to slight agreement (kappa −0.75 to 0.05) for EDI IgM and from slight to perfect agreement (kappa 0.15 to 1.00) for Euroimmun IgG.
Semiquantitative comparison of IgM and IgG ELISA values with the LFA results provided further insight into the performance of each LFA. EDI IgM ELISA normalized optical density (ODn) values between positive and negative LFA results had little variation (Fig. S4), which underscores the poor agreement between EDI IgM and the LFAs. Euroimmun IgG specimen/calibrator (S/C) values had a stronger correlation with LFA results (Fig. S5). However, for a subset of assays, samples with high positive ELISA values had a negative LFA result, suggesting LFA false-negative results.
Sensitivity by duration of infection.
The sensitivity of IgM or IgG reactivity or any reactivity increased with duration of infection. For five LFAs (CoronaChek, DNA Link, Nirmidas, Sensing Self, and TBG) (where we could obtain enough testing kits) and for two ELISAs (Euroimmun IgG and EDI IgM), the sensitivity by duration from symptom onset was evaluated on longitudinal samples from 47 hospitalized patients. Patient characteristics are included in Table S2 in the supplemental material. The sensitivity for both IgM and IgG increased over time up to 20 days after symptom onset (Fig. 3). The median time to seroreactivity was shorter for IgM than for IgG: 7 days (interquartile range [IQR], 5.4 to 9.8 days) for IgM versus 8.2 days (IQR, 6.3 to 11.3 days) for IgG. However, it was not always true that that IgM band appeared before the IgG band. For 13 patients, the IgG bands appeared earlier than or on the same date as the IgM bands. For two of these patients, only IgG was detected regardless of the time of testing. Four others did not obtain a positive result either by LFA or ELISA for the time points evaluated.
FIG 3.
Longitudinal evaluation of analytical performance for four SARS-CoV-2 antibody-based LFAs and two ELISAs. The boxes represent the lower and upper 95% confidence intervals (95% binomial exact CI), and the line inside each box indicates the value determined for each assay at each indicated time range.
DISCUSSION
LFAs for SARS-CoV-2 antibody testing are appealing given their low cost, ease of distribution, and clinical use. However, the performance and reliability of these tests for detection of IgM and IgG against SARS-CoV-2 at different stages of COVID-19 remain unclear, and information about cross-reactivity of these assays toward other viral antibodies is lacking. It is unknown whether this represents a problem with the antigen being used or with the formulation of the assay itself. Using 15 commercially available LFAs, we demonstrated variation in test performance of these assays as well as cross-reactivity with other viral antibodies. Using longitudinal samples from patients with documented cases of COVID-19, the findings indicate optimal sensitivity approximately 3 weeks following onset of symptoms.
The panel of LFAs evaluated had combined levels of sensitivity and specificity for IgM and IgG of between 55% and 98% and between 80% to 100%, respectively. AllTest, Premier Biotech, and Wondfo have been previously evaluated (17, 19, 27–29). Our performance results are similar to previously reported values, except for Wondfo, which had lower sensitivity and specificity (55% [95% CI, 38% to 71%] and 98% [95% CI, 91% to 100%], respectively). It is unclear why the values differed.
Of the tests evaluated, only the CoronaChek, Premier Biotech, and Sensing Self assays did not generate a false-positive response on our panel of 60 samples from individuals known to have been infected with other respiratory infections. Our data demonstrated that samples from patients infected with other coronaviruses (229E, HKU1, NL63, and OC43) are more prone to cross-reactivity than samples from those infected with influenza A, B, or C virus, parainfluenza virus, HIV, rhinovirus, and enterovirus. Cross-reactivity with other viral antibodies has been reported for other SARS-CoV-2-specific IgM and IgG antibody immunoassays (17, 19). Whitman and coworkers performed cross-reactivity controls with 10 POCTs by testing samples from individuals that tested negative for SARS-CoV-2 and/or had other viral and inflammatory illnesses (17). The evaluated POCTs were prone to cross-react against viruses other than SARS-CoV-2, but no consistent pattern was identified.
The immunologic response to SARS-CoV-2 infection begins as early as a couple of days after symptom onset. In our cohort of symptomatic, hospitalized patients diagnosed with COVID-19, seroconversion occurred in 64% of individuals by 14 days, similarly to previous investigations (11, 21, 30–32). IgM levels have been shown to decline after seroconversion and to be almost undetectable by week 7 after the onset of symptoms, while IgG levels persist past week 7 (11, 33). In our study, we also observed an increase in IgM and IgG detection in the first 2 weeks after the onset of symptoms and less seroreactivity to IgM among convalescent plasma donors whose samples were tested approximately a month and a half after diagnosis by RT-PCR. We did observe a sample from a convalescent donor that was IgG negative for all assays evaluated (15 LFAs and Euroimmun IgG ELISA), highlighting that not all infected individuals generate detectable antibody responses.
The limitations to our study included a lack of early infection samples from nonhospitalized patients. Additionally, specificity analysis needs to be performed on hundreds if not thousands of samples to determine factors associated with misclassification and to give better precision of the point estimate. Furthermore, the samples evaluated were from the Baltimore-Washington region of the United States and may not reflect performance of these assays in different parts of the world. Future studies should include samples from different regions of the world where the underlying host genetics and common viral infections differ to determine the robustness of POCT performance. Additionally, studies using testing algorithms that apply different assays in combination and that test different target antigens of the virus should be evaluated, as such methodologies have proven highly effective for testing other infections such as HIV.
The current test for the diagnosis of SARS-CoV-2 infection is RT-PCR. RT-PCR has disadvantages, including cost, lengthy turnaround times, and preanalytical variability. Additionally, the sensitivity of this method declines past the first week after onset of symptoms (31, 34). POCTs could be used in parallel with RT-PCR testing as a supplemental diagnostic tool in patients suspected to have infection who are RT-PCR negative and who are more than 14 days from onset of symptoms. Serologic assays also facilitate population-level monitoring of COVID-19 exposure. Of note, POCTs should be considered supplemental diagnostic tools, not confirmatory tests.
Overall, antibody-based testing shows great promise as an easy and rapid screening method for determining SARS-CoV-2 exposure, if those assays with poor performance are removed from the market. However, comprehensive evaluation of these tests should be performed prior to their clinical implementation. If antibodies to SARS-CoV-2 do provide long-term immunological protection, then POCTs could play a pivotal role in the evaluation of protective immunity. In summary, our report provides insight into the performance of a series of LFAs and into some of the factors capable of influencing their performance (e.g., time of testing, coinfection).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to all of the study participants who donated plasma and to the clinical staff, including Sonali Thapa and Liz Martinez, Mary De’Jarnette, Carlos Aguado, Peggy Iraola, and Jackie Lobien, who collected samples.
The study was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH). Research reported in this publication was supported by the following research awards: from the NIAID, UM1-AI068613, R01AI120938, and R01AI128779; from the National Institute of Biomedical Imaging and Bioengineering, U54EB007958; from the National Heart, Lung, and Blood Institute of the National Institutes of Health, 1K23HL151826-01. The work described here was supported in part by NIAID contract HHSN272201400007C awarded to the Johns Hopkins Center for Influenza Research and Surveillance (JHCEIRS).
E.M.B. is a member of the United States Food and Drug Administration (FDA) Blood Products Advisory Committee.
Any views or opinions that are expressed in this article are ours, based on our own scientific expertise and professional judgment; they do not necessarily represent the views of either the Blood Products Advisory Committee or the formal position of FDA and also do not bind or otherwise obligate or commit either Advisory Committee or the Agency to the views expressed.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gronvall G, Connel N, Kobokovich A, West R, Warmbrod K, Shearer M, Mullen L, Inglesby T. 2020. Developing a national strategy for serology (antibody testing) in the United States. Johns Hopkins Bloomberg School of Public Health, Baltimore, MD. [Google Scholar]
- 3.Clarke W, Sokoll LJ, Rai AJ. 2020. Immunoassays, p 201–214. In Clarke W, Marzinke MA (ed), Contemporary practice in clinical chemistry, 4th ed. Academic Press, New York, NY. [Google Scholar]
- 4.Koczula KM, Gallotta A. 2016. Lateral flow assays. Essays Biochem 60:111–120. doi: 10.1042/EBC20150012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joseph A, Branswell H. 24 April 2020. The results of coronavirus ‘serosurveys’ are starting to be released. Here’s how to kick their tires. Stat. https://www.statnews.com/2020/04/24/the-results-of-coronavirus-serosurveys-are-starting-to-be-released-heres-how-to-kick-their-tires/.
- 6.U.S. Food and Drug Administration. 2020. Independent evaluations of COVID-19 serological tests. FDA. https://open.fda.gov/apis/device/covid19serology/.
- 7.Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, Sun R, Wang Y, Hu B, Chen W, Zhang Y, Wang J, Huang B, Lin Y, Yang J, Cai W, Wang X, Cheng J, Chen Z, Sun K, Pan W, Zhan Z, Chen L, Ye F. 2020. Development and clinical application of a rapid IgM‐IgG combined antibody test for SARS‐CoV‐2 infection diagnosis. J Med Virol 92:1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barna V, Katalin K, Eszter O, Dóra S, Zoltán P, Béla M. 2020. The diagnostic value of rapid anti IgM and IgG detecting tests in the identification of patients with SARS CoV-2 virus infection. Orv Hetil 161:807–812. doi: 10.1556/650.2020.31859. [DOI] [PubMed] [Google Scholar]
- 9.Cassaniti I, Novazzi F, Giardina F, Salinaro F, Sachs M, Perlini S, Bruno R, Mojoli F, Baldanti F, Members of the San Matteo Pavia COVID‐19 Task Force. 2020. Performance of VivaDiag COVID-19 IgM/IgG Rapid Test is inadequate for diagnosis of COVID-19 in acute patients referring to emergency room department. J Med Virol 92:1724–1724. doi: 10.1002/jmv.25800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases. 2020. Interim guidelines for COVID-19 antibody testing. CDC. https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antibody-tests-guidelines.html.
- 11.Long Q-X, Liu B-Z, Deng H-J, Wu G-C, Deng K, Chen Y-K, Liao P, Qiu J-F, Lin Y, Cai X-F, Wang D-Q, Hu Y, Ren J-H, Tang N, Xu Y-Y, Yu L-H, Mo Z, Gong F, Zhang X-L, Tian W-G, Hu L, Zhang X-X, Xiang J-L, Du H-X, Liu H-W, Lang C-H, Luo X-H, Wu S-B, Cui X-P, Zhou Z, Zhu M-M, Wang J, Xue C-J, Li X-F, Wang L, Li Z-J, Wang K, Niu C-C, Yang Q-J, Tang X-J, Zhang Y, Liu X-M, Li J-J, Zhang D-C, Zhang F, Liu P, Yuan J, Li Q, Hu J-L, Chen J, et al. 2020. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 12.Montesinos I, Gruson D, Kabamba B, Dahma H, Van den Wijngaert S, Reza S, Carbone V, Vandenberg O, Gulbis B, Wolff F, Rodriguez-Villalobos H. 2020. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti-SARS-CoV-2 antibodies. J Clin Virol 128:104413. doi: 10.1016/j.jcv.2020.104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aj J, Kuivanen S, Kekäläinen E, Mj A, Loginov R, Vapalahti O, Jarva H, Kurkela S, Lappalainen M. 2020. Performance of six SARS-CoV-2 immunoassays in comparison with microneutralisation. medRxiv 10.1101/2020.05.18.20101618. [DOI] [PMC free article] [PubMed]
- 14.Paradiso AV, De Summa S, Loconsole D, Procacci V, Sallustio A, Centrone F, Silvestris N, Cafagna V, De Palma G, Tufaro A, Garrisi V, Chironna M. 2020. Clinical meanings of rapid serological assay in patients tested for SARS-Co2 RT-PCR. medRxiv 10.1101/2020.04.03.20052183. [DOI] [PMC free article] [PubMed]
- 15.Pan Y, Li X, Yang G, Fan J, Tang Y, Zhao J, Long X, Guo S, Zhao Z, Liu Y, Hu H, Xue H, Li Y. 2020. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J Infect 130:e28–e32. doi: 10.1016/j.jinf.2020.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bond K, Nicholson S, Hoang T, Catton M, Howden B, Williamson D. 2020. Post-market validation of three serological assays for COVID-19. https://www.health.gov.au/sites/default/files/documents/2020/08/post-market-validation-of-three-serological-assays-for-covid-19-final-report_0.pdf.
- 17.Whitman JD, Hiatt J, Mowery CT, Shy BR, Yu R, Yamamoto TN, Rathore U, Goldgof GM, Whitty C, Woo JM, Gallman AE, Miller TE, Levine AG, Nguyen DN, Bapat SP, Balcerek J, Bylsma S, Lyons AM, Li S, Wong AW, Gillis-Buck EM, Steinhart ZB, Lee Y, Apathy R, Lipke MJ, Smith JA, Zheng T, Boothby IC, Isaza E, Chan J, Acenas DD, Lee J, Macrae TA, Kyaw TS, Wu D, Ng DL, Gu W, York VA, Eskandarian HA, Callaway PC, Warrier L, Moreno ME, Levan J, Torres L, Farrington L, Loudermilk R, Koshal K, Zorn KC, Garcia-Beltran WF, Yang D, Astudillo MG, Bernstein BE, Gelfand JA, Ryan ET, Charles RC, Iafrate AJ, Lennerz JK, Miller S, et al. 2020. Test performance evaluation of SARS-CoV-2 serological assays. medRxiv 10.1101/2020.04.25.20074856. [DOI] [PMC free article] [PubMed]
- 18.Klein S, Pekosz A, Park H-S, Ursin R, Shapiro J, Benner S, Littlefield K, Kumar S, Naik HM, Betenbaugh M, Shrestha R, Wu A, Hughes R, Burgess I, Caturegli P, Laeyendecker O, Quinn T, Sullivan D, Shoham S, Redd A, Bloch E, Casadevall A, Tobian A. 2020. Sex, age, and hospitalization drive antibody responses in a COVID-19 convalescent plasma donor population. medRxiv 10.1101/2020.06.26.20139063. [DOI] [PMC free article] [PubMed]
- 19.Lassaunière R, Frische A, Harboe ZB, Nielsen AC, Fomsgaard A, Krogfelt KA, Jørgensen CS. 2020. Evaluation of nine commercial SARS-CoV-2 immunoassays. medRxiv 10.1101/2020.04.09.20056325. [DOI]
- 20.Bloch EM, Shoham S, Casadevall A, Sachais BS, Shaz B, Winters JL, van Buskirk C, Grossman BJ, Joyner M, Henderson JP, Pekosz A, Lau B, Wesolowski A, Katz L, Shan H, Auwaerter PG, Thomas D, Sullivan DJ, Paneth N, Gehrie E, Spitalnik S, Hod E, Pollack L, Nicholson WT, Pirofski L, Bailey JA, Tobian AAR. 2020. Deployment of convalescent plasma for the prevention and treatment of COVID-19. J Clin Invest 130:2757–2765. doi: 10.1172/JCI138745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lou B, Li T, Zheng S, Su Y, Li Z, Liu W, Yu F, Ge S, Zou Q, Yuan Q, Lin S, Hong C, Yao X, Zhang X, Wu D, Zhou G, Hou W, Li T, Zhang Y, Zhang S, Fan J, Zhang J, Xia N, Chen Y. 2020. Serology characteristics of SARS-CoV-2 infection since the exposure and post symptoms onset. medRxiv 10.1101/2020.03.23.20041707. [DOI] [PMC free article] [PubMed]
- 22.Hardick J, Sadiq S, Perelstein E, Peterson S, Rothman R, Gaydos CA. 2014. A case-control study evaluating RT-PCR/ESI-MS technology compared to direct fluorescent antibody and xTAG RVP PCR. Diagn Microbiol Infect Dis 79:187–189. doi: 10.1016/j.diagmicrobio.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu GJ, Kula T, Xu Q, Li MZ, Vernon SD, Ndung'u T, Ruxrungtham K, Sanchez J, Brander C, Chung RT, O'Connor KC, Walker B, Larman HB, Elledge SJ. 2015. Comprehensive serological profiling of human populations using a synthetic human virome. Science 348:aaa0698. doi: 10.1126/science.aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohan D, Wansley DL, Sie BM, Noon MS, Baer AN, Laserson U, Larman HB. 2018. PhIP-Seq characterization of serum antibodies using oligonucleotide-encoded peptidomes. Nat Protoc 13:1958–1978. doi: 10.1038/s41596-018-0025-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landis JR, Koch GG. 1977. The measurement of observer agreement for categorical data. Biometrics 33:159–174. doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
- 26.GraphPad Software Inc. 2020. GraphPad Prism 8.0. GraphPad Software Inc, La Jolla, Ca. [Google Scholar]
- 27.Garcia FP, Perez Tanoira R, Romanyk Cabrera JP, Arroyo Serrano T, Gomez Herruz P, Cuadros Gonzalez J. 2020. Rapid diagnosis of SARS-CoV-2 infection by detecting IgG and IgM antibodies with an immunochromatographic device: a prospective single-center study. medRxiv 10.1101/2020.04.11.20062158. [DOI]
- 28.Bendavid E, Mulaney B, Sood N, Shah S, Ling E, Bromley-Dulfano R, Lai C, Weissberg Z, Saavedra R, Tedrow J, Tversky D, Bogan A, Kupiec T, Eichner D, Gupta R, Ioannidis J, Bhattacharya J. 2020. COVID-19 antibody seroprevalence in Santa Clara County, California. medRxiv 10.1101/2020.04.14.20062463. [DOI] [PMC free article] [PubMed]
- 29.Kontou PI, Braliou GG, Dimou NL, Nikolopoulos G, Bagos PG. 2020. Antibody tests in detecting SARS-CoV-2 infection: a meta-analysis. Diagnostics 10:319. doi: 10.3390/diagnostics10050319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sethuraman N, Jeremiah SS, Ryo A. 2020. Interpreting diagnostic tests for SARS-CoV-2. JAMA 2019:2019–2021. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 31.Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, Wang X, Yuan J, Li T, Li J, Qian S, Hong C, Wang F, Liu Y, Wang Z, He Q, Li Z, He B, Zhang T, Fu Y, Ge S, Liu L, Zhang J, Xia N, Zhang Z. 2020. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis 1:ciaa344. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiang F, Wang X, He X, Peng Z, Yang B, Zhang J, Zhou Q, Ye H, Ma Y, Li H, Wei X, Cai P, Ma W-L. 2020. Antibody detection and dynamic characteristics in patients with COVID-19. Clin Infect Dis 1:1930–1934. doi: 10.1093/cid/ciaa461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao AT, Gao C, Zhang S. 2020. Profile of specific antibodies to SARS-CoV-2: the first report. J Infect 81:147–178. doi: 10.1016/j.jinf.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J. 2020. Variation in false-negative rate of reverse transcriptase polymerase chain reaction–based SARS-CoV-2 tests by time since exposure. Ann Intern Med 173:262–267. doi: 10.7326/M20-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.