The purpose of this study was to detect coronavirus disease 2019 (COVID-19) cases with persistent positive reverse transcription-PCR (RT-PCR) results for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), for which viable virus can be inferred due to the presence of subgenomic (SG) viral RNA, which is expressed only in replicating viruses. RNA remnants purified from diagnostic nasopharyngeal specimens were used as the templates for RT-PCR-specific detection of SG E gene RNA.
KEYWORDS: COVID-19, SARS-CoV-2, subgenomic RNA, persistence
ABSTRACT
The purpose of this study was to detect coronavirus disease 2019 (COVID-19) cases with persistent positive reverse transcription-PCR (RT-PCR) results for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), for which viable virus can be inferred due to the presence of subgenomic (SG) viral RNA, which is expressed only in replicating viruses. RNA remnants purified from diagnostic nasopharyngeal specimens were used as the templates for RT-PCR-specific detection of SG E gene RNA. As controls, we also detected viral genomic RNA for the E gene and/or a human housekeeping gene (RNase P). We assessed the samples of 60 RT-PCR-positive cases with prolonged viral SARS-CoV-2 shedding (24 to 101 days) since the first diagnostic RT-PCR. SG viral RNA was detected in 12/60 (20%) of the persistent cases, 28 to 79 days after the onset of symptoms. The age range of the cases with prolonged viral shedding and the presence of SG RNA was quite wide (40 to 100 years), and the cases were equally distributed between males (42%) and females (58%). No case was HIV positive, although seven were immunosuppressed. According to the severities of the COVID-19 episodes, they were mild (40%), intermediate (20%), and severe (40%). In a percentage of persistent SARS-CoV-2 PCR-positive cases, the presence of actively replicating virus may be inferred, far beyond diagnosis. We should not assume a universal lack of infectiousness for COVID-19 cases with prolonged viral shedding.
INTRODUCTION
The reference standard for coronavirus disease 2019 (COVID-19) diagnosis is based on reverse transcription-PCR (RT-PCR) detection of genomic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA. From the experience accumulated with the systematic use of these RT-PCRs, some authors have reported cases with prolonged viral shedding.
The time between the first positive RT-PCR result for SARS-CoV-2, close to the onset of symptoms, and the first negative RT-PCR result for persistent COVID-19 cases ranges from 3 weeks (1–3) to 45 to 48 days for mild and severe cases, respectively (4). Longer periods have also been communicated (53.5 days on average), with one reported case of 83 days (5).
These findings have raised uncertainty regarding the meaning of prolonged persistent positive RT-PCR results. It is essential to determine whether persistently RT-PCR SARS-CoV-2-positive results are due to remnant genomic RNA or because of the existence of viable, infectious viruses.
Several efforts have been made to clarify this issue, mainly supported based on the possibility of isolating in cell cultures the virus from persistent positive RT-PCR cases and/or determining its cytopathic effect. To date, these are considered indicators of viral viability and, therefore, of contagiousness. The few data available (3, 6–8) indicate that no viruses are isolated from cultures, nor is a cytopathic effect observed from specimens from persistent cases.
An alternative strategy to assess viral viability may be the detection of SARS-CoV-2 subgenomic (SG) mRNAs. These intermediates, required by the virus to express its assembly proteins, are transcribed only in actively infected cells; are not packed into virions; and therefore are produced only when the virus is actively replicating (9, 10).
The purpose of this study was to detect cases with persistent positive RT-PCR results for SARS-CoV-2 for which active viral replication can be inferred due to the presence of SG viral RNA.
MATERIALS AND METHODS
Detection of genomic and subgenomic RNA.
RNA remnants purified (KingFisher; Thermo Fisher Scientific, Waltham, MA, USA) from diagnostic nasopharyngeal specimens (300 μl of UTM (Universal Transport Medium) swabs; Copan, bioMérieux, Marcy-l’Etoile, France) from patients diagnosed (by targeting the open reading frame 1ab [ORF1ab] and N genes using a Thermo Fisher RT-PCR kit) in our institution, a tertiary hospital in Madrid, Spain, were used as the templates.
We used a PCR design described previously (6) for the specific detection of SG E gene RNA (plus strand). We used a leader-specific primer described previously by Wolfel et al. (6), sgLeadSARSCoV2-F (CGATCTCTTGTAGATCTGTTCTC), and the primers and probes targeting sequences downstream of the start codons of the E gene described previously by Corman et al. (11).
The SG RNA RT-PCR assay included 400 nM each primer, 200 nM probe, and one of the three following enzymes (depending on availability): the Superscript III one-step RT-PCR system with Platinum Taq polymerase (Invitrogen, Darmstadt, Germany), one-step RT-PCR polymerase mix (Roche), or the TaqPath COVID-19 CE-IVD RT-PCR kit (Thermo Fisher Scientific). RT-PCR conditions were 15 min at 50°C for reverse transcription followed by 2 min at 95°C (SuperScript), 5 min at 55°C followed by 5 min at 95°C (Roche), or 5 min at 50°C for reverse transcription followed by 20 s at 95°C (TaqPath), followed by 45 cycles of 15 s at 95°C, 30 s at 60°C, and 5 s at 72°C. Reactions were run with a CFX384 real-time PCR detection system (Bio-Rad, Hercules, CA, USA).
As controls, we also detected viral genomic RNA for the E gene (plus strand) and/or a human housekeeping gene (RNase P). For validating the detection of SG RNA, a positive signal must be obtained for both the genomic viral E gene and the human gene. In all reaction mixtures, we included in the same set an SG RNA-positive specimen and an SG RNA-negative clinical specimen, which were reextracted and tested in parallel with the clinical problem specimens.
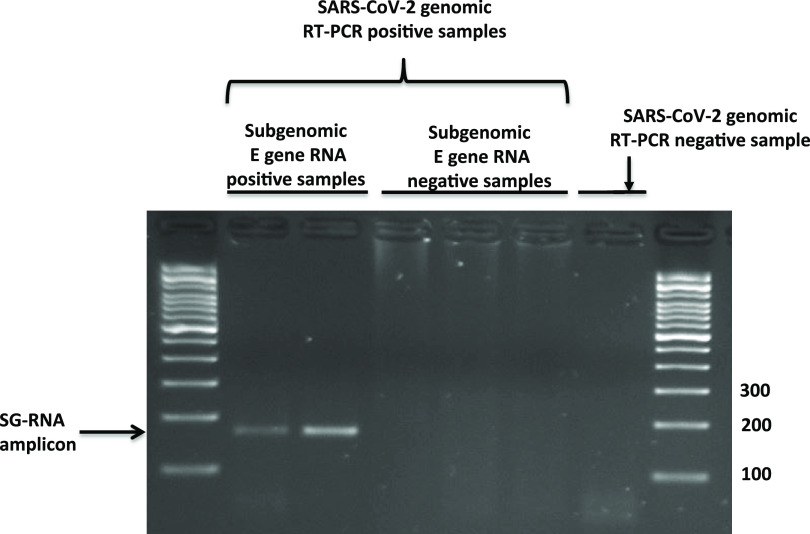
In brief, RT-PCR used to specifically detect SG E gene RNA included a forward primer that anneals with the leader sequence of the E gene and a reverse primer that anneals in the gene sequence. These primers bind to either the genomic viral RNA or the corresponding SG RNA. However, because of the discontinuous transcription of the RNA-dependent RNA polymerase of SARS-CoV-2 when producing SG intermediates (10), the distance between the leader sequence and the inner primer in the SG RNAs is shorter (171 bp) than in the corresponding genomic viral RNAs (26,337 bp; the sgLeadSARSCoV2-F 5′ end anneals at position 44, and the E_Sarbeco_R 5′ end anneals at position 26381). The 26,381-bp product cannot be obtained by PCR. The proper size of the amplicon was checked by agarose gel electrophoresis (Fig. 1).
FIG 1.
Agarose gel electrophoresis indicating the proper amplicon size (171 bp) expected for E gene subgenomic RNA RT-PCR from positive specimens taken close to symptom onset (diagnostic specimens), when active viral replication is expected. A selection of RT-PCR genomic SARS-CoV-2-positive specimens with negative E gene subgenomic RNA RT-PCR results among those taken far beyond diagnosis (persistence specimens) is also shown. Finally, an RT-PCR genomic SARS-CoV-2-negative specimen is included.
The study was approved by the Ethical Research Committee of the Hospital General Universitario Gregorio Marañón (MICRO.HGUGM.2020-017, v3).
RESULTS AND DISCUSSION
We initially analyzed the first nasopharyngeal specimens from a selection of 19 newly diagnosed, at our hospital, RT-PCR-positive cases, for which active viral replication was expected, as was the presence of SG RNA. SG RNA was detected in 15/19 cases. According to the clinical charts of the four cases for which no SG RNA was found, three of them had had previous clinical episodes compatible with COVID-19 (two had SARS-CoV-2-positive serology), and the remaining one had positive serology. This explains the absence of SG RNAs in the four cases and confirms the accuracy of RT-PCR targeting SG RNA to identify cases with true active infection at the beginning of symptom onset.
Next, we assessed the samples of persistent cases: 60 RT-PCR-positive cases with >21 days since the first diagnostic RT-PCR. Forty patients did not require hospitalization; for the 20 patients who were hospitalized, the antiviral scheme approved at our center was dexamethasone at 6 mg/24 h intravenously (i.v.)/orally for 5 to 10 days, and for those with respiratory insufficiency with the need for low-flow oxygen therapy and radiologically confirmed pneumonia and <7 days after the onset of symptoms, remdesivir was given at 200 mg on day 1, followed by 100 mg/24 h i.v. on days 2 and 5.
To the best of our knowledge, the periods from symptom onset for our cases with positive RT-PCR results (24 to 101 days) (Table 1) are the longest reported for COVID-19 (1–5). For most cases (48/60), only genomic E gene viral RNA, and no SG RNA, was detected, consistent with nonreplicating viruses. However, in the remaining 12 persistent cases (20%), SG RNA was detected (28 to 79 days after the onset of symptoms). The size of the amplicons was determined by electrophoresis to confirm the length (171 bp) expected for SG RNA.
TABLE 1.
Detection of genomic and subgenomic viral RNA from diagnostic specimens (patients 1 to 19) and persistent cases (patients 20 to 79)a
| Patient |
CT |
No. of days since diagnosisb |
||
|---|---|---|---|---|
| E gene genomic RNA | E gene SG RNA | RNase P | ||
| 1 | 16.9 | 24.8 | 29.6 | 0 |
| 2 | 24.2 | 30.1 | 23 | 0 |
| 3 | 24.3 | 35 | 19 | 0 |
| 4 | 23.9 | 31.3 | 17.4 | 0 |
| 5 | 25.8 | 31.6 | 15.8 | 0 |
| 6 | 38.2 | — | 16.9 | 0 |
| 7 | 37.6 | — | 20.5 | 0 |
| 8 | 22 | 27.3 | 17.3 | 0 |
| 9 | 21.1 | 25.9 | 20.1 | 0 |
| 10 | 22.1 | 30.1 | 17.8 | 0 |
| 11 | 21.8 | 30.1 | 17.6 | 0 |
| 12 | 20.5 | 28 | 18.9 | 0 |
| 13 | 26.5 | — | 14.3 | 0 |
| 14 | 31.7 | — | 9.3 | 0 |
| 15 | 25.8 | 31.9 | 24.1 | 0 |
| 16 | 23.1 | 30 | 24.4 | 0 |
| 17 | 23.5 | 29.7 | 28.1 | 0 |
| 18 | 25.9 | 32.8 | 26.5 | 0 |
| 19 | 25.9 | 30.8 | 27.2 | 0 |
| 20 | 29.2 | 34 | 17.4 | 37 |
| 21 | 27.7 | 38.4 | 21.3 | 35 |
| 22 | 29.4 | 38.5 | 18.5 | 38 |
| 23 | 26.4 | 33 | 22.9 | 28 |
| 24, sample 1 | 16 | 22 | 25.1 | 60 |
| 24, sample 2 | 15.1 | 21.5 | NA | 66 |
| 24, sample 3 | 17.9 | 22 | 26 | 75 |
| 25 | 27.7 | 33.5 | 22 | 33 |
| 26 | 23.1 | 33.5 | 22.4 | 25 |
| 27 | 24 | 33.4 | NA | 63 |
| 28 | 16.9 | 21.9 | 27.4 | 55 |
| 29 | 27.9 | 40.1 | NA | 79 |
| 30, sample 1 | 25.8 | 33.5 | 21.9 | 29 |
| 30, sample 2 | 33.5 | — | 17 | 34 |
| 31, sample 1 | 18.6 | 26.2 | 26.3 | 44 |
| 31, sample 2 | 38.2 | — | 25.5 | 78 |
| 32 | 32.5 | — | 28.5 | 71 |
| 33 | 29.7 | — | 25.7 | 81 |
| 34 | 32.8 | — | 25.9 | 74 |
| 35 | 31.6 | — | 30.7 | 80 |
| 36 | 36.2 | — | 24.4 | 27 |
| 37 | 37.5 | — | 27 | 47 |
| 38 | 41.7 | — | 21.1 | 51 |
| 39 | 39.4 | — | 21.4 | 24 |
| 40 | 33.4 | — | 20.1 | 54 |
| 41 | 34.3 | — | 20.1 | 24 |
| 42 | 35.2 | — | 17.2 | 42 |
| 43 | 33.6 | — | 20.7 | 39 |
| 44 | 34.8 | — | 20.9 | 52 |
| 45 | 36.1 | — | 20.3 | 44 |
| 46 | 35.5 | — | 29.3 | 35 |
| 47 | 35.4 | — | 17.5 | 51 |
| 48 | 35.6 | — | 22 | 58 |
| 49 | 34.4 | — | 19.4 | 51 |
| 50 | 41.8 | — | 21.5 | 40 |
| 51 | 33.2 | — | 14.9 | 38 |
| 52 | 36.1 | — | 15.8 | 50 |
| 53 | 34.1 | — | 19.8 | 56 |
| 54 | 32.3 | — | 19.3 | 39 |
| 55 | 33.2 | — | 19.3 | 43 |
| 56 | 36.6 | — | 19.3 | 49 |
| 57 | 34.1 | — | 18.4 | 49 |
| 58 | 36.3 | — | 16.5 | 46 |
| 59 | 34 | — | 18.8 | 46 |
| 60 | 33.3 | — | 19.6 | 51 |
| 61 | 34.1 | — | 18.9 | 36 |
| 62 | 36.6 | — | 20.1 | 48 |
| 63 | 32.6 | — | 18.2 | 40 |
| 64 | 34.6 | — | 18.1 | 47 |
| 65 | 27.2 | — | 10.6 | 26 |
| 66 | 24.7 | — | 14.2 | 49 |
| 67 | 32.2 | — | 24.6 | 56 |
| 68 | 30.8 | — | 28.4 | 28 |
| 69 | 32.8 | — | 21.6 | 41 |
| 70 | 36 | — | 18.9 | 30 |
| 71 | 36.6 | — | 18.2 | 40 |
| 72 | 33.4 | — | 17.3 | 36 |
| 73 | 37.6 | — | 17.7 | 22 |
| 74 | 36.4 | — | 16.6 | 37 |
| 75 | 32.3 | — | 24 | 33 |
| 76 | 27.1 | — | 26.4 | 101 |
| 77 | 41.3 | — | 25.1 | 79 |
| 78 | 32.5 | — | 24.9 | 67 |
| 79 | 32.9 | — | 27.3 | 71 |
—, no amplification; NA, not available.
The day of diagnosis was considered the day when the first RT-PCR for SARS-CoV-2 was positive.
Three sequential samples (days 60, 66, and 75) were available for one of the cases. SG RNA was detected in all three samples, with consistent threshold cycle (CT) values (22, 21.5, and 22, respectively), confirming the prolonged presence of actively replicating virus (Table 1). New specimens were available for another two cases, obtained 5 and 34 days after the first specimen in which SG detected; SG RNA was no longer identified. In all cases with detectable SG RNA, CT values for genomic RNA were <30, consistent with the values expected for an active virus.
The age range of subjects with prolonged viral shedding and SG viral RNA was quite wide (40 to 100 years), equally distributed between males (42%) and females (58%). None of the subjects were HIV positive, although seven were immunosuppressed. The severities of the COVID-19 episodes in patients for whom detailed clinical information was available were mild (40%), intermediate (20%), and severe (40%); seven patients were hospitalized, two of whom died (Table 2). It is worth noting that one case with SG RNA at day 25 was asymptomatic.
TABLE 2.
Characteristics of patients with prolonged viral shedding and SG viral RNAa
| Patient | Gender | Age (yrs) | Preexisting condition(s) | Day of symptom onset (no. of days before 1st positive PCR) | Duration of viral shedding (days) | COVID-19 symptom severity (score)b | Hospital admission | Immunosuppression | HIV | COVID-19 serology | Exitus |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 20 | F | 61 | High blood pressure | −5 | 37 | Mild (1 + 2) | No | No | Neg | Pos | No |
| 21 | M | 53 | No | −15 | 35 | Mild (1) | No | No | Neg | Pos | No |
| 22 | F | 62 | No | −5 | 38 | Mild (1 + 2) | No | No | Ned | Pos | No |
| 23 | F | 45 | DM, high blood pressure, morbid obesity, systemic lupus erythematosus | −3 | 28 | Severe (1) | Yes | Yes | Neg | ND | No |
| 24 | M | 47 | Follicular lymphoma | −22 | 75 | Severe (2) | Yes | Yes | Neg | Pos | Yes |
| 25 | F | 81 | Parkinson’s disease, high blood pressure | −7 | 33 | Mild (1) | No | No | Neg | ND | No |
| 26 | F | 100 | No data | 25 | No data | No | No | Neg | ND | No | |
| 27 | M | 56 | Coronary heart disease, high blood pressure, DM | 63 | No symptoms | Yesc | Yes | Neg | ND | No | |
| 28 | M | 74 | Acute myeloid leukemia | −2 | 55 | Severe (2) | Yes | Yes | Neg | Pos | No |
| 29 | F | 40 | Hodgkin’s lymphoma | −2 | 79 | Severe (2) | Yes | Yes | Neg | Neg | No |
| 30 | M | 69 | Squamous cell lung carcinoma, DM, COPD, high blood pressure | −18 | 29 | Intermediate (1) | Yes | Yes | Neg | Pos | Yes |
| 31 | F | 63 | Congenital cardiopathy, follicular lymphoma | −20 | 44 | Intermediate (2) | Yes | Yes | Neg | Neg | No |
M, male; F, female; DM, diabetes mellitus; COPD, chronic obstructive pulmonary disease; Pos, positive; Neg, negative; ND, not determined.
Mild (1), fever, cephalea, anosmia, ageusia, rhinorrhea, odynophagia, and cough (one or several); Mild (1 + 2), plus diarrhea; Intermediate (1), dyspnea, cough, and fever; Intermediate (2), asthenia and general unrest; Severe (1), respiratory failure; Severe (2), severe dyspnea and bilateral pneumonia.
Due to upper gastrointestinal bleeding.
Our data, based on SG RNA detection, indicate that the presence of actively replicating SARS-CoV-2 may be inferred in certain persistent COVID-19 cases, far beyond diagnosis. In a recent study (12), SG RNA was rarely detectable beyond 8 days after the onset of illness; however, we detected it up to 79 days from symptom onset.
Other authors have ruled out the presence of viable virus in cases with prolonged RT-PCR-positive results based on an analysis of specimens in cell culture. In a selection of cases with persistent positive RT-PCR results (up to 21 days), there was no viral growth in samples taken >8 days from onset (3). No virus was recovered in a limited sample of nine cases after 7 days from onset (6), and no virus was recovered from culture in a sample of 108 specimens taken from persistent RT-PCR-positive cases 44 days from onset, on average (7). The sensitivity of culture is limited, and a minimum viral load in the samples is necessary for viral recovery or detection of a cytopathic effect. The virus is not recovered from cell cultures when the CT value of the RT-PCR-positive specimen is >30 (7, 13) or the viral load is <106 copies/ml (6), even when samples were from cases with active infection. The SARS-CoV-2 peak viral load appears near symptom onset, from where it begins a progressive decrease (1–3, 8). Consequently, most cases with prolonged positive RT-PCR results should have a low viral load, which limits the detection of viable viruses in culture.
Our data, using an alternative SG RNA method, show the usefulness of a non-culture-based approach for inferring the existence of actively replicating viruses. SG RNA detection has been used as proof of active viral replication of other coronaviruses (14), and in SARS-CoV-2, SG RNAs were used (6) to infer active virus replication in the throat.
Our results, acknowledging the limited size of our sample, allow us to suggest that a percentage of persistent SARS-CoV-2 PCR-positive cases might still be contagious. Because of the relevance of these findings, we should extend the analysis of persistent cases in other settings, not assuming their universal lack of infectiousness. Our SG RNA determination approach may be used as an alternative tool to assess the relationships between RT-PCR positivity and viral viability in COVID-19.
ACKNOWLEDGMENTS
We declare that there is no conflict of interest.
This study did not receive any specific funding.
The participants in the Gregorio Marañón Microbiology-ID COVID-19 Study Group include the following: Javier Adán-Jiménez, Luis Alcalá, Teresa Aldámiz, Roberto Alonso, Beatriz Álvarez, Ana Álvarez-Uría, Alexi Arias, Luis Antonio Arroyo, Juan Berenguer, Elena Bermúdez, Emilio Bouza, Almudena Burillo, Ana Candela, Raquel Carrillo, Pilar Catalán, Emilia Cercenado, Alejandro Cobos, Cristina Díez, Pilar Escribano, Agustín Estévez, Chiara Fanciulli, Alicia Galar, María Dolores García, Darío García de Viedma, Paloma Gijón, Adolfo González, Helmuth Guillén, Jesús Guinea, Laura Vanessa Haces, Marta Herranz, Martha Kestler, Juan Carlos López, Carmen Narcisa Losada, Marina Machado, Mercedes Marín, Pablo Martín, Pedro Montilla, Zaira Moure, Patricia Muñoz, María Olmedo, Belén Padilla, María Palomo, Francisco Parras, María Jesús Pérez-Granda, Laura Pérez-Lago, Leire Pérez, Paula Pescador, Sandra R Maus, Elena Reigadas, Cristina Rincón, Belén Rodríguez, Sara Rodríguez, Cristina Rodríguez-Grande, Sandra Rodríguez Maus, Adriana Rojas, María Jesús Ruiz-Serrano, Carlos Sánchez, Mar Sánchez, Julia Serrano, Pedro J. Sola Campoy, Francisco Tejerina, Maricela Valerio, María Cristina Veintimilla, Lara Vesperinas, Teresa Vicente, and Sofía de la Villa.
REFERENCES
- 1.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. 2020. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He X, Lau EHY, Wu P, Deng X, Wang J, Hao X, Lau YC, Wong JY, Guan Y, Tan X, Mo X, Chen Y, Liao B, Chen W, Hu F, Zhang Q, Zhong M, Wu Y, Zhao L, Zhang F, Cowling BJ, Li F, Leung GM. 2020. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med 26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 3.Bullard J, Dust K, Funk D, Strong JE, Alexander D, Garnett L, Boodman C, Bello A, Hedley A, Schiffman Z, Doan K, Bastien N, Li Y, Van Caeseele PG, Poliquin G. 22 May 2020. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin Infect Dis doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun J, Xiao J, Sun R, Tang X, Liang C, Lin H, Zeng L, Hu J, Yuan R, Zhou P, Peng J, Xiong Q, Cui F, Liu Z, Lu J, Tian J, Ma W, Ke C. 2020. Prolonged persistence of SARS-CoV-2 RNA in body fluids. Emerg Infect Dis 26:1834–1838. doi: 10.3201/eid2608.201097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li T-Z, Cao Z-H, Chen Y, Cai M-T, Zhang L-Y, Xu H, Zhang J-Y, Ma C-H, Liu Y, Gao L-J, Duan Z-H, Mou D-L, Liang L-C. 9 July 2020. Duration of SARS-CoV-2 RNA shedding and factors associated with prolonged viral shedding in patients with COVID-19. J Med Virol doi: 10.1002/jmv.26280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Muller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brunink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. 2020. Virological assessment of hospitalized patients with COVID-2019. Nature 581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 7.KDCA. 2020. Finding from investigation and analysis of re-positive cases. KDCA, Cheongju, South Korea. [Google Scholar]
- 8.La Scola B, Le Bideau M, Andreani J, Hoang VT, Grimaldier C, Colson P, Gautret P, Raoult D. 2020. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis 39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. 2020. The architecture of SARS-CoV-2 transcriptome. Cell 181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sztuba-Solińska J, Stollar V, Bujarski JJ. 2011. Subgenomic messenger RNAs: mastering regulation of (+)-strand RNA virus life cycle. Virology 412:245–255. doi: 10.1016/j.virol.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, Bleicker T, Brunink S, Schneider J, Schmidt ML, Mulders DGJC, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette J-L, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MPG, Drosten C. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perera R, Tso E, Tsang OTY, Tsang DNC, Fung K, Leung YWY, Chin AWH, Chu DKW, Cheng SMS, Poon LLM, Chuang VWM, Peiris M. 2020. SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg Infect Dis 26:2701–2704. doi: 10.3201/eid2611.203219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou J, Otter JA, Price JR, Cimpeanu C, Garcia DM, Kinross J, Boshier PR, Mason S, Bolt F, Holmes AH, Barclay WS. 8 July 2020. Investigating SARS-CoV-2 surface and air contamination in an acute healthcare setting during the peak of the COVID-19 pandemic in London. Clin Infect Dis doi: 10.1093/cid/ciaa905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hornyak A, Balint A, Farsang A, Balka G, Hakhverdyan M, Rasmussen TB, Blomberg J, Belak S. 2012. Detection of subgenomic mRNA of feline coronavirus by real-time polymerase chain reaction based on primer-probe energy transfer (P-sg-QPCR). J Virol Methods 181:155–163. doi: 10.1016/j.jviromet.2012.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]