Reliable serological tests are required to determine the prevalence of antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and to characterize immunity to the disease in order to address key knowledge gaps in the coronavirus disease 2019 (COVID-19) pandemic. Quantitative suspension array technology (qSAT) assays based on the xMAP Luminex platform overcome the limitations of rapid diagnostic tests and enzyme-linked immunosorbent assays (ELISAs) with their higher precision, dynamic range, throughput, miniaturization, cost-efficiency, and multiplexing capacity.
KEYWORDS: antibody, immunoassay, SARS-CoV-2, COVID-19, IgM, IgA, IgG, Luminex, quantitative suspension array technology, multiplex, spike, RBD, nucleocapsid, coronavirus, sensitivity, specificity, immunity, performance
ABSTRACT
Reliable serological tests are required to determine the prevalence of antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and to characterize immunity to the disease in order to address key knowledge gaps in the coronavirus disease 2019 (COVID-19) pandemic. Quantitative suspension array technology (qSAT) assays based on the xMAP Luminex platform overcome the limitations of rapid diagnostic tests and enzyme-linked immunosorbent assays (ELISAs) with their higher precision, dynamic range, throughput, miniaturization, cost-efficiency, and multiplexing capacity. We developed three qSAT assays for IgM, IgA, and IgG against a panel of eight SARS-CoV-2 antigens, including spike protein (S), nucleocapsid protein (N), and membrane protein (M) constructs. The assays were optimized to minimize the processing time and maximize the signal-to-noise ratio. We evaluated their performances using 128 prepandemic plasma samples (negative controls) and 104 plasma samples from individuals with SARS-CoV-2 diagnosis (positive controls), of whom 5 were asymptomatic, 51 had mild symptoms, and 48 were hospitalized. Preexisting IgG antibodies recognizing N, M, and S proteins were detected in negative controls, which is suggestive of cross-reactivity to common-cold coronaviruses. The best-performing antibody/antigen signatures had specificities of 100% and sensitivities of 95.78% at ≥14 days and 95.65% at ≥21 days since the onset of symptoms, with areas under the curve (AUCs) of 0.977 and 0.999, respectively. Combining multiple markers as assessed by qSAT assays has the highest efficiency, breadth, and versatility to accurately detect low-level antibody responses for obtaining reliable data on the prevalence of exposure to novel pathogens in a population. Our assays will allow gaining insights into antibody correlates of immunity and their kinetics, required for vaccine development to combat the COVID-19 pandemic.
INTRODUCTION
In a globalized world where emerging infectious diseases of broad distribution can put at stake the health and economy of millions of people, there is a need for versatile and reliable serological tools that can be readily applicable to (i) determine the seroprevalence of antibodies against any new pathogen and, more importantly, (ii) characterize the immune response to the disease at the individual and community levels. In the case of the coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), one of the main priorities since the beginning of the epidemics in China by the end of 2019 (1) was to ascertain the percentage of the population that had been exposed to the virus, considering that a substantial number of people could have been asymptomatic (2, 3). The lack of sensitive and specific serological tests early in the COVID-19 pandemic delayed the precise estimation of the burden of infection for the rational implementation of public health measures to control viral spread (4). Furthermore, immunological assays that can measure a high breadth of antibody types and specificities are needed to dissect which are the naturally acquired protective responses and identify correlates of immunity (5). Additionally, when a vaccine becomes available, such assays would be valuable to evaluate the immunogenicity of candidate vaccines and monitor the duration of immunity at the population level (6).
Common tools for antibody studies are (i) rapid diagnostic tests (RDTs) as point-of-care (POC) devices that usually measure either total immunoglobulins or IgG and IgM, qualitatively (7); (ii) traditional enzyme-linked immunosorbent assays (ELISAs) (8) that can quantify different isotypes and subclasses of antibodies against single antigens at a time and that require certain previous expertise, personnel, and equipment; and (iii) chemiluminescence assays (CLIAs), widely used in clinical practice, which are faster and with higher throughput than ELISAs (9). The performance of the best assays externally validated with >100 negative and >100 positive samples (as recommended by the Foundation for Innovative New Diagnostics [FIND]), mostly from symptomatic patients who are known to have higher antibody levels, ranges from 79.7 to 83.1% sensitivity and 99.1 to 100% specificity for RDTs, 7 days after symptom onset (10); 93.1 to 96.6% sensitivity and 86.3 to 96.4% specificity for ELISAs (Euroimmune); and 87.7 to 99.1% sensitivity and 96.1 to 100% specificity for CLIAs (Siemens, DiaSorin, Abbot, and Roche ECLIA), 14 days after symptom onset (11–14).
A number of in-house ELISAs have also been developed in hospital and research laboratories (15), but they have the limitations that (i) a relatively large amount of sample is required, (ii) the large surface area of the individual microplate wells and the hydrophobic binding of capture antibody can lead to nonspecific binding and increased background, and (iii) most ELISAs rely upon enzyme-mediated amplification of signals in order to achieve reasonable sensitivity (16).
An alternative technique that offers the benefits of ELISAs but also a larger dynamic range of antibody quantification and higher sensitivity (16, 17) is based on the xMAP Luminex platform (www.luminexcorp.com/bibliography). Secondary antibodies are labeled with fluorescent phycoerythrin (PE) directly or with biotin that mediates binding to streptavidin R-phycoerythrin (SAPE), which does not depend on an additional colorimetric reaction. The technique has the added value of higher throughput (up to a 384-well-plate format), increased flexibility, and lower cost with the same workflow as ELISAs, particularly if using magnetic MagPlex microspheres. Paramagnetic beads allow automation of the workflow and better reproducibility than with the previous generation of MicroPlex microspheres. Since the beads have the capture antigen covalently bound and immobilized on their much smaller surface area than with passive coating on a 96-well microplate well, reduced sample volumes are required, and nonspecific binding is diminished (18). Furthermore, a chief advantage over ELISAs is the multiplex nature of the assay that allows measuring antibodies to different antigens simultaneously. This increases the probabilities of detecting a positive antibody response due to the heterogeneity of the human response, and therefore, it has a higher sensitivity relevant for identifying seropositive individuals. The Luminex technology, capable of simultaneously measuring antibodies against 50 (MagPix), 80 (Luminex 100/200), and up to 500 (FlexMap3D) different antigens, makes it an invaluable tool for antigen and epitope screening. Finally, its versatility to set up adapted antigen panels makes Luminex an excellent platform to ensure better preparedness for faster responses to future emerging diseases and pandemics.
Here, we report the establishment and validation of three quantitative suspension array technology (qSAT) assays to measure IgM, IgA, and IgG antibodies against eight SARS-CoV-2 antigens based on the adaptation of previous in-house protocols that measured antibodies to other infectious diseases, including malaria (19–22). Due to the need to process a large number of samples with minimal time and cost during a pandemic like the COVID-19 pandemic, we optimized several conditions to reduce the duration of the assays and report them here for the three main isotypes that have proven useful for seroprevalence studies (23).
MATERIALS AND METHODS
Samples.
Positive samples were 104 plasma samples from individuals with SARS-CoV-2 infection confirmed by real-time reverse transcriptase PCR (rRT-PCR). Fifty-five individuals were recruited in a study of health care workers at Hospital Clínic in Barcelona, Spain (HCB); most of them had mild symptoms, 1 of them was hospitalized, and 5 were without symptoms (23). Forty-nine COVID-19 patients were recruited at the Clínica Universidad de Navarra in Pamplona (Spain), of whom 47 had severe symptoms and were hospitalized and 2 had mild symptoms. The time since the onset of symptoms ranged from 0 to 46 days. Positive samples were used as pools of up to 20 sample subsets for assay optimization tests and individually for the assay evaluations. Negative controls were plasma samples from 128 healthy European donors collected before the COVID-19 pandemic and were used individually. The numbers of positive and negative samples were in line with protocol recommendations from FIND.
Ethics.
The samples analyzed in this study received ethical clearance for immunological evaluation and/or inclusion as controls in immunoassays, and the protocols and informed consent forms were approved by the Institutional Review Board (IRB) at HCB (reference numbers CEIC-7455 and HCB/2020/0336) or the Universidad de Navarra (reference number UN/2020/067) prior to study implementation.
Antigens.
The receptor-binding domain (RBD) of the spike (S) glycoprotein of SARS-CoV-2, the leading vaccine candidate target, was selected as the primary antigen to develop the initial qSAT assays because (i) S is one of the most immunogenic surface proteins together with the nucleocapsid protein (N) (24), (ii) the RBD is the fragment of the virus that mediates binding to the host receptor ACE2 in lung cells (25), (iii) antibodies to RBD correlate with neutralizing antibodies (22, 24) that could be associated with protection based on studies of other coronaviruses and animal models (24, 26–28), and (iv) an ELISA based on this same protein has received FDA approval for COVID-19 serology (15). The RBD was from the Krammer laboratory (Mount Sinai, New York, NY, USA) (15), and the S antigen was produced in-house using Chinese hamster ovary (CHO) cells transiently transfected with a plasmid from the Krammer laboratory (15), followed by purification of the recombinant protein from 4-day culture supernatants using nickel affinity chromatography. The multiplex antigen panel was completed with commercial S1 (GenScript Biotech, Netherlands) and S2 (Sino Biologicals, Germany) proteins and in-house-produced N (full-length, N-terminal, and C-terminal constructs) and membrane (M) (full-length construct) recombinant proteins (see methods in the supplemental material).
qSAT assays.
The optimal concentration of protein to be coupled to beads depends on the antigen and needs to be tested with each new lot. The protein antigens (10, 30, and 50 μg/ml) were coupled to magnetic MagPlex 6.5-μm COOH-microspheres from Luminex Corporation (Austin, TX) in reaction mixtures with a maximum of 625,000 beads, at 10,000 beads/μl (19), and left at 4°C overnight (ON) on a rotatory shaker protected from light. After blocking, washing, and counting, antigen-coupled beads were validated by incubating them with serial dilutions of anti-histidine-biotin antibody for antigens with a histidine tag (catalog number ab27025; Abcam). Further protocol details are provided in the supplemental material. Appropriate coupling concentrations for maximum antibody signal were established by titrating IgG and IgM levels in 11 3-fold serial dilution curves of a pool of 20 positive samples. Batches of coupled beads were stored multiplexed at 2,000 beads/μl in PBS-BN (phosphate-buffered saline [Sigma] plus 1% bovine serum albumin [BSA; Biowest] and 0.05% sodium azide [catalog number S8032; Sigma]) at 4°C protected from light until use.
We compared the performances of the assays when samples were incubated with the antigen-coupled beads for 1 h or 2 h at room temperature (RT) in relation to our regular protocol ON at 4°C. Two thousand antigen-coupled beads in 90 μl PBS-BN were added per well to a 96-well μClear flat-bottom plate (catalog number 655096; Greiner Bio-One), together with individual positive and negative plasma controls (range of dilutions of 1/100 to 1/5,000) at a final volume of 100 μl per well. Two blank control wells with beads in PBS-BN were set up in each plate to control for background signal. Plates were incubated on a microplate shaker at 600 rpm protected from light and then washed three times with 200 μl/well of PBS–0.05% Tween 20 using a magnetic manual washer (catalog number 43-285; Millipore). For more accurate IgM measurements, we tested whether diluting samples 1:10 with GullSORB IgG inactivation reagent (Meridian Bioscience) prior to testing for IgM levels could reduce the high responses observed in some negative samples (29). Additionally, we tested the levels of RBD and S antibodies at different plasma dilutions when incubated in a multiplex panel with additional antigen-coupled beads, including S1, S2, M, and N constructs, compared to those obtained in singleplex to check for potential interferences. Definitive plasma dilutions were established with titration experiments in individual positive and negative samples once the final multiplex panel and all assay conditions were selected to account for the different immunogenicities of the viral antigens.
For the secondary antibody reaction, we compared the performance of biotinylated antibodies followed by SAPE versus that of antibodies conjugated directly to PE and at different incubation times (45 versus 30 min). In all cases, each new lot of secondary antibody was titrated for selecting the optimal concentration. For the first option, 100 μl of biotinylated secondary antibody diluted in PBS-BN (anti-human IgG [catalog number B1140] at a 1/1,250 dilution, anti-human IgM [catalog number B1265] at 1/1,000, or anti-human IgA [catalog number SAB3701227] at 1/500; Sigma) was added to all wells and incubated for 45 min at 600 rpm at RT protected from light. Plates were washed three times, 100 μl of SAPE (catalog number 42250; Sigma) diluted 1:1,000 in PBS-BN was added, and the mixture was incubated during 30 min at 600 rpm at RT protected from light. For the second option, 100 μl of PE-secondary antibody diluted in PBS-BN (goat anti-human IgG [catalog number GTIG-001] at 1/400, goat anti-human IgM [catalog number GTIM-001] at 1/200, or goat anti-human IgA [catalog number GTIA-001] at 1/200; Moss, MD, USA) was added to all wells and incubated for 45 or 30 min at 600 rpm at RT protected from light.
Before reading, plates were washed three times, beads were resuspended in 100 μl of PBS-BN, and data were acquired using a Luminex 100/200 analyzer with a 70-μl acquisition volume per well, doublet discriminator (DD) gating 5,000 to 25,000 settings, and the high-photomultiplier tube (PMT) option. Plates could also be kept ON at 4°C, protected from light, and read the next day. At least 50 beads were acquired per antigen and sample. Crude median fluorescence intensities (MFIs) were exported using xPONENT software. The seropositivity threshold (cutoff) was calculated as 10 to the mean plus 3 standard deviations of log10-transformed MFIs of the negative controls for each antibody isotype and antigen.
Performance of the SARS-CoV-2 qSAT assays.
The receiver operating characteristic (ROC) curves, their corresponding areas under the curve (AUC), and the specificities and sensitivities of the IgM, IgA, and IgG assays were established by testing all 104 positive samples from participants diagnosed with SARS-CoV-2 infection, regardless of symptom information and at different periods since the onset of symptoms (7, 14, 21, and 28 days), and 128 negative samples. For IgM, IgA, and IgG assays, the multiplex panel including RBD, S, S1, S2, M, and N antigen constructs was used according to the same procedures as the ones indicated above and after selecting the optimal assay conditions: samples at 1/500 or 1/3,500 dilutions were incubated with antigen-coupled beads for 1 h followed by a 30-min incubation with secondary antibodies conjugated directly to PE.
Data analysis.
ROC curves, AUCs, and sensitivities and specificities were calculated using the predicted values estimated by supervised machine learning random forest (RF) algorithm models. A classification RF algorithm is a supervised machine learning method that is able to predict the class of an observation based on the knowledge that it has previously acquired from the training data. By seeing many observations, with the value of each predictor variable and the classification outcome, the algorithm is trained and is then able to classify a previously unseen observation. It will do so based on the majority of votes from all the classification trees that make up the forest. IgM, IgA, and IgG MFIs for the different antigens or their combinations were the predictors, and the outcome was SARS-CoV-2 positivity or negativity determined by rRT-PCR. Antibody/antigen pairs that did not discriminate between positive and negative samples were excluded from the analysis. Next, the antibody/antigen variables were further down-selected using an RF algorithm, including all negative and positive controls (n = 232) or all negative controls plus positive controls corresponding to each different period since the onset of symptoms: ≥7 days (n = 213), ≥14 days (n = 199), ≥21 days (n = 174), and ≥28 days (n = 152).
Besides classifying, RFs also rank predictor variables according to their importance to explain the outcome variable. The ranking is based on two criteria: mean decrease accuracy and mean decrease Gini. Mean decrease accuracy refers to the accuracy that is lost by removing each predictor from the model. Mean decrease Gini is based on the Gini impurity, which measures how often a randomly chosen observation from the data set used to train the model will be incorrectly classified. Using an important variable to classify the data entails a high decrease in node impurity and, therefore, a high mean decrease in Gini.
Next, different RFs were built to explore all possible variable combinations at the different periods since the onset of symptoms based on the selected variables per period: all samples (top 12 markers), ≥7 days (top 11 markers), ≥14 days (top 11 markers), ≥21 days (top 11 markers), and ≥28 days (top 11 markers). For each model, we calculated the AUC and selected three seropositivity cutoffs aiming at specificities of (i) 100%, (ii) ≥99% and <100%, and (iii) ≥98% and <99% and obtained the corresponding sensitivities. Models with 100% specificity and the highest sensitivities were selected for ROC curve representations. The analysis was carried out using R studio version R-3.5.1 statistical software (30) (the packages used were randomForest [31] and pROC [32]).
RESULTS
The characteristics of SARS-CoV-2 infected participants whose plasma samples have been used in the study, with regard to age, sex, days since rRT-PCR diagnosis, and days since the onset of symptoms, are included in Table 1.
TABLE 1.
Characteristics of individuals from whom positive samples were tested with regard to age, sex, symptoms, days since symptom onset, and days since rRT-PCR diagnosis
| Variablea | Value |
|---|---|
| Continuous variables [median (IQR)] | |
| Age (yrs) | 50 (24.73) |
| Categorical variables [no. (%) of individuals] | |
| Sex | |
| Female | 61 (58.65) |
| Male | 43 (41.35) |
| Presence of symptoms | |
| No | 5 (4.81) |
| Yes | 99 (95.19) |
| Hospitalized | |
| Yes | 48 (46.15) |
| No. of days since onset of symptoms | |
| 0–6 | 13 (13.13) |
| 7–13 | 14 (14.14) |
| 14–20 | 25 (25.25) |
| 21–27 | 22 (22.22) |
| ≥28 | 24 (24.24) |
| No information | 1 (1.01) |
| No. of days since first positive rRT-PCR | |
| 0–6 | 22 (21.15) |
| 7–13 | 24 (23.08) |
| 14–20 | 29 (27.88) |
| ≥21 | 17 (16.35) |
| After sample collection | 5 (4.81) |
| Not available | 7 (6.73) |
IQR, interquartile range.
Optimization of assay conditions.
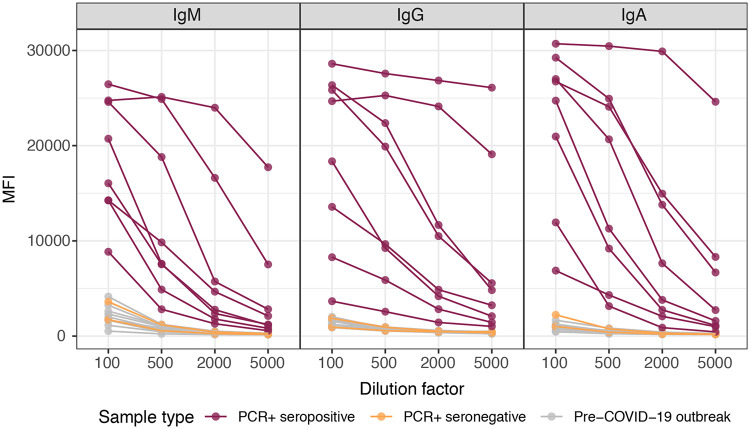
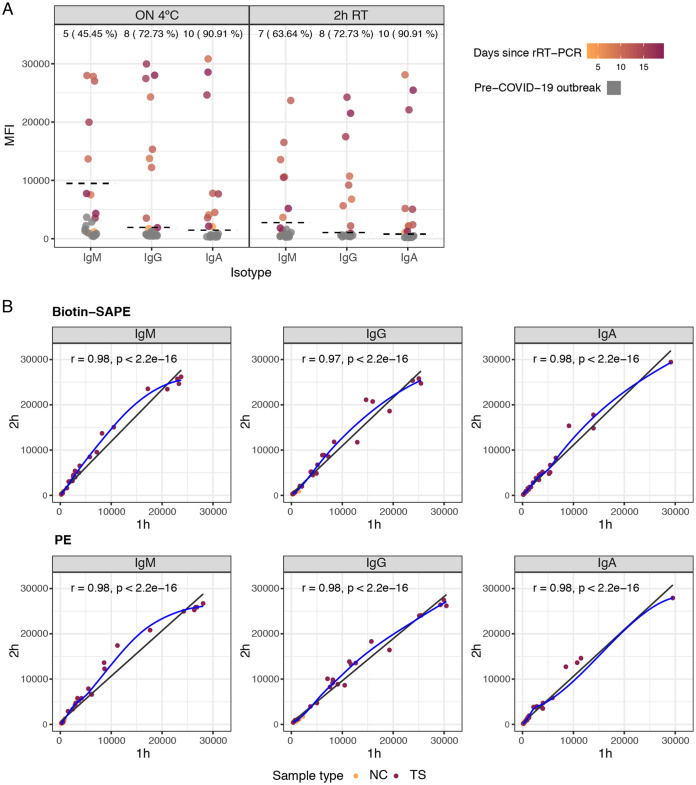
Titration curves did not change substantially between 10, 30, and 50 μg/ml protein, in which case the lower concentration was chosen (see Fig. S1 in the supplemental material). We found that a 1/500 plasma dilution was optimal for quantifying IgM, IgA, and IgG antibodies to RBD in singleplex (Fig. 1), and it was used for subsequent optimizations. Our original standard operating procedures (SOP) established for large seroepidemiological and vaccine studies using Plasmodium falciparum antigens were based on ON incubations of samples at 4°C (20). For COVID-19 serology, we prioritized having faster assays and thus compared the performances of ON incubations at 4°C versus shorter times at RT. Reducing the incubation time to 2 h with the same plasma dilutions did not change the percentage of IgG- or IgA-seropositive samples, and it increased the sensitivity for IgM (Fig. 2A). Although the MFI readings in positive samples generally diminished, the MFI readings in the negative samples were also reduced; i.e., the signal-to-noise ratio was the same or sometimes better, maintaining or increasing the overall proportion of seropositive responses among the positive samples and, thus, the sensitivity. We initially adopted the 2-h incubation time for a first COVID-19 seroprevalence study (23) and subsequently tested shorter incubations, finding that 1 h was noninferior to 2 h of incubation (Fig. 2B). Treatment with GullSORB generally reduced the IgM reactivity in negative controls and increased the signal-to-noise ratio, number of seropositive responses, and sensitivity; therefore, this incubation was adopted for this isotype (Fig. S2).
FIG 1.
Levels (median fluorescence intensity [MFI]) of IgM, IgA, and IgG antibodies to our primary SARS-CoV-2 antigen (RBD) in singleplex using samples from positive and negative individuals at different dilutions (1/100, 1/500, 1/2,000, and 1/5,000) after overnight incubation at 4°C. This served to establish the optimal range of plasma dilutions and to choose the 1/500 dilution for the subsequent assay optimization experiments.
FIG 2.
Levels of IgM, IgA, and IgG antibodies (MFI) to RBD antigen of SARS-CoV-2 in positive and negative plasma samples (1/500 dilution) comparing incubation overnight (ON) at 4°C versus 2 h at room temperature (RT) (A) and 2 h versus 1 h at RT with two different secondary antibodies (B). In panel A, the dashed lines indicate cutoff values; the numbers and percentages of seropositive samples among rRT-PCR-positive samples are shown at the top of the dot plots. In panel B, the blue fitting curve was calculated using the LOESS (locally estimated scatterplot smoothing) method, and the black line was calculated by linear regression. The Spearman test was used to assess the correlations. Biotin-SAPE refers to secondary antibodies conjugated to biotin and streptavidin-phycoerythrin (SAPE), and PE refers to secondary antibodies conjugated with phycoerythrin. NC, negative controls; TS, test samples.
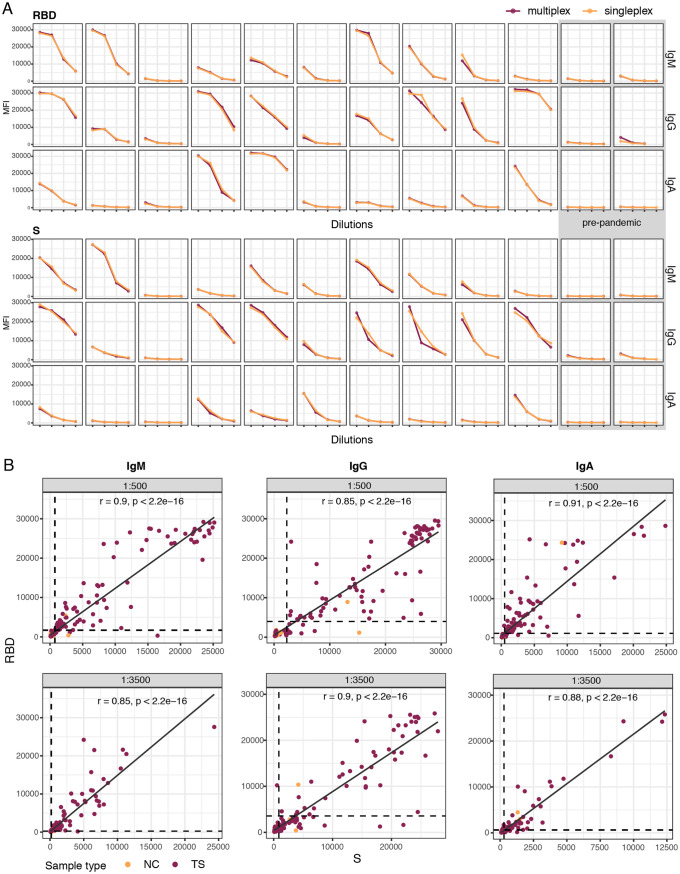
Multiplexing the antigens (8-plex panel) did not significantly decrease the MFI antibody levels to RBD or S compared to singleplex testing (Fig. 3A). Interestingly, there was no evidence of any interference between RBD, S, S1, and S2 antigens despite sharing epitopes within the same multiplex panel. A number of negative prepandemic samples had preexisting antibodies recognizing SARS-CoV-2 proteins for certain isotypes and dilutions: IgG to S1, S2, M, and N constructs and IgA to S1 and the N terminus and C terminus of N (Fig. S2). Furthermore, testing plasma samples against multiple antigens increased the sensitivity of the assay since some individuals who were seronegative or low responders to RBD responded with higher antibodies to S (Fig. 3B).
FIG 3.
Levels of plasma IgM, IgA, and IgG antibodies to the SARS-CoV-2 spike (S) and receptor-binding domain (RBD) primary antigens at different dilutions (1/100, 1/500, 1/2,000, and 1/5,000). (A) Comparison of antibody levels (MFI) in singleplex versus multiplex. The first 10 samples from left to right are from individuals who were positive by rRT-PCR at different times since diagnosis, and the last two samples on the right are from individuals before the COVID-19 pandemic. (B) Correlation of IgG, IgM, and IgA antibody levels against the RBD versus S at different dilutions (1/500 and 1/3,500) showing the benefit of including multiple antigens in the panel to maximize the detection of seropositives. Cutoff values are indicated by dashed lines. The Spearman test was used to assess the correlations. NC, negative controls; TS, test samples.
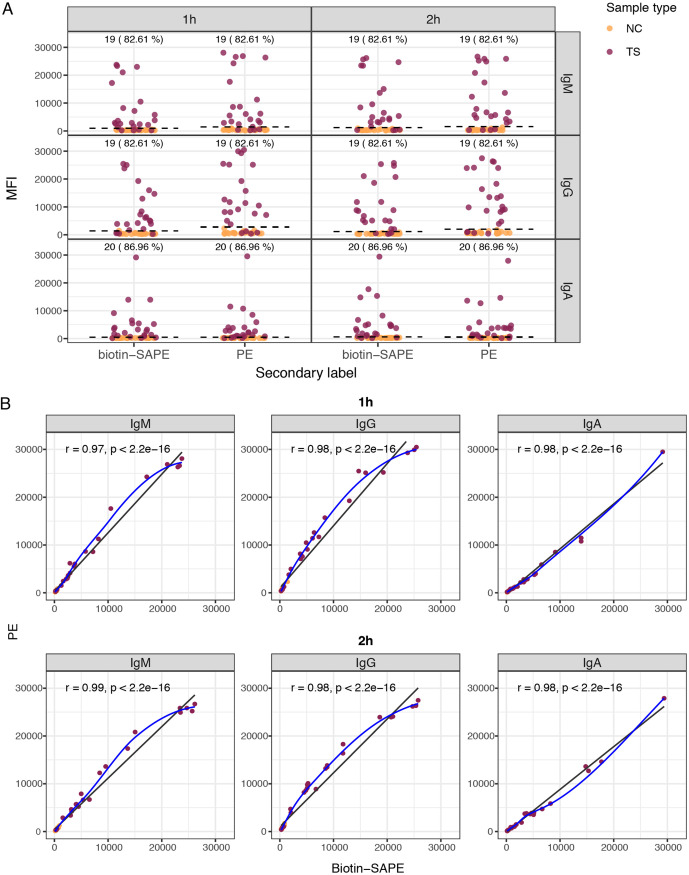
One-step incubation with secondary antibodies conjugated to PE performed as well as a two-step secondary antibody conjugated to biotin followed by SAPE incubation (Fig. 4). A 30-min incubation with the PE-antibody reagent was noninferior to a 45-min incubation (Fig. S3).
FIG 4.
Antibody levels to the RBD using different secondary antibodies and sample incubation times. (A) Levels of IgM, IgA, and IgG antibodies (median fluorescence intensity [MFI]) and percent seropositivity to the RBD among positive controls (burgundy), comparing secondary antibodies conjugated to biotin and streptavidin-phycoerythrin (SAPE) versus PE. Negative controls are in orange. (B) Correlations between antibody levels measured using secondary antibodies conjugated to biotin and SAPE versus PE, for 1-h and 2-h sample incubations. Seropositivity cutoff values are indicated by dashed lines. The numbers and percentages of seropositive samples among the rRT-PCR-positive samples are shown at the top of the dot plots. In panel B, the blue fitting curve was calculated using the LOESS method, and the black line was calculated by linear regression. The Spearman test was used to assess the correlations.
Sensitivity and specificity of the qSAT assays.
We sought the combination of immunoglobulin and antigen responses that yielded the highest specificity (primarily), sensitivity, and AUC to detect seropositive responses. To that end, we built and assessed several RF models, each of them with all the negative controls (n = 128) plus one of the following: (i) all the positive controls (n = 104), (ii) positive controls with ≥7 days since the onset of symptoms (n = 85), (iii) positive controls with ≥14 days since the onset of symptoms (n = 71), (iv) positive controls with ≥21 days since the onset of symptoms (n = 46), or (v) positive controls with ≥28 days since the onset of symptoms (n = 30).
Prior to fitting the RF models, exploratory dot plots were used to select antigens that could discriminate well positive from negative controls and the best plasma dilution for each antigen. M and S1 (IgG and IgA) and the N terminus of N (IgM) were discarded because they had similar responses in negative and positive controls. For RBD and S, samples at a 1/500 dilution for IgG and IgA and samples at a 1/3,500 dilution for IgM gave higher percentages of seropositive responses among the positive controls and thus were selected for the calculations; for N constructs, samples at a 1/3,500 dilution for IgG and IgA performed better except for the C terminus of N, in which IgG was better for samples at a 1/500 dilution.
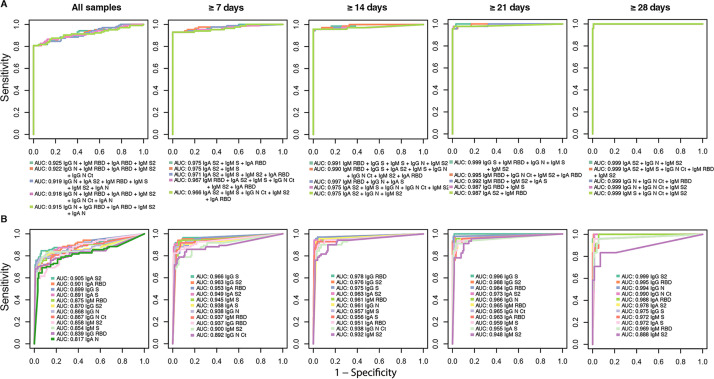
The importance of the predictor variables to explain the outcome variable was ranked by the RFs according to the mean decrease in accuracy and the mean decrease in Gini. We downselected the top 11 to 12 antibody/antigen variables that entailed a greater loss of mean accuracy or Gini impurity in the RF algorithms, which were computed using positive samples from different periods since the onset of symptoms plus the negative controls (Fig. S4). Next, we ran an RF for each possible combination of the downselected top variables for each different period since the onset of symptoms. The resulting RFs were ranked and selected, prioritizing three high-specificity targets (100%, 99%, and 98%). Within each specificity target, the RFs were then sorted by sensitivity and AUC. When targeting a specificity of 100%, the sensitivity of the qSAT assays for samples from participants with a SARS-CoV-2-positive diagnosis with ≥14 days since the onset of symptoms was up to 95.78%, and the AUC was up to 0.977, for the best combinations of antibody/antigen. The top 3 best-performing antibody/antigen signatures for the three different seropositivity thresholds targeting specificities of 100%, 99%, and 98% are shown in Table 2, and their ROC curves with the corresponding AUCs are shown in Fig. 5. High AUCs mean high specificity and high sensitivity and, therefore, a greater predictive capacity of the model. For a set specificity of 100%, the sensitivity for samples from participants with ≥21 days since the onset of symptoms was up to 95.65%, with an AUC of up to 0.999, for the best combinations of antibody/antigen. Using samples from all participants regardless of the time since symptom onset, the sensitivity was up to 80.77%, with an AUC of up to 0.925, for the best combinations. The performance of the qSAT assays to predict positivity using combinations of multiple antibodies/antigens was clearly superior to using single antibody/antigen markers (Fig. 5), as higher AUCs with higher sensitivities for a given specificity target were obtained in the first case, for all different periods since the onset of symptoms. Higher sensitivities were obtained when specificities were set to 98% or 99% than when they were set to 100% (Table 2), reaching sensitivities of 100% for samples from patients with ≥21 or ≥28 days since the onset of symptoms.
TABLE 2.
Sensitivities and specificities of the qSAT antibody assays for negative controls plus either all positive samples or positive samples at different times since the onset of symptoms and different thresholds targeting specificities of 100%, 99%, and 98%a
| Antibody/antigen combination | AUC | % specificity | % sensitivity |
|---|---|---|---|
| All samples | |||
| IgG N + IgM RBD + IgA RBD + IgM S2 | 0.925 | 100 | 80.77 |
| IgG N + IgM RBD + IgA RBD + IgM S2 + IgG N Ct | 0.922 | 100 | 80.77 |
| IgG N + IgA S2 + IgM RBD + IgM S + IgM S2 + IgA N | 0.919 | 100 | 80.77 |
| IgG N + IgA S2 + IgM RBD | 0.920 | 99.22 | 82.69 |
| IgG N + IgA S2 + IgM RBD + IgG S + IgA N + IgA S | 0.919 | 99.22 | 82.69 |
| IgG N + IgM RBD + IgA S | 0.917 | 99.22 | 82.69 |
| IgG N + IgA S2 + IgM RBD | 0.920 | 98.44 | 82.69 |
| IgG N + IgA S2 + IgM RBD + IgG S + IgA S | 0.919 | 98.44 | 82.69 |
| IgG N + IgA S2 + IgM RBD + IgG S + IgA N + IgA S | 0.918 | 98.44 | 82.69 |
| ≥7 days since onset of symptoms | |||
| IgA S2 + IgM S + IgA RBD | 0.975 | 100 | 92.94 |
| IgA S2 + IgM S | 0.975 | 100 | 92.94 |
| IgA S2 + IgM S + IgM S2 + IgA RBD | 0.971 | 100 | 92.94 |
| IgA S2 + IgG S2 + IgM S + IgA RBD | 0.981 | 99.22 | 92.94 |
| IgM RBD + IgA S2 + IgG S + IgM S + IgM S2 + IgA RBD | 0.980 | 99.22 | 92.94 |
| IgM RBD + IgA S2 + IgG S | 0.980 | 99.22 | 92.94 |
| IgA S2 + IgG S + IgM S | 0.983 | 98.44 | 92.94 |
| IgG S2 + IgM S + IgA RBD | 0.983 | 98.44 | 92.94 |
| IgA S2 + IgG S2 + IgM S + IgA RBD | 0.981 | 98.44 | 92.94 |
| ≥14 days since onset of symptoms | |||
| IgM RBD + IgG N + IgA S | 0.977 | 100 | 95.78 |
| IgA S2 + IgM S + IgG N + IgG N Ct + IgM S2 | 0.975 | 100 | 95.78 |
| IgA S2 + IgG N + IgM S2 | 0.975 | 100 | 95.78 |
| IgM RBD + IgG S + IgM S + IgG N | 0.992 | 99.22 | 95.78 |
| IgM RBD + IgG S + IgA S2 + IgM S + IgG N + IgG RBD + IgG N Ct | 0.992 | 99.22 | 95.78 |
| IgM RBD + IgG S + IgA S2 + IgG N + IgM S2 | 0.991 | 99.22 | 95.78 |
| IgG S + IgG RBD + IgG N Ct + IgM S2 | 0.985 | 98.44 | 97.18 |
| IgG S + IgG RBD | 0.984 | 98.44 | 97.18 |
| IgG S + IgA S2 + IgG RBD + IgM S2 + IgA RBD | 0.983 | 98.44 | 97.18 |
| ≥21 days since onset of symptoms | |||
| IgG S + IgM RBD + IgG N + IgM S + IgM S2 | 0.999 | 100 | 95.65 |
| IgM RBD + IgG N Ct + IgM S2 + IgA RBD | 0.995 | 100 | 95.65 |
| IgM RBD + IgM S2 + IgA S | 0.992 | 100 | 95.65 |
| IgG S + IgG RBD + IgM RBD | 0.999 | 99.22 | 97.83 |
| IgG S + IgG RBD + IgM RBD + IgG N + IgM S2 | 0.999 | 99.22 | 97.83 |
| IgG S + IgG RBD + IgM RBD + IgG N + IgM S2 + IgA RBD | 0.999 | 99.22 | 97.83 |
| IgG S + IgG RBD + IgM RBD | 0.999 | 98.44 | 100 |
| IgG S + IgA S2 + IgG RBD + IgM RBD + IgM S2 + IgA RBD + IgA S | 0.999 | 98.44 | 100 |
| IgG S + IgA S2 + IgM RBD + IgM S2 + IgA RBD | 0.999 | 98.44 | 100 |
| ≥28 days since onset of symptoms | |||
| IgA S2 + IgG N + IgM S2 | 0.9997 | 100 | 95.83 |
| IgA S2 + IgM S + IgG N Ct + IgM RBD + IgM S2 | 0.9997 | 100 | 95.83 |
| IgG N + IgG N Ct + IgM RBD | 0.9997 | 100 | 95.83 |
| IgA S2 + IgG N + IgM S2 | 0.9997 | 99.22 | 100 |
| IgA S2 + IgM S + IgG N Ct + IgM RBD + IgM S2 | 0.9997 | 99.22 | 100 |
| IgG N + IgG N Ct + IgM RBD | 0.9997 | 99.22 | 100 |
| IgA S2 + IgG N + IgM S2 | 0.9997 | 98.44 | 100 |
| IgA S2 + IgM S + IgG N Ct + IgM RBD + IgM S2 | 0.9997 | 98.44 | 100 |
| IgG N + IgG N Ct + IgM RBD | 0.9997 | 98.44 | 100 |
The top 3 best-performing signatures for each category are shown. AUC, area under the curve; N Ct, C terminus of N.
FIG 5.
Antibody Luminex assay performance. Shown are receiver operating characteristic (ROC) curves and areas under the curve (AUCs) using samples from prepandemic negative controls plus either all participants with a positive COVID-19 diagnosis or participants with a positive diagnosis at different times since the onset of symptoms. ROC curves and AUCs from different combinations of multiple immunoglobulin isotypes to different antigens with top performances are included in panel A, whereas those of single isotype/antibody markers are included in panel B.
DISCUSSION
We developed three novel multiplex immunoassays for quantifying IgM, IgA, and IgG to eight SARS-CoV-2 protein constructs and evaluated, by machine learning classification algorithms, the performances of several antibody/antigen combinations to detect any positive antibody response to infection, obtaining specificities of 100% and sensitivities of 95.78% (≥14 days since symptom onset) or 95.65% (≥21 days since symptom onset) and very high predictability (AUC of ≥0.98). Our qSAT assays, based on the xMAP technology, provide excellent precision and accuracy and a wide range of detection.
For any given test, there is usually a trade-off between sensitivity and specificity. To evaluate the performances of the assays here, we prioritized specificity over sensitivity for the impact that specificity has on seroprevalence studies. Particularly when the prevalence of infection is low, the positive predictive value of a test strongly relies on a high specificity. For example, in a scenario of a 5% prevalence, a decrease in specificity from 100% to 95% would cause 5% of negative results to be wrongly classified as positive results, giving a false prevalence of 10%, the double of the real prevalence (50% positive predictive value). In a scenario of a 20% prevalence, a 95% specificity would estimate a prevalence of 24% (83% positive predictive value). The lower the prevalence, the higher the impact of a low specificity. For this reason, we highlight the importance of targeting high specificities when dealing with a low prevalence of infection (33). However, other seropositivity thresholds can be used to have a balanced specificity/sensitivity or to maximize sensitivity. For studies aiming at characterizing the kinetics of antibody responses, the immunogenicity of vaccine candidates, or correlates of protection, a cutoff prioritizing sensitivity over specificity would be preferable.
A time period after the onset of symptoms is usually established for these analyses because antibodies take an average of 4 to 14 days since infection to be produced and detected depending on the isotype and test (IgM, 5 to 12 days; IgA, 5 to 12 days; IgG, 4 to 14 days) (34–39). Thus, antibodies are not necessarily expected to be detected in individuals who are acutely infected and diagnosed around the time of plasma collection. Accordingly, when considering all samples, which included samples from 13 individuals with <6 days since the onset of symptoms plus 1 symptomatic individual without information on the number of days since the onset of symptoms and the 5 asymptomatic individuals, the sensitivity was lower (up to 80.77%) at specificities of 100%. However, we detected IgM or IgA as early as 2 days from the onset of symptoms. In fact, since samples were collected in the early days of the COVID-19 pandemic, it is expected that IgM and IgA, which are induced upon primary infection earlier than IgG, could contribute to a higher sensitivity of detection. Most of the best signatures identified included IgM and IgA besides IgG, regardless of the time period since the onset of symptoms, also beyond 28 days. However, over time, the only antibodies that would be expected to remain in blood are IgG due to the decay of IgM and IgA; e.g., IgM levels may become undetectable by the fifth week after symptom onset (40). Therefore, with longer times since infection, serological assays to detect maintenance of antibodies could focus on IgG detection. For diagnosis early in the disease, we recommend using the combinations of antibody/antigen that achieve a cutoff that results in 100% specificity with the highest possible sensitivity and AUC when fitting an RF algorithm with all the negative plus positive controls. Specificity would also prevail over sensitivity for the implications that a false-positive result may have at a personal level. However, antibody assays are not able to discern current from recent infection, and, as mentioned above, the production and detection of antibodies are delayed from the onset of infection.
The sensitivities and specificities of other SARS-CoV-2 Luminex assays evaluated with >100 positive and >100 negative samples have been shown to be comparable to ours. A study with sera from asymptomatic to hospitalized patients, measuring IgG and IgA to several peptides derived from S and N, showed a 99% sensitivity and a 98% specificity (41). Another study with samples from outpatients and hospitalized cases measuring IgG to S1, the RBD, and N reported a 90% sensitivity and a 100% specificity (42). A third study with samples from health care workers and hospitalized patients measuring IgG and IgM to S trimer, the RBD, S1, and S2 estimated a 96% sensitivity and a 99% specificity (43).
A key value of Luminex compared to ELISAs is multiplexing, which allows the capture of a wider breadth of responses, and this is needed because some individuals may not respond to one antigen (e.g., the RBD) but may do so to other antigens (e.g., S or N proteins) (44–46), or responses may change over time. Here, we substantially increased the sensitivity of the assay by combining antibodies and antigens compared to using only one. An added advantage of multiplexing is the reduced usage of sample volumes, resources, and time if antibodies to several antigens are to be evaluated. The possibility of performing miniaturized assays with small amounts of blood is very attractive in pediatric studies, in large field surveys where fingerpick may be more logistically feasible, and for testing special tissues of interest, including mucosal fluids. These combined advantages have a direct impact on the cost of the qSAT assay, which can be less than 1/5 of the cost of the least expensive commercial ELISA and less than 1/16 of that of the most expensive commercial kit. The cost is reduced because there is less protein used due to the smaller surface area and smaller amounts of other materials and reagents. Compared to the commercial xMAP SARS-CoV-2 multiantigen IgG assay, our assay includes 2 additional isotypes and 5 additional antigens, allowing a better study of the immune response and likely being more sensitive. We reduced the dilutions of plasma and titrated the secondary antibody to use minimal amounts of samples and reagents without compromising sensitivity. The economy of scale is improved further when the assays are adapted to the high-throughput FlexMap3D 384-well-plate format, but they are also easily adaptable to the benchtop MagPix 96-well format, which is more affordable and simpler to maintain even in remote laboratory settings.
Positive responses to M, S1, S2, and N antigens were detected in samples collected before the COVID-19 pandemic. The presence of such IgGs has been interpreted as cross-reactivity with human coronaviruses (HCoVs) causing the common cold (47–49). Indeed, higher sequence homology at the protein level between SARS-CoV-2 and HCoV has been reported for N (particularly the N-terminal and central regions), M, and S2 (47, 50, 51). The addition of a commercial S1 protein did not have any added value, and future versions of the assay will test S1 from different sources as this subunit is expected to not cross-react with other betacoronaviruses and be specific for SARS-CoV-2 diagnostics (15, 50). Preexisting SARS-CoV-2-specific T cells have also been reported and attributed to cross-reactivity with HCoVs previously encountered (52, 53). The multiplex nature of the assay will allow testing of this hypothesis in the future with the addition of antigens to the related HCoVs 229E, HKU1, NL63, and OC43 in the same assay panel, by comparing the patterns of antibody reactivity, to address the significance of this in immunity to COVID-19.
The assay performances were excellent, but further testing needs to be performed with longer periods of time since the onset of symptoms, although we expect to maintain high specificity and sensitivity, albeit antibody signatures would be different and based on IgG only. Future studies will include additional positive samples from asymptomatic individuals, who probably have lower antibody levels than mild or severe cases and are rarely included for the validation of commercial kits. In addition, it will be interesting to include negative controls reacting with other coronaviruses or other infections (e.g., malaria) and pathologies known to induce polyclonal responses or rheumatoid factor, which may increase background responses.
In conclusion, we developed 100% specific and fast assays with optimal diagnostic characteristics to assess the seroprevalence of COVID-19. Considering their high sensitivity, these qSAT assays would be suited to identify individuals with low levels of antibodies, such as asymptomatic or immunosuppressed individuals, or long-term decaying antibodies (43). In addition, this approach would be particularly suited to identify hyperimmune donors with very high levels of antibodies and the largest antigenic breadth for immunotherapy. The assays are highly versatile, being easily adaptable to quantify other antibodies like IgE, or IgG and IgA subclasses, and avidity with the use of chaotropic agents, and even functional activity like inhibition of binding to the virus receptor ACE2. The multiplex capabilities also make them ideal for sizeable peptide screenings to accelerate epitope mapping and selection for identifying the fine specificity of immune correlates of protection for vaccine development and would also be applicable in vaccine evaluation when the first candidates reach larger-scale phase 3 clinical trials.
Supplementary Material
ACKNOWLEDGMENTS
We thank the volunteers who donated blood for COVID-19 studies and the clinical and laboratory staff who participated in sample collection and processing. We especially thank ISGlobal colleagues P. Cisteró, R. A. Mitchell, C. Jairoce, S. Alonso, J. Moreno, L. Puyol, C. Chaccour, and those involved in data analysis and/or the recruitment of volunteers at the hospital, J. L. del Pozo, M. Fernández, M. Tortajada, C. Guinovart, S. Sanz, S. Méndez, A. Llupià, E. Chóliz, A. Cruz, S. Folchs, M. Lamoglia, N. Ortega, N. Pey, M. Ribes, N. Rosell, P. Sotomayor, S. Torres, S. Williams, S. Barroso, A. Trilla, and P. Varela. We are grateful to F. Krammer for the donation of RBD and S plasmids, L. Mayer for assistance with literature review, and Wilco de Jager from Luminex for technical advice.
Assay development and sample collection were performed with internal funds from the investigators’ groups and institutions, and the performance analysis received support from FIND. G.M. had the support of the Department of Health, Catalan Government (SLT006/17/00109). J.C. is supported by an SAF2016-76080-R grant from the Spanish Ministry of Economy (AEI/FEDER, UE) to L.I. The development of SARS-CoV-2 reagents was partially supported by NIAID Centers of Excellence for Influenza Research and Surveillance (CEIRS) contract HHSN272201400008C. We acknowledge support from the Spanish Ministry of Science and Innovation through the Centro de Excelencia Severo Ochoa 2019-2023 Program (CEX2018-000806-S) and the Generalitat de Catalunya through the CERCA Program.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Day M. 2020. Covid-19: four fifths of cases are asymptomatic, China figures indicate. BMJ 369:m1375. doi: 10.1136/bmj.m1375. [DOI] [PubMed] [Google Scholar]
- 3.Wilder-Smith A, Teleman MD, Heng BH, Earnest A, Ling AE, Leo YS. 2005. Asymptomatic SARS coronavirus infection among healthcare workers, Singapore. Emerg Infect Dis 11:1142–1145. doi: 10.3201/eid1107.041165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winter AL, Hegde ST. 2020. The important role of serology for COVID-19 control. Lancet Infect Dis 20:758–759. doi: 10.1016/S1473-3099(20)30322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altmann DM, Douek DC, Boyton RJ. 2020. What policy makers need to know about COVID-19 protective immunity. Lancet 395:1527–1529. doi: 10.1016/S0140-6736(20)30985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.den Hartog G, van Binnendijk R, Buisman AM, Berbers GAM, van der Klis FRM. 2020. Immune surveillance for vaccine-preventable diseases. Expert Rev Vaccines 19:327–339. doi: 10.1080/14760584.2020.1745071. [DOI] [PubMed] [Google Scholar]
- 7.Koczula KM, Gallotta A. 2016. Lateral flow assays. Essays Biochem 60:111–120. doi: 10.1042/EBC20150012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engvall E, Perlmann P. 1971. Enzyme-linked immunosorbent assay (ELISA). Quantitative assay of immunoglobulin G. Immunochemistry 8:871–874. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
- 9.Cinquanta L, Fontana DE, Bizzaro N. 2017. Chemiluminescent immunoassay technology: what does it change in autoantibody detection? Auto Immun Highlights 8:9. doi: 10.1007/s13317-017-0097-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cota G, Freire ML, de Souza CS, Pedras MJ, Saliba JW, Faria V, Alves LL, Rabello A, Avelar DM. 8 October 2020. Diagnostic performance of commercially available COVID-19 serology tests in Brazil. Int J Infect Dis doi: 10.1016/j.ijid.2020.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manthei DM, Whalen JF, Schroeder LF, Sinay AM, Li S-H, Valdez R, Giacherio DA, Gherasim C. 9 October 2020. Differences in performance characteristics among four high-throughput assays for the detection of antibodies against SARS-CoV-2 using a common set of patient samples. Am J Clin Pathol doi: 10.1093/AJCP/AQAA200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turbett SE, Anahtar M, Dighe AS, Garcia Beltran W, Miller T, Scott H, Durbin SM, Bharadwaj M, Thomas J, Gogakos TS, Astudillo M, Lennerz J, Rosenberg ES, Branda JA. 5 October 2020. Evaluation of three commercial SARS-CoV-2 serologic assays and their performance in two-test algorithms. J Clin Microbiol doi: 10.1128/JCM.01892-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National SARS-CoV-2 Serology Assay Evaluation Group. 23 September 2020. Performance characteristics of five immunoassays for SARS-CoV-2: a head-to-head benchmark comparison. Lancet Infect Dis doi: 10.1016/S1473-3099(20)30634-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Batra R, Olivieri LG, Rubin D, Vallari A, Pearce S, Olivo A, Prostko J, Nebbia G, Douthwaite S, Rodgers M, Cloherty G. 2020. A comparative evaluation between the Abbott Panbio COVID-19 IgG/IgM rapid test device and Abbott Architect SARS CoV-2 IgG assay. J Clin Virol 132:104645. doi: 10.1016/j.jcv.2020.104645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amanat F, Stadlbauer D, Strohmeier S, Nguyen THO, Chromikova V, McMahon M, Jiang K, Arunkumar GA, Jurczyszak D, Polanco J, Bermudez-Gonzalez M, Kleiner G, Aydillo T, Miorin L, Fierer DS, Lugo LA, Kojic EM, Stoever J, Liu STH, Cunningham-Rundles C, Felgner PL, Moran T, García-Sastre A, Caplivski D, Cheng AC, Kedzierska K, Vapalahti O, Hepojoki JM, Simon V, Krammer F. 2020. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med 26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luminex Corporation. 2010. Overcoming the cost and performance limitations of ELISA with xMAP technology. Tech note 1–4. Luminex Corporation, Austin, TX. [Google Scholar]
- 17.Rizzi G, Zhang YJ, Latek R, Weiner R, Rhyne PW. 2010. Characterization and development of a Luminex-based assay for the detection of human IL-23. Bioanalysis 2:1561–1572. doi: 10.4155/bio.10.68. [DOI] [PubMed] [Google Scholar]
- 18.Carson RT, Vignali DA. 1999. Simultaneous quantitation of 15 cytokines using a multiplexed flow cytometric assay. J Immunol Methods 227:41–52. doi: 10.1016/s0022-1759(99)00069-1. [DOI] [PubMed] [Google Scholar]
- 19.Vidal M, Aguilar R, Campo JJ, Dobaño C. 2018. Development of quantitative suspension array assays for six immunoglobulin isotypes and subclasses to multiple Plasmodium falciparum antigens. J Immunol Methods 455:41–54. doi: 10.1016/j.jim.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ubillos I, Jiménez A, Vidal M, Bowyer PW, Gaur D, Dutta S, Gamain B, Coppel R, Chauhan V, Lanar D, Chitnis C, Angov E, Beeson J, Cavanagh D, Campo JJ, Aguilar R, Dobaño C. 2018. Optimization of incubation conditions of Plasmodium falciparum antibody multiplex assays to measure IgG, IgG(1-4), IgM and IgE using standard and customized reference pools for sero-epidemiological and vaccine studies. Malar J 17:219. doi: 10.1186/s12936-018-2369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ubillos I, Campo JJ, Jiménez A, Dobaño C. 2018. Development of a high-throughput flexible quantitative suspension array assay for IgG against multiple Plasmodium falciparum antigens. Malar J 17:216. doi: 10.1186/s12936-018-2365-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ubillos I, Aguilar R, Sanz H, Jiménez A, Vidal M, Valmaseda A, Dong Y, Gaur D, Chitnis CE, Dutta S, Angov E, Aponte JJ, Campo JJ, Valim C, Harezlak J, Dobaño C. 2018. Analysis of factors affecting the variability of a quantitative suspension bead array assay measuring IgG to multiple Plasmodium antigens. PLoS One 13:e0199278. doi: 10.1371/journal.pone.0199278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Basteiro AL, Moncunill G, Tortajada M, Vidal M, Guinovart C, Jiménez A, Santano R, Sanz S, Méndez S, Llupià A, Aguilar R, Alonso S, Barrios D, Carolis C, Cisteró P, Chóliz E, Cruz A, Fochs S, Jairoce C, Hecht J, Lamoglia M, Martínez MJ, Mitchell RA, Ortega N, Pey N, Puyol L, Ribes M, Rosell N, Sotomayor P, Torres S, Williams S, Barroso S, Vilella A, Muñoz J, Trilla A, Varela P, Mayor A, Dobaño C. 2020. Seroprevalence of antibodies against SARS-CoV-2 among health care workers in a large Spanish reference hospital. Nat Commun 11:3500. doi: 10.1038/s41467-020-17318-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang S, Hillyer C, Du L. 2020. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol 41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen K-Y, Wang Q, Zhou H, Yan J, Qi J. 2020. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181:894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Callow KA, Parry HF, Sergeant M, Tyrrell DAJ. 1990. The time course of the immune response to experimental coronavirus infection of man. Epidemiol Infect 105:435–446. doi: 10.1017/s0950268800048019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bao L, Deng W, Gao H, Xiao C, Liu J, Xue J, Lv Q, Liu J, Yu P, Xu Y, Qi F, Qu Y, Li F, Xiang Z, Yu H, Gong S, Liu M, Wang G, Wang S, Song Z, Zhao W, Han Y, Zhao L, Liu X, Wei Q, Qin C. 2020. Reinfection could not occur in SARS-CoV-2 infected rhesus macaques. bioRxiv 10.1101/2020.03.13.990226. [DOI]
- 28.Liu W, Fontanet A, Zhang P, Zhan L, Xin Z, Baril L, Tang F, Lv H, Cao W. 2006. Two year prospective study of the humoral immune response of patients with severe acute respiratory syndrome. J Infect Dis 193:792–795. doi: 10.1086/500469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kareinen L, Hepojoki S, Huhtamo E, Korhonen EM, Schmidt-Chanasit J, Hedman K, Hepojoki J, Vapalahti O. 2019. Immunoassay for serodiagnosis of Zika virus infection based on time-resolved Förster resonance energy transfer. PLoS One 14:e0219474. doi: 10.1371/journal.pone.0219474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.R Core Team. 2020. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. [Google Scholar]
- 31.Liaw A, Wiener M. 2002. Classification and regression by randomForest. R News 2:18–22. [Google Scholar]
- 32.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, Müller M. 2011. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watson J, Whiting PF, Brush JE. 2020. Interpreting a covid-19 test result. BMJ 369:m1808. doi: 10.1136/bmj.m1808. [DOI] [PubMed] [Google Scholar]
- 34.Long Q, Deng H, Chen J, Hu J, Liu B, Liao P, Lin Y, Yu L, Mo Z, Xu Y, Gong F, Wu G, Zhang X, Chen Y, Li Z, Wang K, Zhang X, Tian W, Niu C, Yang Q, Xiang J, Du H, Liu H, Lang C, Luo X, Wu S, Cui X, Zhou Z, Wang J, Xue C, Li X, Wang L, Tang X, Zhang Y, Qiu J, Liu X, Li J, Zhang D, Zhang F, Cai X, Wang D, Hu Y, Ren J, Tang N, Liu P, Li Q, Huang A. 2020. Antibody responses to SARS-CoV-2 in COVID-19 patients: the perspective application of serological tests in clinical practice. medRxiv doi: 10.1101/2020.03.18.20038018. [DOI]
- 35.Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, Wang X, Yuan J, Li T, Li J, Qian S, Hong C, Wang F, Liu Y, Wang Z, He Q, Li Z, He B, Zhang T, Fu Y, Ge S, Liu L, Zhang J, Xia N, Zhang Z. 28 March 2020. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.To KK-W, Tsang OT-Y, Leung W-S, Tam AR, Wu T-C, Lung DC, Yip CC-Y, Cai J-P, Chan JM-C, Chik TS-H, Lau DP-L, Choi CY-C, Chen L-L, Chan W-M, Chan K-H, Ip JD, Ng AC-K, Poon RW-S, Luo C-T, Cheng VC-C, Chan JF-W, Hung IF-N, Chen Z, Chen H, Yuen K-Y. 2020. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiang F, Wang X, He X, Peng Z, Yang B, Zhang J, Zhou Q, Ye H, Ma Y, Li H, Wei X, Cai P, Ma W-L. 19 April 2020. Antibody detection and dynamic characteristics in patients with COVID-19. Clin Infect Dis doi: 10.1093/cid/ciaa461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qu J, Wu C, Li X, Zhang G, Jiang Z, Li X, Zhu Q, Liu L. 27 April 2020. Profile of IgG and IgM antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis doi: 10.1093/cid/ciaa489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo L, Ren L, Yang S, Xiao M, Chang D, Yang F, Dela Cruz CS, Wang Y, Wu C, Xiao Y, Zhang L, Han L, Dang S, Xu Y, Yang Q, Xu S, Zhu H, Xu Y, Jin Q, Sharma L, Wang L, Wang J. 2020. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis 71:778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao AT, Gao C, Zhang S. 2020. Profile of specific antibodies to SARS-CoV-2: the first report. J Infect 81:147–178. doi: 10.1016/j.jinf.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shrock E, Fujimura E, Kula T, Timms RT, Lee I-H, Leng Y, Robinson ML, Sie BM, Li MZ, Chen Y, Logue J, Zuiani A, McCulloch D, Lelis FJN, Henson S, Monaco DR, Travers M, Habibi S, Clarke WA, Caturegli P, Laeyendecker O, Piechocka-Trocha A, Li J, Khatri A, Chu HY, Villani A-C, Kays K, Goldberg MB, Hacohen N, Filbin MR, Yu XG, Walker BD, Wesemann DR, Larman HB, Lederer JA, Elledge SJ. 29 September 2020. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.den Hartog G, Schepp RM, Kuijer M, GeurtsvanKessel C, van Beek J, Rots N, Koopmans MPG, van der Klis FRM, van Binnendijk RS. 2020. SARS-CoV-2-specific antibody detection for seroepidemiology: a multiplex analysis approach accounting for accurate seroprevalence. J Infect Dis 222:1452–1461. doi: 10.1093/infdis/jiaa479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosado J, Pelleau S, Cockram C, Merkling SH, Nekkab N, Demeret C, Meola A, Kerneis S, Terrier B, Fafi-Kremer S, de Seze J, Dejardin F, Petres S, Longley R, Backovic M, Mueller I, White MT. 2020. Serological signatures of SARS-CoV-2 infection: implications for antibody-based diagnostics. medRxiv 10.1101/2020.05.07.20093963. [DOI] [PMC free article] [PubMed]
- 44.Qiu M, Shi Y, Guo Z, Chen Z, He R, Chen R, Zhou D, Dai E, Wang X, Si B, Song Y, Li J, Yang L, Wang J, Wang H, Pang X, Zhai J, Du Z, Liu Y, Zhang Y, Li L, Wang J, Sun B, Yang R. 2005. Antibody responses to individual proteins of SARS coronavirus and their neutralization activities. Microbes Infect 7:882–889. doi: 10.1016/j.micinf.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Chang Z, Ouyang J, Wei H, Yang R, Chao Y, Qu J, Wang J, Hung T. 2005. Profiles of IgG antibodies to nucleocapsid and spike proteins of the SARS-associated coronavirus in SARS patients. DNA Cell Biol 24:521–527. doi: 10.1089/dna.2005.24.521. [DOI] [PubMed] [Google Scholar]
- 46.Giménez LG, Rojas J, Rojas A, Mendoza J, Camacho AG. 2009. Development of an enzyme-linked immunosorbent assay-based test with a cocktail of nucleocapsid and spike proteins for detection of severe acute respiratory syndrome-associated coronavirus-specific antibody. Clin Vaccine Immunol 16:241–245. doi: 10.1128/CVI.00252-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyer B, Drosten C, Müller MA. 2014. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res 194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khan S, Nakajima R, Jain A, de Assis RR, Jasinskas A, Obiero JM, Adenaiye O, Tai S, Hong F, Milton DK, Davies H, Felgner PL. 2020. Analysis of serologic cross-reactivity between common human coronaviruses and SARS-CoV-2 using coronavirus antigen microarray. bioRxiv 10.1101/2020.03.24.006544. [DOI]
- 49.Che X-Y, Qiu L-W, Liao Z-Y, Wang Y, Wen K, Pan Y-X, Hao W, Mei Y-B, Cheng VCC, Yuen K-Y. 2005. Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. J Infect Dis 191:2033–2037. doi: 10.1086/430355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okba NMA, Muller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, Lamers MM, Sikkema RS, de Bruin E, Chandler FD, Yazdanpanah Y, Hingrat QL, Descamps D, Houhou-Fidouh N, Reusken CBEM, Bosch B-J, Drosten C, Koopmans MPG, Haagmans BL. 2020. SARS-CoV-2 specific antibody responses in COVID-19 patients. medRxiv 10.1101/2020.03.18.20038059. [DOI] [PMC free article] [PubMed]
- 51.Hachim A, Kavian N, Cohen CA, Chin AWH, Chu DKW, Mok CKP, Tsang OTY, Yeung YC, Perera RAPM, Poon LLM, Peiris MJS, Valkenburg SA. 2020. Beyond the spike: identification of viral targets of the antibody response to SARS-CoV-2 in COVID-19 patients. medRxiv 10.1101/2020.04.30.20085670. [DOI]
- 52.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Rydyznski Moderbacher C, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, Marrama D, de Silva AM, Frazier A, Carlin AF, Greenbaum JA, Peters B, Krammer F, Smith DM, Crotty S, Sette A. 2020. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Braun J, Loyal L, Frentsch M, Wendisch D, Georg P, Kurth F, Hippenstiel S, Dingeldey M, Kruse B, Fauchere F, Baysal E, Mangold M, Henze L, Lauster R, Mall M, Beyer K, Roehmel J, Schmitz J, Miltenyi S, Mueller MA, Witzenrath M, Suttorp N, Kern F, Reimer U, Wenschuh H, Drosten C, Corman VM, Giesecke-Thiel C, Sander L-E, Thiel A. 2020. Presence of SARS-CoV-2 reactive T cells in COVID-19 patients and healthy donors. medRxiv 10.1101/2020.04.17.20061440. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.