Neisseria meningitidis and Neisseria gonorrhoeae are pathogenic bacteria that can cause human infections. While N. meningitidis infections are associated with bacterial meningitis and bacteremia, a strain of N. meningitidis, isolated from the urogenital system, has recently been associated with urethritis.
KEYWORDS: Neisseria, urethritis, whole-genome sequencing, antibiotic resistance
ABSTRACT
Neisseria meningitidis and Neisseria gonorrhoeae are pathogenic bacteria that can cause human infections. While N. meningitidis infections are associated with bacterial meningitis and bacteremia, a strain of N. meningitidis, isolated from the urogenital system, has recently been associated with urethritis. As this strain is becoming prominent as an emerging pathogen, it is essential to assess identification tools for N. meningitidis and N. gonorrhoeae urogenital isolates. Consecutive N. meningitidis isolates recovered from urogenital cultures of symptomatic patients with presumptive diagnoses of gonorrhea and a random selection of N. gonorrhoeae isolates recovered from the same population within the same time frame were characterized with routine identification systems, antimicrobial susceptibility testing, and whole-genome sequencing. Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), multilocus sequence typing, 16S rRNA gene sequence, and average nucleotide identity methods accurately identified 95% (18/19) of N. meningitidis and N. gonorrhoeae isolates. With the Aptima Combo 2 CT/NG test, 30% (3/10) of N. meningitidis isolates were misidentified as N. gonorrhoeae, but no misidentifications were found with the Xpert CT/NG nucleic acid amplification test (NAAT). Phylogenetic core genome and single nucleotide polymorphism (SNP)-based grouping analyses showed that urogenital N. meningitidis isolates were highly related and phylogenetically distinct from N. gonorrhoeae and respiratory N. meningitidis isolates but similar to urogenital N. meningitidis isolates from patients with urethritis in the United States. Urogenital N. meningitidis isolates were predominantly azithromycin resistant, while N. gonorrhoeae isolates were azithromycin susceptible. These data indicate that urogenital isolates of N. meningitidis can cause false-positive detections with N. gonorrhoeae diagnostic assays. Misidentification of urogenital N. meningitidis isolates may confound public health-related activities for gonorrhea, and future studies are needed to understand the impact on clinical outcome of N. meningitidis urogenital infection.
INTRODUCTION
Neisseria gonorrhoeae and Neisseria meningitidis are human bacterial pathogens that can occupy different niches in the body (1, 2). N. gonorrhoeae is the causative agent of the sexually transmitted infection gonorrhea, which impacts 78 million people worldwide (3). As an obligate pathogen, N. gonorrhoeae primarily colonizes the genital mucosa and has evolved virulence factors that allow it to survive and evade the host immune system (1). Recently, N. gonorrhoeae has received increased public health attention, and drug-resistant N. gonorrhoeae has been categorized as an urgent threat by the U.S. Centers for Disease Control and Prevention due to resistance to commonly used antibiotics, limiting treatment in patients (4–6). Molecular point-of-care and sample-to-answer assays have been developed to rapidly and accurately identify the presence of N. gonorrhoeae in clinical specimens (7). While the development of these assays is important for patient treatment, there is evidence for sporadic false-positive molecular results due to cross-reactivity between Neisseria species (8).
N. meningitidis is found as a commensal in the respiratory system, and ∼10% of healthy adults and 40% of men who have sex with men (MSM) demonstrate naso/oropharyngeal carriage (2). N. meningitidis is also a leading cause of bacterial meningitis and causes significant morbidity and mortality in children and young adults, with an estimated 1.2 million cases of meningococcal infection per year worldwide (9). The virulence of N. meningitidis is determined by host factors (i.e., complement deficiency) and several virulence genes that facilitate adherence and survival in the respiratory system and invasion of the bloodstream (9).
The genus Neisseria has evolved mechanisms that result in a high frequency of horizontal gene transfer (HGT), both within and between species, with up to 10% of the N. meningitidis genome made up of mobile genetic elements (9, 10). Colocalization of N. meningitidis and N. gonorrhoeae in the urogenital system may result in increased transfer of virulence or antibiotic resistance genes (11). Recent studies have identified a strain of N. meningitidis that has been isolated from the urogenital system and is associated with urethritis (12–16). As this strain becomes a more prominent emerging pathogen in areas with high N. gonorrhoeae infection rates, it is essential to assess the ability of identification tools to discriminate between N. meningitidis and N. gonorrhoeae urogential isolates (13, 14, 17).
Following the implementation of a total laboratory automation system for culture-based microbiology (BD Kiestra total laboratory automation (TLA) system; Beckton Dickinson), the clinical microbiology laboratory at Barnes Jewish Hospital in St. Louis, MO, observed significant increases in the recovery of N. gonorrhoeae and, more recently, N. meningitidis incidentally from urine specimens submitted for routine culture-based testing (18). In this study, we characterize consecutively recovered N. meningitidis and compare them to N. meningitidis urethritis and invasive strains reported elsewhere.
MATERIALS AND METHODS
Clinical isolates and human study approval.
Consecutive Neisseria meningitidis and a random selection of Neisseria gonorrhoeae isolates recovered from March 2018 to March 2019 from clinical specimens submitted for routine testing to the Barnes Jewish Hospital Clinical Microbiology Laboratory in St. Louis, MO, were included in this study. Previous studies from our laboratory found increased recovery of N. gonorrhoeae and N. meningitidis isolates incidentally from urine cultures submitted for routine testing when incubated with the Kiestra total laboratory automation (TLA) (18). Study isolates were deidentified, but patient age, gender, and isolate source were documented. This study was approved by the Human Research Protection Office of Washington University School of Medicine.
Laboratory characterization.
Frozen N. meningitidis and N. gonorrhoeae isolates were subcultured to chocolate (CHC) agar (Hardy Diagnostics, Santa Maria, CA), incubated at 35°C and 5% CO2, and passaged twice prior to additional testing. For phenotypic characterization, 10 µl of a 0.5 McFarland (McF) suspension of each isolate was cross-struck to CHC and modified Thayer-Martin (MTM) (Hardy Diagnostics, Santa Maria, CA) to achieve less subjective interpretation and was quadrant struck to a third CHC plate with 10 µg colistin disk (BD BBL, Sparks, MD). Following incubation at 35°C and 5% CO2 for 18 to 20 h, CFU were enumerated and the colistin zone size to the nearest millimeter was recorded. For biochemical characterization, the RapidID NH system (Remel, Lenexa, KS) was used per the manufacturer’s instructions. Briefly, biochemical strips were inoculated with 3 McF suspensions of each isolate and incubated at 35°C for 4 h in air. Following incubation, biochemical reactions were read and scored, and microcodes were interpreted using the ERIC system to obtain organism identifications. For molecular characterization with matrix-assisted-laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), 2 commercially available systems were utilized, Bruker BioTyper (Bruker, Billerica, MA) and Vitek MS (bioMérieux, Durham, NC). Briefly, single colonies of pure growth were spotted to target slides and overlaid with matrix prior to analysis on each instrument per the manufacturer’s instructions. For N. meningitidis isolates, target slides were spotted with organism and matrix and fully dried inside a biosafety cabinet (BSC) before removal for loading onto the MALDI-TOF MS instrument. For analysis with commercial in vitro diagnostic (IVD) nucleic acid amplification tests (NAATs), both contrived swab and urine specimens of N. meningitidis and N. gonorrhoeae isolates were tested to confirm the lack of matrix-specific effects. Swabs from Aptima vaginal and Xpert vaginal/endocervical specimen collection kits were inserted into a 0.5-McF suspension of each isolate for 10 s, or 0.5-McF isolate suspensions were diluted 10-fold with remnant urine specimens previously determined to be negative for Chlamydia trachomatis and Neisseria gonorrhoeae. Both contrived swab and urine specimens were tested with the Aptima Combo 2 CT/NG (A-CT/NG) on the Tigris GTS system (Hologic, Inc., San Diego, CA) and the Xpert CT/NG (X-CT/NG) assay on the GeneXpert Infinity system (Cepheid, Sunnyvale, CA) per the manufacturer’s instructions.
Antimicrobial susceptibility testing.
Antimicrobial susceptibility testing (AST) of N. meningitidis and N. gonorrhoeae isolates was performed using disk diffusion and gradient diffusion strips, and the MIC and/or disk diffusion zone size was interpreted according to the Clinical and Laboratory Standards Institute (CLSI) M100, 29th edition (19). For N. meningitidis isolates, 0.5 McF suspensions of test isolates were inoculated to Mueller-Hinton Agar with 5% sheep blood and incubated with penicillin, azithromycin, ceftriaxone ETESTs (bioMérieux, Durham, NC) and ciprofloxacin (5 µg), rifampin (5 µg), minocycline (30 µg) and trimethoprim-sulfamethoxazole disks (1.25/23.75 µg) (BD BBL™, Sparks, MD) at 35°C and 5% CO2, for 20–24 h. For N. gonorrhoeae isolates, 0.5-McF suspensions of test isolates were inoculated to GC agar base with 1% defined growth supplement and incubated with penicillin, azithromycin, ceftriaxone Etests (bioMérieux, Durham, NC), and ciprofloxacin (5 µg) and rifampin (5 µg) disks (BD BBL, Sparks, MD) at 35°C and 5% CO2, for 20 to 24 h. MIC doubling dilutions and zone sizes to the nearest millimeter were read with reflected light and interpreted per CLSI M100, 29th edition, guidelines (19). AST categorical results were visualized using pheatmap (R) with color strips used to indicate the source of the isolate, species, and SNP pairwise distance-based grouping.
Whole-genome sequencing.
Total genomic DNA was extracted from cell cultures suspended in 1 ml of deionized water using the QIAamp BiOstic bacteremia DNA kit (Qiagen, Germantown, MD, USA). We quantified the DNA concentration using Qubit double-stranded DNA (dsDNA) assays (Thermo Fisher Scientific). Illumina sequencing libraries were prepared using 5 ng/μl of isolate DNA in a modified Nextera kit protocol (Illumina, San Diego, CA, USA). We then pooled and sequenced libraries on a NextSeq high-output platform (Illumina) to obtain ∼2 million 2 × 150-bp reads. The reads were demultiplexed by barcode, adapters were removed with Trimmomatic v.36, and contaminating human sequences were removed with Deconseq v.4.3 (20, 21). We assembled processed reads into draft genomes using the de novo assembler SPAdes v3.11 (22). The quality of draft genomes was assessed using QUAST v.4.5 and CheckM (23, 24). Assembles were considered to have passed quality control when the assembly length represented in contigs of <1 kb was less than 10%, the number of contigs greater than 500 bp was less than 5,000, completeness was greater than 90%, and contaminated reads were less than 5%. Draft genomes were annotated using Prokka v.1.12 (25).
Genomic taxonomic identification.
Following draft genome assembly, we determined genomic taxonomic identification by average nucleotide identity (ANI), 16S rRNA gene identification, and multilocus sequence typing (MLST). The assembled scaffolds were submitted to the Neisseria multilocus sequence typing website (https://pubmlst.org/neisseria/) to determine MLST and clonal complex (26). For all isolates, 16S rRNA gene sequences were identified using RNAmmer v.1.2 and submitted to the EZ BIoCloud taxonomic database for classification (27, 28). Using ANI analysis, species were determined if the genome in question had >95% ANI with the type genome (N. meningitidis, NM_MC58; N. gonorrhoeae, NG_ref_FA_1090) using dnadiff (29). Pairwise ANI for each isolate was clustered and visualized using pheatmap (R) (30).
Phylogenetic analysis.
To phylogenetically compare isolate sequences, 16S rRNA gene sequences identified by RNAammer were aligned using MUSCLE, and an approximate maximum likelihood tree was built with FastTree (31, 32). FastTree uses a heuristic variant of neighbor joining to construct a rough topology, reduces the length of the tree using a mix of nearest-neighbor interchanges and subtree-prune-regraft moves, and improves the tree with maximum-likelihood rearrangements (32). Branch length precision was rounded to 0.0001 substitutions per site. The output Newick files were visualized and annotated with isolate source using ggtree (R) (33, 34). To compare isolate genomes, .gff files produced by Prokka were used to construct a core genome alignment with Roary v.3.8.0 for N. meningitidis (35). Roary alignments were used to create an approximate maximum likelihood tree with FastTree (32). The output Newick files were visualized and annotated with isolate source using ggtree (R) (33, 34).
Isolate groupings based on SNP pairwise distances.
Snippy v.4.3.8 was used to map forward and reserve reads for N. meningitidis isolates to the N. meningitidis MC58 type strain complete genome assembly (ID) and to call SNPs (36). To determine groupings, we compared pairwise SNP distances between each N. meningitidis isolate pair. Isolates were grouped into perfectly reciprocal groups at every pairwise distance cutoff between N. meningitidis isolates using igraph v.1.2.4.1 as described previously (37). Groupings were visualized with a SNP cutoff of 2,000.
Antibiotic resistance mutation identification and analysis.
Targeted analysis of acquired antibiotic resistance genes/mutations (ARGs) against β-lactams (blaTEM, penA, porA, ponA, mtrR), macrolides/lincosamides/streptogramins (23S rRNA, mtrR), and quinolones (gyrA, parC) was performed as a result of phenotypic AST findings using PointFinder (38). The presence/absence matrix of ARGs was visualized in pheatmap (R). Associated metadata were displayed as a color strip to represent bacterial isolate identification and Aptima CT results. We further validated PointFinder results for key resistance gene mutations using BLASTn with MUSCLE alignment and maximum likelihood tree visualization (see Fig. S3 in the supplemental material).
Data availability.
All assemblies are uploaded to the NCBI under BioProject PRJNA643774.
RESULTS
N. meningitidis urinary isolates can cause false-positive N. gonorrhoeae molecular test result.
Consecutive N. meningitidis isolates and a random selection of N. gonorrhoeae isolates recovered from clinical specimens during the same time period were characterized by phenotypic and molecular methods routinely used to identify Neisseria species in clinical microbiology laboratories, including MALDI-TOF MS and commercial biochemical and molecular tests. Detailed demographic information was not available for these isolates, but limited information including patient age and gender, and isolate source is summarized in Table 1. All urogenital N. meningitidis and N. gonorrhoeae isolates were correctly identified using the Bruker Biotyper and Vitek MS MALDI-TOF MS platforms (Table 1). One Neisseria isolate from a respiratory source (NM12) was incorrectly identified using MALDI-TOF MS and biochemical tests as N. meningitidis (Bruker Biotyper) or Neisseria polysaccharea/N. meningitidis (Vitek MS) and N. gonorrhoeae (RapidID NH). However, this isolate was phenotypically consistent with nonpathogenic Neisseria species with no growth on MTM medium and a zone of inhibition when incubated with a 10-µg colistin disk on solid medium and was identified using whole-genome sequencing (WGS) methods as N. polysaccharea.
TABLE 1.
Neisseria meningitidis and Neisseria gonorrhoeae isolate characterizationa
| Isolate | Age (yrs), gender | Source | Serogroupb | Results from: |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Colistin (10 μg) disk, zone size (mm) | RapID NH | VITEK MS | Bruker Biotyper | Aptima CT/NG | Xpert CT/NG | 16S rRNA ID | MLST CC | ||||
| NM02 | 43, M | Urine | Nongroupable | R, 6 | Nm | Nm | Nm | Neg | Neg | Nm | ST-11 |
| NM03 | 31, M | Urine | ND | R, 6 | Nm | Nm | Nm | Neg | Neg | Nm | No type |
| NM04 | 27, M | Urine | Nongroupable | R, 6 | Nm | Nm | Nm | Ng Posc | Negc | Nm | ST-11 |
| NM05 | 18, F | Urine | ND | R, 6 | Nm | Nm | Nm | Neg | Neg | Nm | ST-11 |
| NM06 | 31, M | Urine | W135 | R, 6 | Nm | Nm | Nm | Neg | Neg | Nm | ST-11 |
| NM07 | 43, M | Urine | Nongroupable | R, 6 | Nm | Nm | Nm | Ng Posc | Negc | Nm | ST-11 |
| NM08 | 23, M | Urine | Nongroupable | R, 6 | Nm | Nm | Nm | Ng Posc | Negc | Nm | ST-11 |
| NM09 | 22, M | Urine | Nongroupable | R, 6 | Mo | Nm | Nm | Negc | Negc | Nm | ST-32 |
| NM10 | 29, M | Urine | ND | R, 6 | Nm | Nm | Nm | Neg | Neg | Nm | ST-11 |
| NM11 | 46, M | Respiratory, Sp | Nongroupable | R, 6 | Nm | Nm | Nm | Neg | Neg | Nm | No type |
| NM12 | 8, F | Respiratory, TA | NA | S, 20 | Ng | Nm/Np | Nm | Negc | Negc | Np | No type |
| NM13 | 21, F | Respiratory, BAL | Nongroupable | R, 6 | Nm | Nm | Nm | Neg | Neg | Nm | ST-11 |
| NM14 | 71, M | Respiratory, BW | B | R, 6 | Mo | Nm | Nm | Negc | Negc | Nm | ST-11 |
| NM15 | 32, M | Genital | Nongroupable | R, 6 | Nm | Nm | Nm | Neg | Neg | Nm | ST-4821 |
| NG ATCC 49226 | NA | ND | R, 6 | Ng | Ng | Ng | Ng Pos | Ng Pos | Ng | No type | |
| NG01 | 35, M | Urine | ND | R, 6 | Ng | Ng | Ng | Ng Pos | Ng Pos | Ng | No type |
| NG02 | 34, M | Urine | ND | R, 6 | Ng | Ng | Ng | Ng Pos | Ng Pos | Ng | No type |
| NG03 | 31, M | Urine | ND | R 6 | Ng | Ng | Ng | Ng Pos | Ng Pos | Ng | No type |
| NG04 | 29, M | Urine | ND | R, 6 | Ng | Ng | Ng | Ng Pos | Ng Pos | Ng | No type |
| NG05 | 23, M | Urine | ND | R, 6 | Ng | Ng | Ng | Ng Pos | Ng Pos | Ng | No type |
Abbreviations: M, male; F, female; Sp, sputum; TA, tracheal aspirate; BAL, bronchoalveolar lavage; BW, bronchial wash; Nm, Neisseria meningitis; Ng, Neisseria gonorrhoeae; Np, Neisseria polysaccharea; Mo, Moraxella osloensis; ND, not done; NA, not applicable; R, resistant; S, susceptible; Pos, positive; Neg, negative for all targets.
Serogroup determination or confirmatory testing performed by Missouri State Public health Laboratory (MSPHL) Jefferson City, MO.
Confirmed by repeat testing in remnant urine matrix.
Organism suspensions of each isolate were also tested with the Aptima CT/NG Combo 2 assay (A-CT/NG) on the Tigris DTS system and Xpert CT/NG (X-CT/NG) on the GeneXpert system. All N. gonorrhoeae isolates were detected by both systems, while all N. meningitidis isolates were not detected by X-CT/NG. Importantly, urinary isolates of N. meningitidis (NM04, NM07, NM08) tested positive for N. gonorrhoeae with A-CT/NG. This result was confirmed with remnant urine specimens spiked with NM04, NM07, NM08, NM09, NM12, and NM14 (Table 1).
Urogenital N. meningitidis classified as N. meningitidis by MLST, 16S rRNA gene classification, and average nucleotide identity.
We performed Illumina whole-genome sequencing (WGS) on all Saint Louis, MO (STL)-collected isolates. After draft genome assembly, scaffolds were submitted to the Neisseria MLST website (https://pubmlst.org/neisseria/) to determine MLST and clonal complex (26). For 18 of 19 Neisseria isolates, MLST species classification agreed with MALDI-TOF MS classification. One respiratory isolate (NM12), which MALDI-TOF MS was unable to classify to a single species, was characterized as N. polysaccharea. MLST clonal complex indicated that 7 of 10 STL-collected urogenital N. meningitidis isolates fell into the sequence type 11 (ST-11) clonal complex (Table 1).
To determine the phylogenetic context of isolates, we downloaded the following series of N. meningitidis genomes from NCBI and PubMLST: 28 N. meningitidis isolates from urinary tract infections (UTIs) in the United States (13, 14), 3 N. meningitidis ST-11 isolates from cases of meningitis in MSM in the United States (39), 29 N. meningitidis ST-11 isolates from a meningitis epidemic in Africa (40), and 8 N. meningitidis isolates from non-ST-11 meningitis cases (Table S1).
Ribosomal RNA (rRNA) classification is used in the Aptima Combo 2 assay, with the specific loci being proprietary (41), and in 16S rRNA gene sequence classification to determine the bacterial operational taxonomic unit (OTU) or amplicon sequence variant (ASV) (28). Thus, we classified and compared 16S rRNA gene sequences across Neisseria isolates. For all STL-collected isolates, 16S rRNA gene sequences were submitted to the EZ BIoCloud taxonomic database for classification (28). 16S rRNA gene classification correlated with MLST for all isolates (Table 1). An approximate maximum likelihood tree with NM12 as the outgroup shows N. gonorrhoeae sequences form a monophyletic clade distinct from N. meningitidis sequences with N. meningitidis that tested positive for N. gonorrhoeae using the A-CT/NG falling within the N. meningitidis sequences (Fig. S1).
Finally, we used ANI for genomic species classification. Species were determined if the genome in question had >95% pairwise ANI with the type genome (Fig. 1). All N. meningitidis or N. gonorrhoeae isolates from urogenital samples that were identified by MALDI-TOF MS and MLST were also identified as N. meningitidis or N. gonorrhoeae, respectively, by ANI. NM12 did not fall above the cutoff for N. meningitidis, N. gonorrhoeae, or N. polysaccharea type strains. Pairwise ANI of all N. meningitidis isolates and select reference N. meningitidis genomes indicated that ST-11 isolates form a distinct cluster with ANI above 99% (Fig. 1). This cluster included all urogenital N. meningitidis isolates for which N. gonorrhoeae was detected by A-CT/NG and one N. meningitidis respiratory isolate, NM13. Thus, MALDI-TOF MS, MLST, 16S rRNA, and ANI agree on classification for 18 of 19 Neisseria isolates.
FIG 1.
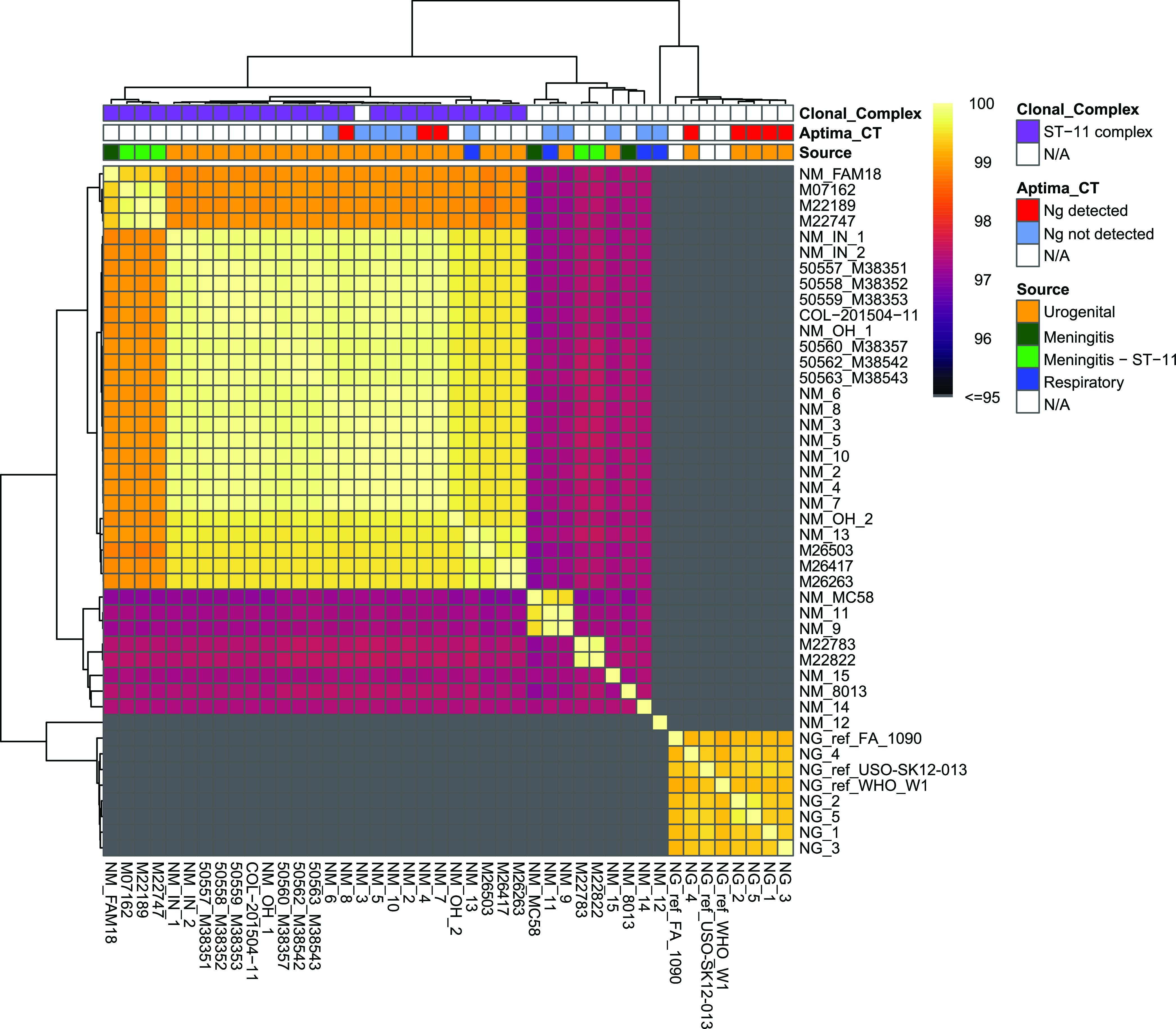
Identification of clinical N. meningitidis (Nm) and N. gonorrhoeae (Ng) by ANI analysis. All N. meningitidis and N. gonorrhoeae isolates from urogenital samples in St. Louis (STL) were characterized by pairwise ANI with all other isolates from STL and a subset of meningitis and ST-11 meningitis genomes. Isolates cluster within 96% ANI for all but one N. gonorrhoeae and N. meningitidis isolate. A single isolate (NM12) from a respiratory sample did not fall above a 96% cutoff for either N. meningitidis or N. gonorrhoeae type strain. Color strips indicate source of isolate, clonal complex identified my PubMLST, and Aptima test results.
N. meningitidis urogenital isolates form a primary lineage that is distinct from ST-11 meningitis isolates.
To determine genomic similarity across N. meningitidis genomes, we used a core genome alignment of 1,057 genes at 95% identity of all N. meningitidis isolates, using NM12 as an outgroup. The phylogenetic tree of this alignment shows that urogenital N. meningitidis isolates primarily fall within a single lineage (Fig. 2). All (3 of 3) STL-collected N. meningitidis isolates for which the A-CT/NG test detected N. gonorrhoeae formed a single clade within STL-collected ST-11 urogenital N. meningitidis isolates, suggesting a recent common ancestor. Of the STL-collected urogenital N. meningitidis isolates, 8/10 cluster together and form a sister clade to other urogenital N. meningitidis isolates. This similarity suggests a single common ancestor for 93% (31 of 33) of urogenital N. meningitidis isolates. Two urogenital STL-collected N. meningitidis isolates, NM09 and NM15, did not cluster with other urogenital isolates, and NM09 was, instead, highly related to a respiratory N. meningitidis isolate. Both NM09 and NM15 isolates were nongroupable using serotyping methods, did not fall into the ST-11 clonal complex, and were misclassified by RapID NH-; NM15 had been misclassified as N. gonorrhoeae and NM09 was misclassified as Moraxella osloensis (Table 1). ST-11 urethritis isolates were a sister clade to a lineage that included one STL-collected respiratory isolate and all 3 of the MSM meningitis isolates. This clade was a sister clade to the African ST-11 meningitis isolates. In contrast to N. meningitidis urinary isolates, N. meningitidis respiratory isolates were highly diverse and distantly related.
FIG 2.
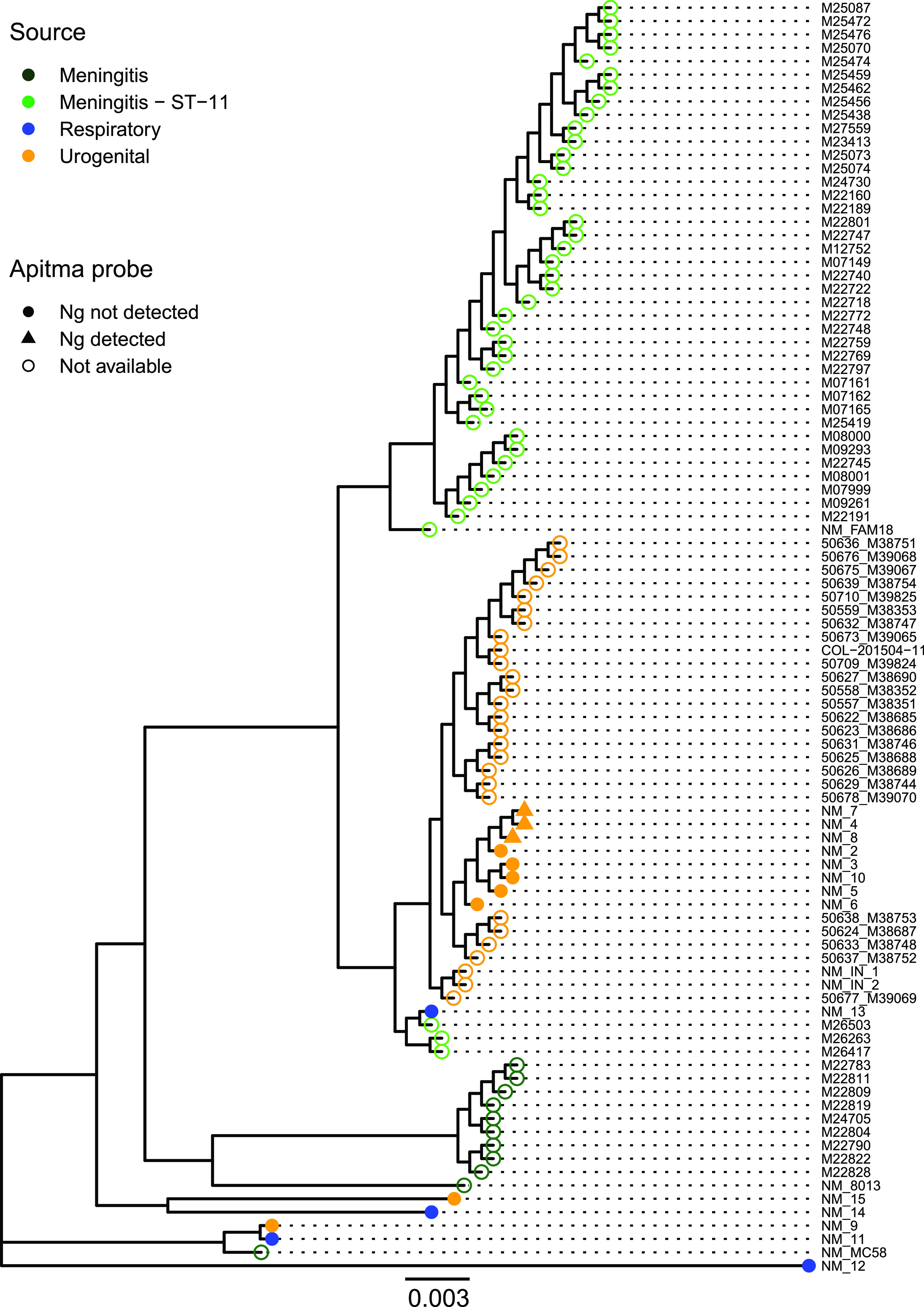
Urogenital N. meningitidis isolates primarily fall within a single, highly related clade. An approximate maximum likelihood tree of core genome alignment of St. Louis and select Neisseria isolates with tree branch lengths of >0.0001 is shown. Two urogenital isolates collected from St. Louis fall outside of this clade and are distantly related to the ST-11 clonal complex. All isolates that tested positive for N. gonorrhoeae fall into a clade with other urogenital N. meningitidis isolates. The source is indicated by color of the tip, and Aptima test results are indicated by the shape of tip. Ng, N. gonorrhoeae.
N. meningitidis urinary isolates are highly related to other urogenital isolates and not respiratory isolates.
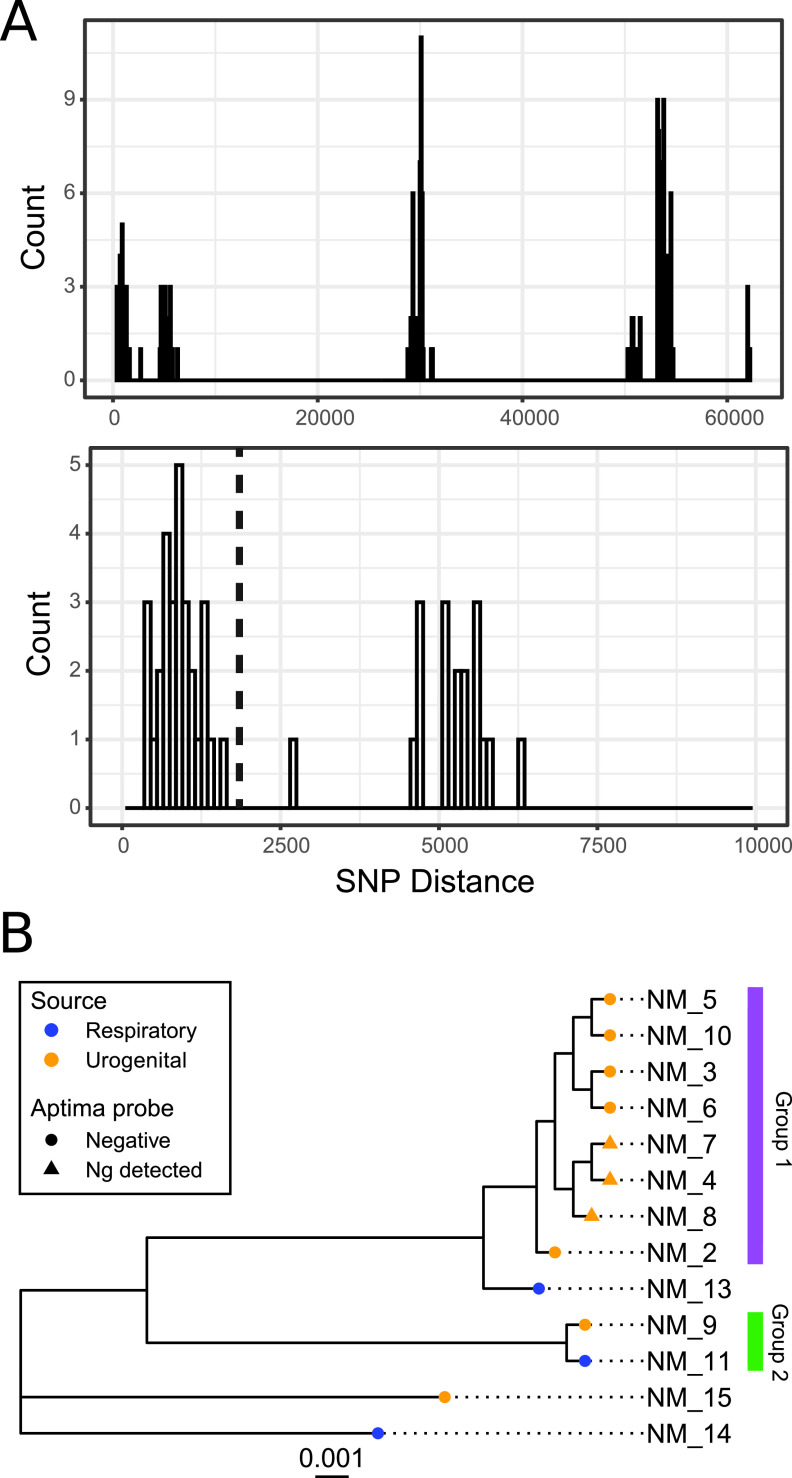
SNP distances across whole genomes have been found to provide higher resolution of phylogenetic distances than core genome comparisons (37). Thus, to further investigate the genomic similarity of STL-collected N. meningitidis isolates, we calculated pairwise SNP distances by mapping quality-filtered reads from N. meningitidis isolates to the N. meningitidis type strain. To find groupings, we used a grouping technique, “clique” (37), on STL-collected N. meningitidis isolates. We compared pairwise SNP distances between N. meningitidis isolate pairs and iterated through each unique SNP distance cutoff to filter the isolate pairwise network list (Fig. 3A). For each cutoff, we found reciprocal groups and recorded the number of groups and isolates per group. Then, groups were defined as complete subgraphs, where each node in the group was connected to every other node in the group. The number of N. meningitidis groups rose initially from 1 to 3 groups as SNP distances increased from 357 to 6,269. Only a single SNP distance of 5,624 SNPs had 4 groups, and immediately after this peak, groups decreased again to 3, with a decline in group size to 1 after 20,000 SNPs. Figure 3A shows the groups, which correspond to a SNP cutoff that includes only highly related N. meningitidis isolates with less than 2,000 SNP distances.
FIG 3.
Urogenital N. meningitidis isolates primarily form a single SNP pairwise distance-based grouping. (A) Histogram of pairwise SNP distances indicates three modes of pairwise distances. The first corresponds to within clade, the second to within species, and the third to between species. We define variant threshold as variant pairwise distances that fall before 2,000 (black line). (B) Groupings are visualized on the N. meningitidis core genome phylogeny. Group 1 includes 8 of 10 urogenital isolates. Group 2 includes a respiratory and a urogenital isolate. The source is indicated by color of the tip, and Aptima test results are indicated by the shape of the tip. Ng, N. gonorrhoeae.
Urogenital N. meningitidis isolates primarily formed a single grouping (Fig. 3B). The first grouping includes 8/10 urogenital isolates and all ST-11 urethritis isolates. This grouping fell entirely with the ST-11 urogenital clade described in the core genome phylogeny (Fig. 2). A second grouping included a respiratory (NM11) and a urogenital (NM09) isolate. These groupings suggest that while ST-11 urethritis isolates are highly related with between 9.5 and 9.6 × 10−4 pairwise SNPs/genome length, not all urogenital isolates fall into the grouping, and one isolate shares high similarity (4.8 × 10−4 pairwise SNPs/genome length) with a respiratory isolate.
N. meningitidis isolates have an antibiotic susceptibility profile distinct from N. gonorrhoeae isolates.
To consider clinical implications of misidentified N. meningitidis isolates, we performed phenotypic AST on all STL-collected isolates. AST was performed against azithromycin, penicillin, ceftriaxone, rifampin, ciprofloxacin, minocycline, trimethoprim-sulfamethoxazole, and colistin. AST profiles varied between N. gonorrhoeae isolates and urogenital N. meningitidis isolates (Fig. 4A). Most N. meningitidis isolates tested nonsusceptible to azithromycin with MIC50/90 of 4 µg/ml (range, 0.5 to 4 μg/ml) compared to N. gonorrhoeae isolates, which were mostly susceptible to azithromycin with MIC50 of 0.125 µg/ml and MIC90 of 2 µg/ml (range, 0.064 to 2 μg/ml). The respiratory isolate, NM012, has a unique AST profile distinct from both N. gonorrhoeae and N. meningitidis isolates in that it tested resistant to azithromycin, penicillin, ciprofloxacin, and trimethoprim-sulfamethoxazole. We also evaluated chromosomal point mutations that may account for antibiotic resistance in Neisseria both by using PointFinder and by individually validating mutations of known interest in specific genes (Table S2, Fig. S3). Point mutations for resistance were primarily shared by species (Fig. S2). One prominent point mutation in N. meningitidis isolates was in nonmosaic penA allele p.N512Y. This point mutation is associated with mosaic penA, which can contribute to decreased susceptibility to expanded-spectrum cephalosporins (42); however, all isolates in this study were ceftriaxone susceptible with MIC of ≤0.016 µg/ml. While all urogenital N. meningitidis isolates and 1/3 respiratory N. meningitidis isolates had this mutation, this mutation was not present in any N. gonorrhoeae isolate. Isolates within the same SNP pairwise distance-based grouping had identical resistance mutation profiles (Fig. S2 and S3, Table S2).
FIG 4.
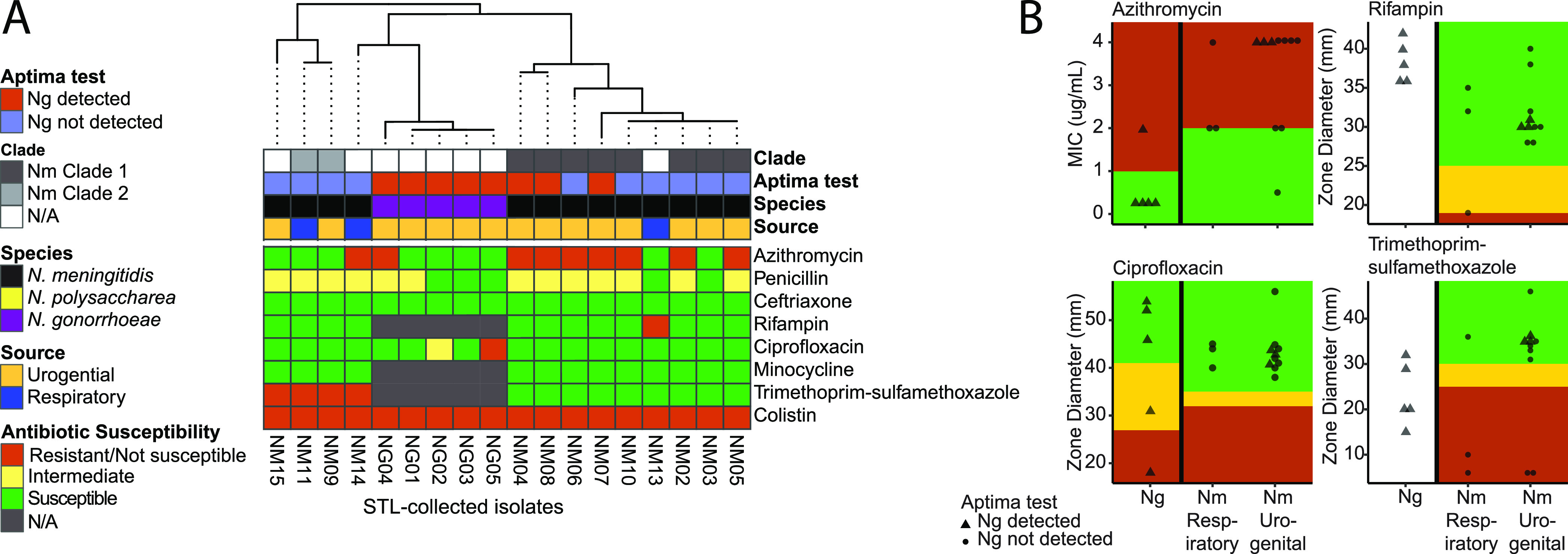
The antibiotic susceptibility profile varies between N. gonorrhoeae isolates and urogenital N. meningitidis isolates. (A) The heatmap of AST profiles for each isolate shows N. meningitidis and N. gonorrhoeae isolates organized by 16S phylogenetic gene tree, with major differences being that N. meningitidis isolates are resistant to azithromycin and intermediate to penicillin, while N. gonorrhoeae isolates are largely susceptible to both antibiotics. Isolate NM12 has a distinct antibiotic susceptibility profile that varies from both N. gonorrhoeae and N. meningitidis isolates. Color strips indicate the source of isolate, species identification by ANI, and SNP pairwise distance-based grouping. (B) Distributions of MIC and zone diameter for N. meningitidis urogenital isolates and N. gonorrhoeae respiratory isolates for azithromycin, rifampin, ciprofloxacin, and trimethoprim-sulfamethoxazole. Ng, N. gonorrhoeae; Nm, N. meningitidis.
DISCUSSION
As N. meningitidis becomes an increasingly recognized pathogen in the urogenital system, accurate species identification of N. meningitidis and N. gonorrhoeae urogenital isolates may be important for clinical care. Thus, it is essential to assess tools used for identification and compare N. meningitidis and N. gonorrhoeae urogenital isolates. In this study, we demonstrate that 30% (3/10) of urogenital N. meningitidis isolates were misidentified as N. gonorrhoeae with the A-CT/NG NAAT and that these urogenital N. meningitidis isolates were predominantly nonsusceptible to azithromycin. We found that specific identification using MALDI-TOF MS, MLST, 16S rRNA gene sequence, and ANI methods was 100% accurate for both urogenital N. meningitidis and N. gonorrhoeae isolates. However, our data indicate that some urogenital isolates of N. meningitidis can cause false-positive detections with N. gonorrhoeae-specific molecular tests and that some commensal Neisseria strains can be identified as N. meningitidis by MALDI-TOF MS.
While NAAT tests such as the Aptima CT/NG Combo 2 or the Xpert CT/NG are the standard of care for detection of N. gonorrhoeae from urine and genital specimens in clinical laboratories, there is evidence that other Neisseria species can cause false-positive N. gonorrhoeae detections (7, 8). A previous report suggested these false-positive results were sporadic and low level, as no isolate tested positive twice in their study (8). In contrast, our findings were not sporadic, as 3 unique urogenital N. meningitidis isolates were detected as N. gonorrhoeae both when tested as pure isolate suspensions in saline and when spiked into urine. The Xpert CT/NG NAAT has two N. gonorrhoeae-specific targets, both of which must be detected to return an N. gonorrhoeae-positive result, while the Aptima CT/NG Combo 2 NAAT targets a region of the 16S rRNA to detect N. gonorrhoeae. Since the exact locus of both the Aptima CT/NG Combo 2 and the Xpert CT/NG tests are proprietary, we are unable to directly test for sequence differences that may result in misidentifications. However, genomic characterization of these isolates demonstrated that N. meningitidis isolates that test falsely positive for N. gonorrhoeae form a distinct clade based on a core genome phylogeny, suggesting a common ancestor and indicating a genomic component rather than random chance or a sporadic error in the NAAT test is responsible for the false-positive N. gonorrhoeae result.
Accurate identification of Neisseria isolates is important, as AST profiles vary between N. gonorrhoeae and N. meningitidis isolates. Though rising rates of reduced susceptibly to azithromycin in N. gonorrhoeae have been reported across the United States (43), we found that the N. gonorrhoeae isolates tested in our study were primarily susceptible to azithromycin (n = 4/5), while urogenital N. meningitidis isolates collected over the same time period as N. gonorrhoeae isolates were more likely to be azithromycin nonsusceptible (n = 7/10). Our observation of largely azithromycin-susceptible N. gonorrhoeae is consistent with a previous study of a larger cohort of N. gonorrhoeae isolates recovered from the same institution that reported that azithromycin nonsusceptibility was rare (<2%) (44). Interestingly, all N. meningitidis isolates that tested falsely positive for N. gonorrhoeae were azithromycin nonsusceptible (n = 3/3). Currently, a single dose of intramuscular ceftriaxone (250 mg) plus a single dose of oral azithromycin (1 g) is the primary treatment recommendation for uncomplicated gonococcal infection (45). Although dual therapy was primarily aimed at treatment of uncomplicated chlamydial coinfections, routine combination therapy may hinder development of antimicrobial resistance in N. gonorrhoeae, particularly in light of increased cephalosporin resistance in the United States (43). Given the rare reports of ceftriaxone-resistant N. meningitidis (46–48), ceftriaxone plus azithromycin dual therapy is likely effective for treatment of urogenital N. meningitidis infection. However, if azithromycin nonsusceptibility is common among urogenital N. meningitidis isolates and in the setting of reduced susceptibility to 3rd-generation cephalosporins, current gonococcal treatment guidelines may be suboptimal for urogenital N. meningitidis infection. Interestingly, despite recent reports of ciprofloxacin-resistant, beta-lactamase-producing N. meningitidis serogroup Y (49), all of the N. meningitidis isolates evaluated in this study were ciprofloxacin and ceftriaxone susceptible (n = 13/13), though most were nonsusceptible to penicillin (n = 10/13); beta-lactamase testing was not performed.
One hypothesis for conflicting identification of N. meningitidis isolates may be increased HGT between N. gonorrhoeae and N. meningitidis. However, our 16S rRNA gene sequence, MLST, and whole-genome analyses indicate that urogenital N. meningitidis isolates are not more similar to N. gonorrhoeae than other N. meningitidis isolates. The majority of urogenital isolates for which we performed genomic analyses (n = 35/37) share a recent common ancestor. This suggests that most cases of N. meningitidis urethritis are due to the spread of urethritis-associated N. meningitidis ST-11 and not due to translocation and subsequent infection of respiratory or meningitis-associated N. meningitidis isolates to the genitourinary tract, consistent with previous reports (11, 15). However, we did observe two instances where urogenital N. meningitidis isolates were not from the ST-11 urethritis clade, and in one case, a respiratory N. meningitidis isolate was highly related to a urinary N. meningitidis isolate. This suggests urogenital N. meningitidis isolates do not derive exclusively from the ST-11 urethritis clade and that transmission between body sites may be possible.
Studies have indicated that invasive N. meningitidis isolates from MSM are associated with colonization of the urethra or rectum (2) and that N. meningitidis urethritis outbreaks are closely related to cases of invasive N. meningitidis in MSM populations, suggesting that urethral colonization may contribute to invasive disease (11). In our study, the sister clade to all ST-11 urethritis isolates included one respiratory isolate and three meningitis-associated isolates from MSM patients. This phylogeny suggests a common ancestor between MSM meningitidis and the origin of urogenital N. meningitidis isolates. However, as this study is focused on urogenital N. meningitidis isolates, only a subset of 54 N. meningitidis meningitis isolates was used for comparison, with a focus on N. meningitidis isolates within ST-11. A more exhaustive study of N. meningitidis meningitis isolates may find additional clades related to the urogenital NM isolates. Further, in this data set, we do not see evidence for closely related urethritis and invasive N. meningitidis isolates. It is possible that increasing the collection and analysis of invasive and urogenital N. meningitidis isolates from meningitis patients may expand further on this issue.
Public health-related activities associated with gonococcal infection, such as contact-tracing and expedited partner therapy, may be indicated and initiated following notification of this reportable infection. However, misidentification of N. meningitidis can confound these activities, particularly if identification of N. gonorrhoeae and/or N. meningitidis is inconsistent across currently available diagnostic tests. Increased vigilance surrounding these (mis)identifications will be required for a more complete understanding of the scope, epidemiology, susceptibility, and clinical outcomes associated with N. meningitidis urogenital infections, as has been previously suggested (50).
Overall, our findings demonstrate that some urogenital N. meningitidis isolates are incorrectly identified as N. gonorrhoeae by the Aptima CT/NG NAAT despite being correctly identified as N. meningitidis by other molecular methods, including MALDI-TOF MS, MLST, 16S rRNA gene sequencing, and ANI analysis. Further, these urogenital N. meningitidis isolates are related to the ST-11 urethritis-associated N. meningitidis isolates from across the United States. These isolates may be recovered from routine urine cultures with increasing frequency, as laboratories transition to automated inoculation and incubation systems. Finally, our studies have found that accurate identification of N. meningitidis and N. gonorrhoeae may be important due to implications for public-health related activities and potential differences in susceptibility profiles. Future studies further describing the scope, epidemiology, clinical course, and outcomes of N. meningitidis-mediated urogenital infection compared to gonococcal infection will be needed to justify strategies to identify and/or differentiate N. meningitidis from N. gonorrhoeae urogenital specimens.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by awards to G.D. through the National Institute of Allergy and Infectious Diseases and the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health (NIH) under award numbers R01AI123394 and R01HD092414, respectively. K.V.S. is supported by the Society for Healthcare Epidemiology of America Research Scholar Award. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
We thank the Edison Family Center for Genome Sciences & Systems Biology at WUSM staff, Eric Martin, Brian Koebbe, Jessica Hoisington-López, and MariaLynn Crosby for technical support in high-throughput sequencing and computing.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Quillin SJ, Seifert HS. 2018. Neisseria gonorrhoeae host adaptation and pathogenesis. Nat Rev Microbiol 16:226–240. doi: 10.1038/nrmicro.2017.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janda WM, Bohnoff M, Morello JA, Lerner SA. 1980. Prevalence and site-pathogen studies of Neisseria meningitidis and N gonorrhoeae in homosexual men. JAMA 244:2060–2064. doi: 10.1001/jama.1980.03310180026026. [DOI] [PubMed] [Google Scholar]
- 3.Newman L, Rowley J, Vander Hoorn S, Wijesooriya NS, Unemo M, Low N, Stevens G, Gottlieb S, Kiarie J, Temmerman M. 2015. Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS One 10:e0143304. doi: 10.1371/journal.pone.0143304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fifer H, Natarajan U, Jones L, Alexander S, Hughes G, Golparian D, Unemo M. 2016. Failure of dual antimicrobial therapy in treatment of gonorrhea. N Engl J Med 374:2504–2506. doi: 10.1056/NEJMc1512757. [DOI] [PubMed] [Google Scholar]
- 5.Eyre DW, Town K, Street T, Barker L, Sanderson N, Cole MJ, Mohammed H, Pitt R, Gobin M, Irish C, Gardiner D, Sedgwick J, Beck C, Saunders J, Turbitt D, Cook C, Phin N, Nathan B, Horner P, Fifer H. 2019. Detection in the United Kingdom of the Neisseria gonorrhoeae FC428 clone, with ceftriaxone resistance and intermediate resistance to azithromycin, October to December 2018. Euro Surveill 24:1900147. https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2019.24.10.1900147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CDC. 2019. Antibiotic resistance threats in the United States, 2019. U.S. Department of Health and Human Services, CDC, Atlanta, GA. https://www.cdc.gov/drugresistance/Biggest-Threats.html. [Google Scholar]
- 7.Tabrizi SN, Unemo M, Golparian D, Twin J, Limnios AE, Lahra M, Guy R, TTANGO Investigators. 2013. Analytical evaluation of GeneXpert CT/NG, the first genetic point-of-care assay for simultaneous detection of Neisseria gonorrhoeae and Chlamydia trachomatis. J Clin Microbiol 51:1945–1947. doi: 10.1128/JCM.00806-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tabrizi SN, Unemo M, Limnios AE, Hogan TR, Hjelmevoll SO, Garland SM, Tapsall J. 2011. Evaluation of six commercial nucleic acid amplification tests for detection of Neisseria gonorrhoeae and other Neisseria species. J Clin Microbiol 49:3610–3615. doi: 10.1128/JCM.01217-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rouphael NG, Stephens DS. 2012. Neisseria meningitidis: biology, microbiology, and epidemiology. Methods Mol Biol 799:1–20. doi: 10.1007/978-1-61779-346-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marri PR, Paniscus M, Weyand NJ, Rendon MA, Calton CM, Hernandez DR, Higashi DL, Sodergren E, Weinstock GM, Rounsley SD, So M. 2010. Genome sequencing reveals widespread virulence gene exchange among human Neisseria species. PLoS One 5:e11835. doi: 10.1371/journal.pone.0011835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Retchless AC, Kretz CB, Chang HY, Bazan JA, Abrams AJ, Norris Turner A, Jenkins LT, Trees DL, Tzeng YL, Stephens DS, MacNeil JR, Wang X. 2018. Expansion of a urethritis-associated Neisseria meningitidis clade in the United States with concurrent acquisition of N. gonorrhoeae alleles. BMC Genomics 19:176. doi: 10.1186/s12864-018-4560-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelley GA, Kelley KS. 2018. Systematic reviews and cancer research: a suggested stepwise approach. BMC Cancer 18:246. doi: 10.1186/s12885-018-4163-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bazan JA, Turner AN, Kirkcaldy RD, Retchless AC, Kretz CB, Briere E, Tzeng YL, Stephens DS, Maierhofer C, Del Rio C, Abrams AJ, Trees DL, Ervin M, Licon DB, Fields KS, Roberts MW, Dennison A, Wang X. 2017. Large cluster of Neisseria meningitidis urethritis in Columbus, Ohio, 2015. Clin Infect Dis 65:92–99. doi: 10.1093/cid/cix215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toh E, Gangaiah D, Batteiger BE, Williams JA, Arno JN, Tai A, Batteiger TA, Nelson DE. 2017. Neisseria meningitidis ST11 complex isolates associated with nongonococcal urethritis, Indiana, USA, 2015–2016. Emerg Infect Dis 23:336–339. doi: 10.3201/eid2302.161434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma KC, Unemo M, Jeverica S, Kirkcaldy RD, Takahashi H, Ohnishi M, Grad YH. 2017. Genomic characterization of urethritis-associated Neisseria meningitidis shows that a wide range of N. meningitidis strains can cause urethritis. J Clin Microbiol 55:3374–3383. doi: 10.1128/JCM.01018-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jannic A, Mammeri H, Larcher L, Descamps V, Tosini W, Phung B, Yazdanpanah Y, Bouscarat F. 2019. Orogenital transmission of Neisseria meningitidis causing acute urethritis in men who have sex with men. Emerg Infect Dis 25:175–176. doi: 10.3201/eid2501.171102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.CDC. 2018. Sexually transmitted disease surveillance 2018: gonorrhea. cdc.gov/std/stats18/gonorrhea.htm.
- 18.Lainhart W, Burnham CA. 2018. Enhanced recovery of fastidious organisms from urine culture in the setting of total laboratory automation. J Clin Microbiol 56 doi: 10.1128/JCM.00546-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute. 2019. Performance standards for antimicrobial susceptibility testing, 29th ed. CLSI, Wayne, PA. [Google Scholar]
- 20.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmieder R, Edwards R. 2011. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS One 6:e17288. doi: 10.1371/journal.pone.0017288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 26.Jolley KA, Bray JE, Maiden MCJ. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J. 2017. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolde R. 2019. pheatmap: pretty heatmaps. R package version 1012. https://cran.r-project.org/web/packages/pheatmap/pheatmap.pdf.
- 31.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2: approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu G, Smith DK, Zhu H, Guan Y, Lam TT‐Y. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 34.Yu G, Lam TT-Y, Zhu H, Guan Y. 2018. Two methods for mapping and visualizing associated data on phylogeny using ggtree. Mol Biol Evol 35:3041–3043. doi: 10.1093/molbev/msy194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seemann T. Snippy: rapid haploid variant calling and core genome alignment. https://github.com/tseemann/snippy.
- 37.D’Souza AW, Potter RF, Wallace M, Shupe A, Patel S, Sun X, Gul D, Kwon JH, Andleeb S, Burnham CD, Dantas G. 2019. Spatiotemporal dynamics of multidrug resistant bacteria on intensive care unit surfaces. Nat Commun 10:4569. doi: 10.1038/s41467-019-12563-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zankari E, Allesoe R, Joensen KG, Cavaco LM, Lund O, Aarestrup FM. 2017. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother 72:2764–2768. doi: 10.1093/jac/dkx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Folaranmi TA, Kretz CB, Kamiya H, MacNeil JR, Whaley MJ, Blain A, Antwi M, Dorsinville M, Pacilli M, Smith S, Civen R, Ngo V, Winter K, Harriman K, Wang X, Bowen VB, Patel M, Martin S, Misegades L, Meyer SA. 2017. Increased risk for meningococcal disease among men who have sex with men in the United States, 2012–2015. Clin Infect Dis 65:756–763. doi: 10.1093/cid/cix438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Retchless AC, Hu F, Ouedraogo AS, Diarra S, Knipe K, Sheth M, Rowe LA, Sangare L, Ky Ba A, Ouangraoua S, Batra D, Novak RT, Ouedraogo Traore R, Wang X. 2016. The establishment and diversification of epidemic-associated serogroup W meningococcus in the African meningitis belt, 1994 to 2012. mSphere 1:e00201-16. doi: 10.1128/mSphere.00201-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hologic. 2020. Aptima Combo 2 assay. 501798 Rev. 007. https://www.hologic.com/sites/default/files/2020-11/501798-IFU-PI_007_01.pdf. Accessed 30 November 2020.
- 42.Tomberg J, Unemo M, Davies C, Nicholas RA. 2010. Molecular and structural analysis of mosaic variants of penicillin-binding protein 2 conferring decreased susceptibility to expanded-spectrum cephalosporins in Neisseria gonorrhoeae: role of epistatic mutations. Biochemistry 49:8062–8070. doi: 10.1021/bi101167x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirkcaldy R, Harvey A, Papp J, Del Rio C, Soge O, Holmes K, Hook E, Kubin G, Riedel S, Zenilman J, Pettus K, Sanders T, Sharpe S, Torrone E. 2016. Neisseria gonorrhoeae antimicrobial susceptibility surveillance: The Gonococcal Isolate Surveillance Project, 27 Sites, United States, 2014. MMWR Surveill Summ 65:1–19. doi: 10.15585/mmwr.ss6507a1. [DOI] [PubMed] [Google Scholar]
- 44.Bailey A, Potter R, Wallace M, Johnson C, Dantas G, Burnham C. 2019. Genotypic and phenotypic characterization of antimicrobial resistance in Neisseria gonorrhoeae: a cross-sectional study of isolates recovered from routine urine cultures in a high-incidence setting. mSphere 4:e00373-19. doi: 10.1128/mSphere.00373-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Workowski KA, Berman S, Centers for Disease Control and Prevention. 2010. Sexually transmitted diseases treatment guidelines, 2010. MMWR Recomm Rep 59:1–110. [PubMed] [Google Scholar]
- 46.Manchanda V, Bhalla P. 2006. Emergence of non-ceftriaxone-susceptible Neisseria meningitidis in India. J Clin Microbiol 44:4290–4291. doi: 10.1128/JCM.01903-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deghmane A, Hong E, Taha M. 2017. Emergence of meningococci with reduced susceptibility to third-generation cephalosporins. J Antimicrob Chemother 72:95–98. doi: 10.1093/jac/dkw400. [DOI] [PubMed] [Google Scholar]
- 48.Alemayehu T, Mekasha A, Abebe T. 2017. Nasal carriage rate and antibiotic susceptibility pattern of Neisseria meningitidis in healthy Ethiopian children and adolescents: a cross-sectional study. PLoS One 12:e0187207. doi: 10.1371/journal.pone.0187207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McNamara LA, Potts C, Blain AE, Retchless AC, Reese N, Swint S, Lonsway D, Karlsson M, Lunquest K, Sweitzer JJ, Wang X, Hariri S, Fox LM, Antimicrobial-Resistant Neisseria meningitidis T. 2020. Detection of ciprofloxacin-resistant, beta-lactamase-producing Neisseria meningitidis serogroup Y isolates: United States, 2019–2020. MMWR Morb Mortal Wkly Rep 69:735–739. doi: 10.15585/mmwr.mm6924a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brooks A, Lucidarme J, Campbell H, Campbell L, Fifer H, Gray S, Hughes G, Lekshmi A, Schembri G, Rayment M, Ladhani S, Ramsay M, Borrow R. 2020. Detection of the United States Neisseria meningitidis urethritis clade in the United Kingdom, August and December 2019: emergence of multiple antibiotic resistance calls for vigilance. Euro Surveill 25:2000375. https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2020.25.15.2000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All assemblies are uploaded to the NCBI under BioProject PRJNA643774.