Acanthamoeba is a free-living amoeba of extensive genetic diversity. It may cause infectious keratitis (IK), which can also be caused by bacteria, fungi, and viruses.
KEYWORDS: Denmark, keratitis, NGS, ocular disease, microbiome, parasite infection
ABSTRACT
Acanthamoeba is a free-living amoeba of extensive genetic diversity. It may cause infectious keratitis (IK), which can also be caused by bacteria, fungi, and viruses. High diagnostic sensitivity is essential to establish an early diagnosis of Acanthamoeba-associated keratitis. Here, we investigated the applicability of next-generation sequencing (NGS)-based ribosomal gene detection and differentiation (16S-18S) compared with specific real-time PCR for the detection of Acanthamoeba. Two hundred DNAs extracted from corneal scrapings and screened by Acanthamoeba-specific real-time PCR were analyzed using an in-house 16S-18S NGS assay. Of these, 24 were positive by specific real-time PCR, of which 21 were positive by the NGS assay. Compared with real-time PCR; the specificity and sensitivity of the NGS assay were 100% and 88%, respectively. Genotypes identified by the NGS assay included T4 (n = 19) and T6 (n = 2). Fungal and bacterial species of potential clinical relevance were identified in 31 of the samples negative for Acanthamoeba, exemplified by Pseudomonas aeruginosa (n = 11), Moraxella spp. (n = 6), Staphylococcus aureus (n = 2), Fusarium spp. (n = 4), and Candida albicans (n = 1). In conclusion, the 16S-18S assay was slightly less sensitive than real-time PCR in detecting Acanthamoeba-specific DNA in corneal scrapings. Robust information on genotypes was provided by the NGS assay, and other pathogens of potential clinical relevance were identified in 16% of the samples negative for Acanthamoeba. NGS-based detection of ribosomal genes in corneal scrapings could be an efficient screening method for detecting nonviral causes of IK, including Acanthamoeba.
INTRODUCTION
Acanthamoeba is a free-living amoeba found primarily in soil and water. The genus exhibits a high degree of genetic diversity, and to date, 20 genotypes have been identified (1). Acanthamoeba can cause infectious keratitis (IK), which can lead to blindness if left untreated (2). The overall disease burden of Acanthamoeba-associated keratitis (AK) remains low, with an estimated prevalence of 1 to 9 per 100,000 according to the Orphanet database (2). However, the incidence has increased dramatically since the first cases of AK were reported in the 1970s (3). This increase is most likely associated with increased clinical awareness, the development of sensitive tests, and an increase in the number of individuals exposed to risk factors.
Acanthamoeba-associated keratitis is often seen in contact lens wearers (4, 5). A recent study from Denmark, which included PCR-based diagnostic data on corneal scrapings tested at our laboratory, revealed that 89% of AK patients were contact lens users, and 49% and 54% had received corticosteroids and treatment for herpes, respectively, before the diagnosis was established (6). In Denmark, an Acanthamoeba-positive test result is typically seen in unilateral cases of IK not responding to treatment with chloramphenicol, ciprofloxacin, and/or tobramycin (https://en.ssi.dk/news/epi-news/2016/no-44---2016).
In general, the incidence ranges from 17 to 70 cases per million individuals wearing contact lenses (3). Other events, such as corneal surgery, trauma, or exposure to contaminated water, have also been associated with AK.
An early diagnosis of AK is critical to ensuring a good prognosis (7). However, since IK can also be caused by bacteria, fungi, and viruses with a significant clinical overlap, establishing a diagnosis requires clinical expertise supported by specialized microbiological diagnostics, e.g., real-time PCR (RT-PCR), confocal microscopy, and/or culture using specialized media (8). This was illustrated in a German multicenter study in which 172 cases of AK were reported. Only 23% were initially diagnosed as AK. Several other causes were suggested: 48% were first attributed to herpes simplex virus, 25% were thought to be of bacterial origin, and 3% were wrongly attributed to fungi (9) While an early diagnosis is critical to the prognosis of AK, an average of 2.8 ± 4.0 months (range, 0 to 23 months) of delay between the onset of symptoms and the diagnosis has been reported (9).
Since IK can be caused by a variety of microorganisms and often reflects a polymicrobial coinfection including fungi and/or bacteria in addition to Acanthamoeba (10, 11), a strategy to address this situation could be to apply next-generation sequencing (NGS) of ribosomal genes in corneal scrapings in order to screen for all nonviral causes of IK in the early phase of the disease.
In this study, we used a recently developed amplicon-based sequencing assay targeting parasites, fungi, and bacteria based on analysis of nuclear ribosomal genes (16S and 18S rRNA) amplified from genomic DNA extracted directly from clinical corneal scrapings (12, 13). We compared the results obtained by a well-established real-time PCR for Acanthamoeba with the detection and differentiation of nuclear ribosomal genes present in corneal scrapings from patients with keratitis in order to (i) evaluate the sensitivity of the 16S-18S assay compared with real-time PCR, (ii) evaluate the robustness of genotyping based on 16S-18S analysis, and (iii) explore the potential of comprehensive 16S-18S analysis in terms of simultaneous detection of clinically relevant fungi, parasites, and bacteria as causative agents of keratitis.
MATERIALS AND METHODS
Sampling and reference data.
A total of 200 corneal scrapings collected in 2015 and 2016 were included in this study. These samples, from patients with suspected IK, had been referred for Acanthamoeba PCR at the Laboratory of Parasitology at the Statens Serum Institut, Copenhagen, Denmark. Clinicians are advised to take representative and sufficiently large samples in sterile saline water. Samples are usually processed within 24 h of sampling. DNAs had been extracted using the QIAamp DNA blood and tissue kit (Qiagen). The TaqMan-based real-time PCR assay for Acanthamoeba spp. in use in our routine diagnostic laboratory was a single-assay modification of the triplex assay previously described by Qvarnström et al. in 2006 (14). All samples had been tested in duplicate by real-time PCR. One water sample (nontemplate; used to test for contamination, including between samples) was included and “DNA-extracted” in each run (12 samples were extracted per run); these water samples were also included in the real-time PCR and NGS assays. Moreover, a water sample is included in the real-time PCR and in the NGS assay. In the real-time PCR, DNA from an Acanthamoeba culture (kindly provided by Marianne Lebbad, Public Health Agency of Sweden, Solna, Sweden) was included as a positive control. No positive controls were included in the NGS assay, since this is a microbe profiling assay and was not developed as a diagnostic assay. Both real-time PCR-positive (n = 24) and -negative (n = 176) samples were included in this study for validation of the 16S-18S assay. Two of the PCR-positive samples were weakly positive, which means that only one of the two duplicates yielded a signal, and this signal reflected a threshold cycle (CT) value of 40 or more.
16S-18S assay: NGS-based detection and differentiation of nuclear ribosomal genes.
Amplification of nuclear ribosomal genes was performed as described previously (13, 15, 16). Briefly, the three following primer sets were chosen for 18S rRNA genes: G3F1/G3R1 (GCCAGCAGCCGCGGTAATTC/ACATTCTTGGCAAATGCTTTCGCAG), G4F3/G4R3 (CAGCCGCGGTAATTCCAGCTC/GGTGGTGCCCTTCCGTCAAT), and G6F1/G6R1 (TGGAGGGCAAGTCTGGTGCC/ACGGTATCTGATCGTCTTCGATCCC). G3 and G6 primers target the V3-V4 hypervariable regions, and G4 targets the V3-V5 hypervariable region of the 18S rRNA gene. For 16S rRNA gene amplification, we used a modified version of the published universal prokaryotic primer set 341F/806R, targeting the V3-V4 hypervariable regions (17). The forward primer, 341F3, had three additional nucleotides attached in the 5′ end (ACTCCTAYGGGRBGCASCAG), and the reverse primer, 806R5, had five additional nucleotides attached in the 5′ end (AGCGTGGACTACNNGGGTATCTAAT).
For each primer pair, the rRNA gene was amplified using a short PCR setup as follows: initial denaturation at 95°C for 3 min was followed by 20 cycles of 95°C (30 s for 16S rRNA; 1 min for 18S rRNA), 60°C for 1 min, and 72°C for 30 s; final elongation was carried out at 72°C (7 min for 16S rRNA; 4 min for 18S rRNA). PCR was performed in a 25-μl volume, using the Extract-N-Amp PCR ReadyMix (Sigma-Aldrich, St. Louis, MO, USA) with 0.4 μM each primer and 2 μl of the template. This PCR is referred to as PCR1. The products from PCR1 were prepared for sequencing by a second PCR (PCR2) using the same PCR program. PCR2 attached an adaptor A, an index i5, and a forward sequencing primer site (FSP) to the 5′ ends of the amplicons and an adaptor B, an index i7, and a reverse sequencing primer site (RSP) to the 3′ ends of the amplicons (Fig. 1). Hence, four PCR products were generated for each sample. DNA was quantified using the Quant-IT high-sensitivity double-stranded DNA (dsDNA) assay kit (Thermo Fisher Scientific), and PCR2 products were pooled in equimolar amounts from the individual samples into the pooled amplicon library (PAL). Undesirable DNA amplicons were removed from the PAL by Agencourt AMPure XP bead (Beckman Coulter)-based purification in a two-step process. First, DNA fragments of <300 nucleotides (nt) were removed by use of 24 μl AMPure beads per 10 μl PAL, according to the manufacturer’s protocol, and were eluted in 40 μl Tris-EDTA (TE) buffer. Second, large DNA fragments of >1 kbp were removed by 16 μl AMPure beads per 10 μl AM1 (DNA resulting from the first AMPure bead treatment). The resulting AMPure bead-purified PAL was designated bPAL. The bPAL was diluted to its final concentration of 11.5 pM DNA with a 0.001 N NaOH concentration and was used for sequencing on the Illumina MiSeq desktop sequencer (Illumina Inc., San Diego, CA, USA). The library was sequenced with the 500-cycle MiSeq reagent kit, V2, in a 2 × 250-nt setup (Illumina Inc., San Diego, CA, USA).
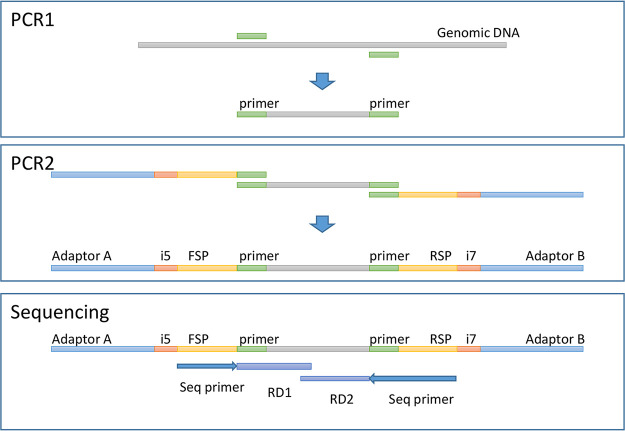
FIG 1.
(Top) PCR1. Shown is a diagram of the PCR of the gene target area with genomic DNA as the template. Genomic template DNA was subjected to PCR-based amplification with one primer set targeting prokaryotes and three primer sets targeting eukaryotes. Each PCR was run in parallel. (Center) PCR2. Shown is a diagram of the attachment of the required elements to amplicons for MiSeq sequencing. The products from PCR1 were used as templates for the adaptor PCR, where adaptors, bar codes, and sequencing primer-binding sites were added. This was performed in parallel for each of the four primer sets. (Bottom) Sequencing. Shown is a diagram of the regions sequenced by MiSeq as RD1 and RD2. See the text for details.
Data analysis was performed using BION. The pipeline accepts raw sequences and includes steps for demultiplexing, primer extraction, sampling, sequence- and quality-based trimming and filtering, dereplication, clustering, chimera checking, identification of similarities to reference data, and taxonomic mapping and formatting. Nonoverlapping paired reads were allowed for analysis.
Identification of Acanthamoeba genotypes.
BION automatically assigns a species name to Acanthamoeba-specific ribosomal DNA (rDNA) sequences. According to Martín-Pérez et al. (18), Acanthamoeba genotypes can be robustly grouped based on 18S rDNA sequences. Clustered FASTA files representing Acanthamoeba-specific DNA sequences were downloaded from the BION server and aligned with representative reference sequences downloaded from GenBank. The work by Martín-Pérez and colleagues (18) was used to inform the selection of reference sequences. The alignment was manually edited, and phylogenetic analysis of the edited alignment used the neighbor-joining algorithm as implemented in MEGA7 (19).
Identification of organisms of potential clinical relevance.
A multitude of organisms have been reported to be involved in IK, including organisms commonly found on the skin (e.g., Staphylococcus and Candida species). Therefore, in the absence of a reference methodology, we reasoned that it would be useful, for a preliminary set of observations, to define a cutoff for the detection of organisms of potential clinical importance using a standardized proportion rather than a standardized absolute number of sequences, given the vast range in sequence read output (see Results).
Therefore, in Acanthamoeba-negative samples where ≥50% of the total output reflecting nonvertebrate DNA (i.e., nonhost DNA) could be assigned to a bacterial or fungal genus/species, this genus/species was considered to be of potential clinical relevance. This threshold reflected a conservative approach and was based on the rationale that if at least half of the sequences produced for any given sample could be attributed to a single microorganism, the likelihood that this organism was involved in the keratitis might be considered substantial.
Ethical considerations.
This study complied with ASM's Ethical Guidelines (https://journals.asm.org/content/ethical-guidelines). Although the study involved DNA from human clinical samples (corneal scrapings), human DNA was not subjected to analysis. Moreover, the samples were anonymized prior to analysis by the 16S-18S assay. Hence, informed consent for publication was not deemed necessary, since patients could not be identified from any material in this article.
Data availability.
Acanthamoeba-specific sequences produced by the NGS assay in this study have been submitted to GenBank under accession numbers MT919356 to MT919376.
RESULTS
16S-18S sequence read output.
16S and 18S rRNA gene data were generated from all samples with a range of 3,909 to 381,110 reads per sample (median, 81,848 reads; interquartile range [IQR], 48,219 to 106,487 reads) (see Fig. S1 in the supplemental material). Verterbrate DNA (i.e., host DNA) represented a significant proportion of all sequences generated (median, 42,548 sequences; IQR, 24,861 to 64,661 sequences). Three to 74 genera were identified in each sample.
Ability of the 16S-18S assay to identify Acanthamoeba-positive samples and usefulness of the sequences obtained for genotyping.
A total of 24 samples had been scored Acanthamoeba positive by real-time PCR; of these, 2 samples were considered weakly positive for Acanthamoeba. By the 16S-18S assay, Acanthamoeba rDNA was detected in 21 samples, all of which had been scored positive by real-time PCR (specificity, 100%). Meanwhile, the 16S-18S assay failed to detect Acanthamoeba DNA in three of the samples positive by real-time PCR, two of which were weakly positive (sensitivity, 88%). None of the samples identified as Acanthamoeba negative by real-time PCR were found positive by the 16S-18S assay.
For 16 samples, a result at the species level was returned by BION. The species identified were Acanthamoeba palestinensis (n = 2), Acanthamoeba hatchetti (n = 3), Acanthamoeba polyphaga (n = 5), Acanthamoeba castellani (n = 4), and Acanthamoeba mauritaniensis (n = 2). The remaining Acanthamoeba spp. (n = 5) could be identified only to the genus level. Based on phylogenetic analysis, a total of 19 sequences reflected genotype T4, and 2 sequences reflected genotype T6.
Overall mapping of bacterial DNA in the samples.
The 16S rRNA gene sequences were analyzed at various taxonomic levels, ranging from phylum to species. The dominant phylum was Proteobacteria, which, on average, represented 67.2% of bacterial reads (range, 0 to 100%). Firmicutes (mean, 16%; range, 0 to 100%), Actinobacteria (mean, 10.3%; range, 0 to 72.1%), and Bacteroidetes (mean, 5.0%; range, 0 to 59.4%) were also highly represented. Other phyla, such as Acidobacteria (mean, <0.1%; range, 0 to 3.0%), Fusobacteria (mean, 0.6%; range, 0 to 21.4%), Deinococcus-Thermus (mean, 0.3%; range, 0 to 9.3%), Chlamydiae (mean, <0.1%; range, 0 to 76.2%), Spirochaetes (mean, <0.1%; range, 0 to 3.4%), Tenericutes (mean, <0.1%; range, 0 to 10.1%), Thermatogae (mean, <0.1%; range, 0 to 1.18%), and Verrucomicrobia (mean, <0.1; range, 0 to 2.8%), were present, but in smaller proportions. The remaining bacterial reads from a diverse set of phyla contributed very few reads (mean, 0.2%; range, 0 to 9.8%). The most common bacterial genera were Pseudomonas, Acinetobacter, Propionibacterium, and Streptococcus, but their relative proportions differed greatly between the samples.
Organisms potentially causing nonviral IK detected in Acanthamoeba-negative samples.
In addition to Acanthamoeba, several other pathogens that could be involved in the development of IK were detected (Table 1). Fungal and bacterial species of potential clinical relevance were identified in 31 of the samples negative for Acanthamoeba; these included Pseudomonas aeruginosa (n = 11), Moraxella spp. (n = 6), Staphylococcus aureus (n = 2), Fusarium spp. (n = 4), and Candida albicans (n = 1). All these accounted for ≥50% of the total sequence output reflecting nonvertebrate DNA (i.e., nonhost DNA) in Acanthamoeba-negative samples.
TABLE 1.
Organisms detected in Acanthamoeba-negative samples and reflecting at least 50% of sequence reads
| Microbial genus/speciesa | No. of samples in which the organism was observed | Possible interpretationb |
|---|---|---|
| Pseudomonas spp. | 12 (P. aeruginosa, 11; P. fragi, 1) | C |
| Moraxella spp. | 6 (M. catarrhalis, 4; M. nonliquefaciens, 2) | C |
| Saccharomyces cerevisiae | 5 | S |
| Fusarium spp. | 4 | C |
| Malassezia globosa | 4 | S |
| Streptococcus spp. | 4 (S. mitis, 2; S. dysgalactiae, 1; S. pneumoniae, 1) | C |
| Staphylococcus aureus | 2 | C |
| Candida albicans | 1 | C |
| Eikenella corrodens | 1 | C |
| Enterococcus faecium | 1 | C |
| Total | 40 | C (n = 31), S (n = 9) |
Listed according to frequency.
S, more likely skin contamination; C, more likely clinical relevance.
DISCUSSION
In this study, we examined the feasibility of the 16-18S assay as a diagnostic tool for the detection of Acanthamoeba-specific DNA in corneal scrapings received due to infectious keratitis (IK).
Our data demonstrate that the 16-18S assay was able to detect Acanthamoeba-specific DNA with a sensitivity of 88% and a specificity of 100% relative to Acanthamoeba-specific real-time PCR. As an added benefit, the Acanthamoeba sequences obtained by the 16S-18S assay served both to confirm the presence of Acanthamoeba-specific DNA and also enabled genotyping of the strains based on the sequence output analyzed by BION. Finally, we developed an automated algorithm that identified organisms represented by at least 50% of the total sequence read count in the sample as potentially clinically relevant nonviral causes of IK, and 31 (18%) of the Acanthamoeba-negative samples (n = 176) were positive for a clinically relevant bacterium or fungus.
Reports on NGS-based approaches to detecting and differentiating organisms involved in IK are still very limited. Li et al. applied metagenomics to a very limited set of paraffin-embedded samples (n = 16) (20). Prashanthi et al. used amplicon-based NGS of the internal transcribed spacer (ITS) region to characterize alterations in the ocular surface fungal microbiome in fungal keratitis (21). So far, however, no studies have been published on amplicon-based NGS of ribosomal rRNA genes from bacteria, fungi, and parasites in corneal scrapings.
The primers used in the 16S-18S assay have very limited specificity compared with the primers (and probe) used in the real-time PCR assay, and the fact that the 16S-18S assay was able to detect 21 out of 24 Acanthamoeba-positive samples is quite remarkable, especially considering the fact that 2 of the 24 samples scored positive by real-time PCR were categorized as “weakly positive,” so the 16S-18S assay identified 21 out of 22 clearly positive samples (95%). No other microorganism that could explain IK was found in these samples using the 16S-18S assay. Unfortunately, we did not have any clinical information on the samples, and it is not known to us whether the two faintly positive samples came from patients who had already been receiving Acanthamoeba-targeted treatment prior to sampling, which could explain the very weak signals. None of the three samples testing weakly positive by real-time PCR and negative by the 16S-18S assay were positive for other organisms relevant to IK by 16S-18S analysis; however, since data from local clinical microbiology laboratories were not available for analysis in the present study, we could not investigate whether these samples were from patients with IK potentially suffering from viral infections. The sensitivity of the real-time PCR used as a reference in the present study remains unknown. However, about 15% of all samples referred for Acanthamoeba PCR and tested by this real-time PCR in Denmark are positive. The ability to establish a diagnosis of AK correctly based on analysis of corneal scrapings relies not only on sufficiently sensitive microbiological methods but also on the ability to sample appropriately.
Establishing a diagnosis of AK is very difficult under normal circumstances, especially in the later stages of the disease, when very few trophozoites are present and the disease is dominated by cysts. It remains to be determined if the sensitivity of this analysis is high enough for routine diagnostics or whether it will remain a supplement to the specific RT-PCR. It is possible that the assay could prove useful in the initial stage of IK, when trophozoites are more abundant than they are in later stages.
Organisms from >75% of samples (16 of 21) were identified to the species level, providing useful information on the pathogens. Although it is encouraging that the microbiome platform enables species identification, this information should not stand alone but needs to be supplemented by genotype determination. Table 2 summarizes data from similar studies reflecting Acanthamoeba genotypes identified mainly in corneal scrapings, but also in corneal/nasal swabs, contact lenses, and contact lens solutions. As can be seen, T4 is the by far the most common genotype detected, accounting for >83% of the cases, followed in prevalence by T3, which accounts for 6% of the cases. As suggested by the distribution of data in Table 2, the data obtained in the present study add support to the claim that T4 is by far the most common genotype involved in human AK (22). Of note, we identified T6, a genotype rarely seen in AK, in two corneal scrapings. Walochnik and colleagues identified the T6 genotype when analyzing a “hypervirulent” strain of Acanthamoeba from a contact lens-wearing patient with keratitis whose case was managed in Austria (23). It remains unclear whether the genotypes involved in AK differ in terms of clinical course/severity and susceptibility to treatment. Given the rarity of the infection, multicenter prospective studies to investigate this question would be appropriate.
TABLE 2.
Selection of studies showing the distribution of Acanthamoeba genotypes identified in Acanthamoeba-positive samples
| Reference or source | Country | Sample material(s) | No. of samples analyzed | No. of samples with Acanthamoeba genotype: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T3 | T4 | T5 | T6 | T9 | T10 | T11 | T13 | T15 | ||||
| Behera et al. (24) | India | Cornea | 18 | 16 | 2 | |||||||
| Rocha-Cabrera et al. (25) | Spain | Cornea | 17 | 17 | ||||||||
| Omaña-Molina et al. (26) | Mexico | Cornea | 2 | 1 | 1 | |||||||
| Montalbano Di Filippo et al. (27) | Italy | Cornea (n = 12), lens solution (n = 19), swab (n = 24) | 55 | 11 | 33 | 1 | 10 | |||||
| Arnalich-Montiel et al. (28) | Spain | Cornea | 17 | 2 | 14 | 1 | ||||||
| Grün et al. (29) | Germany | Contact lens | 1 | 1 | ||||||||
| Cabello-Vilchez et al. (30) | Peru | Nasal swabs | 21 | 20 | 1 | |||||||
| Takaoka-Sughihara et al. (31) | Japan | Cornea | 6 | 6 | ||||||||
| Lorenzo-Morales et al. (32) | Spain | Cornea | 1 | 1 | ||||||||
| Abe and Kimata (33) | Japan | Cornea | 7 | 7 | ||||||||
| Sharifi et al. (34) | Sweden | Cornea (n = 10), contact lens (n = 1) | 11 | 1 | 8 | 1 | 1 | |||||
| Zhao et al. (35) | China | Cornea | 14 | 14 | ||||||||
| Ledee et al. (36) | USA | Cornea | 37 | 36 | 1 | |||||||
| Yera et al. (37) | France | Cornea | 9 | 1 | 7 | 1 | ||||||
| Gatti et al. (38) | Italy | Cornea | 15 | 15 | ||||||||
| Hajialilo et al. (39) | Iran | Cornea | 17 | 14 | 1 | 2 | ||||||
| Present study | Denmark | Cornea | 21 | 19 | 2 | |||||||
| Total | 269 | 16 | 227 | 1 | 2 | 1 | 2 | 7 | 1 | 12 | ||
In this study, we applied a strict criterion for determining which of the organisms detected could be considered potential contributors to the development of IK. Hence, only bacterial or fungal species that made up >50% of nonvertebrate sequence reads were included. Still, quite a few genera/species of potential clinical relevance were noticed. P. aeruginosa was detected in 11 samples and S. aureus in 2; both are well-known causes of IK. Several other bacterial species that could potentially be causes of IK were detected. Unsurprisingly, these bacteria were mainly bacterial commensals found in the upper airways and as part of the oropharyngeal environment, and the clinical significance of these findings should be evaluated on a case-to-case basis, just as it should be confirmed using conventional culturing. We did, however, adopt a very conservative algorithm to ensure that the bacterium identified was found at levels that warranted further clinical follow-up. This was also true for fungal species; we found four samples containing Fusarium spp. and one sample containing C. albicans, all of which are potential causes of keratitis and could be of clinical significance.
The availability and cost of this assay is also a concern that needs to be taken into account. In our setup, this assay requires 17.5 technician working hours and 6 molecular bioinformatics working hours. Downstream analysis of the NGS assay output has been automatized so that the freely available software BION annotates the sequence results automatically to taxonomic units, significantly reducing the amount of work related to sequence read analysis.
These findings are encouraging because they demonstrate the ability of the assay to detect and differentiate microorganisms that are usually found using conventional methods. Due to ethical considerations and limitations, this study could not include data from local clinical microbiology laboratories to confirm our findings and validate our algorithm. We suggest developing a prospective study where data from the 16S-18S assay can be compared with data from routine clinical microbiology analyses and other investigations used to establish a diagnosis of AK (such as confocal microscopy), since this will be crucial in determining the feasibility of this platform as a frontline screening tool for nonviral causes of IK. The microbiome platform is not dependent on viable bacteria or fungi in order to determine their presence, and it is conceivable that this analysis could provide new/additional information. It is not unusual for a patient to have been prescribed topical antibiotics prior to sampling for keratitis, and a diagnostic approach that is independent of the viability of pathogens could be of great value. Further studies comparing the 16S-18S platform to standard culturing in this setting would be of great value.
Since it is based on small-subunit (SSU) rRNA genes and not ITS regions, the 16S-18S assay does not have sufficient discriminatory power to identify molds to the species level, and subsequent analyses will be needed to make a full identification. The same issue pertains to Streptococcus spp., for which further testing will be needed for identification to the species level.
In conclusion, our study confirmed that the 16S-18S assay is able to detect the presence of Acanthamoeba spp. with a sensitivity of 88% and a specificity of 100%. The assay was able to provide valuable information on Acanthamoeba genotypes. Furthermore, the assay was able to detect bacterial and fungal pathogens potentially involved in IK; however, further studies are needed to ascertain the sensitivity of this analysis.
Supplementary Material
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Fuerst PA. 2014. Insights from the DNA databases: approaches to the phylogenetic structure of Acanthamoeba. Exp Parasitol 145:S39–S45. doi: 10.1016/j.exppara.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 2.Lorenzo-Morales J, Khan NA, Walochnik J. 2015. An update on Acanthamoeba keratitis: diagnosis, pathogenesis and treatment. Parasite 22:10. doi: 10.1051/parasite/2015010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lorenzo-Morales J, Martín-Navarro CM, López-Arencibia A, Arnalich-Montiel F, Piñero JE, Valladares B. 2013. Acanthamoeba keratitis: an emerging disease gathering importance worldwide? Trends Parasitol 29:181–187. doi: 10.1016/j.pt.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Seal DV, Kirkness CM, Bennett HG, Peterson M, Keratitis Study Group. 1999. Acanthamoeba keratitis in Scotland: risk factors for contact lens wearers. Cont Lens Anterior Eye 22:58–68. doi: 10.1016/S1367-0484(99)80004-6. [DOI] [PubMed] [Google Scholar]
- 5.Sauer A, Bourcier T, French Study Group for Contact Lenses Related Microbial Keratitis. 2011. Microbial keratitis as a foreseeable complication of cosmetic contact lenses: a prospective study. Acta Ophthalmol 89:e439–e442. doi: 10.1111/j.1755-3768.2011.02120.x. [DOI] [PubMed] [Google Scholar]
- 6.Nielsen SE, Ivarsen A, Hjortdal J. 2020. Increasing incidence of Acanthamoeba keratitis in a large tertiary ophthalmology department from year 1994 to 2018. Acta Ophthalmol 98:445–448. doi: 10.1111/aos.14337. [DOI] [PubMed] [Google Scholar]
- 7.Maycock NJR, Jayaswal R. 2016. Update on Acanthamoeba keratitis: diagnosis, treatment, and outcomes. Cornea 35:713–720. doi: 10.1097/ICO.0000000000000804. [DOI] [PubMed] [Google Scholar]
- 8.Lakhundi S, Siddiqui R, Khan NA. 2017. Pathogenesis of microbial keratitis. Microb Pathog 104:97–109. doi: 10.1016/j.micpath.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 9.Daas L, Szentmáry N, Eppig T, Langenbucher A, Hasenfus A, Roth M, Saeger M, Nölle B, Lippmann B, Böhringer D, Reinhard T, Kelbsch C, Messmer E, Pleyer U, Roters S, Zhivov A, Engelmann K, Schrecker J, Zumhagen L, Thieme H, Darawsha R, Meyer-Ter-Vehn T, Dick B, Görsch I, Hermel M, Kohlhaas M, Seitz B. 2015. The German Acanthamoeba keratitis register: initial results of a multicenter study. Ophthalmologe 112:752–763. (In German.) doi: 10.1007/s00347-014-3225-7. [DOI] [PubMed] [Google Scholar]
- 10.Raghavan A, Baidwal S, Vijayaraghavan P, Rajeswari S, Rajaraman R, Venkatapathy N, Menon S, Rammohan R. 2018. Diagnostic evaluation of co-occurrence of Acanthamoeba and fungi in keratitis. Cornea 37:227–234. doi: 10.1097/ICO.0000000000001441. [DOI] [PubMed] [Google Scholar]
- 11.Buchele MLC, Wopereis DB, Casara F, de Macedo JP, Rott MB, Monteiro FBF, Bazzo ML, Spada FDR, Dos Santos JI, Caumo KS. 2018. Contact lens-related polymicrobial keratitis: Acanthamoeba spp. genotype T4 and Candida albicans. Parasitol Res 117:3431–3436. doi: 10.1007/s00436-018-6037-x. [DOI] [PubMed] [Google Scholar]
- 12.Ring HC, Thorsen J, Saunte DM, Lilje B, Bay L, Riis PT, Larsen N, Andersen LO, Nielsen HV, Miller IM, Bjarnsholt T, Fuursted K, Jemec GB. 2017. The follicular skin microbiome in patients with hidradenitis suppurativa and healthy controls. JAMA Dermatol 153:897–905. doi: 10.1001/jamadermatol.2017.0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krogsgaard LR, Andersen LO, Johannesen TB, Engsbro AL, Stensvold CR, Nielsen HV, Bytzer P. 2018. Characteristics of the bacterial microbiome in association with common intestinal parasites in irritable bowel syndrome. Clin Transl Gastroenterol 9:161. doi: 10.1038/s41424-018-0027-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qvarnström Y, Visvesvara GS, Sriram R, da Silva AJ. 2006. Multiplex real-time PCR assay for simultaneous detection of Acanthamoeba spp., Balamuthia mandrillaris, and Naegleria fowleri. J Clin Microbiol 44:3589–3595. doi: 10.1128/JCM.00875-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gogarten JF, Calvignac-Spencer S, Nunn CL, Ulrich M, Saiepour N, Nielsen HV, Deschner T, Fichtel C, Kappeler PM, Knauf S, Müller-Klein N, Ostner J, Robbins MM, Sangmaneedet S, Schülke O, Surbeck M, Wittig RM, Sliwa A, Strube C, Leendertz FH, Roos C, Noll A. 2020. Metabarcoding of eukaryotic parasite communities describes diverse parasite assemblages spanning the primate phylogeny. Mol Ecol Resour 20:204–215. doi: 10.1111/1755-0998.13101. [DOI] [PubMed] [Google Scholar]
- 16.Stensvold CR, Lebbad M, Hansen A, Beser J, Belkessa S, O’Brien Andersen L, Clark CG. 2020. Differentiation of Blastocystis and parasitic archamoebids encountered in untreated wastewater samples by amplicon-based next-generation sequencing. Parasite Epidemiol Control 9:e00131. doi: 10.1016/j.parepi.2019.e00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu Y, Lee C, Kim J, Hwang S. 2005. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol Bioeng 89:670–679. doi: 10.1002/bit.20347. [DOI] [PubMed] [Google Scholar]
- 18.Martín-Pérez T, Criado-Fornelio A, Martínez J, Blanco MA, Fuentes I, Pérez-Serrano J. 2017. Isolation and molecular characterization of Acanthamoeba from patients with keratitis in Spain. Eur J Protistol 61:244–252. doi: 10.1016/j.ejop.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 19.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Breitwieser FP, Lu J, Jun AS, Asnaghi L, Salzberg SL, Eberhart CG. 2018. Identifying corneal infections in formalin-fixed specimens using next generation sequencing. Invest Ophthalmol Vis Sci 59:280–288. doi: 10.1167/iovs.17-21617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prashanthi GS, Jayasudha R, Chakravarthy SK, Padakandla SR, SaiAbhilash CR, Sharma S, Bagga B, Murthy SI, Garg P, Shivaji S. 2019. Alterations in the ocular surface fungal microbiome in fungal keratitis patients. Microorganisms 7:309. doi: 10.3390/microorganisms7090309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maciver SK, Asif M, Simmen MW, Lorenzo-Morales J. 2013. A systematic analysis of Acanthamoeba genotype frequency correlated with source and pathogenicity: T4 is confirmed as a pathogen-rich genotype. Eur J Protistol 49:217–221. doi: 10.1016/j.ejop.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Walochnik J, Haller-Schober E, Kölli H, Picher O, Obwaller A, Aspöck H. 2000. Discrimination between clinically relevant and nonrelevant Acanthamoeba strains isolated from contact lens-wearing keratitis patients in Austria. J Clin Microbiol 38:3932–3936. doi: 10.1128/JCM.38.11.3932-3936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behera HS, Panda A, Satpathy G, Bandivadekar P, Vanathi M, Agarwal T, Nayak N, Tandon R. 2016. Genotyping of Acanthamoeba spp. and characterization of the prevalent T4 type along with T10 and unassigned genotypes from amoebic keratitis patients in India. J Med Microbiol 65:370–376. doi: 10.1099/jmm.0.000234. [DOI] [PubMed] [Google Scholar]
- 25.Rocha-Cabrera P, Reyes-Batlle M, Martín-Navarro CM, Dorta-Gorrín A, López-Arencibia A, Sifaoui I, Martínez-Carretero E, Piñero JE, Martín-Barrera F, Valladares B, Lorenzo-Morales J. 2015. Detection of Acanthamoeba on the ocular surface in a Spanish population using the Schirmer strip test: pathogenic potential, molecular classification and evaluation of the sensitivity to chlorhexidine and voriconazole of the isolated Acanthamoeba strains. J Med Microbiol 64:849–853. doi: 10.1099/jmm.0.000103. [DOI] [PubMed] [Google Scholar]
- 26.Omaña-Molina M, Vanzzini-Zago V, Hernandez-Martinez D, Gonzalez-Robles A, Salazar-Villatoro L, Ramirez-Flores E, Oregon-Miranda E, Lorenzo-Morales J, Martinez-Palomo A. 2016. Acanthamoeba genotypes T3 and T4 as causative agents of amoebic keratitis in Mexico. Parasitol Res 115:873–878. doi: 10.1007/s00436-015-4821-4. [DOI] [PubMed] [Google Scholar]
- 27.Montalbano Di Filippo M, Santoro M, Lovreglio P, Monno R, Capolongo C, Calia C, Fumarola L, D’Alfonso R, Berrilli F, Di Cave D. 2015. Isolation and molecular characterization of free-living amoebae from different water sources in Italy. Int J Environ Res Public Health 12:3417–3427. doi: 10.3390/ijerph120403417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arnalich-Montiel F, Lumbreras-Fernández B, Martín-Navarro CM, Valladares B, Lopez-Velez R, Morcillo-Laiz R, Lorenzo-Morales J. 2014. Influence of Acanthamoeba genotype on clinical course and outcomes for patients with Acanthamoeba keratitis in Spain. J Clin Microbiol 52:1213–1216. doi: 10.1128/JCM.00031-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grün A-L, Stemplewitz B, Scheid P. 2014. First report of an Acanthamoeba genotype T13 isolate as etiological agent of a keratitis in humans. Parasitol Res 113:2395–2400. doi: 10.1007/s00436-014-3918-5. [DOI] [PubMed] [Google Scholar]
- 30.Cabello-Vílchez AM, Martín-Navarro CM, López-Arencibia A, Reyes-Batlle M, González AC, Guerra H, Gotuzzo E, Valladares B, Piñero E, Lorenzo-Morales J. 2014. Genotyping of potentially pathogenic Acanthamoeba strains isolated from nasal swabs of healthy individuals in Peru. Acta Trop 130:7–10. doi: 10.1016/j.actatropica.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 31.Takaoka-Sugihara N, Yamagami S, Yokoo S, Matsubara M, Yagita K. 2012. Cytopathic effect of Acanthamoeba on human corneal fibroblasts. Mol Vis 18:2221–2228. [PMC free article] [PubMed] [Google Scholar]
- 32.Lorenzo-Morales J, Morcillo-Laiz R, Martín-Navarro CM, López-Vélez R, López-Arencibia A, Arnalich-Montiel F, Maciver SK, Valladares B, Martínez-Carretero E. 2011. Acanthamoeba keratitis due to genotype T11 in a rigid gas permeable contact lens wearer in Spain. Cont Lens Anterior Eye 34:83–86. doi: 10.1016/j.clae.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 33.Abe N, Kimata I. 2010. Genotyping of Acanthamoeba isolates from corneal scrapings and contact lens cases of Acanthamoeba keratitis patients in Osaka. Jpn J Infect Dis 63:299–301. [PubMed] [Google Scholar]
- 34.Sharifi N, Botero-Kleiven S, Ohman D, Barragan A, Winiecka-Krusnell J. 2010. Genotypic characterization of Acanthamoeba spp. causing ocular infections in Swedish patients: identification of the T15 genotype in a case of protracted keratitis. Scand J Infect Dis 42:781–786. doi: 10.3109/00365548.2010.490563. [DOI] [PubMed] [Google Scholar]
- 35.Zhao G, Sun S, Zhao J, Xie L. 2010. Genotyping of Acanthamoeba isolates and clinical characteristics of patients with Acanthamoeba keratitis in China. J Med Microbiol 59:462–466. doi: 10.1099/jmm.0.016667-0. [DOI] [PubMed] [Google Scholar]
- 36.Ledee DR, Iovieno A, Miller D, Mandal N, Diaz M, Fell J, Fini ME, Alfonso EC. 2009. Molecular identification of T4 and T5 genotypes in isolates from Acanthamoeba keratitis patients. J Clin Microbiol 47:1458–1462. doi: 10.1128/JCM.02365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yera H, Zamfir O, Bourcier T, Ancelle T, Batellier L, Dupouy-Camet J, Chaumeil C. 2007. Comparison of PCR, microscopic examination and culture for the early diagnosis and characterization of Acanthamoeba isolates from ocular infections. Eur J Clin Microbiol Infect Dis 26:221–224. doi: 10.1007/s10096-007-0268-6. [DOI] [PubMed] [Google Scholar]
- 38.Gatti S, Rama P, Matuska S, Berrilli F, Cavallero A, Carletti S, Bruno A, Maserati R, Di Cave D. 2010. Isolation and genotyping of Acanthamoeba strains from corneal infections in Italy. J Med Microbiol 59:1324–1330. doi: 10.1099/jmm.0.019786-0. [DOI] [PubMed] [Google Scholar]
- 39.Hajialilo E, Behnia M, Tarighi F, Niyyati M, Rezaeian M. 2016. Isolation and genotyping of Acanthamoeba strains (T4, T9, and T11) from amoebic keratitis patients in Iran. Parasitol Res 115:3147–3151. doi: 10.1007/s00436-016-5072-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Acanthamoeba-specific sequences produced by the NGS assay in this study have been submitted to GenBank under accession numbers MT919356 to MT919376.