Abstract
Background
Missing MRI sequences represent an obstacle in the development and use of deep learning (DL) models that require multiple inputs.
Purpose
To determine if synthesizing brain MRI scans using generative adversarial networks (GANs) allows for the use of a DL model for brain lesion segmentation that requires T1-weighted images, postcontrast T1-weighted images, fluid-attenuated inversion recovery (FLAIR) images, and T2-weighted images.
Materials and Methods
In this retrospective study, brain MRI scans obtained between 2011 and 2019 were collected, and scenarios were simulated in which the T1-weighted images and FLAIR images were missing. Two GANs were trained, validated, and tested using 210 glioblastomas (GBMs) (Multimodal Brain Tumor Image Segmentation Benchmark [BRATS] 2017) to generate T1-weighted images from postcontrast T1-weighted images and FLAIR images from T2-weighted images. The quality of the generated images was evaluated with mean squared error (MSE) and the structural similarity index (SSI). The segmentations obtained with the generated scans were compared with those obtained with the original MRI scans using the dice similarity coefficient (DSC). The GANs were validated on sets of GBMs and central nervous system lymphomas from the authors’ institution to assess their generalizability. Statistical analysis was performed using the Mann-Whitney, Friedman, and Dunn tests.
Results
Two hundred ten GBMs from the BRATS data set and 46 GBMs (mean patient age, 58 years ± 11 [standard deviation]; 27 men [59%] and 19 women [41%]) and 21 central nervous system lymphomas (mean patient age, 67 years ± 13; 12 men [57%] and nine women [43%]) from the authors’ institution were evaluated. The median MSE for the generated T1-weighted images ranged from 0.005 to 0.013, and the median MSE for the generated FLAIR images ranged from 0.004 to 0.103. The median SSI ranged from 0.82 to 0.92 for the generated T1-weighted images and from 0.76 to 0.92 for the generated FLAIR images. The median DSCs for the segmentation of the whole lesion, the FLAIR hyperintensities, and the contrast-enhanced areas using the generated scans were 0.82, 0.71, and 0.92, respectively, when replacing both T1-weighted and FLAIR images; 0.84, 0.74, and 0.97 when replacing only the FLAIR images; and 0.97, 0.95, and 0.92 when replacing only the T1-weighted images.
Conclusion
Brain MRI scans generated using generative adversarial networks can be used as deep learning model inputs in case MRI sequences are missing.
© RSNA, 2021
Online supplemental material is available for this article.
See also the editorial by Zhong in this issue.
An earlier incorrect version of this article appeared online. This article was corrected on April 12, 2021.
Summary
Brain MRI scans synthesized using generative adversarial networks can be used as inputs in an existing deep learning model, thus allowing for their application in case MRI data are missing.
Key Results
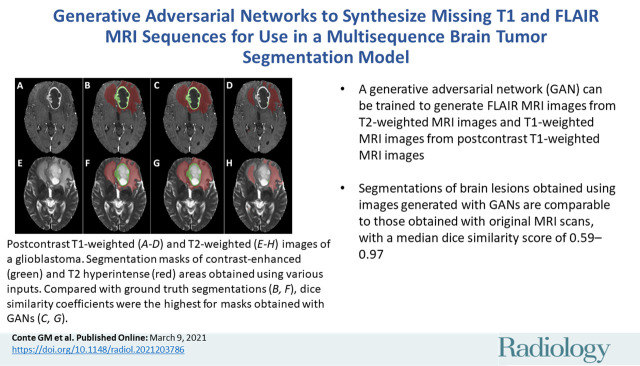
■ A generative adversarial network (GAN) can be trained to generate fluid-attenuated inversion recovery MRI scans from T2-weighted MRI scans and T1-weighted MRI scans from postcontrast T1-weighted MRI scans.
■ Segmentations of brain lesions obtained using images generated with GANs are comparable to those obtained with original MRI scans, with a median dice similarity score of 0.59–0.97.
Introduction
Manual segmentation of brain tumors is a time-consuming and tedious task that has low reproducibility between tracers (1). To address this issue, many research groups have developed automatic segmentation models using deep learning (DL) (2–7). Brain tumor segmentation models often rely on multimodal MRI inputs that are difficult to obtain because of differences in the acquisition protocols among institutions, time constraints, and/or the presence of image artifacts. Thus, sequences are often missing from MRI examinations.
Different strategies can be used during model training and inference to handle missing sequences. During training, models could be designed to produce inference in the absence of an input parameter where the missing data are represented by a fixed value (eg, 0). Although compelling, this approach has drawbacks, as follows: (a) it is only applicable during training and might not be used for inference if the model was not trained to support inference in this manner and (b) representing all combinations of model input data will increase the data set size exponentially as the number of inputs increases; this will in turn increase the size of the model required to achieve good results, and the computational resources and/or time required for training (8). Alternatively, missing sequences could be substituted with synthetic artificial data during training and/or inference, which represents a best guess at the missing value. This approach decouples the handling of the missing data from model training.
Generative adversarial networks (GANs) (9,10) are a relatively new type of DL model that have received much attention because of their ability to generate synthetic images. GANs are trained using two neural networks—a generator and a discriminator. The generator learns to create data that resemble examples contained within the training data set, and the discriminator learns to distinguish real examples from the ones created by the generator. The two networks are trained together until the generated examples are indistinguishable from the real examples. GANs have found many applications in medical imaging (11–16).
In our study, we evaluated the feasibility of using GANs to generate missing MRI sequences for multi-input pretrained models. The aim of our study was to determine if synthesizing brain MRI scans using GANs allows for the use of a DL model for brain lesion segmentation that requires T1-weighted images, postcontrast T1-weighted images, fluid-attenuated inversion recovery (FLAIR) images, and T2-weighted images.
Materials and Methods
Training, Validation, and Testing of Data Sets
For this retrospective study, we used both publicly available MRI scans and MRI scans collected at our institution. The inclusion criteria were a histologic diagnosis of glioblastoma (GBM) or central nervous system lymphoma (CNSL) and availability of precontrast T1-weighted, T2-weighted, and FLAIR MRI scans and postcontrast T1-weighted MRI scans.
The public data set was the Multimodal Brain Tumor Image Segmentation Benchmark (BRATS) 2017 data set (https://www.med.upenn.edu/sbia/brats2017/data.html) (210 examinations) (17–19), which included precontrast T1-weighted, T2-weighted, and FLAIR images and postcontrast T1-weighted images of GBMs obtained at multiple institutions. The 210 BRATS examinations were randomly split into training, validation, and test sets, resulting in 135, 33, and 42 examinations for a total of 20 925, 5115, and 6510 images for each image type, respectively.
The MRI scans from our institution were selected from a retrospectively curated database of GBMs and CNSLs collected between 2011 and 2019.
We obtained institutional review board approval with a waiver of informed consent and Health Insurance Portability and Accountability Act authorization for the use of the data sets from our institution.
Training a GAN Model to Create Missing MRI Sequences
We developed a GAN to create missing T1-weighted and FLAIR images for use in a brain tumor segmentation model that required multiple MRI series (T1-weighted, postcontrast T1-weighted, T2-weighted, and FLAIR). To assess the relative performance of GAN, we simulated missing data and compared model results generated using GAN-generated missing data and other common approaches to handle missing data with control (all real data).
We trained two GAN models to generate missing MRI sequences (Fig 1, A and B)—one that generated precontrast T1-weighted images from postcontrast T1-weighted images (T1 model) and one that generated FLAIR images from T2-weighted images (FLAIR model). GAN models were trained using the pix2pix framework, a conditional GAN designed to perform image-to-image translation (20). Before training, all MRI volumes were resized to 256 × 256 dimensions, and MRI intensities were normalized to (–1, 1) range.
Figure 1:
Generative adversarial network training process. A, T1 model training. Generator learns to obtain precontrast T1-weighted images from postcontrast T1-weighted (T1 Gd) images, section by section. Discriminator is trained to distinguish generated images (gT1) from original images (T1). At each iteration, generator uses discriminator output to synthetize images that more closely resemble real data. B, Fluid-attenuated inversion recovery (FLAIR) model training. Training of FLAIR model follows same steps as T1 model, except generator learns to obtain FLAIR images from T2-weighted images and discriminator is trained to distinguish generated FLAIR (gFLAIR) images from original T2-weighted images. C, After training, generators of the two models are used to obtain full volumes of gT1 images from postcontrast T1-weighted images and gFLAIR images from T2-weighted images of test set. Images are generated section by section and are then used to reconstruct complete volumes.
During training, the GAN models received two paired sections as input, one from the source domain and one from the target domain. For the T1 model, the source domain was the postcontrast T1-weighted images and the target domain was the precontrast T1-weighted images, whereas for the FLAIR model, the source domain was the T2-weighted images and the target domain was the FLAIR images. GAN-generated precontrast T1-weighted images and GAN-generated FLAIR images are shown in Fig 1, C.
Each model was trained for 300 epochs using the scheme described in the study by Isola et al (20). The discriminator and the generator were trained alternately for one gradient descent step, and the discriminator loss was halved to slow down its training process against the generator. The final loss function was a combination of entropy and least absolute deviation, or L1, loss. The weights were updated using minibatch stochastic gradient descent with a batch size of one, an Adam optimizer with a learning rate of 0.0002, and momentum parameters of β1 = 0.5 and β2 = 0.999. Data augmentation consisted of random cropping and horizontal (left to right) flipping.
The models were implemented with Python software (version 3.5; Python Software Foundation) and TensorFlow software (version 2.0; Google Brain). Model training and predictions were performed with an NVIDIA V100 card with 32 GB of memory. The code for the pix2pix model is available at https://github.com/giemmecci/pix2pixRAD.
Quantitative Evaluation of the Generated Images
Differences in the MRI signal intensities of the GAN-generated images and the original images were assessed using the mean squared error (MSE) (21) and the structural similarity index (SSI) (22). For the MSE, a value closer to 0 indicated better image quality, whereas for the SSI, a value closer to 1 indicated better similarity. Compared with the MSE, the SSI was more indicative of the human perceived similarity because it took into account the structural, contrast, and luminance information between two images (23,24).
Quantitative Evaluation of Segmentations
To quantify the ability of GAN-generated images to function as surrogates for images from missing MRI sequences (precontrast T1 sequence and/or FLAIR sequence), we used a publicly available DL brain tumor segmentation model developed outside our institution (25,26). We selected this model for its comprehensive training set, its ease of implementation, and its good performance (27). In brief, the model was trained by others on three data sets (3220 MRI scans in 1450 patients with brain tumors) from different institutions, and it required four MRI scan types (T1-weighted, postcontrast T1-weighted, T2-weighted, and FLAIR images) as input to generate two distinct segmentation masks—one for the contrast-enhanced regions and one for the areas of T2 and/or FLAIR hyperintensities in the tumor. The model was a modified U-Net (28) and is available at https://github.com/NeuroAI-HD/HD-GLIO.
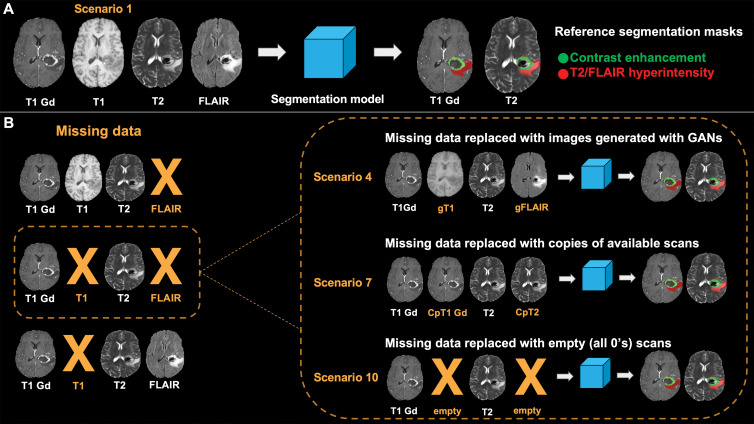
We tested the effects of using GAN-generated images, replacement images, and null input (Fig 2) as strategies to replace missing MRI sequences and determined the relative effect of each strategy with reference to the reference standard model segmentation (no missing data) (Fig 3). Specifically, we simulated 10 scenarios as follows: (a) all original images, which were the reference standard; (b) GAN-generated T1-weighted images were used instead of the original T1-weighted image; (c) GAN-generated FLAIR images were used instead of FLAIR images; (d) both GAN-generated T1-weighted images and GAN-generated FLAIR images were used instead of T1-weighted and FLAIR images, respectively; (e) a postcontrast T1-weighted image was copied and used as a precontrast T1 image; (f) a T2-weighted image was copied and used as a FLAIR image; (g) both postcontrast T1-weighted and T2-weighted images were copied and used as precontrast T1-weighted and FLAIR images, respectively; (h) an image of all 0s (null) was supplied as a precontrast T1-weighted image; (i) an image of all 0s (null) was supplied as a FLAIR image; and (j) both precontrast T1-weighted and FLAIR images were replaced with images of all 0s. We compared model-derived segmentations (T2 abnormal component, the contrast-enhanced component, and the whole lesion) using the dice similarity coefficient (DSC) with reference to the first scenario, as follows:
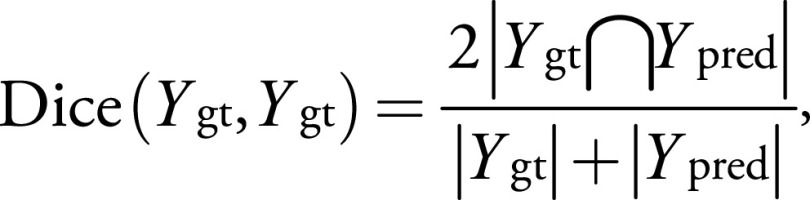 |
Figure 2:
Experimental scenarios. FLAIR = fluid-attenuated inversion recovery, T1 Gd = postcontrast T1-weighted image. A, Scenario 1 represents ideal scenario in which all MRI sequences required as inputs of segmentation model are available. Segmentations obtained in this scenario are used as reference in comparisons with segmentations obtained in other scenarios. B, Scenarios 2–4 are depicted in which missing data are replaced with images generated with generative adversarial networks (GANs) (in orange). gFLAIR = generated FLAIR image, gT1 = generated T1-weighted image. C, Scenarios 5–7 are depicted in which missing data are replaced with copies of other available scans. Copy of postcontrast T1-weighted image (CpT1 Gd) is used instead of precontrast T1-weighted image, and copy of T2-weighted image (CpT2) is used instead of FLAIR image (in orange). D, Scenarios 8–10 are depicted in which missing data are not replaced, and segmentation model is used without all required MRI sequences.
Figure 3:
Lesion segmentation. T1 Gd = postcontrast T1-weighted image, FLAIR = fluid-attenuated inversion recovery. A, Segmentation model requires four MRI sequences (postcontrast T1-weighted, precontrast T1-weighted, T2-weighted, and FLAIR imaging) as inputs to obtain masks of contrast-enhanced areas and areas of T2 and FLAIR hyperintensity in lesions. Masks obtained with original MRI scans serve as reference in comparison with those obtained in different scenarios. B, In case both precontrast T1-weighted and FLAIR images are missing, we simulated three different scenarios. Both precontrast T1-weighted and FLAIR images are replaced with those generated with generative adversarial networks (GANs) (scenario 4), replaced with copies of available MRI scans (scenario 7), or are left empty (scenario 10). Segmentations obtained in these scenarios are compared with those obtained using only original scans. A similar approach was used when only FLAIR images or T1-weighted images were missing (not shown). CpT1 Gd = copy of postcontrast T1-weighted image, CpT2 = copy of T2-weighted image, gFLAIR = FLAIR image generated with GANS, gT1 = T1-weighted image generated with GANS.
where Ygt were the segmentations obtained using the original MRI scans that served as ground truth and Ypred were the segmentations obtained in the scenarios where we simulated missing MRI sequences. The DSC ranged from 0 (no overlap) to 1 (perfect overlap).
GAN Generalization
To assess how well the GANs generalized to independent data sets, we tested them on two data sets from our institution—one from GBMs (n = 26) and one from CNSLs (n = 21). We repeated scenarios 1 through 7 (Fig 2); we did not test the use of blank images because previous analyses did not show consistent superior performance.
To evaluate the potential benefits of retraining the models that included data from our institution, we retrained the original models, adding 20 GBMs (different from the 26 used as a test set) to the training set; we referred to these models with the suffix “retrained-Inst.” To compare the advantage of retraining the models using more data from the BRATS data set, we randomly selected 20 of the 42 volumes originally reserved as a test set and retrained the original model; we referred to these models with the suffix “retrained-BRATS.” We then repeated the predictions on the two internal test sets and compared them with the ones obtained with the original models in terms of MSE, SSI, and DSC.
Evaluating Time Required to Perform Image Segmentation Error Correction
To assess how the use of GANs could affect the clinical routine in terms of the amount of time needed to correct image segmentation errors, we asked two radiologists (M.B. and F.S., with 3 and 5 years of experience in neuroradiology, respectively), who were blinded to the aim of the study and the origin of the images, to independently correct the segmentation of 20 examinations randomly selected from the BRATS test set obtained from scenarios 1, 4, and 7 (Fig 2), for a total of 60 masks, and measured the time needed to perform the corrections. To avoid any bias, the segmentations were presented to the radiologists in a random order.
Statistical Analysis
Statistics were computed with Prism 8 software (GraphPad Software). All analyses were performed with nonparametric tests. We used the Mann-Whitney test to compare the MSE and SSI between the T1 models and the FLAIR models, and we used the Friedman test to compare MSE, SSI, and DSC values obtained in the different scenarios we simulated and to compare the time required by the two radiologists to correct the segmentations. We used the Dunn test to correct for multiple comparisons when comparing the MSE and SSI obtained with the original and the retrained models, the DSC obtained in all scenarios we simulated (Fig 2), and the times for the radiologists’ corrections for scenarios 1, 4, and 7 (Fig 2). P < .05 indicated a statistically significant difference.
Results
Patient Characteristics
All MRI scans from the BRATS 2017 data set were included in the study (n = 210).
The database from our institution included preoperative MRI scans in 150 patients with GBMs and in 91 patients with CNSLs. We excluded 104 GBMs and 70 CNSLs because of incomplete examinations (Fig E1 [online]). The final numbers of GBMs and CNSLs included from our institution were 46 (mean patient age, 58 years ± 11; 27 men [59%] and 19 women [41%]) and 21 (mean patient age, 67 years ± 13; 12 men [57%] and nine women [43%]) (Table E1 [online]), respectively. We refer to the data from our institution as internal sets.
Quantitative Evaluation of the Generated Images
Table 1 shows the median and 25th and 75th percentiles of the MSE and the SSI for all test sets. For both the T1 model and the FLAIR model, the lowest MSE and the highest SSI were obtained with the BRATS test set (median MSE of the T1 model = 0.005 and median MSE of the FLAIR model = 0.004, P < .001; median SSI of the T1 model = 0.92 and median SSI of the FLAIR model = 0.92, P < .001) (Table 1, Fig E2 [online]). For the internal GBM and CNSL test sets, the T1 models outperformed the FLAIR models in all scenarios, with a median MSE ranging from 0.006 to 0.013 and a median SSI ranging from 0.82 to 0.88 (Table 1, Fig E3 [online]). Retraining the models resulted in different MSE and SSI, except for the MSE of the FLAIR models retrained using internal and BRATS data for both test sets, the SSI of the original FLAIR model versus the model retrained with additional BRATS data in the CNSLs test set, and the SSI of the FLAIR models retrained using internal and BRATS data in the GBMs test set (Tables 1, E2 [online]; Fig E4 [online]).
Table 1:
MSE and SSI Values for Test Sets
Quantitative Evaluation of Segmentations
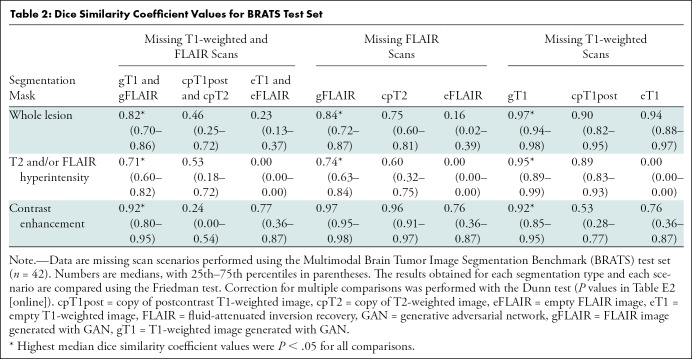
BRATS test set.—Table 2 shows the medians and 25th and 75th percentiles of the DSC values obtained with the BRATS test set (n = 42). The segmentations obtained with the GAN-generated images had the highest median DSC values in all scenarios, ranging from 0.71 to 0.97, with significant differences for all comparisons except for the segmentation of the contrast-enhanced portion of the tumor obtained by substituting the original FLAIR image using either the generated FLAIR image or a copy of the T2-weighted image (DSC for generated FLAIR image = 0.97 and DSC for copy of T2-weighted image = 0.96, P = .98) (Tables 2, E3 [online]; Fig E5 [online]).
Table 2:
Dice Similarity Coefficient Values for BRATS Test Set
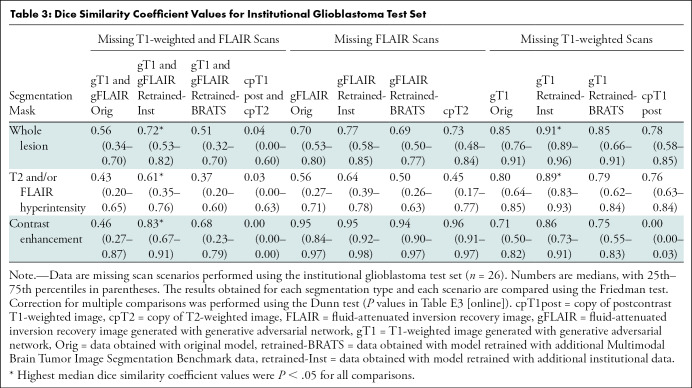
Internal test sets.—Tables 3 and 4 show the medians and 25th and 75th percentiles of the DSC values obtained with the two internal test sets. Overall, the segmentations obtained with the GAN-generated images had the highest median DSC values, except for two comparisons (median DSC ranged from 0.59 to 0.97) (Tables 3, 4, E4 [online], E5 [online]; Fig E6 [online]).
Table 3:
Dice Similarity Coefficient Values for Institutional Glioblastoma Test Set
Table 4:
Dice Similarity Coefficient Values for Institutional CNSL Test Sets
For the GBM test set (n = 26), retraining the models using internal data improved the performance in all scenarios except for one (Tables 3, E4 [online]; Fig E6 [online]). For the CNSL test set (n = 21), retraining the models ensured a better performance in the segmentation task, but the advantage of using additional internal data or additional BRATS data is less clear because both approaches improved the quality of the segmentation masks similarly (Tables 4, E5 [online]; Fig E6 [online]).
Evaluating Time Required to Perform Image Segmentation Error Correction
Correcting the segmentation masks obtained with GAN-generated images required less time than the use of copies of existing scans (Figs 4, E7 [online]; Tables E6, E7 [online]). For the first radiologist, the total time required for the corrections obtained using only the original MRI scan, the generated MRI scan, and copies of existing MRI scans was 266 minutes, 396 minutes, and 650 minutes, respectively. For the second radiologist, the total time was 106 minutes, 287 minutes, and 426 minutes, respectively.
Figure 4:
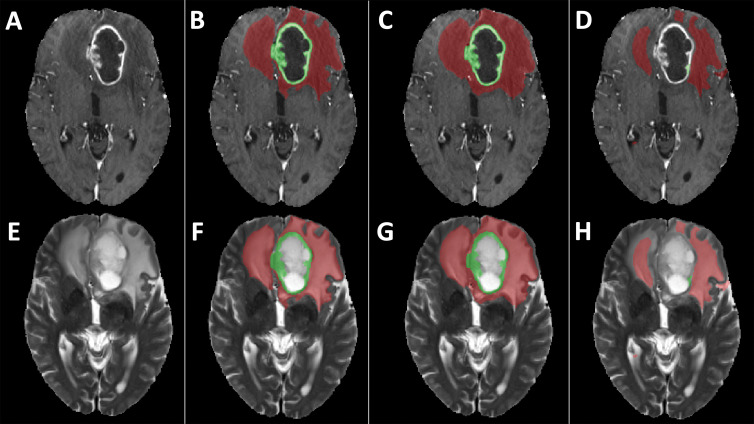
Examples of segmentation masks. A–D, Postcontrast T1-weighted images and, E–H, T2-weighted images of glioblastoma from Multimodal Brain Tumor Segmentation Challenge test set. Segmentation masks of contrast-enhanced area (green) and T2 hyperintense area (red) obtained, B, F, with segmentation model using only original scans as inputs (scenario 1), C, G, using images generated with generative adversarial networks (GANs) (scenario 4), and, D, H, using copies of other scans (scenario 7). Use of only empty scans (scenario 10) failed to produce any masks (not shown). When compared with ground truth segmentations (B, F), dice similarity coefficient for whole segmentation masks obtained with GANs (C, G) was 0.88, whereas dice similarity coefficient obtained with copies of other available scans (D, H) was 0.56.
Discussion
The development and application of deep learning (DL) models often rely on the availability of multimodal MRI inputs. In practice, MRI examinations often miss one or more sequences because of differences in the acquisition protocols and/or the presence of image artifacts. Thus, missing MRI sequences represent an obstacle for the development of de novo DL models and for the application of existing ones.
In this study, we demonstrated that GANs are an effective approach to generating missing MRI sequences to use for a multisequence convolutional neural network model for brain tumor segmentation. Specifically, we demonstrated that two GANs trained on public data using the pix2pix architecture (20), which uses only one sequence as input, successfully synthesized precontrast T1 and FLAIR sequences starting from postcontrast T1-weighted and T2-weighted images, respectively (median MSE ranged from 0.004 to 0.103 and median SSI ranged from 0.76 to 0.92). The synthetized images were used as inputs to apply an already trained segmentation model to the test set from the public data set and the one from our institution, obtaining a median DSC ranging from 0.71 to 0.97 for the public test set and from 0.43 to 0.95 for the test sets from our institution (Figs 5, 6). A relative decrease in the models’ performance on the external validation sets, here the data sets from our institution, could be expected, given the different scanners used to acquire the MRI scans. We demonstrated that adding a small data set from our institution to the original training set and retraining the models exerted a beneficial effect on the models’ performance (DSC ranged from 0.59 to 0.97). This finding suggests that institutions with limited data to train DL models can leverage the existence of comprehensive, public data sets to perform the initial training and then finetune their models using data from their institutions to maximize the models’ performance.
Figure 5:
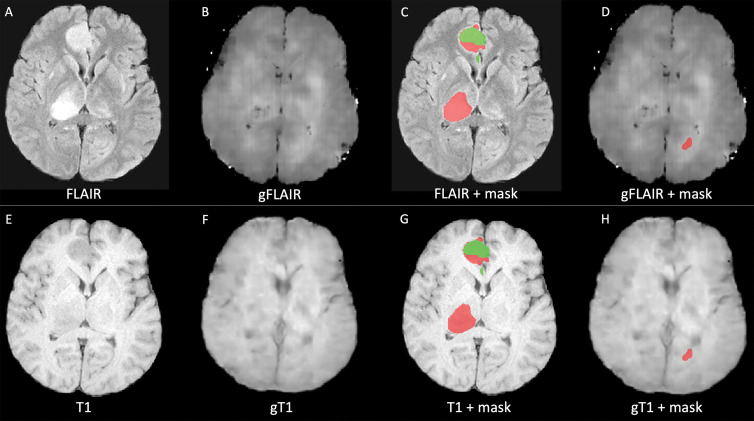
Examples of low-quality segmentation. A, Original fluid-attenuated inversion recovery (FLAIR) MRI scan, B, FLAIR MRI scan generated with generative adversarial network (GAN) (gFLAIR), E, T1-weighted MRI scan, and, F, T1-weighted MRI scan generated with GAN (gT1) created by our models. D, H, Segmentation of lesion obtained with generated FLAIR image and generated T1-weighted image instead of original MRI scans is different from, C, G, segmentation obtained using only original MRI scans that served as reference, with dice similarity coefficient of 0.23. Red indicates segmentation of T2 and/or FLAIR hyperintensity. Green area indicates segmentation of enhanced area of lesion.
Figure 6:
Examples of high-quality segmentation. A, Original fluid-attenuated inversion recovery (FLAIR) scan, B, FLAIR MRI scan generated with generative adversarial network (GAN) (gFLAIR), E, T1-weighted MRI scan, and, F, T1-weighted MRI scan generated with GAN (gT1) created by our models. D, H, Segmentation of lesion obtained with generated FLAIR image and generated T1 image instead of original MRI scans is similar to, C, G, segmentation obtained using only original MRI scans that served as reference, with dice similarity coefficient of 0.87. Red indicates segmentation of T2 and/or FLAIR hyperintensity. Green indicates segmentation of enhanced area of lesion.
Although measures like MSE, SSI, and DSC offer quantitative feedback of the quality of the generated images, it could be difficult to estimate their clinical value. For example, a low DSC could indicate that the model is including in the segmentation a large area that is clearly identifiable as an error by a human. For this reason, we also assessed the impact of using images generated with GANs in a clinical scenario, showing that they reduced the time radiologists needed to spend correcting the segmentation masks (for GANs-derived images vs copies of other sequences, the time was 396 minutes vs 650 minutes, respectively, for the first radiologist and 287 minutes vs 426 minutes for the second radiologist).
Other groups have explored the use of GANs as a possible solution for missing brain MRI data (14–16). Sharma et al (14) designed a multi-input, multi-output network that combines information from four MRI sequences and synthesizes the missing sequences in a single forward pass. They were able to generate all missing sequences in the scenarios where either one, two, or three of the four sequences were missing. Lee et al (15) trained a GAN to investigate what MRI contrasts can be effectively reproduced by generative networks and evaluated the quality of the generated images using them as inputs for a brain tumor segmentation model as we did in our study. Moreover, in line with the study by Sharma et al (14), Lee et al (15) also reported a decrease in performance when trying to synthesize postcontrast T1-weighted images; other studies successfully generated postcontrast T1-weighted images using networks different from GANs (29,30). In a preprint article, Li et al (16) trained a multimodal GAN to synthetize double inversion recovery and postcontrast T1-weighted images using FLAIR, T1-weighted, and T2-weighted images.
We have expanded on these works, confirming our findings with a larger and diverse test set, totaling 89 patients (8010 single MRI sections) from different institutions and disease types. This provides evidence that our model generalized well. There is an urgent need to demonstrate generalizability of DL models. Recently, the American College of Radiology and the RSNA cosigned a letter addressed to the U.S. Food and Drug Administration, underlying how ensuring models’ generalizability should be a required step during evaluations (31).
Our results show that combining publicly available DL models, public data sets, and GANs allows for a broader adoption of DL models in the medical community. For example, Yan and colleagues (11) proved how GANs can improve the generalizability of a segmentation model in MRI scans obtained using equipment from different manufacturers.
Our study had limitations. First, we did not generate all possible MRI sequences; others have proved the limits of GANs in synthesizing MRI sequences, especially postcontrast T1-weighted images (14,15). Second, we validated GAN performance with a single DL model for brain tumor segmentation (25,26). Third, the generalizability of the study may be limited due to our relatively small data set because we included only two tumor types, and it is possible that our results might not generalize to other diseases. Nevertheless, to our knowledge, ours is one of the largest and most comprehensive test sets published so far on this topic, including more than one tumor type. Fourth, we only tested the pix2pix model (20). Although it is possible that other approaches, such as a multi-input multi-output model (14–16), might improve performance, it was out of the scope of this study to test and compare different GANs.
Our results suggested that GANs are an effective approach to synthesize missing brain MRI data for multi-input convolutional neural networks. It is likely that we will soon witness an increase in the number of publicly available DL models because of the availability of software that makes model implementation easy in everyday practice (32,33). However, missing data might limit the broad implementation of these models. Our study proved that GANs can be a possible solution to this issue.
In the future, we plan to assess the validity of our approach using different deep learning models trained for purposes other than image segmentation (eg, classification) and other imaging types. Moreover, a fundamental step toward the implementation of images generated with generative adversarial networks in clinical practice will be to assess the biologic impact that these images have on tasks such as differential diagnosis or imaging follow-up.
Research reported in this publication was supported by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health under award number R01NS113803. The content is solely the responsibility of the Authors and does not necessarily represent the official views of the National Institutes of Health.
Disclosures of Conflicts of Interest: G.M.C. disclosed no relevant relationships. A.D.W. disclosed no relevant relationships. D.C.V. disclosed no relevant relationships. K.A.P. disclosed no relevant relationships. J.C.C. disclosed no relevant relationships. M.B. disclosed no relevant relationships. F.S. disclosed no relevant relationships. D.H.L. disclosed no relevant relationships. R.B.J. disclosed no relevant relationships. W.O.T. Activities related to the present article: institution received grant from National Institutes of Health. Activities not related to the present article: has research grant pending with Mallinckrodt Pharmaceuticals. Other relationships: disclosed no relevant relationships. J.E.E.P. Activities related to the present article: institution received grant from National Institutes of Health. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. B.J.E. disclosed no relevant relationships.
Abbreviations:
- BRATS
- Multimodal Brain Tumor Image Segmentation Benchmark
- CNSL
- central nervous system lymphoma
- DL
- deep learning
- DSC
- dice similarity coefficient
- FLAIR
- fluid-attenuated inversion recovery
- GAN
- generative adversarial network
- GBM
- glioblastoma
- MSE
- mean squared error
- SSI
- structural similarity index
References
- 1.Weltens C, Menten J, Feron M, et al. Interobserver variations in gross tumor volume delineation of brain tumors on computed tomography and impact of magnetic resonance imaging. Radiother Oncol 2001;60(1):49–59. [DOI] [PubMed] [Google Scholar]
- 2.Wang G, Li W, Ourselin S, Vercauteren T. Automatic Brain Tumor Segmentation Based on Cascaded Convolutional Neural Networks With Uncertainty Estimation. Front Comput Neurosci 2019;13:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Havaei M, Davy A, Warde-Farley D, et al. Brain tumor segmentation with Deep Neural Networks. Med Image Anal 2017;35:18–31. [DOI] [PubMed] [Google Scholar]
- 4.Kamnitsas K, Ledig C, Newcombe VFJ, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal 2017;36:61–78. [DOI] [PubMed] [Google Scholar]
- 5.Pereira S, Pinto A, Alves V, Silva CA. Brain Tumor Segmentation Using Convolutional Neural Networks in MRI Images. IEEE Trans Med Imaging 2016;35(5):1240–1251. [DOI] [PubMed] [Google Scholar]
- 6.Sun L, Zhang S, Chen H, Luo L. Brain Tumor Segmentation and Survival Prediction Using Multimodal MRI Scans With Deep Learning. Front Neurosci 2019;13:810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang K, Beers AL, Bai HX, et al. Automatic assessment of glioma burden: a deep learning algorithm for fully automated volumetric and bidimensional measurement. Neuro Oncol 2019;21(11):1412–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolesnikov A, Beyer L, Zhai X, et al. Big Transfer (BiT): General Visual Representation Learning. http://arxiv.org/abs/1912.11370. Published 2019. Accessed September 4, 2020. [Google Scholar]
- 9.Goodfellow I, Pouget-Abadie J, Mirza M, et al. Generative Adversarial Nets. In: Ghahramani Z, Welling M, Cortes C, Lawrence ND, Weinberger KQ, eds. Adv Neural Inf Process Syst 27. Curran Associates, Inc, 2014; 2672–2680. http://papers.nips.cc/paper/5423-generative-adversarial-nets.pdf. [Google Scholar]
- 10.Erickson BJ, Cai J. Magician’s Corner: 5. Generative Adversarial Networks. Radiol Artif Intell 2020;2(2):e190215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan W, Huang L, Xia L, et al. MRI Manufacturer Shift and Adaptation: Increasing the Generalizability of Deep Learning Segmentation for MR Images Acquired with Different Scanners. Radiol Artif Intell 2020;2(4):e190195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagiwara A, Otsuka Y, Hori M, et al. Improving the quality of synthetic FLAIR images with deep learning using a conditional generative adversarial network for pixel-by-pixel image translation. AJNR Am J Neuroradiol 2019;40(2):224–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcadent S, Hofmeister J, Preti MG, Martin SP, Van De Ville D, Montet X. Generative Adversarial Networks Improve the Reproducibility and Discriminative Power of Radiomic Features. Radiol Artif Intell 2020;2(3):e190035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma A, Hamarneh G. Missing MRI Pulse Sequence Synthesis Using Multi-Modal Generative Adversarial Network. IEEE Trans Med Imaging 2020;39(4):1170–1183. [DOI] [PubMed] [Google Scholar]
- 15.Lee D, Moon WJ, Ye JC. Assessing the importance of magnetic resonance contrasts using collaborative generative adversarial networks. Nat Mach Intell 2020;2(1):34–42. [Google Scholar]
- 16.Li H, Paetzold JC, Sekuboyina A, et al. DiamondGAN: Unified multi-modal generative adversarial networks for MRI sequences synthesis. Lect Notes Comput Sci 2019;11767:795–803. https://arxiv.org/abs/1904.12894. Accessed November 25, 2020. [Google Scholar]
- 17.Menze BH, Jakab A, Bauer S, et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging 2015;34(10):1993–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bakas S, Reyes M, Jakab A, et al. Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge. http://arxiv.org/abs/1811.02629. Published 2018. Accessed June 9, 2020. [Google Scholar]
- 19.Bakas S, Akbari H, Sotiras A, et al. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data 2017;4:170117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isola P, Zhu JY, Zhou T, Efros AA, Research BA. Image-to-Image Translation with Conditional Adversarial Networks. https://github.com/phillipi/pix2pix. Accessed February 18, 2020.
- 21.Pedregosa F, Grisel O, Weiss R, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res 2011;12:2825–2830. http://scikit-learn.sourceforge.net. Accessed July 12, 2020. [Google Scholar]
- 22.van der Walt S, Schönberger JL, Nunez-Iglesias J, et al. scikit-image: image processing in Python. PeerJ 2014;2(1):e453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, Bovik AC. Mean squared error: Love it or leave it? A new look at Signal Fidelity Measures. IEEE Signal Proc Mag 2009;26(1):98–117. [Google Scholar]
- 24.Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process 2004;13(4):600–612. [DOI] [PubMed] [Google Scholar]
- 25.Kickingereder P, Isensee F, Tursunova I, et al. Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: a multicentre, retrospective study. Lancet Oncol 2019;20(5):728–740. [DOI] [PubMed] [Google Scholar]
- 26.Isensee F, Jäger PF, Kohl SAA, Petersen J, Maier-Hein KH. Automated Design of Deep Learning Methods for Biomedical Image Segmentation. https://arxiv.org/abs/1904.08128. Published 2019. Accessed June 9, 2020. [DOI] [PubMed] [Google Scholar]
- 27.Ghaffari M, Sowmya A, Oliver R. Automated Brain Tumor Segmentation Using Multimodal Brain Scans: A Survey Based on Models Submitted to the BraTS 2012-2018 Challenges. IEEE Rev Biomed Eng 2020;13:156–168. [DOI] [PubMed] [Google Scholar]
- 28.Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. Lect Notes Comput Sci 2015;9351:234–241. [Google Scholar]
- 29.Gong E, Pauly JM, Wintermark M, Zaharchuk G. Deep learning enables reduced gadolinium dose for contrast-enhanced brain MRI. J Magn Reson Imaging 2018;48(2):330–340. [DOI] [PubMed] [Google Scholar]
- 30.Kleesiek J, Morshuis JN, Isensee F, et al. Can Virtual Contrast Enhancement in Brain MRI Replace Gadolinium?: A Feasibility Study. Invest Radiol 2019;54(10):653–660. [DOI] [PubMed] [Google Scholar]
- 31.ACR and RSNA Comments - FDA WS - Autonomous Imaging AI. 2020. [Google Scholar]
- 32.Philbrick KA, Weston AD, Akkus Z, et al. RIL-Contour: a Medical Imaging Dataset Annotation Tool for and with Deep Learning. J Digit Imaging 2019;32(4):571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mehrtash A, Pesteie M, Hetherington J, et al. DeepInfer: Open-Source Deep Learning Deployment Toolkit for Image-Guided Therapy. Proc SPIE Int Soc Opt Eng 2017;10135:101351K. [DOI] [PMC free article] [PubMed] [Google Scholar]