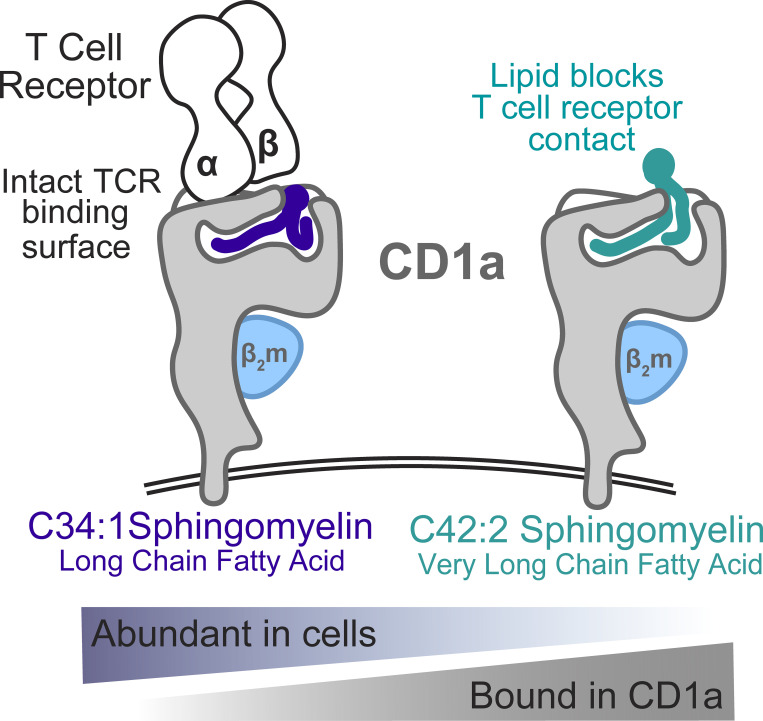
CD1a is expressed on Langerhans cells, where it presents lipid antigens to activate T cells. However, skin cells also produce a natural lipid that binds CD1a and blocks T cell response as a mechanism to down-regulate autoreactivity.
Abstract
We optimized lipidomics methods to broadly detect endogenous lipids bound to cellular CD1a proteins. Whereas membrane phospholipids dominate in cells, CD1a preferentially captured sphingolipids, especially a C42, doubly unsaturated sphingomyelin (42:2 SM). The natural 42:2 SM but not the more common 34:1 SM blocked CD1a tetramer binding to T cells in all human subjects tested. Thus, cellular CD1a selectively captures a particular endogenous lipid that broadly blocks its binding to TCRs. Crystal structures show that the short cellular SMs stabilized a triad of surface residues to remain flush with CD1a, but the longer lipids forced the phosphocholine group to ride above the display platform to hinder TCR approach. Whereas nearly all models emphasize antigen-mediated T cell activation, we propose that the CD1a system has intrinsic autoreactivity and is negatively regulated by natural endogenous inhibitors selectively bound in its cleft. Further, the detailed chemical structures of natural blockers could guide future design of therapeutic blockers of CD1a response.
Graphical Abstract
Introduction
CD1 proteins are monomorphic antigen-presenting molecules that bind self and foreign cellular lipids for display to T cells (Calabi and Milstein, 1986; Salio et al., 2014; Van Rhijn et al., 2015). Newly synthesized CD1 proteins fold in the endoplasmic reticulum, where they capture lipids and traffic as CD1–lipid complexes through the secretory pathway (Briken et al., 2002; Kang and Cresswell, 2002; Park et al., 2004) to the cell surface and bind to αβ and γδ TCRs (Borg et al., 2007; Luoma et al., 2013; Uldrich et al., 2013). Among human CD1 antigen-presenting molecules, only CD1a lacks any known reinternalization motif in its cytoplasmic tail (Sugita et al., 1999). Thus, more so than CD1b, CD1c, and CD1d, which efficiently traffic to the endolysosomal network to capture exogenous lipids, CD1a is thought to acquire self lipids in the secretory pathway for presentation to T cells (Moody and Cotton, 2017).
In agreement with the hypothesis that self lipids dominate in the control of CD1a-mediated T cell response, the frequency of CD1a autoreactive T cells in the blood is higher than for T cells that recognize CD1b, CD1c, and CD1d when measured by activation or limiting dilution assays (de Jong et al., 2010; de Lalla et al., 2011). CD1a autoreactive T cells colonize human skin, where CD1a tetramer-binding T cells are ∼100-fold more frequent than CD1b tetramer-binding T cells and account for ∼1% of total T cells (Cotton et al., 2021). Likewise, CD1a proteins are expressed at high density in skin, where they are found on myeloid dendritic cells and at particularly high density on Langerhans cells (LCs; Dougan et al., 2007). Self-antigens for CD1a are extracted from skin in larger amounts than from other tissues, and CD1a autoreactive T cell clones respond to lipids that normally accumulate in skin, including squalene, which is the main component of sebum, as well as wax esters and free fatty acids (de Jong et al., 2014).
The abundance of CD1a autoreactive T cells, CD1a, and CD1a-presented antigens raises basic questions about possible mechanisms of inhibition of CD1a autoreactivity, which might otherwise result in autoimmune disease. Indeed, recent studies suggest that CD1a autoreactive T cells are present at higher levels in human skin involved with atopic dermatitis and psoriasis compared with skin from healthy donors or noninvolved skin (Cheung et al., 2016; Subramaniam et al., 2016). Allergens that contain phospholipase activity generate lysolipids that contribute to T intradermal cell response (Bourgeois et al., 2015; Cheung et al., 2016; Subramaniam et al., 2016). Further, human CD1a transgenic mice display markedly augmented skin immune responses to the contact dermatitis antigen urushiol and the contact sensitizer imiquimod, a widely used model of psoriasiform skin inflammation (Kim et al., 2016).
CD1a has the smallest cleft among the four types of human CD1 antigen-presenting molecules (Moody et al., 2005) and is composed of two named pockets: A′ and F′. The first CD1a–lipid structure solved was CD1a–sulfatide: the sphingosine chain resides in the A′ pocket, with the acyl chain in the F′ pocket and the sulfo-sugar protruding through the F′ portal (Zajonc et al., 2003). Subsequent CD1a–lipid structures show that lipopeptides (Zajonc et al., 2005), lysophospholipids (Birkinshaw et al., 2015), urushiol (Kim et al., 2016), and farnesol (Nicolai et al., 2020) each show distinct positioning within CD1a. These structural studies raise questions about the extent of the natural diversity of lipid ligands, suggesting that CD1a might bind to chemically diverse lipids or, at the extreme, nearly every type of endogenous cellular lipid.
In this study, we used comparative mass spectrometry (MS) of lipids bound to CD1a to ask a fundamental question, which to our knowledge has not been addressed previously. Do CD1a proteins randomly bind nearly all types of lipids present in the cell in which it is expressed, or does the size and shape of the CD1a cleft guide capture specific classes of cellular lipids? By comparing the spectrum of lipids bound in CD1a to all lipids present in the cell, we found that sphingolipids, especially certain molecular subspecies of sphingomyelins (SMs), were markedly over-represented in CD1a eluents. The preferentially captured SMs are normally expressed in cells and tissues that express CD1a, and they are potent and broadly acting blockers of CD1a–TCR binding. The detailed molecular mechanism of TCR blockade was solved through three crystal structures of CD1a proteins bound to SMs that do or do not have inhibitory properties.
CD1a T cell autoreactivity is particularly common in blood (de Jong et al., 2010; de Lalla et al., 2011) and skin-derived (Cotton et al., 2021) T cells, and a recent study showed how polyclonal T cell autoreactivity to CD1a can occur through lipid-independent contact of TCRs on the surface of CD1a (Cotton et al., 2021). Opposite to most views that emphasize antigen-driven T cell activation, here we used an unbiased cell screen to discover endogenous lipids that are selectively captured by CD1a proteins in cells and provide information about how their structures confer binding to CD1a and act broadly as a dominant negative control of polyclonal TCR binding.
Results
Lipidomic analysis of cells versus CD1a
We used three parallel MS approaches to detect ligands bound to CD1 proteins expressed in cells. Nano-electrospray ionization–MS shotgun MS provides a highly sensitive method to broadly but nonquantitatively detect ionizable ligands eluted from CD1 proteins recoverable from cells (Cox et al., 2009; Huang et al., 2011; Joyce et al., 1998; Yuan et al., 2009). In contrast to shotgun methods, preseparation of compounds by HPLC–time of flight (ToF)–MS limits ionization cross-suppression by chemically dissimilar ions, allowing sensitive and quantitative detection even on a microscale needed for detection of trace lipids eluted from proteins. The high mass resolution of ToF-MS allows reliable assignment of chemical formulas (Cotton et al., 2021; Wun et al., 2018). Normal-phase chromatography separates lipid classes, where retention time (RT) correlates with polarity, and RT comparisons to lipid standards are possible (Layre et al., 2011). Last, quadrupole ToF (QToF)–MS detects fragments corresponding to functional groups and neutral loss of components that allow identification of key unknowns.
We recently demonstrated that CD1a complexes formed in cells with endogenous lipids can be sensitively detected and are heterogeneous (Cotton et al., 2021). However, it is currently unknown if CD1a randomly surveys membrane and other lipids present in the cells in which it is expressed or instead has mechanisms for selective capture of specialized lipid classes. Therefore, we performed comparative lipidomics of all ionizable lipids in chloroform and methanol extracts of HEK293T cells compared with the cohort of lipids eluted from CD1a proteins expressed in this cell line. CD1a proteins carrying endogenous lipids (CD1a-endo) from HEK293T cells were previously shown to bind TCRs, so both CD1a protein folding and capture of a cohort of ligands with physiological T cell reactivity were validated (Cotton et al., 2021). Prior studies also indicated that detergent extraction of transmembrane CD1 proteins displaces lipid ligands from CD1 clefts, a problem that is bypassed by engineering transmembrane domain-truncated constructs that traverse the secretory pathway for lipid capture in detergent-free conditions (Cox et al., 2009; Huang et al., 2011; Yuan et al., 2009). Originally developed to profile unlimited amounts of lipids present from cells (Layre et al., 2011), new microscale methods of normal-phase HPLC broadly separate and detect the lipids present in eluents of 20–50 µg of CD1 protein (Cotton et al., 2021; Wun et al., 2018). Multiple ligands in CD1 eluates can be detected over a broad dynamic range with low rates of false positivity compared with eluents of MHC proteins (de Jong et al., 2014; Huang et al., 2011).
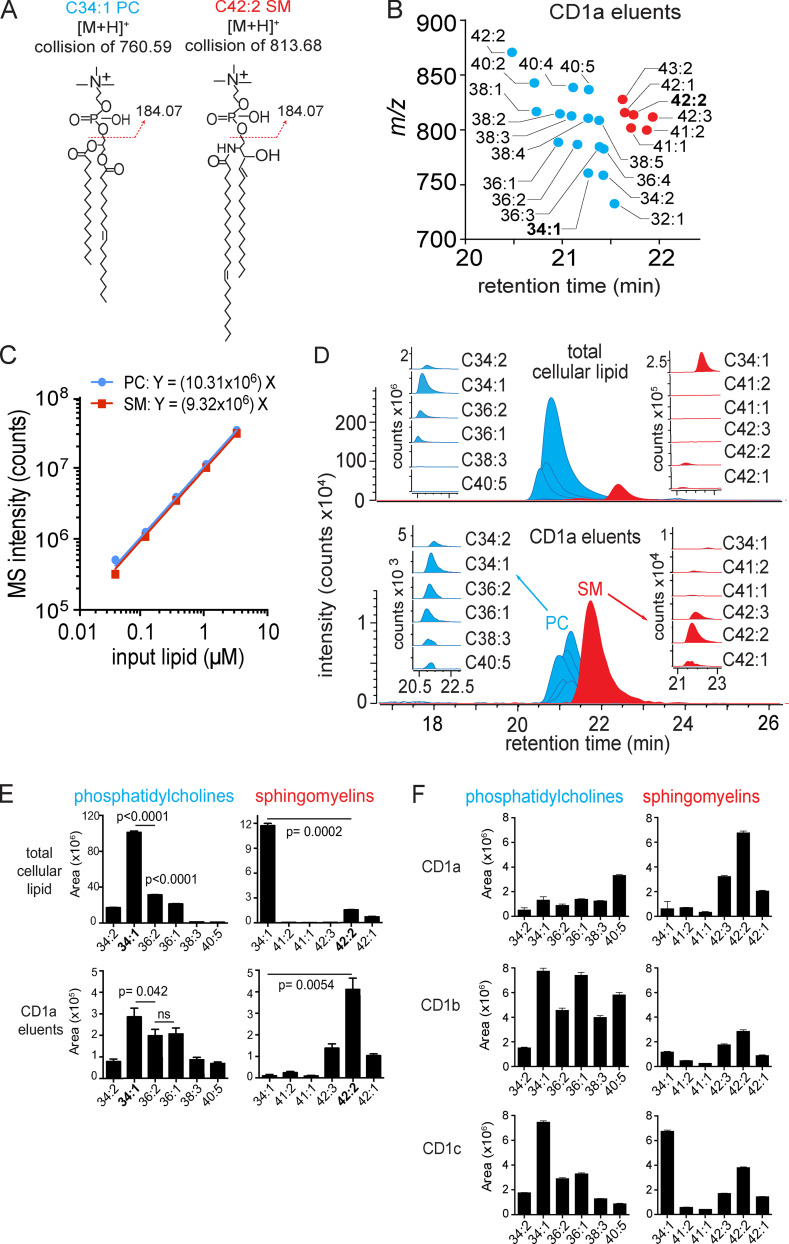
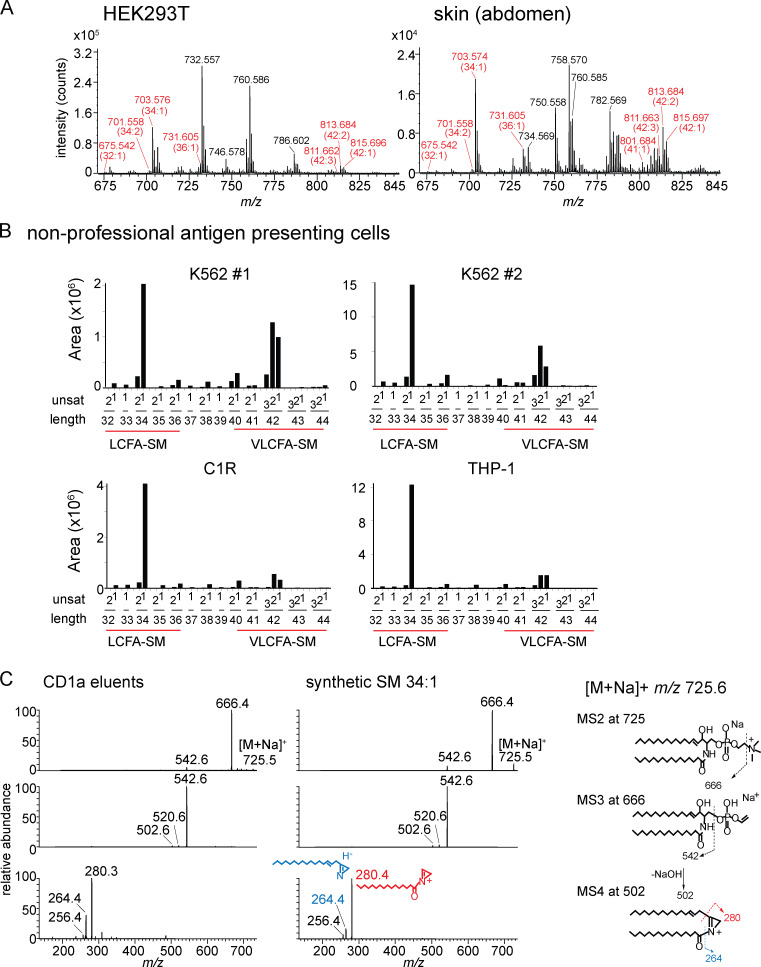
We compared total lipids from HEK293T cells to the spectrum of endogenous lipids eluted from CD1a, focusing on the most intense ions. We tentatively identified lipids when their elution time and m/z values matched those of authentic standards for sulfatides, ceramides, hexosyl ceramides, diacylglycerols, phosphatidylinositols, SMs, and phosphatidylcholines (PCs) as detailed previously (Cotton et al., 2021). Then, key compounds were confirmed using collision-induced dissociation MS and coelution of standards. For example, the ion at m/z 760.59 matched the expected mass of PC, whose two fatty acyl units combined for 34 methylene groups and one unsaturation (34:1 PC). Another ion of m/z 813.68 matched the expected mass of SM, where the combined fatty acyl and sphingosine base contained 42 methylene units and two unsaturations (42:2 SM; Fig. 1 A).
Figure 1.
Targeted HPLC-MS analysis of CD1 monomer eluents and cellular lipids. (A) Collision-induced dissociation–MS analysis for CD1a ligands matching the expected m/z of 34:1 PC and 42:2 SM identified the phosphocholine groups, allowing assignment of the overall length and unsaturation states of the alkyl chains. The position and Z or E stereochemistry are inferred from known lipid structures but cannot be established by MS. (B) Chain length and saturation variants identified within the same lipid class have equivalent RTs that match those of SM and PC standards. The m/z values allowed deduction of the combined chain length of the acyl chain and sphingosine units that vary by an integer number of methylene units (X) or unsaturations (Y), shown as X:Y. We identified 16 molecular variants of PC (blue points) and six SMs (red points), which were seen in two datasets analyzed. (C) Response factors for SM and PC were highly similar and nearly linear based on MS intensity measured as a function of the mass input for two synthetic standards, 34:1 PC AND 42:2 SM. (D) Mass chromatograms of the six most abundant PC and SM family members eluted from the CD1a monomer (bottom) and in the total lipid extract from matched CD1a-producing HEK293T.TPM cells (top). (E) PCs and SMs quantified as integrated area under the curve (counts) for each lipid chain variant detected in triplicate (± SEM) at a diagnostic m/z value and RT window. P values were calculated using Welch’s corrected t test. (F) PCs and SMs, eluted from CD1a, CD1b, and CD1c protein monomers, quantified as integrated area under the curve for each lipid chain variant detected in triplicate at a diagnostic m/z value and RT window. Error bars indicate SD from the mean. For D and E, results are representative of three experiments, and for F, results are representative of more than three experiments with interexperimental replication shown in Fig. S1. [M+H]+, mass of the molecular ion plus a proton adduct.
PC and SM are structurally related lipids that both contain phosphocholine head groups and showed the diagnostic fragment of phosphocholine (m/z 184.07; Fig. 1 A). Next, we counted and assigned the molecular variants within each lipid class based on total chain length and unsaturations. Considering ions released from CD1a that nearly coelute with 34:1 PC (20.4 min–21.5 min), we deduced 15 additional PC variants ranging from C34 to C42 with up to five unsaturations (Fig. 1 B, blue). Using the same approach, we identified five additional SMs eluting from CD1a that nearly coeluted with 42:2 SM (Fig. 1 B, red). The names, formulas, and experimental mass errors for the 22 distinct PCs and SMs are detailed in Table S1. Overall, mass-based detection revealing molecular heterogeneity of chain length and lipid unsaturation within each class was detected.
Quantitation of cellular lipids on the microscale with MS
To quantitate lipids in cells or eluted from CD1a, we determined the MS response factors for injections of PC and SM standards lipids (Fig. 1 C). The signal was directly related and highly correlated to the mass injected for PC and SM up to 10 million counts, and the response factors were quite similar, as expected based on their equivalent head groups, which typically control ionization efficiency (Fig. 1 C). The nearly linear relationship and equivalent response factors for PC and SM create a situation in which MS intensity measurements, quantitated from the area under the HPLC-MS curve, approximate the relative molar amount of compounds, allowing ranking of compounds as a measure of relative abundance (Fig. 1 D, insets). In cells, the most abundant PC species, 34:1 PC, was approximately eightfold more intense than the most abundant 34:1 SM (Fig. 1 D, upper). This finding is in agreement with prior reports, where PC is a major structural component of membrane bilayers, and SM is a less-abundant lipid with specialized roles in lipid rafts, signaling, and other processes (Fahy et al., 2005; van Meer, 2005).
CD1a-bound lipids versus cellular lipids
Comparison of triplicate lipidomic profiles from HEK293T cells to lipid eluents of CD1a proteins expressed in HEK293T cells generated two striking findings. First, whereas PCs dominated in cells, SM species showed much higher signals in CD1a eluents, representing an inversion of the PC/SM ratio (Fig. 1 D, red and blue). This strong effect was quantitatively validated by separately determining chromatogram areas for the six most abundant molecular species in each class in triplicate, allowing for statistical comparisons (Fig. 1 E). Second, the SMs bound to CD1a skewed toward longer and more unsaturated alkyl chains compared with the cellular pool of SMs (Fig. 1, D and E). Among cellular SMs, a shorter 34:1 SM species was predominant, with the ion chromatogram area measuring 7.5-fold more than 42:2 SM, which was scarcely detectable (P = 0.0002; Fig. 1 D, inset, red). Conversely, in CD1a eluents, 34:1 SM was so low in intensity that it was not initially picked up by the automated peak-picking software. By manually searching for this specific mass and integrating the peak area, we found that 34:1 SM was detectable only at trace levels: 44-fold lower than 42:2 SM in CD1a eluents (P = 0.0054). Overall CD1a selectivity for 42:2 was clearly evident, such that the molecule was scarcely detectable in total cells but was the most abundant molecule eluting from CD1a among the PCs and SMs tested (Fig. 1 E). Considering the PC:SM inversion and the SM length and saturation preference together, relative enrichment was conservatively estimated to exceed 100-fold. Both a C8 longer chain length and a second unsaturation correlated with preferential capture.
Extraction of cellular lipids is a simple, one-step process that provides quantitative yield. In contrast, the necessary design and handling of CD1 proteins before generating lipid eluents is more complex and might have influenced the spectrum of lipids detected. For example, longer chain lipids might remain preferentially bound to CD1a proteins during extracellular protein purification. However, whereas CD1a preferentially captured and released longer SMs, the profiles of PCs seen in whole cells and on CD1a were similar, suggesting that CD1a protein purification was not causing bias for long chain length in a general way (Fig. 1 D, blue inset; and Fig. 1 E). Further, we analyzed capture patterns in triplicate from three independently produced batches of group 1 CD1 proteins in HEK293T cell lines, CD1a, CD1b, and CD1c (Fig. 1 F). This second CD1a protein batch confirmed prior findings, as SMs were greater than PCs, and 42:2 SM was the most abundant CD1a-associated SM species, followed by 42:1 and 42:3 SM variants. In CD1b and CD1c, detectable skewing toward longer SMs was seen, but it was much weaker than for CD1a, and the short-chain 34:1 SM remained the predominant SM released from CD1c.
Lipid elution profiles from three CD1a preparations with a different protein production system or cell type (HEK293S, HEK293T), which were generated in different laboratories, confirmed the SM predominance and the strong skewing to longer 42:2 SM (Fig. S1). In one preparation, somewhat higher 34:1 SM eluted from CD1a, but this effect did not reproduce in a second experiment using the same proteins or in a separately prepared CD1a batch (Fig. S1). Overall, the selective 42:2 SM capture was reproducible among cell lines with independently engineered CD1 proteins and purification methods in two laboratories. The chain length effect was only found for the particular combination of CD1a and SM, and it was strong, where sphingolipids bound preferentially compared with phospholipids, and among SMs, one unsaturation in the alkyl chain and C8 added length correlated with increased capture. We interpret the over-representation of lipids in the CD1a eluent as preferential capture or retention in CD1a proteins traversing the secretory pathway.
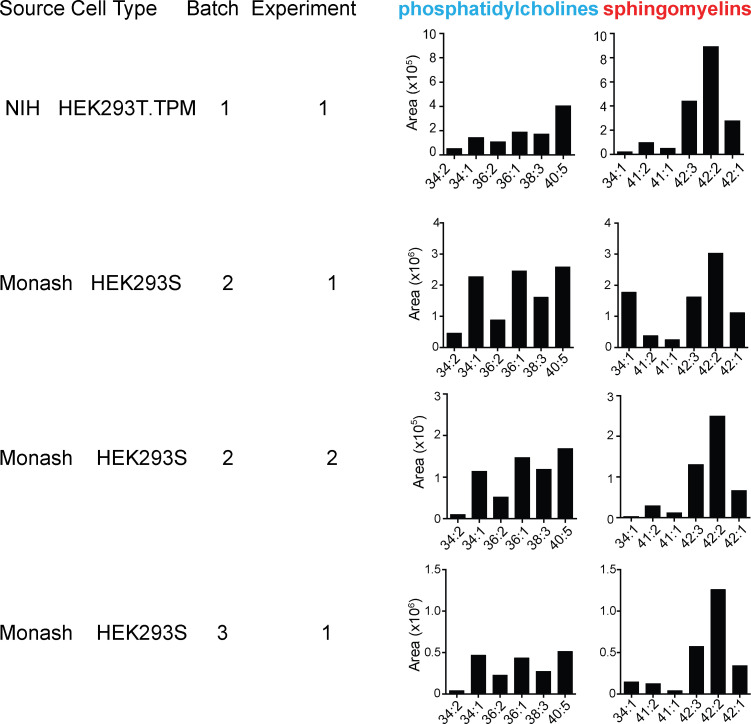
Figure S1.
Detailed SM profiles from HEK293 cells. CD1a proteins generated in HEK293S or HEK293T.TPM cells were treated with chloroform and methanol, with eluents subject to analysis by positive ion mode HPLC-MS, where the combined length (X) and unsaturations (Y), shown as X:Y, were deduced based on the detected m/z values of natural SMs nearly coeluting with an SM standard. CD1a constructs and proteins were generated at the NIH tetramer facility or at Monash University (Monash).
Molecular analysis of natural and synthetic SMs
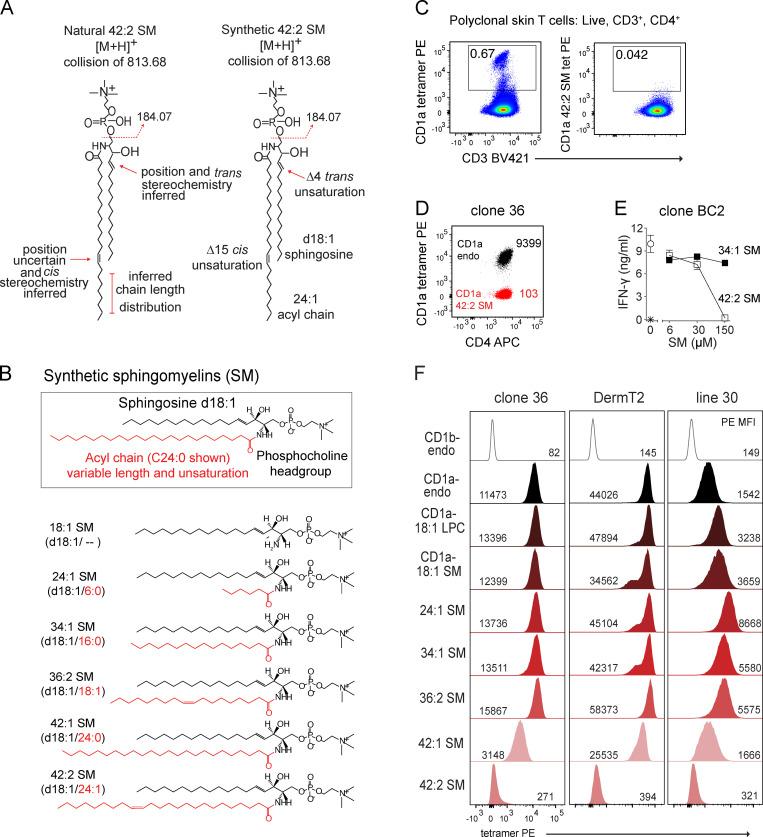
To determine the molecular basis of SM capture by CD1a, we assembled a panel of synthetic SMs, which differed only in alkyl chain length and saturation. Electrospray ionization–MS provides high sensitivity for the microanalysis of ligands, and it could reliably establish molecular formulae (Fig. 1) but not the complete structures of SM species (Fig. 2 A). For example, the total number of methylene units and unsaturations in the combined sphingosine and alkyl chains can be reliably inferred, but assigning the position and stereochemistry of unsaturations is indirect and relies on analogies to known compounds. Sphingosine, also known as D-erythro-sphingosine or (E)-sphing-4-enine in reference to the trans unsaturation at the fourth carbon (abbreviated d18:1), is the most common sphingoid base in mammalian cells, and so the sphingosine base unsaturation is somewhat reliably inferred (Merrill, 2011). The true location of the fatty acyl unsaturation is unknown and might be varied. Also, for any single ion predicting a total length of two chains is C42, the actual composition might contain isobaric lipids, where incremental increases in the sphingosine base are matched to shorter fatty acyl units and vice versa (Fig. 2 A). In contrast, the actual length as well as the position and stereochemistry of unsaturations in pure synthetic molecules is known. Accordingly, we studied synthetic SMs with the d18:1 Δ4 trans unsaturated sphingosine base (18:1 SM) with varied amide-linked fatty acids up to C24:0 (42:1 SM) or C24:1 cis unsaturation at the indicated position (42:2 SM; Fig. 2, A and B).
Figure 2.
Lipid chain length and unsaturation determine CD1a tetramer binding. (A) CD1a-eluted natural versus synthetic 42:2 SMs are shown with regard to known or inferred aspects of structure. (B) Synthetic SM variants differ in the length and saturation of the fatty acyl unit (red). (C) Polyclonal skin T cells stained with CD1a tetramer (tet) or tetramer treated with 42:2 SM, showing results that are representative of two experiments. (D) CD1a autoreactive skin T cell clone 36 staining with CD1a-endo or CD1a–42:2 SM, with results shown being representative of three experiments. Insets indicate tetramer MFI. (E) IFN-γ ELISA of clone BC2 T cells (star indicates T cells only) exposed to plate-bound CD1a treated with the synthetic SM, with results representative of two experiments. (F) CD1a–SM tetramer staining of skin T cell lines DermT2, Line 30, and Line 36. Cells were pregated (live, CD3+, CD4+ Autofluoresence[FITC]neg), and histograms were normalized to mode. [M+H], mass of the molecular ion plus a proton adduct.
SM influence on CD1a–TCR binding
We considered the immunological implications of selective capture of long-chain unsaturated SMs on T cell activation. Sphingolipids, like the natural killer T cell (NKT) agonist α-galactosylceramide (Kawano et al., 1997), are antigens in the CD1d system, but to our knowledge SM has not been prominently identified as a CD1-restricted T cell antigen, and in one study, SM was able to block activation of a CD1a autoreactive clone (de Jong, 2015). To search for 42:2 SM–specific T cells, we tetramerized 42:2 SM–treated CD1a monomers and stained polyclonal skin-derived T cells (Fig. 2 C). CD3+CD4+ T cells staining a control CD1a tetramer that carried endogenous lipids (CD1a-endo) derived from the cellular expression system for CD1 was readily detectable (0.67%) as a discrete, brightly staining population. 42:2 SM treatment of CD1a-endo tetramers blocked rather than augmented this staining. Recent studies have found that CD1a-endo tetramers frequently and reproducibly stain T cells expressing TCRs that bind to the outer surface of CD1a but ignore the carried lipids, so are said to be CD1a specific (Birkinshaw et al., 2015; Cotton et al., 2021). Here, treatment of CD1a tetramers with 42:2 SM strongly blocked staining of both bright and dim tetramer+ cells (Fig. 2 C). This result in one subject was inconsistent with the idea that 42:2 SM might be an immunodominant antigen and pointed to a possible blocking function, which was tested further.
Prior screening for CD1a antigens in vitro had hinted at SM as an antagonist of CD1a-mediated T cell activation (de Jong et al., 2014), a result that was confirmed and broadened in recent studies of polyclonal NKT cells recognizing CD1d (Melum et al., 2019). Therefore, we next considered that 42:2 SM might act as an inhibitor of CD1a-autoreactive T cell clones in which the CD1a-autoreactive TCRs are known: BC2 (de Jong et al., 2014) and clone 36 (Cotton et al., 2021). Clone 36 stains brightly with the CD1a-endo tetramer, and treating CD1a with synthetic 42:2 SM reduced tetramer staining intensity (mean fluorescence intensity [MFI]) by 90-fold (Fig. 2 D). Furthermore, when we treated plate-bound CD1a proteins with synthetic 34:1 SM or 42:2 SM and co-cultured with the CD1a-autoreactive T cell clone BC2 (Fig. 2 E; de Jong et al., 2014), increasing concentrations of synthetic 42:2 SM reduced T cell IFN-γ production. This effect was unlikely to be attributable to general toxicity because it was not seen with 34:1 SM, and the plate assay uses a pulse format in which antigens, although used at high concentrations, are not coincubated with T cells. Overall, three observations with synthetic SMs of known structure suggested T cell inhibition.
Molecular fine mapping of CD1a–TCR blockade
Sphingosines can be signaling molecules that might have blocked functional T cell response through general effects on APCs, CD1a, or T cell viability. However, the inhibition of CD1a tetramer staining and T cell activation by plate-bound CD1a both pointed toward a specific mechanism whereby SM binds CD1a and blocks TCR contact. The cellular basis of sphingolipid capture by CD1a remained unknown and might have occurred through delivery of sphingolipids to CD1a or lipid-loading cofactors, like saposins and microsomal triglyceride transfer proteins, which play key roles in endogenous lipid loading, especially for CD1d in late endosomes (Brozovic et al., 2004; Dougan et al., 2005; Kang and Cresswell, 2004; Zhou et al., 2004). However, tetramer loading with exogenous lipids onto CD1a is independent of cellular cofactors. Therefore, the nearly identical patterns of endogenous (Fig. 1) and exogenous (Fig. 2) loading based on lipid saturation and chain length provided a hint that the patterns were determined by intrinsic properties of CD1a proteins. We therefore hypothesized that long chain length and unsaturation directly promote the efficiency of CD1a–SM complex formation.
To test this hypothesis, CD1a monomers were treated with candidate blockers ranging from C34 to C42 SM with one or two unsaturations, tetramerized, and used to stain human T cell lines with partially characterized or known TCRs: clone 36, DermT2 (TRBV2*01 TRBJ1-5*01, TRAV8-6 TRAJ20*01), and line 30 (>90% Vβ22+; Fig. 2, B andF). These three T cell lines were chosen for their low (line 30; MFI = 1,542), intermediate (clone 36; MFI = 11,473), or high (Derm T2; MFI = 44,026) staining with CD1a-endo tetramers, creating a range of baseline values from which each line might show differing sensitivity to increased or decreased staining with alternatively loaded tetramers (Fig. 2 F). We used two controls: CD1b-endo tetramers, which are not expected to bind to CD1a autoreactive lines, and CD1a tetramers treated with lysophosphatidylcholine (LPC), which is a small C18 ligand that does not generally block CD1a-specific TCRs (Birkinshaw et al., 2015; de Jong, 2015) or CD1c–TCR interactions (Wun et al., 2018).
CD1a tetramers formed with shorter-chain lipids, including LPC, 18:1 SM, 24:1 SM, 34:1 SM, and 36:2 SM, stained lines at a similar or slightly higher MFI than the heterogeneously loaded CD1a-endo tetramers (Fig. 2 F). For line 30, the LPC and smaller SMs up to C36 in length clearly augmented staining compared with CD1a-endo. In contrast, both 42:1 and 42:2 SM ligands blocked CD1a staining for all lines tested. The doubly unsaturated 42:2 SM blocked tetramer binding 50–100-fold, nearly to the background levels seen with CD1b tetramers. The molecular patterns of tetramer staining blockade matched those conferring CD1a capture in cells: a C6–8 increment of increased chain length and a second unsaturation separately conferred increased biological effects, again suggesting that these features of natural SMs are binding determinants for CD1a. The opposite biological effects of short- and long-chain SMs could have occurred either as a differential extent of binding to CD1a or as formation of CD1a–SM complexes that are structurally distinct in ways that affect TCR binding.
Structural basis of CD1a presenting different classes of SM
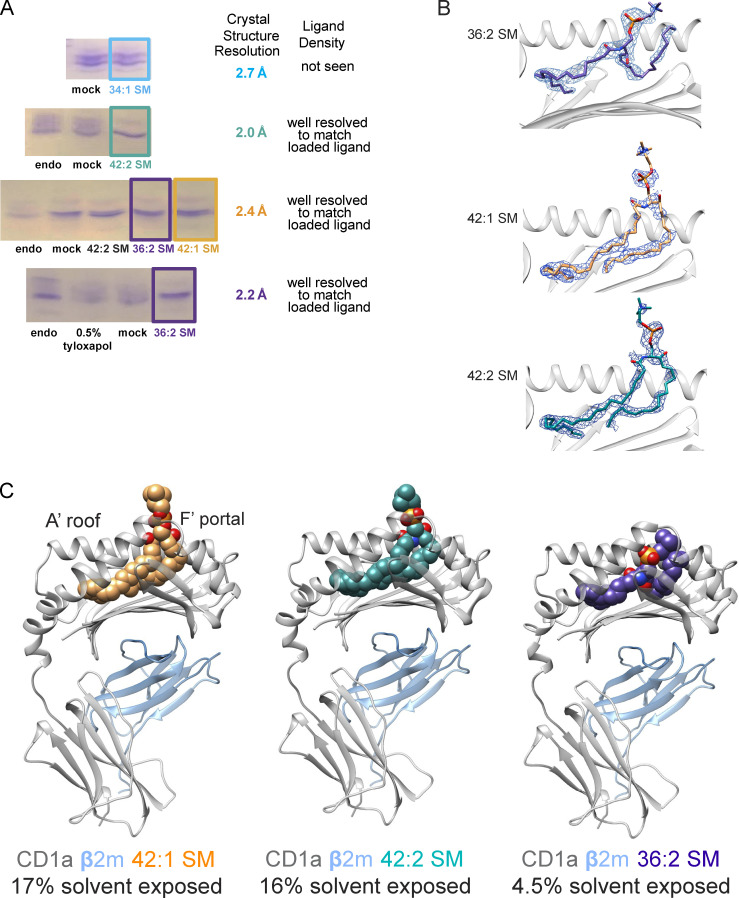
To determine how different SMs might fine-tune CD1a–lipid complex architecture, we aimed to solve binary crystal structures of a CD1a bound to 34:1 SM, 36:2 SM, 42:1 SM, and 42:2 SM. Our initial attempts to load 34:1 SM onto CD1a proved challenging: crystals formed, but they diffracted to 2.7 Å resolution with no clearly defined electron density corresponding to a single lipid in the binding pocket, which is consistent with either low occupancy of a single lipid or occupancy with diverse lipids from the expression system. (Fig. 3 A and data not shown). Thus, the poor in vitro loading efficiency of exogenous 34:1 SM again matches patterns seen in capture of endogenous cellular SMs (Fig. 1), whereby the 34:1 SM showed the highest MS intensity among SM species in cells, yet was the least abundant form eluted from recombinant cellular CD1a proteins (Fig. 1).
Figure 3.
Loading and structural elucidation of CD1a–SM complexes with altered fatty acyl chain length. (A) Deglycosylated CD1a carrying HEK293T cell–derived lipids (endo) was treated with CHAPS detergent (mock) or different species of SM and resolved on isoelectric focusing gels. In case of 36:2 SM, endogenous lipids were “washed out” using 0.5% tyloxapol before SM loading. Results are typical for two or more experiments. (B) 2Fo-Fc electron density maps of the ligands in the cleft are contoured at σ of 0.7 Å. (C) Overview of the binary structures of CD1a/β2m heterodimer (gray and blue, respectively) bound to 42:1 SM (orange), 42:2 SM (teal), or 36:2 SM (purple).
Isoelectric focusing (Fig. 3 A) of CD1a treated with 36:2 SM, 42:1 SM, or 42:2 SM showed markedly reduced band heterogeneity, suggesting high loading efficiency into CD1a. All three binary complexes crystallized, leading to crystal structures of CD1a–42:1 SM, CD1a–42:2 SM, and CD1a–36:2 SM, resolved to 2.4 Å, 2.0 Å, and 2.1 Å resolution, respectively (Fig. 3, B and C; andTable S2). In all three structures, we observed a clear unbiased electron density in the cleft of CD1a, leading to the unambiguous assignment of the lipid ligand position. The CD1a cleft has two pockets, A′ and F′, which connect to the outer surface of CD1a via the F′ portal. The internal volume of CD1a measured from prior crystal structures is 1660 ± 220 Å3 and corresponds to a chain length capacity of ~C34–C39 (Zajonc et al., 2005; Zajonc et al., 2003). Therefore, the combined fatty acyl and sphingosine chains in these three SM molecules of C36–42 plus the molecular volume of the choline head group were expected to just fill or perhaps overfill the CD1a cleft and protrude through the F′ portal.
Indeed, for C42:1 SM and C42:2 SM complexes, 16% and 17%, respectively, of the ligand protrudes and is solvent exposed on the outer surface of CD1a. In contrast, the 36:2 SM is sequestered within CD1a so that only 4% is solvent exposed (Fig. 3 C). The simplest structural explanation might be that the C6 length increment creates protrusion on a size basis of otherwise similarly seated molecules, pushing the phosphocholine groups through the F′ portal toward the surface. However, while there is some size effect contributing to the protrusion, comparative analysis of the C42 and C36 CD1a–SM structures show marked differences in lipid seating within CD1a, revealing both lipid and CD1a remodeling after binding different SMs.
An ionic platform stabilizing CD1a
Both 42:1 SM and 42:2 SM bound to CD1a in a manner that is similar to each other and to the mechanism observed previously for CD1a–SM (Protein Data Bank: 4X6F; Birkinshaw et al., 2015). Both lipid tails enter the F′ pocket and run in parallel until the shorter, C18 sphingosine unit ends near V12, while the longer C24 acyl tail turns around F70 and terminates deep inside the A′ pocket (Fig. 4 A). Both chains fully occupy the cleft and establish an extensive network of mostly hydrophobic interactions with apolar residues of CD1a (Table S3).
Figure 4.
Remodeling of the CD1a cleft and surface with short- and long-chain SMs. (A) Side view of CD1a–42:2 SM shows the acyl and sphingosine chains lying parallel in the A′ pocket, where the fatty acyl unit encircles an internal structure known as the A′ pole (Moody et al., 2005) formed by F70 and V12, with the position of the unsaturation in the 42:2 SM shown with a cyan arrow. W14 at the bottom of the A′ pocket floor is flipped toward the F′ pocket. (B) In contrast, 36:2 SM is seated 7 Å lower within the CD1a cleft, and its two chains are oriented antiparallel. W14 is bent toward the A′ pocket so that it lies between the acyl chain in the A′ pocket and the sphingosine chain in the F′ pocket. Unlike CD1a–42:2 SM, R76 and R73 stabilize the phosphate group to bend the SM back toward the cleft. The hydrogen bonds and electrostatic interactions are marked with a dashed line. (C) Superimposed top-down views of the CD1a roof for CD1a–42:1 SM and CD1a–42:2 SM complexes, indicating positioning of CD1a F′ portal adjacent residues R73, R76, and E154, in which R76 and the SM head group orient vertically to protrude above the CD1a platform for CD1–42:2 SM. (D) For the CD1a–36:2 SM complex, R76 and the phosphate in the choline group of SM are positioned near the plane of the CD1a platform.
However, for 36:2 SM, the sphingosine and fatty acyl chains orient nearly antiparallel: here, the acyl chain turns around F70 and anchors deep inside the A′ pocket near V28 (Fig. 4 B). Its reduced length results in a clear movement of the phosphate group, which in this case sits ∼7 Å deeper in the F′ pocket compared with that of the 42 SMs, significantly decreasing the remaining volume of the binding cleft (Fig. 4 B and overlay). Consequently, the sphingosine unit cannot adopt similar orientation to the one observed in CD1a–42:2 and CD1a–42:1 SM, as it would sterically clash against F70. Instead, it induces remodeling at the bottom of the pocket. Namely, the side chain of W14 flips 180° and orientates toward V12, creating an open space that accommodates a segment of the sphingosine chain, which then sharply turns toward the F′ portal, where it is further coordinated by a stretch of apolar residues (Fig. 4 B), similar to the previously solved sulfatide-bound CD1a structure (Zajonc et al., 2003). The orientation of the lipid tail within the F′ portal area is unlikely to have an important role in the CD1a–TCR interaction because it is enclosed within CD1a.
As compared to 42:2 SM and 42:1 SM, the shorter acyl chain in 36:2 SM positions the phosphate group ∼7 Å deeper into CD1a (Fig. 4, A and B). This lower position buries the phosphocholine group of SM within CD1a, where the phosphate group makes ionic interactions with R73 and R76, as well as a hydrogen bond between the oxygen on the acyl chain and R73 (Fig. 4 B). These three charge–charge interactions create a stable network, locking 36:2 SM more deeply inside CD1a within the neck of the portal to the F′ pocket. The smaller alkyl chains do not merely allow lower seating of the whole lipid, but instead these three defined interactions bend the phosphocholine head group back toward the CD1a platform so that it runs parallel to and below the presumed CD1a–TCR contact surface (Fig. 4 B).
In contrast, the longer C24 acyl chains in 42:1 SM and 42:2 SM push these ligands higher in the cleft so R73 does not interact with SM, and the oxygen on the SM sphingosine chain forms a hydrogen bond with R76 (Fig. 4 A). Therefore, the phosphocholine unit protrudes substantially through the F′ portal so that 16–17% of its surface area is solvent exposed. Also, this higher position disrupts the R73-E154-R76 salt bridge and the margin of the A′ roof (Fig. 4, A–C). The bulky phosphate group is positioned close to the side chain of E154 and forces R76 to orient vertically. This kind of lipid-induced molecular arrangement was previously proposed to negatively impact the recognition of CD1a by the BK6 TCR and illustrates the situation in which an inhibitory ligand renders a surface epitope of CD1a unavailable to TCR binding (Birkinshaw et al., 2015). In contrast, when CD1a harbors the shorter 36:2 SM ligand, the A′ roof integrity is maintained (Fig. 4 D): the lipid does not significantly impact the surface of CD1a or extend above CD1a, which would then allow the autoreactive TCRs to dock, as suggested by the permissive ligand scenario.
Role of acyl chain unsaturation
Next, we focused on the acyl chain Δ15 cis unsaturation present in 42:2 SM but absent from 42:1 SM, as this unsaturation increased capture by CD1a but blocked CD1a–SM staining of T cell lines by SM (Fig. 2 F). Both 42:1 and 42:2 SM have essentially identical seating. An observable difference is that the electron density of the acyl moiety is slightly weaker in the case of 42:1 SM, which might point toward a higher degree of flexibility, as expected for saturated versus unsaturated fatty acid chains. The unsaturation is sequestered deeply within the A′ pocket and distant from any TCR contact surface, so its strong effect in augmenting tetramer blockade is unlikely to involve TCR contact (Fig. 4 A, cyan arrow). Instead, the cis unsaturation bends the lipid to curve around the outer toroidal wall of the A′ pocket to encircle the A′ pole, formed by F70 and V12. Thus, the cis unsaturation provides a covalent constraint that could plausibly stabilize the bent lipid and could thereby plausibly increase binding and account for the increased biological effects of 42:2 SM (Fig. 1, Fig. 2, and Fig. 3 A).
Analysis of SMs in cell lines and human skin
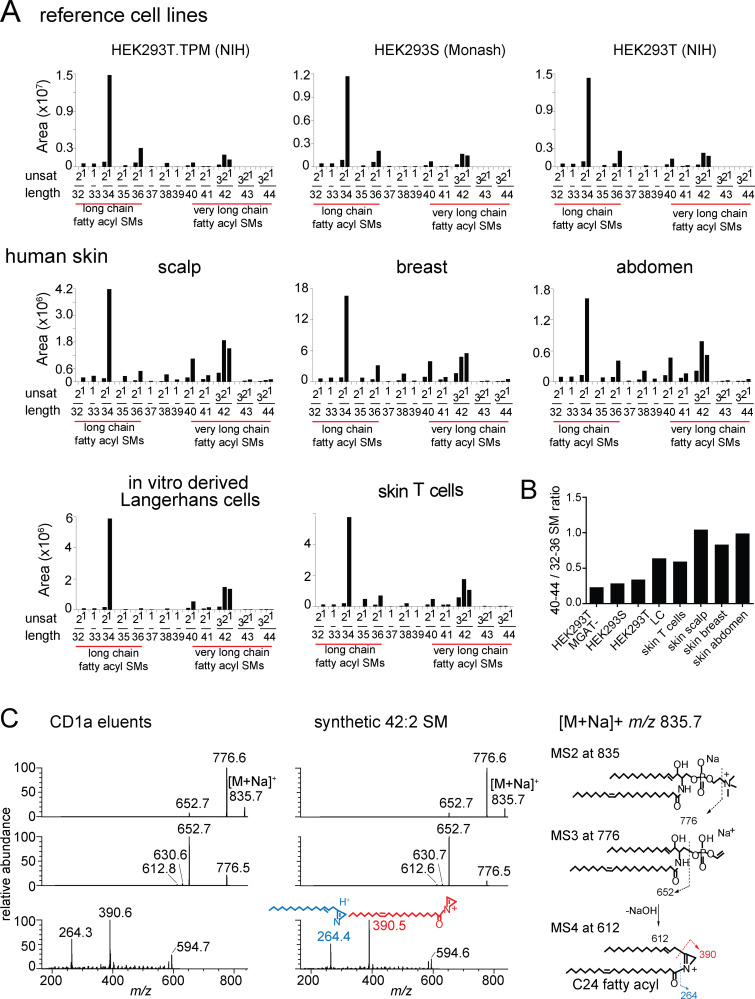
Next, we asked whether the lipid blockers (Fig. 1 and Fig. S2) also exist in cell types with roles in CD1a autoreactivity. CD1a is expressed in the thymus and on dermal dendritic cells, but the highest CD1a expression is seen in skin on epidermal LCs (Furue et al., 1992). Further, CD1a autoreactive T cells are enriched in healthy (Cotton et al., 2021) and diseased human skin (Cheung et al., 2016; Jarrett and Ogg, 2016; Jarrett et al., 2016; Subramaniam et al., 2016). Mass spectra (Fig. S2 A) were converted to SM profiles, where the overall number of CH2 units and unsaturation were reported for model cell lines used in the study of CD1a autoreactivity (HEK293 cells, C1R cells, K562 cells, and THP-1 cells; Fig. S2 B). We also studied skin T cells, in vitro–derived LCs (Bourgeois et al., 2015), and full-thickness skin biopsies as more physiologically relevant cell types. We separately detected all SMs in each cell type (Fig. 5 A and Fig. S2) and calculated the ratio of total 42 SMs to 34 SMs (42/34 ratios; Fig. 5 B), which correlates with expected blocking potential. 42/34 SM ratios from three genetically modified HEK293 sublines (Fig. 5 A) were <1 and similar in magnitude to one another and to our prior results (Fig. 2). Otherwise, 42/34 SM ratios varied by cell type, with higher ratios in skin-resident cell types, including T cells, in vitro–generated LCs, and skin from breast, abdomen, and scalp (Fig. 5 B; and Fig. S2, A and B). Thus, the inhibitor is broadly expressed among cells and tissues and appears to be overexpressed in skin, where CD1a is also present.
Figure S2.
Total lipid extracts from the indicated sources were analyzed by HPLC-MS in the positive ion mode. (A and B) SM ion (red) intensity values were matched to the deduced SM variant (A), where the overall chain length (CH2 units) and unsaturations present in both chains are indicated in (B). (C) MS/MS/MS of 34:1 SM from CD1a eluents and an authentic standard detected ion corresponding to the alkyl chains present in the sphingosine base (blue) and the fatty acyl unit (red), allowing assignment of a C16 fatty acid (m/z 280.4) as the predominant species. Thus, most chain-length variations in SMs are accounted for by the fatty acyl unit. Unsat, number of unsaturations in both chains.
Figure 5.
Length distribution of SMs in cells and skin. (A) Mass spectra of lipids (shown in Fig. S2) derived from HEK293 cells and human skin are shown as MS intensity values for each of 30 possible chain length and saturation variants of SM, where length is the number of methylene units in the fatty acyl and sphingosine chains, and unsat is the number of unsaturations in both chains. (B) The 42/34 ratio is calculated based on the sum of intensity values of C42 SMs divided by the sum of intensity values of C34 SMs for each cell or tissue. Because C42 SMs are inhibitory and C34 SMs are weakly activating for T cells, higher ratios predict stronger inhibitory functions of SM profiles. (A and B) Results are representative of two experiments. (C) Collision-induced dissociation of natural 42:2 SM from HEK293T identified the neutral loss of the choline head group and, through chain cleavage products, the length of the sphingosine as predominantly C24:1 (m/z 390.5) acyl chain and C18:1 sphingosine chain in the synthetic standard and CD1a-eluted molecule. Similar analysis of the 34:2 SM demonstrated a C18 sphingosine chain and a C16 fatty acyl unit (Fig. S2). Thus, chain length variation is determined mainly or exclusively in the fatty acyl unit, so that 42 SMs are formed from VLCFAs and 34 SMs and 36 SMs are made from LCFAs. The position and Z or E stereochemistry are inferred from known lipid structures but cannot be established by MS.
Inhibitory ligands are very long–chain fatty acyl SMs
The chain-length profiles and the higher-expression in skin sources suggested a specific biochemical origin of inhibitory lipids via biosynthesis pathways involving very long–chain fatty acids (VLCFAs). Lipids are normally produced in alkane series, whereby each species typically differs by two CH2 units, but here, the profiles show a C6–8 gap in length between inhibitory and noninhibitory SMs (Fig. 1, Fig. 2, and Fig. 3). These observations could be explained if all SMs contain the same kind of d18:1 sphingosine unit. Then, the 34:1, 34:2, 36:1, 36:2, and related weak agonist SMs would be composed of LCFAs, which are C16–18 (Sassa and Kihara, 2014), whereas the inhibitory 42 SMs would be composed of VLCFAs. VLCFAs are typically C24 in length and are made by distinct fatty acyl elongase isoforms that show tissue-specific expression, including overexpression in healthy (Ohno et al., 2010) skin and altered expression in diseased skin (Tawada et al., 2014).
To directly determine fatty acyl length, collisional MS of natural 34:1 SM and 42:2 SM was completed. We identified the d18:1 sphingosine chain in ester linkage with a fragment (Hsu and Turk, 2000) derived from VLCFAs (Fig. 5 C) or LCFAs (Fig. S2 C), respectively, which ruled in this hypothesis. Specifically, MS4 of 42:2 SM eluted from CD1a matched the spectrum of the 42:2 standard with a d18:1 sphingosine chain, with a key ion of m/z 390.5 directly identifying the C24:1 VLCFA-derived fragment (Fig. 5 C). Therefore, we no longer refer to agonists or antagonists as short or long in general terms; instead, antagonists could be formally defined as VLCFA-SMs, and the short agonistic molecules are LCFA-SMs (Fig. 5 A).
Blockade among polyclonal skin-derived CD1a autoreactive T cells
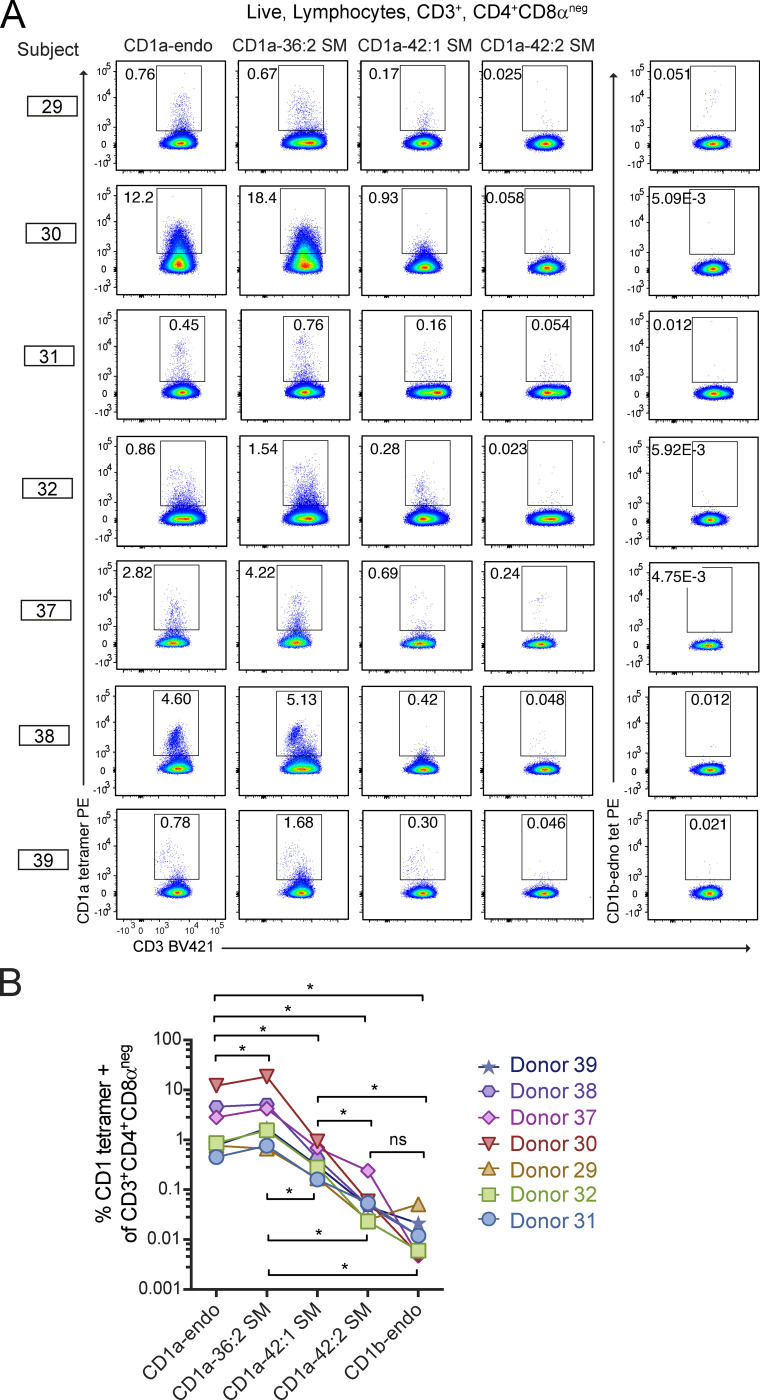
Having seen blockade of tetramer staining in T cell lines (Fig. 2), we next asked if VLCFA and LCFA SMs have effects on polyclonal CD1a autoreactive T cells from seven unrelated donors measured ex vivo. This experiment was possible based on the implementation of a 3-dimensional culture system, whereby viable T cells passively crawl into collagen matrices in the presence of T cell growth factors (Clark et al., 2006a, 2006b; Cotton et al., 2021). We focused on CD4+ T cells based on their high rates of CD1a autoreactivity measured previously (Cotton et al., 2021; de Jong et al., 2014). As with the individual lines (Fig. 2 F), untreated CD1a-endo tetramers achieved an intermediate intensity of staining on 0.1% to 13% of T cells from each donor, allowing assessment of whether SMs augment or block CD1a–TCR interaction at the polyclonal level.
Regardless of whether each individual started with high or low rates of CD1a-endo binding to T cells, all seven donors showed similar effects with SM treatment. The 36:2 SM–treated CD1a tetramers stained a greater frequency of CD3+CD4+CD8aneg cells than did CD1a-endo tetramers in six of seven subjects (P = 0.0313; Fig. 6). Prior measurements demonstrated the presence of >90 mixed lipids with agonist and blocking properties in untreated CD1a–endo complexes (Cotton et al., 2021), so this finding was consistent with short-chain SM displacement of endogenous CD1a ligands with a weak agonist, such that net TCR-CD1a binding was increased. Similar to results from T cell lines (Fig. 2 F), 42:1 SM treatment provided strong but partial blockade of staining among polyclonal cells, detecting cells at a frequency ∼10-fold lower than CD1a–36:2 SM tetramers. Strikingly, CD1a–42:2 SM tetramer staining was blocked nearly to baseline levels of CD1b-endo staining (P = 0.0156) in all seven individuals (Fig. 6, A and B), implicating 42:2 SM as a strong and broadly acting inhibitor of CD1a interactions with diverse CD1a autoreactive TCRs.
Figure 6.
CD1a–SM tetramer staining of polyclonal skin T cells. (A and B) Polyclonal T cells from seven donors were stained with CD1a or CD1b tetramers treated with the indicated lipids and shown as flow cytometry plots (A) and as summary data and statistics (B). Individual patients were tested on different days using the same method. *, P < 0.05; Wilcoxon matched-pairs signed rank test.
Discussion
The classical understanding of αβ T cell function emphasizes antigen-mediated T cell activation. Namely, T cell response occurs through TCR contact with an exposed region of the peptide, lipid, or small molecule that generates an epitope on MHC, CD1, or MR1 proteins, respectively (Borg et al., 2007; Garboczi et al., 1996; Patel et al., 2013). Antigenic epitopes are formed in specific situations and interact with a very small percentage of TCRs, so the default outcome of TCR encounter with most cellular antigen complexes is no activation. However, for CD1a autoreactive T cells, lipid-derived epitopes are not required because TCRs frequently bind on the unliganded “roof” of CD1a without contacting carried lipids (Birkinshaw et al., 2015; Cotton et al., 2021; Cotton et al., 2018; Van Rhijn et al., 2015). Accordingly, increasing evidence points to binding or activation as the default outcome of contact of CD1a with CD1a-autoreactive TCRs, especially in human skin, where both CD1a-autoreactive T cells and CD1a+ LCs are common (Cheung et al., 2016; Cotton et al., 2021; de Jong et al., 2010; de Lalla et al., 2011; Jarrett et al., 2016; Subramaniam et al., 2016). Thus, whereas prior models have emphasized antigen-regulated “off until on” mechanisms, studies of CD1a autoreactivity increasingly point to an “on until off” mode of regulation (Cotton et al., 2018). Emerging studies suggest a similar situation may occur for autoreactive CD1c-restricted T cells (Wun et al., 2018).
This new perspective supports the search for constitutively active inhibitory mechanisms for CD1a autoreactive T cells, especially since most approaches have discovered CD1 ligands using add-back assays where antigenic lipids induce T cell activation. Using MS as an unbiased method for detection of CD1 ligands without regard to their activating properties, we identified endogenous cellular lipids that are bound to CD1a in cells. We found that CD1a does not randomly survey the pool of self lipids, but rather specifically captures sphingolipids, especially VLCFA-SMs, in preference to membrane phospholipids. These preferred ligands broadly inhibit CD1a–TCR interactions among polyclonal T cells. The detection of nonactivating ligands raises comparisons to class II invariant chain peptide (Cresswell et al., 1990; Teyton et al., 1990), which is an endogenous peptide ligand that binds newly folded MHC II proteins, protecting their grooves from premature capture of endogenous self-peptides before arrival in the endosomal network (Cresswell, 1994).
Unlike class II invariant chain peptide, 42:2 SM does not universally occupy clefts in a cohort of CD1a-endo complexes and therefore does not completely block TCR binding to CD1a. In this and prior studies (Birkinshaw et al., 2015; Cotton et al., 2021; de Jong et al., 2014), the spectrum of cellular CD1a-endo complexes includes weak agonists and blockers (including 34:1 SM and 36:2 SM) that cause moderate activation and, when formed into tetramers, generates moderately bright staining of large numbers of polyclonal CD1a-reactive T cells, ranging from 0.1% to 13% of skin T cells. Thus, we propose that 42:2 SM and potentially other yet-to-be-discovered blockers do not “protect” all CD1a clefts, but instead act constitutively to generate a mid-range set point for CD1a–TCR interactions from which the mixtures of natural lipid agonists and inhibitors in tissues can tune T cell activation. Our model diverges fundamentally from antigen-driven off until on activation models because it emphasizes negative regulation and the spectrum of ligands present, rather than rare, strongly activating antigens.
We are unaware of other chemically defined lipid blockers that are selectively captured by CD1 proteins from the pool of self lipids or broadly acting blockers that act on a constitutive basis in cells. However, a related mechanism is the active regulation of nonstimulatory lipids so that altered cellular lipid levels can influence T cell response. In the CD1d system, deletion of glycolipid degradative enzymes (Gadola et al., 2006), including acid sphingomyelinase that degrades SM, induces overexpression of nonantigenic lipids in ways that strongly inhibit both NKT development and activation (Melum et al., 2019). Although our data point to constitutive capture of 42:2 SM, its regulated expression could likewise influence CD1a autoreactive T cells. Although we could readily detect 42:2 SMs in all cells tested, including cells and tissues that normally express CD1a, the ratio of inhibitory to activating SMs was highest in LCs and skin-derived cells. Among SMs, the switch from low to high capture and from weak T cell agonism to strong inhibition both derive from the C6–8-length increment that distinguishes natural LCFAs from VLCFAs.
Thus, incorporation of VLCFAs into SM could regulate the T cell response
Fatty acyl chain length is controlled by regulated and tissue-specific expression of individual isoenzymes in the elongation of very long (ELOVL) fatty acid family (Ohno et al., 2010). ELOVL1 and ELOVL4 prefer the longer substrates needed for VLCFA biosynthesis. VLCFAs are enriched in skin, where they likely account for the skin-specific expression of VLCFA-SMs observed here, and VLCFAs contribute to maintaining water retention and barrier function (Ohno et al., 2010; Sassa and Kihara, 2014). Further, ELOVL enzymes are biologically regulated. ELOVL1 and VLCFAs are down-regulated by IFN-γ in vitro in human keratinocytes and cultured epidermis, as well as in psoriatic skin disease and atopic dermatitis (Tawada et al., 2014), and ELOVL3 is down-regulated in psoriasis (Gudjonsson et al., 2010). Also, VLCFAs are abundant in the stratum corneum (Ohno et al., 2010; Sassa and Kihara, 2014). Thus, the regulation and microlocalization of 42:2 SM inhibitors in cells and tissues could control CD1a response during immune response or in monitoring skin integrity.
CD1a has a broad roof structure that allows TCRs to sit down on the surface of CD1a (Birkinshaw et al., 2015), ignoring lipids in the cleft (Cotton et al., 2021). This molecular mechanism predicts that lipids binding in the cleft is not sufficient to block TCR binding to the CD1a roof. Therefore, it is notable that CD1a cocrystalized with 42:1 and 42:2 SM shows two specific mechanisms to block roof-specific autoreactive T cells. The longer acyl chains position R76 and the phosphocholine unit high above the CD1a platform, allowing steric interference of TCRs. Also, the phosphate in SM disrupts the R73-E154-R76 salt bridge to alter the intrinsic roof structure. These two effects, which were not seen for 36:2 SM, alter the surface of CD1a near the center of the platform, which can account for the observed blockade of polyclonal roof-specific TCRs (Cotton et al., 2021). Further, CD1a–SM crystal structures provide plausible mechanisms for the observed preferential 42:2 SM capture, whereby the longer chains favor parallel insertion into the A′ pocket, and the Δ15 cis unsaturation promotes curvature around the A′ pole. The cis unsaturation is predicted to provide a covalent constraint to stabilize the bent lipid conformation, which could promote retention of 42:2 SM in CD1a. These two mechanisms, discovered with endogenous lipids, provide a chemical basis for future design of exogenous lipids that could be used as therapeutic antagonists of CD1a based on altered chain length and unsaturation. Lipid blockers could be applied topically on the skin in psoriasis, contact dermatitis, and other diseases linked to CD1a autoreactivity (Bourgeois et al., 2015; Cheung et al., 2016; Jarrett et al., 2016; Kim et al., 2016; Nicolai et al., 2020; Subramaniam et al., 2016).
Finally, these results may explain the mechanism of action of recently identified small lipids that act as natural agonists for CD1a-reactive T cells. These include skin-derived squalene and wax esters (de Jong et al., 2014), poison ivy–derived urushiol (Kim et al., 2016), and lysolipid products of endogenous phospholipases (Bourgeois et al., 2015; Cheung et al., 2016; Jarrett et al., 2016; Subramaniam et al., 2016), as well as farnesol and benzyl cinnamate, which act as contact allergens (Nicolai et al., 2020), and the 36:2 SMs shown here. For lysoPC, urushiol, farnesol, and 36:2 SM, binary crystal structures show that agonistic lipids are seated fully inside CD1a (Birkinshaw et al., 2015; Kim et al., 2016; Nicolai et al., 2020), so they cannot likely contact TCRs. Capture of these exogenous lipids would necessarily displace endogenous CD1a-bound blockers, leading to T cell activation, and increased endogenous short-chain SMs could also promote activation. Here, we observed that the intermediate level of CD1a tetramer–TCR binding was augmented by treating CD1a with 34 SM, 36 SM, or lysoPC or blocked with long-chain 42 SMs. Thus, agonistic properties of CD1a ligands can be increasingly understood not as generating epitopes on the outside of CD1a but instead as displacing inhibitors to free up the surface of the CD1a protein for TCR binding. If “antigens” are molecules that contact TCRs, then these natural lipid agonists are not antigens, but instead are displacers of dominant negative blockers.
Materials and methods
Human subjects
Discarded skin from cosmetic surgeries was used for T cell assays and analysis of skin lipids and was obtained through the Human Skin Disease Resource Center at Harvard Medical School and Brigham and Women’s Hospital under approved protocols by Partners Institutional Review Board. Human peripheral blood mononuclear cells were obtained from leukoreduction collars provided by Brigham and Women’s Hospital.
Recovery of skin T cells by three-dimensional culture
Skin T cells were recovered after culture for 21–28 d on three-dimensional cell foam growth matrices (Cytomatrix) seeded with collagen I (Thermo Fisher Scientific; 354236). Skin T cell culture media (Iscove’s modified Dulbecco’s medium, 10–20% FCS, L-glutamine, penicillin-streptomycin, and 2-mercaptoethanol) was supplemented with IL-2 (Brigham and Women’s Hospital or Peprotech) and recombinant human IL-15 (10 ng/ml; Peprotech; #200-15) as described (Clark et al., 2006b; Cotton et al., 2021).
Loading CD1a monomers with defined lipids for tetramers
Lipids stored in chloroform and methanol were transferred to new borosilicate glass tubes, dried under nitrogen gas, and reconstituted to 400 µM in Tris-buffered saline (TBS), pH 8.0, 0.5% CHAPS (3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate hydrate) buffer by sonication in a 37°C water bath for ∼1 h. Lipid-buffer sonicates or a buffer-only control was transferred to 1.5-ml Eppendorf tubes on a 37°C heat block to which CD1a monomer was added to a final concentration of 0.2 µg/µl and incubated for 2 h at 37°C, then overnight at room temperature. Loaded monomers were stored at 4°C and used for tetramer assembly.
Lipid sources
The following synthetic lipids were obtained from Avanti: C34:1 PC (850475), C18:1 Lyso-PC (845875), C18:1 Lyso-SM (860600), C24:1 SM (860582), C34:1 SM (860584), C36:2 SM (860587), C42:1 SM (860592), and C42:2 SM (860593).
CD1a recombinant expression and purification for crystallization studies
CD1a was expressed in HEK293S GnTI− (lacking N-acetylglucosaminyltransferase I activity) cells and purified at Monash University (Birkinshaw et al., 2015) or from HEK293T or HEK293T.TPM cells from the National Institutes of Health (NIH) Tetramer Facility (unpublished data). TPM is engineered to express the Escherichia coli BirA enzyme and has CRISPR knockouts of the TAP2 and MGAT1 genes. The MGAT1 knockout renders it deficient in the GNT-I protein that is also lacking in the HEK293S line (Cotton et al., 2021). Following endoglycosidase H (New England BioLabs) and thrombin (Sigma) treatment, the purified CD1a was loaded with C42:2 SM, C42:1 SM, and C36:2 SM (Avanti) dissolved in TBS/0.5% CHAPS (Sigma). CD1a was incubated overnight at pH 5.5 with a 40-fold molar excess of each lipid. The detergent and unbound SMs were removed by ion exchange chromatography (MonoQ 10/100 GL, glass; GE Healthcare). Because the initially obtained crystals of CD1a–36:2 SM did not diffract, probably due to sample heterogeneity, we modified the loading procedure. First, CD1a was incubated overnight at low pH with 0.5% tyloxapol (Sigma) to remove endogenous lipids. The detergent was removed by gel filtration chromatography (Superdex 200; GE Healthcare). In a second step, the tyloxapol-treated CD1a was loaded overnight with 40-fold excess of SM 36:2 dissolved in TBS/0.5% CHAPS, and the mixture was separated by ion exchange chromatography (MonoQ 10/100 GL; GE Healthcare).
Crystallization, structure determination, and refinement of CD1a–SM complexes
Seeds from binary CD1a-antigen crystals (Kim et al., 2016) were used to grow crystals of the CD1a–36:2 SM, CD1a–42:1 SM, and CD1a–42:2 SM binary complexes in 20–25% polyethylene glycol 1500/10% DL-Malic acid, MES monohydrate, Tris buffer at pH 5–6. The crystals were flash-frozen, and data were collected at the MX1 and MX2 beamlines (Australian Synchrotron). Data were processed with XDS (Kabsch, 2010) and were scaled with SCALA from the CCP4 program suite (Winn et al., 2011). Upon successful phasing by molecular replacement using PHASER (McCoy et al., 2007) and the CD1a-farnesol structure as the search model (Nicolai et al., 2020), the electron densities of the ligands were clear in respective unbiased electron density maps. Initial rigid body refinement was performed using phenix.refine (Afonine et al., 2012), and iterative model improvement was performed using COOT (Emsley et al., 2010) and phenix.refine. The quality of the structure was confirmed at the Research Collaboratory for Structural Bioinformatics Protein Data Bank Data Validation and Deposition Services website. Molecular graphics were created with University of California San Francisco–Chimera (Pettersen et al., 2004). The data and refined coordinates were deposited in the Protein Data Bank under accession nos. 7KOZ (CD1a–36:2 SM), 7KP0 (CD1a–42:1 SM), and 7KP1 (CD1a–42:2 SM).
HPLC-MS analysis
CD1a, CD1b, and CD1c proteins were produced in HEK293S cells at Monash University or HEK293T.TPM at the NIH Tetramer Facility. For lipid elution, 20–200 µg of CD1 protein was extracted in 15-ml glass tubes using chloroform, methanol, and water (Bligh and Dyer, 1959). The organic phase was recovered and dried under nitrogen gas. Eluent residue was redissolved in chloroform-methanol, normalized to 20 µM based on input protein and stored at −20°C. We injected 20 µl for Q-ToF HPLC-MS−positive ion mode analysis using an Agilent 6530 Accurate-Mass Q-TOF and 1260 series HPLC system with a normal phase Inertsil Diol column (150 mm × 2 mm; GL Sciences), running at 0.15 ml/min as described (Huang et al., 2011). For the CD1a lipidomics, the protein was produced in HEK293T.TPM cell line and subject to lipid elution in triplicate, typically using 80 µg. However, when less protein was available, comparisons were normalized to the absolute mass of protein. For lipid extraction from cells, CD1a-transduced HEK293T.TPM cells were matched to the CD1a monomer batches used for CD1a lipidomics, tetramer staining, and sorting. Cells were grown up in DMEM 20% fetal calf serum according to tetramer core facility culture conditions. Cells were washed twice with PBS. Cell pellets were extracted successively with chloroform-methanol (at a ratio of 1/2 for 2 h and 2/1 for 1 h). Successive extractions were combined and dried under nitrogen gas, and lipids were weighed and stored at 1 mg/ml in chloroform-methanol at −20°C. For cellular lipidomic analysis, HEK293T cells (NIH Tetramer Facility), skin samples (Brigham and Women’s Hospital Dermatology), C1R cells (de Jong et al., 2014), K562 cells (de Jong et al., 2010), skin T cells (Cotton et al., 2021), and in vitro derived LCs (Bourgeois et al., 2015) were obtained or prepared from the indicated sources. Cells or tissues were extracted as above in triplicate for HPLC-MS analysis with 0.25 mg/ml lipid concentration.
Online supplemental material
Fig. S1 provides detailed SM profiles from HEK293 cells. Fig. S2 provides detailed SM profiles from cells and tissues. Table S1 provides HPLC-MS–based identification of PCs and SMs from CD1a. Table S2 shows the data collection and refinement statistics for CD1a–SM complexes. Table S3 provides residues with contacts between CD1a and SMs.
Supplementary Material
provides HPLC-MS–based identification of PCs and SMs from CD1a.
shows the data collection and refinement statistics for CD1a–SM complexes.
provides residues with contacts between CD1a and SMs.
Acknowledgments
We thank the staff at the Australian Synchrotron for assistance with data collection.
This work was supported by the Wellcome Trust Collaborative Award (G. Ogg, J. Rossjohn, G.S. Besra, D.B. Moody), the Medical Research Council (G. Ogg and MR/R001154/1 and MR/S000542/1 to G.S. Besra), the National Institute for Health Research Oxford Biomedical Research Centre (G. Ogg), an Australian Research Council Laureate Fellowship (J. Rossjohn), Comprehensive Research Network (G. Ogg), The Gillian Reny Stepping Strong Center for Trauma (D.P. Orgill), and the National Institutes of Health (R01 AR048632 to D.B. Moody; R01 AR074037 to A. De Jong; P30AR069625 and R01AI127654 to R.A. Clark). J.D. Altman is supported by National Institutes of Health contract HHSN272201300006C. G.S. Besra acknowledges support in the form of a Personal Research Chair from Mr. James Bardrick.
Author contributions: J. Rossjohn and D.B. Moody initiated the project and provided oversight. R.N. Cotton, T.-Y. Cheng, Y.-L. Chen, A. de Jong, and I. Van Rhijn performed immunology and lipidomics experiments. M. Wegrecki and J. Le Nours performed protein chemistry and structural biology experiments. J.D. Altman and R. Willis designed and produced proteins for tetramer experiments and lipid elution. T.-Y. Cheng designed and performed the lipid elution experiments. G.S. Besra and N. Veerapen planned and designed lipid ligands. D.P. Orgill, B. Pomahac, S.G. Talbot, and R.A. Clark provided skin samples and T cell purification methods. J. Le Nours, J.D. Altman, I. Van Rhijn, R.A. Clark, G. Ogg, J. Rossjohn, and D.B. Moody conceived and oversaw the immunology experiments. R.N. Cotton, M. Wegrecki, T.-Y. Cheng, J. Rossjohn, and D.B. Moody wrote the manuscript with input from all authors.
References
- Afonine, P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., and Adams P.D.. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68:352–367. 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkinshaw, R.W., Pellicci D.G., Cheng T.Y., Keller A.N., Sandoval-Romero M., Gras S., de Jong A., Uldrich A.P., Moody D.B., Godfrey D.I., and Rossjohn J.. 2015. αβ T cell antigen receptor recognition of CD1a presenting self lipid ligands. Nat. Immunol. 16:258–266. 10.1038/ni.3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh, E.G., and Dyer W.J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917. 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- Borg, N.A., Wun K.S., Kjer-Nielsen L., Wilce M.C., Pellicci D.G., Koh R., Besra G.S., Bharadwaj M., Godfrey D.I., McCluskey J., and Rossjohn J.. 2007. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 448:44–49. 10.1038/nature05907 [DOI] [PubMed] [Google Scholar]
- Bourgeois, E.A., Subramaniam S., Cheng T.Y., De Jong A., Layre E., Ly D., Salimi M., Legaspi A., Modlin R.L., Salio M., et al. 2015. Bee venom processes human skin lipids for presentation by CD1a. J. Exp. Med. 212:149–163. 10.1084/jem.20141505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briken, V., Jackman R.M., Dasgupta S., Hoening S., and Porcelli S.A.. 2002. Intracellular trafficking pathway of newly synthesized CD1b molecules. EMBO J. 21:825–834. 10.1093/emboj/21.4.825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brozovic, S., Nagaishi T., Yoshida M., Betz S., Salas A., Chen D., Kaser A., Glickman J., Kuo T., Little A., et al. 2004. CD1d function is regulated by microsomal triglyceride transfer protein. Nat. Med. 10:535–539. 10.1038/nm1043 [DOI] [PubMed] [Google Scholar]
- Calabi, F., and Milstein C.. 1986. A novel family of human major histocompatibility complex-related genes not mapping to chromosome 6. Nature. 323:540–543. 10.1038/323540a0 [DOI] [PubMed] [Google Scholar]
- Cheung, K.L., Jarrett R., Subramaniam S., Salimi M., Gutowska-Owsiak D., Chen Y.L., Hardman C., Xue L., Cerundolo V., and Ogg G.. 2016. Psoriatic T cells recognize neolipid antigens generated by mast cell phospholipase delivered by exosomes and presented by CD1a. J. Exp. Med. 213:2399–2412. 10.1084/jem.20160258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, R.A., Chong B., Mirchandani N., Brinster N.K., Yamanaka K., Dowgiert R.K., and Kupper T.S.. 2006a. The vast majority of CLA+ T cells are resident in normal skin. J. Immunol. 176:4431–4439. 10.4049/jimmunol.176.7.4431 [DOI] [PubMed] [Google Scholar]
- Clark, R.A., Chong B.F., Mirchandani N., Yamanaka K.-I., Murphy G.F., Dowgiert R.K., and Kupper T.S.. 2006. b. A novel method for the isolation of skin resident T cells from normal and diseased human skin. J. Invest. Dermatol. 126(5):1059–1070. 10.1038/sj.jid.5700199 [DOI] [PubMed] [Google Scholar]
- Cotton, R.N., Shahine A., Rossjohn J., and Moody D.B.. 2018. Lipids hide or step aside for CD1-autoreactive T cell receptors. Curr. Opin. Immunol. 52:93–99. 10.1016/j.coi.2018.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton, R.N., Cheng T., Wegrecki M., Le Nours J., Orgill D.G., Pomahac B., Talbot S.G., Willis R.A., Altman J.D., de Jong A., et al. 2021. Human skin is colonized by T cells that recognize CD1a independently of lipid. J. Clin. Invest. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, D., Fox L., Tian R., Bardet W., Skaley M., Mojsilovic D., Gumperz J., and Hildebrand W.. 2009. Determination of cellular lipids bound to human CD1d molecules. PLoS One. 4:e5325. 10.1371/journal.pone.0005325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresswell, P. 1994. Assembly, transport, and function of MHC class II molecules. Annu. Rev. Immunol. 12:259–291. 10.1146/annurev.iy.12.040194.001355 [DOI] [PubMed] [Google Scholar]
- Cresswell, P., Avva R.R., Davis J.E., Lamb C.A., Riberdy J.M., and Roche P.A.. 1990. Intracellular transport and peptide binding properties of HLA class II glycoproteins. Semin. Immunol. 2:273–280. [PubMed] [Google Scholar]
- de Jong, A. 2015. Activation of human T cells by CD1 and self-lipids. Immunol. Rev. 267:16–29. 10.1111/imr.12322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong, A., Peña-Cruz V., Cheng T.Y., Clark R.A., Van Rhijn I., and Moody D.B.. 2010. CD1a-autoreactive T cells are a normal component of the human αβ T cell repertoire. Nat. Immunol. 11:1102–1109. 10.1038/ni.1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong, A., Cheng T.Y., Huang S., Gras S., Birkinshaw R.W., Kasmar A.G., Van Rhijn I., Peña-Cruz V., Ruan D.T., Altman J.D., et al. 2014. CD1a-autoreactive T cells recognize natural skin oils that function as headless antigens. Nat. Immunol. 15:177–185. 10.1038/ni.2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lalla, C., Lepore M., Piccolo F.M., Rinaldi A., Scelfo A., Garavaglia C., Mori L., De Libero G., Dellabona P., and Casorati G.. 2011. High-frequency and adaptive-like dynamics of human CD1 self-reactive T cells. Eur. J. Immunol. 41:602–610. 10.1002/eji.201041211 [DOI] [PubMed] [Google Scholar]
- Dougan, S.K., Salas A., Rava P., Agyemang A., Kaser A., Morrison J., Khurana A., Kronenberg M., Johnson C., Exley M., et al. 2005. Microsomal triglyceride transfer protein lipidation and control of CD1d on antigen-presenting cells. J. Exp. Med. 202:529–539. 10.1084/jem.20050183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan, S.K., Kaser A., and Blumberg R.S.. 2007. CD1 expression on antigen-presenting cells. Curr. Top. Microbiol. Immunol. 314:113–141. [DOI] [PubMed] [Google Scholar]
- Emsley, P., Lohkamp B., Scott W.G., and Cowtan K.. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy, E., Subramaniam S., Brown H.A., Glass C.K., Merrill A.H. Jr., Murphy R.C., Raetz C.R., Russell D.W., Seyama Y., Shaw W., et al. 2005. A comprehensive classification system for lipids. J. Lipid Res. 46:839–861. 10.1194/jlr.E400004-JLR200 [DOI] [PubMed] [Google Scholar]
- Furue, M., Nindl M., Kawabe K., Nakamura K., Ishibashi Y., and Sagawa K.. 1992. Epitopes for CD1a, CD1b, and CD1c antigens are differentially mapped on Langerhans cells, dermal dendritic cells, keratinocytes, and basement membrane zone in human skin. J. Am. Acad. Dermatol. 27:419–426. 10.1016/0190-9622(92)70211-W [DOI] [PubMed] [Google Scholar]
- Gadola, S.D., Silk J.D., Jeans A., Illarionov P.A., Salio M., Besra G.S., Dwek R., Butters T.D., Platt F.M., and Cerundolo V.. 2006. Impaired selection of invariant natural killer T cells in diverse mouse models of glycosphingolipid lysosomal storage diseases. J. Exp. Med. 203:2293–2303. 10.1084/jem.20060921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garboczi, D.N., Ghosh P., Utz U., Fan Q.R., Biddison W.E., and Wiley D.C.. 1996. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 384:134–141. 10.1038/384134a0 [DOI] [PubMed] [Google Scholar]
- Gudjonsson, J.E., Ding J., Johnston A., Tejasvi T., Guzman A.M., Nair R.P., Voorhees J.J., Abecasis G.R., and Elder J.T.. 2010. Assessment of the psoriatic transcriptome in a large sample: additional regulated genes and comparisons with in vitro models. J. Invest. Dermatol. 130:1829–1840. 10.1038/jid.2010.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, F.F., and Turk J.. 2000. Structural determination of sphingomyelin by tandem mass spectrometry with electrospray ionization. J. Am. Soc. Mass Spectrom. 11:437–449. 10.1016/S1044-0305(99)00150-6 [DOI] [PubMed] [Google Scholar]
- Huang, S., Cheng T.Y., Young D.C., Layre E., Madigan C.A., Shires J., Cerundolo V., Altman J.D., and Moody D.B.. 2011. Discovery of deoxyceramides and diacylglycerols as CD1b scaffold lipids among diverse groove-blocking lipids of the human CD1 system. Proc. Natl. Acad. Sci. USA. 108:19335–19340. 10.1073/pnas.1112969108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett, R., and Ogg G.. 2016. Lipid-specific T cells and the skin. Br. J. Dermatol. 175(Suppl 2):19–25. 10.1111/bjd.14908 [DOI] [PubMed] [Google Scholar]
- Jarrett, R., Salio M., Lloyd-Lavery A., Subramaniam S., Bourgeois E., Archer C., Cheung K.L., Hardman C., Chandler D., Salimi M., et al. 2016. Filaggrin inhibits generation of CD1a neolipid antigens by house dust mite-derived phospholipase. Sci. Transl. Med. 8:325ra18. 10.1126/scitranslmed.aad6833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce, S., Woods A.S., Yewdell J.W., Bennink J.R., De Silva A.D., Boesteanu A., Balk S.P., Cotter R.J., and Brutkiewicz R.R.. 1998. Natural ligand of mouse CD1d1: cellular glycosylphosphatidylinositol. Science. 279:1541–1544. 10.1126/science.279.5356.1541 [DOI] [PubMed] [Google Scholar]
- Kabsch, W. 2010. Xds. Acta Crystallogr. D Biol. Crystallogr. 66:125–132. 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, S.J., and Cresswell P.. 2002. Regulation of intracellular trafficking of human CD1d by association with MHC class II molecules. EMBO J. 21:1650–1660. 10.1093/emboj/21.7.1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, S.J., and Cresswell P.. 2004. Saposins facilitate CD1d-restricted presentation of an exogenous lipid antigen to T cells. Nat. Immunol. 5:175–181. 10.1038/ni1034 [DOI] [PubMed] [Google Scholar]
- Kawano, T., Cui J., Koezuka Y., Toura I., Kaneko Y., Motoki K., Ueno H., Nakagawa R., Sato H., Kondo E., et al. 1997. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 278:1626–1629. 10.1126/science.278.5343.1626 [DOI] [PubMed] [Google Scholar]
- Kim, J.H., Hu Y., Yongqing T., Kim J., Hughes V.A., Le Nours J., Marquez E.A., Purcell A.W., Wan Q., Sugita M., et al. 2016. CD1a on Langerhans cells controls inflammatory skin disease. Nat. Immunol. 17:1159–1166. 10.1038/ni.3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layre, E., Sweet L., Hong S., Madigan C.A., Desjardins D., Young D.C., Cheng T.Y., Annand J.W., Kim K., Shamputa I.C., et al. 2011. A comparative lipidomics platform for chemotaxonomic analysis of Mycobacterium tuberculosis. Chem. Biol. 18:1537–1549. 10.1016/j.chembiol.2011.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoma, A.M., Castro C.D., Mayassi T., Bembinster L.A., Bai L., Picard D., Anderson B., Scharf L., Kung J.E., Sibener L.V., et al. 2013. Crystal structure of Vδ1 T cell receptor in complex with CD1d-sulfatide shows MHC-like recognition of a self-lipid by human γδ T cells. Immunity. 39:1032–1042. 10.1016/j.immuni.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy, A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., and Read R.J.. 2007. Phaser crystallographic software. J. Appl. Cryst. 40:658–674. 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melum, E., Jiang X., Baker K.D., Macedo M.F., Fritsch J., Dowds C.M., Wang J., Pharo A., Kaser A., Tan C., et al. 2019. Control of CD1d-restricted antigen presentation and inflammation by sphingomyelin. Nat. Immunol. 20:1644–1655. 10.1038/s41590-019-0504-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill, A.H. Jr. 2011. Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem. Rev. 111:6387–6422. 10.1021/cr2002917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody, D.B., and Cotton R.N.. 2017. Four pathways of CD1 antigen presentation to T cells. Curr. Opin. Immunol. 46:127–133. 10.1016/j.coi.2017.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody, D.B., Zajonc D.M., and Wilson I.A.. 2005. Anatomy of CD1-lipid antigen complexes. Nat. Rev. Immunol. 5:387–399. 10.1038/nri1605 [DOI] [PubMed] [Google Scholar]
- Nicolai, S., Wegrecki M., Cheng T.Y., Bourgeois E.A., Cotton R.N., Mayfield J.A., Monnot G.C., Le Nours J., Van Rhijn I., Rossjohn J., et al. 2020. Human T cell response to CD1a and contact dermatitis allergens in botanical extracts and commercial skin care products. Sci. Immunol. 5:eaax5430. 10.1126/sciimmunol.aax5430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, Y., Suto S., Yamanaka M., Mizutani Y., Mitsutake S., Igarashi Y., Sassa T., and Kihara A.. 2010. ELOVL1 production of C24 acyl-CoAs is linked to C24 sphingolipid synthesis. Proc. Natl. Acad. Sci. USA. 107:18439–18444. 10.1073/pnas.1005572107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.J., Kang S.J., De Silva A.D., Stanic A.K., Casorati G., Hachey D.L., Cresswell P., and Joyce S.. 2004. Lipid-protein interactions: biosynthetic assembly of CD1 with lipids in the endoplasmic reticulum is evolutionarily conserved. Proc. Natl. Acad. Sci. USA. 101:1022–1026. 10.1073/pnas.0307847100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, O., Kjer-Nielsen L., Le Nours J., Eckle S.B., Birkinshaw R., Beddoe T., Corbett A.J., Liu L., Miles J.J., Meehan B., et al. 2013. Recognition of vitamin B metabolites by mucosal-associated invariant T cells. Nat. Commun. 4:2142. 10.1038/ncomms3142 [DOI] [PubMed] [Google Scholar]
- Pettersen, E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., and Ferrin T.E.. 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Salio, M., Silk J.D., Jones E.Y., and Cerundolo V.. 2014. Biology of CD1- and MR1-restricted T cells. Annu. Rev. Immunol. 32:323–366. 10.1146/annurev-immunol-032713-120243 [DOI] [PubMed] [Google Scholar]
- Sassa, T., and Kihara A.. 2014. Metabolism of very long-chain Fatty acids: genes and pathophysiology. Biomol. Ther. (Seoul). 22:83–92. 10.4062/biomolther.2014.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam, S., Aslam A., Misbah S.A., Salio M., Cerundolo V., Moody D.B., and Ogg G.. 2016. Elevated and cross-responsive CD1a-reactive T cells in bee and wasp venom allergic individuals. Eur. J. Immunol. 46:242–252. 10.1002/eji.201545869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita, M., Grant E.P., van Donselaar E., Hsu V.W., Rogers R.A., Peters P.J., and Brenner M.B.. 1999. Separate pathways for antigen presentation by CD1 molecules. Immunity. 11:743–752. 10.1016/S1074-7613(00)80148-X [DOI] [PubMed] [Google Scholar]
- Tawada, C., Kanoh H., Nakamura M., Mizutani Y., Fujisawa T., Banno Y., and Seishima M.. 2014. Interferon-γ decreases ceramides with long-chain fatty acids: possible involvement in atopic dermatitis and psoriasis. J. Invest. Dermatol. 134:712–718. 10.1038/jid.2013.364 [DOI] [PubMed] [Google Scholar]
- Teyton, L., O’Sullivan D., Dickson P.W., Lotteau V., Sette A., Fink P., and Peterson P.A.. 1990. Invariant chain distinguishes between the exogenous and endogenous antigen presentation pathways. Nature. 348:39–44. 10.1038/348039a0 [DOI] [PubMed] [Google Scholar]
- Uldrich, A.P., Le Nours J., Pellicci D.G., Gherardin N.A., McPherson K.G., Lim R.T., Patel O., Beddoe T., Gras S., Rossjohn J., and Godfrey D.I.. 2013. CD1d-lipid antigen recognition by the γδ TCR. Nat. Immunol. 14:1137–1145. 10.1038/ni.2713 [DOI] [PubMed] [Google Scholar]
- van Meer, G. 2005. Cellular lipidomics. EMBO J. 24:3159–3165. 10.1038/sj.emboj.7600798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rhijn, I., Godfrey D.I., Rossjohn J., and Moody D.B.. 2015. Lipid and small-molecule display by CD1 and MR1. Nat. Rev. Immunol. 15:643–654. 10.1038/nri3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn, M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., et al. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67:235–242. 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wun, K.S., Reijneveld J.F., Cheng T.Y., Ladell K., Uldrich A.P., Le Nours J., Miners K.L., McLaren J.E., Grant E.J., Haigh O.L., et al. 2018. T cell autoreactivity directed toward CD1c itself rather than toward carried self lipids. Nat. Immunol. 19:397–406. 10.1038/s41590-018-0065-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, W., Kang S.J., Evans J.E., and Cresswell P.. 2009. Natural lipid ligands associated with human CD1d targeted to different subcellular compartments. J. Immunol. 182:4784–4791. 10.4049/jimmunol.0803981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajonc, D.M., Elsliger M.A., Teyton L., and Wilson I.A.. 2003. Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 A. Nat. Immunol. 4:808–815. 10.1038/ni948 [DOI] [PubMed] [Google Scholar]
- Zajonc, D.M., Crispin M.D., Bowden T.A., Young D.C., Cheng T.Y., Hu J., Costello C.E., Rudd P.M., Dwek R.A., Miller M.J., et al. 2005. Molecular mechanism of lipopeptide presentation by CD1a. Immunity. 22:209–219. 10.1016/j.immuni.2004.12.009 [DOI] [PubMed] [Google Scholar]
- Zhou, D., Cantu C. III, Sagiv Y., Schrantz N., Kulkarni A.B., Qi X., Mahuran D.J., Morales C.R., Grabowski G.A., Benlagha K., et al. 2004. Editing of CD1d-bound lipid antigens by endosomal lipid transfer proteins. Science. 303:523–527. 10.1126/science.1092009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
provides HPLC-MS–based identification of PCs and SMs from CD1a.
shows the data collection and refinement statistics for CD1a–SM complexes.
provides residues with contacts between CD1a and SMs.