Abstract
Aims:
The ability to predict response to treatment remains a key unmet need in psoriatic disease. We conducted a systematic review of studies relating to biomarkers associated with response to treatment in either psoriasis vulgaris (PsV) or psoriatic arthritis (PsA).
Methods:
A search was conducted in PubMed, Embase and the Cochrane library from their inception to 2 September 2020, and conference proceedings from four major rheumatology conferences. Original research articles studying pre-treatment biomarker levels associated with subsequent response to pharmacologic treatment in either PsV or PsA were included.
Results:
A total of 765 articles were retrieved and after review, 44 articles (22 relating to PsV and 22 to PsA) met the systematic review’s eligibility criteria. One study examined the response to methotrexate, one the response to tofacitinib and all the other studies to biologic disease-modifying antirheumatic drugs (DMARDs). Whilst several studies examined the HLA-C*06 allele in PsV, the results were conflicting. Interleukin (IL)-12 serum levels and polymorphisms in the IL-12B gene show promise as biomarkers of treatment response in PsV. Most, but not all, studies found that higher baseline levels of C-reactive protein (CRP) were associated with a better clinical response to treatment in patients with PsA.
Conclusion:
Several studies have identified biomarkers associated with subsequent response to treatment in psoriatic disease. However, due to the different types of biomarkers, treatments and outcome measures used, firm conclusions cannot be drawn. Further validation is needed before any of these biomarkers translate to clinical practice.
Keywords: biological therapy, DMARD, drug response biomarkers, psoriatic arthritis, psoriasis, therapeutics
Introduction
Psoriasis is a common inflammatory skin disorder. The prevalence of psoriasis vulgaris (PsV), the most common form of psoriasis, is about 2%,1 and up to 30% of these patients develop psoriatic arthritis (PsA) – a chronic inflammatory condition that affects the joints, entheses and axial skeleton.
In the past 15 years, several effective biologic disease-modifying antirheumatic drugs (bDMARDs) and targeted synthetic DMARDs (tsDMARDs) have been licensed for the treatment of psoriatic disease. However, these treatments are either only partially or not effective for some patients. The best-designed, phase III randomised controlled trials (RCTs) in patients with PsA have been those conducted with bDMARDs and more recently with tsDMARDs. Less than 60% of patients achieve the primary outcome measure of an American College of Rheumatology 20% (ACR20) response, with approximately 40% and 20%, respectively, reaching harder targets of ACR50 or ACR70.2–5 The < 60% ACR20 response rate, which is a minimal disease response measure, means of course that > 40% do not respond. Additionally, patients can exhibit discordant responses for their different manifestations of psoriatic disease with, for example, treatment targeting interleukin (IL)-17 resulting in sometimes dramatic improvements in skin psoriasis while features of peripheral arthritis may show little or no response. Trying to identify which drug to prescribe for which patient can be challenging and clinicians often use an individual’s clinical features and history of previous drug response as the best guide to treatment choice. This can result in patients cycling through several therapies before finding one that is effective for them, with this period of non-response contributing to disease progression and poor outcomes. bDMARDs are occasionally associated with serious adverse events, most commonly infection,6 and their high cost compared to conventional synthetic DMARDs (csDMARDs) must also be considered. The application of precision and stratified medicine is therefore needed, whereby psoriatic patients most likely to respond to different bDMARDs and tsDMARDs can be identified, thereby justifying their additional toxicity and cost.
The objectives of this systematic review were to identify studies of biomarkers associated with response to treatment in (i) PsV and (ii) PsA.
Methods
A protocol was developed in advance and contained eligibility criteria, information sources, search strategy and study selection. Our study aligns with ‘The PRISMA Statement for Reporting Systematic Reviews and Meta-Analyses of Studies That Evaluate Health Care Interventions’.7
Inclusion criteria
We included cohort studies, case-control studies and RCTs that examined the relationship in patients with PsV or PsA between biomarker concentration prior to drug commencement and subsequent treatment response. The following types of biomarkers were included: genetic, serum, cellular, urine, synovial tissue and skin tissue.
Exclusion criteria
The following were exclusion criteria: studies with patients under 18 years of age; studies using clinical, radiological, or stool biomarkers; and studies examining response to non-pharmacologic treatments.
Searches
The initial search was performed on 18 June 2018 and was repeated on 2 September 2020 to capture the most up to date published information possible. The following medical literature electronic databases were searched: PubMed, Embase, and Cochrane Central Register of Controlled Trials (CENTRAL). The following MeSH, EMTree or key terms were used: biomarkers; psoriatic arthritis; psoriasis; DMARD; biologic; antirheumatic agent. Conference proceedings were also searched for potential inclusion, including: American College of Rheumatology (ACR) annual meeting (2015–2019); European League Against Rheumatism (EULAR) annual congress (2015–2019); British Society for Rheumatology (BSR) annual conference (2015–2019); and Group for Research and Assessment of Psoriasis and Psoriatic Arthritis (GRAPPA) annual meeting (2015–2019).
Study selection
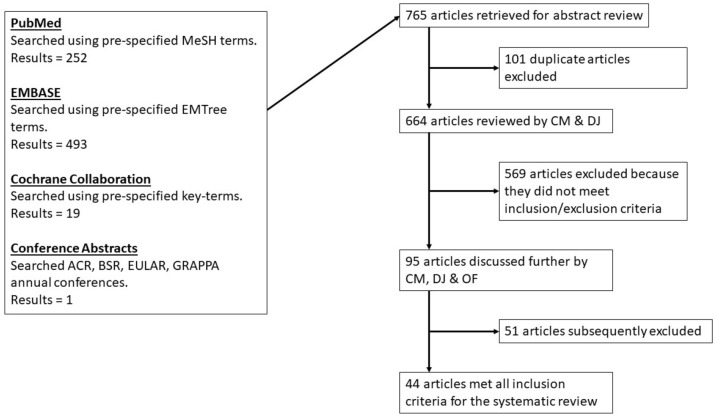
All search results were assessed independently by two reviewers (CM, DJ) for potential inclusion. Where there was a difference of opinion, the full article was discussed by the two reviewers with a third reviewer (OF) to reach a consensus. Figure 1 details the process of article selection.
Figure 1.
Systematic review search algorithm.
ACR, American College of Rheumatology; BSR, British Society for Rheumatology; EULAR, European League Against Rheumatism; GRAPPA, Group for Research and Assessment of Psoriasis and Psoriatic Arthritis.
Outcome measurements of treatment response accepted included objective measurements such as changes in psoriasis area severity index (PASI), disease activity score (DAS)28 and an ACR20 response, but also included patient reported outcomes such as EuroQol score and health assessment questionnaire (HAQ) score.
Results
The searches identified 765 articles; 101 duplicate articles were excluded, and of the 664 remaining unique articles, 569 were excluded because they did not meet the inclusion and exclusion criteria. Of the remaining 95 articles a further 51 were excluded, for example, if the citations failed to match the study design, outcome or population of interest. A total of 44 articles met all eligibility criteria (Table 1): 32 were full-length articles in peer-reviewed international journals and 12 were abstracts from peer-reviewed international conferences.
Table 1.
Characteristics of studies included in the systematic review.
| References | PsV/PsA | Country | No. of subjects | Study design |
|---|---|---|---|---|
| Chicharro et al.19 | PsV | Spain | 33 | Prospective, single centre |
| De Keyser et al.11 | PsV | Belgium, the Netherlands | 137 | Prospective, multicentre |
| Dand et al.8 | PsV | UK | 1326 | Retrospective, multicentre |
| Ovejero-Benito et al.15 | PsV | Spain | 95 | Prospective, single centre |
| Prieto-Pérez et al.10 | PsV | Spain | 144 | Prospective, single centre |
| Ovejero-Benito et al.16 | PsV | Spain | 78 | Prospective, single centre |
| Lu et al.26 | PsV | China | 43 | Prospective, single centre |
| Masouri et al.9 | PsV | Greece | N/A | Retrospective, single centre |
| Nishikawa et al.17 | PsV | Japan | 65 | Prospective, multicentre |
| Tan et al.22 | PsV | US | N/A | Prospective, multicentre |
| Lima et al.27 | PsV | Brazil | 38 | Prospective, single centre |
| Hoffman et al.29 | PsV | Germany | 146 | Retrospective, single centre |
| Kivelevitch et al.18 | PsV | US | 35 | Prospective, single centre |
| Lembo et al.21 | PsV | Italy | 16 | Prospective, single centre |
| Ryan et al.12 | PsV | US | 138 | Retrospective, multicentre |
| Strober et al.24 | PsV | US | 152 | Prospective, multicentre |
| Gedebjerg et al.20 | PsV | Denmark | 18 | Prospective, single centre |
| Jokai et al.30 | PsV | Hungary | 38 | Prospective, single centre |
| Shimauchi et al.28 | PsV | Japan | 28 | Retrospective, single centre |
| Chiu et al.13 | PsV | Taiwan | 102 | Prospective, single centre |
| Gulliver et al.14 | PsV | Canada | 45 | Retrospective, single centre |
| Kanelleas et al.25 | PsV | Greece | 41 | Prospective, single centre |
| Alivernini et al.36 | PsA | Italy | 12 | Prospective, single centre |
| David et al.31 | PsA | UK | 128 | Prospective, multicentre |
| Hellman et al.47 | PsA | Sweden | 20 | Prospective, multicentre |
| Mascia et al.32 | PsA | Italy | 70 | Prospective, single centre |
| Ørnbjerg et al.40 | PsA | Multinational | 7975 | Retrospective, multicentre |
| Siebert et al.46 | PsA | UK, US | 1069 | Retrospective, multicentre |
| Song et al.45 | PsA | US | 142 | Prospective, multicentre |
| Ovejero-Benito et al.33 | PsA | Spain | 20 | Prospective, single centre |
| Scrivo et al.41 | PsA | Italy | 149 | Prospective, single centre |
| Muramatsu et al.48 | PsA | Japan | 29 | Prospective, single centre |
| Ademowo et al.37 | PsA | Ireland | 10 | Retrospective, single centre |
| Collins et al.38 | PsA | Ireland | 32 | Prospective, multicentre |
| Fabris et al.34 | PsA | Italy | 74 | Prospective, single centre |
| Murdaca et al.35 | PsA | Italy | 57 | Prospective, single centre |
| Chandran et al.49 | PsA | Canada | 40 | Prospective, single centre |
| Wagner et al.50 | PsA | Multinational | 100 | Prospective, multicentre |
| Chimenti et al.51 | PsA | Italy | 55 | Prospective, single centre |
| Marotta et al.52 | PsA | Canada | 24 | Prospective, single centre |
| Pontifex et al.39 | PsA | Ireland | 25 | Prospective, single centre |
| Pedersen et al.42 | PsA | Denmark | 17 | Prospective, single centre |
| Gratacos et al.43 | PsA | Spain | 69 | Prospective, multicentre |
| Kristensen et al.44 | PsA | Sweden | 261 | Prospective, multicentre |
PsA, psoriatic arthritis; PsV, psoriasis vulgaris; UK, United Kingdom; US, United States.
Biomarkers associated with treatment response in PsV
The 22 articles describing biomarkers associated with treatment response in PsV are shown in Table 2.
Table 2.
Studies evaluating biomarkers predictive of treatment response in PsV.
| Reference | Outcome measure | Treatment | Biomarker | Outcome |
|---|---|---|---|---|
| Chicharro et al.19 | PASI | TNFi, anti-IL-12/IL-23, anti-IL-17 | miRNA in lesional and non-lesional psoriatic skin | Baseline expression of miRNA-146a in non-lesional skin and miRNA-135b in lesional skin were related to response to treatment |
| De Keyser et al.11 | PASI | UST | HLA-C*06 allele | No statistically significant difference in clinical response between HLA-C*06 positive and HLA-C*06 negative patients |
| Dand et al.8 | PASI90 | ADA, UST | HLA-C*06:02 allele | HLA-C*06:02-negative patients were significantly more likely to respond to ADA than UST |
| Ovejero-Benito et al.15 | PASI75 | ADA, IFX | Genetic polymorphisms | Association between polymorphisms in IVL, IL-12B, NFKBIA, ZNF816A and SLC9A8 genes and treatment response |
| Prieto-Perez et al.10 | PASI75 | TNFi | Genetic polymorphisms | Association between polymorphisms in PGLYR4, ZNF816A, CTNNA2, IL12B, MAP3K1 and HLA-C genes and treatment response |
| Ovejero-Benito et al.16 | PASI75 | ETN | Genetic polymorphisms | Association between polymorphisms in HLA-B/MICA, MAP3K1, PTTG1, ZNF816A genes and response to ETN |
| Lu et al.26 | PASI75 | ETN | Serum cytokines | Baseline IL-12 serum level was a significant factor affecting the clinical response to ETN |
| Masouri et al.9 | PASI | TNFi, UST | Genetic polymorphisms | Rs10484554, a genetic polymorphism in the HLA-C gene showed an association with a good response to TNFi agents but not to UST |
| Nishikawa et al.17 | PASI | ADA, IFX | Genetic polymorphisms (GWAS) | Reported on the 10 SNPs showing the strongest association with response to TNFi treatment |
| Tan et al.22 | PASI75 | TOF | CRP | Baseline CRP was not predictive of treatment response |
| Lima et al.27 | NA | NA | Serum chemokines (CXCL9, CXCL10 and CXCL16) | Levels of serum chemokines do not predict treatment response |
| Hoffman et al.29 | LOR, SSE | ADA, ETN | Anti-dsDNA concentration | Low baseline anti-dsDNA concentrations associated with better outcomes in ADA therapy |
| Kivelevitch et al.18 | PASI | ADA, UST | DEGs | 57 DEGs differentiated UST responders from non-responders |
| Lembo et al.21 | PASI | ADA, ETN, EFZ | MCP-1 levels in plasma and skin | MCP-1 levels not found as a predictor of disease response |
| Ryan et al.12 | PASI75 | ADA, ETN | HLA-C, KIR, VDR genotypes | None of the genotypes examined were predictive of treatment response |
| Strober et al.24 | PASI | ADA | CRP | Baseline CRP was not associated with change in PASI |
| Gedebjerg et al.20 | PASI | UST | mRNA expression in skin | IL-20, IL-21 and p40 mRNA expression in lesional psoriatic skin were upregulated in non-responders compared to responders |
| Jokai et al.30 | PASI | ADA, ETN, IFX | CLA | Responders showed (not significantly) lower initial CLA expression than relapsing patients |
| Shimauchi et al.28 | PASI75 | IFX, ADA, UST | Serum IL-22 & VEGF | Baseline levels of serum IL-22 and VEGF were not significantly different between responders and non-responders |
| Chiu et al.13 | PASI50 | ALC, EFZ, ETN, UST | HLA-B & HLA-C alleles | HLA-C*06 status did not affect PASI 50 response |
| Gulliver et al.14 | PASI75 | ALC | Genetic polymorphisms (GWAS) | HLA-C*06 did not predict response to alefacept |
| Kanelleas et al.25 | PASI75 | ETN | Inflammatory markers | No significant difference at baseline between responders and non-responders |
ADA, adalimumab; ALC, alefacept; CLA, cutaneous lymphocyte-associated antigen; CRP, C-reactive protein; DEG, differentially expressed gene; dsDNA, double stranded DNA; EFZ, efalizumab; ETN, etanercept; GWAS, genome-wide association study; HLA, human leucocyte antigen; IFX, infliximab; IL, interleukin; KIR, killer immunoglobulin receptor; LOR, loss of response; MCP, monocyte chemoattractant protein; miRNA, microRNA; mRNA, messenger RNA; NA, not available; PASI, psoriasis area and severity index; PsV, psoriasis vulgaris; SNP, single nucleotide polymorphism; SSE, serious side effect; TNFi, TNF inhibitors; TOF, tofacitinib; UST, ustekinumab; VDR, vitamin D receptor; VEGF, vascular endothelial growth factor.
Ten articles examined the potential role of genetic polymorphisms and specific human leukocyte antigen (HLA) alleles as biomarkers of treatment response. Three of these studies reported associations between response and the HLA-C*06 haplotype. Using a national psoriasis registry, Dand et al. examined genotype data on 1326 patients.8 They reported that HLA-C*06:02-negative patients were significantly more likely to respond at all time points to the tumour necrosis factor-alpha inhibitor (TNFi), adalimumab, than to ustekinumab, which blocks the p40 subunit common to both IL-12 and IL-23 cytokines. They found no evidence that an interaction between the ERAP1 genotype and HLA-C*06:02 could provide a more effective predictive biomarker than HLA-C*06:02 alone. Masouri et al. found that rs10484554, a single nucleotide polymorphism (SNP) in the HLA-C gene, showed an association with a good response to TNF is but not to ustekinumab, while rs151823 and rs26653 SNPs in the ERAP1 gene showed associations with a good response to ustekinumab therapy.9 The study by Prieto-Pérez et al. studied 173 polymorphisms in an effort to establish an association with response to TNFi therapy.10 A multivariable analysis showed an association between polymorphisms in several genes including HLA-C.
Other studies have not found an association between the HLA-C gene and treatment response. De Keyser et al. examined the relationship between the presence of the HLA-C*06 haplotype and subsequent response to ustekinumab.11 They found no statistically significant difference in clinical response between HLA-C*06 positive and HLA-C*06 negative patients. Ryan et al. compared the frequencies of HLA-C, killer cell immunoglobulin like receptor (KIR) and vitamin D receptor (VDR) genes in responders and non-responders to etanercept or adalimumab in patients with severe chronic plaque psoriasis.12 None of the HLA-C, KIR or VDR genotypes examined were predictive of treatment response. A case-control study of 199 Chinese patients with PsV found that the presence of certain HLA-C*06 haplotypes was not predictive of treatment response to etanercept, ustekinumab, efalizumab or alefacept.13 Gulliver et al. conducted a retrospective study and identified 45 patients with psoriasis who had been treated with alefacept.14 They found that the presence of certain HLA-C*06 haplotypes was not predictive of response to treatment in PsV.
Three studies reported associations between other non-HLA polymorphisms and response to TNFi treatment. Ovejero-Benito et al. performed two studies investigating response to monoclonal antibody treatment and etanercept, respectively.15,16 Multivariable analyses showed five SNPs, in IVL, IL-12B, NFKBIA, ZNF816A and SLC9A8 genes, to be associated with achieving PASI75 response after 3 months of either adalimumab or infliximab. Multivariable analyses showed an association between polymorphisms in HLA-B/MICA, MAP3K1, PTTG1 and ZNF816A genes and the response to etanercept at 3 months. A genome-wide association study (GWAS) of 65 Japanese psoriasis patients reported on 10 SNPs, mapping to the SPEN, JAG2, MACC1, GUCY1B3, PDE6A, CDH23, SHOC2, LOC728724, ADRA2A and KCNIP1 genes, showing association with TNFi treatment response.17 The authors also examined 68 SNPs that had previously been reported to be associated with response to TNFi treatment. Only one, rs11096957, mapping to the toll-like receptor (TLR) 10 gene was associated with treatment response.
Kivelevitch et al. examined differentially expressed genes using microarray analysis in 35 patients treated with either adalimumab or ustekinumab.18 They found 57 differentially expressed genes, 14 upregulated and 43 downregulated, that differentiated ustekinumab responders from non-responders. The most significant differences in responders compared with non-responders were upregulation of HLA-DRB4 and carbohydrate metabolism pathways, and downregulation of tetrahydrobiopterin synthesis.
Three studies described either chemokine, microRNA (miRNA) or gene expression levels in lesional psoriatic skin. Chicharro et al. reported on expression of miRNAs in psoriatic skin at baseline and their associations with subsequent response to biologic therapy.19 They found that expression of miRNA-146a in non-lesional skin and miRNA-135b in lesional skin were related to improvement after 3 months of treatment. Gedebjerg et al.20 measured messenger RNA (mRNA) expression of various genes in skin biopsies by quantitative polymerase chain reaction. A total of 18 adult patients with moderate-to-severe chronic plaque psoriasis were included in the study and all patients were treated with ustekinumab. IL-20, IL-21 and p40 mRNA expression were significantly upregulated by factors of 2.7, 2.4 and 2.3, respectively, among non-responders compared with responders. Lembo et al. studied monocyte chemoattractant protein-1 (MCP-1) plasma levels in psoriatic patients seeking an association between plasma and cutaneous MCP-1 expression and response to biological drugs.21 They also performed lesional skin biopsies in five patients treated with TNFi. They did not find an association between baseline MCP-1 levels and subsequent response to treatment.
The potential role of inflammatory markers as predictors of treatment response was examined in three studies. Tan et al. examined data from the Oral-treatment Psoriasis Trial (OPT) Pivotal 1 phase 3 study on the use of tofacitinib, a Janus kinase (JAK) inhibitor, for the treatment of psoriasis.22,23 Baseline C-reactive protein (CRP) was not associated with PASI75 response. Similarly, Strober et al. reported that baseline levels of CRP were not associated with subsequent change in PASI in patients treated with adalimumab who had a suboptimal response to previous therapies.24 Kanelleas et al. reported similar results.25 They found that neither baseline levels of high sensitivity (hs) -CRP, nor ESR were associated with subsequently achieving a PASI75 response in patients treated with etanercept.
The remaining five studies on PsV examined levels of serum cytokines, chemokines, anti-double stranded (ds)DNA antibodies and cutaneous lymphocyte-associated antigen (CLA). Lu et al. measured baseline levels of IL-6, IL-12, IL-17A, IL-23 and TNF-α in patients with moderate to severe psoriasis before commencing on etanercept therapy.26 They reported that baseline IL-12 serum levels were significantly higher in responders compared with non-responders (p = 0.03). Lima et al. measured serum levels of CXCL9, CXCL10 and CXCL16 and the frequencies of CD4+CXCR3+ T lymphocytes through ELISA and flow cytometry, respectively.27 They found systemic levels of chemokine ligands unable to predict response to treatment. Shimauchi et al. examined serum levels of IL-22 and vascular endothelial growth factor (VEGF),28 but found them unable to predict response to treatment with ustekinumab or TNFi. Hoffman et al. measured baseline anti-dsDNA antibody concentrations in patients undergoing treatment with adalimumab.29 They found patients with lower baseline anti-dsDNA concentrations responded better. Lastly, a study by Jokai et al. examined a potential role for CLA as a predictor of response to TNFi therapy.30 They reported baseline CLA expression was not significantly different between those who responded to treatment and those who relapsed over a 24-week period.
Biomarkers associated with treatment response in PsA
The 22 articles describing biomarkers predictive of treatment response in PsA are shown in Table 3.
Table 3.
Studies evaluating biomarkers predictive of treatment response in PsA.
| Reference | Outcome measure | Treatment | Biomarker | Outcome |
|---|---|---|---|---|
| Alivernini et al.36 | MDA | MTX | Synovial CD3+ cells | Patients who reached MDA status at 6 months had lower baseline CD3+ cell immunohistochemistry scores |
| David et al.31 | DAS28 | bDMARD | HLA-B*27 allele | HLA-B*27 status was not associated with treatment response |
| Hellman et al.47 | MDA, DAPSA, ACR20/50/70 | ADA | HA in skin and serum | Higher levels of HA in serum associated with higher overall disease activity after 12 weeks of treatment |
| Mascia et al.32 | PsARC, ACR20 | TNFi | Genetic polymorphisms | SNP-29 predicts response to TNFi |
| Ørnbjerg et al.40 | DAPSA28 remission | TNFi | CRP | Normal CRP at baseline decreased the probability of DAPSA28 remission at 6 months |
| Siebert et al.46 | ACR20, PASI75 | GUS, UST | IL-17A, IL-17F, CRP | Baseline levels of proteins measured not associated with treatment response to UST. Baseline IL-17F modestly associated with ACR20 response to GUS |
| Song et al.45 | ACR20, PASI75 | GUS | CRP, SAA, slCAM1, svCAM1, IL-17A, IL-17F, IL-22 | None of the baseline proteins measured were associated with treatment response |
| Ovejero-Benito et al.33 | Improvement in Arthritis, EuroQol | ADA, ETN, IFX | Genetic polymorphisms | Association between polymorphisms in the TNFAIP3 gene and treatment response |
| Scrivo et al.41 | Achievement of MDA | GOL | hs-CRP | A higher baseline hs-CRP value and the absence of comorbidities were predictive factors for achieving MDA at 6 months |
| Muramatsu et al.48 | DAS28-CRP | IFX, ADA, UST | Serum IL-6 levels | Baseline serum IL-6 levels not statistically different between good responders and poor responders to treatment |
| Ademowo et al.37 | DAS28-CRP | ADA | Synovial tissue proteins | Panel of 57 proteins predictive of response to treatment (AUC of 0.76) |
| Collins et al.38 | DAS28 | TNFi | Synovial tissue proteins | 25 proteins differentially expressed between good and poor responders |
| Fabris et al.34 | Survival of first TNFi agent | TNFi | Genetic polymorphisms | TNFα -308A allele and IL-6 -174GG homozygosis resulted as independent biomarkers predicting survival of the first TNFi therapy |
| Murdaca et al.35 | ACR 20/50/70; DAS28; HAQ | ADA, ETN, IFX | Genetic polymorphisms | TNFα gene polymorphisms at −308 and −238 not associated with response to TNFi treatment. SNP +489 A/A genotype associated with response to ADA |
| Chandran et al.49 | SJC, TJC, PASI | ADA, ETN, IFX, GOL | MMP-3 | Baseline level of MMP-3 was independently associated with treatment response |
| Wagner et al.50 | ACR20; DAS28-CRP; PASI75 | GOL | 92 serum biomarkers | Pyridinoline, adiponectin, PAP and factor VII were identified as a panel of markers having the potential to be predictive of ACR20 response |
| Chimenti et al.51 | DAS28 | ETN, ADA | Complement C3 | Higher baseline C3 levels were associated with non-response |
| Marotta et al.52 | SJC68; PASI, CRP, ESR, DAS28, ACR50 | ADA | 14-3-3 eta serum protein | Baseline 14-3-3 eta titres were predictive of an ACR50 response |
| Pontifex et al.39 | DAS28 | ANR, ETN | CD3+ T cells (synovium & peripheral blood) | Baseline levels of CD3+ T cells were not predictive of treatment response |
| Pedersen et al.42 | VAS-pain; PGA; 28 joint count | ADA, ETN, IFX | CRP, IL-6, VEGF, YKL-40, MMP-3, total aggrecan | Baseline levels of serum CRP and MMP3 and plasma IL-6 and VEGF were all higher in responders compared to non-responders |
| Gratacos et al.43 | ACR50 | IFX | ESR, CRP | High CRP values were independently associated with a good therapeutic response |
| Kristensen et al.44 | TNFi survival | ADA, ETN, IFX | ESR, CRP | Higher baseline CRP levels associated with drug survival |
ACR, American college of rheumatology; ADA, adalimumab; ANR, anakinra; AUC, area under the curve; bDMARD, biologic DMARD; CAM, cell adhesion molecule; CRP, C-reactive protein; DAPSA, disease activity in psoriatic arthritis; DAS, disease activity score; ESR, erythrocyte sedimentation rate; ETN, etanercept; GOL, golimumab; GUS, guselkumab; HA, hyaluronan; HAQ, health assessment questionnaire; HLA, human leucocyte antigen; hs-CRP, high sensitivity CRP; IFX, infliximab; IL, interleukin; MDA, minimal disease activity; MMP, matrix metalloprotease; MTX, methotrexate; PASI, psoriasis area and severity index; PGA, patient global assessment; PsA, psoriatic arthritis; PsARC, psoriatic arthritis response criteria; SAA, serum amyloid A; SJC, swollen joint count; SNP, single nucleotide polymorphism; TJC, tender joint count; TNFi, TNF inhibitor; UST, ustekinumab; VAS, visual analogue score; VEGF, vascular endothelial growth factor.
Five studies investigated HLA alleles and other genetic polymorphisms in responders and non-responders. David et al. examined whether the presence of HLA-B*27 is a predictor of treatment response to biologics in PsA,31 but concluded it was not associated with EULAR good response or DAS28 improvement. Mascia et al. aimed to identify genetic variants in the TNF-α genomic region able to predict therapeutic response to TNFi therapy.32 They found a significant association between SNP29, located between the lymphotoxin alpha (LTA) and TNF genes, with the response to TNFi treatment. Ovejero-Benito et al. examined 10 polymorphisms located in genes related to TNF in 20 PsA patients treated with TNFi therapy.33 rs6920220 and rs610604 mapping the TNFAIP3 gene showed a significant association with an improvement EuroQol score after 3 months of treatment. Fabris et al. reported that the TNFα-308A allele as well as the presence of IL6-174GG homozygosity were independent biomarkers predicting survival of the first TNFi therapy in patients with spondyloarthritis,34 some of whom had PsA. Murdaca et al.35 investigated the role of SNPs in the TNFα gene in the response to TNFi therapy. The +489A allele showed a statistically non-significant trend for association with response to treatment with etanercept. Alleles −308 and −238 did not influence the clinical outcome of PsA patients treated with TNFi.
Four studies examined potential synovial tissue biomarkers for predicting response to treatment. Alivernini et al. examined synovial tissue biopsies using immunohistochemistry (IHC) in DMARD naive PsA patients prior to them commencing methotrexate (MTX).36 They reported a lower IHC score of CD3+ T-cells in patients reaching minimal disease activity (MDA) status at 6 months compared to those not achieving this outcome. Two of these studies utilised an unbiased proteomic analysis approach by using mass spectrometry to report levels of synovial tissue proteins. Ademowo et al. described a biomarker panel of 57 proteins confirmed to be predictive of treatment response with an area under the curve of 0.76.37 Collins et al. reported 25 synovial tissue proteins that were differentially expressed between good responders and poor responders to TNFi therapy.38 Another study, by Pontifex et al., quantified cellular markers including CD3+ T-cells but found baseline levels were not predictive of treatment response.39
A number of studies examined the association between inflammatory markers at baseline and subsequent response to treatment with bDMARDs. Five studies reported that a higher baseline level of CRP was associated with better treatment response or treatment continuation. Ørnbjerg et al. reported on data from nearly 8000 PsA patients in 13 European registries commencing on first TNFi.40 Using a multivariate model, they found a normal CRP at baseline decreased the probability of DAPSA28 remission at 6 months. Scrivo et al. reported higher levels of hs-CRP predicted MDA achievement after 6 months of treatment with golimumab.41 A study by Pedersen et al. of patients treated with TNFi therapy reported that compared with non-responders, responders had higher baseline CRP, IL-6, VEGF and MMP-3, whereas no difference was seen in YKL-40 or total aggrecan.42 Similarly, Gratacos et al. found that high CRP levels at the start of treatment were independently associated with a good therapeutic response to infliximab.43 In a study by Kristensen et al., drug persistence was used as a surrogate of treatment response.44 They reported that high CRP levels at TNFi initiation were associated with better overall drug survival. Conversely, other studies did not find an association between baseline levels of CRP and subsequent treatment response. Song et al. measured CRP, serum amyloid A (SAA), soluble cell adhesion molecules (sICAM1, sVCAM1) and Th17 effector cytokines (IL17A, IL17F and IL22) at baseline in patients subsequently treated with guselkumab.45 They did not identify an association between baseline protein levels and subsequent clinical response. Siebert et al. examined baseline levels of CRP, Il17A and IL17F in patients treated with either ustekinumab or guselkumab.46 While none of the baseline levels of evaluated cytokines were associated with clinical response to ustekinumab, baseline levels of Il17F in patients treated with guselkumab were modestly associated with ACR20 response at week 24.
The remaining six studies identified other candidate biomarkers of treatment response. In a prospective clinical study, Hellman et al. measured skin inflammation, serum hyaluronan (HA) and molecular mass of HA in patients subsequently treated with adalimumab.47 Patients with elevated HA values had more retained swollen joints and higher overall disease activity after 12 weeks of treatment. Muramatsu et al. found that baseline serum IL-6 levels were statistically not significantly different between good and poor responders to biologic treatment.48 Chandran et al. studied 10 soluble biomarkers in patients commencing TNFi treatment but found only baseline level of MMP-3 to be associated with responder status.49 Notably, they found no association between hs-CRP and treatment response. In a prospectively planned biomarker sub-study, Wagner et al. examined baseline levels of 92 biomarkers in 100 patients from the GO-REVEAL trial examining the response of patients with PsA to golimumab.50 Pyridinoline, adiponectin, prostatic acid phosphate and factor VII were identified as a panel of markers having the potential to be predictive of ACR20 response. As in the study by Chandran et al., baseline CRP levels were not associated with any of the clinical outcomes. Chimenti et al. examined baseline levels of complement, CRP and ESR.51 They found that higher baseline C3 levels were associated with non-response to TNFi therapy. Neither CRP nor ESR were associated with treatment response. Lastly, Marotta et al. reported baseline titres of 14-3-3 eta serum protein were predictive of an ACR50 response in patients with PsA treated with adalimumab.52
Discussion
This review reports several different types of biomarkers that have been shown to be associated with treatment response in psoriatic disease. Of the 22 PsA studies, 21 involved bDMARD therapy; 13 were limited to TNFi therapy only, while two studies involved TNFi therapy as well as another agent, either ustekinumab or anakinra. One of the studies on PsV involved the use of a tsDMARD, tofacitinib. The other studies on PsV involved biologic therapy, predominantly TNFis, although six studies did involve ustekinumab treatment. The majority of the studies assessed outcomes after 12 to 28 weeks, which is a reasonable period of time after which to assess response to treatment. One limitation of the data is that only one of the studies reviewed included patients on a csDMARD and further studies exploring biomarkers following use of csDMARDs would be valuable.36
A significant limitation to the majority of studies in this review was the number of subjects enrolled. Only 14 of the 44 studies had at least 100 subjects, while only 4 had at least 200. In smaller studies, particularly where less stringent outcome measures were used, the numbers of non-responders tended to be low, making it difficult to identify statistically significant associations due to the higher standard error.
Another factor that made it more difficult to compare results from different studies was the number of different outcome measures used. All of the studies on PsV used change in PASI, most commonly PASI75, as an outcome measure. In contrast, a number of different measures were used to assess outcome in PsA, reflecting the heterogenous nature of the disease. A DAS28 score was the most common outcome measure used, while ACR20/50/70, drug persistence, MDA and patient global assessment (PGA) were among the other measures used. Some of these outcome measures are more achievable than others. For example, MDA is a much stricter criteria for response to treatment than ACR20. The adoption of standardised, widely used outcome measures remains a challenge in PsA.
While some studies assessed response to one treatment only, a number of studies included patients treated with different therapies, sometimes acting on different molecular pathways, for example, TNFα inhibition and IL-12/23 inhibition. Mechanistically, it is likely that a biomarker is predictive of response to one specific class of treatment but not another, due to the immune axis being altered. Therefore, it is difficult to interpret analyses where pooling of patients treated with different classes of agents occurred.
These reasons may partially explain some of the seemingly inconsistent results reported. For example, of the six studies that investigated associations between the HLA-C gene and treatment response in PsV, three studies reported associations, while the other three did not. It must be noted that these studies included patients treated with different bDMARDs. Eight studies either focused primarily on, or included, CRP as a possible predictor of treatment response in PsA. Five studies reported higher baseline levels of CRP being associated with better response to treatment, whereas three studies did not. Notably, different outcome measures were used in all five studies where associations were shown. None of the three studies that examined the relationship between CRP and subsequent treatment response in PsV identified any association.
The most significant limitation of research in this field that has been identified by this systematic review is the lack of validation of results in independent cohorts. None of the potential biomarkers identified in this systematic review have been validated in larger independent cohorts. Validation of biomarkers in well defined, prospective cohorts is necessary before they can be developed into clinical tests that can be used on a routine basis.
In PsV, studies examining potential associations between genetic polymorphisms and treatment response gave some of the most promising results. This topic is a good candidate for prioritization for further research, with stratification of patients by type of bDMARD therapy received more likely to uncover meaningful associations.
In PsA, the relationship between CRP and subsequent response to bDMARD therapy is potentially of significant clinical use. The five studies that showed a positive relationship between higher levels of CRP at baseline and a good therapeutic response all related to the use of TNFis, while the more recent large study by Siebert et al. which did not show a similar association related to bDMARDs which block the IL12/23 pathway. Further study in this area is needed.
The ability to predict response to treatment remains a key unmet need in psoriatic disease. While many of the studies included in this review show promise, their results need to be validated before they can be developed into routine clinically useful tests.
Acknowledgments
We would like to thank Diarmuid Stokes for his guidance on constructing the database searches. DRJ and HJ acknowledge that this research was supported by the NIHR Cambridge Biomedical Research Centre (BRC-1215-20014) and Cambridge Arthritis Research Endeavour (CARE) [The views expressed are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care].
Footnotes
Conflict of interest statement: The authors declare that there is no conflict of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Funding is acknowledged from the UCD Wellcome Institutional Strategic Support Fund, which was financed jointly by University College Dublin and the SFI-HRB-Wellcome Biomedical Research Partnership (ref 204844/Z/16/Z). DRJ’s research time is supported by the Cambridge Arthritis Research Endeavour (CARE; charity number 802862).
ORCID iDs: Hannah Jethwa  https://orcid.org/0000-0003-1640-7350
https://orcid.org/0000-0003-1640-7350
Oliver M. FitzGerald  https://orcid.org/0000-0002-6607-6070
https://orcid.org/0000-0002-6607-6070
Contributor Information
Conor Magee, The Conway Institute for Biomolecular Research, University College Dublin, Dublin, Ireland; Department of Rheumatology, St. Vincent’s University Hospital, Dublin, Ireland.
Hannah Jethwa, Department of Rheumatology, Imperial College London NHS Trust, London, UK.
Oliver M. FitzGerald, The Conway Institute for Biomolecular Research, University College Dublin, Dublin, Ireland Department of Rheumatology, St. Vincent’s University Hospital, Dublin, Ireland.
Deepak R. Jadon, Department of Rheumatology, Cambridge University Hospitals NHSFT, Hills Road, Cambridge, CB2 0QQ, UK.
References
- 1. Langley R, Krueger G, Griffiths C. Psoriasis: epidemiology, clinical features, and quality of life. Ann Rheum Dis 2005; 64: ii18–ii23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mease PJ, Kivitz AJ, Burch FX, et al. Etanercept treatment of psoriatic arthritis: safety, efficacy, and effect on disease progression. Arthritis Rheum 2004; 50: 2264–2272. [DOI] [PubMed] [Google Scholar]
- 3. Mease P, Fleischmann R, Deodhar AA, et al. Effect of certolizumab pegol on signs and symptoms in patients with psoriatic arthritis: 24-week results of a phase 3 double-blind randomised placebo-controlled study (rapid-psa). Ann Rheum Dis 2014; 73: 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kavanaugh A, McInnes I, Mease P, et al. Golimumab, a new human tumor necrosis factor α antibody, administered every four weeks as a subcutaneous injection in psoriatic arthritis: twenty-four–week efficacy and safety results of a randomized, placebo-controlled study. Arthritis Rheum 2009; 60: 976–986. [DOI] [PubMed] [Google Scholar]
- 5. McInnes IB, Sieper J, Braun J, et al. Efficacy and safety of secukinumab, a fully human anti-interleukin-17a monoclonal antibody, in patients with moderate-to-severe psoriatic arthritis: a 24-week, randomised, double-blind, placebo-controlled, phase ii proof-of-concept trial. Ann Rheum Dis 2014; 73: 349–356. [DOI] [PubMed] [Google Scholar]
- 6. Dommasch ED, Abuabara K, Shin DB, et al. The risk of infection and malignancy with tumor necrosis factor antagonists in adults with psoriatic disease: a systematic review and meta-analysis of randomized controlled trials. J Am Acad Dermatol 2011; 64: 1035–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liberati A, Altman DG, Tetzlaff J, et al. The prisma statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med 2009; 6: e1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dand N, Duckworth M, Baudry D, et al. HLA-c* 06: 02 genotype is a predictive biomarker of biologic treatment response in psoriasis. J Allergy Clin Immunol 2019; 143: 2120–2130. [DOI] [PubMed] [Google Scholar]
- 9. Masouri S, Stefanaki I, Ntritsos G, et al. A pharmacogenetic study of psoriasis risk variants in a greek population and prediction of responses to anti-TNF-α and anti-IL-12/23 agents. Mol Diagn Ther 2016; 20: 221–225. [DOI] [PubMed] [Google Scholar]
- 10. Prieto-Pérez R, Solano-López G, Cabaleiro T, et al. New polymorphisms associated with response to anti-TNF drugs in patients with moderate-to-severe plaque psoriasis. Pharmacogenomics J 2018; 18: 70–75. [DOI] [PubMed] [Google Scholar]
- 11. De Keyser E, Busard CI, Lanssens S, et al. Clinical consequences of antibody formation, serum concentrations, and HLA-Cw6 status in psoriasis patients on ustekinumab. Ther Drug Monit 2019; 41: 634–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ryan C, Kelleher J, Fagan MF, et al. Genetic markers of treatment response to tumour necrosis factor-alpha inhibitors in the treatment of psoriasis. Clin Exp Dermatol 2014; 39: 519–524. [DOI] [PubMed] [Google Scholar]
- 13. Chiu HY, Huang PY, Jee SH, et al. HLA polymorphism among Chinese patients with chronic plaque psoriasis: subgroup analysis. Br J Dermatol 2012; 166: 288–297. [DOI] [PubMed] [Google Scholar]
- 14. Gulliver W, Pope A, Baker KA, et al. Determination of genetic markers for responsiveness to alefacept. J Am Acad Dermatol 2011; 64: AB163. [Google Scholar]
- 15. Ovejero-Benito MC, Prieto-Pérez R, Llamas-Velasco M, et al. Polymorphisms associated with adalimumab and infliximab response in moderate-to-severe plaque psoriasis. Pharmacogenomics 2018; 19: 7–16. [DOI] [PubMed] [Google Scholar]
- 16. Ovejero-Benito MC, Prieto-Pérez R, Llamas-Velasco M, et al. Polymorphisms associated with etanercept response in moderate-to-severe plaque psoriasis. Pharmacogenomics 2017; 18: 631–638. [DOI] [PubMed] [Google Scholar]
- 17. Nishikawa R, Nagai H, Bito T, et al. Genetic prediction of the effectiveness of biologics for psoriasis treatment. J Dermatol 2016; 43: 1273–1277. [DOI] [PubMed] [Google Scholar]
- 18. Kivelevitch D, Sharma M, Mansouri B, et al. Immune signatures of psoriasis. Br J Dermatol 2014; 171: e114. [Google Scholar]
- 19. Chicharro P, Rodríguez-Jiménez P, Llamas-Velasco M, et al. Expression of miR-135b in psoriatic skin and its association with disease improvement. Cells 2020; 9: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gedebjerg A, Johansen C, Kragballe K, et al. IL-20, IL-21 and p40: potential biomarkers of treatment response for ustekinumab. Acta Derm Venereol 2013; 93: 150–155. [DOI] [PubMed] [Google Scholar]
- 21. Lembo S, Capasso R, Balato A, et al. MCP-1 in psoriatic patients: effect of biological therapy. J Dermatolog Treat 2014; 25: 83–6. [DOI] [PubMed] [Google Scholar]
- 22. Tan H, Valdez H, Wolk R, et al. Early clinical response as a predictor of subsequent response to tofacitinib treatment: results from two phase 3 studies of subjects with moderate to severe plaque psoriasis. J Am Acad Dermatol 2016; 74: AB244. [DOI] [PubMed] [Google Scholar]
- 23. Papp KA, Krueger JG, Feldman SR, et al. Tofacitinib, an oral janus kinase inhibitor, for the treatment of chronic plaque psoriasis: long-term efficacy and safety results from 2 randomized phase-III studies and 1 open-label long-term extension study. J Am Acad Dermatol 2016; 74: 841–850. [DOI] [PubMed] [Google Scholar]
- 24. Strober BE, Poulin Y, Teller C, et al. Changes in C-reactive protein in patients with moderate-to-severe psoriasis switched to adalimumab therapy after suboptimal response to etanercept, methotrexate or phototherapy. J Eur Acad Dermatol Venereol 2014; 28: 1701–1706. [DOI] [PubMed] [Google Scholar]
- 25. Kanelleas A, Liapi C, Katoulis A, et al. The role of inflammatory markers in assessing disease severity and response to treatment in patients with psoriasis treated with etanercept. Clin Exp Dermatol 2011; 36: 845–850. [DOI] [PubMed] [Google Scholar]
- 26. Lu J, Tang S, Xie S, et al. The potential of IL-12 in predicting clinical response to etanercept treatment in patients with psoriasis. Int J Clin Exp Med 2016; 9: 23519–23524. [Google Scholar]
- 27. Lima XT, Oliveira RTD, Braga FG, et al. Circulating levels of chemokines in psoriasis. Autoimmunity 2015; 48: 57–60. [DOI] [PubMed] [Google Scholar]
- 28. Shimauchi T, Hirakawa S, Suzuki T, et al. Serum interleukin-22 and vascular endothelial growth factor serve as sensitive biomarkers but not as predictors of therapeutic response to biologics in patients with psoriasis. J Dermatol 2013; 40: 805–812. [DOI] [PubMed] [Google Scholar]
- 29. Hoffmann JH, Knoop C, Enk AH, et al. Baseline anti-dsDNA concentrations and previous treatments predict response to adalimumab and etanercept: a retrospective investigation of 146 psoriasis patients. J Dermatol Sci 2014; 76: 180–185. [DOI] [PubMed] [Google Scholar]
- 30. Jokai H, Szakonyi J, Kontar O, et al. Cutaneous lymphocyte-associated antigen as a novel predictive marker of TNF-alpha inhibitor biological therapy in psoriasis. Exp Dermatol 2013; 22: 221–223. [DOI] [PubMed] [Google Scholar]
- 31. David T, Jani M, Bowes J, et al. Is HLA-B27 a predictor of treatment response to biologics in psoriatic arthritis? Rheumatology (Oxford) 2019; 58: iii128. [Google Scholar]
- 32. Mascia E, Orrù S, Mathieu A, et al. Genetic variants in the TNF-alpha region: a novel biomarker of clinical response to anti TNF-alpha drugs in psoriatic arthritis patients. J Psoriasis Psoriatic Arthritis 2019; 4: 99. [Google Scholar]
- 33. Ovejero-Benito MC, Muñoz-Aceituno E, Reolid A, et al. Polymorphisms associated with anti-TNF response in psoriatic arthritis. Basic Clin Pharmacol Toxicol 2018; 123: 89. [Google Scholar]
- 34. Fabris M, Quartuccio L, Fabro C, et al. The TNF alpha -308 and the IL-6 -174 promoter polymorphisms associate with effective anti-TNF alpha treatment in seronegative spondyloarthritis. Ann Rheum Dis 2015; 74: 282. [DOI] [PubMed] [Google Scholar]
- 35. Murdaca G, Gulli R, Spano F, et al. TNF-α gene polymorphisms: association with disease susceptibility and response to anti-TNF-α treatment in psoriatic arthritis. J Invest Dermatol 2014; 134: 2503–2509. [DOI] [PubMed] [Google Scholar]
- 36. Alivernini S, Bruno D, Tolusso B, et al. Differential synovial tissue biomarkers among psoriatic arthritis and rheumatoid factor/anti-citrulline antibody-negative rheumatoid arthritis. Arthritis Res Ther 2019; 21: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ademowo OS, Hernandez B, Collins E, et al. Discovery and confirmation of a protein biomarker panel with potential to predict response to biological therapy in psoriatic arthritis. Ann Rheum Dis 2016; 75: 234–241. [DOI] [PubMed] [Google Scholar]
- 38. Collins ES, Butt AQ, Gibson DS, et al. A clinically based protein discovery strategy to identify potential biomarkers of response to anti-TNF-α treatment of psoriatic arthritis. Proteomics Clin Appl 2016; 10: 645–662. [DOI] [PubMed] [Google Scholar]
- 39. Pontifex EK, Gerlag DM, Gogarty M, et al. Change in CD3 positive T-cell expression in psoriatic arthritis synovium correlates with change in DAS28 and magnetic resonance imaging synovitis scores following initiation of biologic therapy–a single centre, open-label study. Arthritis Res Ther 2011; 13: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ørnbjerg LM, Georgiadis S, Jacobsson L, et al. Predictors of DAPSA28 remission at 6 months in bio-naive patients with psoriatic arthritis starting a TNF inhibitor in clinical practice-results from the eurospa collaboration. Arthritis Rheumatol 2019; 71. [Google Scholar]
- 41. Scrivo R, Giardino A, Salvarani C, et al. An italian observational prospective study on predictors of clinical response to golimumab at 6 months in patients with active psoriatic arthritis. Ann Rheum Dis 2018; 77: 383. [PubMed] [Google Scholar]
- 42. Pedersen SJ, Hetland ML, Sørensen IJ, et al. Circulating levels of interleukin-6, vascular endothelial growth factor, YKL-40, matrix metalloproteinase-3, and total aggrecan in spondyloarthritis patients during 3 years of treatment with TNFα inhibitors. Clin Rheumatol 2010; 29: 1301–1309. [DOI] [PubMed] [Google Scholar]
- 43. Gratacos J, Casado E, Real J, et al. Prediction of major clinical response (ACR50) to infliximab in psoriatic arthritis refractory to methotrexate. Ann Rheum Dis 2007; 66: 493–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kristensen LE, Gulfe A, Saxne T, et al. Efficacy and tolerability of anti-tumour necrosis factor therapy in psoriatic arthritis patients: results from the south Swedish arthritis treatment group register. Ann Rheum Dis 2008; 67: 364–349. [DOI] [PubMed] [Google Scholar]
- 45. Song Q, Loza MJ, Leander K, et al. Reduction of serum IL17f and IL22 by IL23p19 blockade with guselkumabis associated with improvement in joint symptoms in psoriatic arthritis. J Clin Rheumatol 2019; 25: S66. [Google Scholar]
- 46. Siebert S, Loza MJ, Song Q, et al. Ustekinumab and guselkumab treatment results in differences in serum IL17a, IL17f and CRP levels in psoriatic arthritis patients: a comparison from ustekinumab ph3 and guselkumab Ph2 programs. Ann Rheum Dis 2019; 78: 293.30559252 [Google Scholar]
- 47. Hellman U, Engström-Laurent A, Larsson A, et al. Hyaluronan concentration and molecular mass in psoriatic arthritis: biomarkers of disease severity, resistance to treatment, and outcome. Scand J Rheumatol 2019; 48: 284–293. [DOI] [PubMed] [Google Scholar]
- 48. Muramatsu S, Kubo R, Nishida E, et al. Serum interleukin-6 levels in response to biologic treatment in patients with psoriasis. Mod Rheumatol 2017; 27: 137–141. [DOI] [PubMed] [Google Scholar]
- 49. Chandran V, Shen H, Pollock RA, et al. Soluble biomarkers associated with response to treatment with tumor necrosis factor inhibitors in psoriatic arthritis. J Rheumatol 2013; 40: 866–871. [DOI] [PubMed] [Google Scholar]
- 50. Wagner CL, Visvanathan S, Elashoff M, et al. Markers of inflammation and bone remodelling associated with improvement in clinical response measures in psoriatic arthritis patients treated with golimumab. Ann Rheum Dis 2013; 72: 83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chimenti MS, Perricone C, Graceffa D, et al. Complement system in psoriatic arthritis: a useful marker in response prediction and monitoring of anti-TNF treatment. Clin Exp Rheumatol 2012; 30: 23–30. [PubMed] [Google Scholar]
- 52. Marotta A, Van Kuijk AW, Maksymowych WP, et al. 14-3-3 eta is a modifiable serum biomarker that marks adalimumab response in psoriatic arthritis. Arthritis Rheumatol 2012; 64: S247. [Google Scholar]