Abstract
Purpose:
To apply our convolutional neural network (CNN) algorithm to predict neoadjuvant chemotherapy (NAC) response using the I-SPY TRIAL breast MRI dataset.
Methods:
From the I-SPY TRIAL breast MRI database, 131 patients from 9 institutions were successfully downloaded for analysis. First post-contrast MRI images were used for 3D segmentation using 3D slicer. Our CNN was implemented entirely of 3 × 3 convolutional kernels and linear layers. The convolutional kernels consisted of 6 residual layers, totaling 12 convolutional layers. Dropout with a 0.5 keep probability and L2 normalization was utilized. Training was implemented by using the Adam optimizer. A 5-fold cross validation was used for performance evaluation. Software code was written in Python using the TensorFlow module on a Linux workstation with one NVidia Titan X GPU.
Results:
Of 131 patients, 40 patients achieved pCR following NAC (group 1) and 91 patients did not achieve pCR following NAC (group 2). Diagnostic accuracy of our CNN two classification model distinguishing patients with pCR vs non-pCR was 72.5 (SD ± 8.4), with sensitivity 65.5% (SD ± 28.1) and specificity of 78.9% (SD ± 15.2). The area under a ROC Curve (AUC) was 0.72 (SD ± 0.08).
Conclusion:
It is feasible to use our CNN algorithm to predict NAC response in patients using a multi-institution dataset.
1. Introduction
Neoadjuvant chemotherapy (NAC) is commonly used to reduce the size of breast tumors before surgery and improve clinical outcomes. Pathological complete response (pCR), i.e., the absence of residual invasive disease in the breast or nodes, is used as a measure of efficacy of NAC. Achieving pCR means better survival for patients [1] and a higher likelihood of benefiting from breast-conserving surgery instead of a full mastectomy [2]. Several large randomized NAC trials have demonstrated pCR to be a potential marker for clinical efficacy as there is a significant correlation between patients who achieved a pCR and improved disease-free survival and overall survival [3,4].
Magnetic resonance imaging (MRI) has become a valuable modality in the evaluation of NAC response and has led to identification of potential imaging-based biomarkers with successful incorporation into the clinical trial setting [5]. One such trial that evaluated MRI based data to predict pCR is the I-SPY TRIAL (Investigation of Serial Studies to Predict Your Therapeutic Response With Imaging and Molecular Analysis). ISPY 1 was a collaboration of the National Cancer Institute Specialized Programs of Research Excellence, the American College of Radiology Imaging Network; the Cancer and Leukemia Group B; and the National Cancer Institute Center for Biomedical Informatics and Information Technology. This multi-institution breast cancer study has made breast MRI imaging dataset publicly available for research.
Deep learning, the use of artificial neural networks applied to learning tasks, presents an efficient solution for complex imaging problems that were otherwise constrained by a limited set of pre-defined features. By employing an abundance of training datasets and processing power, a deep neural network can be trained to identify new imaging features in an unsupervised manner that can identify several different disease states. An example of such a method, convolutional neural networks (CNN) have shown great promise and have already been applied in areas of pathology and radiology. CNNs possess the potential to discover unique multi-scale features highly predictive of response beyond the limits of a handful of explicitly defined radiomic features.
Previously, we have developed CNN algorithms for various classification tasks using breast MRI imaging dataset yielding reasonable diagnostic performance [6–8]. Our studies are limited due to dataset compiled from a single institution. The use of external dataset composed of multiple institutions becomes invaluable to further validate CNN algorithms. The purpose of this study is to apply our CNN algorithm to the publicly available I-SPY TRIAL dataset to predict NAC response.
2. Materials and methods
The ISPY TRIAL protocol was HIPAA-compliant and the informed consent process were approved by the American College of Radiology Institutional Review Board and local-site institutional review boards. Women with invasive breast cancer of 3 cm or greater undergoing NACT with an anthracycline-based regimen, with or without a Taxane, were enrolled between May 2002 and March 2006. MRI data was collected as described by Hylton et al. [5]. From the ISPY TRIAL Breast MRI public database, 131 cases collected from 9 different institutions in the United States were successfully downloaded for this study.
2.1. Data Pre-processing
After the breast MRIs were obtained and anonymized, a fellowship trained breast imaging radiologist with over 10 years of experience subsequently reviewed and segmented the tumor identified on the first post contrast MRI images. The volumes and segmentations were resized to a slice size of 256 × 256 using bicubic and nearest-neighbor inter-polation respectively. The background and chest were zeroed using chest wall and air masks, and contrast limited adaptive histogram normalization of the MRI volumes was performed on each phase separately. Two 3D volumes of each image were generated; the first baseline volume consisted of a 32×32×16×3 pixel bounding cube centered on each tumor; the second normalized box was based on a bounding cube size calculated from the radius of the segmentation mask and subsequently resized to 32×32×16×3 pixels. The last dimension of the volumes represented each of the 3 dynamic phases as a separate channel.
The network was trained on two-dimensional input slices of each patient. During each iteration, one of the 16 slices each from one of the two volumes (the normalized and baseline volume) were used. The volume used was randomly selected and the slice used was based on a random normalized distribution centered on the center slice. Each slice was randomly rotated, flipped, cropped and contrast adjusted. A randomly generated Poisson noise array was added to the slice to simulate the effect of random imaging noise. The slice was then resized to 32×32×3 and used as input to the network.
2.2. CNN architecture
Similar to our previously developed CNN algorithm [6–8], CNN was implemented entirely of 3 × 3 convolutional kernels and linear layers (Fig. 3). The convolutional kernels consisted entirely of 6 residual layers [9], totaling 12 convolutional layers. Feature maps were down-sampled using strided convolutions. Dropout with a 0.5 keep probability and L2 normalization was utilized. Training was implemented by using the Adam optimizer described by Dozat et al. [10–12] which performs parameter wise momentum augmented training. Network weights were initialized randomly.
Fig. 3.
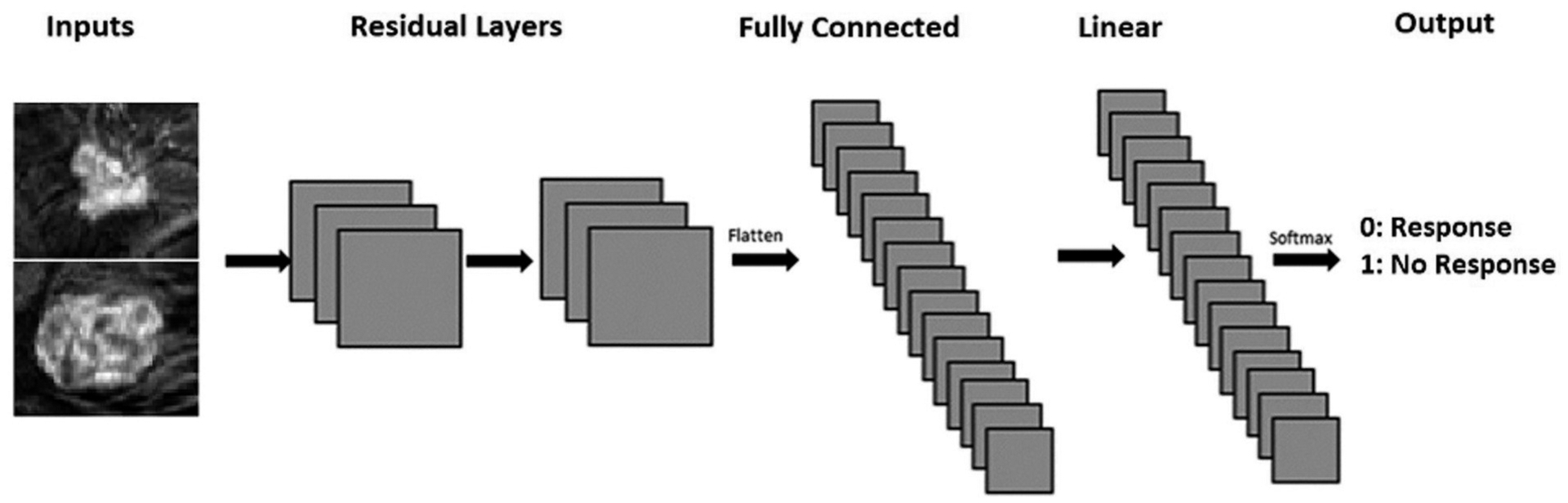
Network architecture: The neural network was custom built and trained with random initializations. It consisted of 12 convolutional layers set up as 6 residual nodes and 2 linear layers before an output layer predicting two classes. Inputs were 32 × 32 pixels.
A 5-fold cross validation was utilized for performance evaluation using 80% (n = 105) for training and 20% (n = 26) for validation. Software code for this study was written in Python using the TensorFlow library v1.13. Experiments and CNN training were performed on a Linux workstation with dual NVIDIA Titan X Pascal GPUs with 12 GB on chip memory, an i7 CPU and 48 GB RAM.
3. Results
The average age of patient was 48.3 years old (SD 9.0). Patients identified themselves as Caucasian (n = 101), African American (20), Asian (6), Native Hawaiian/Pacific Islander (1), American Indian/Alaskan Native (0), Multiple race (1) and N/A (2). Tumor receptor status was: Er + (n = 70), Pr + (55), HR+ (76), Her2+ (34), and triple negative (35).
Of 131 patients, 40 patients achieved pCR following NAC (group 1) and 91 patients did not achieve pCR following NAC (group 2) comprising of the two classification groups in this study (Figs. 1 and 2).
Fig. 1.
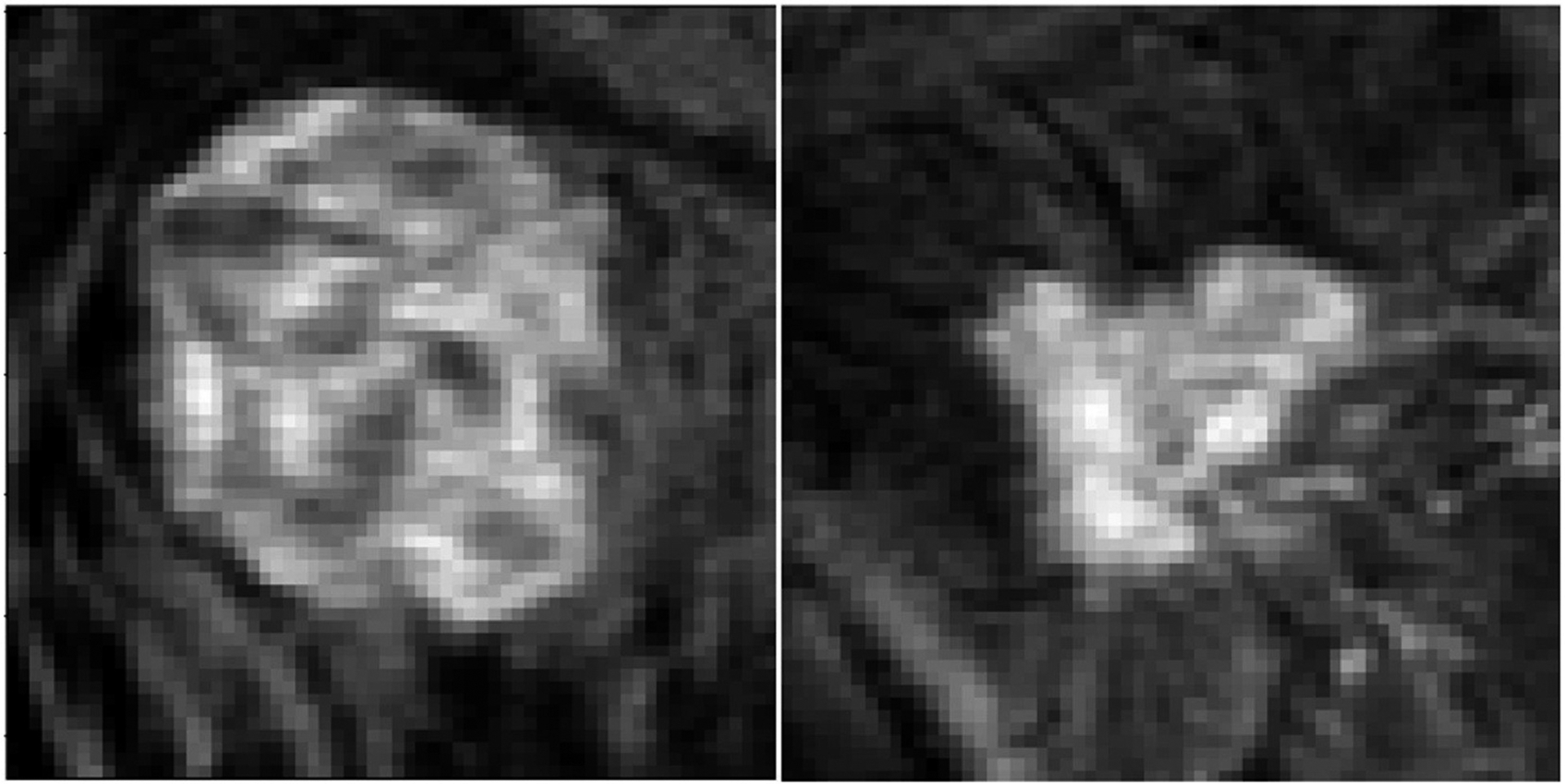
Example images from tumors that responded to neoadjuvant therapy.
Fig. 2.
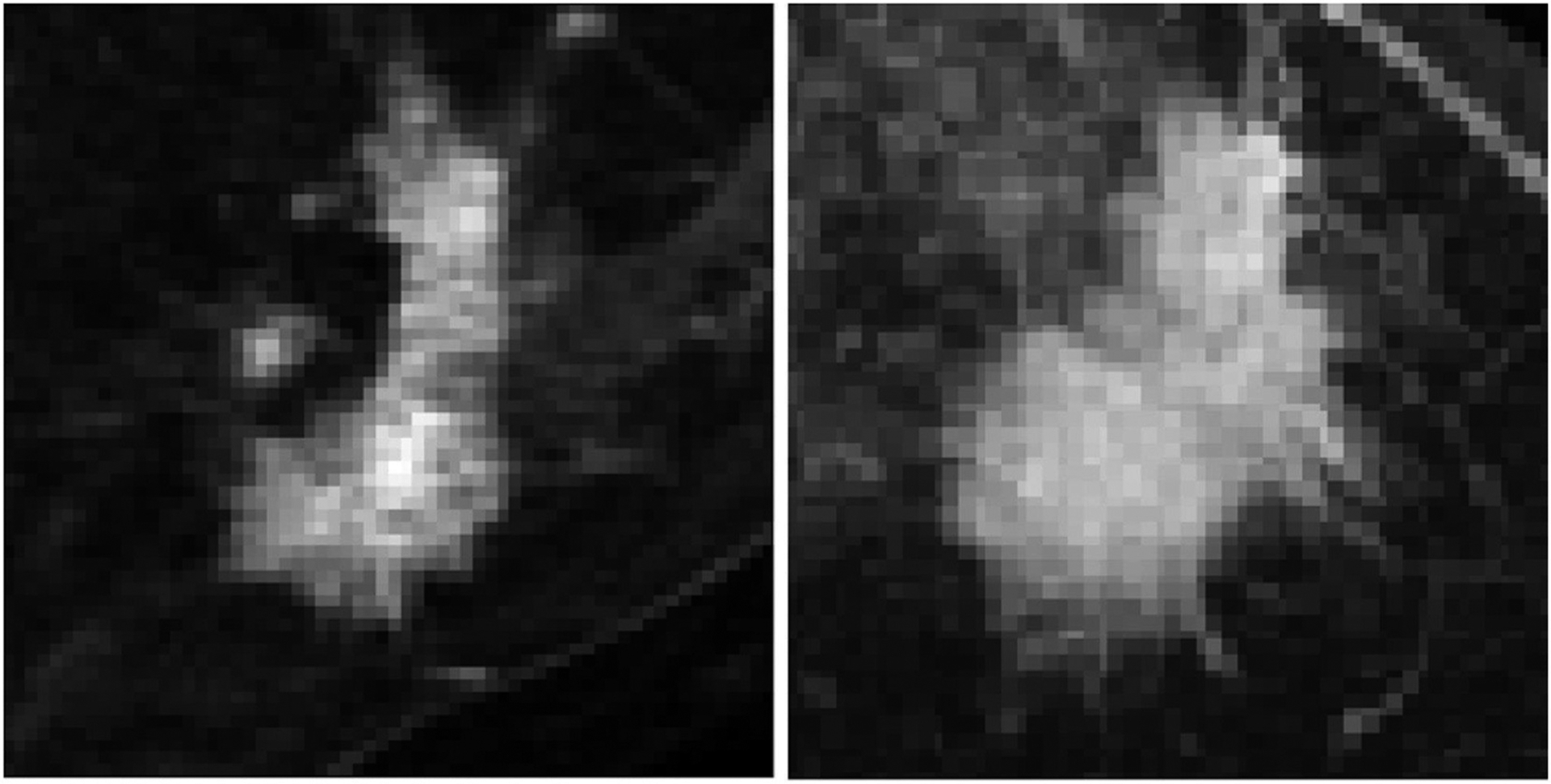
Example images from tumors that did not respond to neoadjuvant therapy.
Diagnostic accuracy of our CNN classification model distinguishing patients with pCR vs non- pCR was 72.5 (SD ± 8.4), with sensitivity 65.5% (SD ± 28.1) and specificity of and 78.9% (SD ± 15.2). The area under a ROC Curve (AUC) was 0.72 (SD ± 0.08).
4. Discussion
Utilizing a publicly available breast MRI dataset from the ISPY-Trial, our CNN algorithm was able to achieve an overall accuracy of 72.5% in predicting patients with pCR following NAC. Our results indicate that it is feasible to use our CNN algorithm on an MRI dataset composed of studies performed from 9 different institutions. There were criteria to keep the MRI protocol consistent among different institutions participating in this trial such as plane of acquisition, temporal resolution timing and spatial resolution parameters [5]. However, it is not possible to keep all the parameters the same across multiple intuitions with inherent variabilities unique to each institution producing some-what heterogeneous set of MRI images. Using this type of dataset from multiple institutions is an important step towards the goal of generalizability of this technology and for further clinical validation [13].
Other groups have also applied advanced imaging techniques prediction of breast cancer response to NAC treatment. Quantitative radiomics extracts data from routine medical imaging and analyzes complex imaging features, unperceivable to the human eye [14]. Radiomics of breast cancer using MRI works on the principle of analyzing various intrinsic features including dynamic contrast enhancement (DCE) kinetics, which often define tumor heterogeneity, to predict NAC response [15,16]. In 2015, Aghaei et al. looked at quantitative kinetic imaging features to predict NAC therapy response from the pre-treatment MRI scan of 68 cancer patients [15]. In the pool of 39 imaging features tested, 10 yielded relatively higher classification performance with the areas under receiver operating characteristic curve (AUC) values ranging from 0.61 to 0.78. A study by Cain et al. used pre-treatment MRI performed in 288 patients to predict response to NAC using a multivariate machine learning-based model [16]. A comprehensive set of 529 radiomic features was extracted from each patient’s pre-treatment MRI and was tested for predictive potential. The AUC values for predicting pCR in a subset of Triple Negative and HER+ patients were significant (0.707, 95% CI 0.582–0.833, p < 0.002).
These previously mentioned studies have shown promising results in breast MRI data analysis. However, these methods are dependent on feature engineering, which require domain knowledge to build feature extractors, which simplify the complex data and create more comprehensible patterns to be applied in algorithms. These methods are limited in their function, as they are dependent on human extraction of crucial features. More recently, a subset of machine learning named CNN that has made great strides in medical imaging analysis. Compared to traditional machine learning, which is dependent on human extracted features, artificial neural networks depend on the curated input data and allow the computer to construct predictive statistical models through increasingly complex layers and self-optimization in an automated way [17]. Using CNN, Ravichandran et al. evaluate 166 breast cancer patients to predict pCR from a pre-treatment MRI tumor dataset [18]. Their CNN algorithm achieved an area under the curve (AUC) of 0.77 and an overall accuracy of 82%. Inclusion of clinical variables improved response prediction with an AUC of 0.85 and an accuracy of 85%. Our group also used a CNN based algorithm in a study of 141 patients to predict whether the patient would achieve a complete pathologic response using a pre-treatment MRI data yielding an overall mean accuracy of 88% in three-class prediction of NAC7.
A CNN algorithm that allows automated extraction of features from the input data are crucial to the defined problem domain using medical images. This process improves its ability to study the input features in an end-to-end manner, using complex, stacked hidden layers to predict a desired output. Therefore, CNN feature extraction is not a variable with each new MRI and thus may perform better than traditional machine approach using feature engineering [17].
Despite these promising results, the use of machine learning and breast MRIs for early prediction of NAC treatment response requires further clinical validation. Published studies that have explored this approach, have been retrospective, single-institutional, and have included relatively small number of patients. Thus, the motivation for this study was to apply CNN algorithm to test the predictive potential of classifying patents with pCR using a database composed of breast MRIs from multiple institutions.
In the 2012 ACRIN 6657 TRIAL by Hylton et al. [5], MRI data from 216 women was analyzed for both pCR and residual cancer burden (RCB). They reported AUC differences between MRI volume and clinical size predictors at 3 time points, respectively, were 0.14, 0.09, and 0.02 for prediction of pCR. AUC for pCR was predicted to be 0.73. They concluded that MRI findings are a stronger predictor of pCR to NACT than clinical assessment, with the greatest advantage observed with the use of volumetric measurement of tumor response early in treatment. Similarly, the study under the ACRIN 6657 TRIAL by Scheel et al. [21] showed data from 138 women to determine the accuracy of preoperative measurements for detecting pathologic complete response (CR) and assessing residual disease after neoadjuvant chemotherapy (NACT) in patients with locally advanced breast cancer. In their study, MRI had the highest accuracy for detecting pathologic CR for all lesions and non-mass enhancement (NME) (AUC = 0.76 and 0.84, respectively). Longest diameter by MRI and longest diameter by clinical examination showed moderate ability for detecting pathologic CR for multiple masses (AUC = 0.78 and 0.74), and longest diameter by MRI and longest diameter by mammography showed moderate ability for detecting pathologic CR for tumors without DCIS (AUC = 0.74 and 0.71). In subjects with residual disease, longest diameter by MRI exhibited the strongest association with pathology size for all lesions and single masses (r = 0.33 and 0.47). They reported that MRI is more accurate than by mammography and clinical examination for preoperative assessment of tumor residua after NACT and may improve surgical planning. Our study using a novel CNN approach yielded similar diagnostic performance of AUC = 0.72 without the need for size measurements which can be prone to subjective bias. In addition, with the larger training data, there is potential for the performance of our CNN algorithm to improve.
A number of large randomized trials have shown that achieving pCR after NAC for locally advanced breast cancer not only aids in predicting patient mortality but also reduces patient morbidity by allowing for less invasive surgery [3,19]. Timely and accurate identification of patients who will respond to NAC based on pre-treatment MRI could substantially improve treatment guidance in the neoadjuvant setting. Ruling out treatment resistance would enable de-escalation of toxic therapeutic measures that have little benefit. There is potential great benefit of a clinical tool such as our CNN algorithm that may be used to accurately predict NAC treatment response in patients.
There are few limitations of our study. Despite being multi-institutional, the dataset was relatively small in size. It has been shown that the performance of a CNN has been shown to increase logarithmically with increasing datasets [20]. In addition, because CNN is a type of artificial neural network and is considered an end-to-end process, as it does not clearly reveal the reasoning behind the final result in a deterministic manner. To better understand the intuition behind the predictions of a neural network is an ongoing area of research. Furthermore, potential of combining clinical information such as molecular subtypes of cancer and the results of our CNN algorithm in order to further improve overall prediction model is under investigation. Lastly, further clinical validation of our CNN algorithm is planned to be performed prospectively with randomization [20].
5. Conclusion
It is feasible to use our CNN algorithm to predict NAC response in patients using the I-SPY TRIAL dataset from multiple institutions. Our CNN algorithm has the potential to impact clinical management in patients with locally advanced breast cancer, including the opportunity to direct appropriate therapy in non-responders, minimize toxicity from ineffective therapies, and facilitate the upfront use of novel targeted treatment in the neoadjuvant setting.
Acknowledgement
We would like to acknowledge and thank World Gold Council for research support.
Footnotes
Work originated from Columbia University Medical Center. No disclosures. No conflict of interest.
References
- [1].Kong X, Moran MS, Zhang N, Haffty B, Yang Q. Meta-analysis confirms achieving pathological complete response after neoadjuvant chemotherapy predicts favourable prognosis for breast cancer patients. Eur J Cancer 2011;47(14):2084–90. [DOI] [PubMed] [Google Scholar]
- [2].Cho JH, Park JM, Park HS, Park S, Kim SI, Park BW. Oncologic safety of breast-conserving surgery compared to mastectomy in patients receiving neoadjuvant chemotherapy for locally advanced breast cancer. J Surg Oncol 2013;108(8):531–6. [DOI] [PubMed] [Google Scholar]
- [3].Cortazar P, Zhang L, Untch M, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014;384(9938):164–72. [DOI] [PubMed] [Google Scholar]
- [4].Gianni L, Pienkowski T, Im YH, et al. 5-year analysis of neoadjuvant pertuzumab and trastuzumab in patients with locally advanced, inflammatory, or early-stage HER2-positive breast cancer (NeoSphere): a multicentre, open-label, phase 2 randomised trial. Lancet Oncol 2016;17(6):791–800. [DOI] [PubMed] [Google Scholar]
- [5].Hylton NM, Blume JD, Bernreuter WK, et al. Locally advanced breast cancer: MR imaging for prediction of response to neoadjuvant chemotherapy–results from ACRIN 6657/I-SPY TRIAL. Radiology. 2012;263(3):663–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ha R, Chang P, Mutasa S, et al. Convolutional neural network using a breast MRI tumor dataset can predict Oncotype dx recurrence score. J Magn Reson Imaging 2019;49(2):518–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ha R, Chin C, Karcich J, et al. Prior to initiation of chemotherapy, can we predict breast tumor response? Deep learning convolutional neural networks approach using a breast MRI tumor dataset. J Digit Imaging 2019;32(5):693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ha R, Mutasa S, Karcich J, et al. Predicting breast Cancer molecular subtype with MRI dataset utilizing convolutional neural network algorithm. J Digit Imaging 2019;32(2):276–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Paper presented at: Proceedings of the IEEE conference on computer vision and pattern recognition. 2016. [Google Scholar]
- [10].Dozat T Incorporating Nesterov momentum into Adam. 2016.
- [11].Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:14126980. 2014. [Google Scholar]
- [12].Nesterov Y Gradient methods for minimizing composite functions. Mathematical Programming 2013;140(1):125–61. [Google Scholar]
- [13].Abramson RG, Li X, Hoyt TL, et al. Early assessment of breast cancer response to neoadjuvant chemotherapy by semi-quantitative analysis of high-temporal resolution DCE-MRI: preliminary results. Magn Reson Imaging 2013;31(9):1457–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kumar V, Gu Y, Basu S, et al. Radiomics: the process and the challenges. Magn Reson Imaging 2012;30(9):1234–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Aghaei F, Tan M, Hollingsworth AB, Qian W, Liu H, Zheng B. Computer-aided breast MR image feature analysis for prediction of tumor response to chemotherapy. Med Phys 2015;42(11):6520–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cain EH, Saha A, Harowicz MR, Marks JR, Marcom PK, Mazurowski MA. Multivariate machine learning models for prediction of pathologic response to neoadjuvant therapy in breast cancer using MRI features: a study using an independent validation set. Breast Cancer Res Treat 2019;173(2):455–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–44. [DOI] [PubMed] [Google Scholar]
- [18].Ravichandran K, Braman N, Janowczyk A, Madabhushi A. A deep learning classifier for prediction of pathological complete response to neoadjuvant chemotherapy from baseline breast DCE-MRI. Vol 10575: SPIE. 2018. [Google Scholar]
- [19].Weis JA, Miga MI, Yankeelov TE. Three-dimensional image-based mechanical modeling for predicting the response of breast Cancer to neoadjuvant therapy. Comput Methods Appl Mech Eng 2017;314:494–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sun C, Shrivastava A, Singh S, Gupta A. Revisiting unreasonable effectiveness of data in deep learning era. Paper presented at: Proceedings of the IEEE international conference on computer vision. 2017. [Google Scholar]
- [21].Scheel JR, Kim E, Partridge SC, et al. MRI, clinical examination, and mammography for preoperative assessment of residual disease and pathologic complete response after neoadjuvant chemotherapy for breast Cancer: ACRIN 6657 trial. AJR Am J Roentgenol 2018;210(6):1376–85. [DOI] [PMC free article] [PubMed] [Google Scholar]


