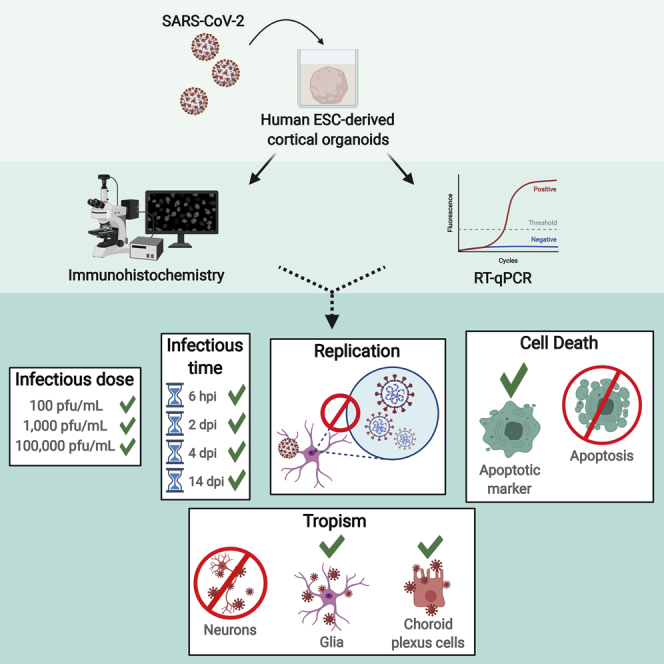
Summary
Coronavirus disease 2019 (COVID-19) patients have manifested a variety of neurological complications, and there is still much to reveal regarding the neurotropism of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Human stem cell-derived brain organoids offer a valuable in vitro approach to study the cellular effects of SARS-CoV-2 on the brain. Here we used human embryonic stem cell-derived cortical organoids to investigate whether SARS-CoV-2 could infect brain tissue in vitro and found that cortical organoids could be infected at low viral titers and within 6 h. Importantly, we show that glial cells and cells of the choroid plexus were preferentially targeted in our model, but not neurons. Interestingly, we also found expression of angiotensin-converting enzyme 2 in SARS-CoV-2 infected cells; however, viral replication and cell death involving DNA fragmentation does not occur. We believe that our model is a tractable platform to study the cellular effects of SARS-CoV-2 infection in brain tissue.
Keywords: SARS-CoV-2, COVID-19, cortical organoid, neurological symptoms, human embryonic stem cells, neurotropism, glial cells
Graphical abstract
Highlights
-
•
SARS-CoV-2 infects human cortical organoids at low viral loads within 6 h
-
•
Replication of SARS-CoV-2 does not occur within 4 days post infection
-
•
SARS-CoV-2 preferentially targets glia and choroid plexus cells in vitro
-
•
Cleaved caspase-3 co-localizes with SARS-CoV-2 in cells at 14 days post infection
In this article, Hsieh and colleagues investigated the neurotropism of the virus responsible for COVID-19, SARS-CoV-2. Using human ESC-derived brain organoids, this study found that SARS-CoV-2 can infect these organoids at low viral loads, but does not replicate within 4 days post infection. Specifically, SARS-CoV-2 preferentially targeted glial cells and cells of the choroid plexus in this model.
Introduction
With the emergence of the novel coronavirus disease 2019 (COVID-19) global pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a wide range of symptoms have been observed in infected patients. Fever, respiratory symptoms, and muscle pain were among the initial indicators of COVID-19, but, more recently, up to one-third of patients have manifested neurological complications (Helms et al., 2020; Mao et al., 2020; Poyiadji et al., 2020; Taherifard and Taherifard, 2020). Neurological symptoms range from milder indicators, such as nausea, headache, loss of smell and/or taste, to the more severe encephalopathy, seizures, stroke, and Guillain-Barré syndrome (Chen et al., 2020b; Helms et al., 2020; Mao et al., 2020; Poyiadji et al., 2020; Sedaghat and Karimi, 2020; Virani et al., 2020). All of these are suggestive of the neurotropism of SARS-CoV-2 (Baig et al., 2020; Conde Cardona et al., 2020). In addition, other coronaviruses have been shown to invade the brain (Arbor et al., 2000; Bleau et al., 2015; Hao et al., 2019; McCray et al., 2007; Zhao et al., 2015) and SARS-CoV-2 RNA has been detected in brain biopsies from fatal cases of COVID-19 (Puelles et al., 2020). The entry receptor for SARS-CoV-2, angiotensin-converting enzyme 2 (ACE2), is expressed throughout the brain (Chen et al., 2020a), although at lower levels compared with lung and other tissues (Li et al., 2020), so it is important to investigate (1) if the brain is susceptible to infection by SARS-CoV-2, (2) if the brain is a viable site of replication, and (3) what the consequential effects of infection are on the brain.
Due to the inaccessibility of human brain tissue for research purposes, 3D in vitro models have proved to be a valuable tool for studying the cellular and molecular effects of a neurological SARS-CoV-2 infection. These models allow for human-specific studies of particular regions and cell types within the brain. Previous studies using brain organoids to investigate SARS-CoV-2 tropism have resulted in conflicting reports regarding infectivity; some have shown that SARS-CoV-2 does not readily infect neurons (Pellegrini et al., 2020), while others suggest that the virus does, but cannot replicate within them (Mesci et al., 2020; Ramani et al., 2020). Another study demonstrated infection in both neurons and neural progenitors with subsequent replication (Ramani et al., 2020; Zhang et al., 2020). SARS-CoV-2 tropism in other cell types of the brain has been investigated, such as glia and cells of the choroid plexus (ChP) (Pellegrini et al., 2020). These studies report infection of the cells of the ChP, but not glial cells (Jacob et al., 2020; Pellegrini et al., 2020). Notably, these studies employed a wide range of viral titers and multiplicity of infection (MOI) (Zhang et al., 2020). Because SARS-CoV-2 tropism in the CNS remains largely unclear, it is imperative to identify the cellular targets of SARS-CoV-2 and the infectious dose at which brain tissue is susceptible.
It is estimated that the infectious dose for SARS-CoV-2 may be as low as a few hundred particles (Beggs, 2020); therefore, we tested organoids for infection using viral titers from 102 to 105 plaque-forming units (PFU)/mL, approximating MOI of 0.00001–0.01. We found infection of cortical organoids at titers as low as 102 PFU/mL. Consistent with studies from Pellegrini et al., (2020) and Jacob et al., (2020), we saw little to no infection of neurons but moderate infection of ChP cells. Interestingly, however, we found that SARS-CoV-2 localized to cells that expressed the glial marker glial fibrillary acidic protein (GFAP) and the astrocytic marker aldehyde dehydrogenase 1 family member L1 (ALDH1L1), as well as cells that co-expressed GFAP and nestin, indicative of radial glia progenitor cells. This study marks the first demonstration of the glial tropism of SARS-CoV-2.
Results
SARS-CoV-2 infects human cortical organoids at low viral titers
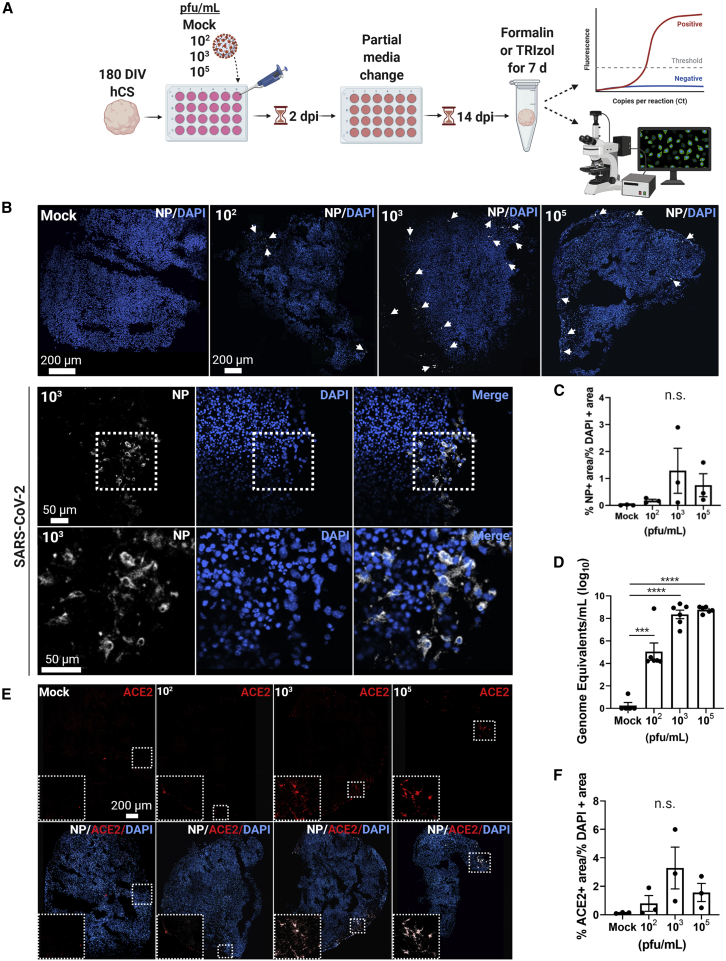
To investigate the ability of SARS-CoV-2 to infect neural cell types, we exposed 180 days in vitro (DIV) human embryonic stem cell (hESC)-derived cortical organoids to SARS-CoV-2 at three viral titers, plus a mock infection group. The organoids were maintained for a total of 14 days, with media changes every 4–5 days, then fixed (Figure 1A). During this period, we monitored the organoids daily with bright field microscopy to look for morphology changes, alterations in media color, and/or external cellular integrity. Infection was first confirmed using immunohistochemistry (IHC) to detect the nucleocapsid protein (NP) of SARS-CoV-2 (Figures 1B and 1C); this protein is the most abundant in coronaviruses and tends to be more immunogenic than other structural proteins (Shi et al., 2003; Tan et al., 2004). Quantitative real-time PCR was also conducted to assess the number of viral particles in each organoid and to determine the level of infection relative to the viral titer (Figure 1D). We found that infection of cortical organoids occurred at titers as low as 102 PFU/mL through positive SARS-CoV-2 NP signal and detection of SARS-CoV-2 genome equivalents (GE) from RT-qPCR (Figures 1B–1D).
Figure 1.
SARS-CoV-2 infects cortical organoids at various viral titers
(A) The experimental schematic.
(B) Representative confocal images from IHC of 180 DIV hESC-derived cortical organoids confirm the presence and localization of SARS-CoV-2 NP (white) in infection groups. Nuclei are stained in DAPI. White arrows indicate regions of NP clusters.
(C) Quantification of viral infection by percentage NP-positive area over percentage DAPI-positive area.
(D) Normalized number of GE/mL show significant differences among groups. Significant statistical differences are indicated among samples (p < 0.05). N = 6 organoids from one independent experiment.
(E) Representative confocal images show ACE2 expression and co-localization to NP.
(F) ACE2 quantification by percentage ACE2-postive area over percentage DAPI-positive area. N = 3 organoids from one independent experiment. Each data point in IHC images represents an average of three sections from three separate organoids.
We then investigated whether SARS-CoV-2 NP co-localized with the putative entry receptor, ACE2 (Figure 1E). IHC data revealed that NP-positive cells also expressed ACE2, and, interestingly, that ACE2 expression appeared to increase with higher viral load (Figures 1E and 1F). Although there was not a significant increase in ACE2, we observed a trend suggesting upregulation. Together, these results indicate that SARS-CoV-2 does infect human brain tissue, and that cortical organoids are a suitable model for evaluating infection.
SARS-CoV-2 does not replicate in cortical organoids within 4 days post infection
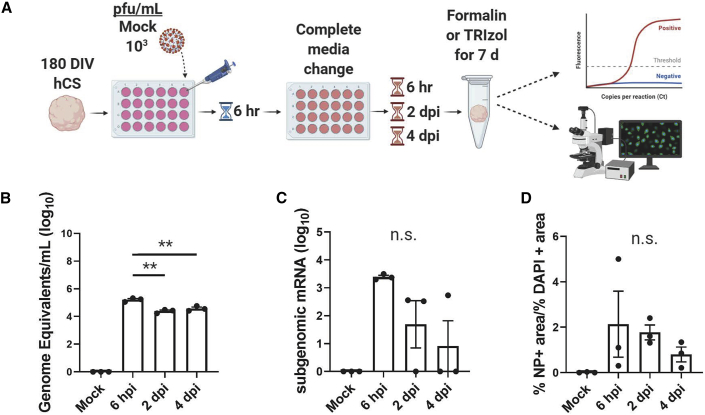
While previous experiments with 102 PFU/mL showed infection, we performed subsequent experiments with 103 PFU/mL to increase the number of infected cells for downstream analysis. We conducted a time-course assay to determine whether infection of brain tissue could occur within a shorter and more physiologically relevant time span (Figure 2A). One-hundred and eighty DIV hESC-derived cortical organoids were exposed to 103 PFU/mL of SARS-CoV-2 for 6 h. A complete media change was conducted to remove any suspended viral particles. After fixation, IHC and RT-qPCR confirmed detection of SARS-CoV-2 in the organoids 6 h post infection (hpi) compared with a mock infection control (Figure 2).
Figure 2.
SARS-CoV-2 does not replicate in cortical organoids within 4 dpi
(A) Time course experimental schematic.
(B) RT-qPCR of organoids shows that viral load decreases after 6 hpi. Significant statistical differences are indicated among samples (p < 0.05). N = 3 organoids from one independent experiment.
(C) Subgenomic mRNA levels show decreasing viral transcription after 6 hpi (p < 0.05). N = 3 organoids from one independent experiment.
(D) Quantification of percentage NP-positive area over percentage DAPI-positive area from IHC shows a decrease in NP detection. N = 3 organoids from one independent experiment.
To determine if viral replication was occurring, we continued incubation of the remaining organoids for either 2 or 4 days post infection (dpi), then harvested them for analysis. RT-qPCR data shows that viral RNA levels slightly decreased at 2 dpi and plateaued at 4 dpi compared with 6 hpi, suggesting that replication was not occurring. To confirm this, we conducted RT-qPCR to detect subgenomic viral mRNA. Subgenomic mRNA is not contained within virions but is transcribed only in infected cells, indicating active viral replication (Wölfel et al., 2020); therefore, we compared subgenomic mRNA levels with the genomic RNA levels of each group. The levels of subgenomic mRNA decreased after 6 hpi, indicating that viral transcription can occur early after infection but slows down by 2 dpi (Figure 2C) as viral load plateaus (Figure 2B). In addition, the level of NP detection decreased after 6 hpi (Figure 2D). Together, these results suggest that, while SARS-CoV-2 can infect cortical organoids, SARS-CoV-2 replication does not occur within 4 dpi.
Glial cells and ChP cells are targeted by SARS-CoV-2
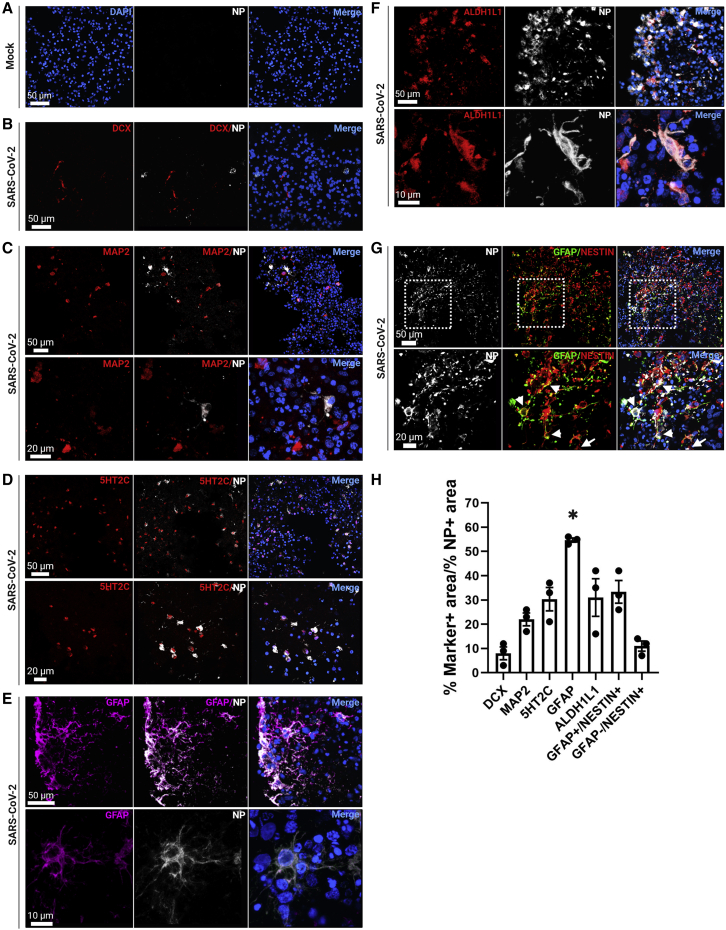
We then identified which cell types within our organoids were susceptible to SARS-CoV-2 infection. First, we investigated whether neurons were vulnerable to infection using doublecortin (DCX) as a marker for immature neurons and MAP2 as a marker for mature neurons. We found that SARS-CoV-2 only minimally infected neurons in cortical organoids, with little to no co-localization of DCX with NP, and low co-localization of MAP2 with NP (Figures 3B, 3C, and 3H). Since previous literature has found significant infection in ChP cells (Jacob et al., 2020; Pellegrini et al., 2020), we next determined the level of ChP infection using the marker 5-HT2C. We observed modest co-localization with NP, consistent with other studies (Figures 3D and 3H).
Figure 3.
SARS-CoV-2 targets glial cells and ChP cells in cortical organoids
(A) Representative confocal images of mock infected cortical organoids.
(B) Images show little to no co-localization of DCX and NP within cortical organoids.
(C) MAP2 modestly co-localizes with NP.
(D) 5-HT2C showed moderate co-localization with NP.
(E) NP highly co-localized with GFAP.
(F) Moderate co-localization of ALDH1L1 with NP.
(G) Little to no NP co-localization is seen in GFAP-negative nestin-positive cells (arrows), but there is moderate co-localization with cells that co-express GFAP and nestin (arrowheads).
(H) Quantification shows low NP co-localization with neuronal markers DCX, MAP2, and GFAP-negative, nestin-positive cells. NP and GFAP co-localization is significant, and there is moderate co-localization with ChP marker 5-HT2C, astrocytic marker ALDH1L1, and radial glial progenitor markers co-expressed nestin and GFAP. Significant statistical differences are indicated among samples (p < 0.05). N = 3 sections from three organoids from two independent experiments.
We then used GFAP as a marker for glial cells. SARS-CoV-2 NP co-localized with GFAP-expressing cells at significant levels (Figures 3E and 3H). To classify the infected glial cells, we first evaluated the expression level of an astrocytic marker, ALDH1L1, and found moderate co-localization with NP (Figures 3F and 3H). To determine the identity of non-astrocytic GFAP-expressing cells, we then looked at co-expression of GFAP with nestin to mark radial glia progenitor cells. We found that NP also co-localized moderately with cells expressing both GFAP and nestin (Figures 3G and 3H). Our IHC data also showed nestin-positive cells that were GFAP negative, which is indicative of neural progenitors (Figure 3F). Quantification of NP in these cells revealed that there was also very little to no infection of neural progenitors (Figure 3H). These data suggest that SARS-CoV-2 selectively infects glial cells, including astrocytes and radial glial progenitors, and ChP cells, but only modestly infects neurons and neural progenitors.
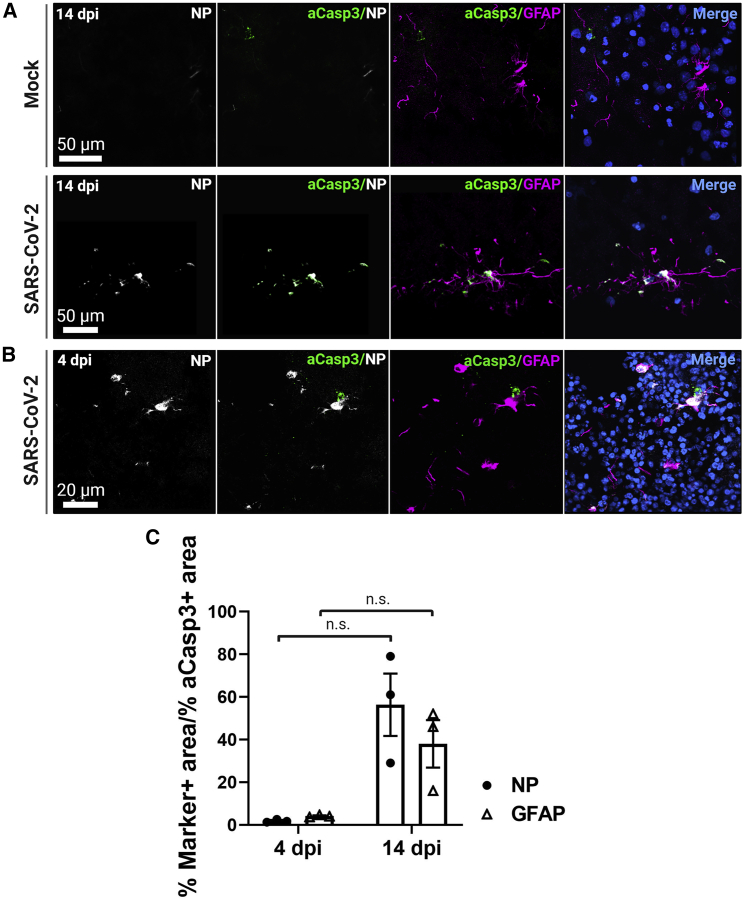
Apoptosis marker cleaved caspase-3 co-localizes with SARS-CoV-2 NP at 14 dpi
We next sought to determine the cellular effects of a SARS-CoV-2 infection. We used the marker cleaved caspase-3 (Asp175) (aCasp3) to indicate apoptotic activity in infected cells. IHC of aCasp3 expression shows that cells demonstrating viral infection also expressed this marker at 14 dpi, compared with the mock infection group (Figure 4A). In addition, expression of aCasp3 co-localized with GFAP expression at 14 dpi (Figures 4A and 4C). We also evaluated aCasp3 expression at 4 dpi and saw little to no co-localization with either NP or GFAP expression (Figures 4B and 4C). Interestingly, no DNA fragmentation was detected at either time point. These results suggest that SARS-CoV-2-infected cells may express markers of apoptosis but may not be undergoing cell death as characterized by DNA fragmentation.
Figure 4.
Apoptosis marker cleaved caspase-3 co-localizes with NP at 14 dpi
(A) Representative confocal images of infected cells show co-localization of NP with cleaved caspase-3 (Asp175) (aCasp3) 14 dpi. In addition, aCasp3 co-localizes with GFAP. DNA fragmentation is not seen.
(B) There is low co-localization of aCasp3 with NP or GFAP 4 dpi.
(C) Quantification of aCasp3 co-localization with NP or GFAP at 4 and 14 dpi. Each point represents the mean of three sections from three organoids. Significant statistical differences are indicated among samples (p < 0.05). N = 3 organoids from two independent experiments.
Discussion
Using hESC-derived cortical organoids allowed us to examine cellular effects of in vitro brain tissue infection by SARS-CoV-2. With NP staining and qPCR, we showed that SARS-CoV-2 can infect cortical organoids at titers as low as 102 PFU/mL (~MOI = 0.00001), although replication does not occur within 4 dpi. We also confirmed that the virus can infect these organoids within 6 h of initial exposure. Previous reports of SARS-CoV-2 infection in brain organoids have been contradictory, and the neurotropism is unresolved. Here, we reveal a previously unreported preference of SARS-CoV-2 to glial cells within our organoids, specifically astrocytes and radial glial progenitor cells. Astrocytes are important for neuron survival and maintenance of the blood-brain barrier (Abbott, 2002; Abbott et al., 2006; Domingues et al., 2016), and preferential targeting of astrocytes by SARS-CoV-2 may offer an additional entry route into the brain. Other studies have reported only modest infection of glial cells in their models but have found that SARS-CoV-2 selectively targets cells of the ChP (Jacob et al., 2020; Pellegrini et al., 2020). In our study, we found moderate infection of ChP cells, similar to the level of glial cell infection.
Furthermore, we investigated the co-localization of NP to the putative entry receptor for SARS-CoV-2, ACE2. We found that NP highly co-localized with ACE2, and, interestingly, we observed a trend of increasing ACE2 expression with increased viral load. The lack of statistical significance may be a result of the limited number of organoids used in this study. Future studies involving more organoids would be interesting but this is beyond the scope of the current work. Other studies have found that ACE2 is upregulated after infection, particularly in tissues where ACE2 normally has low expression (Chua et al., 2020; Garvin et al., 2020; Zamorano Cuervo and Grandvaux, 2020).
Our protocols differed from previous studies in several ways that may account for the discrepancy between results. We utilized a protocol adapted from Birey et al. (2017) to generate cortical organoids, whereas organoids in other studies represented different regions of the brain. Secondly, GFAP-expressing cells constitute ~20% of the organoids after 180 DIV, but this percentage is much lower at earlier time points (Paşca et al., 2015). Since the organoids used in our study were 180 DIV, this may explain why we have seen more astrocyte infection than other studies that used younger organoids. In addition, our organoids were derived from embryonic stem cells rather than induced pluripotent stem cells, which may also contribute to variability between studies (Jacob et al., 2020; Mesci et al., 2020; Ramani et al., 2020; Zhang et al., 2020). Our finding that SARS-CoV-2 does not have tropism for neurons but does infect cells of the ChP is consistent with other studies, however (Jacob et al., 2020; Pellegrini et al., 2020).
We also found co-expression of aCasp3 with SARS-CoV-2 NP in cells, suggestive of increased cleaved caspase-3 activity 14 dpi but not 4 dpi. Furthermore, these cells did not display DNA fragmentation. Further studies using a higher number of organoids is still needed to determine significance. Previous studies have found that activation of caspase-3 can be associated with growth stimulation under certain conditions (Huang et al., 2011; Liu et al., 2015). Thus, the lack of DNA fragmentation may indicate that infected cells are not undergoing cell death, but rather that SARS-CoV-2 may have the potential to induce proliferation of infected cells. Further investigation is necessary to uncover the role of activated caspase-3 in infected cells, and the extent and mechanisms of viral infection-induced cell death in cortical organoids.
Infection of glia by SARS-CoV-2 marks a significant peril, because glia are essential for normal brain function. One neurological manifestation of COVID-19 is the demyelinating form of Guillain-Barré syndrome (Domingues et al., 2016; Hadden et al., 1998; Helms et al., 2020), which is largely associated with astrocyte dysfunction (Domingues et al., 2016; Sher et al., 2019). In all, our study suggests a mechanism for neurological complications in COVID-19 patients: that SARS-CoV-2 preferentially targets glial cells in a way that does not result in the cells' death.
Experimental procedures
SARS-CoV-2 infection
To infect organoids, we used viral titers rather than MOI due to the variability in organoid size and cell count. Each 180 DIV hESC-derived organoid consisted of an average of 10,000,000 cells, according to our cell count, which approximates an MOI of 0.00001–0.01 when using 102 to 105 PFU/mL of SARS-CoV-2 per organoid. The optimal number of viral particles used to infect cortical organoids was determined by comparing different plaque-forming units of virus and was stopped at 103 PFU/mL based on data from the first experiment. The number of viral particles used to infect cortical organoids was 103 PFU/mL for all experiments thereafter. Cortical organoids were maintained in a 24-well plate at 37°C with 5% CO2. Each well contained one organoid and 1 mL of neural medium. To exclude effects not induced by SARS-CoV-2, the control organoids were treated with only neural medium (mock infection). For the time point assays, the medium was removed from the organoids at 6 hpi and replaced with fresh neural medium. The organoids were then harvested at their respective time points.
Statistical analysis
All data are shown as mean ± SEM. Differences were considered statistically significant when p < 0.05 and are signified by ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001, and not statistically significant when p > 0.05, signified by n.s. In all cases, the stated n value represents individual organoids. No statistical methods were used to pre-determine sample sizes. For IHC, compared samples were processed in parallel, and the same settings and laser power were used for confocal microscopy. To compare the mean ± SEM values from the mock infected control and infected organoids, an unpaired two-tailed Student's t test was used with Welch's correction when the F-test indicated significant differences between the variances of both groups. All analyses were conducted using GraphPad Prism software (version 8.3.0).
Quantitative analysis of immunostained sections
Quantification was conducted using Leica LAX software and ImageJ Fiji (NIH). To analyze the amount of marker expression in cortical organoids, we used ImageJ Fiji software to trace and calculate the total DAPI-positive area in three immunostained sections per organoid, then determined the percentage of each marker-positive area out of the total DAPI-positive area. We used ImageJ Fiji software to calculate co-localization by quantifying percentage marker-positive area over percentage NP-positive area or percentage cleaved caspase-3-positive area. The number of organoids used for marker quantification is included in the figure legends.
Author contributions
C.M. contributed to the concept, design, experimentation, analysis of data, and wrote the manuscript. H.S. contributed to experimental design, analysis, and technical expertise. M.G. collected data for RT-qPCR and analysis. R.C. contributed to the experimental design, manuscript review, and provided resources and financial support. J.H. helped design and conceptualize experiments, helped write and edit the manuscript, and provided financial support.
Acknowledgments
We thank Jimena Caballero and Carmen Bartley for technical assistance and Aline McKenzie for manuscript editing. Some figures were created with BioRender.com. This work was supported in part by the Stem Cell Core at the University of Texas at San Antonio and by grants from the NIH (R01NS093992, R01NS113516, R01NS089770, and R21AG066496 (to J.H.) and the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation and the Semmes Foundation (to J.H.).
Published: May 11, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.01.016.
Supplemental information
References
- Abbott N.J. Astrocyte-endothelial interactions and blood-brain barrier permeability. J. Anat. 2002;200:629–638. doi: 10.1046/j.1469-7580.2002.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott N.J., Rönnbäck L., Hansson E. Astrocyte-endothelial interactions at the blood-brain barrier. Nat. Rev. Neurosci. 2006;7:41–53. doi: 10.1038/nrn1824. [DOI] [PubMed] [Google Scholar]
- Arbour N., Day R., Newcombe J., Talbot P.J. Neuroinvasion by human respiratory coronaviruses. J. Virol. 2000;74:8913–8921. doi: 10.1128/jvi.74.19.8913-8921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baig A.M., Khaleeq A., Ali U., Syeda H. Evidence of the COVID-19 virus targeting the CNS: tissue distribution, host-virus interaction, and proposed neurotropic mechanisms. ACS Chem. Neurosci. 2020;11:995–998. doi: 10.1021/acschemneuro.0c00122. [DOI] [PubMed] [Google Scholar]
- Beggs C.B. Is there an airborne component to the transmission of COVID-19?: a quantitative analysis study. medRxiv. 2020 [Google Scholar]
- Birey F., Andersen J., Makinson C.D., Islam S., Wei W., Huber N., Fan H.C., Metzler K.R.C., Panagiotakos G., Thom N. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545:54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleau C., Filliol A., Samson M., Lamontagne L. Brain invasion by mouse hepatitis virus depends on impairment of tight junctions and beta interferon production in brain microvascular endothelial cells. J. Virol. 2015;89:9896–9908. doi: 10.1128/JVI.01501-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Wang K., Yu J., Chen Z., Wen C., Xu Z. The spatial and cell-type distribution of SARS-CoV-2 receptor ACE2 in human and mouse brain. bioRxiv. 2020 doi: 10.3389/fneur.2020.573095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Wu D., Chen H., Yan W., Yang D., Chen G., Ma K., Xu D., Yu H., Wang H. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:m1091. doi: 10.1136/bmj.m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F., Debnath O., Thürmann L., Kurth F., Kazmierski J. Cross-talk between the airway epithelium and activated immune cells defines severity in COVID-19. medRxiv. 2020 doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- Conde Cardona G., Quintana Pájaro L.D., Quintero Marzola I.D., Ramos Villegas Y., Moscote Salazar L.R. Neurotropism of SARS-CoV 2: mechanisms and manifestations. J. Neurol. Sci. 2020;412:116824. doi: 10.1016/j.jns.2020.116824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues H.S., Portugal C.C., Socodato R., Relvas J.B. Oligodendrocyte, astrocyte, and microglia crosstalk in myelin development, damage, and repair. Front. Cell Dev. Biol. 2016;4:71. doi: 10.3389/fcell.2016.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin M.R., Alvarez C., Miller J.I., Prates E.T., Walker A.M., Amos B.K., Mast A.E., Justice A., Aronow B., Jacobson D. A mechanistic model and therapeutic interventions for COVID-19 involving a RAS-mediated bradykinin storm. eLife. 2020;9:e59177. doi: 10.7554/eLife.59177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadden R.D., Cornblath D.R., Hughes R.A., Zielasek J., Hartung H.P., Toyka K.V., Swan A.V. Electrophysiological classification of Guillain-Barré syndrome: clinical associations and outcome. Plasma exchange/sandoglobulin Guillain-barré syndrome trial group. Ann. Neurol. 1998;44:780–788. doi: 10.1002/ana.410440512. [DOI] [PubMed] [Google Scholar]
- Hao X.Y., Lv Q., Li F.D., Xu Y.F., Gao H. The characteristics of hDPP4 transgenic mice subjected to aerosol MERS coronavirus infection via an animal nose-only exposure device. Anim. Model. Exp. Med. 2019;2:269–281. doi: 10.1002/ame2.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms J., Kremer S., Merdji H., Clere-Jehl R., Schenck M., Kummerlen C., Collange O., Boulay C., Fafi-Kremer S., Ohana M. Neurologic features in severe SARS-CoV-2 infection. N. Engl. J. Med. 2020;382:2268–2270. doi: 10.1056/NEJMc2008597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Li F., Liu X., Li W., Shi W., Liu F.F., O'Sullivan B., He Z., Peng Y., Tan A.C. Caspase 3-mediated stimulation of tumor cell repopulation during cancer radiotherapy. Nat. Med. 2011;17:860–866. doi: 10.1038/nm.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F., Pather S.R., Huang W.K., Zhang F., Wong S.Z.H., Zhou H., Cubitt B., Fan W., Chen C.Z., Xu M. Human pluripotent stem cell-derived neural cells and brain organoids reveal SARS-CoV-2 neurotropism predominates in choroid plexus epithelium. Cell Stem Cell. 2020;27:937–950.e9. doi: 10.1016/j.stem.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.Y., Li L., Zhang Y., Wang X.S. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect. Dis. Poverty. 2020;9:45. doi: 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., He Y., Li F., Huang Q., Kato T.A., Hall R.P., Li C.Y. Caspase-3 promotes genetic instability and carcinogenesis. Mol. Cell. 2015;58:284–296. doi: 10.1016/j.molcel.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L., Jin H., Wang M., Hu Y., Chen S., He Q., Chang J., Hong C., Zhou Y., Wang D. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 2020;77:1–9. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray P.B., Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesci P., Macia A., Saleh A., Martin-Sancho L., Yin X., Snethlage C., Avansini S., Chanda S.K., Muotri A. Sofosbuvir protects human brain organoids against SARS-CoV-2. bioRxiv. 2020 [Google Scholar]
- Paşca A.M., Sloan S.A., Clarke L.E., Tian Y., Makinson C.D., Huber N., Kim C.H., Park J.Y., O'Rourke N.A., Nguyen K.D. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L., Albecka A., Mallery D.L., Kellner M.J., Paul D., Carter A.P., James L.C., Lancaster M.A. SARS-CoV-2 infects the brain choroid plexus and disrupts the blood-CSF barrier in human brain organoids. Cell Stem Cell. 2020;27:951–961.e5. doi: 10.1016/j.stem.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyiadji N., Shahin G., Noujaim D., Stone M., Patel S., Griffith B. COVID-19-associated acute hemorrhagic necrotizing encephalopathy: imaging features. Radiology. 2020;296:E119–E120. doi: 10.1148/radiol.2020201187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles V.G., Lütgehetmann M., Lindenmeyer M.T., Sperhake J.P., Wong M.N., Allweiss L., Chilla S., Heinemann A., Wanner N., Liu S. Multiorgan and renal tropism of SARS-CoV-2. N. Engl. J. Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani A., Müller L., Ostermann P.N., Gabriel E., Abida-Islam P., Müller-Schiffmann A., Mariappan A., Goureau O., Gruell H., Walker A. SARS-CoV-2 targets cortical neurons of 3D human brain organoids and shows neurodegeneration-like effects. bioRxiv. 2020 doi: 10.15252/embj.2020106230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedaghat Z., Karimi N. Guillain Barre syndrome associated with COVID-19 infection: a case report. J. Clin. Neurosci. 2020;76:233–235. doi: 10.1016/j.jocn.2020.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher A.A., Glover K.K.M., Coombs K.M. Zika virus infection disrupts astrocytic proteins involved in synapse control and axon guidance. Front. Microbiol. 2019;10:596. doi: 10.3389/fmicb.2019.00596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Yi Y., Li P., Kuang T., Li L., Dong M., Ma Q., Cao C. Diagnosis of severe acute respiratory syndrome (SARS) by detection of SARS coronavirus nucleocapsid antibodies in an antigen-capturing enzyme-linked immunosorbent assay. J. Clin. Microbiol. 2003;41:5781–5782. doi: 10.1128/JCM.41.12.5781-5782.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taherifard E., Taherifard E. Neurological complications of COVID-19: a systematic review. Neurol. Res. 2020:1–8. doi: 10.1080/01616412.2020.1796405. [DOI] [PubMed] [Google Scholar]
- Tan Y.J., Goh P.Y., Fielding B.C., Shen S., Chou C.F., Fu J.L., Leong H.N., Leo Y.S., Ooi E.E., Ling A.E. Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin. Diagn. Lab. Immunol. 2004;11:362–371. doi: 10.1128/CDLI.11.2.362-371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virani A., Rabold E., Hanson T., Haag A., Elrufay R., Cheema T., Balaan M., Bhanot N. Guillain-Barré syndrome associated with SARS-CoV-2 infection. IDCases. 2020;20:e00771. doi: 10.1016/j.idcr.2020.e00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Zamorano Cuervo N., Grandvaux N. ACE2: evidence of role as entry receptor for SARS-CoV-2 and implications in comorbidities. eLife. 2020;9:e61390. doi: 10.7554/eLife.61390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.Z., Chu H., Han S., Shuai H., Deng J., Hu Y.F., Gong H.R., Lee A.C., Zou Z., Yau T. SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res. 2020:1–4. doi: 10.1038/s41422-020-0390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G., Jiang Y., Qiu H., Gao T., Zeng Y., Guo Y., Yu H., Li J., Kou Z., Du L. Multi-organ damage in human dipeptidyl peptidase 4 transgenic mice infected with Middle East respiratory syndrome-coronavirus. PLoS One. 2015;10:e0145561. doi: 10.1371/journal.pone.0145561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.