Summary
SARS-CoV-2 infection is associated with lower blood oxygen levels, even in patients without hypoxia requiring hospitalization. This discordance illustrates the need for a more unifying explanation as to whether SARS-CoV-2 directly or indirectly affects erythropoiesis. Here, we show significantly enriched CD71+ erythroid precursors/progenitors in the blood circulation of COVID-19 patients. We found that these cells have distinctive immunosuppressive properties. In agreement, we observed a strong negative correlation between the frequency of these cells with T and B cell proportions in COVID-19 patients. The expansion of these CD71+ erythroid precursors/progenitors was negatively correlated with the hemoglobin levels. A subpopulation of abundant erythroid cells, CD45+ CD71+ cells, co-express ACE2, TMPRSS2, CD147, and CD26, and these can be infected with SARS-CoV-2. In turn, pre-treatment of erythroid cells with dexamethasone significantly diminished ACE2/TMPRSS2 expression and subsequently reduced their infectivity with SARS-CoV-2. This provides a novel insight into the impact of SARS-CoV-2 on erythropoiesis and hypoxia seen in COVID-19 patients.
Keywords: COVID-19, CD71+ erythroid cells, RBCs, SARS-CoV-2, dexamethasone, Erythroid precursors/progenitors
Highlights
-
•
Expansion of erythroid precursors and progenitors in COVID-19 infection
-
•
Erythroid precursors/progenitors suppress adaptive immune responses
-
•
Erythroid precursors/progenitors get infected with SARS-CoV-2
-
•
Dexamethasone treatment reduces the infectivity of these cells with SARS-CoV2
In this article Elahi and colleagues show that COVID-19 infection is associated with significant expansion of erythroid precursors/progenitors in the blood of patients. These cells suppress patients' immune response against SARS-CoV-2 infection. Also, they express ACE2 and TMPRSS2 and get infected with SARS-CoV-2. In turn, pre-treatment with dexamethasone significantly diminishes ACE2/TMPRSS2 expression and subsequently reduces their infectivity with SARS-CoV-2.
Introduction
SARS-CoV-2 infection manifests as a spectrum from asymptomatic or mild symptoms to moderate and severe disease (Guan et al., 2020). A subgroup will become critically ill and develop acute respiratory distress syndrome, a clinical phenomenon characterized by the development of bilateral infiltrates and hypoxemia (Thompson, 2017), often accompanied with septic shock and organ failure (Xu et al., 2020; Zhou et al., 2020).
The pathogenesis of SARS-CoV2 is being delineated rapidly; however, the causes of hypoxia have remained elusive. It has been shown that SARS-CoV-2 infection is initiated by the viral surface spike glycoprotein (S protein) (Walls et al., 2020) binding to the angiotensin-converting enzyme 2 (ACE2) for cell entry (Hoffmann et al., 2020). Subsequently, the S protein gets cleaved by the transmembrane protease serine 2 (TMPRSS2) (Hoffmann et al., 2020). It appears that SARS-CoV-2 not only gains initial entry through ACE2 but also downregulates cell surface ACE2 expression such that this enzyme cannot exert its protective role (Kuba et al., 2005). Downregulation of ACE2 in the respiratory tract is linked to neutrophil infiltration in response to liposaccharide (Sodhi et al., 2018) and may result in angiotensin II accumulation and lung injury, as has been reported in animal models of respiratory virus infections (Gu et al., 2016; Yang et al., 2014). In addition to the respiratory tract, ACE2 expression has been reported in intestinal epithelial cells, endothelial cells, renal tubules, cerebral neurons, and possibly immune cells (e.g., alveolar monocytes/macrophages) (Lukassen et al., 2020). Reduced numbers of T, B, and natural killer cells in the peripheral blood of COVID-19 patients has been reported, especially in those with severe disease (Cao, 2020; Huang et al., 2020; Xu et al., 2020). Despite increased levels of granulocyte macrophage colony-stimulating factor, a decline in the proportion of monocytes, eosinophils, and basophils has been reported (Cao, 2020). In contrast to what occurs in peripheral blood, higher neutrophil recruitment in the lungs has been associated with disease severity (Cao, 2020). Despite the frequency of hypoxia, the impact of SARS-CoV-2 infection on erythropoiesis has received limited attention. Preliminary modeling reports have suggested that SARS-CoV-2 may inhibit heme metabolism and induce hemoglobin denaturation (Cavezzi et al., 2020). As such, hemoglobin alteration may compromise oxygen-carrying capacity of red blood cells (RBCs) in COVID-19 patients resulting in hypoxia. The entry receptor, ACE2, has been confirmed in RBCs by proteomic studies (D'Alessandro et al., 2017). This suggests that RBCs might be targeted by the SARS-CoV-2 virus, although they cannot support viral replication. RBCs can be directly invaded by pathogens (e.g., in malaria), resulting in hemolysis (McCullough, 2014). In support of this concept, structural protein damage and changes in RBC membrane lipids have been reported in COVID-19 patients (Thomas et al., 2020). In addition to ACE2, SARS-CoV-2 invades host cells via CD147 (Wang et al., 2020), a known RBC receptor for Plasmodium falciparum (Crosnier et al., 2011). Finally, CD26 was reported to interact with the SARS-CoV-2 spike (Li et al., 2020), which is involved in stress hematopoiesis (Broxmeyer et al., 2012). In light of the above, it is possible that SARS-CoV-2 directly or indirectly invades RBCs. Hence, depletion of RBCs by SARS-CoV-2 may result in stress erythropoiesis as a compensatory mechanism to meet the oxygen supply, resulting in the abundance of erythroid precursors in the blood.
Erythroid precursors are defined as CD71+ erythroid cells (CECs) co-expressing CD71 (the transferrin receptor) and CD235a (glycophorin A, erythroid lineage marker) in humans, and CD71 and TER119 in mice (Elahi, 2014, 2019; Elahi et al., 2013). CECs are a heterogeneous population of erythroid progenitors and precursors with a wide range of immunosuppressive and/or immunomodulatory properties (Elahi and Mashhouri, 2020). We and others have reported that CECs compromise innate and adaptive immune responses against infections and tumors due to their immunosuppressive properties (Dunsmore et al., 2017; Elahi et al., 2013; Namdar et al., 2017; Shahbaz et al., 2018; Yang et al., 2020; Zhao et al., 2018). In addition, it has been shown that CECs can harbor infective HIV particles and that the binding of HIV to CD235a mediates HIV trans-infection to CD4+ T cells (Namdar et al., 2019). However, whether these cells can be the target of SARS-CoV-2 remains to be explored.
In light of the above, we investigated the frequency and functionality of CECs in different groups of COVID-19 patients.
Results
Study population
Among 128 patients included in this study, 40 were critically ill patients (age 76.2 ± 13.2 years) admitted to the intensive care unit (ICU), whom we defined as having severe disease. Forty-eight individuals (age 62.3 ± 17.5) were hospitalized on a hospital ward >5 days with moderate disease and the remaining 40 patients had mild disease requiring less than 5 days in hospital (age 60 ± 15.4). ICU patients were older and 72.5% male (29/40) while non-ICU patients were 62.5% male (55/88). The mean age average for men and women were (68.6 ± 15.5) and (62.8 ± 21.4), respectively. Patient age ranged from 17 to 95 years. Fifteen healthy individuals were recruited as negative controls (age 48 ± 14.2).
COVID-19 infection results in the expansion of CECs in the peripheral blood
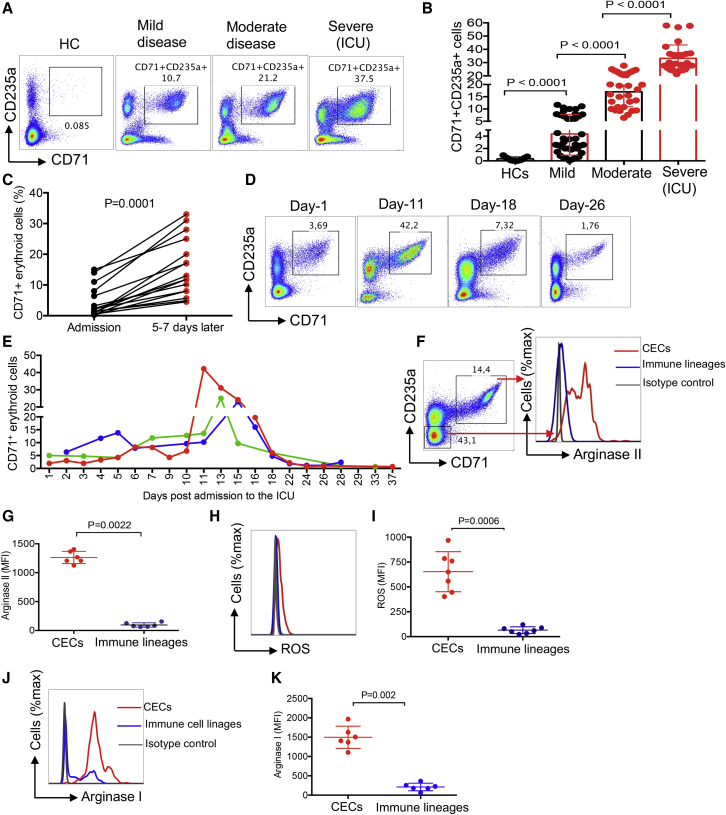
In this study, we observed that COVID-19 infection results in the expansion of CECs in the peripheral blood of patients compared with healthy controls (HCs). Of note, patients with severe COVID-19 disease had significantly higher percentages of CECs in peripheral blood compared with those with a moderate or mild disease (Figures 1A, 1B, and S1A) and CECs were very low or absent in HCs (Figures 1A and 1B). As the disease progressed over time, the CECs expanded in peripheral blood (Figure 1C). We monitored the frequency of CECs over the entire disease course in three patients admitted to hospital. As shown in Figures 1D and 1E, CECs expanded gradually after hospitalization but increased rapidly with the development of critical illness and eventually declined as patients no longer had detectable virus. The erythroid nature of these expanded cells in COVID-19 patients was confirmed by the elevated expression level of the erythroid transcriptional factor, GATA1 gene (Figure S1B). We also quantified the mRNA level of 5-aminolevulinate synthase-2 as a key downstream target of GATA1 (Zhang et al., 2017) and a wide range of associated enzymes with heme biosynthesis in CECs, including hydroxymethylbilane synthase, uroporphyrinogen III synthase, uroporphyrinogen decarboxylase, ferrochelatase, aminolevulinic acid dehydratase, protoporphyrinogen oxidase, and coproporphyrinogen-III oxidase (Figure S1C). As we have reported elsewhere (Dunsmore et al., 2018a; Elahi, 2019; Elahi et al., 2013; Elahi and Mashhouri, 2020; Namdar et al., 2019), CECs are a heterogeneous population of erythroid precursors/progenitors and possibly reticulocytes. CECs express significantly a greater level of the transferrin receptor (CD71) compared with immune cell lineages (Figures S1D and S1E). Besides, the intensity of CD71 varied among CECs (Figures S1F and S1G), and those with the highest CD71 expression level were enriched with CD45-expressing CECs (Figure S1H), suggesting CD45+ CECs as early erythroid progenitors. As the intensity of CD71 expression was reduced, the percentages of the CD45+ subpopulation declined in each subpopulation (Figure S1H).
Figure 1.
Expansion of CECs in COVID-19 patients is associated with disease progression
(A and B) (A) Representative plots and (B) cumulative data of the percentages of CECs in the PBMCs from COVID-19 patients with mild, moderate, and severe disease versus healthy controls (HCs).
(C) Data showing the percentage of CECs in PBMCs from patients at the time of admission to hospital and 5–7 days later.
(D and E) (D) Representative plots and (E) cumulative data of longitudinal changes in the frequency of CECs in three patients admitted to the ICU, each line represents one patient.
(F and G) (F) Representative plots and (G) cumulative data of arginase II expression in CECs compared with immune cell lineages.
(H and I) (H) Representative plots and (I) cumulative data of ROS expression in CECs compared with other immune cell lineages in COVID-19 patients.
(J and K) (J) Representative plots and (K) cumulative data of arginase I expression in CECs compared with immune cell lineages (total PBMCs without CECs) in COVID-19 patients. Each point represents data from an individual/patient. Bar, mean ± 1 standard error.
We also observed an increase in the quantity of lightweight RBCs (CD235a+ CD71− cells) in the blood of patients while their presence in HCs was negligible (Figures S1I and S1J). Although characterizing the properties of these expanded CD235+ CD71− cells was out of the scope of our study, we defined them as lightweight RBCs because they remained on the top Ficoll gradient. We reasoned that the underlying mechanism for the expansion of CECs in COVID-19 patients might be related to dysregulated activity of hematopoietic stem and progenitor cells. We first examined interleukin-33 (IL-33) levels in plasma since increased levels of IL-33 may inhibit the differentiation of CECs to RBCs (Swann et al., 2020). However, we did not observe any detectable level of IL-33 in our patients. These observations suggest that SARS-CoV-2 infection influences erythropoiesis.
CECs express arginase II, arginase I, and reactive oxygen species to mediate immunosuppression
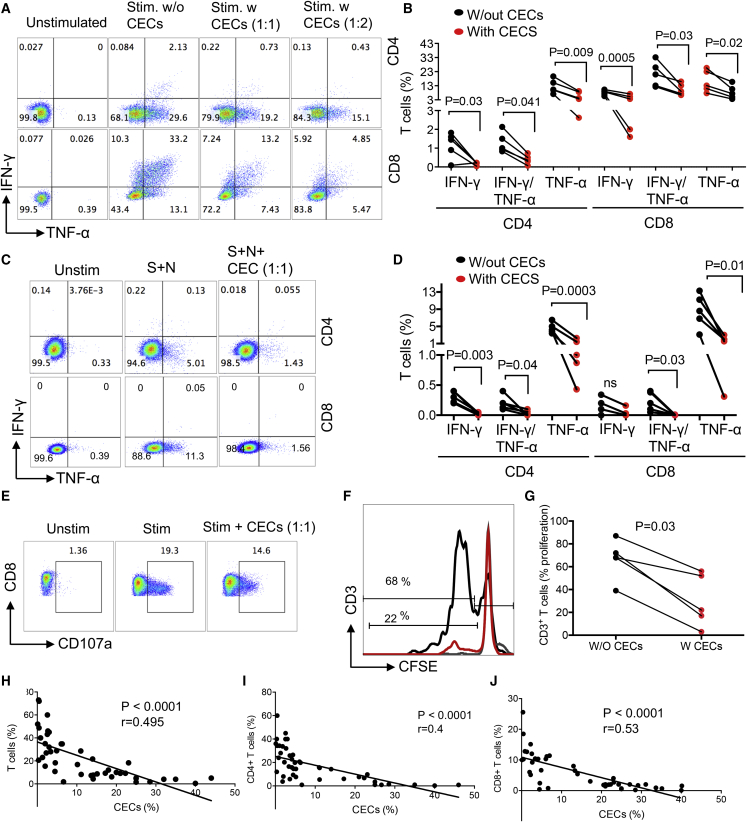
Due to their immunomodulatory properties, the pathological abundance of CECs during disease progression can have immunological consequences (Elahi, 2014, 2019; Elahi and Mashhouri, 2020). To better understand the biological properties of these expanded CECs in COVID-19 patients, we subjected them to immunological phenotyping. In contrast to other reports (Delyea et al., 2018; Shahbaz et al., 2018), CECs in COVID-19 patients expressed negligible amount of PDL-1/PDL-2 but expressed the V-domain immunoglobulin suppressor of T cell activation (Figure S1K). In agreement with our previous reports in other models (Dunsmore et al., 2017; Elahi et al., 2013), we found that CECs express significantly higher amounts of arginase II (Figures 1F and 1G) and reactive oxygen species (ROS) (Figures 1H and 1I) compared with other immune cell lineages, similar to what has been described for their counterparts in human newborns (Elahi et al., 2020b), HIV (Namdar et al., 2019), and cancer (Zhao et al., 2018). As a comparison, we measured the expression of ROS in CECs from HIV patients (Figures S2A and S2B). For the very first time, we also detected expression of arginase I in CECs of COVID-19 patients (Figures 1J and 1K). These observations guided us to investigate their immunosuppressive properties in vitro. CECs isolated from the peripheral blood mononuclear cells (PBMCs) that were >95% pure (Figure S2C) co-cultured with PBMCs at ratios of 1:1 or 1:2. The selected ratio reflects the minimal physiological relevance of T cells to CECs in the peripheral blood of COVID-19 patients. Because in most patients percentages of CECs were 3- to 5-fold higher than T cells, the CECs significantly suppressed cytokine production (e.g., tumor necrosis factor alpha [TNF-α] and interferon-γ [IFN-γ]) by both CD4+ and CD8+ T cells when stimulated with anti-CD3/CD28 in vitro (Figures 2A and 2B), and with overlapping peptide pools covering the main SARS-CoV-2 structural protein spike (S) and nucleocapsid (N) (Figures 2C and 2D). Of note, antigen-specific response was dominated by TNF-α but not IFN-γ (Figures 2C and 2D). The CECs had a similar immunosuppressive effect on the capacity of CD8+ T cells to degranulation in response to viral peptide stimulation, as measured using CD107a (Figures 2E and S2D). In agreement with previous reports in other models (Dunsmore et al., 2018a; Zhao et al., 2018), CECs significantly inhibited T cell proliferation following stimulation of PBMCs with SARS-CoV-2 peptides in vitro (Figures 2F and 2G). This was supported in vivo by the negative correlation between the percentages of CECs and CD3+ (Figure 2H), CD4+ (Figure 2I), and CD8+ T cells (Figure 2J) in COVID-19 patients. We also observed an inverse correlation between CECs and the frequency of antibody-secreting cells (plasmablasts) in COVID-19 (Figures S2E and S2F).
Figure 2.
CECs exhibit immunosuppressive properties and their abundance may compromise adaptive immunity
(A and B) (A) Representative plots and (B) cumulative data of IFN-γ and TNF-α expression in CD4 and CD8 T following stimulation (Stim.) of PBMCs with anti-CD3/CD28 antibodies for 6 h with (w) or without (w/out) CECs at 1:1 or 1:2 ratios.
(C and D) (C) Representative plots and (D) cumulative data of IFN-γ and TNF-α expression in CD4 and CD8 T cells following stimulation of PBMCs with S and N peptides of SARS-CoV-2 (2 μg/mL) for 6 h with (w) or without (w/out) CECs at the indicated ratio.
(E) Representative plots of CD107a in stimulated CD8 T cells with S and N peptides of SARS-CoV-2 for 6 h in the absence or presence of CECs (1:1 ratio).
(F and G) (F) Representative plots and (G) cumulative data showing proliferation of CD3 T cells as measured by CFSE dilution without (w/o) or with (w) CECs at 1:1 ratio following stimulation with S and N peptides of SARS-CoV-2 (2 μg/mL) for 3 days.
(H–J) (H) The correlation of total T cells, (I) CD4 T cells, and (J) CD8 T cells with the percentage of CECs in the PBMCs from COVID-19 patients. Selected patients were those having >15% CECs in their PBMCs. Each point represents data from one patient.
These observations demonstrated the immunosuppressive properties of CECs in COVID-19 patients, potentially resulting in the impairment of both T and B cell effector functions.
Progenitor CECs express SARS-CoV-2 receptor, ACE2
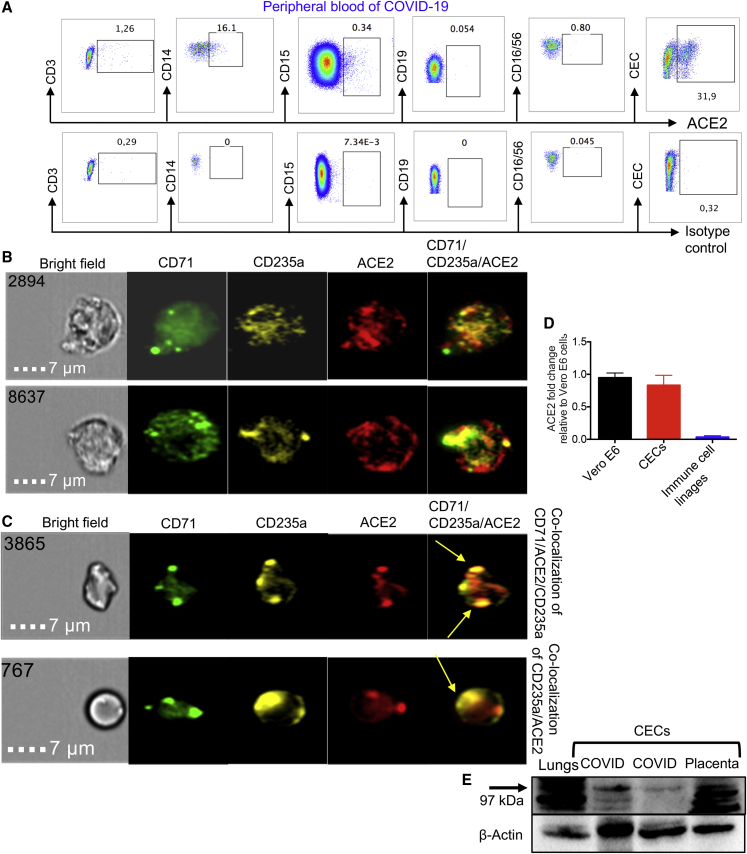
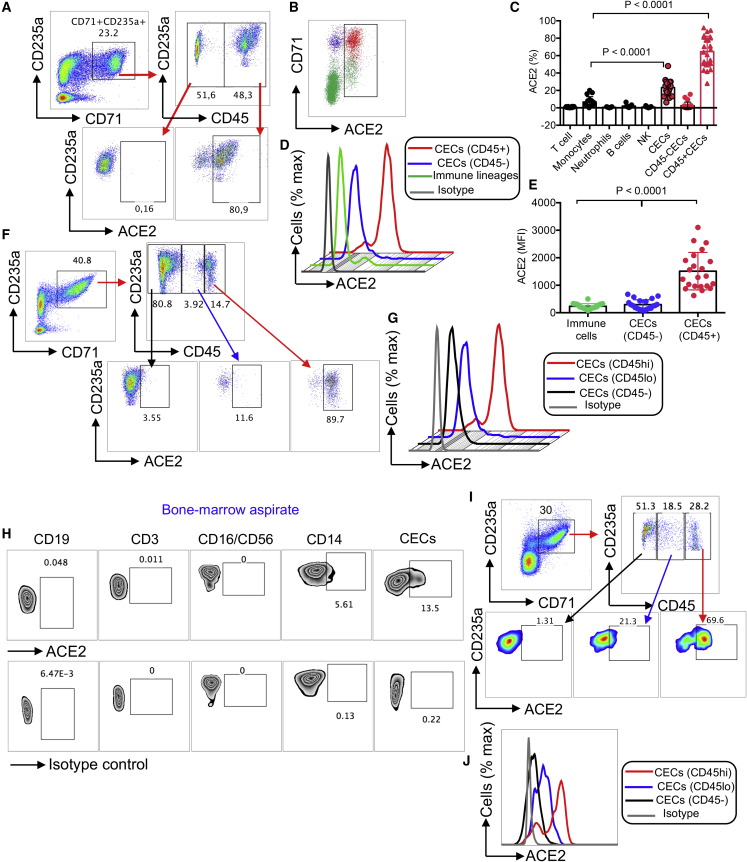
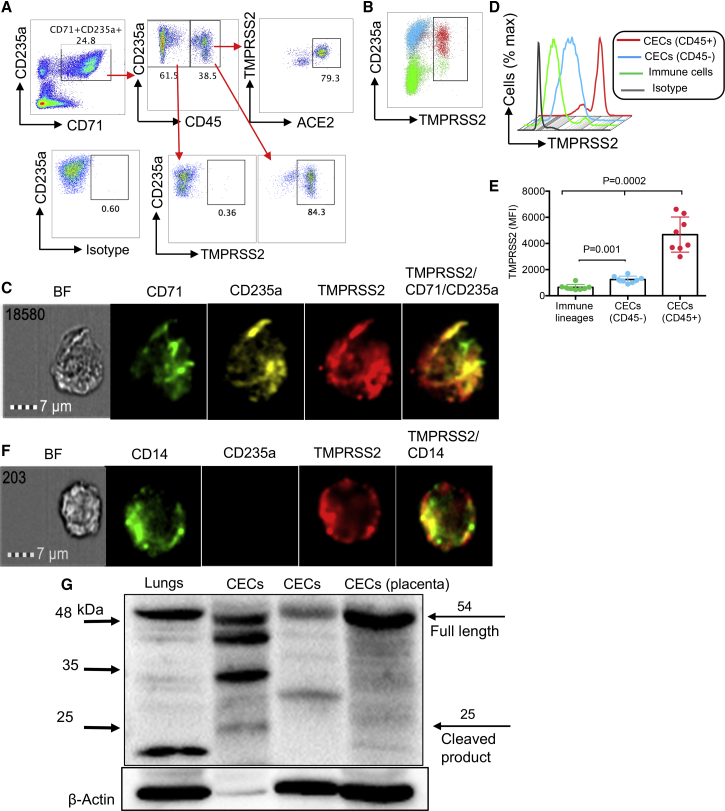
We have recently reported that HIV can both reside in CECs and that CECs can trans-infect CD4+ T cells (Namdar et al., 2017). We analyzed ACE2 expression on CECs compared with immune cell subsets, and found that CECs were the dominant cells expressing ACE2, followed by monocytes (Figure 3A). Similar observations were made by image stream analysis (Figures 3B and S2G), and co-localization of ACE2 with CD71 and/or CD235a was noted (Figures 3C and S2H). These observations were further re-confirmed by qPCR (Figure 3D) and western blotting. The expression of ACE2 in different tissues/organs in mice was first confirmed (Figure S3A). Then, we examined CECs isolated from the blood of COVID-19 patients (Figure S3B), the placental tissues of humans (Figure 3E), immune cell lineages, and VeroE6 cells as positive control for ACE2 expression (Figure S3C). Next, we identified ACE2-expressing CECs as erythroid progenitors that express CD45 (Figures 4A–4C), using the gating strategy in Figure S3D. CD45+ CECs appeared to be the major ACE2-expressing cells in the peripheral blood of COVID-19 patients, while other immune cells express negligible level of ACE2, except monocytes (Figures 3A and 4C). Importantly, the expression and intensity of ACE2 was also significantly elevated on CD45+ CECs compared with CD45− CECs and other immune cell lineages (Figures 4D and 4E). In particular, the intensity of ACE2 was substantially higher in CD45hiCECs compared with their counterparts with lower CD45 expression (Figures 4F and 4G). Nevertheless, the percentage of ACE2-expressing CECs varied during the course of disease (Figure S3E).
Figure 3.
CECs possess surface expression of ACE2
(A) Plots showing the expression of ACE2 on CD3 T cells, CD14 monocytes, CD15 neutrophils, CD19 B cells, CD16CD56 NK cells, and CECs in PBMCs from COVID-19 patients compared with the isotype control antibody.
(B and C) (B) Image stream plots showing the expression of ACE2 on CECs, and (C) the co-localization of ACE2 with CD71/CD235a on CECs in PBMCs from COVID-19 patients.
(D) Fold regulation of ACE2 mRNA in CECs and immune cell lineages from PBMCs from six COVID-19 patients, three with moderate and three with severe disease, compared with VeroE6 cells as quantified by qPCR.
(E) Western blot data showing the presence of ACEs protein in a mouse lung tissue, CECs from PBMCs from two COVID-19 patients and CECs (∼1 × 106 cells) isolated from a human placenta.
Figure 4.
CD45+ CECs from the peripheral blood of COVID-19 patients and human bone marrow exhibit the highest intensity of ACE2 expression compared with other immune cells
(A) Plots showing the expression of ACE2 on CD45− CECs versus CD45+ CECs from PBMCs from a COVID-19 patient.
(B) Percentage of ACE2 surface expression on total CECs and immune cell lineages in PBMCs from a COVID-19 patient.
(C) Cumulative data of the percentage of ACE2 expressing cells among different immune cell lineages and CECs in PBMCs from COVID-19 patients.
(D and E) (D) Representative plots and (E) cumulative data of the intensity of ACE2 expression in CD45+ CECs, CD45− CECs, and immune cell lineages measured by mean fluorescence intensity (MFI) in PBMCs from COVID-19 patients.
(F) Representative plots of the percentage of ACEs expression in CD45−, CD45lo, and CD45hiCECs in PBMCs from COVID-19 patients.
(G) Plot showing the intensity of ACE2 expression in CD45hi, CD45lo, and CD45− CECs compared with isotype control in PBMCs from COVID-19 patients.
(H) Plots showing the expression of ACE2 on CD19 B cells, CD3 T cells, CD16CD56 NK cells, CD14 monocytes, and CECs compared with the isotype control antibody in the bone marrow aspirates of a healthy adult.
(I) Representative plots of the percentage of ACEs expression in CD45−, CD45lo, and CD45hiCECs of the bone marrow.
(J) Histogram plot of the intensity of ACE2 expression in CD45hi, CD45lo, and CD45− CECs of the bone marrow compared with isotype control. Each point represents data from one patient. Bar, mean ± 1 standard error.
Next, we decided to quantify the expression level of CD45 and ACE2 in CECs from the human bone marrow. We found that CECs were the most dominant ACE2-expressing cell type followed by monocytes (Figures 4H and 4I). Similar to the peripheral blood CECs, the bone marrow CD45+ hiCECs had the highest intensity of ACE2 (Figures 4I and 4J). Also, we found that CD45+ CECs possess the highest levels of CD147 expression when compared with other immune cells from COVID-19 patients (Figures S3F and S3G). Finally, we found a higher expression of CD26 on CECs compared with RBCs (Figures S3H and S2I) and CD45+ CECs had higher surface CD26 expression compared with CD45− CECs (Figure S3J). Since SARS-CoV-2 infection might lead to damage and lysis of these cells, we also measured soluble ACE2 in the plasma of COVID-19 patients. Interestingly, we found significantly higher levels of plasma ACE2 in patients with more moderate/severe disease than those with mild disease (Figure S4A). As the disease progressed, plasma ACE2 levels also increased (Figure S4B). Longitudinal analysis in ICU patients also indicated a gradual increase in the plasma ACE2 with clinical progression (Figure S4C). Overall, these observations showed that CD45+ CECs were the dominant ACE2/CD147-expressing cells.
CD45+ CECs also express SARS-CoV-2 co-receptor, TMPRSS2
Recent evidence indicates that viral entry into target cells depends not only on the binding of the S protein to ACE2 but also requires S protein priming by the cellular serine protease TMPRSS2 (Hoffmann et al., 2020). Thus, we found that CD45+ CECs both express TMPRSS2 (Figures 5A, 5B, and S4D) and co-express ACE2 and TMPRSS2 (Figure 5A) as confirmed by image stream analysis (Figures 5C and S4E–S4G). The intensity of TMPRSS2 was significantly greater on CD45+ CECs compared with CD45− CECs and other immune cell lineages (Figures 5D and 5E). Of note, a small subset of the immune cell lineages, mainly CD14 monocytes, also express TMPRSS2 (Figures 5F and S4H), which were further re-confirmed by western blotting. Initially, we confirmed the expression of full-length (54 kDa) and the cleavage fragment (25 kDa) of TMPRSS2 in mice tissues compared with the positive control (human colorectal adenocarcinoma grade II cell line) (Figure S5A); however, the cleaved product appeared to be smaller than 25 kDa in mice tissues. Western blot confirmed the expression of TMPRSS2 in CECs isolated from COVID-19 patients and human placental tissues (Figure 5G). Interestingly, when the same number of cell lysate was loaded, TMPRSS2 protein level was higher in CECs compared with immune cell linages (CECs-) (Figure S5B). Of note, β-actin levels appeared to be lower in CECs compared with other immune cells. Similarly, bone marrow CECs also possess the surface expression of TMPRSS2 (Figure S5C). Taken together, these results confirm the presence/surface expression of TMPRSS2 and its co-expression with ACE2 in CECs.
Figure 5.
CD45+ CECs from the peripheral blood of COVID-19 patients are the dominant TMPRSS2-expressing cells
(A) Plots showing the expression of ACE2 on CD45− CECs versus CD45+ CECs and the co-expression of ACE2/TMPRSS2 on CD45+ CECs in PBMCs from a COVID-19 patient.
(B) Percent TMPRSS2 surface expression on total CECs and immune cell lineages in PBMCs from a COVID-19 patient.
(C) Image stream plots of TMPRSS2 expression on CECs from PBMCs from a COVID-19 patient.
(D and E) (D) Histogram plot and (E) cumulative data of the intensity of TMPRSS2 expression in CD45+ CECs, CD45− CECs and immune cell lineages measured by MFI in PBMCs from COVID-19 patients.
(F) Image stream plots of TMPRSS2 expression on CD14 monocytes in PBMCs from a COVID-19 patient.
(G) Western blot data showing the presence of TMPRSS2 protein in a mouse lungs tissue, CECs isolated from PBMCs from two COVID-19 patients and CECs (∼1 × 106 cells) isolated from a human placenta tissue. Each point represents data from one patient. Bar, mean ± 1 standard error.
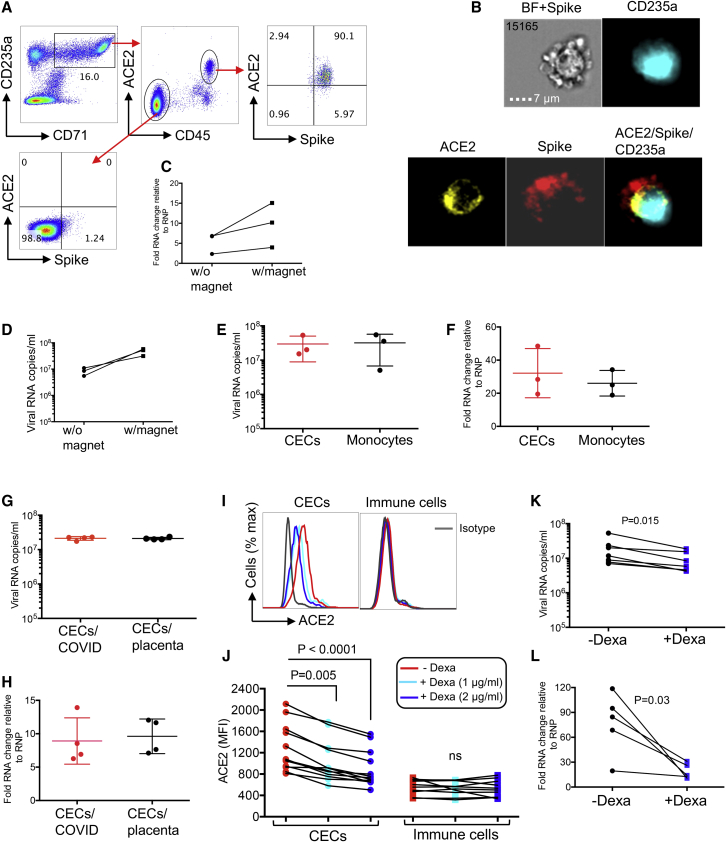
CECs get infected with SARS-CoV-2 and dexamethasone reduces their infectivity in vitro
In light of ACE2 and TMPRSS2 expression/co-expression on CECs, we hypothesized that these cells could get infected by SARS-CoV-2. To test this hypothesis, we found that the spike binding domain (conjugated by a fluorescent dye) bound ACE2 on CD45+ CECs, whereas it did not bind CD45− CECs (Figure 6A). This was further confirmed by image stream analysis (Figure 6B and S5D). In image stream analysis, we first gated on CD235a+ T cells (Figure S5E) in area less than 300 (Figure S5F) to avoid larger cells, such as central macrophages. Furthermore, we did not observe the binding of spikes with cells that lacked ACE2 (e.g., T cells) (Figure S5G). The ability of CECs from COVID-19 patients to be infected by SARS-CoV-2 using the traditional approach versus magnetofection was tested as we have reported for HIV-1 (Elahi et al., 2012, 2016). We were able to detect viral RNA produced in cell culture supernatants as well as viral RNA in the cells 24 h after infection using both methods, but the magnetofection was more efficient (Figures 6C and 6D). While viral RNA levels were not high compared with VeroE6 cells, they were >2 logs higher than the final wash after infection and infectious SARS-CoV2 was produced by these cells. Since it has been recently shown that monocytes can be infected with SARS-CoV2 (Pence, 2020), we compared the amount of infection in CECs with that in monocytes. We found that the amount of viral RNA in CECs and produced by CECs was similar to monocytes (Figures 6E and 6F). To examine the infectivity of CECs obtained from a SARS-CoV2-naive source we infected CECs isolated from human placenta with SARS-CoV2, because placenta is physiologically enriched with CECs (Delyea et al., 2018; Dunsmore et al., 2018a, 2018b; Gomez-Lopez et al., 2016), possessing ACE2 and TMPRSS2 (Figures 3D and 5G). Similar to CECs from COVID-19 patients, placental CD45+ CECs had substantial surface expression of ACE2 (Figures S6A and S6B) and TMPRSS2 (Figure S6C). We found that, similar to CECs from COVID-19 patients, CECs from placenta contained viral RNA after infection and secreted viral RNA into the cell culture supernatants (Figures 6G and 6H). These results indicate that CECs can be directly infected by SARS-CoV2.
Figure 6.
CECs allow SARS-CoV-2 infection, but this can be reversed in part by dexamethasone
(A) Plots showing co-expression of spike binding domain with ACE2 on CD45+ CECs but not CD45− CECs from PBMCs from a COVID-19 patient.
(B) Image stream plots of spike protein interaction/binding with ACE2 on CECs in PBMCs from a COVID-19 patient; bright field (BF) plus spike staining (BF + Spike).
(C and D) (C) Cellular RNA level in CECs and (D) viral RNA copies in culture supernatants of CECs without (w/o) or with (w) magnetofection with SARS-CoV-2.
(E) Viral RNA copies in the culture supernatant of total CECs or monocytes of COVID-19 patients infected with SARS-CoV-2 and measured by qPCR 24 h after infection.
(F) Cellular RNA changes relative to the housekeeping gene (RNP) in cell pellet of CECs and monocytes of COVID-19 patients infected with SARS-CoV-2 and measured by qPCR 24 h after infection.
(G) Viral RNA copies in culture supernatants of total CECs of COVID-19 patients versus CECs of the placental tissues from healthy deliveries 24 h after infection with SARS-CoV-2.
(H) Cellular fold RNA changes relative to the housekeeping gene (RNP) in cell pellets of total CECs from COVID-19 patients or the placental tissues from healthy deliveries 24 h after infection with SARS-CoV-2.
(I and J) (I) Histogram plots and (J) cumulative data of the expression of ACE2 on total CECs and immune cells lineages of COVID-19 patients treated with or without dexamethasone (1 or 2 μg/mL) overnight.
(K) Viral RNA copies in culture supernatants of CECs either untreated or treated with dexamethasone (2 μg/mL) for 24 h before infection with SARS-CoV-2 as measured 24 h after infection.
(L) Cellular fold RNA changes relative to the housekeeping gene (RNP) in cell pellets of CECs either untreated or treated with dexamethasone (2 μg/mL) for 24 h before infection with SARS-CoV-2 as measured 24 h after infection. Each point represents data from cells of one patient, infection studies are representative of three independent experiments. Bar, mean ± 1 standard error.
Erythroid progenitors possess a glucocorticoid receptor that enhances the response to erythropoietin (Epo) (Golde et al., 1976). Glucocorticoids, such as dexamethasone are known to aid in the treatment of Epo-resistant anemia by stimulating self-renewal of progenitors (Flygare et al., 2011; Lee et al., 2015). Since multiple studies reported that COVID-19 patients and primate models of SARS-CoV-2 infection are generally anemic (Chen et al., 2020; Munster et al., 2020), and little is known about the mechanism associated with the therapeutic effects of dexamethasone in severely ill COVID-19 patients (Horby et al., 2020), we reasoned that the enhanced maturation of expanded CECs in severe disease might be one molecular mechanism for the lower mortality rate in patients receiving dexamethasone. Similar to other observations, we noted low hemoglobin (Hgb) levels in our cohort, particularly in those with severe disease (Figure S6D), and more importantly we found a negative correlation between the frequency of CECs in the peripheral blood with Hgb concentrations (Figure S6E). Then we examined the effect of dexamethasone in mice. Since young mice possess a higher frequency of CECs in their spleens compared with adults (Shahbaz et al., 2018), we treated young mice (15 days) with dexamethasone (1 μg/g body weight intraperitoneally) and 2 days later collected their spleens for the quantification of CECs. A significant reduction in the frequency of CECs in treated versus control animals was observed (Figures S6F and S6G), suggesting that dexamethasone enhances the maturation of CECs to mature RBCs. We then examined whether CECs obtained from the peripheral blood of COVID-19 patients displayed the same phenomena. We treated total PBMCs and/or isolated CECs with 1 and 2 μg/mL dexamethasone overnight (Okoye et al., 2020). We observed that such treatment enhanced the maturation of CECs, which resulted in the downregulation of ACE2 in a dose-dependent manner (Figures 6I and 6J), but did not affect the ACE2 expression on other immune cells lineages (Figures 6I and 6J). We also found downregulation of TMPRSS2 expression on CECs following treatment with dexamethasone (Figure S6H). However, atorvastatin, despite its immunomodulatory properties (Elahi et al., 2016; Okoye et al., 2017), did not impact ACE2 expression (Figure S6I). Since dexamethasone downregulated the expression of ACE2, we reasoned it might also reduce the permissibility of CECs to SARS-CoV-2 infection. When we pre-treated CECs with 2 μg/mL dexamethasone for 24 h before SARS-CoV2 infection, there was a significant reduction in the viral RNA in cell culture supernatant and intracellular viral RNA compared with untreated cells (Figures 6K and 6L). Importantly, this was not the case for monocytes when treated with dexamethasone (Figures S6J and S6K). Taken together, these observations indicate that CECs can be infected by SARS-CoV-2.
Discussion
We identified significant expansion of CECs in the peripheral blood of COVID-19 patients, particularly in those with moderate or severe disease. Interestingly, we found that the expansion of CECs was associated with disease severity and ICU admission. CECs have a wide range of immunomodulatory properties and via cell:cell interactions or soluble mediators, such as transforming growth factor β, ROS, and arginase II, and can suppress effector T cell and B cell functions (Dunsmore et al., 2018a; Elahi, 2019; Elahi and Mashhouri, 2020; Namdar et al., 2017; Zhao et al., 2018). Consistent with previous reports in the context of infection (e.g., HIV) and cancer (Dunsmore et al., 2017; Namdar et al., 2019; Zhao et al., 2018), CECs from COVID-19 patients exhibited substantial expression of arginase I/II and ROS. These capabilities not only enabled CECs to exert a global immunosuppression effects on T cells but they also significantly impaired cytokine production, proliferation, and degranulation capacities of antigen-specific T cells in vitro. These observations were further confirmed in vivo by the presence of a negative correlation between the frequency of T cells with the percentages of CECs in the peripheral blood of COVID-19 patients. Therefore, the pathological abundance of CECs in COVID-19 patients may in part explain the mechanism underlying a wide range of changes in the frequency and functionality of different immune cells in these patients (Huang et al., 2020; Xu et al., 2020). One striking observation was that CECs from COVID-19 patients appear to have a different membrane structure. For example, measuring arginase II activity in human CECs has been technically impossible as CECs are lysed when exposed to the fixation/permeabilization buffer required for intracellular staining. Surprisingly, this was not the case for CECs isolated from COVID-19 patients and, indeed, these CECs were resistant to permeabilization. This suggests that SARS-CoV-2 infection or the immunopathology associated with COVID-19 disease can modify RBCs and/or CEC structural components, as COVID-19 infection is associated with RBC structural protein damage and modifications in RBC membrane lipids (Thomas et al., 2020). As such, the abundance of these unwanted guests and the possible altered structural proteins following infection with SARS-CoV-2 may contribute to other serious complications (such as thromboembolic and coagulopathic events) commonly observed in COVID-19 patients (Giannis et al., 2020). The role of RBC morphology and deformability in clot formation has been widely studied (Aleman et al., 2014; Byrnes and Wolberg, 2017). Although thrombosis is likely multifactorial in nature, involving vasculature, platelets, and dysfunctional RBCs, it will be of interest to determine if expanded CECs, like their older siblings, are involved within thrombi in COVID-19 patients. The severely low oxygen saturations observed in critically ill patients (Aggarwal et al., 2020), particularly without substantial damage to the lungs (Henry, 2020) suggests that SARS-CoV-2 may affect oxygenation via paths unrelated to pulmonary function. Elevated cytokines (e.g., IL-1, IL-6, and TNF-α) may influence erythropoiesis (Sarkar et al., 2015) and increase the permissibility of RBCs to oxidant stress-induced lysis in COVID-19 patients (Thomas et al., 2020). In light of the above, there is the possibility that SARS-CoV-2 infection directly or indirectly invades RBCs in the periphery or the bone marrow resulting in their enhanced lysis and anemia (Chen et al., 2020; Mitra et al., 2020; Munster et al., 2020). In agreement with other reports, we observed low Hgb levels in COVID-19 patients, particularly in those with severe disease. In this scenario, we expect stress erythropoiesis and subsequently abundant CECs in the periphery due to passive incontinence of hematopoietic cells from the bone marrow (Elahi and Mashhouri, 2020; Johns and Christopher, 2012). Our results further confirmed that lower Hgb level was associated with increased proportion of CECs in the blood circulation. Although the lack of organelles in RBCs precludes viral survival/replication, this might not be true for CECs. Recently, we reported that HIV can reside and possibly replicate within CECs (Namdar et al., 2019). In support of this hypothesis, we identified CECs as the major ACE2- and TMPRSS2-expressing cells. Furthermore, the receptor-like tyrosine phosphatase CD45 is expressed on all nucleated hematopoietic cells, including erythroid progenitors (Shim et al., 2020) and gets downregulated as erythroid progenitors mature into RBCs (Shim et al., 2020). The expression of CD45 and CD235a on the erythroid progenitors have been controversial. For example, several studies have reported the expression of CD235a on pro-erythroblasts (Dzierzak and Philipsen, 2013; Edelman et al., 1986; Fukuda and Fukuda, 1981). In contrast, another study reported the lack of CD235a on pro-erythroblasts (van Lochem et al., 2004). Similarly, although CD45 is considered to be a leukocyte marker, its expression on both burst-forming unit-erythroid and colony-forming unit-erythroid cells has been reported (Li et al., 2014). However, a report suggested low expression and absence of CD45 expression on pro-erythroblast and erythroblasts, respectively (van Lochem et al., 2004). Our data demonstrate the presence of a subpopulation of CECs as CD71+ CD235a+ CD45+ cells in COVID-19 patients. We found CD45+ CECs were the dominant ACE2/TMPRSS2-expressing cells compared with their CD45− CEC counterparts. In this setting, we speculated that erythroid progenitors in the bone marrow should express ACE2 and TMPRSS2. Indeed, this was the case and once again we observed CD45+ CECs as the dominant cells in terms of ACE2 and TMPRSS2 expression in the human bone marrow. In addition, we found that CECs from COVID-19 patients, particularly CD45+ CECs, possess the highest intensity of CD147/CD26 expression, another SARS-CoV-2 receptor (Wang et al., 2020). Taken together, these observations suggest that CECs could be an attractive target for SARS-CoV-2. More importantly, CECs, in particular their CD45+ progenitors possess nuclei and other organelles that can support viral survival and replication. Therefore, we first confirmed the binding of the SARS-CoV-2 spike with ACE2 on the surface of CD45+ CECs. These observations led us to determine if CECs can get infected with SARS-CoV-2 and, more importantly, whether the virus replicates in these cells. We found that the infectivity of CECs to SARS-CoV-2 was comparable with monocytes. Despite the similar level of viral infection in these two cell subsets, we believe CECs are more permissible to infection compared with monocytes for multiple reasons. (1) CECs are the most abundant cells in the peripheral blood of COVID-19 patients, especially those with a moderate/severe disease. (2) CECs contain the highest ACE2, CD147, and TMPRSS2 expression. (3) Given a lower proportion of nucleated CECs, but that similar viral RNA copies were observed in the culture supernatant and cell pellet of CECs and monocytes, this suggests more efficient viral infectivity/replication in CECs. The CD45+ CEC population comprises approximately 20%–80% of CECs, while monocytes are 100% nucleated and can support viral replication. Thus, the infectivity of RBC progenitors to SARS-CoV-2 infection may explain one potential mechanism for the observed hypoxia in COVID-19 patients. As such, higher percentages of CECs in these severely ill patients is indicative of stress hematopoiesis. In line with our findings, anemia has been considered as an independent risk factor related to severe COVID-19 illness (Tao et al., 2020; Zhou et al., 2020). We hypothesize that this phenomenon might be due to the elimination of infected/damaged CECs by lysis or/and phagocytosis. However, further studies are required to confirm these observations by detecting viral proteins or infective viral particles in CECs of COVID-19 patients.
On the other hand, the immunosuppressive properties of CECs may be beneficial to COVID-19 patients since hyper-inflammation and cytokine storm is associated with disease severity (Huang et al., 2020). As such, CECs might appear protective at the early stage of disease to prevent a robust innate immune response (Elahi, 2020). Nevertheless, expansion of CECs in the peripheral blood of COVID-19 patients coincides with the disease progression, which is the time for the induction of an efficient adaptive immune response. Therefore, the absence of CECs at the early stage of disease deprives the host from their highly desired immunosuppressive properties but instead their appearance later can compromise T cell effector functions and antibody production. The uncontrolled inflammatory response can itself damage the lungs via the excessive release of proteases, ROS, and pro-inflammatory cytokines (Tay et al., 2020). In agreement with this concept, several immunosuppressive strategies are recommended for the treatment of COVID-19 patients. Although the administration of systemic corticosteroids for COVID-19 patients was initially not supported by the WHO guidelines (Huang et al., 2020), several trials are under way for the efficacy of such treatment options (Tay et al., 2020). In fact, a recent randomized clinical trial has shown that dexamethasone reduces deaths by one-third in patients on ventilators and by one-fifth in those receiving oxygen without invasive ventilation (Horby et al., 2020). In addition to the anti-inflammatory function of dexamethasone, it influences hematopoiesis and promotes the maturation of erythroid cells (Narla et al., 2011). This may be supported by our observation that dexamethasone reduced ACE2/TMPRSS2 expression by enhanced CEC maturation. In this respect, dexamethasone-mediated downregulation of ACE2/TMPRSS2 may explain the reduced susceptibility of CECs to SARS-CoV-2 infection. Although we were unable to determine the exact underlying mechanism of ACE2/TMPRSS2 downregulation following treatment of CECs with dexamethasone, there are two possibilities. (1) Dexamethasone enhances maturation of CECs and subsequently downregulates ACE2/TMPSS2. (2) It modifies the plasma membrane of CECs (Zingariello et al., 2019). Our observations support the former and knowledge gained from this study may illuminate the pivotal role CECs play in COVID-19 pathogenesis. In addition, this study provides mechanistic rationale for the clinical use of dexamethasone in COVID-19 patients, particularly in those with severe disease. It is therefore, tempting to speculate that immunosuppressive drugs might be harmful when given in the induction phase of immune response. However, considering the massive expansion of CECs in COVID-19 patients, targeting these cells with medications, such as dexamethasone might be beneficial rather than detrimental for the patient. It appears that dexamethasone may not only attenuate the hyperactive immune response but also protect CECs from the virus, enhancing their maturation and possibly preventing hypoxia. This concept merits further investigations in larger cohorts.
Given the high surface expression of ACE2 and TMPRSS2 on placental CECs and their permissibility to ARS-CoV-2 infection, the possibility of vertical viral transmission seems possible. Also, CECs become abundant in the peripheral blood of pregnant women at the later stage of pregnancy (Delyea et al., 2018; Dunsmore et al., 2018a, 2018b), which in part may support the reported viremia and placental transmission of SARS-CoV-2 (Vivanti et al., 2020). This was challenged due to negligible co-expression of ACE2 and TMPRSS2 by placental cell types (Pique-Regi et al., 2020). However, mRNA expression does not necessarily correlate with the protein expression pattern. More importantly, the expression of ACE2 and TMPRSS2 on CECs as the major cells in placenta was not studied in this report. Thus, the abundance of CECs expressing ACE2 and TMPRSS2 in the peripheral blood of pregnant mothers and placenta tissue suggests transplacental transmission of SARS-CoV-2 infection.
We are aware of the study limitations. For example, we would have liked to perform additional infection assays (e.g., drug titration, time points, blood group). Another area of intense interest that requires further attention is correlating the plasma antibody levels with the frequency of CECs. A larger sample size would also be required to evaluate whether dexamethasone treatment reduces the viral load in patients (e.g., lungs), reduces the frequency of CECs, and if it influences the expression of ACE2 in different tissues.
Experimental procedures
Human sample collection and processing
Blood samples were collected from hospitalized COVID-19 patients in different hospitals in Edmonton, Alberta. All COVID-19 patients were SARS-CoV-2-positive by qRT-PCR assay specific for viral RNA-dependent RNA polymerase and envelope transcripts detected using a nasopharyngeal swab. The Human Research Ethics Board (HREB) at the University of Alberta approved the study (Pro00099502). Waiver of consent was obtained by the HREB for those patients admitted to the ICU but a verbal consent was required from all other patients. Wet consent was not required due to logistics and the possible risk of viral transmission. The Health Research Ethics Board of Alberta (HREBA no. CC-17-0307) approved bone marrow aspirates. Also, the research ethics boards at the University of Alberta approved blood collection from HCs (no. Pro00063463) and placenta tissues (no. Pro00046080). Written informed consent was obtained from bone marrow donors, healthy individuals, and pregnant mothers. Fresh PBMCs or bone marrow aspirates were isolated over Ficoll-Hypaque gradients. For CECs or mock depletion samples were stained using anti-CD71 out of CD235a+ cells or isotype control biotin-conjugated antibody and fractioned using streptavidin-linked magnetic beads (Miltenyi Biotec) according to our previous reports (Dunsmore et al., 2017, 2018a; Elahi et al., 2013; Namdar et al., 2019). Normally, the isolated CECs have a purity >95% (Figures S2C and S3B).
Animal studies
BALB/c mice were used for studying the effects of dexamethasone on CECs. The research ethics boards at the University of Alberta approved these studies (AUP00001021).
Antibodies and flow cytometry
The list of antibodies and flow cytometry methods according to our previous protocols (Dunsmore et al., 2017; Namdar et al., 2019) are provided in the supplemental information.
Co-culture and stimulation
For in vitro intracellular cytokine staining, PBMCs were cultured and stimulated with anti-CD3/CD28 in RPMI medium supplemented with 10% FBS for 6 h in the presence or absence of CECs according to our previous report (Namdar et al., 2017) and our previous protocols (Dunsmore et al., 2018a; Elahi et al., 2011).
ELISA
IL-33 (Novus Biologicals) and ACE2 DuoSet ELISA (R&D) were performed on frozen plasma samples of patients and HCs according to the manufacturing protocol.
Western blot analyses
Protein samples were separated by electrophoresis on either 7%, 17%, or 4%–15% gradient polyacrylamide gels and then transferred to polyvinylidene fluoride membranes. Further details are provided in the supplemental information.
Virus infection and quantification
SARS-CoV-2 stocks were pre-incubated with ViroMag transduction reagent (OZ Biosciences) as reported elsewhere (Elahi et al., 2012, 2016; Namdar et al., 2019) and further details are provided in the supplemental information.
Gene expression analysis
RNA isolation and qPCR were conducted according to our published data (Elahi et al., 2020a; Shahbaz et al., 2018) with further information in the supplemental information.
Statistical analysis
Statistical comparisons between various groups were performed by using t test and Mann- Whitney tests (as appropriate) using GraphPad Prism software. Also, differences were evaluated using one-way ANOVA followed by Tukey's test for multiple comparisons. Correlation analysis was performed using the Spearman test. Results are expressed as mean ± SEM. p < 0.05 was considered statistically significant.
Author contributions
S.S. performed most of the experiments and analyzed the data. L.X. performed some of the experiments and analyzed the data. M.O. contributed to patient recruitment. W.S., as a clinician scientist (in critical care medicine and infectious disease), identified and recruited patients for the study. J.S. performed viral infection experiments. M.J. provided assistance and guided viral infection studies. D.L.T. advised on infection experiments. O.O. performed all the western blotting studies. S.E. conceptualized, designed, secured funding and resources, performed some of studies, analyzed the data, designed the figures, supervised all of the research, and wrote the manuscript. All authors revised and edited the manuscript.
Acknowledgments
This work was supported by a grant from FASTGRANT.com, the Canadian Institutes of Health Research (CIHR), Grant number MM1-174901, and a grant from the Li Ka Shing Institute of Virology, University of Alberta, Canada.
Published: May 11, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.04.001.
Supplemental information
References
- Aggarwal S., Garcia-Telles N., Aggarwal G., Lavie C., Lippi G., Henry B.M. Clinical features, laboratory characteristics, and outcomes of patients hospitalized with coronavirus disease 2019 (COVID-19): early report from the United States. Diagnosis (Berl) 2020;7:91–96. doi: 10.1515/dx-2020-0046. [DOI] [PubMed] [Google Scholar]
- Aleman M.M., Walton B.L., Byrnes J.R., Wolberg A.S. Fibrinogen and red blood cells in venous thrombosis. Thromb. Res. 2014;133:S38–S40. doi: 10.1016/j.thromres.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broxmeyer H.E., Hoggatt J., O'Leary H.A., Mantel C., Chitteti B.R., Cooper S., Messina-Graham S., Hangoc G., Farag S., Rohrabaugh S.L. Dipeptidylpeptidase 4 negatively regulates colony-stimulating factor activity and stress hematopoiesis. Nat. Med. 2012;18:1786–1796. doi: 10.1038/nm.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes J.R., Wolberg A.S. Red blood cells in thrombosis. Blood. 2017;130:1795–1799. doi: 10.1182/blood-2017-03-745349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020;20:269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavezzi A., Troiani E., Corrao S. COVID-19: hemoglobin, iron, and hypoxia beyond inflammation. A narrative review. Clin. Pract. 2020;10:1271. doi: 10.4081/cp.2020.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosnier C., Bustamante L.Y., Bartholdson S.J., Bei A.K., Theron M., Uchikawa M., Mboup S., Ndir O., Kwiatkowski D.P., Duraisingh M.T. Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum. Nature. 2011;480:534–U158. doi: 10.1038/nature10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alessandro A., Dzieciatkowska M., Nemkov T., Hansen K.C. Red blood cell proteomics update: is there more to discover? Blood Transfus-Italy. 2017;15:182–187. doi: 10.2450/2017.0293-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delyea C., Bozorgmehr N., Koleva P., Dunsmore G., Shahbaz S., Huang V., Elahi S. CD71(+) erythroid suppressor cells promote fetomaternal tolerance through arginase-2 and PDL-1. J. Immunol. 2018;200:4044–4058. doi: 10.4049/jimmunol.1800113. [DOI] [PubMed] [Google Scholar]
- Dunsmore G., Bozorgmehr N., Delyea C., Koleva P., Namdar A., Elahi S. Erythroid suppressor cells compromise neonatal immune response against Bordetella pertussis. J. Immunol. 2017;199:2081–2095. doi: 10.4049/jimmunol.1700742. [DOI] [PubMed] [Google Scholar]
- Dunsmore G., Koleva P., Ghobakhloo N., Sutton R.T., Ambrosio L., Meng X., Hotte N., Nguyen V., Madsen K.L., Dieleman L.A. Lower abundance and impaired function of CD71+ erythroid cells in inflammatory bowel disease patients during pregnancy. J. Crohns Colitis. 2018;13:230–244. doi: 10.1093/ecco-jcc/jjy147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunsmore G., Koleva P., Sutton R.T., Ambrosio L., Huang V., Elahi S. Mode of delivery by an ulcerative colitis mother in a case of twins: immunological differences in cord blood and placenta. World J. Gastroenterol. 2018;24:4787–4797. doi: 10.3748/wjg.v24.i42.4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzierzak E., Philipsen S. Erythropoiesis: development and differentiation. Cold Spring Harb Perspect. Med. 2013;3:a011601. doi: 10.1101/cshperspect.a011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman P., Vinci G., Villeval J.L., Vainchenker W., Henri A., Miglierina R., Rouger P., Reviron J., Breton-Gorius J., Sureau C. A monoclonal antibody against an erythrocyte ontogenic antigen identifies fetal and adult erythroid progenitors. Blood. 1986;67:56–63. [PubMed] [Google Scholar]
- Elahi S. New insight into an old concept: role of immature erythroid cells in immune pathogenesis of neonatal infection. Front. Immunol. 2014;5:376. doi: 10.3389/fimmu.2014.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahi S. Neglected cells: immunomodulatory roles of CD71(+) erythroid cells. Trends Immunol. 2019;40:181–185. doi: 10.1016/j.it.2019.01.003. [DOI] [PubMed] [Google Scholar]
- Elahi S. Neonatal and children's immune system and COVID-19: biased immune tolerance versus resistance strategy. J. Immunol. 2020;205:1990–1997. doi: 10.4049/jimmunol.2000710. [DOI] [PubMed] [Google Scholar]
- Elahi S., Dinges W.L., Lejarcegui N., Laing K.J., Collier A.C., Koelle D.M., McElrath M.J., Horton H. Protective HIV-specific CD8+ T cells evade Treg cell suppression. Nat. Med. 2011;17:989–995. doi: 10.1038/nm.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahi S., Ertelt J.M., Kinder J.M., Jiang T.T., Zhang X., Xin L., Chaturvedi V., Strong B.S., Qualls J.E., Steinbrecher K.A. Immunosuppressive CD71+ erythroid cells compromise neonatal host defence against infection. Nature. 2013;504:158–162. doi: 10.1038/nature12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahi S., Mashhouri S. Immunological consequences of extramedullary erythropoiesis: immunoregulatory functions of CD71+ erythroid cells. Haematologica. 2020;105:1478–1483. doi: 10.3324/haematol.2019.243063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahi S., Niki T., Hirashima M., Horton H. Galectin-9 binding to Tim-3 renders activated human CD4+ T cells less susceptible to HIV-1 infection. Blood. 2012;119:4192–4204. doi: 10.1182/blood-2011-11-389585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahi S., Shahbaz S., Houston S. Selective upregulation of CTLA-4 on CD8+ T cells restricted by HLA-B∗35Px renders them to an exhausted phenotype in HIV-1 infection. Plos Pathog. 2020;16 doi: 10.1371/journal.ppat.1008696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahi S., Vega-López M.A., Herman-Miguel V., Ramírez-Estudillo C., Mancilla-Ramírez J., Motyka B., West L., Oyegbami O. CD71+ erythroid cells in human neonates exhibit immunosuppressive properties and compromise immune response against systemic infection in neonatal mice. Front. Immunol. 2020;11:597433. doi: 10.3389/fimmu.2020.597433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahi S., Weiss R.H., Merani S. Atorvastatin restricts HIV replication in CD4+ T cells by upregulation of p21. Aids. 2016;30:171–183. doi: 10.3389/fimmu.2020.597433. [DOI] [PubMed] [Google Scholar]
- Flygare J., Rayon Estrada V., Shin C., Gupta S., Lodish H.F. HIF1alpha synergizes with glucocorticoids to promote BFU-E progenitor self-renewal. Blood. 2011;117:3435–3444. doi: 10.1182/blood-2010-07-295550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M., Fukuda M.N. Changes in cell surface glycoproteins and carbohydrate structures during the development and differentiation of human erythroid cells. J. Supramol Struct. Cell Biochem. 1981;17:313–324. doi: 10.1002/jsscb.380170403. [DOI] [PubMed] [Google Scholar]
- Giannis D., Ziogas I.A., Gianni P. Coagulation disorders in coronavirus infected patients: COVID-19, SARS-CoV-1, MERS-CoV and lessons from the past. J. Clin. Virol. 2020;127:104362. doi: 10.1016/j.jcv.2020.104362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golde D.W., Bersch N., Cline M.J. Potentiation of erythropoiesis in vitro by dexamethasone. J. Clin. Invest. 1976;57:57–62. doi: 10.1172/JCI108269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Lopez N., Romero R., Xu Y., Miller D., Unkel R., T C.M., Frascoli M., Hassan S.S. Umbilical cord CD71+ erythroid cells are reduced in neonates born to women in spontaneous preterm labor. Am. J. Reprod. Immunol. 2016;76:280–284. doi: 10.1111/aji.12556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Xie Z., Li T., Zhang S., Lai C., Zhu P., Wang K., Han L., Duan Y., Zhao Z. Angiotensin-converting enzyme 2 inhibits lung injury induced by respiratory syncytial virus. Sci. Rep. 2016;6:19840. doi: 10.1038/srep19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C. Clinical characteristics of coronavirus disease 2019 in China. New Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry B.M. COVID-19, ECMO, and lymphopenia: a word of caution. Lancet Resp Med. 2020;8:E24. doi: 10.1016/S2213-2600(20)30119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horby P., Lim W.S., Emberson J., Mafham M., Bell J., Linsell L., Staplin N., Brightling C., Ustianowski A., Elmahi E. Effect of dexamethasone in hospitalized patients with COVID-19: preliminary report. medRxiv. 2020 2020.2006.2022.20137273. [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns J.L., Christopher M.M. Extramedullary hematopoiesis: a new look at the underlying stem cell niche, theories of development, and occurrence in animals. Vet. Pathol. 2012;49:508–523. doi: 10.1177/0300985811432344. [DOI] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.Y., Gao X., Barrasa M.I., Li H., Elmes R.R., Peters L.L., Lodish H.F. PPAR-alpha and glucocorticoid receptor synergize to promote erythroid progenitor self-renewal. Nature. 2015;522:474–477. doi: 10.1038/nature14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Hale J., Bhagia P., Xue F., Chen L., Jaffray J., Yan H., Lane J., Gallagher P.G., Mohandas N. Isolation and transcriptome analyses of human erythroid progenitors. Bfu-e Cfu-e. Blood. 2014;124:3636–3645. doi: 10.1182/blood-2014-07-588806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang Z.D., Yang L., Lian X.Y., Xie Y., Li S., Xin S.Y., Cao P.F., Lu J.H. The MERS-CoV receptor DPP4 as a candidate binding target of the SARS-CoV-2 spike. Iscience. 2020;23:101160. doi: 10.1016/j.isci.2020.101160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., Winter H., Meister M., Veith C., Boots A.W. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. Embo J. 2020;39:e105114. doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J. RBCs as targets of infection. Hematol-am Soc. Hemat. 2014:404–409. doi: 10.1182/asheducation-2014.1.404. [DOI] [PubMed] [Google Scholar]
- Mitra A., Dwyre D.M., Schivo M., Thompson G.R., 3rd, Cohen S.H., Ku N., Graff J.P. Leukoerythroblastic reaction in a patient with COVID-19 infection. Am. J. Hematol. 2020;95:999–1000. doi: 10.1002/ajh.25793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V.J., Feldmann F., Williamson B.N., van Doremalen N., Perez-Perez L., Schulz J., Meade-White K., Okumura A., Callison J., Brumbaugh B. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namdar A., Dunsmore G., Shahbaz S., Koleva P., Xu L., Jovel J., Houston S., Elahi S. CD71(+) erythroid cells exacerbate HIV-1 susceptibility, mediate trans-infection, and harbor infective viral particles. MBio. 2019;10 doi: 10.1128/mBio.02767-19. e02767–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namdar A., Koleva P., Shahbaz S., Strom S., Gerdts V., Elahi S. CD71+ erythroid suppressor cells impair adaptive immunity against Bordetella pertussis. Sci. Rep. 2017;7:7728. doi: 10.1038/s41598-017-07938-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narla A., Dutt S., McAuley J.R., Al-Shahrour F., Hurst S., McConkey M., Neuberg D., Ebert B.L. Dexamethasone and lenalidomide have distinct functional effects on erythropoiesis. Blood. 2011;118:2296–2304. doi: 10.1038/s41598-017-07938-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoye I., Namdar A., Xu L., Crux N., Elahi S. Atorvastatin downregulates co-inhibitory receptor expression by targeting Ras-activated mTOR signalling. Oncotarget. 2017;8:98215–98232. doi: 10.18632/oncotarget.21003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoye I.S., Xu L., Walker J., Elahi S. The glucocorticoids prednisone and dexamethasone differentially modulate T cell function in response to anti-PD-1 and anti-CTLA-4 immune checkpoint blockade. Cancer Immunol. Immunother. 2020;69:1423–1436. doi: 10.1007/s00262-020-02555-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pence B.D. Severe COVID-19 and aging: are monocytes the key? Geroscience. 2020;42:1051–1061. doi: 10.1007/s11357-020-00213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pique-Regi R., Romero R., Tarca A.L., Luca F., Xu Y., Alazizi A., Leng Y., Hsu C.D., Gomez-Lopez N. Does the human placenta express the canonical cell entry mediators for SARS-CoV-2? eLife. 2020;9:e58716. doi: 10.7554/eLife.58716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar M., Rajta P.N., Khatana J. Anemia in chronic obstructive pulmonary disease: prevalence, pathogenesis, and potential impact. Lung India. 2015;32:142–151. doi: 10.4103/0970-2113.152626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbaz S., Bozorgmehr N., Koleva P., Namdar A., Jovel J., Fava R.A., Elahi S. CD71+VISTA+ erythroid cells promote the development and function of regulatory T cells through TGF-beta. Plos Biol. 2018;16 doi: 10.1371/journal.pbio.2006649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim Y.A., Campbell T., Weliwitigoda A., Dosanjh M., Johnson P. Regulation of CD71(+)TER119(+) erythroid progenitor cells by CD45. Exp. Hematol. 2020;86:53–66.e1. doi: 10.1016/j.exphem.2020.05.005. [DOI] [PubMed] [Google Scholar]
- Sodhi C.P., Wohlford-Lenane C., Yamaguchi Y., Prindle T., Fulton W.B., Wang S., McCray P.B., Jr., Chappell M., Hackam D.J., Jia H. Attenuation of pulmonary ACE2 activity impairs inactivation of des-Arg(9) bradykinin/BKB1R axis and facilitates LPS-induced neutrophil infiltration. Am. J. Physiol. Lung Cell Mol Physiol. 2018;314:L17–L31. doi: 10.1152/ajplung.00498.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swann J.W., Koneva L.A., Regan-Komito D., Sansom S.N., Powrie F., Griseri T. IL-33 promotes anemia during chronic inflammation by inhibiting differentiation of erythroid progenitors. J. Exp. Med. 2020;217:e20200164. doi: 10.1084/jem.20200164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z.Y., Xu J., Chen W., Yang Z.T., Xu X.M., Liu L., Chen R.W., Xie J.Y., Liu M.Y., Wu J.Y. Anemia is associated with severe illness in COVID-19: a retrospective cohort study. J. Med. Virol. 2020;93:1478–1488. doi: 10.1002/jmv.26444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Renia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T., Stefanoni D., Dzieciatkowska M., Issaian A., Nemkov T., Hill R.C., Francis R.O., Hudson K.E., Buehler P.W., Zimring J.C. Evidence for structural protein damage and membrane lipid remodeling in red blood cells from COVID-19 patients. medRxiv. 2020;19:4455–4469. doi: 10.1021/acs.jproteome.0c00606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B.T. Acute respiratory distress syndrome in another 50 years: historical footnote or persistent malady? Curr. Opin. Crit. Care. 2017;23:1–3. doi: 10.1097/MCC.0000000000000383. [DOI] [PubMed] [Google Scholar]
- van Lochem E.G., van der Velden V.H., Wind H.K., te Marvelde J.G., Westerdaal N.A., van Dongen J.J. Immunophenotypic differentiation patterns of normal hematopoiesis in human bone marrow: reference patterns for age-related changes and disease-induced shifts. Cytometry B Clin. Cytom. 2004;60:1–13. doi: 10.1002/cyto.b.20008. [DOI] [PubMed] [Google Scholar]
- Vivanti A.J., Vauloup-Fellous C., Prevot S., Zupan V., Suffee C., Do Cao J., Benachi A., De Luca D. Transplacental transmission of SARS-CoV-2 infection. Nat. Commun. 2020;11:3572. doi: 10.1038/s41467-020-17436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292 e286. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Chen W., Zhou Y.-S., Lian J.-Q., Zhang Z., Du P., Gong L., Zhang Y., Cui H.-Y., Geng J.-J. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv. 2020;5:283. doi: 10.1101/2020.03.14.988345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Gu H., Zhao Z., Wang W., Cao B., Lai C., Yang X., Zhang L., Duan Y., Zhang S. Angiotensin-converting enzyme 2 (ACE2) mediates influenza H7N9 virus-induced acute lung injury. Sci. Rep. 2014;4:7027. doi: 10.1038/srep07027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Shivakumar P., Kinder J.M., Way S.S., Donnelly B., Mourya R., Luo Z., Bezerra J.A. Regulation of bile duct epithelial injury by hepatic CD71+ erythroid cells. JCI Insight. 2020;5 doi: 10.1172/jci.insight.135751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang J., An W., Wan Y., Ma S., Yin J., Li X., Gao J., Yuan W., Guo Y. Intron 1 GATA site enhances ALAS2 expression indispensably during erythroid differentiation. Nucleic Acids Res. 2017;45:657–671. doi: 10.1093/nar/gkw901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., He R., Long H., Guo B., Jia Q., Qin D., Liu S.Q., Wang Z., Xiang T., Zhang J. Late-stage tumors induce anemia and immunosuppressive extramedullary erythroid progenitor cells. Nat. Med. 2018;24:1536–1544. doi: 10.1038/s41591-018-0205-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingariello M., Bardelli C., Sancillo L., Ciaffoni F., Genova M.L., Girelli G., Migliaccio A.R. Dexamethasone predisposes human erythroblasts toward impaired lipid metabolism and renders their ex vivo expansion highly dependent on plasma lipoproteins. Front. Physiol. 2019;10:281. doi: 10.3389/fphys.2019.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.