Abstract
Classifying ground-glass lung nodules (GGNs) into atypical adenomatous hyperplasia (AAH), adenocarcinoma in situ (AIS), minimally invasive adenocarcinoma (MIA), and invasive adenocarcinoma (IAC) on diagnostic CT images is important to evaluate the therapy options for lung cancer patients. In this paper, we propose a joint deep learning model where the segmentation can better facilitate the classification of pulmonary GGNs. Based on our observation that masking the nodule to train the model results in better lesion classification, we propose to build a cascade architecture with both segmentation and classification networks. The segmentation model works as a trainable preprocessing module to provide the classification-guided ‘attention’ weight map to the raw CT data to achieve better diagnosis performance. We evaluate our proposed model and compare with other baseline models for 4 clinically significant nodule classification tasks, defined by a combination of pathology types, using 4 classification metrics: Accuracy, Average F1 Score, Matthews Correlation Coefficient (MCC), and Area Under the Receiver Operating Characteristic Curve (AUC). Experimental results show that the proposed method outperforms other baseline models on all the diagnostic classification tasks.
Keywords: pulmonary ground glass opacity nodules, classification, automatic segmentation, joint training, deep learning
1. Introduction
Lung cancer is one of the leading causes of cancer-related deaths in the world. The Lung Cancer Staging Project of International Association for the Study of Lung Cancer (IASLC) [1] showed that there is a significant decrease in survival rate as tumor size increases, indicating that early detection and diagnosis is very effective to reduce the death of patients due to lung cancer. Therapy options for treatment of lung cancer also depend on the type of lung nodules. To address the problem of accurate classification of lung nodules, the IASLC, the American Thoracic Society (ATS), and the European Respiratory Society (ERS) proposed a new classification scheme for lung adenocarcinoma classification [2] in 2011, according to which Ground-glass nodules (GGNs) are classified as atypical adenomatous hyperplasia (AAH), adenocarcinoma in situ (AIS), minimally invasive adenocarcinoma (MIA), or invasive adenocarcinoma (IAC) based on the size of the lesion and the presence of a solid component on pathology analysis (as shown in Fig. 1). The new classification has a significant impact on patient therapy options and follow-up because prognosis varies widely among the different pathologic subtypes [3, 4]. It has been shown in recent studies that patients with early-stage AIS and MIA have a disease-free survival rate of almost 100%, while patients with IACs have a disease-free survival rate of 60–70% [5, 6, 7, 8], necessitating the need for accurate classification of GGNs for planning the therapy option and the extent of resection.
Figure 1:
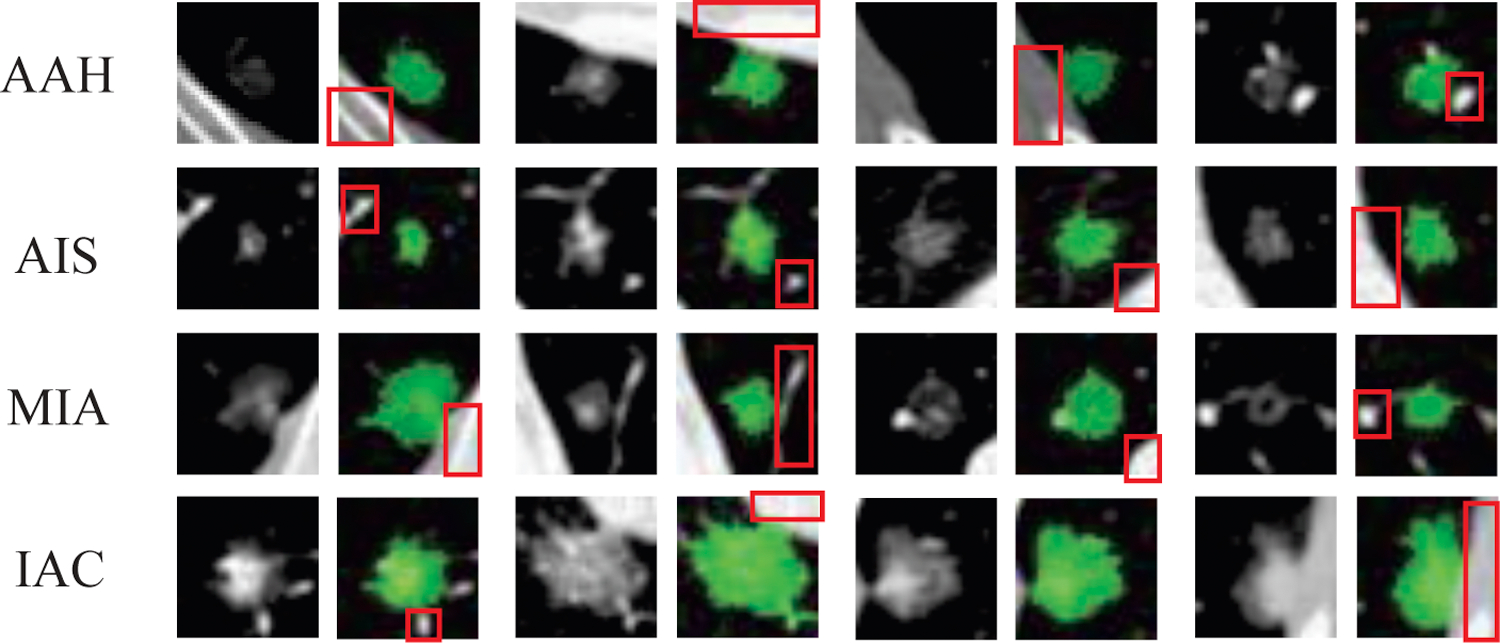
Subtypes of lung nodules - AAH, AIS, MIA and IAC. Green mask is the segmentation mask annotated by experts. Red box contains some interference area for the nodule detection and classification, such as blood vessel, chest wall and rib.
Traditional computer-aided diagnosis (CAD) methods utilize various feature extraction protocols to quantify the appearance of nodules on diagnostic computed tomography (CT) images, and machine learning algorithms have been employed to classify the nodules. Statnikov et al. [9] make a comprehensive comparison of random forests and support vector machines on 22 diagnostic and prognostic cancer datasets. In our previous paper [10], we extract 57 quantified heterogeneity metrics of GGNs and use them to train the SVM to learn and predict the lesion type. In a recent work [11], as many as 1117 features are extracted from 3D nodule CTs. Although these works have achieved impressive performance, extracting appropriate nodule features is very time-consuming and laborious, and these hand-craft features may not be enough for high-level tasks.
Recent deep learning methods can greatly reduce the difficulty of feature extraction and outperforms the methods of hand-crafted feature engineering. With the power of automatic representation learning and end-to-end training, deep learning has achieved remarkable success in several important computer vision tasks, such as image classification [12, 13, 14, 15], object detection [16, 17], and segmentation [18, 19, 20, 21, 22]. Recently, such methods have been extensively applied to medical image analysis [23], especially to the diagnosis of pulmonary adenocarcinoma [24, 25, 26, 27, 28].
Despite different network architectures exploited in these works, one common characteristic among these deep learning models is that they are trained with the raw CT volume or data patch cropped from raw CT volume. Due to the proximity of the nodule to the chest wall or blood vessels passing through the nodule (as shown in the red boxes in Fig. 1), these additional structures on the CT images may result in misclassification of the nodules, leading to errors in diagnosis and therapy. To tackle this problem, deep learning networks with auxiliary tasks, such as detection [24] and segmentation [25], have been integrated with the classification task. These approaches have built a two-branch architecture for the classification and auxiliary tasks respectively, and train the model in a multi-task learning setting. Such two-branch architecture networks solve the classification problem at the ‘feature level’ since the auxiliary task will enforce the network to focus on the nodule region and less on the surrounding structures during the feature extraction step. However, due to the uncertainty and randomness of deep learning training, there is no guarantee that the interference from the surrounding structures will be eliminated.
In this paper, we utilize the segmentation information to aid in the classification of lung nodules. We observe that when we use the expertly segmented CT volumes to training the classification model, the performance is much better than that trained with raw CT volumes. However, creating segmentation maps manually is very time-consuming and of high cost. Thus we propose to first train a segmentation model with the annotated CT data to automatically segment the nodule from raw CT data, which is then used to train the classification model. Motivated by other assembled models [29, 30], we propose to build a cascade architecture with both segmentation and classification models, where the segmentation model is used as a trained data preprocessing module for the classification model. The output of the segmentation model can be considered as the ‘attention’ weight map applied to the data indicating the importance of different regions for the classification task. We jointly fine-tune the whole model to further improve the performance. Since we use the segmentation map directly to mask the background region of raw CT data, the segmentation and classification tasks are done at the ‘data level’. Our model can provide a better trade-off between the classification performance and the cost of data preparation.
We have evaluated our method on 4 classification tasks, defined by different combinations of pathology types. First, we merge AAH and AIS into one class and classify nodules as AAH+AIS, MIA, or IAC. The first two classes are combined because AAH nodules are too few in clinical practice and are usually considered as benign [25]. Second, we classify indolent nodules (AAH+AIS) and invasive nodules (MIA+IAC), which is of great clinical significance since the two types of nodules usually require different treatment plans [2]. Third, we specifically differentiate IAC from other 3 non-IAC (AAH+AIS+MIA) types, as patients with IAC nodules have a much lower disease-free survival rate and require more aggressive surgical treatment. Finally, we differentiate between AIS and MIA because these two types are similar in appearance and very difficult to distinguish even by expert clinicians.
We compare our method with other baselines using 4 classification metrics: Accuracy, Average F1 Score, Matthews Correlation Coefficient (MCC), and Area Under the Receiver Operating Characteristic Curve (AUC, only applied to 2-class problems). Experimental results show that applying segmentation information to the original data in the data level can help improve the performance, and the data-level method behaves more stable than the feature-level method.
The main contributions of this work are 1) Utilization of the segmentation annotation at the data-level for the classification of pulmonary nodules, 2) Training of a segmentation network to mask the input CT data, 3) A cascade architecture consisting of the segmentation and classification models with joint training, 4) Extensive experiments with a large dataset and comparison with a series of baseline models to demonstrate the effectiveness. We also conduct a detailed analysis and explanation on the experimental results and their impact on clinical decision support for surgeons. We believe our paper makes a valuable contribution to the clinical field of classifying GGOs for determining the optimal therapy and the resection margins for sub-lobar resections.
The rest part of this paper is organized as follows: in Section 2 we describe our cascade architecture model consisting of the classification and segmentation models, and the way to assemble them. In Section 3 we present the experimental setting, data, results, and analysis. In Section 4 we present the conclusions and discuss our future work.
2. Method
In this section, we first describe in detail the classification and segmentation networks. Then, we introduce the entire architecture of the cascade model.
2.1. Classification Model
The classification model is composed of 8 convolution blocks, 3 downsampling blocks, and 3 fully-connected blocks, as shown in Fig. 2a. Each convolution block consists of a 3D convolution layer with kernel size 3 × 3 × 3 and stride size [1,1,1], followed by batch normalization [31] layer and ReLU as the nonlinear activation function. Output channels of the 8 convolution blocks are 16, 16, 32, 32, 64, 64, 256 and 256, respectively. After every 2 convolution blocks, we downsample the feature map by max-pooling of scale [2,2,2]. The output of the last convolution block is reshaped and input to 3 fully-connected (fc) blocks. Output dimension of the first two fc blocks is 256 and the nonlinear activation function is ReLU. Output dimension of the last fc block is equal to the number of nodule pathology types and softmax function is exploited as the activation function. The classification model outputs the probability vector indicating the probability that the input data belongs to each nodule type. Dropout [32] with zero rate of 0.1 is exploited after the first two fc blocks to avoid overfitting. Training is performed by minimizing the cross-entropy loss Lcla between the model output and true class label in one-hot form as follows:
| (1) |
where and are the cth element of ground-truth class label and model prediction, respectively, N is the number of training samples.
Figure 2:
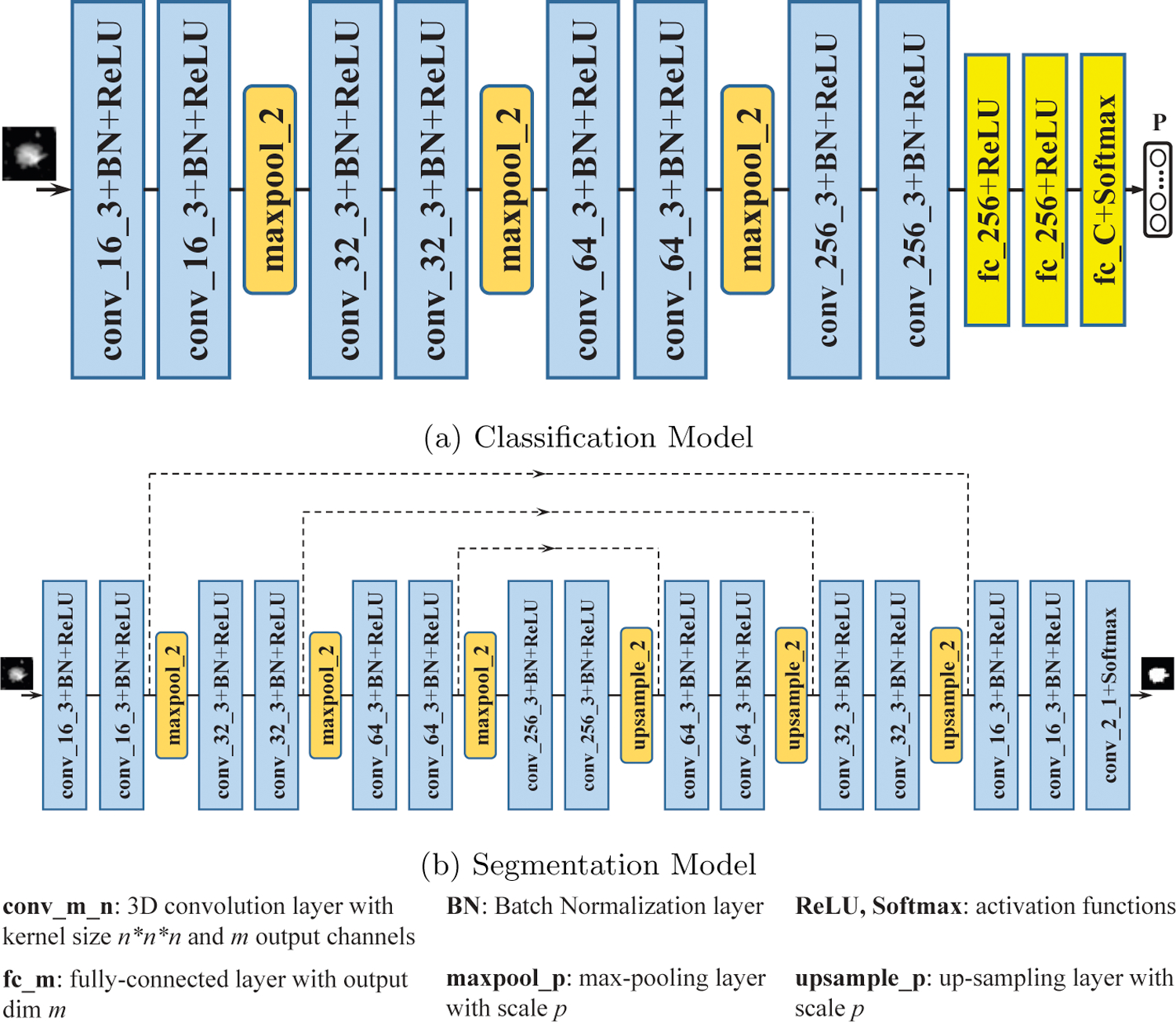
The classification model (a) is 3D CNN containing 8 convolution blocks, 3 max-pooling layers and 3 fully-connected blocks. The segmentation model (b) is a 3D U-Net with 15 convolution blocks, 3 max-pooling layers and 3 up-sampling layers. Dilated-convolution is exploited in the segmentation model to increase the receptive field.
2.2. Segmentation Model
We build a 3D U-Net [20] architecture for automatic lung nodule segmentation, as shown in Fig. 2b. The segmentation model consists of 15 3D convolution blocks, with 8 in the encoder (contracting) path and 7 in the decoder (expanding) path. Each convolution block contains a 3D convolution layer with kernel size 3 × 3 × 3, which is followed by a batch normalization layer and ReLU as the nonlinear activation function except for the last block where kernel with size 1 × 1 × 1 and softmax function are used. Dilated convolution is exploited in the segmentation model to increase the receptive field thus making full use of spatial context. Dilation factors are set as [1,1,2,2,2,2,4,4,2,2,2,2,1,1,1] in the 15 convolution layers, respectively. The stride and padding are chosen accordingly to make the size of the output feature identical to that of the input. There are 3 max-pooling and 3 up-sampling layers of scale [2,2,2] in the encoder and decoder paths. The feature map before each max-pooling layer in the encoder path is skip-connected and concatenated to the corresponding feature map after the up-sampling layer in the decoder path. The last convolution layer uses the channel-wise softmax function to output a dense segmentation map with C channels (with background included as 1 channel). Dice loss [21] Lseg is used for the segmentation model training, and is given as:
| (2) |
where and are the ith element of ground-truth and model predicted nodule segmentation map, respectively, n is a small value used for numerical stability and N is the number of training samples.
2.3. Assembling Classification and Segmentation Model
Existing deep learning models for classifying pulmonary nodules are trained with raw CT volume or data patch cropped from raw CT volume, as is shown in Fig. 3a. The input CT volume contains not only the lung nodule but also other interference regions, such as blood vessels, chest wall, and rib. The existence of these regions could introduce erroneous features in the training and test data, which will lead to misclassification of the lung nodules and poor generalizability of the model.
Figure 3:
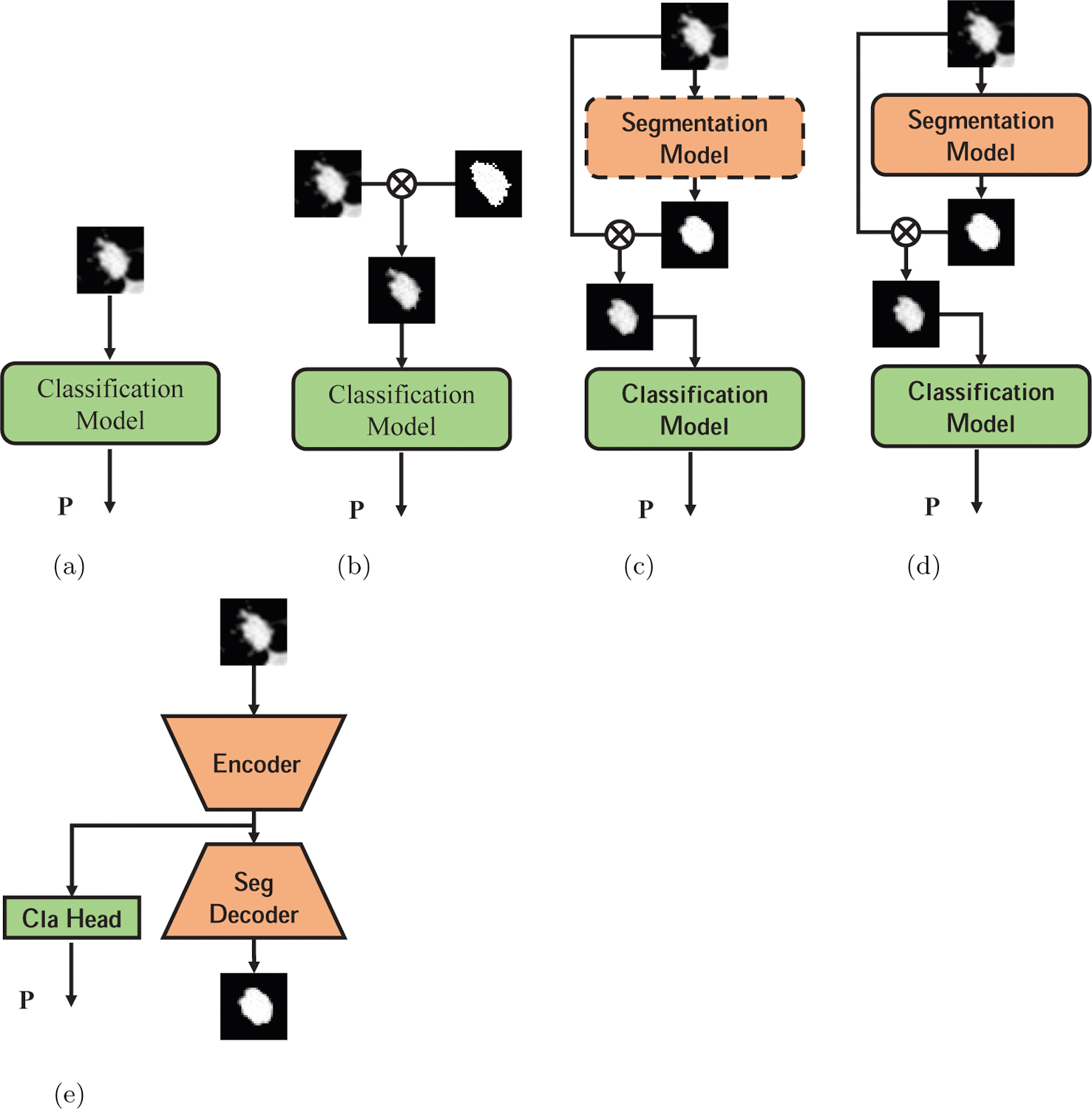
Different deep learning models used in this paper. (a) Train classification model with raw CT data. (b) Train classification model with data masked by ground-truth segmentation. (c) Train segmentation model first, then train classification model with data masked by automatically generated segmentation map, dash rectangle means the model parameters are fixed. (d) Jointly train classification model and segmentation model. (e) Multi-task learning model with segmentation and classification.
In this paper, we utilize the segmentation mask to remove the interference information. The segmentation map of the nodule annotated by an expert clinician is used to mask the background in the original CT data, which is performed as follows:
| (3) |
where draw denotes original CT data, s denotes the binary segmentation map, 0 corresponds to the background region and 1 corresponds to the nodule region, and denotes element-wise (Hadamard) product. We observe that when we use the masked data to train and evaluate the classification model, its performance is significantly better than that of the model trained with the original data, as shown in Fig. 3b. However, this method requires manual lung nodule segmentation by an expert before diagnosis, which is time-consuming and tedious. Based on this observation and motivated by other assembled models [29, 14, 30], we propose to build a cascade model with both the segmentation and classification networks. We first train a segmentation model with the provided segmentation map and then freeze its parameters. The output of the segmentation model is a volume with values between 0 and 1 indicating the probability that each pixel belongs to the nodule region. We use it, in the same way, to mask the original data and use the masked data to train and evaluate the classification model, as shown in Fig. 3c. Finally, we jointly fine-tune the whole model by minimizing the weighted sum of Dice loss of segmentation network and cross-entropy loss of classification network as follows (see Fig. 3d):
| (4) |
In the joint model, the output of segmentation model can be considered as the ‘attention’ weight map applied to the data, indicating the importance of different regions. Through joint training, the segmentation model will be trained to not only provide an accurate segmentation map of a nodule but also focus more on the region that is more useful for the classification task (such as the regions that are more discriminative between different types), making it easier to train the classification model.
The idea of our method is similar to that of [24, 25], as we use additional supervisory information to assist in the training of the classification model. Existing works build a two-branch architecture for the classification task and the auxiliary task respectively and train the model in a multi-task learning setting. The auxiliary task will enforce the network to focus on the nodule and less on the surrounding anatomy in the feature extraction step, therefore attempting to improve the classification accuracy at the ‘feature level’. However, in this paper, we apply the segmentation map directly to raw CT data by Hadamard product to reduce the influence of background region for the classification problem, therefore working at the ‘data level’ to improve the classification.
In this paper, we also build a network with two branches for classification and segmentation respectively for comparison. We use the same U-Net architecture shown in Fig. 2b for segmentation and add a branch after the bottleneck convolution layer (the 8th convolution layer shown in Fig. 2b for classification. The classification branch consists of 3 fully-connected blocks similar to the architecture shown in Fig. 2a. The multi-branch model is trained by minimizing the sum of classification and segmentation loss, see Fig. 3e.
3. Experiments
3.1. Data Collection and Preprocessing
In this study, non-contrast enhanced CT images of the patient before surgery are collected, with the mean interval between the CT examination and surgery of 13 days. The CT volume is acquired with the patients in the supine position, covering the area from the top to the base of the lung, including the chest wall and axillary fossa. The chest CT imaging is performed using 4 scanners: GE Discovery CT750 HD, 64-slice LightSpeed VCT (GE Medical Systems), Somatom Definition flash and Somatom Sensation-16 (Siemens Medical Solutions). The scan parameters were: section width, 1.25 mm; reconstruction interval, 1.25 mm; tube voltage, 120 kV; tube current, 100 – 200 mAs; pitch, 0.75 – 1.5; collimation, 1 – 1.25 mm; display FOV, 28 × 28 cm to 36 × 36 cm; matrix size, 512 × 512; and pixel size, 0.55 – 0.7 mm, respectively. All CT volumes are reconstructed with a medium sharp reconstruction algorithm with a thickness of 1 – 1.25 mm.
3D Slicer [33] (version 4.8.0), a medical image processing and navigation software, is used to segment the volume of interest (VOI) of the nodules. Segmentation is performed by one experienced radiologist and then confirmed by another. Large vessels and bronchioles are excluded as much as possible from the volume of the nodule. The lung CT data is first loaded into the Slicer software in DICOM (Digital Imaging and Communications in Medicine) format for segmentation, and then the images with VOI information are converted to NII format for the next step of processing. Each segmented nodule is given a specific pathology label (one of AAH, AIS, MIA, and IAC), according to the detailed pathology report post surgical resection of the nodule.
A total of 740 CT volumes of subcentimeter nodules with nodule segmentation maps and pathology labels are collected for experiments. To keep the physical meaning behind the voxel identical over all the cases, we first resampled the data to 0.7mm×0.7mm×1.0mm spacing. The CT volumes and ground-truth segmentation maps are resampled with trilinear and nearest-neighbor interpolation methods respectively. We then cropped a cubic patch of size 32 × 32 × 32 around the center of the lung nodule indicated by the expertly annotated segmentation map. To reduce the ambiguity of the grayscale distribution in the data, intensities are first clipped between [−1024, 400] and normalized to the range of [−1,1] by
| (5) |
3.2. Deep Learning Model Setting
The distribution of the 740 nodules is: 32 AAH, 193 AIS, 335 MIA, and 180 IAC nodules. We randomly select 70% samples of each category for training and the rest 30% for testing. In total, we get 515 training samples and 225 test samples. Detailed numbers of data for training and testing of each category are listed in Table 1. In the training dataset, we randomly choose 20% as the validation set. In other words, we have 56%, 14%, and 30% for training, validation, and testing. We carefully adjust part samples of each set to make sure each set contains nodules from different patients.
Table 1:
Number for training and test cases for each category.
| Training | Testing | Total | |
|---|---|---|---|
| AAH | 21 | 11 | 32 |
| AIS | 135 | 58 | 193 |
| MIA | 234 | 101 | 335 |
| IAC | 125 | 55 | 180 |
| Total | 515 | 225 | 740 |
To avoid overfitting, we perform several ways of data augmentation: 1) flipping the volume by a random axis with 50% probability, 2) rotating the volume around a random axis by 90 increments, 3) adding Gaussian noise. For the full use of training data, augmentation is performed on the fly during the training process.
We use Adam optimizer [34] to train all the models. For the training of segmentation and classification models, the initial learning rate is set to 0.0001 and is reduced by a factor of 0.8 if the model performance on the test dataset doesn’t increase in 10 epochs. For joint fine-tuning, the initial learning rate is set to 0.00001 and is reduced by 0.5 every 30 epochs. The maximum number of training epochs for the segmentation model, the classification model, and joint fine-tuning is 150, 120, and 120, respectively, and training is stopped if the maximum training epoch is reached or the performance doesn’t increase within 50 epochs. To avoid overfitting, dropout [32] with zero rate of 0.1 is used for the first two fc blocks of the classification model and L2 regularization with weight 0.0001 is applied to all models. No dropout is used in the segmentation model. The parameters of the weight are randomly initialized by the He method [35] and bias is initialized to zero, except for the last convolution layer of the segmentation model, where we follow the initialization in [36] to tackle the imbalance in the number of pixels between the lung nodule and background region. During training, we use a mini-batch of 24 volumes and after each training epoch, we evaluate the model on the test dataset. The best test result is recorded. For each model, we repeatedly ran the experiments for 5 times and the best results are reported.
3.3. Evaluation Metrics
We applied our method on 4 classification tasks, defined by different combinations of pathology subtypes:
1). AAH+AIS, MIA, IAC:
We merge AAH and AIS into one class and classify AAH+AIS, MIA, and IAC nodules. This is because AAH nodules are relatively less compared to the other three subtypes and the trained model may not work well on this category, which will influence the evaluation of the model’s overall performance. Further, merging the two subtypes is still reasonable from a clinical point of view [2, 25].
2). AAH+AIS, MIA+IAC:
We classify indolent nodules (AAH+AIS) and invasive nodules (MIA+IAC) in this task. This is of great clinical significance since the two types of nodules usually require completely different treatment options [2].
3). AAH+AIS+MIA, IAC:
We differentiate IAC from other 3 non-IAC (AAH+AIS+MIA) types in this task, as patients with IAC nodules have a much lower disease-free survival rate and thus require more aggressive surgical treatment and subsequent adjuvant chemotherapy treatment [2, 25].
4). AIS, MIA:
In this task, we differentiate between AIS and MIA types. As mentioned above, these two types of nodules require different treatment plans. However, on the preoperative CT images, they are similar in appearance and difficult to distinguish, even by expert radiologists.
We evaluate our method and compare it with other baselines using several metrics: Accuracy, Average F1 Score (AveF1), and Matthews Correlation Coefficient (MCC). For the two-category task, we also use the Receiver Operating Characteristic Curve and calculate the area under the curve (AUC).
3.4. Results and Discussion
We evaluate 5 different models, as shown in Fig. 3, for the 4 classification tasks using the 4 metrics. Note that the weight factor of the joint training model is set to 1 here and the effect of different values is evaluated in the following part. In order to prove the generalizability of our method, we have conducted experiments using two different data splits. The results of the two data splits are listed in Table 2 and 3 respectively. From the results, we can see that when we use the ground-truth nodule segmentation map to mask the data and then use them to train the classification model, the performance is much better than that of the model trained with the original data for all 4 metrics (see columns (a) and (b) under each metric). Using the segmentation mask for the nodule helped improve the accuracy of classifying the nodules. The model trained with the data masked by generated segmentation map performs better than that trained with original data but is not as good as the expertly annotated segmentation map (see column (c) under each metric). This is understandable because the automatic segmentation is not as accurate as the expert segmentation. Accurate segmentation of the lung nodule will extract all the nodule features. The segmentation map generated by the U-Net model is not as accurate as the ground-truth, thus some nodule features are missing or additional interference information from the surrounding anatomy is included. This is the reason why the model trained with generated segmentation masked data performs worse. In most cases, jointly fine-tuning the segmentation and classification model together can further improve the performance. Although there exist some exceptions (such as the AUC of task ‘AIS, MIA’ of data split 1 and the ‘MCC’ of task ‘AAH+AIS, MIA+IAC’ of split 2), such situations are very few, and the degradation of performance is not obvious (see column (d) under each metric). The multi-task learning model with segmentation can help improve the classification task, but the improvement is not as large and stable as our model (see column (e) under each metric). The 3-class confusion matrices of different models are shown in Fig. 4 and ROC curves of the 3 two-class tasks are shown in Fig. 5.
Table 2:
Statistical Results of Different Models on Different Metrics with Data Split 1. (a) Train classification model with raw CT data. (b) Train classification model with data masked by ground-truth segmentation. (c) Train classification model with data masked by automatically generated segmentation map (ours1). (d) Jointly train classification model and segmentation model (ours2). (e) Multi-task learning model with segmentation and classification.
| Accuracy(%) | AveF1(%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (a) | (b) | (c)(ours1) | (d)(ours2) | (e) | (a) | (b) | (c)(ours1) | (d)(ours2) | (e) | |
| 1) | 55.73±0.34 | 59.11±0.33 | 57.78±0.24 | 58.67±0.27 | 57.33±0.51 | 55.19±0.35 | 58.88±0.38 | 58.04±0.31 | 58.30±0.31 | 56.72±0.46 |
| 2) | 74.22±0.37 | 78.22±0.31 | 76.89±0.25 | 78.22±0.27 | 74.33±0.31 | 74.27±0.31 | 78.62±0.37 | 75.69±0.29 | 77.53±0.25 | 72.06±0.42 |
| 3) | 82.22±0.31 | 83.56±0.35 | 82.67±0.27 | 82.67±0.27 | 81.33±0.45 | 80.74±0.31 | 82.27±0.39 | 81.12±0.25 | 81.32±0.25 | 80.74±0.41 |
| 4) | 67.29±0.51 | 72.32±0.38 | 71.70±0.31 | 72.30±0.30 | 67.67±0.33 | 67.29±0.43 | 72.95±0.35 | 70.34±0.35 | 72.22±0.21 | 65.78±0.49 |
| MCC(×10−1) | AUC(×10−1) | |||||||||
| (a) | (b) | (c)(ours1) | (d)(ours2) | (e) | (a) | (b) | (c)(ours1) | (d)(ours2) | (e) | |
| 1) | 3.259±0.057 | 3.561±0.054 | 3.415±0.037 | 3.430±0.035 | 3.238±0.047 | - | - | - | - | - |
| 2) | 4.121±0.039 | 5.128±0.031 | 4.467±0.041 | 4.828±0.037 | 4.239±0.045 | 7.441±0.031 | 8.140±0.038 | 7.943±0.039 | 8.129±0.031 | 7.483±0.045 |
| 3) | 4.692±0.033 | 5.134±0.031 | 4.819±0.025 | 4.852±0.021 | 4.548±0.038 | 7.852±0.031 | 8.044±0.036 | 7.884±0.039 | 7.998±0.041 | 7.802±0.046 |
| 4) | 3.000±0.039 | 4.051±0.039 | 3.931±0.021 | 4.029±0.021 | 3.120±0.048 | 6.557±0.023 | 7.542±0.035 | 7.437±0.031 | 7.412±0.034 | 6.809±0.048 |
Table 3:
Statistical Results of Different Models on Different Metrics with Data Split 2. (a) Train classification model with raw CT data. (b) Train classification model with data masked by ground-truth segmentation. (c) Train classification model with data masked by automatically generated segmentation map (ours1). (d) Jointly train classification model and segmentation model(ours2). (e) Multi-task learning model with segmentation and classification.
| Accuracy(%) | AveF1(%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (a) | (b) | (c)(ours1) | (d)(ours2) | (e) | (a) | (b) | (c)(ours1) | (d)(ours2) | (e) | |
| 1) | 54.67±0.31 | 57.78±0.36 | 56.00±0.25 | 54.67±0.21 | 52.67±0.41 | 52.90±0.31 | 57.74±0.37 | 54.40±0.21 | 53.79±0.21 | 51.17±0.41 |
| 2) | 72.00±0.33 | 74.67±0.36 | 72.89±0.29 | 73.78±0.21 | 72.56±0.31 | 70.31±0.31 | 73.86±0.25 | 71.00±0.21 | 71.48±0.23 | 70.75±0.46 |
| 3) | 82.67±0.31 | 84.00±0.34 | 83.11±0.36 | 83.11±0.36 | 82.22±0.41 | 81.20±0.33 | 82.67±0.33 | 81.98±0.24 | 81.70±0.24 | 82.10±0.36 |
| 4) | 64.78±0.41 | 71.07±0.33 | 67.92±0.29 | 67.29±0.35 | 65.41±0.35 | 64.33±0.31 | 69.10±0.32 | 64.69±0.27 | 66.33±0.29 | 64.94±0.34 |
| MCC | AUC | |||||||||
| (a) | (b) | (c)(ours1) | (d)(ours2) | (e) | (a) | (b) | (c)(ours1) | (d)(ours2) | (e) | |
| 1) | 2.697±0.056 | 3.418±0.049 | 2.743±0.047 | 2.750±0.041 | 2.731±0.057 | - | - | - | - | - |
| 2) | 3.204±0. | 4.033±0.031 | 3.232±0.029 | 3.210±0.026 | 3.217±0.037 | 7.052±0.036 | 7.469±0.035 | 7.157±0.031 | 7.154±0.025 | 7.057±0.041 |
| 3) | 4.819±0.033 | 5.263±0.033 | 5.018±0.029 | 4.978±0.024 | 4.821±0.031 | 7.921±0.043 | 8.106±0.044 | 7.942±0.047 | 8.008±0.041 | 7.934±0.055 |
| 4) | 2.351±0.031 | 3.393±0.030 | 2.530±0.029 | 2.657±0.027 | 2.281±0.031 | 6.072±0.025 | 6.882±0.025 | 6.591±0.021 | 6.567±0.027 | 6.148±0.037 |
Figure 4:
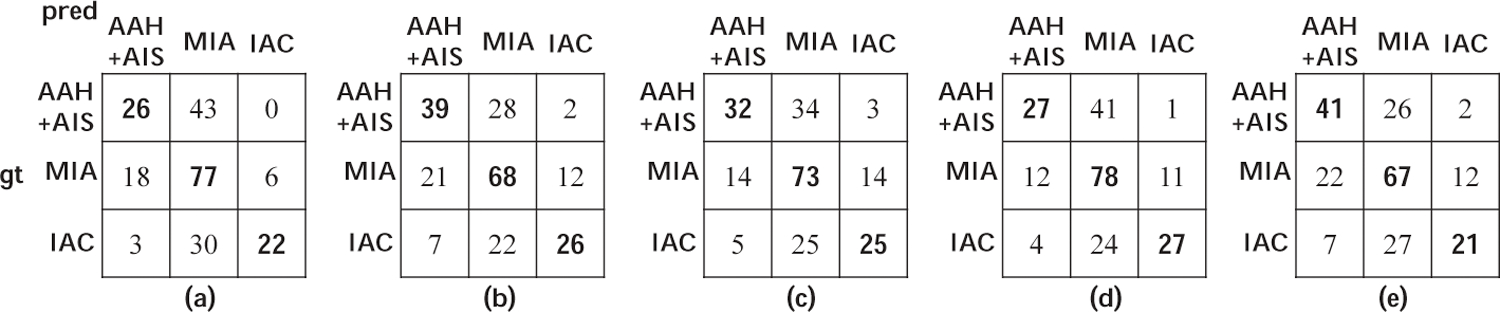
Confusion matrices of the task ‘AAH+AIS, MIA, IAC’ of 5 different models in data oplit 1. Element (i, j) means the number of cases that belong to the class i but are identified as class j.
Figure 5:
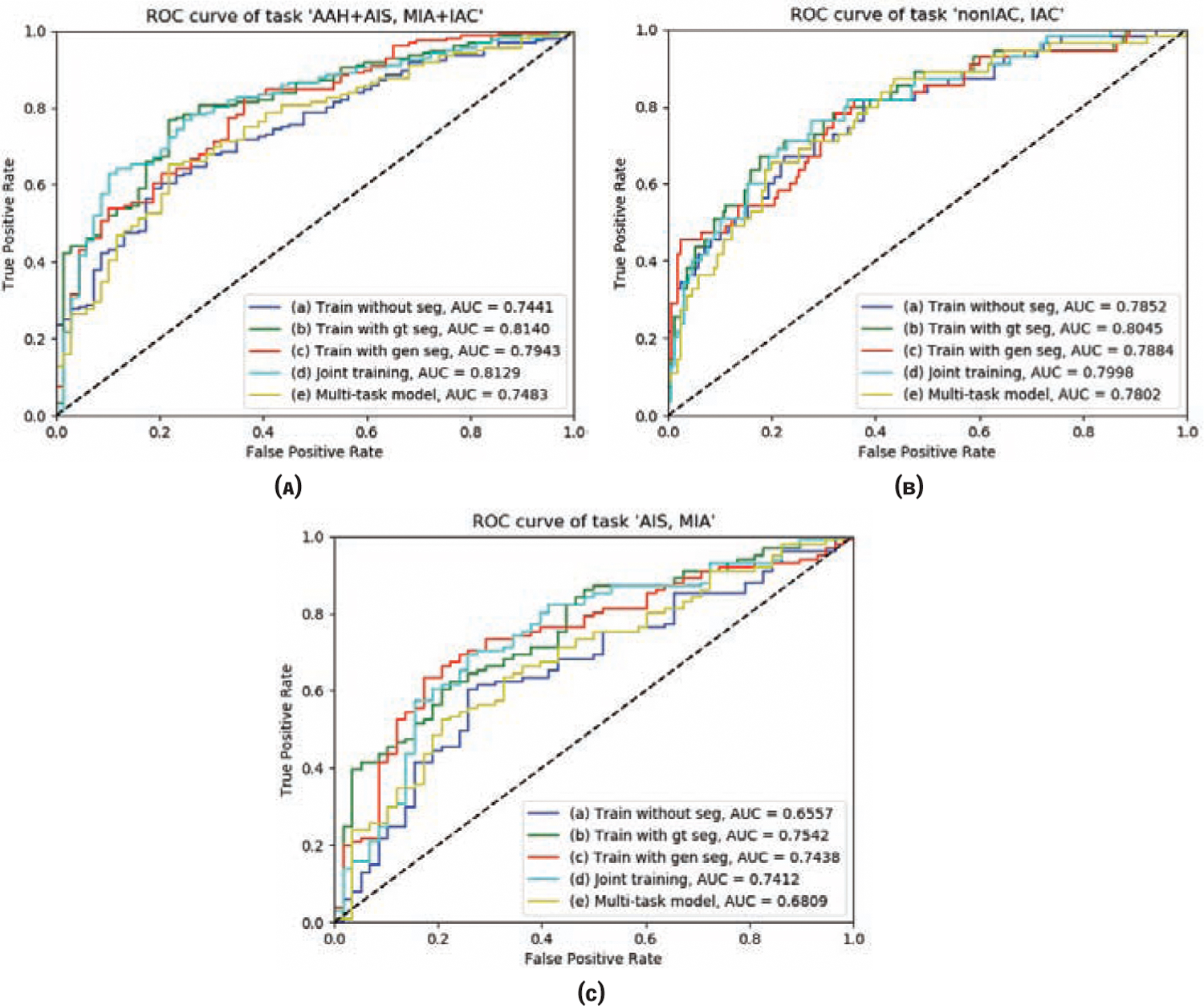
ROC curves of 5 different models of the 3 two-class task: ‘AAH+AIS, MIA+IAC’, ‘AAH+AIS+MIA(nonIAC), IAC’ and ‘AIS, MIA’.
To further prove the effectiveness of our proposed method, we conduct another comparative experiment. We first train a classifier with manually segmented CT data. Then we train an automatic segmentation model. Finally, we apply the classifier to the output of the segmentation model. We follow the same training setting and evaluate on the same 2 data splits for a fair comparison. The results are listed in Table 4. This method can also provide better results than those of the model trained with raw CT data. In most cases, however, our model still performs the best.
Table 4:
Statistical results of applying classifier trained with manually segmented data to automatically segmented data.
| split1 | Accuracy(%) | AveF1(%) | MCC | AUC |
|---|---|---|---|---|
| 1) | 57.67±0.31 | 57.91±0.30 | 3.399±0.045 | - |
| 2) | 76.57±0.34 | 75.22±0.34 | 4.385±0.030 | 7.914±0.031 |
| 3) | 82.47±0.31 | 81.05±0.37 | 4.823±0.025 | 7.815±0.025 |
| 4) | 71.25±0.31 | 70.13±0.30 | 3.857±0.029 | 7.432±0.021 |
| split2 | Accuracy(%) | AveF1(%) | MCC | AUC |
| 1) | 55.85±0.36 | 54.19±0.35 | 2.717±0.049 | - |
| 2) | 72.97±0.24 | 70.90±0.24 | 3.215±0.031 | 7.132±0.023 |
| 3) | 83.01±0.33 | 81.67±0.26 | 5.015±0.039 | 7.967±0.034 |
| 4) | 67.55±0.39 | 64.27±0.35 | 2.507±0.029 | 6.574±0.031 |
To evaluate the effect of the weight factor λ in our joint training model, we test 5 different values, which are 0.5, 0.7, 1.0, 1.3, and 1.5. We only evaluate on the first classification task (AAH+AIS and MIA+IAC) with data split 1 for simplicity. The results are listed in Table 5. We can see that the variation between different values of weight is not obvious, meaning that our proposed method is not very sensitive to the weight within a certain range.
Table 5:
Statistical results of our joint training model (d) on the task 2) ‘AAH+AIS and MIA+IAC’ with different values of weight λ.
| λ | Accuracy(%) | AveF1(%) | MCC | AUC |
|---|---|---|---|---|
| 0.5 | 78.09±0.31 | 77.33±0.30 | 4.817±0.030 | 8.115±0.029 |
| 0.7 | 78.17±0.25 | 77.42±0.24 | 4.823±0.030 | 8.122±0.031 |
| 1.0 | 78.22±0.27 | 77.53±0.25 | 4.828±0.037 | 8.129±0.031 |
| 1.3 | 78.20±0.25 | 77.51±0.25 | 4.828±0.035 | 8.128±0.030 |
| 1.5 | 78.24±0.29 | 77.54±0.25 | 4.830±0.035 | 8.130±0.033 |
We also compare with a traditional method proposed in [10], which collects 248 lung volumes for experiments and exploits the SVM as the classifier. We evaluate our model with the same dataset and on the same 3 tasks as [10] for a fair comparison. The results are listed in Table 6. It can be seen that our model consistently outperforms the traditional method as well as the radiologists.
Table 6:
Comparative results of 3 different tasks with radiologists and traditional SVM in Accuracy(%)
To explore the relationship between the segmentation and classification tasks in the joint training, we illustrate key segmentation results in Fig. 6. Here, we highlight the classification task of ‘AAH+AIS, MIA+IAC’ as an example. The green mask is the ground-truth segmentation while the red and blue masks are the automatic segmentation maps generated by the U-Net before and after joint training, respectively. The Dice and confidence scores of classification for each case are also given under each sample. From Fig. 6, we can see that the confidence scores of classification for all samples increases, which indicates the effectiveness of the joint training. In the cascade model, the output of the segmentation model can be considered as the ‘attention’ weight map applied to the data, indicating the importance of different regions for the classification. However, not all features are useful for the classification task. For example, some features are shared by all the nodule categories. These features are not discriminative and will not contribute to the training of the classification model. Through joint training, the segmentation model will be trained to focus more on the discriminative region of the nodule and ignore the region with a similar appearance between different nodule types that are less useful for classification. In this case, the Dice score after joint training will decrease. On the other hand, the joint training will also help the segmentation model find some region that is important for classification but is not correctly segmented before joint training. In this case, the Dice score will increase.
Figure 6:

Relationship between the segmentation and classification results of joint training (Take task ‘AAH+AIS, MIA+IAC’ as an example). For each case, the first image is the original data. The green mask is the ground-truth segmentation. The red mask and blue mask are the automatic segmentation map generated by the U-Net before joint training and after joint training, respectively. The Dice score and confidence score of classification of each case are also given.
4. Conclusion and Future Work
In this paper, we find that the best classification result is obtained when the segmentation map is used as a mask to remove the interference information around the nodule on the CT volume. When we train a segmentation model with the provided segmentation maps and use its output to mask the original data, the classification model trained with such data still performs better than the model trained without the segmentation mask. Using this insight, we build a cascade architecture with both segmentation and classification networks, and jointly fine-tune the whole model. The cascade model in the data level performs better and more stable than the multi-task learning model. The limitation of our method is that the performance of the segmentation model can’t be maintained after the joint training, as is shown above. This makes our method not suitable for the situation where both good segmentation and classification are required. So, in the future, we will explore more advanced methods in the multi-task learning field to aggregate the two tasks to obtain better results in both of them. Since we may consider the data masked by ground-truth segmentation map and generated segmentation map as the source data domain and target domain, we can also combine our method with transfer learning or domain adaptation techniques, which will also be part of our future work.
Acknowledgements
This project is supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health through Grant Numbers P41EB015898 and R01EB025964, and China Scholarship Council (CSC). Unrelated to this publication, Jayender Jagadeesan owns equity in Navigation Sciences,Inc. He is a co-inventor of a navigation device to assist surgeons in tumor excision that is licensed to Navigation Sciences. Dr. Jagadeesan’s interests are reviewed and are managed by BWH and Partners Healthcare in accordance with their conflict of interest policies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Rami-Porta R, Bolejack V, Crowley J, Ball D, Kim J, Lyons G, et al. , The iaslc lung cancer staging project: proposals for the revisions of the t descriptors in the forthcoming eighth edition of the tnm classification for lung cancer, Journal of Thoracic Oncology 10 (7) (2015) 990–1003. [DOI] [PubMed] [Google Scholar]
- [2].Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, et al. , International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma, Journal of thoracic oncology 6 (2) (2011) 244–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Travis WD, Brambilla E, Van Schil P, Scagliotti GV, Huber RM, Sculier J-P, et al. , Paradigm shifts in lung cancer as defined in the new iaslc/ats/ers lung adenocarcinoma classification (2011). [DOI] [PubMed]
- [4].Travis WD, Brambilla E, Riely GJ, New pathologic classification of lung cancer: relevance for clinical practice and clinical trials, Journal of clinical oncology 31 (8) (2013) 992–1001. [DOI] [PubMed] [Google Scholar]
- [5].Watanabe S.-i, Watanabe T, Arai K, Kasai T, Haratake J, Urayama H, Results of wedge resection for focal bronchioloalveolar carcinoma showing pure ground-glass attenuation on computed tomography, The Annals of thoracic surgery 73 (4) (2002) 1071–1075. [DOI] [PubMed] [Google Scholar]
- [6].Vazquez M, Carter D, Brambilla E, Gazdar A, Noguchi M, Travis WD, et al. , Solitary and multiple resected adenocarcinomas after ct screening for lung cancer: histopathologic features and their prognostic implications, Lung Cancer 64 (2) (2009) 148–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Borczuk AC, Qian F, Kazeros A, Eleazar J, Assaad A, Sonett JR, et al. , Invasive size is an independent predictor of survival in pulmonary adenocarcinoma, The American journal of surgical pathology 33 (3) (2009) 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yim J, Zhu L-C, Chiriboga L, Watson HN, Goldberg JD, Moreira AL, Histologic features are important prognostic indicators in early stages lung adenocarcinomas, Modern pathology 20 (2) (2007) 233. [DOI] [PubMed] [Google Scholar]
- [9].Statnikov A, Wang L, Aliferis CF, A comprehensive comparison of random forests and support vector machines for microarray-based cancer classification, BMC bioinformatics 9 (1) (2008) 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Li M, Narayan V, Gill RR, Jagannathan JP, Barile MF, Gao F, et al. , Computer-aided diagnosis of ground-glass opacity nodules using open-source software for quantifying tumor heterogeneity, American Journal of Roentgenology 209 (6) (2017) 1216–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gong J, Liu J.-y., Hao W, Nie S.-d., Wang S, Peng W, Computer-aided diagnosis of ground-glass opacity pulmonary nodules using radiomic features analysis, Physics in Medicine & Biology. [DOI] [PubMed]
- [12].Krizhevsky A, Sutskever I, Hinton GE, Imagenet classification with deep convolutional neural networks, in: Advances in neural information processing systems, 2012, pp. 1097–1105.
- [13].Simonyan K, Zisserman A, Very deep convolutional networks for large-scale image recognition, arXiv preprint arXiv:1409.1556.
- [14].He K, Zhang X, Ren S, Sun J, Deep residual learning for image recognition, in: Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 770–778. [Google Scholar]
- [15].Huang G, Liu Z, Van Der Maaten L, Weinberger KQ, Densely connected convolutional networks, in: Proceedings of the IEEE conference on computer vision and pattern recognition, 2017, pp. 4700–4708. [Google Scholar]
- [16].Ren S, He K, Girshick R, Sun J, Faster r-cnn: Towards real-time object detection with region proposal networks, in: Advances in neural information processing systems, 2015, pp. 91–99. [DOI] [PubMed]
- [17].Redmon J, Divvala S, Girshick R, Farhadi A, You only look once: Unified, real-time object detection, in: Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 779–788. [Google Scholar]
- [18].Long J, Shelhamer E, Darrell T, Fully convolutional networks for semantic segmentation, in: Proceedings of the IEEE conference on computer vision and pattern recognition, 2015, pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- [19].Chen L-C, Papandreou G, Kokkinos I, Murphy K, Yuille AL, Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs, IEEE transactions on pattern analysis and machine intelligence 40 (4) (2017) 834–848. [DOI] [PubMed] [Google Scholar]
- [20].Ronneberger O, Fischer P, Brox T, U-net: Convolutional networks for biomedical image segmentation, in: International Conference on Medical image computing and computer-assisted intervention, Springer, 2015, pp. 234–241. [Google Scholar]
- [21].Milletari F, Navab N, Ahmadi S-A, V-net: Fully convolutional neural networks for volumetric medical image segmentation, in: 2016 Fourth International Conference on 3D Vision (3DV), IEEE, 2016, pp. 565–571. [Google Scholar]
- [22].Jiang J, Hu Y-C, Liu C-J, Halpenny D, Hellmann MD, Deasy JO, et al. , Multiple resolution residually connected feature streams for automatic lung tumor segmentation from ct images, IEEE transactions on medical imaging 38 (1) (2018) 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. , A survey on deep learning in medical image analysis, Medical image analysis 42 (2017) 60–88. [DOI] [PubMed] [Google Scholar]
- [24].Wang S, Wang R, Zhang S, Li R, Fu Y, Sun X, et al. , 3d convolutional neural network for differentiating pre-invasive lesions from invasive adenocarcinomas appearing as ground-glass nodules with diameters 3 cm using hrct, Quantitative imaging in medicine and surgery 8 (5) (2018) 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhao W, Yang J, Sun Y, Li C, Wu W, Jin L, Yang Z, Ni B, Gao P, Wang P, et al. , 3d deep learning from ct scans predicts tumor invasiveness of subcentimeter pulmonary adenocarcinomas, Cancer research 78 (24) (2018) 6881–6889. [DOI] [PubMed] [Google Scholar]
- [26].Zhao W, Yang J, Ni B, Bi D, Sun Y, Xu M, et al. , Toward automatic prediction of egfr mutation status in pulmonary adenocarcinoma with 3d deep learning, Cancer medicine. [DOI] [PMC free article] [PubMed]
- [27].Wang J, Chen X, Lu H, Zhang L, Pan J, Bao Y, Su J, Qian D, Feature-shared adaptive-boost deep learning for invasiveness classification of pulmonary subsolid nodules in ct images, Medical Physics 47 (4) (2020) 1738–1749. [DOI] [PubMed] [Google Scholar]
- [28].Gong J, Liu J, Hao W, Nie S, Zheng B, Wang S, Peng W, A deep residual learning network for predicting lung adenocarcinoma manifesting as ground-glass nodule on ct images, European Radiology (2019) 1–9. [DOI] [PubMed]
- [29].Chen H, Qi X, Yu L, Heng P-A, Dcan: deep contour-aware networks for accurate gland segmentation, in: Proceedings of the IEEE conference on Computer Vision and Pattern Recognition, 2016, pp. 2487–2496. [Google Scholar]
- [30].Zhang Y, Miao S, Mansi T, Liao R, Task driven generative modeling for unsupervised domain adaptation: Application to x-ray image segmentation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer, 2018, pp. 599–607. [Google Scholar]
- [31].Ioffe S, Szegedy C, Batch normalization: Accelerating deep network training by reducing internal covariate shift, in: International Conference on Machine Learning, 2015, pp. 448–456. [Google Scholar]
- [32].Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R, Dropout: a simple way to prevent neural networks from overfitting, The journal of machine learning research 15 (1) (2014) 1929–1958. [Google Scholar]
- [33].Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin J-C, Pujol S, et al. , 3d slicer as an image computing platform for the quantitative imaging network, Magnetic resonance imaging 30 (9) (2012) 1323–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kingma DP, Ba J, Adam: A method for stochastic optimization, arXiv preprint arXiv:1412.6980.
- [35].He K, Zhang X, Ren S, Sun J, Delving deep into rectifiers: Surpassing human-level performance on imagenet classification, in: Proceedings of the IEEE international conference on computer vision, 2015, pp. 1026–1034. [Google Scholar]
- [36].Lin T-Y, Goyal P, Girshick R, He K, Dollár P, Focal loss for dense object detection, in: Proceedings of the IEEE international conference on computer vision, 2017, pp. 2980–2988. [Google Scholar]


