Abstract
An RNA-dependent RNA polymerase ribozyme that was highly optimized through in vitro evolution for the ability to copy a broad range of template sequences exhibits promiscuity toward other nucleic acids and nucleic acid analogues, including DNA, threose nucleic acid (TNA), and arabinose nucleic acid (ANA). By operating on various RNA templates, the ribozyme catalyzes multiple successive additions of DNA, TNA, or ANA monomers, although with reduced efficiency compared to RNA monomers. The ribozyme can also copy DNA or TNA templates to complementary RNAs, and to a lesser extent it can operate when both the template and product strands are composed of DNA, TNA, or ANA. These results suggest that polymerase ribozymes, which are thought to have replicated RNA genomes during the early history of life, could have transferred RNA-based genetic information to and from DNA, enabling the emergence of DNA genomes prior to the emergence of proteins. In addition, genetic systems based on nucleic acid-like molecules, which have been proposed as precursors or contemporaries of RNA-based life, could have been operated upon by a promiscuous polymerase ribozyme, thus enabling the evolutionary transition between early genetic systems.
Keywords: origins of life, polymerase, ribozyme, reverse transcriptase, RNA world, XNA
Graphical Abstract

All extant life relies on two chemically distinct nucleic acids, namely, DNA and RNA, to store and transfer genetic information, and it utilizes polymerase proteins to copy information between these two polymers. This system is widely thought to have evolved from an ancestral life form containing RNA genomes that were copied by an RNA-dependent RNA polymerase ribozyme.1,2 It has been speculated that other nucleic acid-like molecules may have preceded or even competed with RNA.2−4 Evolutionary transitions between these genetic systems would have required the emergence of mechanisms for information exchange between the different nucleic acid polymers, enabled by polymerase enzymes with the ability to catalyze this exchange. Evolution by natural selection proceeds most efficiently by enhancing or modifying an existing activity. Thus, the promiscuity of polymerase enzymes toward alternative compositions of the template and newly synthesized strands could have facilitated the evolutionary transition between early genetic systems.
Although modern polymerase proteins are typically highly specific for either RNA or DNA, structural and phylogenetic studies suggest that they have swapped substrate and template specificity in the past.5 Similar results have been achieved through targeted mutagenesis and directed evolution6,7 and have expanded polymerase function to accommodate a variety of non-natural xenobiological nucleic acids (XNAs).8,9 The transition from RNA to DNA genomes may have preceded the invention of genetically encoded proteins, and this surely would have been the case for any transition that occurred during the era of RNA-based life.3,4,10 For such early transitions between genetic systems, the polymerases themselves would have been composed of nucleic acids or nucleic acid-like molecules.
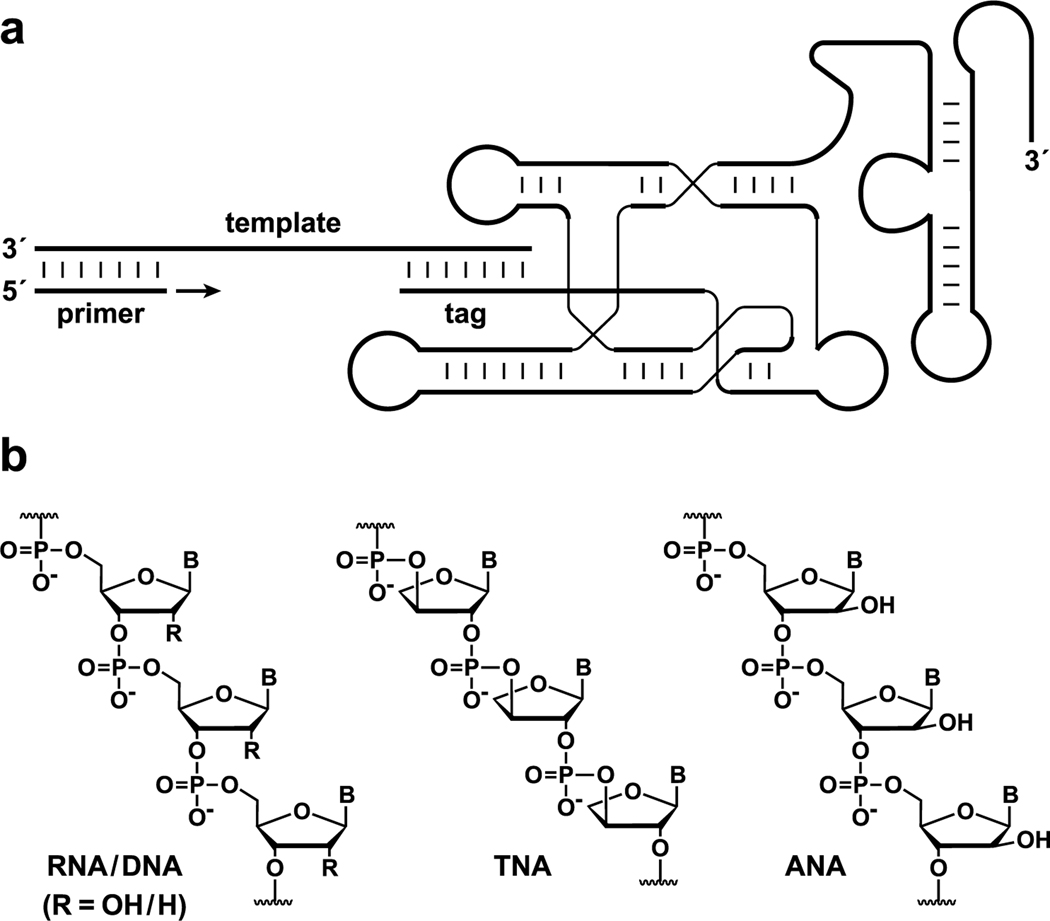
Although no polymerase ribozyme has been found in extant biology, a variety of RNA polymerase ribozymes have been discovered using in vitro evolution.11,12 The most active of these, the class I polymerase (Figure 1a), catalyzes the RNA-templated synthesis of RNA using ribonucleoside 5′-triphosphate (rNTP) substrates.11 This ribozyme has undergone extensive evolutionary optimization and is able to generate long RNA products in a largely sequence-general manner.13−15 Although never selected to operate with substrates or templates other than RNA, the class I polymerase can tolerate some modifications of either the nucleobase or sugar−phosphate backbone for the first nucleotide incorporation step.16,17 However, early versions of the polymerase are unable to add multiple successive nucleotide analogues, nor can they tolerate the presence of even a few DNA residues in the template.18
Figure 1.
RNA-catalyzed polymerization of RNA, DNA, TNA, or ANA. (a) Secondary structure of the complex formed by the ribozyme, template, and primer. The template consists of a primer binding site, the region copied by the polymerase, and the region that binds the processivity tag at the 5′ end of the ribozyme. (b) Composition of nucleic acids and nucleic acid analogues used in this study.
More recent versions of the class I polymerase were optimized not just for catalytic rate but also for the ability to tolerate a wide variety of sequence contexts, enabling the RNA-catalyzed synthesis of complex functional RNAs and the exponential amplification of short RNAs.15 These improvements extend beyond the activities that were directly targeted by selection, with the most active published variant of the enzyme, termed “24−3”, found to have promiscuous activity for reverse transcribing an RNA template to a complementary DNA product using deoxyribonucleoside 5′-triphosphate (dNTP) substrates.19 Under optimal conditions, the 24−3 polymerase can synthesize DNAs that contain more than 30 residues, incorporating all four dNTPs. However, this enzyme was found to be incapable of catalyzing the forward transcription of DNA to complementary RNA.
The present study sought to explore the catalytic promiscuity of the polymerase ribozyme toward other nucleic acid-like polymers, in both the template and complementary product. Attention was directed toward nucleic acid analogues of potential relevance to the origins of life that are based on either threose or arabinose sugars (TNA or ANA, respectively; Figure 1b). RNA, DNA, TNA, and ANA all can engage in Watson−Crick base pairing as either homo- or heteroduplexes.20,21 The tetrose sugar of TNA is compositionally simpler than the pentose of RNA, whereas the arabinose of ANA is an epimer of ribose. Both could have emerged through prebiotic chemistry or during the early history of life.
RESULTS AND DISCUSSION
The class I polymerase ribozyme has undergone many generations of in vitro evolution, resulting in highly robust RNA-dependent RNA polymerase activity.11,13−15 The 24−3 variant of the polymerase was recently shown to function as an RNA-dependent DNA polymerase, albeit with substantially reduced activity compared to RNA polymerization.19 Subsequent to that work, an even more active form of the polymerase, termed “38−6”, was isolated following 14 additional rounds of in vitro evolution in which the ribozyme was selected for the ability to synthesize other functional ribozymes (Horning and Joyce, unpublished). The 38−6 enzyme differs from 24−3 by the acquisition of 14 mutations (for sequences, see Table S1).
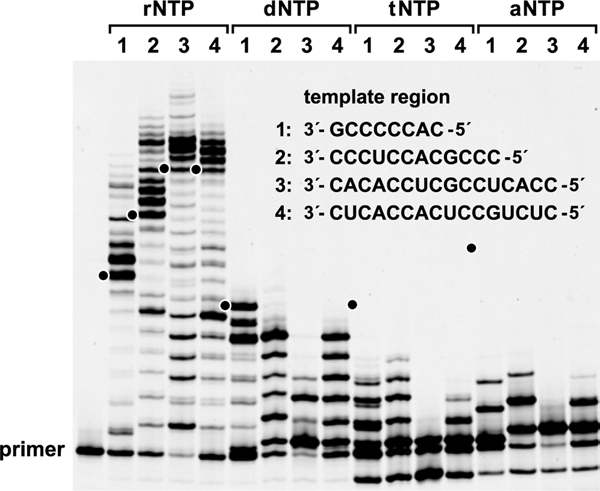
The 38−6 polymerase variant was tested for its ability to copy RNA templates to complementary DNA, TNA, or ANA products. 38−6 catalyzed multiple successive additions of these three NTP analogues (Figure 2). In each case, the polymerase extended a 15-nucleotide RNA primer, bound to a complementary RNA template that also contained a templating region of variable sequence and a region that forms Watson−Crick pairs with the “processivity tag” at the 5′ end of the polymerase (Figure 1a).14 As expected from its evolutionary history, the polymerase performs best with rNTPs, readily extending the primer across the entire templating region, and even copying the portion of the template that binds the processivity tag, displacing that base-paired interaction. As was seen with the 24−3 polymerase,19 the 38−6 variant has lower activity with dNTPs compared to rNTPs, generating full-length products only for template 1, which is the shortest and most C-rich. Activity with threofuranosyl-nucleoside 3′-triphosphates (tNTPs) and arabino-nucleoside 5′-triphosphates (aNTPs) is further reduced, with only a few residues added and a propensity to stall at A residues in the template.
Figure 2.
RNA-catalyzed polymerization of rNTPs, dNTPs, tNTPs, or aNTPs on various RNA templates (1−4). The sequence of the template region to be copied by the polymerase is shown. Reaction conditions: 100 nM polymerase, 100 nM template, 80 nM RNA primer, 0.5 mM each xNTP, 200 mM MgCl2, pH 8.3, 17 °C, 24 h. Black dots indicate the expected position of full-length products resulting from copying the entire templating region.
The four different polymeric products have distinct mobilities in a denaturing polyacrylamide gel (Figure 2), indicative of their distinct chemical composition. To confirm the identity of the TNA and ANA extension products, reactions were performed using a shorter RNA template (5) that required the addition of three successive G residues (Figure 3a). For both TNA and ANA, four product bands were seen in the gel. Three of these bands had progressively slower mobility, likely corresponding to the addition of one, two, or three G residues. The fourth band had faster mobility than the starting primer, and no such band was seen in the reactions with either rNTPs or dNTPs (Figure 2).
Figure 3.
Extension of an RNA primer (p) on a complementary RNA template by addition of up to three tGTP (blue) or aGTP (red) residues. (a) Gel separation of the products corresponding to one, two, or three additions, as well as faster-migrating material (c) that was identified as the primer with a terminal 2′,3′-cyclic phosphate. (b) Summed LC/MS mass spectra from the two extension reactions. Reaction conditions: 200 nM polymerase, 200 nM template 5, 160 nM primer, 1 mM either tGTP or aGTP, 200 mM MgCl2, pH 8.3, 17 °C, 24 h.
The ensemble of products from the TNA and ANA reactions were analyzed by liquid chromatography mass spectrometry (LC/MS; Figure 3b). For TNA, the calculated and observed masses for the addition products were: tG, 5999.0 calcd, 5998.4 found; (tG)2, 6314.1 calcd, 6313.7 found; (tG)3, 6629.3 calcd, 6628.8 found. For ANA, the calculated and observed masses were: aG, 6029.0 calcd, 6028.8 found; (aG)2, 6374.2 calcd, 6373.8 found; (aG)3, 6719.4 calcd, 6718.8 found. For both the TNA and ANA reactions, the product with faster mobility had an observed mass of 5745.7 Da. This product likely corresponds to an RNA primer with a terminal 2′,3′-cyclic phosphate, which has a calculated mass of 5745.7 Da. Such material could be produced by a two-step reaction, involving first the addition of a G residue to the 3′ end of the primer, then excision of the newly added residue via attack of the vicinal 2′-hydroxyl on the newly formed phosphodiester.
Some polymerase proteins have a proofreading mechanism that relies on a separate exonuclease domain, which competes with the polymerase active site to bind the 3′ end of the primer, and which excises a mis-incorporated nucleotide that would otherwise stall the polymerase.5 The presumed exonuclease activity of the ribozyme that is seen with TNA and ANA, but not RNA or DNA polymerization, may reflect either a distinct site or reorganization of the polymerase active site to enable this activity in certain contexts. Such an activity, if further optimized by evolution, may have had selective advantage for RNA-based life by enabling the synthesis of RNA homopolymers, despite the presence of nucleotide analogues in the local environment.
The accuracy of information transfer between an RNA template and DNA, TNA, or ANA products was assessed using template 1 and various combinations of xNTPs. Providing xCTP alone would be expected to allow only a single nucleotide addition, which is the case for all three polymeric products (Figure S1). Providing xGTP alone should not allow any nucleotide addition, but in fact a single mismatched G is added in all cases, followed by five additional templated G residues for DNA and two additional templated G residues for TNA and ANA. When both xCTP and xGTP are provided, the complementary sequence 5′-CGGGGG-3′ is generated in near quantitative yield for DNA, whereas ANA extension progresses no further than 5′-CGG-3′. The situation with TNA is more complicated, because either C or G is added at the first position, followed by three more G residues. In that case, the tetrameric addition products are dominated by the complementary sequence, but the shorter products contain either C or G at the first position. When all four xNTPs are provided, the pattern is similar, except that for DNA the extension reaction continues through the entire sequence 5′-CGGGGGAC-3′.
To further investigate the fidelity of RNA-templated DNA polymerization, a template (6) that contains all four nucleotides was used to generate DNA products with the expected sequence 5′-GCGAGGAGTG-3′. The 10-nucleotide addition product was gel purified and sequenced, both as an ensemble and by examining cloned individuals (Figure S2). In both cases, the majority of products had the correct sequence, with an average fidelity of 97% per nucleotide position, calculated from the sequences of 23 cloned individuals.
Preservation of hereditary information between alternative genetic systems requires copying that information in both the forward and reverse directions between the genetic polymers. Thus, the ability of the 38−6 polymerase to tolerate alteration of both the template and product strands was investigated. Chimeric templates were prepared, where both the primer binding site and the region complementary to the processivity tag were composed of RNA, flanking an 8-nucleotide templating region having the sequence 3′-CC(U/T)CACAC-5′ and composed of RNA (r7), DNA (d7), or TNA (t7).
For each of these templates, primer extension reactions were performed using rNTPs, dNTPs, tNTPS, or aNTPs (Figure 4). As seen previously, the reaction with an RNA template was most efficient, even extending into the region of the processivity tag. The reaction with an RNA template was progressively less efficient when using dNTPs (six nucleotides added), tNTPs (four added), or aNTPs (three added). When either a DNA or TNA template was used, the polymerase exhibited robust activity with rNTPs, generating full-length products. A lower level of activity was seen with dNTPs on either a DNA or TNA template, and still lower activity was seen when either tNTPs or aNTPs were used. Nonetheless, the polymerase ribozyme is able to catalyze multiple nucleotide additions for all 12 combinations of template and product. This high degree of catalytic promiscuity of the 38−6 ribozyme toward alternative backbone compositions had not been seen for earlier, less optimized versions of the class I polymerase.
Figure 4.
RNA-catalyzed polymerization of rNTPs, dNTPs, tNTPs, or aNTPs on an RNA (r7), DNA (d7), or TNA (t7) template. Reaction conditions: 100 nM polymerase, 100 nM template, 80 nM primer, 0.5 mM each xNTP, 200 mM MgCl2, pH 8.3, 17 °C, 24 h.
The high level of activity of the 38−6 polymerase in the DNA-templated synthesis of RNA appears to conflict with the earlier observation that the 24−3 polymerase cannot perform this reaction.19 In those previous experiments, the template strand was composed entirely of DNA, including the region that binds the processivity tag of the ribozyme. To further investigate this issue, a variant of the d7 template was constructed, where the primer-binding region was composed of DNA, and the region that binds the processivity tag was composed of RNA, which exhibited only a modest decrease in DNA-dependent RNA polymerase activity. If, however, both of these regions were composed of DNA, then all activity was lost (Figure S3a). Tests with the 24−3 polymerase variant showed the same pattern of behavior (Figure S3b).
The inability to form a productive complex between a DNA template and RNA processivity tag may simply be due to the lower stability of a DNA•RNA heteroduplex compared to RNA•RNA homoduplex.22 To investigate this hypothesis, the stability of the interaction between ribozyme and template was increased by increasing either the length or the GC content of the pairing region. Either approach resulted in DNA-dependent RNA polymerase activity equivalent to that seen with an RNA•RNA homoduplex in this region (Figure S3). The calculated Tm for the RNA•RNA homoduplex is 22.6 °C, and for the original form of the DNA•RNA heteroduplex it is 3.5 °C, whereas the predicted Tm values for the two stabilized forms are 34.5 and 25.2 °C, respectively.23
With a stabilized processivity tag interaction, the 38−6 polymerase was able to synthesize complementary RNAs from a variety of all-DNA templates (Figure 5a). In each case, the polymerase generated full-length products, surpassing the yield of DNA polymerization with comparable RNA templates.19 The stabilized processivity tag also enables TNA-templated polymerization to occur (Figure 5b). With various all-TNA templates, an RNA primer was extended to yield full-length RNA products, although the yields with TNA templates were lower than those seen with corresponding DNA templates.
Figure 5.
RNA-catalyzed polymerization of RNA on all-DNA or all-TNA templates. (a) Polymerization on DNA templating regions having the sequence 3′-GC5AC-5′ (d1), 3′-GC5ACGC5TC-5′ (d8), 3′-CGCTCCTCACACACAC-5′ (d9), or 3′-GC5ACGC5TCGC5-ACGC5TC-5′ (d10). (b) Polymerization on DNA or TNA templating regions having the sequence 3′-CCTCACGC-5′ (d11 or t11) or that of d8 or d9 (similarly for t8 or t9). Reaction conditions: 100 nM polymerase, 100 nM template, 80 nM primer, 4 mM each rNTP, 200 mM MgCl2, pH 8.3, 17 °C, 24 h.
The ability of an RNA-dependent RNA polymerase to operate with DNA, TNA, or ANA requires its active site to accommodate alternate backbone structures in both the template and product strands. The heteroduplex formed between RNA and each of the other three polymers resembles the A-form helix of an RNA homoduplex, thus easing the requirement for recognition by a polymerase that has been optimized for RNA.24,25 However, when both the template and product strands are not RNA, the geometry can deviate from A-form, as is the case for DNA homoduplexes, which tend to adopt the B-form geometry. Local differences in structure may also pose difficulties for recognition by the polymerase, for example, due to the loss of 2′-hydroxyl contacts in the minor groove of the template-primer complex, which have been observed in structural studies of an earlier version of the RNA polymerase.26 Replacing some of those RNA residues with DNA resulted in decreased catalytic activity.18 Similarly, the alternative orientation of the 2′-hydroxyl in ANA and the lack of a vicinal hydroxyl in TNA may cause local structural perturbations.
Despite its exclusive optimization for activity as an RNA-templated RNA polymerase, the most active variants of the class I polymerase ribozyme exhibit promiscuous activity toward distinct backbone structures in both the template and product strands. Earlier forms of the polymerase were highly sensitive to alteration of the NTP substrates16,18 but also highly restricted in their behavior as an RNA polymerase, strongly preferring to operate on unstructured C-rich templates.15 The more advanced 24−3 and 38−6 variants are the result of extensive evolution for improved activity with a broad range of RNA template sequences. The enhanced tolerance of these enzymes to a variety of RNA templates appears to have increased their tolerance to alternative backbones. The promiscuous activities of the improved polymerases, even without having selected directly for these properties, suggests that the polymerases have enhanced capacity to bind the primer-template complex and, thus, may be more tolerant to the loss of interactions that are specific to RNA. A correlation between catalytic robustness and substrate promiscuity is not unprecedented,27 having been seen in both natural and in vitro evolution of other enzymes, including polymerase proteins.6,7
The 38−6 polymerase has much lower activity in the RNA-templated synthesis of TNA or ANA compared to DNA. The sugar pucker of DNA tends to be more flexible than that of TNA or ANA, potentially allowing DNA to adopt the RNA-like C2′-endo conformation within the polymerase active site.25 Likewise, the DNA-templated polymerization of DNA is less efficient than the DNA-templated polymerization of RNA, which may be due to the greater difficulty for a DNA homoduplex to adopt an A-form geometry. DNA polymerase proteins induce DNA to adopt an A-form within the active site,28 but the polymerase ribozyme may not bind tightly enough to the primer-template complex to achieve the same result. The present form of the polymerase has far lower activity than what would be needed to support the replication of DNA, let alone TNA or ANA. In each case, substantial evolutionary optimization would be required to improve binding of the primer-template complex, achieve greater sequence generality, and operate in a substrate-specific manner with a particular set of xNTPs.
CONCLUSIONS
Transitions between genetic systems can proceed in either a continuous manner or through a genetic takeover. The former may enable the preservation of genetic information across the transition, if there is a way to copy the ordering of subunits from the old genetic material to the new. In a genetic takeover, an entirely different replication machinery emerges and replaces the previous genetic system.29 There is potential selective advantage to a continuous transition, if traits that conferred fitness in the prior system can benefit the successor. The two systems need not have genetic material of similar composition for this to be the case, so long as there is a means to “translate” information from the old system to the new one. However, it likely would have been simpler to “transcribe” information from the prior genetic material to a successor that has similar composition, as is thought to have occurred during the transition from RNA to DNA genomes. Furthermore, if the transcription machinery derives from the replication machinery, a continuous transition becomes more plausible.
The class I polymerase has an evolutionary history of fewer than 100 generations spanning only 25 years, whereas natural polymerase proteins have a history that dates back at least to the last universal common ancestor. However, the class I polymerase recapitulates a key function of prior RNA-based life, which is the RNA-templated polymerization of RNA. This ribozyme, in the absence of direct selection pressure to operate on alternative polymers, has promiscuous activity toward other nucleic acid-like polymers. This finding suggests that an RNA-based organism could have evolved polymerase activity for related genetic polymers, potentially enabling transfer of genetic information to such polymers as an elaboration of the RNA polymerase machinery. Further in vitro evolution studies could be used to determine the capacity for improving these promiscuous activities to achieve the RNA-catalyzed replication of other plausible genetic materials.
EXPERIMENTAL SECTION
Materials.
The sequences of nucleic acids used in this study are listed in Table S1. Polymerase ribozymes were prepared by in vitro transcription of double-stranded DNA templates that were generated by polymerase chain reaction (PCR) from corresponding plasmid DNA. Primers and chimeric templates were prepared by solid-phase synthesis using an Expedite 8909 DNA/RNA synthesizer, with reagents and phosphoramidites purchased from Glen Research, with the exception of the chimeric RNA/TNA template t7, which was prepared by the DNA Core Facility at the University of Utah using TNA phosphoramidites that were chemically synthesized in the Chaput laboratory.30 RNA templates were prepared by in vitro transcription of synthetic DNAs, which were purchased from IDT. DNA templates were obtained from the same source. TNA templates were prepared by in vitro transcription of DNA templates using Kod-RI TNA polymerase, as described previously.31 All primers, templates, and ribozymes were purified by denaturing polyacrylamide gel electrophoresis (PAGE) and ethanol precipitation prior to use. NTPs were purchased from Sigma-Aldrich, dNTPs from Denville Scientific, and aNTPs from Tri-Link Biotechnologies. tNTPs were chemically synthesized as described previously.32 TURBO DNase I, Superscript IV reverse transcriptase, and streptavidin C1 Dynabeads all were from ThermoFisher. Universal miRNA Cloning Linker and thermostable 5′-App DNA/RNA ligase were from NEB.
In Vitro Transcription.
RNA templates were prepared by in vitro transcription of 0.5 μM single-stranded DNA that had previously been annealed with 0.5 μM of a synthetic oligodeoxynucleotide encoding the second strand of the T7 RNA polymerase promoter. Transcription was performed in a mixture containing 15 U/μL T7 RNA polymerase, 0.002 U/μL inorganic pyrophosphatase, 5 mM each NTP, 25 mM MgCl2, 2 mM spermidine, 10 mM DTT, and 40 mM Tris (pH 8.0), which was incubated at 37 °C for 2 h. The DNA then was digested by adding 0.1 U/μL TURBO DNase I and continuing the incubation for 1 h. Polymerase ribozymes were transcribed from fully double-stranded DNA templates (20 mg/mL) that were obtained by PCR amplification of plasmid DNA encoding either the 24−3 or the 38−6 ribozyme. Ribozymes with altered processivity tags were prepared in the same manner, using a different PCR primer (Table S1).
Preparation of TNA Templates by Primer Extension.
The DNA template and DNA primer (for sequences, see Table S2) were mixed at a concentration of 1 μM each in ThermoPol buffer (NEB), heated to 95 °C for 5 min, then annealed at 24 °C. Extension was performed using 1 μM Kod RI TNA polymerase and 100 μM each tNTP at 55 °C for 3 h. The products were diluted with 95% formamide and 25 mM ethylenediaminetetraacetic acid (EDTA), and then the DNA-TNA chimeric material was purified by denaturing PAGE, recovered from the gel by electroelution, exchanged into water using a YM-30 microcentrifuge device, and lyophilized. To remove the DNA primer, the material was resuspended in 10 mM MgCl2, 40 mM Tris (pH 8.4), and 0.0625 U/L snake venom phosphodiesterase I (Sigma-Aldrich) and then incubated at 37 °C overnight. The nuclease was removed by phenol/chloroform extraction, and the TNA was desalted using a YM-30 microcentrifuge device.
RNA-Catalyzed Polymerization.
Unless otherwise noted, all RNA-catalyzed polymerization reactions were performed using 100 nM ribozyme, 100 nM template, and 80 nM primer. The RNA primer contained both a fluorescein label and biotin moiety at its 5′ end. The ribozyme, template, and primer first were heated at 80 °C for 2 min, then cooled to 17 °C over 5 min and added to the reaction mixture, which also contained 200 mM MgCl2, 0.05% TWEEN20, and 50 mM Tris (pH 8.3). Reactions comparing various xNTPs used 0.5 mM each NTP, dNTP, tNTP, or aNTP, whereas reactions comparing DNA and TNA templates used 4 mM each NTP. Polymerization was performed at 17 °C and quenched by adding 250 mM EDTA. The biotinylated primers and extended products were captured on streptavidin C1 Dynabeads, washed twice with alkali (25 mM NaOH, 1 mM EDTA, and 0.05% TWEEN20), once with TE-urea (1 mM EDTA, 0.05% TWEEN20, 10 mM Tris (pH 8.0), and 8 M urea), and then eluted with 98% formamide and 10 mM EDTA at 95 °C for 10 min. The reaction products were analyzed by denaturing PAGE.
Analysis of Polymerization Products.
Products for analysis by LC/MS were prepared using 200 nM ribozyme, 200 nM template, and 160 nM primer. The reactions were performed as described above, using 1 mM tGTP or aGTP at pH 8.3 for 24 h. After purification using streptavidin C1 Dynabeads, 5% of the reaction products were analyzed by denaturing PAGE, while the remainder were ethanol-precipitated, dissolved in 1 mM EDTA, 0.05% TWEEN20, and 10 mM Tris (pH 8.0), and used for LC/MS analysis, which was performed by Novatia LLC using 40 pmol of purified material. Analyses were performed by electrospray ionization LC/MS on the Oligo HTCS platform, which achieves a mass accuracy of 0.01−0.02%.
Products for DNA sequencing were prepared using template 6 and the four dNTPs, as described above. The 10-nucleotide addition product was separated by PAGE, excised from the gel, ethanol-precipitated, and ligated to the Universal miRNA Cloning Linker using thermostable 5′-App DNA/RNA ligase according to the manufacturer’s protocol. The ligated product was reverse-transcribed using Superscript IV reverse transcriptase, PCR-amplified, and purified by agarose gel electrophoresis. This material was used directly for ensemble sequencing. For clonal sequencing, the amplified DNA was cloned into Escherichia coli using the TOPO-TA cloning kit (ThermoFisher), and the cells were grown at 37 °C for 16 h on lysogeny broth (LB) agar plates containing 50 μg/mL kanamycin. Individual colonies were transferred to 3 mL of LB media containing 50 μg/mL kanamycin and grown at 37 °C for 16 h. Plasmid DNA was recovered using the PureLink Quick Plasmid Miniprep Kit (ThermoFisher). Both the ensemble and clonal materials were sequenced by Eton Bioscience.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Grant No. 80NSSC17K0462 from NASA and Grant No. 287624 from the Simons Foundation. The authors gratefully acknowledge the assistance of B. Samanta and A. Ngor for preparation of DNA and TNA templates, respectively.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACSPublicationswebsite at DOI: 10.1021/acssyn-bio.9b00044.
Two tables listing sequences of nucleic acid materials used in this study. Three figures, one showing RNA-templated polymerization with various combinations of xNTPs, a second describing the fidelity of RNA-templated DNA polymerization, and a third comparing DNA-dependent RNA polymerase activity for various strengths of binding interaction between ribozyme and template (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Gilbert W. (1986) The RNA world. Nature 319, 618. [Google Scholar]
- (2).Joyce GF (2002) The antiquity of RNA-based evolution. Nature 418, 214–221. [DOI] [PubMed] [Google Scholar]
- (3).Joyce GF, Schwartz AW, Miller SL, and Orgel LE (1987) The case for an ancestral genetic system involving simple analogues of the nucleotides. Proc. Natl. Acad. Sci. U. S. A. 84, 4398–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Joyce GF (2012) Toward an alternative biology. Science 336, 307–308. [DOI] [PubMed] [Google Scholar]
- (5).Joyce CM, and Steitz TA (1994) Function and structure relationships in DNA polymerases. Annu. Rev. Biochem. 63, 777–822. [DOI] [PubMed] [Google Scholar]
- (6).Gao G, Orlova M, Georgiadis MM, Hendrickson WA, and Goff SP (1997) Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proc. Natl. Acad. Sci. U. S. A. 94, 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Ong JL, Loakes D, Jaroslawski S, Too K, and Holliger P. (2006) Directed evolution of DNA polymerase, RNA polymerase and reverse transcriptase activity in a single polypeptide. J. Mol. Biol. 361, 537–550. [DOI] [PubMed] [Google Scholar]
- (8).Pinheiro VB, Taylor AI, Cozens C, Abramov M, Renders M, Zhang S, Chaput JC, Wengel J, Peak-Chew S-Y, McLaughlin SH, Herdewijn P, and Holliger P. (2012) Synthetic genetic polymers capable of heredity and evolution. Science 336, 341–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Yu H, Zhang S, and Chaput JC (2012) Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat. Chem. 4, 183–187. [DOI] [PubMed] [Google Scholar]
- (10).Smith JM, and Szathmary E. (1997) The Major Transitions in Evolution, Oxford University Press, Oxford. [Google Scholar]
- (11).Johnston WK, Unrau PJ, Lawrence MS, Glasner ME, and Bartel DP (2001) RNA-catalyzed RNA polymerization: accurate and general RNA-templated primer extension. Science 292, 1319–1325. [DOI] [PubMed] [Google Scholar]
- (12).Sczepanski JT, and Joyce GF (2014) A cross-chiral RNA polymerase ribozyme. Nature 515, 440–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Zaher HS, and Unrau PJ (2007) Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA 13, 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Wochner A, Attwater J, Coulson A, and Holliger P. (2011) Ribozyme-catalyzed transcription of an active ribozyme. Science 332, 209–212. [DOI] [PubMed] [Google Scholar]
- (15).Horning DP, and Joyce GF (2016) Amplification of RNA by an RNA polymerase ribozyme. Proc. Natl. Acad. Sci. U. S. A. 113, 9786–9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Attwater J, Tagami S, Kimoto M, Butler K, Kool ET, Wengel J, Herdewijn P, Hirao I, and Holliger P. (2013) Chemical fidelity of an RNA polymerase ribozyme. Chem. Sci. 4, 2804–2814. [Google Scholar]
- (17).Samanta B, Horning DP, and Joyce GF (2018) 3′-End labeling of nucleic acids by a polymerase ribozyme. Nucleic Acids Res. 46, No. e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Mü UF., and Bartel DP (2003) Substrate 2′-hydroxyl groups required for ribozyme-catalyzed polymerization. Chem. Biol. 10, 799–806. [DOI] [PubMed] [Google Scholar]
- (19).Samanta B, and Joyce GF (2017) A reverse transcriptase ribozyme. eLife 6, No. e31153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).ning K-U, Scholz P, Guntha S, Wu X, Krishnamurthy R, and Eschenmoser A. (2000) Chemical etiology of nucleic acid structure: the α-threofuranosyl-(3′→2′) oligonucleotide system. Science 290, 1347–1351. [DOI] [PubMed] [Google Scholar]
- (21).Noronha AM, Wilds CJ, Lok C-N, Viazovkina K, Arion D, Parniak MA, and Damha MJ (2000) Synthesis and biophysical properties of arabinonucleic acids (ANA): circular dichroic spectra, melting temperatures, and ribonuclease H susceptibility of ANA•RNA hybrid duplexes. Biochemistry 39, 7050–7062. [DOI] [PubMed] [Google Scholar]
- (22).Lesnik EA, and Freier SM (1995) Relative thermodynamic stability of DNA, RNA, and DNA:RNA hybrid duplexes: relationship with base composition and structure. Biochemistry 34, 10807−10815. [DOI] [PubMed] [Google Scholar]
- (23).Owczarzy R, Tataurov AV, Wu Y, Manthey JA, McQuisten KA, Almabrazi HG, Pedersen KF, Lin Y, Garretson J, McEntaggart NO, Sailor CA, Dawson RB, and Peek AS (2008) IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. 36, W163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Anosova I, Kowal EA, Sisco NJ, Sau S, Liao J-Y, Bala S, Rozners E, Egli M, Chaput JC, and Van Horn WD (2016) Structural insights into conformation differences between DNA/TNA and RNA/TNA chimeric duplexes. ChemBioChem 17, 1705–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Denisov AY, Noronha AM, Wilds CJ, Trempe J-F, Pon RT, Gehring K, and Damha MJ (2001) Solution structure of an arabinonucleic acid (ANA)/RNA duplex in a chimeric hairpin: comparison with 2′-fluoro-ANA/RNA and DNA/RNA hybrids. Nucleic Acids Res. 29, 4284–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Shechner DM, Grant RA, Bagby SC, Koldobskaya Y, Piccirilli JA, and Bartel DP (2009) Crystal structure of the catalytic core of an RNA-polymerase ribozyme. Science 326, 1271–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Khersonsky O, and Tawfik DS (2010) Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu. Rev. Biochem. 79, 471–505. [DOI] [PubMed] [Google Scholar]
- (28).Steitz TA (1998) Structural biology: a mechanism for all polymerases. Nature 391, 231–232. [DOI] [PubMed] [Google Scholar]
- (29).Cairns-Smith AG (1982) Genetic Takeover and the Mineral Origins of Life, Cambridge University Press, Cambridge. [Google Scholar]
- (30).Sau SP, Fahmi NE, Liao J-Y, Bala S, and Chaput JC (2016) A scalable synthesis of α-L-threose nucleic acid monomers. J. Org. Chem. 81, 2302–2307. [DOI] [PubMed] [Google Scholar]
- (31).Dunn MR, Otto C, Fenton KE, and Chaput JC (2016) Improving polymerase activity with unnatural substrates by sampling mutations in homologous protein architectures. ACS Chem. Biol. 11, 1210–1219. [DOI] [PubMed] [Google Scholar]
- (32).Sau SP, and Chaput JC (2017) A gram-scale HPLC-free synthesis of TNA triphosphates using an iterative phosphorylation strategy. Org. Lett. 19, 4379–4382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.