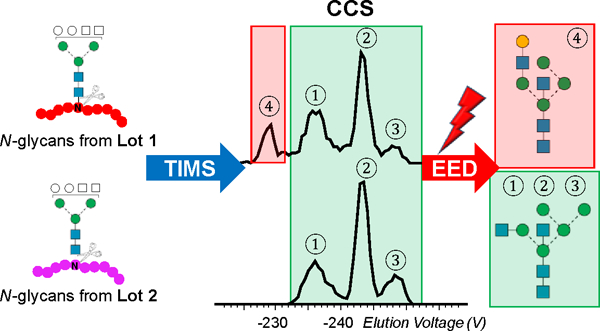
Abstract
Ion mobility-mass spectrometry (IM-MS) has become a powerful tool for glycan structural characterization due to its ability to separate isomers and provide collision cross section (CCS) values that facilitate structural assignment. However, IM-based isomer analysis may be complicated by the presence of multiple gas-phase conformations of a single structure that not only increases difficulty in isomer separation but can also introduce the possibility for misinterpretation of conformers as isomers. Here, the ion mobility behavior of several sets of isomeric glycans, analyzed as their permethylated derivatives, in both non-reduced and reduced forms, was investigated by gated-trapped ion mobility spectrometry (G-TIMS). Notably, reducing-end reduction, commonly performed to remove anomerism-induced chromatographic peak splitting, did not eliminate the conformational heterogeneity of permethylated glycans in the gas phase. At a mobility resolving power of ~100, 14 out of 22 structures showed more than one conformation. These results highlight the need to use IMS devices with high mobility resolving power for better separation of isomers and to acquire additional structural information that can differentiate isomers from conformers. On-line electronic excitation dissociation (EED) MS/MS analysis of isomeric glycan mixtures following G-TIMS separation showed that EED can generate isomer-specific fragments while producing nearly identical tandem mass spectra for conformers, thus allowing confident identification of isomers with minimal evidence of any ambiguity resulting from the presence of conformers. G-TIMS EED MS/MS analysis of N-linked glycans released from ovalbumin revealed that several mobility features previously thought to arise from isomeric structures were conformers of a single structure. Finally, analysis of ovalbumin N-glycans from different sources showed that the G-TIMS EED MS/MS approach can accurately determine the batch-to-batch variations in glycosylation profiles at the isomer level, with confident assignment of each isomeric structure.
Graphical Abstract
INTRODUCTION
Mammalian glycans are constructed with a small alphabet of monosaccharide building blocks, yet they exhibit a high degree of structural heterogeneity, as each monosaccharide residue has multiple potential linkage sites and each glycosidic linkage may occur in one of the two anomeric configurations. For a given oligosaccharide composition, changes in the sequence and the connectivity between residues yield topological and linkage isomers, whereas variations in the stereochemistry of the constituent monosaccharides and the resulting glycosidic linkages produce epimeric and anomeric isomers. Because subtle differences in the glycan structures can lead to significant changes in their biological functions, establishing the structure-function relationship for each glycan entails the development of analytical methods that can differentiate and accurately characterize isomeric structures.
Recent advances in the glycomics field have been largely propelled by the development of mass spectrometry (MS)-based methods for glycan structural characterization. MS offers high detection sensitivity and specificity, and tandem mass spectrometry (MS/MS) provides detailed structural information. In MS/MS, precursor ions are selected based on their m/z values, but co-isolation of isomeric species can give rise to chimeric tandem mass spectra that are prone to misinterpretation. For glycan mixture analysis, MS is often used in conjunction with orthogonal separation methods, such as high-performance liquid chromatography (HPLC) and capillary electrophoresis (CE). Isomer separation has been demonstrated on hydrophilic interaction liquid chromatography (HILIC)-MS,1–3 reversed-phase (RP)LC-MS,4–6 porous graphitic carbon (PGC)LC-MS,7–10 and CE-MS.11,12 Since the retention time of a glycoform is influenced by many external factors, including the column, solvents, analytical gradient, and temperature, glycan standards are often used to establish an elution ladder profile, in glucose units (gu), to facilitate determination of the relative position of elution for each glycan. Structural assignment can then be made by comparing the gu and m/z values of each glycan to those in the database. Reliable identification requires high reproducibility in chromatographic separation, but this is not always achievable, especially for nanoscale LC. Further, confident identification can only be made for previously reported structures that are included in the database.
Ion mobility-mass spectrometry (IM-MS) has recently emerged as a powerful alternative for glycan separation and identification.13–24 Ion mobility spectrometry (IMS)25 separates ions based on their size, shape, mass and charge, and can differentiate isomers with different gas-phase conformations. The measured mobility of an ion may be used to calculate its collision cross section (CCS) from kinetic theory.26 The CCS value is an intrinsic property of an ion (conformation) and provides additional information that aids structural assignment. An online CCS database, GlycoMob, was recently initiated for glycomics. GlycoMob currently contains over 1,200 reference CCS values, including the dextran ladder and N-glycans released from several glycoprotein standards, as well as their fragments. CCS values are listed for both positively- and negatively-charged, native glycans, with a variety of charge carriers.16,23,27 Another recent study reported CCS values for reduced and permethylated N-glycans in their protonated forms.28 Despite this evidence of progress, the construction of a comprehensive glycan CCS library remains challenging because of the common presence of glycan isomers in nature, and the limited access to high-quality glycan standards.29,30 A majority of reported CCS values for glycans were measured on commercial IM-MS instruments with traveling wave IMS (TWIMS) or linear drift tube (DT)-IMS devices. The limited mobility resolving power offered by these instruments, typically ranging from 30 to 60, is often insufficient for isomer resolution, but the presence of isomers is frequently inferred by their asymmetrical arrival time distributions (ATDs). However, a mixture of isomers with only small differences in their CCS values can also produce symmetrical, albeit broadened, ATDs. It is thus advantageous to perform IM-MS analysis on instruments with higher mobility resolving power, such as Trapped Ion Mobility Spectrometry (TIMS), Structures for Lossless Ion Manipulations Serpentine Ultra-long Path with Extended Routing (SLIM SUPER) IM-MS, and cyclic Ion Mobility (cIM) spectrometry.31–33 These instruments can achieve a resolving power of 250 or higher for substantially improved isomer separation.34,35
An inherent confounding factor to IM-MS characterization of isomeric mixtures is the presence of multiple conformations for each isomer. This is particularly evident when a high-resolution IM device is used for separation. For identification of each individual isomer, the appearance of multiple peaks can be beneficial, serving as the conformational fingerprint for more confident identification. For isomeric mixture analysis, however, this conformational multiplicity leads to more complex mobility profiles, and increases the difficulty of isomer separation. A common cause for glycan conformational heterogeneity is the variation in the reducing-end anomeric configuration. A recent cIM study of cellopentaose revealed the presence of three conformations, possibly corresponding to its open-chain form and α,β anomers, based on the heavy oxygen isotope labeling result.33 Ring-opening at the reducing-end is commonly employed to remove anomerism-induced chromatographic peak splitting,36 and thus it seemed worthwhile to investigate whether the gas-phase conformational heterogeneity of glycans can be simplified by reduction prior to analysis.
Since a given glycan composition can produce multiple peaks in its ATD due to the presence of isomers, conformers, or both, differentiation of isomers from conformers is crucial for accurate isomer characterization by IM-MS analysis. Although identification may be made by comparing the observed mobility profile with those in the database, current databases contain CCS values of only a small number of structures, often without clear isomer separation, and typically measured on instruments having low mobility-based resolution. As a complement to IM measurements, the conformational distribution of putative glycan structures and their corresponding CCS values may be calculated by molecular dynamics (MD) simulation. MD modeling could assist the interpretation of experimental results and is particularly useful for predicting structures of gas-phase glycan ions.19,37–39 Although it is usually impractical to consider all possible structures computationally, structural assignments may be validated by orthogonal methods, such as tandem mass spectrometry. Harvey et al. demonstrated that glycan isomers could be assigned by negative-ion collision-induced dissociation (nCID) MS/MS following TWIMS separation.13,40,41 Although nCID can generate diagnostic fragments for isomer differentiation, it generally does not produce structural details sufficient for complete structural determination. Additionally, some diagnostic fragments are not consistently observed across different CID modes.42 For isomers with partially overlapped mobility profiles, extracted fragment ion drift time distributions (XFIDTDs) may be used to differentiate glycan isomers. This approach was used by Clemmer et al. to detect the presence of four Man7 isomers, though definitive structural assignment was challenging due to the sparsity of cross-ring fragments from CID and the overlaps between ATDs of isomers and conformers.18 A similar approach was also used by Harvey et al. to identify isomers with overlapping ATDs. Meanwhile, Ujma et al. recently showed that CID of three IM components of cellopentaose produced fragments of substantially different abundances, which complicated structural assignments.33
These early studies suggest that isomer analysis would benefit greatly from the development of IM-MS/MS methods using IMS devices with higher mobility resolving powers and/or more efficient and definitive fragmentation methods. We have shown that electronic excitation dissociation (EED) can generate far richer fragmentation data than CID for detailed glycan structural characterization.43–46 We have demonstrated successful coupling of EED MS/MS to selective accumulation (SA)-TIMS for characterization of simple binary glycan mixtures of tetrasaccharides, each exhibiting a single conformation.46 Analysis of glycans from biological sources presents a far greater challenge, as they are more heterogeneous, have higher structural complexity, and may exist in multiple conformations. Furthermore, the extended ion storage time during SA-TIMS analysis can result in significant ion heating and fragmentation, as well as preferential loss of ions with high m/z values. By reducing the ion accumulation time in the TIMS device, the recently introduced gated-TIMS (G-TIMS) provides a broader m/z operating range and allows softer ion transmission, generating results comparable to other IMS platforms.47 We have also used G-TIMS, in conjunction with negative electron transfer dissociation (NETD) MS/MS analysis, to characterize and quantify isomeric glycosaminoglycan mixtures.43 In the present study, we use G-TIMS MS to investigate the effect of reduction on the conformation of permethylated glycans. The potential of G-TIMS EED MS/MS for differentiation of isomers from conformers is explored using several sets of isomeric glycan standards. The utility of the G-TIMS EED MS/MS approach for accurate isomer identification from complex glycan mixtures is demonstrated herein by analyzing released N-glycans from chicken ovalbumin.
EXPERIMENTAL
Materials and sample preparation.
Sources of chemicals used in this study and description of the sample preparation procedure can be found in the Supporting Information. All glycan standards and released N-glycans were permethylated, either directly or after deutero-reduction, according to protocols described in detail elsewhere.48,49
G-TIMS MS and G-TIMS EED MS/MS analyses.
All experiments were carried out on a 12T solariX Fourier-transform ion cyclotron resonance (FTICR) mass spectrometer equipped with a prototype G-TIMS device and a static nanoESI source (Bruker Daltonics, Billerica, MA, USA). For ESI analysis, permethylated glycans were dissolved to a concentration of 5–10 pmol/µL in a solution consisting of 50/50 (v/v) acetonitrile/water and 250 µM sodium acetate. The principle of the G-TIMS operation has been described in detail elsewhere.47,50 Briefly, ions were trapped both radially and axially in the TIMS funnel; radially, by an RF potential of 220 Vpp, and axially, by an axially varying electric field gradient (EFG) at positions where their drift velocity matched the carrier gas flow velocity. Following a 9-ms trapping event, the strength of the axial electric field was gradually decreased by ramping the TIMS analyzer entrance potential from -300 V to 50 V, to allow elution of ions in the order of increasing mobilities. When a downstream ion gate was pulsed open, it allowed only ions of selected mobility to pass through and accumulate in the collision cell. The drift gas (nitrogen) pressure in the TIMS tunnel was around 2.6 mBar. A mixture of fluorophosphazene standards (Agilent Technologies, Santa Clara, CA, USA) was used for mass and mobility calibration.51 The reduced mobility (K0) and CCS values from TIMS measurements were calculated as described previously.44,52 Briefly, a calibration curve was generated by a linear fit of the 1/K0 values of fluorophosphazene calibrants as measured by DT-IMS against their elution voltages, allowing calculation of the K0 value of each analyte based on its measured elution voltage. The CCS value of an ion can be calculated from its K0 value using Equation 1,
| (Equation 1) |
where q is the charge of the analyte ion, kB is the Boltzmann constant, T is the temperature, N0 is the neutral number density at standard temperature and pressure, ma and mb are the masses of the analyte and bath gas molecule, respectively.
For G-TIMS EED MS/MS analysis, ions were selected based on mobility via G-TIMS and mass via a quadrupole before being externally accumulated in the collision cell. Accumulated ions were then transferred to the ICR cell where they underwent EED with irradiation of 16- to 19-eV electrons for 350 ms.
Data Analysis.
All spectra were processed by DataAnalysis 4.4 (Bruker, Bremen, Germany), and interpreted manually, assisted by GlycoWorkbench, with a typical mass accuracy of <1 ppm.53 Fragments were annotated according to the Domon and Costello nomenclature, as illustrated in Supporting Scheme S1.54
RESULTS AND DISCUSSION
Effect of reduction on the gas-phase conformation of sodiated and permethylated glycans
Cyclization of a monosaccharide residue creates a new chiral center that may exhibit two possible anomeric configurations, denoted as α and β-anomers, depending on their stereochemistry relative to the anomeric reference carbon. Although the anomeric configuration within larger glycans is fixed upon formation of each glycosidic bond, the reducing end can still interconvert between its two potential anomeric forms via an open-ring intermediate. Such anomerization can be eliminated by chemical conversion of the reducing end into a static open form. A recent study showed that the phosphate adducts ([M+H2PO4]–) of reduced native glycans produced more symmetrical ATDs than their unreduced counterparts, but detailed conformational information could not be obtained due to the limited mobility resolution of the IMS device employed.55 Here we studied the conformational heterogeneity of permethylated glycans, since permethylation is often performed to stabilize labile residues, improve ionization and fragmentation efficiencies, and facilitate spectral interpretation.56 We then evaluated the effect of chemical reduction upon this heterogeneity. Figure 1 shows the structures of the glycans discussed in this study, including both linear and branched glycan standards, and putative structures of N-glycans with a composition of H5N4 from chicken ovalbumin, where H and N denote hexose and N-acetylhexosamine, respectively.
Figure 1.
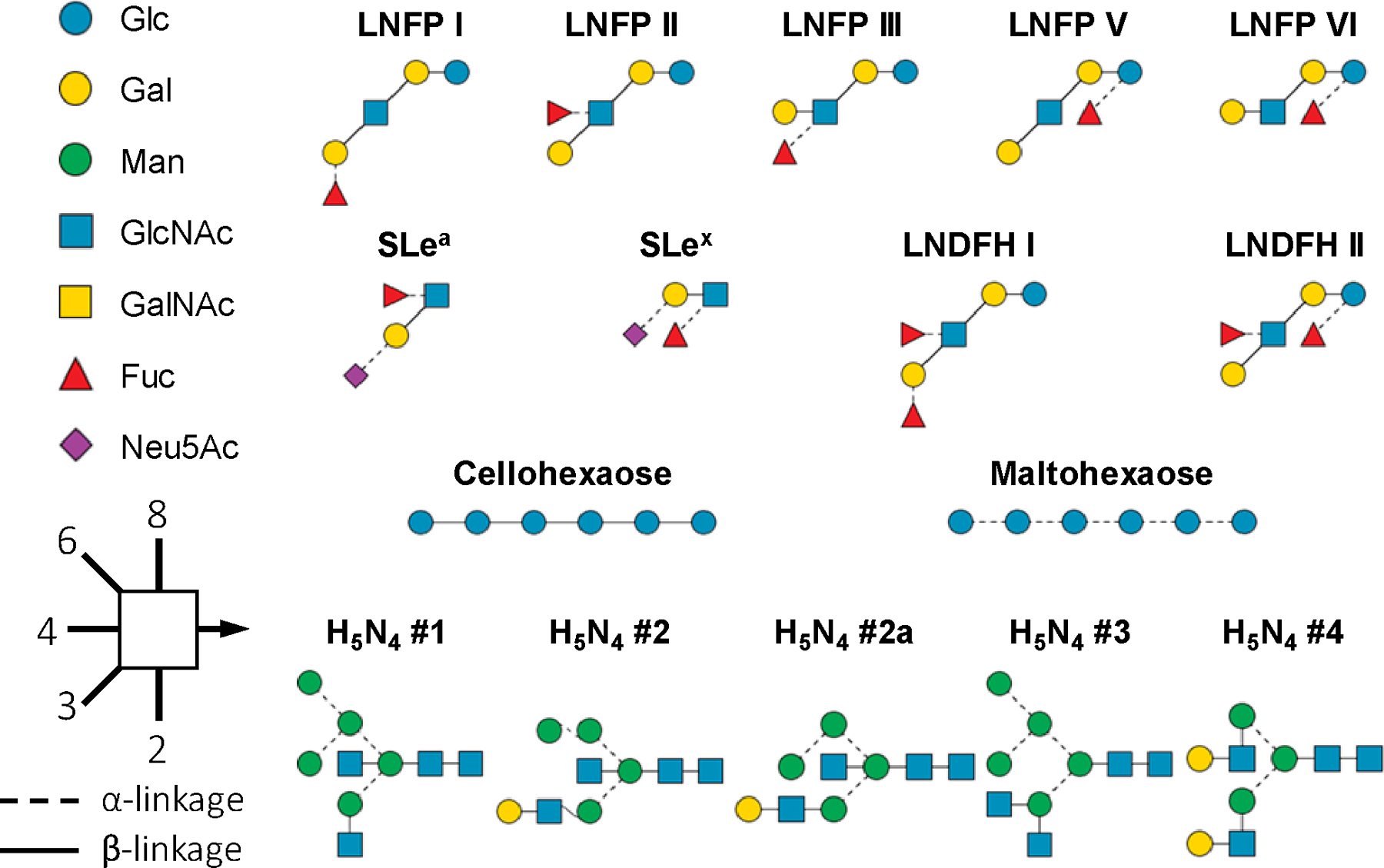
SNFG (symbol nomenclature for glycans) and representations of the structures of glycans discussed in the text.
The TIMS device was first calibrated with a mixture of fluorinated phosphazenes, and showed a mobility resolving power of 92 to 196 (Supplementary Figure S1). The actual peak width observed for each glycan varies depending on its mobility and conformational distribution, and typically indicates a resolution around 100. The gas-phase glycan conformation is influenced by the type and number of charge carriers. Here the investigation is focused on singly sodiated species. The extracted ion mobiligrams (EIMs) of non-reduced glycans are shown in the left panels of Figures 2 and S2. Among the five pentasaccharide isomers studied, the first three, LNFP I, II, and III, each produced only one major conformation, whereas LNFP V and VI each generated two (Figure 2a-e). The primary structural difference between these sets of isomers, LNFP I-III and LNFP V/VI, is the position of branching, with LNFP V/VI having the branch at the reducing end. In contrast, the EIMs recorded for the two linear hexasaccharide isomers included in this study, maltohexaose and cellohexaose, each showed only one major peak (Supplementary Figure S2a, b). From this initial comparison, it might appear that the anomeric configuration could have a stronger influence on the conformation of structures that have a branch near the reducing end, but this trend cannot be generalized. For example, LNDFH II produced one major peak, despite the presence of branching at its reducing end, whereas LNDFH I gave rise to two major peaks without the reducing-end branching (Figure S2c, d). Similarly, although both are branched at the reducing end, sLea produced multiple conformations, whereas sLex exhibited only one major peak in its EIM (Figure S2e, f).
Figure 2.
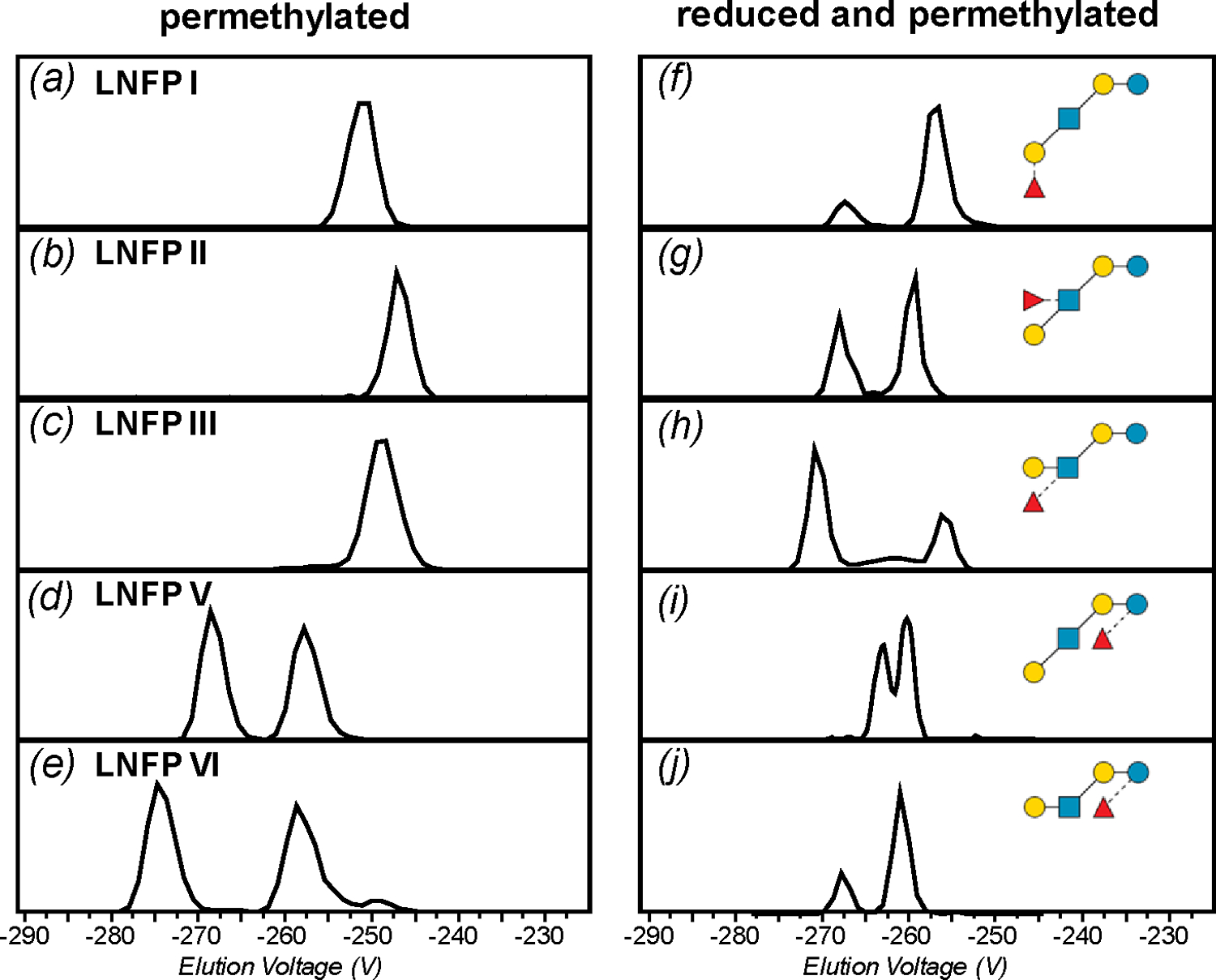
Extracted ion mobiligrams (EIMs) of permethylated (a-e) and reduced and permethylated (f-j) LNFP I, II, III, V, and VI, [M+Na]+, respectively.
Many of the non-reduced glycans studied here showed multiple conformations in the gas phase, and we therefore next examined whether reduction can simplify their mobility profiles. The EIMs of reduced and permethylated glycans are shown in the right panels in Figures 2 and S2. The number of major conformers for each isomer and their measured G-TIMSCCSN2 and K0 values are summarized in Table S1. Surprisingly, a majority of the reduced glycans produced more peaks than their non-reduced counterparts, with none showing a decrease in the number of conformations upon reduction. This is in sharp contrast with the previous report that reduced glycan-phosphate adducts exhibited a lower degree of conformational heterogeneity.55 Such a difference may be rationalized by recognizing that, in metal-cationized permethylated glycans, which no longer bear any free OH or NH groups, intramolecular hydrogen bonding is minimal, and the interplay between stabilization by metal binding and the steric tension caused by maximizing metal coordination becomes the main determinant for conformational stability. After the reducing-end is converted to an open-ring structure by chemical reduction, the glycan gains a higher degree of conformational flexibility for metal binding, potentially leading to more diverse gas-phase structures. Although further studies are needed to understand the effect of reduction on glycan conformations, the results presented here clearly showed that the presence of anomers is not the only factor contributing to the gas-phase glycan conformational diversity. Because there is no universal way to eliminate glycan conformational heterogeneity, there is a pressing need to develop IM-MS methods that can distinguish isomers from conformers, and assign the correct structures to each.
G-TIMS EED MS/MS analysis of LNFP II/III mixtures
We have previously demonstrated that isomeric glycans can be separated by SA-TIMS and subsequently identified by EED MS/MS. However, even with the higher mobility resolution offered by TIMS, some isomers remain unresolved. For example, the measured CCS values of singly sodiated and permethylated LNFP II and III are 319.3 Å2 and 321.7 Å2, respectively, requiring a mobility resolving power of around 300 for baseline separation. Here, G-TIMS analyses were carried out at a moderate resolving power of around 100 to ensure sufficient sensitivity for on-line EED MS/MS analysis, and thus only partially resolved peaks were observed for these two isomers (Figure 3a, top panel). Figure 3b and 3c show the EED spectra acquired at the leading edge of the first peak and the trailing edge of the second peak. In both cases, EED generated complete series of B, C, Y, Z, and 1,5X ions, allowing easy reconstruction of the glycan topology. Several linkage-specific fragments were also produced. The LNFP III-specific 3,5A2, C2/Z3β, and Z3α•-Ac ions (labeled in red) are observed in much higher abundance in Figure 3b, whereas the LNFP II-specific 1,3A2, C2/Z3α, and Z3β•-Ac ions (labeled in green) are more abundantly present in Figure 3c. Proposed mechanisms for the formation of C/Z and Z•-Ac ions and their utility in linkage analysis were discussed in detail previously.4 Here, the EIMs of the diagnostic fragments, e.g. C2/Z3β and C2/Z3α ions (Figure 3a, bottom panels), may be used to assign the two peaks to LNFP III (lower mobility) and LNFP II (higher mobility), respectively. This assignment is also consistent with the elution order of LNFP II and III observed during the G-TIMS analysis of individual standards (Figure 2).
Figure 3.
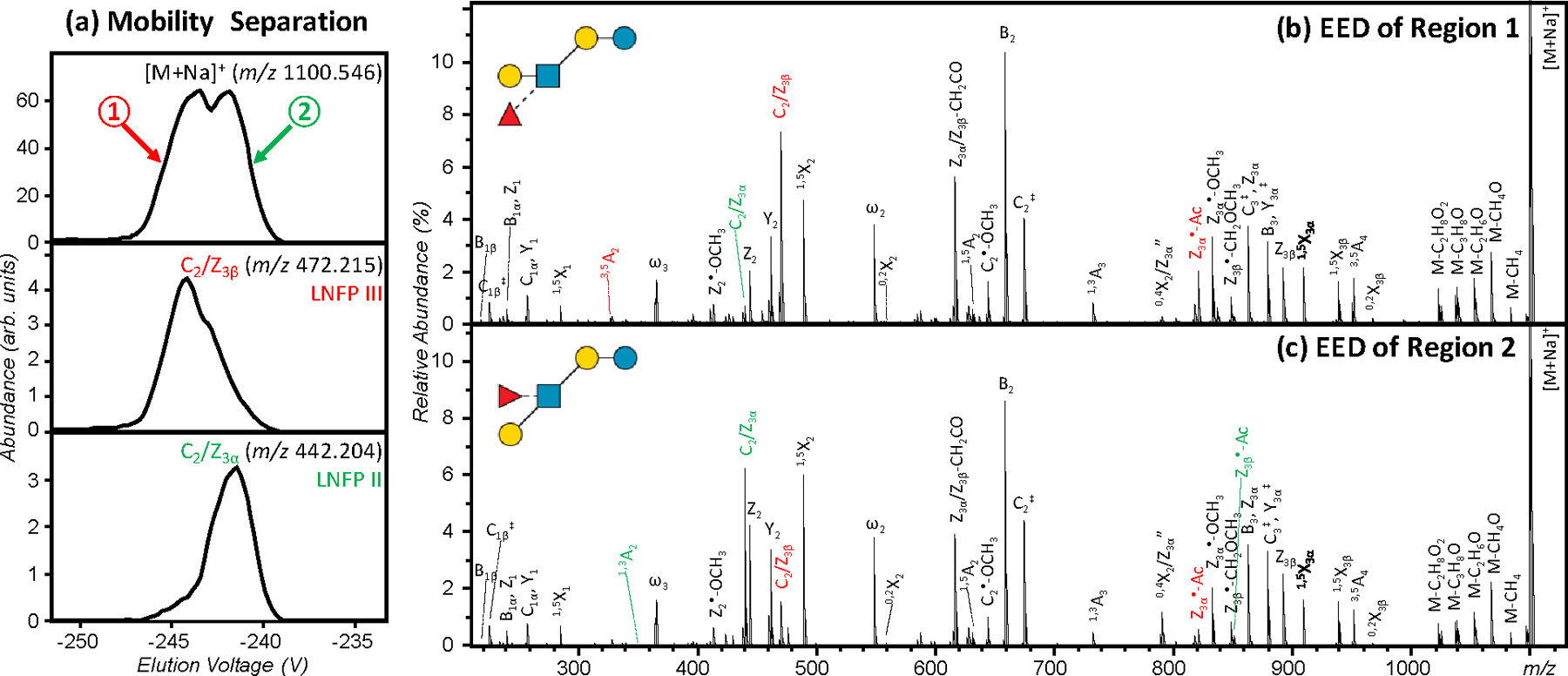
G-TIMS EED MS/MS analysis of a mixture of permethylated LNFP II and III. (a) EIMs of the [M+Na]+ precursor ions and the fragment ions at m/z 442.204 and m/z 472.215; (b, c) EED spectra acquired at two elution voltages marked in (a). Isomer-diagnostic fragments are labeled in color.
Reduction is a common derivatization strategy for glycan analysis to eliminate anomerism-induced chromatographic splitting, or to introduce an isotope label to aid spectral interpretation or relative quantification. Although, as described above, we determined that reduction does not simplify the mobility profiles of metal-adducted permethylated glycans, its wide utilization argues that it is still important to explore the utility of G-TIMS-EED MS/MS for the analysis of reduced and permethylated glycan isomers. For a mixture of reduced and permethylated LNFP II and III, three major peaks were observed in the EIM of the sodiated precursor (Figure 4a). The EED spectra acquired at the rising and falling edges of the first peak and at the apex of the second and third peaks are shown in Figure 4b-e. In all four spectra, complete series of Z-, Y-, and 1,5X-triplets were observed, and the glycan topology can be deduced with confidence. In Figure 4d, the presence of a high-abundance C2/Z3α ion suggests the location of the alpha branch (the non-reducing-end galactose residue) at the C3 position of the GlcNAc residue, consistent with the structure of LNFP II. In Figure 4e, the presence of a high-abundance C2/Z3β ion instead places the beta branch (the fucose residue) at the C3 position, consistent with the structure of LNFP III. Additional fragments characteristic of LNFP II (e.g. 0,4X2/Z3αʺ) and LNFP III (e.g. Z3α•-Ac) are labeled in green and red, respectively. The EIM of the C2/Z3α (Figure 4a, middle panel) contains another component near the falling edge of the first peak, with its EED spectrum (Figure 4c) resembling spectra 4d (LNFP II). Similarly, the EIM of the C2/Z3β ion contains a second component near the rising edge of the first peak, with its EED spectrum (Figure 4b) resembling spectra 4e (LNFP III). The EIMs of these two diagnostic fragments also resemble the mobility profiles of the reduced and permethylated LNFP II and III, respectively (Figure 2g/2f), further confirming the structural assignment. These results show that glycan isomers and conformers may be differentiated based on their EED fragmentation patterns, with conformers producing similar EED spectra, and isomers generating diagnostic fragments for each structure.
Figure 4.
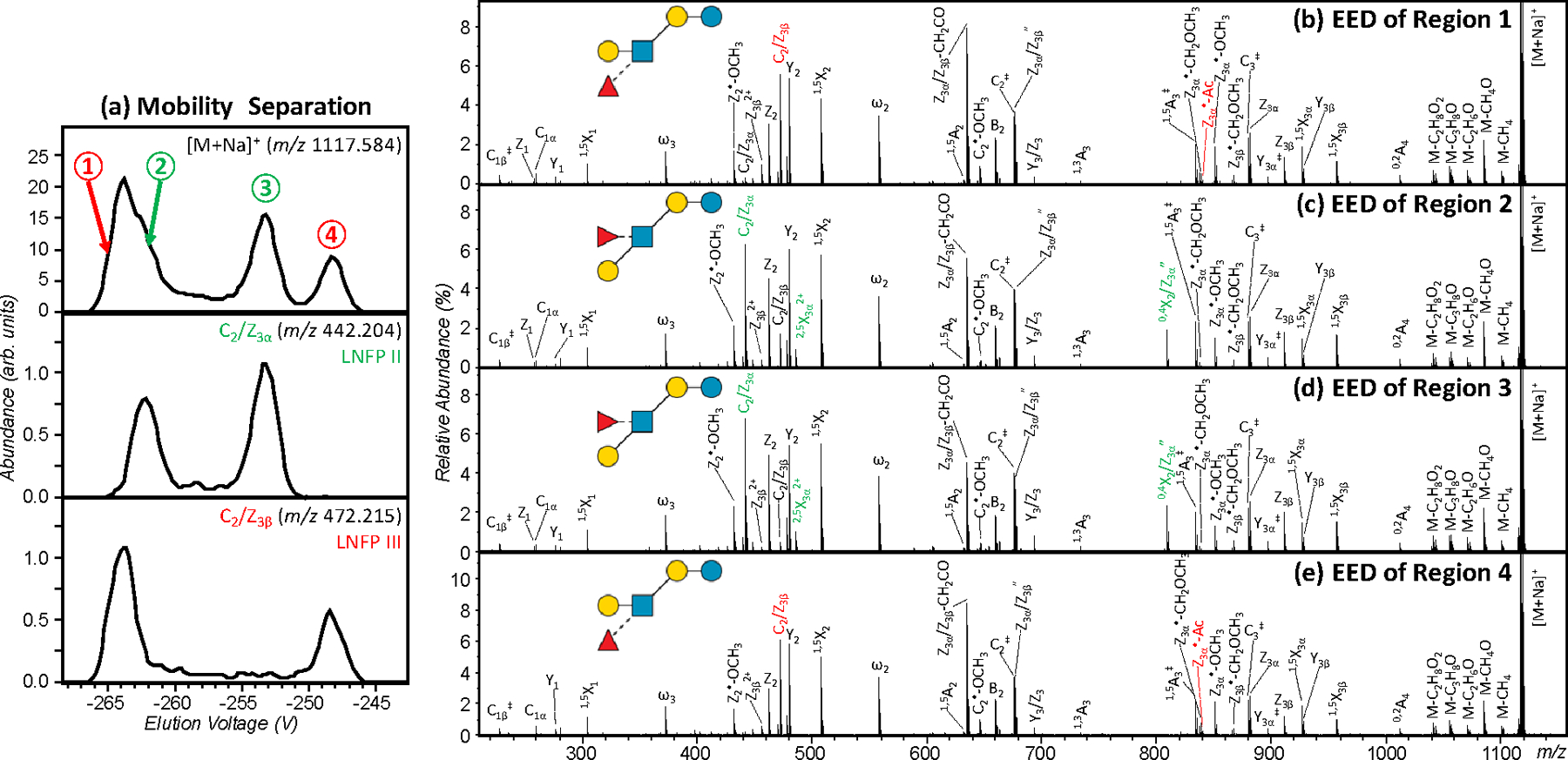
G-TIMS EED MS/MS analysis of a mixture of reduced and permethylated LNFP II and III. (a) EIMs of the [M+Na]+ precursor ions at m/z 1117.584 and two fragment ions at m/z 442.204 and m/z 472.215; (b-e) EED spectra acquired at four elution voltages marked in (a). Isomer-diagnostic fragments are labeled in color.
The discussion thus far has been focused on the analysis of the LNFP II and III binary mixture, as they were the only pair of isomers among five LNFP structures that cannot be resolved by C18 RPLC.4 Based on the measured CCS value of each LNFP isomer (Table S1), an estimated mobility resolving power of at least 500 is required to resolve all conformers of these five LNFP structures, but this is currently unobtainable on our instrument. Supporting Figure S3 shows the G-TIMS EED MS/MS analysis result of the quinary LNFP mixture. Here, even with a moderate resolving power of around 100, differentiation of overlapping mobility profiles could already be achieved by EED MS/MS. For example, the partially resolved LNFP II and III peaks may be identified by the EIMs of the LNFP II-diagnostic C2/Z3α fragment at m/z 442.205 and the LNFP III-diagnostic C2/Z3β fragment at m/z 472.215. For very complex mixtures, off-line LC fractionation can be performed first, to reduce the sample complexity, and subsequent high-resolution IM-EED MS/MS analysis may be utilized to further separate and characterize isomers that are not resolved by LC, while providing valuable CCS fingerprints to facilitate analyte identification.
Structures of H5N4 N-glycans released from ovalbumin
To demonstrate the utility of the G-TIMS EED MS/MS method for isomer analysis, we next applied it to characterize N-glycans released from chicken ovalbumin. Glycans with the H5N4 composition were chosen as the subject of this study, as several isomeric structures have been proposed in the literature for H5N4, yet no consensus exists among different reports.19,41,57–59
The IMS behavior of the H5N4 glycans was first studied by Plasencia and coworkers using a drift-tube instrument. Three partially separated peaks were observed in the ATD of the doubly sodiated, non-reduced and permethylated precursor ion, and assigned to three H5N4 isomers, 1, 2, and 3, based on the molecular modeling results. No tandem MS analysis was performed to validate these assignments, and the authors acknowledged that ambiguity may arise because the IMS-separated features could also correspond to multiple conformations of the same isomer.19 The authors’ choice of these structures for MD simulations was likely influenced by earlier reports suggesting their presence based on MALDI-MS/MS analyses of glycan mixtures.57,58 However, without prior isomer separation or further investigation by MSn analysis, errors can occur when assigning structures from tandem mass spectra of a mixture of isomers. Note that these putative structural assignments, with linkages assumed, or left unassigned, were based on pre-existing biological knowledge.
Here, with improved mobility resolving power, G-TIMS separation of the permethylated H5N4 glycans from chicken ovalbumin produced three baseline-resolved peaks for the doubly sodiated precursor (Figure 5a). Although the measured G-TIMSCCSN2 values (Figure 5d) cannot be directly compared to the DTCCSHe values reported by Plasencia et al., the general shape of the EIM of [H5N4+2Na]2+, m/z 1046.5115, including the number of features, their spacing and relative abundances, closely resembles the ATD of the same precursor in the earlier study. The EED spectra acquired at the peak of elution for these three features are remarkably similar to one another (Figure 6a-c). A bisected hybrid structure, H5N4 isomer 1, may be assigned, based on the following C-type fragments: C1α (m/z 259.1151), C1β or C1γ (m/z 300.1417), C2α (m/z 667.3143), C2β (m/z 504.2412), C3‡ (m/z 1563.7514, 1+, and 794.3789, 2+), and C4 (m/z 1810.8933, 1+, and 916.9407, 2+), along with other supporting B, Z, Y, and 1,5X ions. The presence of a prominent C3/ Z3β ion at m/z 1084.5143 supports a HexNAc1→3Hex branch at the core mannose, whereas the observation of a 0,4A3 ion at m/z 709.3249 suggests the location of the Hex3 branch at the 6-antenna. The 6-branch may be further determined as a branched tri-saccharide with one hexose located at the 3-position based on an abundant C2α/Z4α′ ion at m/z 431.1887, and the other at the 6 position based on the presence of a 0,4A2α ion at m/z 301.1256. Moreover, the observation of a 1,3A3 ion at m/z 560.2674, and a 3,5A3 ion at m/z 982.4821, in addition to the 0,4A3 and C3/ Z3β ions discussed above, suggests the presence of a bisecting HexNAc at the C4 position of the core mannose. In contrast, no glycosidic fragments that would have been unique to the H5N4 isomer 2, e.g. C2β(Hex2) and C3α(Hex2HexNAc), or the H5N4 isomer 3, e.g. C2α(HexHexNAc2), were present in any of these three EED spectra. Thus, the three IM features observed for the H5N4 glycans likely correspond to three conformers of the H5N4 isomer 1, rather than the three different isomers previously suggested. This assignment was confirmed by G-TIMS EED MS/MS analysis of the bisected hybrid H5N4 standard, which produced nearly identical EIM and EED spectra (Figures 5b and S4).
Figure 5.
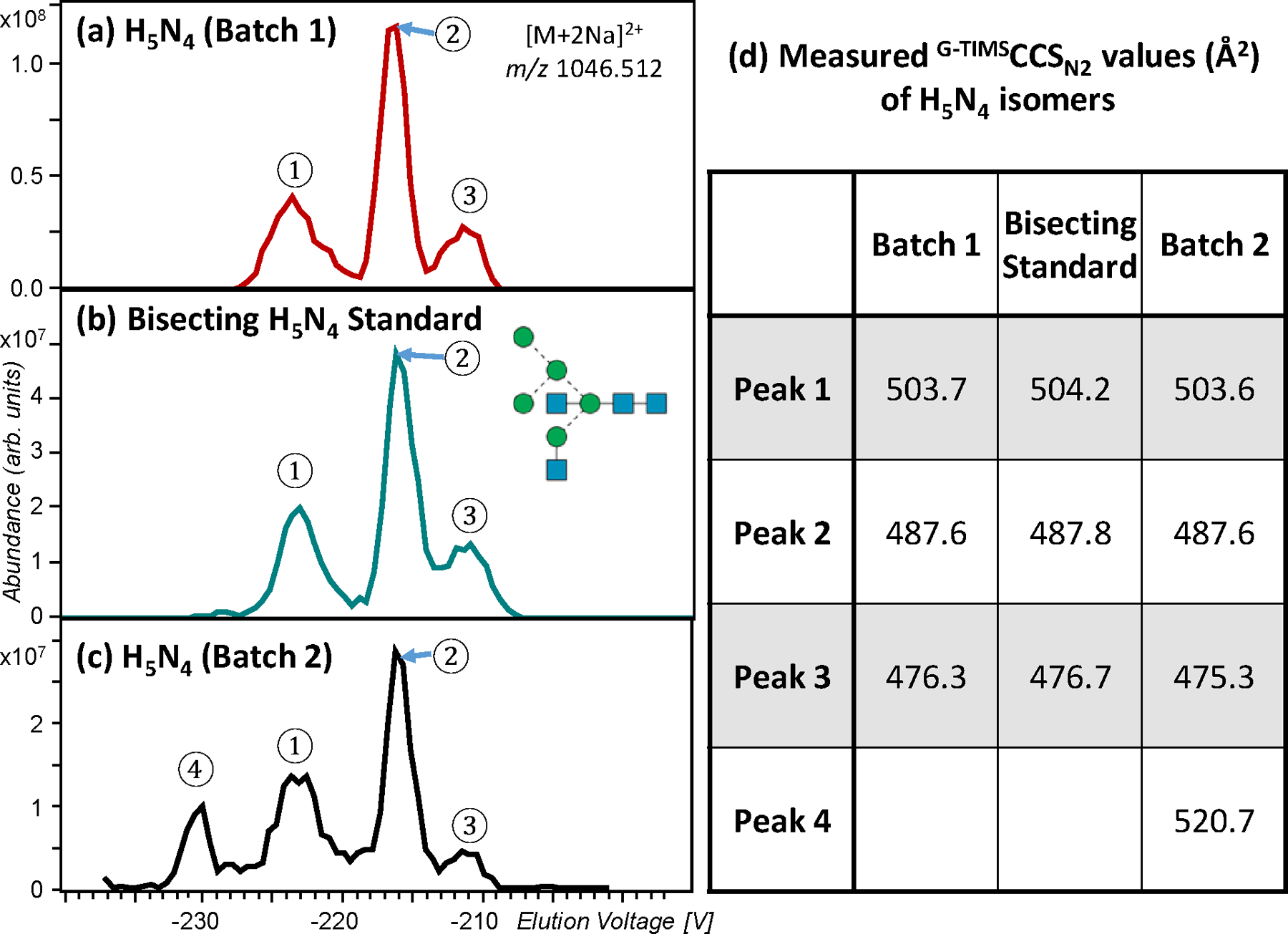
G-TIMS EED MS/MS analysis of permethylated H5N4 glycans. (a-c) EIMs of the H5N4 glycans ([M+2Na]2+, m/z 1046.5122) released from chicken ovalbumin Batch 1, the bisected GlcNAc glycan standard (H5N4 isomer #1 in Figure 1), and H5N4 released from chicken ovalbumin Batch 2, respectively. (d) CCS values of H5N4 isomers measured by G-TIMS.
Figure 6.
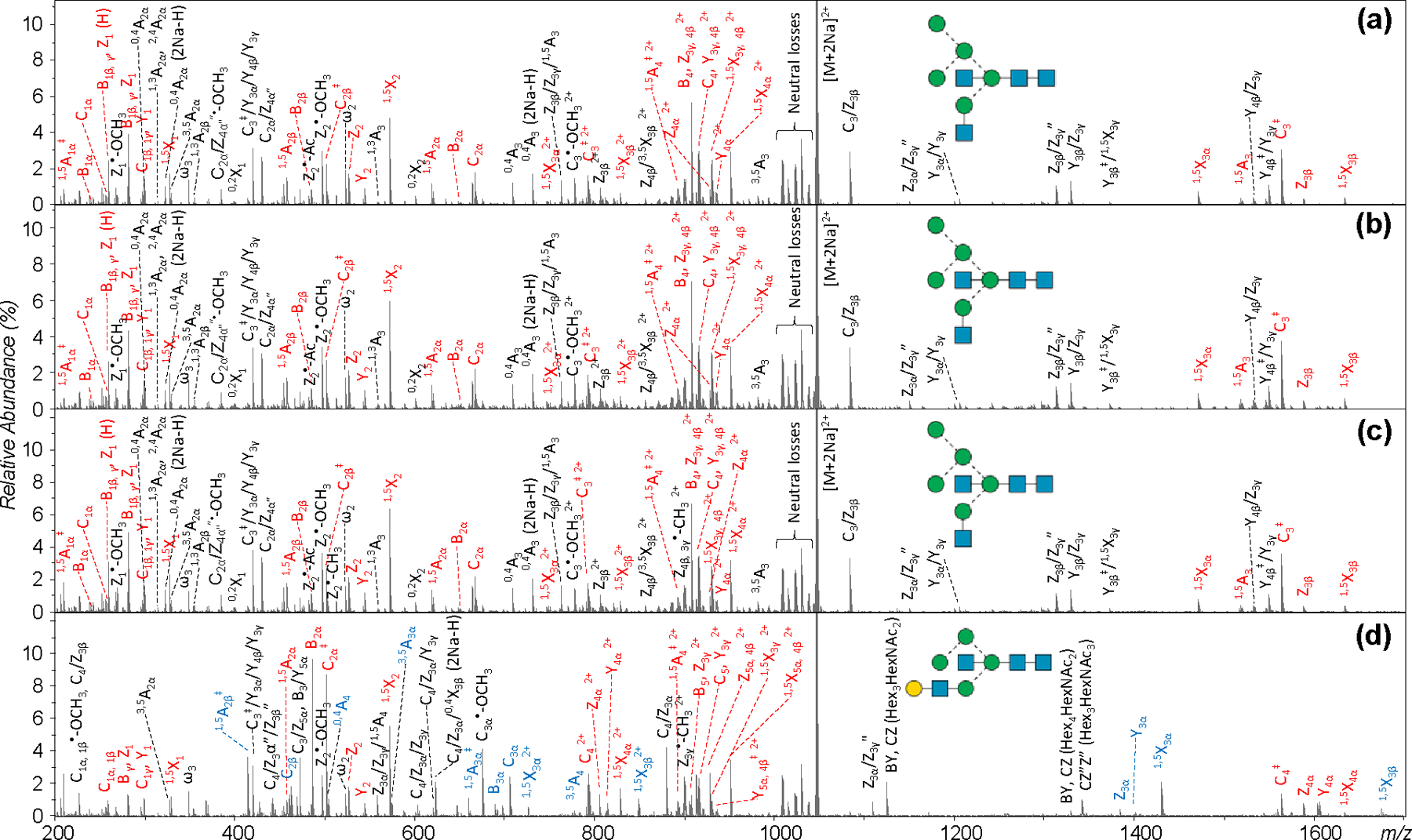
(a-c) EED spectra of H5N4 ([M+2Na]2+, m/z 1046.512) from chicken ovalbumin Batch 1, acquired at the positions marked as 1, 2, and 3 in Figure 5a, respectively. (d) The EED spectrum of H5N4 from chicken ovalbumin Batch 2, acquired at position 4 in Figure 5c. Fragments discussed in the text are labeled in color.
It was recently shown that anomers and the open-ring form of one glycan structure can give rise to multiple mobility features with dissimilar CID fragmentation patterns.33 These IM features arise from a single isomer because it can interconvert in solution and in the gas phase upon activation,60 yet differences in their CID tandem mass spectra may be mistaken as evidence for the presence of structural isomers. To examine whether the gas-phase conformations of permethylated glycans can similarly influence their fragmentation behaviors, CID MS/MS analysis was performed on the three conformers of the sodiated and permethylated H5N4 structure 1 (Figure 5a). As shown in Figure S5, CID spectra of these conformers did not differ as much as those of sodium-bound, native glycans.33 This is perhaps not surprising since the most dissimilar CID spectrum was generated by the open-ring form, which does not exist in unreduced and permethylated glycans. Without the ability to perform IMS/IMS analysis, it was difficult to correlate these changes in fragment ion abundances to the reducing-end configuration. Given that conformational heterogeneity was prevalent in reduced and permethylated glycans, these IM features may also have resulted from other structural variables, such as changes in the sodium coordination sites. Nonetheless, some CID fragments still exhibited significant differences in their relative abundances among three conformers (Figure S6), and the CID spectra of conformers appeared to differ more than their corresponding EED spectra (Figure 6). Figure S7 shows a comparison of the EED and CID cleavage maps. Whereas EED produced a complete series of glycosidic cleavages and an abundance of cross-ring and secondary fragments that enabled detailed structural elucidation, CID did not generate a sufficient number of fragments for either topology or linkage determination.
Recently, Harvey et al. performed an IM-MS/MS study on hybrid and complex glycans from chicken ovalbumin, using a TWIMS-TOF MS instrument. With limited mobility resolution, a single symmetrical mobility peak was observed for the H5N4 glycans, either in sodiated forms ([M+ Na]+ and [M+ 2Na]2+), or as a phosphate adduct, [M+H2PO4]-; this result provided little evidence for the presence of more than one structure.41 The negative CID spectrum of the phosphate adduct following the IM separation revealed structure 1 as the major H5N4 isomer. However, they also observed several minor fragments that could not have been produced by structure 1, but were consistent with structures 2 and 4. The CID fragmentation data were insufficient to define these structures, and the assignment of one minor component to structure 4 was based on the match of its CCS value to that of the biantennary G2 glycan from another glycoprotein standard. Structure 2 was further excluded as the ATD profiles of the characteristic fragments appeared at an earlier arrival time than the main feature (structure 1), and this was inconsistent with previous calculations19 showing that structure 2 has a considerably larger CCS value than structure 1.
Harvey et al. suggested that the difference in findings between the Plasencia study and theirs may have resulted from the different composition of the samples used. This is to be expected as batch-to-batch variation in the glycosylation profiles can result from changes in the expression systems, cell culture conditions, and downstream processing. In the biopharmaceutical industry, such variability must be controlled and defined as glycosylation of therapeutic proteins influences their efficacy, pharmacokinetics, and immunogenicity. The Harvey study suggested that IM-MS/MS can be a valuable tool for characterization of glycosylation changes at the structural level. However, because of the lower mobility resolution and limited fragmentation data, structural assignments could only be made by comparing the measured CCS values to known glycan standards, and the fragment ATD shift to MD simulation results. Thus, assignment accuracy relies on the availability of glycan standards, and the thoroughness and accuracy of the MD simulation.
With a higher mobility resolving power and a fragmentation method capable of delivering more structural details and differentiating isomers from conformers, the G-TIMS-EED MS/MS approach could provide a more reliable method for monitoring lot-to-lot glycosylation changes. To test this hypothesis, we performed G-TIMS-EED MS/MS analysis on N-glycans released from a different batch of chicken ovalbumin. Interestingly, a fourth peak (Peak 4 in Figure 5c) was observed in the EIM of the [H5N4+2Na]2+ precursor from this sample. Figure S8 shows the EED spectra of peaks 1–3 in Figure 5c, which are identical to those shown in Figure S4, thus confirming the presence of structure 1 as the major H5N4 isomer in batch 2 as well. The EED spectrum of peak 4 (Figure 6d), however, was drastically different. Figure 6d contains the following C-ion series: C1α or C1β (m/z 259.1152), C1γ (m/z 300.1417), C2β (m/z 463.2149), C2α‡ (m/z 502.2258), C3α‡ (m/z 706.3263), C4‡ (m/z 1563.7511, 1+, and 794.3784, 2+), and C5 (m/z 916.9411, 2+), as well as other supporting B, Z, Y, and 1,5X ions that are consistent with the presence of structure 2. The bisecting GlcNAc can be confirmed based on the observation of a 0,4A4 ion at m/z 505.2255, along with a 3,5A4 ion at m/z 778.3831, and a 1,3A4 ion at m/z 764.3686. The observation of a 1,3A4 ion and a high-abundance C4/Z3α ion at m/z 880.4148 suggests the presence of a Gal-GlcNAc-Man 3-antenna. Note that the glycosidic fragments from the G2 glycan (structure 4) are a subset of those from structure 2, thus its presence (in addition to structure 2) cannot be ruled out based on glycosidic cleavages alone. However, the absence of any G2-specific cross-ring fragments, such as 0,4A4 at m/z 750.3519 and 0,4A4 (2Na-H) at m/z 772.3339 (Figure S9, zoomed-in regions of Figure 6d, and Figure S10, EED spectrum of the G2 standard), may be used to exclude its presence in peak 4. In contrast, the C2β(Hex2) ion and 0,4A3β ion at m/z 505.2255 are unique to structure 2, and cannot be produced by structures 1, 3, or 4. Further, peak 4 eluted at a higher voltage (larger CCS value) than peaks 1–3 from structure 1, and this is consistent with the Plasencia report showing that structure 2 has the largest calculated CCS value among structures 1, 2, and 3, whereas Harvey et al. showed that structure 4 has a smaller CCS than structure 1. The 6-branch may be determined as Man1→3Man based on the absence of a 0,4A2β ion at m/z 301.1258 and the presence of a 1,3A2β ion at m/z 315.1415 along with a C2/Z4β ion at m/z 227.0890. Similarly, the presence of a 3,5A3α ion at m/z 574.2833 and the absence of a 0,4A3α ion at m/z 546.2521 and a 1,3A3α ion at m/z 560.2677 suggested that the GlcNAc residue is connected to the 3-arm mannose via a 1→4 linkage. This, along with the presence of a 3,5A2α ion at m/z 329.1571 may be used to determine the 3-branch as Gal1→4GlcNAc1→4Man. Thus, all linkages in the putative H5N4 structure 2 can be determined, and structure 2a may be assigned to the minor H5N4 isomer seen in Batch 2. Although the glycan standard for structure 2a was not commercially available, we showed here that peak 4 could be confidently assigned on the basis of an abundance of supporting glycosidic, cross-ring and secondary fragments produced by EED MS/MS, and further substantiated by its elution order relative to structure 1. The EED cleavage map of structure 2a is shown in Figure S11.
CONCLUSIONS
Our investigation on the ion mobility behavior of several sets of isomeric glycans showed that conformational multiplicity is common in permethylated glycans, regardless of the reducing-end status. The apparent lack of a universal strategy to eliminate conformational heterogeneity advocates the use of IMS devices with higher mobility resolving power for better isomer resolution and fragmentation methods that can distinguish conformers from isomers. Enhanced mobility resolution achieved by G-TIMS not only allows better isomer separation, but also provides conformational fingerprints that could improve the confidence of CCS-based glycan identification. EED offers two major advantages for isomer analysis over CID, in that it generates extensive fragmentation that facilitates detailed structural elucidation and isomer identification, and its fragmentation behavior is largely independent of the gas-phase conformation, thus allowing differentiation of conformers and isomers. Results from the present and previous IM-MS studies of the H5N4 glycans from chicken ovalbumin clearly demonstrated the necessity of high mobility resolution and efficient fragmentation for accurate isomer analysis, and the power of the G-TIMS EED MS/MS approach for characterization of the batch-to-batch variation of protein glycosylation at the structural level, beyond changes in glycan compositions.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grants R01 GM232675 (to CL), and R24 GM134210 and S10 RR025082 (to CEC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agency.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Details about materials and sample preparation; Table S1, measured CCS and K0 values of the glycan standards; Scheme S1, Domon-Costello nomenclature for glycan fragmentation; Figure S1-S11, supporting figures as noted in the text; Table S2-3, lists of assigned peaks in the EED spectra of the two H5N4 glycan isomers from chicken ovalbumin.
References
- (1).Mancera-Arteu M; Giménez E; Barbosa J; Sanz-Nebot V Identification and characterization of isomeric N-glycans of human alfa-acid-glycoprotein by stable isotope labelling and ZIC-HILIC-MS in combination with exoglycosidase digestion Anal. Chim. Acta 2016, 940, 92–103. [DOI] [PubMed] [Google Scholar]
- (2).Zhao J; Li S; Li C; Wu S-L; Xu W; Chen Y; Shameem M; Richardson D; Li H Identification of low abundant isomeric N-glycan structures in biological therapeutics by LC/MS Anal. Chem 2016, 88, 7049–7059. [DOI] [PubMed] [Google Scholar]
- (3).Mancera-Arteu M; Giménez E; Barbosa J; Peracaula R; Sanz-Nebot V Zwitterionic-hydrophilic interaction capillary liquid chromatography coupled to tandem mass spectrometry for the characterization of human alpha-acid-glycoprotein N-glycan isomers Anal. Chim. Acta 2017, 991, 76–88. [DOI] [PubMed] [Google Scholar]
- (4).Tang Y; Wei J; Costello CE; Lin C Characterization of Isomeric Glycans by Reversed Phase Liquid Chromatography-Electronic Excitation Dissociation Tandem Mass Spectrometry J. Am. Soc. Mass. Spectrom 2018, 29, 1295–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Zhou S; Hu Y; Mechref Y High‐temperature LC‐MS/MS of permethylated glycans derived from glycoproteins Electrophoresis 2016, 37, 1506–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Vreeker GCM; Wuhrer M Reversed-phase separation methods for glycan analysis Anal. Bioanal. Chem 2017, 409, 359–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Huang Y; Zhou S; Zhu J; Lubman DM; Mechref Y LC-MS/MS isomeric profiling of permethylated N-glycans derived from serum haptoglobin of hepatocellular carcinoma (HCC) and cirrhotic patients Electrophoresis 2017, 38, 2160–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Pabst M; Altmann F Influence of electrosorption, solvent, temperature, and ion polarity on the performance of LC-ESI-MS using graphitic carbon for acidic oligosaccharides Anal. Chem 2008, 80, 7534–7542. [DOI] [PubMed] [Google Scholar]
- (9).Costello CE; Contado-Miller JM; Cipollo JF A Glycomics Platform for the Analysis of Permethylated Oligosaccharide Alditols J. Am. Soc. Mass. Spectrom 2007, 18, 1799–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Zhou S; Huang Y; Dong X; Peng W; Veillon L; Kitagawa DA; Aquino AJ; Mechref Y Isomeric separation of permethylated glycans by porous graphitic carbon (PGC)-LC-MS/MS at high temperatures Anal. Chem 2017, 89, 6590–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Zhuang Z; Starkey JA; Mechref Y; Novotny MV; Jacobson SC Electrophoretic Analysis of N-Glycans on Microfluidic Devices Anal. Chem 2007, 79, 7170–7175. [DOI] [PubMed] [Google Scholar]
- (12).Snyder CM; Zhou X; Karty JA; Fonslow BR; Novotny MV; Jacobson SC Capillary electrophoresis–mass spectrometry for direct structural identification of serum N-glycans J. Chromatogr. A 2017, 1523, 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Harvey DJ; Scarff CA; Edgeworth M; Struwe WB; Pagel K; Thalassinos K; Crispin M; Scrivens J Travelling-wave ion mobility and negative ion fragmentation of high-mannose N-glycans J. Mass Spectrom 2016, 51, 219–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Harvey DJ; Scarff CA; Edgeworth M; Crispin M; Scanlan CN; Sobott F; Allman S; Baruah K; Pritchard L; Scrivens JH Travelling wave ion mobility and negative ion fragmentation for the structural determination of N‐linked glycans Electrophoresis 2013, 34, 2368–2378. [DOI] [PubMed] [Google Scholar]
- (15).Both P; Green AP; Gray CJ; Šardzík R; Voglmeir J; Fontana C; Austeri M; Rejzek M; Richardson D; Field RA; Widmalm G; Flitsch SL; Eyers CE Discrimination of epimeric glycans and glycopeptides using IM-MS and its potential for carbohydrate sequencing Nat. Chem 2013, 6, 65. [DOI] [PubMed] [Google Scholar]
- (16).Pagel K; Harvey DJ Ion Mobility–Mass Spectrometry of Complex Carbohydrates: Collision Cross Sections of Sodiated N-linked Glycans Anal. Chem 2013, 85, 5138–5145. [DOI] [PubMed] [Google Scholar]
- (17).Zheng X; Zhang X; Schocker NS; Renslow RS; Orton DJ; Khamsi J; Ashmus RA; Almeida IC; Tang K; Costello CE; Smith RD; Michael K; Baker ES Enhancing glycan isomer separations with metal ions and positive and negative polarity ion mobility spectrometry-mass spectrometry analyses Anal. Bioanal. Chem 2017, 409, 467–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Zhu F; Lee S; Valentine SJ; Reilly JP; Clemmer DE Mannose7 Glycan Isomer Characterization by IMS-MS/MS Analysis J. Am. Soc. Mass. Spectrom 2012, 23, 2158–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Plasencia MD; Isailovic D; Merenbloom SI; Mechref Y; Clemmer DE Resolving and assigning N-linked glycan structural isomers from ovalbumin by IMS-MS J. Am. Soc. Mass. Spectrom 2008, 19, 1706–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Struwe WB; Benesch JL; Harvey DJ; Pagel K Collision cross sections of high-mannose N-glycans in commonly observed adduct states – identification of gas-phase conformers unique to [M − H]− ions Analyst 2015, 140, 6799–6803. [DOI] [PubMed] [Google Scholar]
- (21).Huang Y; Dodds ED Discrimination of Isomeric Carbohydrates as the Electron Transfer Products of Group II Cation Adducts by Ion Mobility Spectrometry and Tandem Mass Spectrometry Anal. Chem 2015, 87, 5664–5668. [DOI] [PubMed] [Google Scholar]
- (22).Williams JP; Grabenauer M; Holland RJ; Carpenter CJ; Wormald MR; Giles K; Harvey DJ; Bateman RH; Scrivens JH; Bowers MT Characterization of simple isomeric oligosaccharides and the rapid separation of glycan mixtures by ion mobility mass spectrometry Int. J. Mass spectrom 2010, 298, 119–127. [Google Scholar]
- (23).Hofmann J; Struwe WB; Scarff CA; Scrivens JH; Harvey DJ; Pagel K Estimating Collision Cross Sections of Negatively Charged N-Glycans using Traveling Wave Ion Mobility-Mass Spectrometry Anal. Chem 2014, 86, 10789–10795. [DOI] [PubMed] [Google Scholar]
- (24).Seo Y; Andaya A; Leary JA Preparation, Separation, and Conformational Analysis of Differentially Sulfated Heparin Octasaccharide Isomers Using Ion Mobility Mass Spectrometry Anal. Chem 2012, 84, 2416–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Mosely JA; Smith MJ; Prakash AS; Sims M; Bristow AW Electron-induced dissociation of singly charged organic cations as a tool for structural characterization of pharmaceutical type molecules Anal. Chem 2011, 83, 4068–4075. [DOI] [PubMed] [Google Scholar]
- (26).Revercomb HE; Mason EA Theory of plasma chromatography/gaseous electrophoresis Review Anal. Chem 1975, 47, 970–983. [Google Scholar]
- (27).Struwe WB; Pagel K; Benesch JLP; Harvey DJ; Campbell MP GlycoMob: an ion mobility-mass spectrometry collision cross section database for glycomics Glycoconj. J 2016, 33, 399–404. [DOI] [PubMed] [Google Scholar]
- (28).Glaskin RS; Khatri K; Wang Q; Zaia J; Costello CE Construction of a Database of Collision Cross Section Values for Glycopeptides, Glycans, and Peptides Determined by IM-MS Anal. Chem 2017, 89, 4452–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Chen Z; Glover MS; Li L Recent advances in ion mobility–mass spectrometry for improved structural characterization of glycans and glycoconjugates Curr. Opin. Chem. Biol 2018, 42, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Wiederschain GY Essentials of glycobiology Biochem. (Moscow) 2009, 74, 1056–1056. [Google Scholar]
- (31).Adams KJ; Montero D; Aga D; Fernandez-Lima F Isomer separation of polybrominated diphenyl ether metabolites using nanoESI-TIMS-MS Int. J. Ion Mobil. Spectrom 2016, 19, 69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Deng L; Webb IK; Garimella SVB; Hamid AM; Zheng X; Norheim RV; Prost SA; Anderson GA; Sandoval JA; Baker ES; Ibrahim YM; Smith RD Serpentine Ultralong Path with Extended Routing (SUPER) High Resolution Traveling Wave Ion Mobility-MS using Structures for Lossless Ion Manipulations Anal. Chem 2017, 89, 4628–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Ujma J; Ropartz D; Giles K; Richardson K; Langridge D; Wildgoose J; Green M; Pringle S Cyclic Ion Mobility Mass Spectrometry Distinguishes Anomers and Open-Ring Forms of Pentasaccharides J. Am. Soc. Mass. Spectrom 2019, 30, 1028–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Nagy G; Attah IK; Garimella SVB; Tang K; Ibrahim YM; Baker ES; Smith RD Unraveling the isomeric heterogeneity of glycans: ion mobility separations in structures for lossless ion manipulations Chem. Commun 2018, 54, 11701–11704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Leyva D; Tose LV; Porter J; Wolff J; Jaffé R; Fernandez-Lima F Understanding the structural complexity of dissolved organic matter: isomeric diversity Faraday Discuss 2019, 218, 431–440. [DOI] [PubMed] [Google Scholar]
- (36).Ritamo I; Räbinä J; Natunen S; Valmu L Nanoscale reversed-phase liquid chromatography–mass spectrometry of permethylated N-glycans Anal. Bioanal. Chem 2013, 405, 2469–2480. [DOI] [PubMed] [Google Scholar]
- (37).Struwe WB; Baldauf C; Hofmann J; Rudd PM; Pagel K Ion mobility separation of deprotonated oligosaccharide isomers – evidence for gas-phase charge migration Chem. Commun 2016, 52, 12353–12356. [DOI] [PubMed] [Google Scholar]
- (38).Mucha E; Stuckmann A; Marianski M; Struwe WB; Meijer G; Pagel K In-depth structural analysis of glycans in the gas phase Chemical Science 2019, 10, 1272–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Re S; Watabe S; Nishima W; Muneyuki E; Yamaguchi Y; MacKerell AD; Sugita Y Characterization of Conformational Ensembles of Protonated N-glycans in the Gas-Phase Sci. Rep 2018, 8, 1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Harvey DJ; Seabright GE; Vasiljevic S; Crispin M; Struwe WB Isomer Information from Ion Mobility Separation of High-Mannose Glycan Fragments J. Am. Soc. Mass. Spectrom 2018, 29, 972–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Harvey DJ; Scarff CA; Edgeworth M; Pagel K; Thalassinos K; Struwe WB; Crispin M; Scrivens JH Travelling‐wave ion mobility mass spectrometry and negative ion fragmentation of hybrid and complex N‐glycans J. Mass Spectrom 2016, 51, 1064–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Ashwood C; Lin C-H; Thaysen-Andersen M; Packer NH Discrimination of isomers of released N-and O-Glycans using diagnostic product ions in negative ion PGC-LC-ESI-MS/MS J. Am. Soc. Mass. Spectrom 2018, 29, 1194–1209. [DOI] [PubMed] [Google Scholar]
- (43).Yu X; Jiang Y; Chen Y; Huang Y; Costello CE; Lin C Detailed glycan structural characterization by electronic excitation dissociation Anal. Chem 2013, 85, 10017–10021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Pu Y; Ridgeway ME; Glaskin RS; Park MA; Costello CE; Lin C Separation and identification of isomeric glycans by selected accumulation-trapped ion mobility spectrometry-electron activated dissociation tandem mass spectrometry Anal. Chem 2016, 88, 3440–3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Yu X; Huang Y; Lin C; Costello CE Energy-Dependent Electron Activated Dissociation of Metal-Adducted Permethylated Oligosaccharides Anal. Chem 2012, 84, 7487–7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Hong P; Sun H; Sha L; Pu Y; Khatri K; Yu X; Tang Y; Lin C GlycoDeNovo–an Efficient Algorithm for Accurate de novo Glycan Topology Reconstruction from Tandem Mass Spectra J. Am. Soc. Mass. Spectrom 2017, 28, 2288–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Ridgeway ME; Wolff JJ; Silveira JA; Lin C; Costello CE; Park MA Gated trapped ion mobility spectrometry coupled to fourier transform ion cyclotron resonance mass spectrometry Int. J. Ion Mobil. Spectrom 2016, 19, 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Ciucanu I; Kerek F A simple and rapid method for the permethylation of carbohydrates Carbohydr. Res 1984, 131, 209–217. [Google Scholar]
- (49).Ciucanu I; Costello CE Elimination of Oxidative Degradation during the per-O-Methylation of Carbohydrates J. Am. Chem. Soc 2003, 125, 16213–16219. [DOI] [PubMed] [Google Scholar]
- (50).Wei J; Wu J; Tang Y; Ridgeway ME; Park MA; Costello CE; Zaia J; Lin C Characterization and Quantification of Highly Sulfated Glycosaminoglycan Isomers by Gated-Trapped Ion Mobility Spectrometry Negative Electron Transfer Dissociation MS/MS Anal. Chem 2019, 91, 2994–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Stow SM; Causon TJ; Zheng X; Kurulugama RT; Mairinger T; May JC; Rennie EE; Baker ES; Smith RD; McLean JA; Hann S; Fjeldsted JC An Interlaboratory Evaluation of Drift Tube Ion Mobility–Mass Spectrometry Collision Cross Section Measurements Anal. Chem 2017, 89, 9048–9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Hernandez DR; DeBord JD; Ridgeway ME; Kaplan DA; Park MA; Fernandez-Lima F Ion dynamics in a trapped ion mobility spectrometer Analyst 2014, 139, 1913–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Ceroni A; Maass K; Geyer H; Geyer R; Dell A; Haslam SM GlycoWorkbench: A Tool for the Computer-Assisted Annotation of Mass Spectra of Glycans J. Proteome Res 2008, 7, 1650–1659. [DOI] [PubMed] [Google Scholar]
- (54).Domon B; Costello CE A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates Glycoconj. J 1988, 5, 397–409. [Google Scholar]
- (55).Harvey DJ; Abrahams JL Fragmentation and ion mobility properties of negative ions from N-linked carbohydrates: Part 7. Reduced glycans Rapid Commun. Mass Spectrom 2016, 30, 627–634. [DOI] [PubMed] [Google Scholar]
- (56).Morelle W; Michalski J-C Analysis of protein glycosylation by mass spectrometry Nat. Protoc 2007, 2, 1585–1602. [DOI] [PubMed] [Google Scholar]
- (57).Lattova E; Perreault H; Krokhin O Matrix-assisted laser desorption/ionization tandem mass spectrometry and post-source decay fragmentation study of phenylhydrazones of N-linked oligosaccharides from ovalbumin J. Am. Soc. Mass. Spectrom 2004, 15, 725–735. [DOI] [PubMed] [Google Scholar]
- (58).Lattová E; Snovida S; Perreault H; Krokhin O Influence of the labeling group on ionization and fragmentation of carbohydrates in mass spectrometry J. Am. Soc. Mass. Spectrom 2005, 16, 683–696. [DOI] [PubMed] [Google Scholar]
- (59).Jiao J; Zhang H; Reinhold VN High Performance IT-MS Sequencing of Glycans (Spatial Resolution of Ovalbumin Isomers) Int. J. Mass spectrom 2011, 303, 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Spengler B; Dolce JW; Cotter RJ Infrared laser desorption mass spectrometry of oligosaccharides: fragmentation mechanisms and isomer analysis Anal. Chem 1990, 62, 1731–1737. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


