Summary
Endothelial cells (ECs) from the small intestine, colon, liver, and heart have distinct phenotypes and functional adaptations that are dependent on their physiological environment. Gut ECs adapt to low oxygen, heart ECs to contractile forces, and liver ECs to low flow rates. Isolating high-purity ECs in sufficient quantities is crucial to study their functions. Here, we describe protocols combining magnetic and fluorescent activated cell sorting for rapid and reproducible EC purification from four adult murine tissues.
For complete details on the use and execution of these protocols, please refer to Kalucka et al. (2020).
Subject areas: Cell isolation, Single Cell, Flow Cytometry/Mass Cytometry
Graphical abstract

Highlights
-
•
Protocols allowing for simultaneous isolation of murine ECs from different tissue
-
•
Rapid and efficient isolation of ECs from the small intestine, colon, heart, and liver
-
•
Combination of magnetic and fluorescent activated cell sorting
-
•
High purity and quality of isolated murine endothelial cells evident from scRNA-seq
Endothelial cells (ECs) from the small intestine, colon, liver, and heart have distinct phenotypes and functional adaptations that are dependent on their physiological environment. Gut ECs adapt to low oxygen, heart ECs to contractile forces, and liver ECs to low flow rates. Isolating high-purity ECs in sufficient quantities is crucial to study their functions. Here, we describe protocols combining magnetic and fluorescent activated cell sorting for rapid and reproducible EC purification from four adult murine tissues.
Before you begin
The protocols were established using 8-week-old male C57BL6/J mice purchased from Charles River (strain code: 632). All experimental procedures for the establishment and application of the protocols were done under approval by the Institutional Animal Ethics Committee of the KU Leuven (Belgium); protocol number P012/2018.
The protocols described below provide the details on EC isolation from one tissue at a time. However, it is possible to isolate cells from multiple organs of the same mouse. In that case, we recommend to perform transcardial perfusion with ice-cold PBS at a perfusion rate of 2 mL/minute for 5 min, followed by additional perfusion with digestion buffer: Supplemented KnockOutTM DMEM-medium with 0.1% (w/v) collagenase I (Thermo Fisher Scientific, Cat#17018029), 0.1% (w/v) collagenase II (Thermo Fisher Scientific, Cat#17101015) and 7.5 μg/mL DNase I (Sigma-Aldrich, Cat#D4527-10KU) at a perfusion rate of 2 mL/minute for 5 min. The purpose of this step is to remove blood from the blood vessels and replace it with the digestion buffer to ensure efficient digestion. Additionally, if cells from multiple tissues are isolated simultaneously from the same mouse, we suggest to assign one person per single organ isolation.
The cells have been sorted using the BD FACSAria™ III sorter. Considering EC fragility, the following settings have been used: nozzle size - 100 μm, pressure - 20 psi. To maximize sorted EC purity and efficiency, we have been using the 4-way purity and sorting ECs on flow rate 1, respectively.
The optical paths used per fluorochrome:
-
1.
FITC - Laser: Blue 488; Detector 502 LP; Filter set up: 530/30 BP
-
2.
PE-Cy7 - Laser: Yellow-Green 561; Detector 735 LP; Filter set up: 780/60 BP
-
3.
eFluor450 - Laser: Violet 407; Filter set up: 450/40 BP
-
4.
PE - Laser: Yellow-Green 561; Filter set up: 582/15 BP
The settings are tailored for EC sorting with the BD FACSAria™ III sorter and have to be optimized specifically to the sorter of choice.
Of note, the protocols are based on magnetic bead enrichment by magnetic bead for ECs (CD31) and fluorescent activated cell (FACS) sorting. The listed dilution of antibodies might vary between lots and manufacturers/vendors. Therefore, we highly recommend to optimize these parameters for each antibody before the usage.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti-mouse CD31 FITC (clone 390) | Thermo Fisher Scientific | Cat#11-0311-82 RRID: AB_465012 |
| Rat anti-mouse/human CD11b-PE (clone M1/70) | BioLegend | Cat#101208 RRID: AB_312791 |
| Rat anti-mouse CD45 PE-Cy7 (clone 30-F11) | Thermo Fisher Scientific | Cat#25-0451-82 RRID: AB_2734986 |
| Chemicals, peptides, and recombinant proteins | ||
| Dithiothreitol (DTT) | Sigma-Aldrich | Cat#10197777001 |
| DNase I | Sigma-Aldrich | Cat#D4527-10KU |
| EDTA | Sigma-Aldrich | Cat#ED2P-500G |
| Fixable Viability Dye eFluor™ 450 | Thermo Fisher Scientific | Cat#65-0863-18 |
| Sodium pyruvate | Thermo Fisher Scientific | Cat#11360070 |
| Antibiotic-antimycotic | Thermo Fisher Scientific | Cat#15240062 |
| Bovine serum albumin (BSA Fraction V) | Sigma-Aldrich | Cat#10735086001 |
| Collagenase type I | Thermo Fisher Scientific | Cat#17018029 |
| Collagenase type II | Thermo Fisher Scientific | Cat#17101015 |
| Collagenase type IV | Worthington Biochemical | Cat#LS004188 |
| Dispase | Thermo Fisher Scientific | Cat#17105-041 |
| Endothelial cell growth factor supplements (ECGS/Heparin) | PromoCell | Cat#C-30120 |
| Fetal bovine serum (FBS) | Thermo Fisher Scientific | Cat#16000044 |
| Hank's Balanced Salt Solution (HBSS) | Thermo Fisher Scientific | Cat#14025092 |
| KnockOutTM DMEM | Thermo Fisher Scientific | Cat#10829018 |
| MEM NEAA | Thermo Fisher Scientific | Cat#11140035 |
| Penicillin/streptomycin | Thermo Fisher Scientific | Cat#15140122 |
| Phosphate buffered saline (DPBS) | Thermo Fisher Scientific | Cat#14190094 |
| Critical commercial assays | ||
| CD31 MicroBeads, mouse | Miltenyi Biotec | Cat#130-097-418 |
| Deposited data | ||
| RNA-sequencing raw and analyzed data mouse EC | (Kalucka et al., 2020) | ArrayExpress: E-MTAB-8077 |
| Experimental models: organisms/strains | ||
| C57BL6/J male mice | Charles River | Strain code: 632 |
| Software and algorithms | ||
| BIOMEX | (Taverna et al., 2020) | https://www.vibcancer.be/software-tools/BIOMEX |
| Cell Ranger; version 2.2.0 | 10× Genomics | tenx, RRID:SCR_01695 |
| FindClusters (Seurat package; version 2.3.4) | (Satija et al., 2015) | (Seurat; RRID: SCR_016341) |
| flashpcaR; version 2.0 | (Abraham et al., 2017) | https://github.com/gabraham/flashpca/releases |
| FlowJo (version 8.8.6) | https://www.flowjo.com | (FlowJo, RRID: SCR_008520) |
| NormalizeData (Seurat package; version 2.3.4) | (Satija et al., 2015) | (Seurat; RRID: SCR_016341) |
| Rtsne package; version 0.15 | (van der Maaten, 2008) | https://cran.r-project.org/web/packages/Rtsne/index.html |
| Seurat FindVariableGenes | (Satija et al., 2015) | (Seurat; RRID: SCR_016341) |
| Other | ||
| 40 μm Cell strainer | Sigma-Aldrich | Cat#CLS431750-50EA |
| 100 μm Cell strainer | Sigma-Aldrich | Cat#CLS431752-50EA |
| BD FACSAria™ III sorter | BD Biosciences | N/A |
| Centrifuge tube, conical, HDPE CentriStar™, PP, 15 mL | VWR | Cat#734-1867 |
| Centrifuge tube, conical, HDPE CentriStar™, PP, 50 mL | VWR | Cat#734-1869 |
| gentleMACS™ C Tubes | Miltenyi Biotec | Cat#130-093-237 |
| gentleMACS™ Octo Dissociator | Miltenyi Biotec | Cat#130-095-937 |
| gentleMACS™ Dissociator | Miltenyi Biotec | Cat#130-093-235 |
| Insyte-WTM 18G 1.3 × 48 mm | BD Vialon™ | Cat#381346 |
| LS columns | Miltenyi Biotec | Cat#130-042-401 |
| MACS MultiStand | Miltenyi Biotec | Cat#130-042-303 |
| Multipurpose centrifuge and microcentrifuge | N/A | N/A |
| Perfusion pump: Perfusor® fm (MFC) | B. Braun Malaysia | N/A |
| Surgical Scalpel Blade No 10 | Swann-Morton | Cat#0201 |
| Syringe 1 mL (without needle) | HSW HENKE-JECT® | Cat#8300014579 |
| Syringe Pump Harvard Apparatus | Harvard Apparatus | Cat#PHD 22/2000 |
| QuadroMACS™ Separator | Miltenyi Biotec | Cat#130-091-051 |
Materials and equipment
Media and buffers
The following media/buffers are required.
CRITICAL: Protocols are tissue-specific, please read the instructions carefully before proceeding with the isolations.
-
•PBS-based Wash Buffer 1 (necessary for all tissues) containing:
- 0.5% (w/v) BSA (BSA Fraction V, Sigma-Aldrich, Cat#10735086001)
- 2 mM EDTA (Sigma-Aldrich, Cat#ED2P-500G) in PBS (Thermo Fisher Scientific, Cat#14190-094)
| Reagent | Final concentration | Amount |
|---|---|---|
| BSA | 0.5% | 2.5 g |
| EDTA | 2 mM | 0.2 mL (from 5 M pre-prepared stock solution, according to manufacturer's instructions) |
| PBS | - | 499.8 mL |
-
•For digestion of colon prepare PBS-based Wash Buffer 2 containing:
- 3% (v/v) FBS (Thermo Fisher Scientific, Cat#16000044)
- 10 mM EDTA (Sigma-Aldrich, Cat#ED2P-500G) in PBS (Thermo Fisher Scientific, Cat#14190-094)
| Reagent | Final concentration | Amount |
|---|---|---|
| FBS | 3% (v/v) | 15 mL |
| EDTA | 10 mM | 1 mL (from 5 M pre-prepared stock solution according to manufacturer's instructions) |
| PBS | - | 484 mL |
Note: Wash Buffer 1 and 2 can be prepared in advance and stored for up to 3 days at 4°C.
-
•Supplemented KnockOutTM DMEM-medium (necessary for all tissues) containing:
- KnockOutTM DMEM-medium (Thermo Fisher Scientific, Cat#10829018)
- 1% (v/v) Penicillin/Streptomycin (Thermo Fisher Scientific, Cat#15140122)
- 2× Antibiotic-Antimycotic (Thermo Fisher Scientific, Cat#15240062)
- 1 mM Sodium Pyruvate (Thermo Fisher Scientific, Cat#11360070)
- 1× MEM Non-Essential Amino Acids Solution (MEM-NEAA) (Thermo Fisher Scientific, Cat#11140035)
- 1× Endothelial Cell Growth Factor supplements (ECGS/ Heparin) (PromoCell, Cat#C-30120).
| Reagent | Final concentration | Amount |
|---|---|---|
| KnockOutTM DMEM-medium | - | 473 mL |
| Penicillin/Streptomycin | 1% (v/v) | 5 mL |
| Antibiotic-Antimycotic | 2× | 10 mL |
| Sodium Pyruvate | 1 mM | 5 mL |
| MEM Non-Essential Amino Acids Solution | 1× | 5 mL |
| Endothelial Cell Growth Factor supplements (ECGS/ Heparin) | 1× | 2 mL (one vial) |
Note: Please check specific protocols for additional details. Supplemented KnockOutTM DMEM-medium can be stored in a sterile manner at 4°C, up to several months. However, the digestion enzymes (Collagenases and DNase) have to be added freshly to the digestion medium shortly before the start of the isolation procedures.
Note: ECGS/Heparin is added to the media to sustain EC viability. If there is any indication that the growth factors included in ECGS might affect isolated cells and alter the results according to the designed experiments, these can be left out.
Equipment
The following equipment was used for the described protocols. Please check the protocols for specific instructions. For details about the equipment and alternatives please, check the section “Equipment and reagent alternatives”.
-
•
gentleMACS™ Octo Dissociator (Miltenyi Biotec, Cat#130-095-937)
-
•
gentleMACS™ Dissociator (Miltenyi Biotec, Cat#130-093-235)
-
•
MACS MultiStand (Miltenyi Biotec, Cat#130-042-303)
-
•
QuadroMACS™ Separator (Miltenyi Biotec, Cat#130-091-051)
-
•
Water bath or incubator adjusted to 37°C (see specific protocols for details)
-
•
Multipurpose- and Micro-centrifuge (see specific protocols for details)
-
•
BD FACSAria™ III sorter
-
•
Perfusion pump: Perfusor® fm (MFC) - B. Braun Malaysia or Harvard Apparatus PHD 22/2000
Equipment and reagent alternatives
-
•
GentleMACS™ Dissociator (Miltenyi Biotec, Cat#130-093-235) colon tissue (step 8b and heart tissue (step 12f) can be replaced by gentleMACS™ Octo Dissociator (Miltenyi Biotec, Cat#130-095-937). The protocol for enzymatic digestion of small intestine tissue (step 3b) requires gentleMACS™ Octo Dissociator (Miltenyi Biotec, Cat#130-095-937).
-
•
MACS® Columns contain a matrix composed of superparamagnetic spheres, which are covered with a cell-friendly coating. When the column is placed in a MACS Separator, the spheres amplify the magnetic field by 10.000-fold. LS columns (Miltenyi Biotec, Cat#130-042-401) are designed for positive selection and depletion of strongly magnetically labeled cells and its loading capacity for labeled cells is up to 1×108 and for total cells is up to 2×109). LS columns (Miltenyi Biotec, Cat#130-042-401) can be used with the QuadroMACS™ Separator (Miltenyi Biotec, Cat#130-091-051) or the MidiMACS™ Separator (Miltenyi Biotec, Cat#130-042-302). Miltenyi Biotec also offers alternative columns such as MS columns (Miltenyi Biotec, Cat#130-042-201) designed for positive selection and depletion of strongly magnetically labeled cells with different capacity than LS columns (for MS column: labeled cells: up to 1×107 and total cells: up to 2×108); LD columns (Miltenyi Biotec, Cat#130-042-901; designed for depletion of even weakly labeled cells) or AutoMACS columns (Miltenyi Biotec, Cat#130-021-101; designed both for positive and negative selection). However, using other columns than the ones mentioned in the protocols may require further optimization. Additionally, non-column based magnetic isolations methods are available (e.g., from STEMCELL Technologies) that could be optimized and used as alternatives for depletion and enrichment purposes.
-
•
For the protocol of dissection and preparation of colon tissue (step 7b) and intestine tissue (step 2b) for enzymatic digestion, we used the protecting insert from Insyte-WTM 18G 1.3 × 48 mm (BD Vialon™, Cat#381346); however other protecting inserts with the same parameters can be used.
-
•In the above protocols we suggest to use the following reagents:
- Antibodies: CD31 (PECAM-1) Monoclonal Antibody (390), e.g.,: CD31-FITC (Thermo Fisher Scientific, Cat#11-0311-82; 1:300), CD45 Monoclonal Antibody (30-F11), e.g.,: CD45-PE-Cyanine7 (Thermo Fisher Scientific, Cat#25-0451-82; 1:700), CD11b Monoclonal Antibody (M1/70), e.g.,: CD11b-PE (BioLegend, Cat#101208) (1:500).
- Viability Dye, e.g.,: eFluor 450 (Thermo Fisher Scientific, Cat#65-0863-18; 1:1000).
Different antibodies with similar properties (antigen and clone of the antibody), or with different clonality, conjugated with fluorochrome of choice can be used. Please note that staining efficiency depends on the clonality, fluorochrome, vendor and/or the LOT number of the antibody as well as on the sorter used for FACS. We recommend performing optimization of the staining panel and antibody titration.
-
•
The BD FACSAria™ III sorter was used to perform fluorescent activated sorting. However, any other sorter that allows to sort the cells at the required optical paths can be used. Please, note that the settings of sorting are specific for the brand and model of the sorter and optimization of the procedure always needs to be performed.
-
•
A perfusion pump Perfusor® fm (MFC) - B. Braun Malaysia or Harvard Apparatus PHD 22/2000 was used to perform the transcardial perfusion, however, any other perfusion pump that allows the setting described in the protocols can be used.
-
•
Digestion efficiency may depend on the vendor and/or the LOT number of the enzymes used. We recommend to test all newly purchased reagents before performing final experiments.
-
•
Please note that, depending on the intended further use of the isolated ECs, working in a laminar flow hood and applying sterile lab-practice may be necessary. For example, if the cells will be used for cell culture purposes, all the reagents for isolation should be kept and used in a sterile manner and we would recommend to perform the EC isolation under a laminar flow cabinet to avoid any external contamination. However, if sterility of the purified ECs is not necessary after the isolation (e.g., nucleic acid isolation, for sequencing, Western blotting, cell staining) the protocols can be performed outside the laminar flow cabinet.
Step-by-step method details
Small intestine endothelial cell isolation
Timing: ∼6 h 30 min
For preparation of small intestine digestion buffer
Timing: 45 min
For dissection and preparation of intestine tissue for enzymatic digestion
Timing: 2 h
For enzymatic digestion of intestine tissue
Timing: 1 h
For endothelial cell enrichment using CD31 murine MicroBeads
Timing: 45 min
For FACS
Timing: 1–2 h
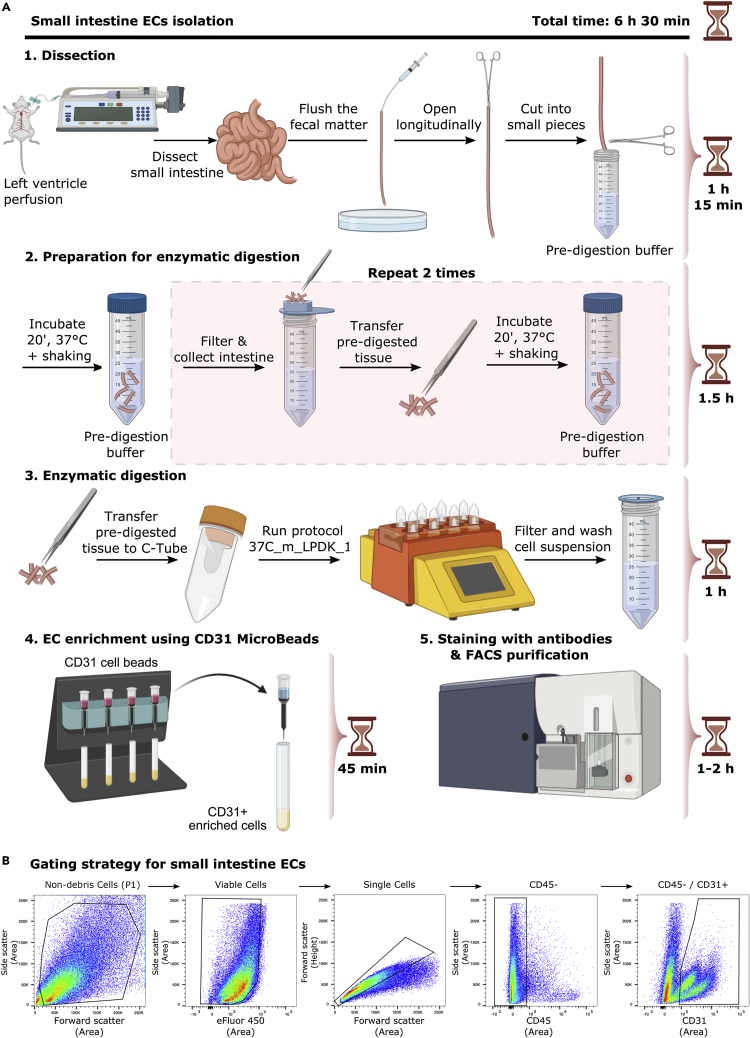
Figure 1A shows a detailed scheme of small intestine ECs isolation.
-
1.Preparation of small intestine digestion buffer
-
a.Right before isolation prepare small intestine digestion buffer containing:
-
i.Supplemented KnockOutTM DMEM-medium (see details in section ‘materials and equipment’)
-
ii.0.1% (w/v) collagenase I
-
iii.0.25% (w/v) collagenase IV
-
iv.0.25 U/mL Dispase
-
v.7.5 μg/mL DNAse I
Reagent Final concentration Amount Supplemented KnockOutTM DMEM-medium - 8.925 mL Collagenase I 0.1% (w/v) 10 mg Collagenase IV 0.25% (w/v) 25 mg Dispase 0.25 U/mL 1 mL (from 2.5 U/mL stock solution prepared according to manufacturer's instructions) DNAse I 7.5 μg/mL 75 μL (from 1 mg/mL stock solution prepared according to manufacturer's instructions)
-
i.
-
b.Right before isolation prepare pre-digestion buffer:
-
i.HBSS (Ca2+, Mg2+)
-
ii.1.5 mM DTT
-
iii.5% (v/v) FBS
-
iv.5 mM EDTA
Reagent Final concentration Amount HBSS (Ca2+, Mg2+) - 475 mL DTT 1.5 mM 0.23 g FBS 5% (v/v) 25 mL EDTA 0.5 mM 0.5 mL (from 5 M pre-prepared stock solution according to manufacturer's instructions)
-
i.
-
c.Prepare cold HBSS (Ca2+, Mg2+).
-
d.Store the small intestine digestion buffer and pre-digestion buffer (freshly prepared) at 4°C until needed.
- 10 mL of small intestine digestion buffer suffices for 3 adult murine small intestines. 50 mL of pre-digestion buffer suffices for 1 adult murine small intestine.
-
a.
-
2.Dissection and preparation of intestine tissue for enzymatic digestion
-
a.Before dissecting the small intestine perform transcardial perfusion via the left ventricle with ice-cold PBS at a perfusion rate of 2 mL/minute for 5 min.
-
b.Flush the fecal matter from the dissected small intestine with ice-cold HBSS using a 20 mL syringe and an elastic needle (use protecting insert from Insyte-WTM 18G 1.3 × 48 mm (BD Vialon™, Cat#381346)).Note: Keep the samples on ice during the dissections.
-
c.Open the small intestine longitudinally.
-
d.Cut intestine into very small pieces (4 mm2) and place them in the 50 mL conical tube (1 intestine per conical tube).
-
e.Add 25 mL ice-cold pre-digestion buffer – invert 10–15 times, then incubate with 37°C water bath for 20 min.
-
i.Shake the tube vigorously by hand every 5 min.
-
i.
-
f.Vortex for 10 seconds.
-
g.Filter the content of the conical tube through a 100 μm cell strainer.
-
h.Collect undigested intestines from the strainer and place them into a new 50 mL conical tube containing 20 mL of pre-digestion buffer and incubate at 37°C water bath for 20 min. Shake the tube vigorously by hand every 5 min.
-
i.Vortex for 10 seconds.
-
j.Filter the content of the conical tube through a 100 μm cell strainer.
-
k.Collect undigested intestines from the strainer and place them into a new 50 mL conical tube containing 20 mL of HBSS and incubate at 37°C for 20 min.
-
l.Filter the content of the tube through a 100 μm cell strainer.
-
a.
-
3.Enzymatic digestion of intestine tissue
-
a.Collect undigested small intestines from the strainer and put them in the gentleMACS C tubes (Miltenyi Biotec, Cat#130-093-237) containing pre-warmed at 37°C digestion buffer (3 mL per gentleMACS C tube).
 CRITICAL: Make sure the tube is closed tightly.
CRITICAL: Make sure the tube is closed tightly. -
b.Place the gentleMACS C tubes in the gentleMACS™ Octo Dissociator (Miltenyi Biotec, Cat#130-095-937). Run protocol 37C_m_LPDK_1 (pre-programmed by manufacturer).
-
c.Stop the reaction by adding 5 mL of Wash Buffer 1.
-
d.Filter cell suspension through a 100 μm cell strainer to remove leftover undigested tissue fragments into the 50 mL conical tube.
 CRITICAL: Keep the pass-through suspension in the 50 mL conical tube.
CRITICAL: Keep the pass-through suspension in the 50 mL conical tube. -
e.Wash the strainer by rinsing it with the additional 10 mL of Wash Buffer 1. Collect the pass-through suspension to the same conical tube.
-
f.Centrifuge cells at 300 g for 10 min.
-
g.Carefully remove the supernatant.Note: The supernatant contains chyle therefore should have a milky/cloudy appearance.
-
h.Resuspend pellet in 5 mL of Wash Buffer 1 and centrifuge at 300 g for 5 min.Note: If supernatant is not clear, repeat washing step 3e.
-
a.
-
4.Endothelial cell enrichment using CD31 murine MicroBeads
-
a.Resuspend the pellet in an appropriate amount of Wash Buffer 1 and add the appropriate volume of CD31 MicroBeads (Miltenyi Biotec, Cat#130-097-418) to the suspension according to the manufacturer's instructions.Note: We suggest to resuspend the cells in 90 μL of Wash Buffer 1, determine the cell number and follow the manufacturer's instructions (https://www.miltenyibiotec.com/US-en/products/cd31-microbeads-mouse.html?countryRedirected=1#gref) i.e., for up to 1 × 107 total cells, resuspend the cells in 90 μL of Wash Buffer 1 and add 10 μL of CD31 MicroBeads. If fewer than 1 × 107 cells are available use the same volumes as indicated above. When working with higher cell numbers than 1 × 107 cells, scale up all reagent volumes and total volumes accordingly.
-
b.Mix and incubate for 15 min at 4°C.Note: Process samples fast in order to avoid non-specific binding of beads at 20°C–22°C.
-
c.Wash cells by adding 3 mL of Wash Buffer 1 and centrifuge at 300 g for 5 min at 4°C.
-
i.During the centrifugation step prepare collection tubes and LS columns (Miltenyi Biotec, Cat#130-042-401) according to the manufacturer’s instructions (https://www.miltenyibiotec.com/US-en/products/ls-columns.html#gref) i.e., place the column in the suitable magnetic separator and prepare the column by rinsing with 3 mL of Wash Buffer.
-
i.
-
d.Remove the supernatant and resuspend the pellet in 0.5 mL of Wash Buffer 1.
-
e.Apply the cell suspension onto the prepared LS column through a 40 μm cell strainer to remove potential cell aggregates in order to prevent clogging of the column.
-
f.Wash the LS column 3 times with 3 mL Wash Buffer 1 adding buffer each time once the column reservoir is empty.Note: The eluent (CD31-negative fraction) from step 4f can be used to prepare controls for FACS analysis or can be discarded if not further needed.
-
g.Remove the LS column from the separator and place it onto a new 15 mL conical collection tube.
-
h.Pipette 5 mL Wash Buffer 1 onto the LS column. Immediately flush out the fraction containing the magnetically labeled cells (CD31-positive fraction) by firmly applying the plunger supplied with the column.
-
a.
-
5.
FACS
CRUCIAL: Prepare all required controls for flow cytometry analysis beforehand i.e., unstained cell (US) control, viability control (VB), Fluorescence Minus One (FMO) control, isotype controls (optional), as well as the compensation controls (one for each fluorophore) for proper set up of the cytometer.Note: We recommend to use the cells from the final step of the isolation as controls (US, VB, FMO, isotype controls). However, if the cell number is limited, one can use the cells from, e.g.,: CD31-negative fraction, to prepare the controls. For compensation controls we would recommend to use compensation beads (e.g., OneComp eBeads™ Compensation Beads, Thermo Fisher Scientific, Cat#01-1111-41) to ensure clear fluorescence signal and sufficient compensation.-
a.Centrifuge the cell suspension at 300 g for 5 min, remove the supernatant.
-
b.Resuspend the pellet in 0.5 mL Wash Buffer 1-based staining solution containing:
-
i.CD31 (PECAM-1) Monoclonal Antibody (390), e.g.,: CD31-FITC (Thermo Fisher Scientific, Cat#11-0311-82; 1:300).
-
ii.CD45 Monoclonal Antibody (30-F11), e.g.,: CD45-PE-Cyanine7 (Thermo Fisher Scientific, Cat#25-0451-82; 1:700).
-
iii.Viability Dye, e.g.,: eFluor 450 (Thermo Fisher Scientific, Cat#65-0863-18; 1:1000).
-
i.
-
c.Stain the cells for 30 min at 4°C min in the dark.
-
d.Add 3 mL of Wash Buffer 1 and centrifuge the stained cells at 300 g for 5 min at 4°C, remove the supernatant.
-
e.Resuspend the pellet in Wash Buffer 1 (based on the pellet size between 200 μL and 0.5 mL) and proceed with FACS.
-
f.Sort viable, CD45-, CD31+ cells to collection tubes.
-
a.
Note: To ensure high viability of the ECs, sort the ECs to collection medium (10% (v/v) FBS in PBS, Thermo Fisher Scientific, Cat#A38401) in an Eppendorf (∼ 200 μL) or 15 mL conical tube (∼ 2 mL).
Figure 1.
EC isolation from mouse small intestine
(A) Detailed scheme illustrating isolation of ECs from small intestine.
(B) Representative FACS plots for the gating strategy to sort ECs from small intestine based on sorting live/CD45-/CD31+ cells.
Figure 1B shows representative FACS plots and gating strategy of viable, CD45-, CD31+ small intestine ECs.
Colon endothelial cell isolation
Timing: ∼5 h 15 min
For preparation of colon digestion buffer
Timing: 45 min
For dissection and preparation of colon tissue for enzymatic digestion
Timing: 45 min
For digestion of colon tissue
Timing: 1 h
For endothelial cell enrichment using CD31 murine MicroBeads
Timing: 45 min
For FACS
Timing: 1–2 h
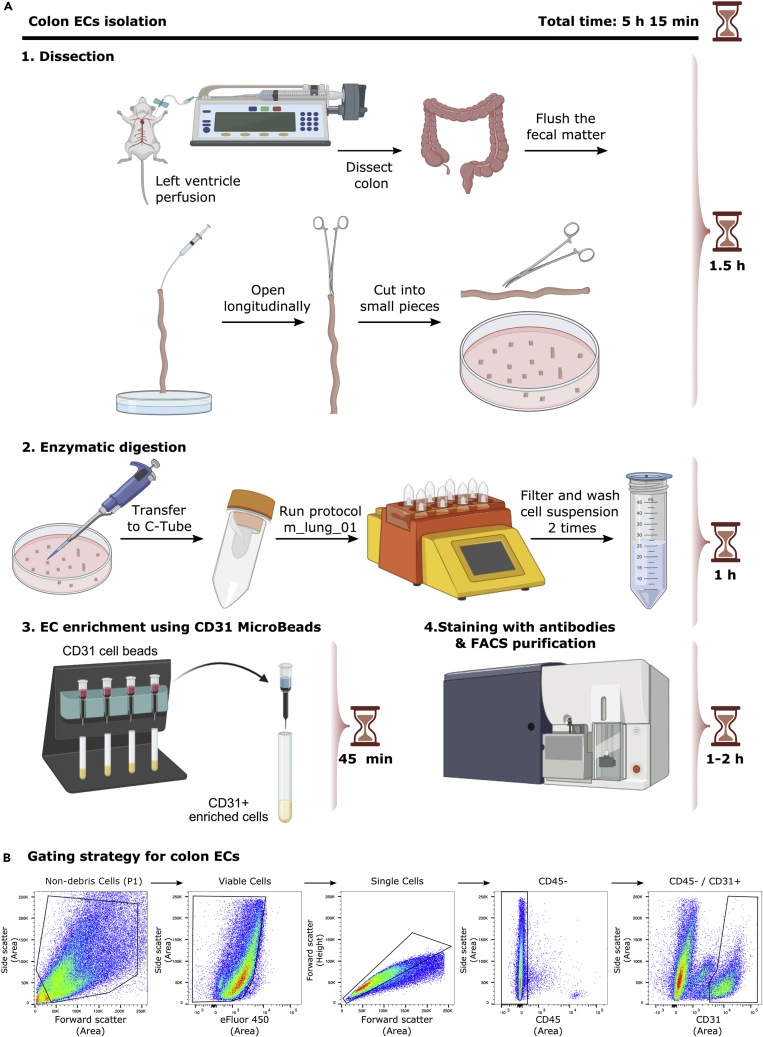
Figure 2A shows a detailed scheme of colon ECs isolation.
-
6.Preparation of colon digestion buffer
-
a.Right before isolation prepare colon digestion buffer containing:
-
i.Supplemented KnockOutTM DMEM-medium (see details in section ‘materials and equipment’)
-
ii.0.1% (w/v) collagenase I
-
iii.0.25% (w/v) collagenase IV
-
iv.0.625 U/mL Dispase
-
v.7.5 μg/mL DNAse I
Reagent Final concentration Amount Supplemented KnockOutTM DMEM-medium - 7.425 mL Collagenase I 0.1% (w/v) 10 mg Collagenase IV 0.25% (w/v) 25 mg Dispase 0.625 U/mL 2.5 mL (from 2.5 U/mL stock solution) DNAse I 7.5 μg/mL 75 μL (from 1 mg/mL stock solution)
-
i.
-
b.Prepare PBS-based Wash Buffer 2 containing:
-
i.3% (v/v) FBS in PBS
-
ii.10 mM EDTA
Reagent Final concentration Amount FBS 3% (v/v) 15 mL EDTA 10 mM 1 mL (from 5 M pre-prepared stock solution according to manufacturer's instructions) PBS - 484 mL
-
i.
-
c.Store the (freshly prepared) colon digestion buffer and Wash Buffer 2 at 4°C until they are needed.
-
a.
Figure 2.
EC isolation from mouse colon
(A) Detailed scheme illustrating isolation of ECs from colon.
(B) Representative FACS plots for the gating strategy to sort ECs from colon based on sorting live/CD45-/CD31+ cells.
10 mL of colon digestion buffer suffices for 3 adult murine colons.
-
7.Dissection and preparation of colon tissue for enzymatic digestion
-
a.Before dissecting the colon, perform transcardial perfusion via the left ventricle with ice-cold PBS at a perfusion rate of 2 mL/minute for 5 min.
-
b.Flush the fecal matter from the dissected colon using a 20 mL syringe and an elastic needle (use protecting insert from Insyte-WTM 18G 1.3 × 48 mm (BD Vialon™, Cat#381346)) with ice-cold PBS.Note: Keep the samples on ice during the dissections.
-
c.Open the colon longitudinally.Note: For easier manipulations you can fix the colon using pins or syringe needles at both ends.
-
d.Use a sterile scalpel blade to swiftly scrape out the mucus from the colon inner surface.
-
e.Place the colon on the Petri dish and cut it into very small pieces (as small as possible), place them into a 50 mL conical tube (3 colons/conical tube with 10 mL of colon digestion buffer).Note: The pieces should pass through a cut P1000 tip (approx. 0.8 mm).
-
f.Rinse the Petri dish with an additional 4 mL of colon digestion buffer, and transfer cell suspension to gentleMACS C tube (Miltenyi Biotec, Cat#130-093-237) using a P1000 micropipette.
-
a.
CRITICAL: Make sure the tube is closed tightly.
-
8.Digestion of colon tissue
-
a.Incubate cell suspension in gentle MACS C tube for 15 min at 37°C.
-
i.Shake the tube vigorously by hand every 5 min to ensure rapid tissue dissociation.
-
i.
-
b.Place tubes in gentleMACS™ Dissociator (Miltenyi Biotec, Cat#130-093-235):
-
i.Run m_lung_01 protocol (pre-programmed by manufacturer).
-
i.
-
c.Stop the enzymatic reaction by adding 15 mL of Wash Buffer 2.
-
d.Filter the content using a 100 μm cell strainer into a 50 mL conical tube to remove larger tissue fragments and mucus.
-
e.Divide the filtered cells over two 15 mL conical tubes and centrifuge both cell suspensions at 300 g for 7 min.
-
f.Remove the supernatant and resuspend the pellets in 5 mL of Wash Buffer 2.
-
g.Pool previously divided samples.
-
h.Filter the content using a 40 μm cell strainer into a new 50 mL conical tube to remove leftover tissue fragments and mucus, then transfer content to a 15 mL conical tube.
-
i.Centrifuge cell suspension at 300 g for 5 min.
-
j.Remove the supernatant and wash the pellet with 5 mL Wash Buffer 2.
-
k.Centrifuge cell suspension at 300 g for 5 min, remove the supernatant.
-
a.
-
9.Endothelial cell enrichment using CD31 murine MicroBeads
-
a.Resuspend the pellet in an appropriate amount of Wash Buffer 1 and add the appropriate volume of CD31 MicroBeads (Miltenyi Biotec, Cat#130-097-418) to the suspension according to the manufacturer's instructions.Note: We suggest to resuspend the cells in 90 μL of Wash Buffer 1, determine the cell number and follow the manufacturer's instructions (https://www.miltenyibiotec.com/US-en/products/cd31-microbeads-mouse.html?countryRedirected=1#gref) i.e., for up to 1 × 107 total cells, resuspend the cells in 90 μL of Wash Buffer 1 and add 10 μL of CD31 MicroBeads. If fewer than 1 × 107 cells are available use the same volumes as indicated above. When working with higher cell numbers than 1 × 107 cells, scale up all reagent volumes and total volumes accordingly.
-
b.Mix and incubate for 15 min at 4°C.Note: Process samples fast in order to avoid non-specific binding of beads at 20°C–22°C.
-
c.Wash the cells by adding 3 mL of Wash Buffer 2 and centrifuge at 300 g for 5 min at 4°C.During the centrifugation step prepare collection tubes and LS columns (Miltenyi Biotec, Cat#130-042-401) according to the manufacturer's instructions (https://www.miltenyibiotec.com/US-en/products/ls-columns.html#gref) i.e., place the column in the suitable magnetic separator and prepare the column by rinsing with 3 mL of Wash Buffer.
-
d.Remove the supernatant and resuspend the pellet in 0.5 mL of Wash Buffer 2.
-
e.Apply the cell suspension onto the prepared LS column through a 40 μm cell strainer to remove potential cell aggregates in order to prevent clogging of the column.
-
f.Wash the LS column 3 times with 3 mL Wash Buffer 2 adding buffer each time once the column reservoir is empty.Note: The eluent (CD31-negative fraction) from step 9f can be used to prepare controls for FACS analysis or can be discarded if not further needed.
-
g.Remove the LS column from the separator and place it onto a new 15 mL conical collection tube.
-
h.Pipet 5 mL Wash Buffer 1 onto the LS column. Immediately flush out the fraction containing the magnetically labeled cells (CD31-positive fraction) by firmly applying the plunger supplied with the column.
-
a.
-
10.
FACS
CRUCIAL: Prepare all required controls for flow cytometry analysis beforehand i.e., unstained cell (US) control, viability control (VB), Fluorescence Minus One (FMO) control, isotype controls (optional), as well as the compensation controls (one for each fluorophore) for proper set up of the cytometer.Note: We recommend to use the cells from the final step of the isolation as controls (US, VB, FMO, isotype controls). However, if the cell number is limited, one can use the cells from, e.g.,: CD31-negative fraction, to prepare the controls. For compensation controls we would recommend to use compensation beads (e.g., OneComp eBeads™ Compensation Beads, Thermo Fisher Scientific, Cat#01-1111-41) to ensure clear fluorescence signal and sufficient compensation.-
a.Centrifuge the cell suspension at 300 g for 5 min, remove the supernatant.
-
b.Resuspend the pellet in 0.5 mL Wash Buffer 1-based staining solution containing:
-
i.CD31 (PECAM-1) Monoclonal Antibody (390), e.g.,: CD31-FITC (Thermo Fisher Scientific, Cat#11-0311-82; 1:300).
-
ii.CD45 Monoclonal Antibody (30-F11), e.g.,: CD45-PE-Cyanine7 (Thermo Fisher Scientific, Cat#25-0451-82; 1:700).
-
iii.Viability Dye, e.g.,: eFluor 450 (Thermo Fisher Scientific, Cat#65-0863-18; 1:1000).
-
i.
-
c.Stain the cells for 30 min at 4°C in the dark.
-
d.Add 3 mL of Wash Buffer 1 and centrifuge the stained cells at 300 g for 5 min at 4°C, remove the supernatant.
-
e.Resuspend the pellet in Wash Buffer 1 (based on the pellet size between 200 μL and 0.5 mL) and proceed with FACS.
-
f.Sort viable, CD45-, CD31+ cells to collection tubes.
-
a.
Note: To ensure high viability of the ECs, sort the ECs to collection medium (10% (v/v) FBS in PBS, Thermo Fisher Scientific, Cat#A38401) in an Eppendorf (∼ 200 μL) or 15 mL conical tube (∼ 2 mL).
Figure 2B shows representative FACS plots and gating strategy of viable, CD45-, CD31+ colon ECs.
Heart endothelial cell isolation
Timing: ∼4h 15 min
For preparation of heart digestion buffer
Timing: 30 min
For dissection and digestion of heart tissue
Timing: 1 h
For endothelial cell enrichment using CD31 murine MicroBeads
Timing: 45 min
For FACS
Timing: 1–2 h
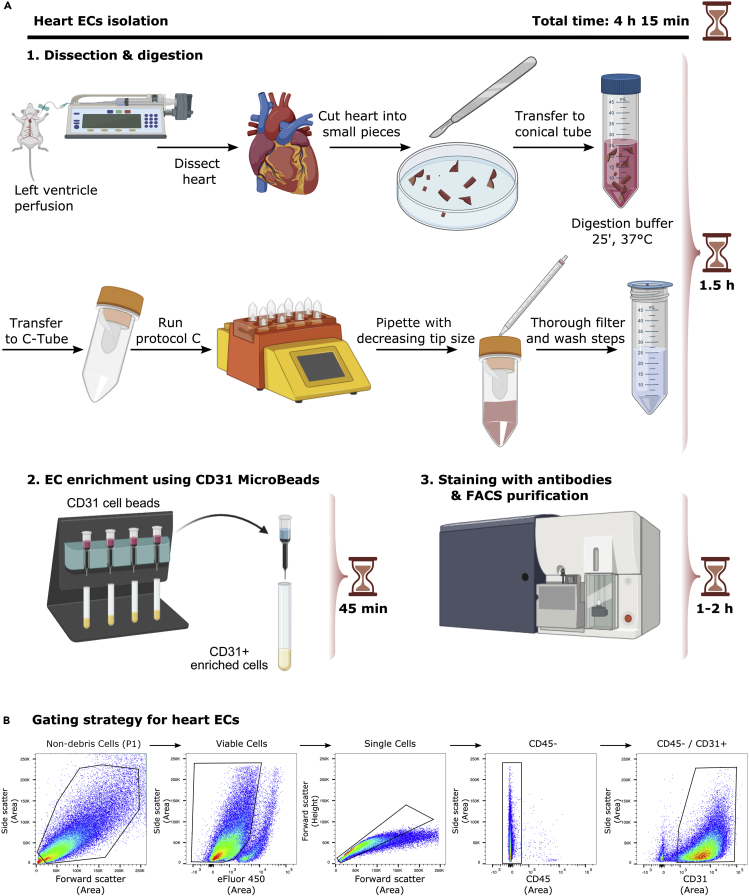
Figure 3A shows a detailed scheme of heart ECs isolation.
-
11.Preparation of heart digestion buffer
-
a.Right before isolation prepare heart digestion buffer containing:
-
i.Supplemented KnockOutTM DMEM-medium (see details in section ‘materials and equipment’)
-
ii.0.1% (w/v) collagenase II
-
iii.0.25% (w/v) collagenase IV
-
iv.7.5 μg/mL DNAse I
Reagent Final concentration Amount Supplemented KnockOutTM DMEM-medium - 8.925 mL Collagenase II 0.1% (w/v) 10 mg Collagenase IV 0.25% (w/v) 25 mg Dispase 0.25 U/mL 1 ml (from 2.5 U/mL stock solution prepared according to manufacturer's instructions) DNAse I 7.5 μg/mL 75 μL (from 1 mg/mL stock solution prepared according to manufacturer's instructions)
-
i.
-
b.Store the heart digestion buffer (freshly prepared) at 4°C until it is needed.
- 5 mL of heart digestion buffer suffices for 3 adult murine hearts.
-
a.
-
12.Dissection and digestion of heart tissue
-
a.Before dissecting the heart, perform transcardial perfusion via the left ventricle with ice-cold PBS at a perfusion rate of 2 mL/minute for 5 min.
-
b.Harvest the hearts and cut them with scalpel blade into small pieces (approx. 2 mm2).
-
c.Pool all pieces in a 50 mL conical tube containing 5 mL of heart digestion buffer.Note: Keep the samples on ice during the dissections.
-
d.Incubate the sample in digestion buffer in a 37°C water bath for 25 min.
-
i.Shake the tube vigorously by hand every 5-10 min for faster tissue dissociation.
-
i.
-
e.Transfer the mix into a gentleMACS C tube (Miltenyi Biotec, Cat#130-093-237).
 CRITICAL: Make sure the tube is closed tightly.
CRITICAL: Make sure the tube is closed tightly. -
f.Place tubes in gentleMACS™ Dissociator (Miltenyi Biotec, Cat#130-093-235):
-
i.Run protocol C (pre-programmed by manufacturer).
-
i.
-
g.Pipet the cell suspension up and down using serological pipets with a decreasing tip size (e.g., 25 mL > 5 mL > 10 mL) until there are no big tissue clumps left and then stop the enzymatic reaction by adding 25 mL of Wash Buffer 1.
-
h.Filter cell suspension through a 100 μm cell strainer to remove larger tissue fragments.
-
i.Centrifuge the cell suspension at 300 g for 7 min.
-
j.Collect supernatant into a new 50 mL conical tube and centrifuge again at 300 g for 5 min, remove the supernatant.
-
k.Resuspend the pellet obtained in step 12i in 5 mL of Wash Buffer 1 and filter through a 40 μm cell strainer to remove leftover tissue fragments. Wash the conical tube and the filter with an additional 4 mL of Wash Buffer 1.
-
l.Resuspend pellet of obtained in step 12j in 4 mL of Wash Buffer 1 and filter the cell suspension through the 40 μm cell strainer used previously.
-
m.Pool both fractions and transfer to a 15 mL conical tube.
-
n.Centrifuge at 300 g for 5 min, remove the supernatant.
-
a.
-
13.Endothelial cell enrichment using CD31 murine MicroBeads
-
a.Resuspend the pellet in an appropriate amount of Wash Buffer 1 and add the appropriate volume of CD31 MicroBeads (Miltenyi Biotec, Cat#130-097-418) to the suspension according to the manufacturer's instructions.Note: We suggest to resuspend the cells in 90 μL of Wash Buffer 1, determine the cell number and follow the manufacturer's instructions (https://www.miltenyibiotec.com/US-en/products/cd31-microbeads-mouse.html?countryRedirected=1#gref) i.e., for up to 1 × 107 total cells, resuspend the cells in 90 μL of Wash Buffer 1 and add 10 μL of CD31 MicroBeads. If fewer than 1 × 107 cells are available use the same volumes as indicated above. When working with higher cell numbers than 1 × 107 cells, scale up all reagent volumes and total volumes accordingly.
-
b.Mix and incubate for 15 min at 4°C.Note: Process samples fast in order to avoid non-specific binding of beads at 20°C–22°C.
-
c.Wash the cells by adding 3 mL of Wash Buffer 1 and centrifuge at 300 g for 5 min at 4°C.
-
i.During the centrifugation step prepare collection tubes and LS (Miltenyi Biotec, Cat#130-042-401) columns according to the manufacturer’s instructions (https://www.miltenyibiotec.com/US-en/products/ls-columns.html#gref) i.e., place the column in the suitable magnetic separator and prepare the column by rinsing with 3 mL of Wash Buffer.
-
i.
-
d.Remove the supernatant and resuspend the pellet in 0.5 mL of Wash Buffer 1.
-
e.Apply the cell suspension onto the prepared LS column through a 40 μm cell strainer to prevent clogging of the column.
-
f.Wash the LS column 3 times with 3 mL Wash Buffer 1, adding buffer each time once the column reservoir is empty.Note: The eluent (CD31-negative fraction) from step 13f can be used to prepare controls for FACS analysis or can be discarded if not further needed.
-
g.Remove the LS column from the separator and place it onto a new 15 mL conical collection tube.
-
h.Pipet 5 mL Wash Buffer 1 onto the LS column. Immediately flush out the fraction containing the magnetically labeled cells by firmly applying the plunger supplied with the column.
-
a.
-
14.
FACS
CRUCIAL: Prepare all required controls for flow cytometry analysis beforehand i.e., unstained cell (US) control, viability control (VB), Fluorescence Minus One (FMO) control, isotype controls (optional), as well as the compensation controls (one for each fluorophore) for proper set up of the cytometer.Note: We recommend to use the cells from the final step of the isolation as controls (US, VB, FMO, isotype controls). However, if the cell number is limited, one can use the cells from, e.g.,: CD31-negative fraction, to prepare the controls. For compensation controls we would recommend to use compensation beads (e.g., OneComp eBeads™ Compensation Beads, Thermo Fisher Scientific, Cat#01-1111-41) to ensure clear fluorescence signal and sufficient compensation.-
a.Centrifuge the cell suspension at 300 g for 5 min, remove the supernatant.
-
b.Resuspend the pellet in 0.5 mL Wash Buffer 1-based staining solution containing:
-
i.CD31 (PECAM-1) Monoclonal Antibody (390), e.g.,: CD31-FITC (Thermo Fisher Scientific, Cat#11-0311-82; 1:300).
-
ii.CD45 Monoclonal Antibody (30-F11), e.g.,: CD45-PE-Cyanine7 (Thermo Fisher Scientific, Cat#25-0451-82; 1:700).
-
iii.Viability Dye, e.g.,: eFluor 450 (Thermo Fisher Scientific, Cat#65-0863-18; 1:1000).
-
i.
-
c.Stain the cells for 30 min at 4°C in the dark.
-
d.Add 3 mL of Wash Buffer 1 and centrifuge the stained cells at 300 g for 5 min at 4°C, remove the supernatant.
-
e.Resuspend the pellet in Wash Buffer 1 (based on the pellet size between 200 μL and 0.5 mL) and proceed with FACS.
-
f.Sort viable, CD45-, CD31+ cells to collection tubes.
-
a.
Note: To ensure high viability of the ECs, sort the ECs to collection medium (10% (v/v) FBS in PBS, Thermo Fisher Scientific, Cat#A38401) in an Eppendorf (∼ 200 μL) or 15 mL conical tube (∼ 2 mL).
Figure 3.
EC isolation from mouse heart
(A) Detailed scheme illustrating isolation of ECs from heart.
(B) Representative FACS plots for the gating strategy to sort ECs from heart based on sorting live/CD45-/CD31+ cells.
Figure 3B shows representative FACS plots and gating strategy of viable, CD45-, CD31+ heart ECs.
Liver endothelial cell isolation
Timing: ∼4 h 15 min
For preparation of liver digestion buffer
Timing: 30 min
For dissection and digestion of liver tissue
Timing: 1 h
For endothelial cell enrichment using CD31 murine MicroBeads
Timing: 45 min
For FACS
Timing: 1–2 h
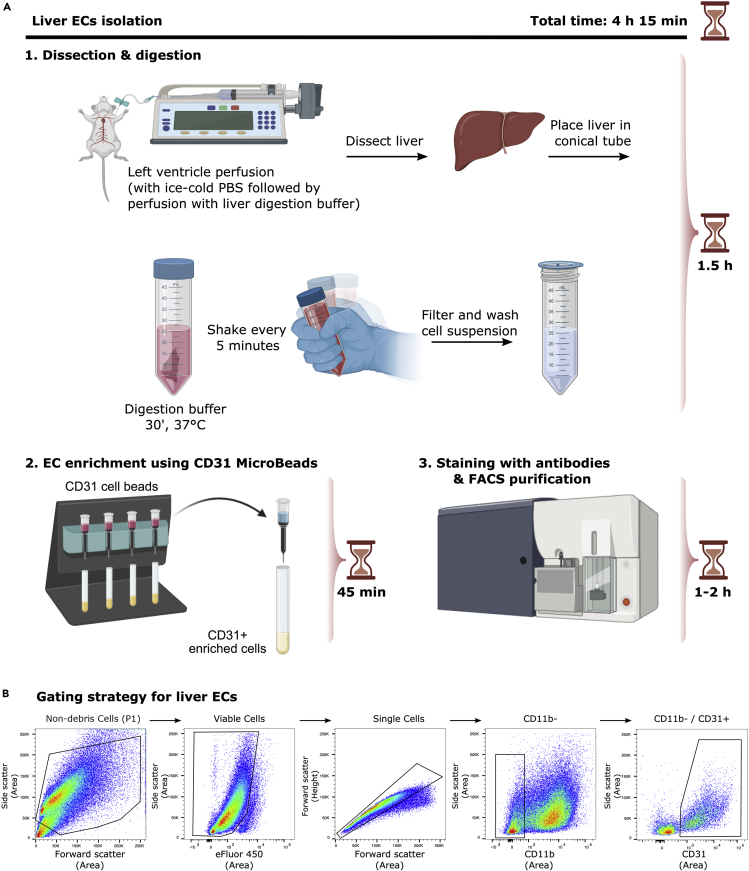
Figure 4A shows a detailed scheme of liver ECs isolation.
-
15.Preparation of liver digestion buffer
-
a.Right before isolation prepare liver digestion buffer containing:
-
i.Supplemented KnockOutTM DMEM-medium (see details in section materials and equipment’)
-
ii.0.1% (w/v) collagenase I
-
iii.0.1% (w/v) collagenase II
-
iv.0.25 U/mL Dispase
-
v.7.5 μg/mL DNAse I
Reagent Final concentration Amount Supplemented KnockOutTM DMEM-medium - 13.39 mL Collagenase I 0.1% (w/v) 15 mg Collagenase II 0.1% (w/v) 15 mg Dispase 0.25 U/mL 1.5 mL (from 2.5 U/mL stock solution prepared according to manufacturer's instructions) DNAse I 7.5 μg/mL 112.5 μL (from 1 mg/mL stock solution prepared according to manufacturer's instructions)
-
i.
-
b.Store the freshly prepared liver digestion buffer at 4°C until needed.
-
a.
-
15
mL of liver digestion buffer suffices for 1 adult murine liver.
-
16.Dissection and digestion of liver tissue
-
a.Before dissecting the liver perform transcardial perfusion via the left ventricle with ice-cold PBS followed by additional perfusion with liver digestion buffer at a perfusion rate of 2 mL/minute for 5 min.
-
b.Transfer a single liver into a 50 mL conical tube containing 5 mL of liver digestion buffer.
-
c.Incubate the sample in the liver digestion buffer in a 37°C water bath for 30 min.
-
i.Shake the tube vigorously by hand every 10 min for faster tissue dissociation.
-
i.
-
d.Mix the sample by pipetting up and down using a 10 mL serological pipette.
-
e.Stop the digestion by adding 8 mL of Wash Buffer 1.
-
f.Mix and filter the cell suspension through a 100 μm cell strainer to remove undigested tissue fragments.
-
g.Centrifuge cell suspension at 300 g for 7 min.
-
h.Remove the supernatant carefully with a pipette.
-
i.Repeat washing step by adding 5 mL of Wash Buffer 1 and centrifuge cell suspension at 300 g for 5 min.
-
j.Remove the supernatant carefully.
-
a.
CRITICAL: The supernatant should be clear after these washing steps. If the supernatant is still turbid/hazy, perform an additional washing step.
-
17.Endothelial cell enrichment using CD31 murine MicroBeads
-
a.Resuspend the pellet in an appropriate amount of Wash Buffer 1 and add the appropriate volume of CD31 MicroBeads (Miltenyi Biotec, Cat#130-097-418) to the suspension according to the manufacturer's instructions.Note: We suggest to resuspend the cells in 90 μL of Wash Buffer 1, determine the cell number and follow the manufacturer's instructions (https://www.miltenyibiotec.com/US-en/products/cd31-microbeads-mouse.html?countryRedirected=1#gref) i.e., for up to 1 × 107 total cells, resuspend the cells the cells in 90 μL of Wash Buffer 1 and add 10 μL of CD31 MicroBeads. If fewer than 1 × 107 cells are available use the same volumes as indicated above. When working with higher cell numbers than 1 × 107 cells, scale up all reagent volumes and total volumes accordingly.Note: In case the sample is too viscous or contains clumps, filter through 70 μm cell strainer (do not filter if not necessary).
-
b.Mix well and incubate for 20 min at 4°C.Note: Process samples fast in order to avoid non-specific binding of beads at 20°C–22°C.
-
c.Wash cells by adding 3 mL of Wash Buffer 1 and centrifuge at 300 g for 5 min at 4°C.
-
i.During the centrifugation step prepare collection tubes and LS columns (Miltenyi Biotec, Cat#130-042-401) according to the manufacturer's instructions (https://www.miltenyibiotec.com/US-en/products/ls-columns.html#gref) i.e., place the column in the suitable magnetic separator and prepare the column by rinsing with 3 mL of Wash Buffer.
-
i.
-
d.Remove the supernatant and resuspend the pellet in 0.5 mL of Wash Buffer 1.
-
e.Apply the cell suspension onto the prepared LS column through a 40 μm cell strainer to prevent clogging of the column.
-
f.Wash the LS column 3 times with 3 mL Wash Buffer 1 adding buffer each time once the column reservoir is empty.Note: The eluent (CD31-negative fraction) from step 17f can be used to prepare controls for FACS analysis or can be discarded if not further needed.
-
g.Remove the LS column from the separator and place it onto a new 15 mL conical collection tube.
-
h.Pipet 5 mL Wash Buffer 1 onto the LS column. Immediately flush out the fraction containing the magnetically labeled cells (CD31-positive fraction) by firmly applying the plunger supplied with the column.
-
a.
-
18.
FACS
CRUCIAL: Prepare all required controls for flow cytometry analysis beforehand i.e., unstained cell (US) control, viability control (VB), Fluorescence Minus One (FMO) control, isotype controls (optional), as well as the compensation controls (one for each fluorophore) for proper set up of the cytometer.Note: We recommend to use the cells from the final step of the isolation as controls (US, VB, FMO, isotype controls). However, if the cell number is limited, one can use the cells from, e.g.,: CD31-negative fraction, to prepare the controls. For compensation controls we would recommend to use compensation beads (e.g., OneComp eBeads™ Compensation Beads, Thermo Fisher Scientific, Cat#01-1111-41) to ensure clear fluorescence signal and sufficient compensation.-
a.Centrifuge the cell suspension at 300 g for 5 min and remove the supernatant.
-
b.Resuspend the pellet in 0.5 mL Wash Buffer 1-based staining solution containing:
-
i.CD11b Monoclonal Antibody (M1/70), e.g.,: CD11b-PE (BioLegend, Cat#101208) (1:500).
-
ii.CD31 (PECAM-1) Monoclonal Antibody (390), e.g.,: CD31-FITC (Thermo Fisher Scientific, Cat#11-0311-82; 1:300).
-
iii.CD45 Monoclonal Antibody (30-F11), e.g.,: CD45-PE-Cyanine7 (Thermo Fisher Scientific, Cat#25-0451-82; 1:700).
-
iv.Viability Dye, e.g.,: eFluor 450 (Thermo Fisher Scientific, Cat#65-0863-18; 1:1000).
-
i.
-
c.Stain the cells for 30 min at 4°C in the dark.
-
d.Add 3 mL of Wash Buffer 1 and centrifuge the stained cells at 300 g for 5 min at 4°C, remove the supernatant.
-
e.Resuspend the pellet in Wash Buffer 1 (based on the pellet size between 200 μL and 0.5 mL) and proceed with FACS.
-
f.Sort viable, CD11b-, and CD31+ cells to collection tubes.
-
a.
Note: To ensure high viability of the ECs, sort the ECs to collection medium (10% (v/v) FBS in PBS, Thermo Fisher Scientific, Cat#A38401) in an Eppendorf (∼ 200 μL) or 15 mL conical tube (∼ 2 mL).
Figure 4.
EC isolation from mouse liver
(A) Detailed scheme illustrating isolation of ECs from liver.
(B) Representative FACS plots for the gating strategy to sort ECs from liver based on sorting live/CD11b-/CD31+ cells.
Figure 4A shows representative FACS plots and gating strategy of viable, CD11b-, CD31+ liver ECs.
Expected outcomes
The described protocols from several murine organs (small intestine, colon, heart, and liver) provide consistent and reproducible method for isolation of high purity blood EC. The protocols involve mechanical and enzymatic digestion, CD31 MicroBeads enrichment and fluorescence-activated cell sorting. The final content of isolated ECs in the cell suspension, after digestion and depletion/enrichment with MicroBeads, and before FACS sorting (in % of the total number of events recorded during FACS sorting) varies from around 4% for small intestine; 6.5% for colon; 10% for liver to around 45.6% for heart. The viability of the sorted ECs measured immediately after FACS varies from 41% for small intestine to 92% for liver. For details about the flow cytometry analysis and viability see Table S1. Figures 1, 2, 3, and 4 show representative FACS plots and EC gating strategy.
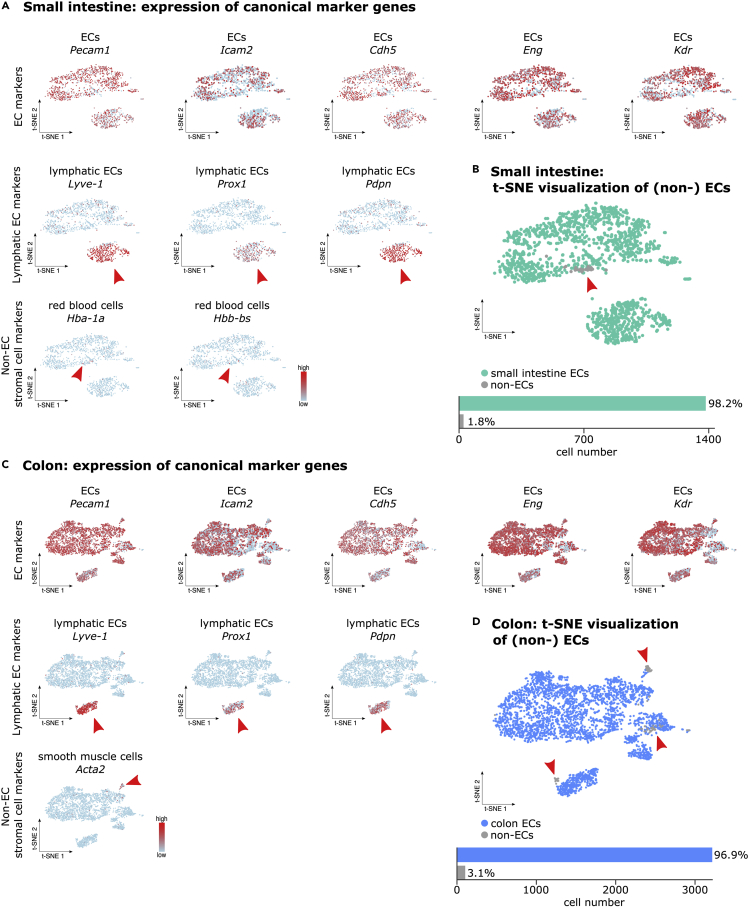
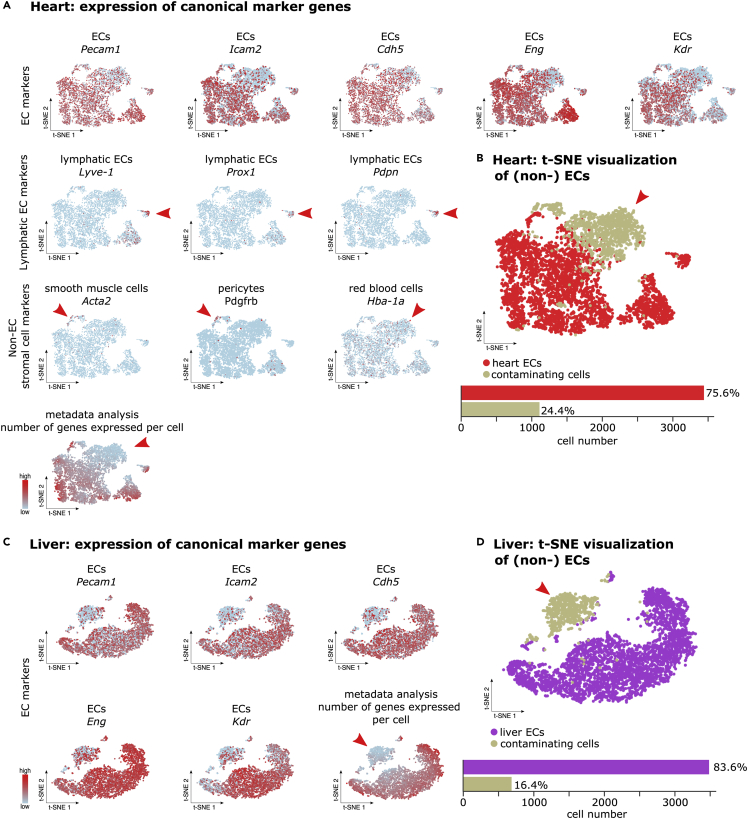
To confirm the quality and purity of isolated ECs we have used the scRNA-seq data generated from ECs isolated during construction of the “Single-Cell Transcriptome Atlas of Murine Endothelial Cells” (Kalucka et al., 2020). The clusters containing EC and “non-EC clusters” were annotated based on the expression of known EC and non-EC marker genes, including Pecam1 and Cdh5 (vascular ECs), Prox1 and Lyve-1 (lymphatic ECs), Col1a1 (fibroblasts), Hba-a1, Hba-a2, Hbb-bs (red blood cells), Pdgfrb (pericytes) and Acta2 (smooth muscle cells). The ECs from small intestine and colon had very high purity (98.2% and 96.9% respectively), and the 1.8% and 3.1% remaining ECs were derived from red blood cells and smooth muscle cells (Figure 5; Table S1). The ECs had high and consistent expression of know endothelial cell markers like CD31 (Pecam1), VE-cadherin (Cdh5), ICAM2 (Icam2), Endoglin/CD105 (Eng) and VEGFR2 (Kdr) (Figure 5). Cells sequenced from heart contained 75.6% of high-quality pure ECs (Figures 6A and 6B; Table S1). In the cluster composed of contaminating cells (24.4%) only a small fraction of cells expressed smooth muscle cell and red blood cell markers. The remaining cells in this cluster didn’t show expression of endothelial cell markers (Figures 6A and 6B). Isolation of liver ECs resulted in 83.6% of high-quality pure ECs. 16.4% of isolated ECs were in silico identified as contaminating ECs due low expression of classical EC markers and low number of total gene expression per cell (Figures 6C and 6D; Table S1).
Figure 5.
Characterization of the small intestine and colon EC sample purity
(A) t-SNE (t-distributed stochastic neighbor embedding) visualization of scRNA-seq analyses on ECs isolated from mouse small intestine showing representative EC and non-EC gene markers expression. Red arrowheads are pointing at cells highly expressing the marker gene. Color scale: red, high expression; blue, low expression.
(B) Top: t-SNE visualization of small intestine (non-) ECs color coded per condition. Red arrowheads are pointing at non-ECs. Bottom: bar plot illustrating the quantification of EC and non-ECs.
(C) t-SNE visualization of scRNA-seq analyses on ECs isolated from mouse colon showing representative EC and non-EC gene markers expression.Red arrowheads are pointing at cells highly expressing the marker gene. Color scale: red, high expression; blue, low expression.
(D) Top: t-SNE visualization of colon (non-) ECs color coded per condition. Red arrowheads are pointing at non-ECs. Bottom: bar plot illustrating the quantification of EC and non-ECs.
Figure 6.
Characterization of the heart and liver EC sample purity
(A) t-SNE visualization of scRNA-seq analyses on ECs isolated from mouse heart showing representative EC and non-EC gene markers expression and t-SNE visualization of the number of genes expressed per cell. Red arrowheads are pointing at cells highly expressing the marker gene (or cells with low gene number expression). Color scale: red, high expression; blue, low expression.
(B) Top: t-SNE visualization of heart (non-) ECs color coded per cell type. Red arrowheads are pointing at non-ECs. Bottom: bar plot illustrating the quantification of EC and non-ECs.
(C) t-SNE visualization of scRNA-seq analyses on ECs isolated from mouse liver showing representative EC and non-EC gene markers expression and t-SNE visualization of the number of genes expressed per cell. Red arrowheads are pointing at cells highly expressing the marker gene (or cells with low gene number expression). Color scale: red, high expression; blue, low expression.
(D) Top: t-SNE visualization of liver (non-) ECs color coded per cell type. Red arrowheads are pointing at non-ECs. Bottom: bar plot illustrating the quantification of EC and non-ECs
Quantification and statistical analysis
A brief overview of data processing and in silico EC Selection (related to the Figures 5 and 6)
-
1.
Generate gene expression matrices using the CellRanger software (10× Genomics).
-
2.
Aggregate sample data using CellRanger software, and process raw data further in R (version 3.4.4).
-
3.Perform the following quality control steps on the pooled tissue datasets:
- a.genes expressed by fewer than 10 cells or with a row average of < 0.002 should not be considered and therefore removed;
- b.cells that expressed fewer than 300 genes (low quality), and cells that expressed over 4,000 genes (potential doublets) should be excluded from further analysis;
- c.cells in which over 10% of unique molecular identifiers (UMIs) were derived from the mitochondrial genome should be removed.
-
4.
Normalize the data using the NormalizeData function as implemented in the Seurat package (Satija et al., 2015).
-
5.
Cluster the cells per organ prior to in silico EC selection for each organ separately.
-
6.
First, for EC selection, identify highly variable genes using the Seurat FindVariableGenes function (mean lower threshold = 0.0125, mean higher threshold = 8, dispersion threshold = 0.5.
-
7.
Auto-scale the data (using highly variable genes only) and summarize by principal component analysis (PCA) using the flashPCA package (Abraham et al., 2017)
-
8.
Visualize the data using t-Distributed Stochastic Neighbor Embedding (t-SNE, Rtsne package; top 8 principal components (PCs)) (van der Maaten, 2008).
-
9.
Perform graph-based clustering to cluster cells according to their gene expression profile using the FindClusters function in Seurat (clustering resolution = 1, k-nearest neighbors = 10).
-
10.
Annotate EC clusters based on the expression of known EC and non-EC marker genes, including Pecam1 and Cdh5 (vascular ECs), Prox1 and Lyve-1 (lymphatic ECs), Col1a1 (fibroblasts), Hba-a1, Hba-a2, Hbb-bs (red blood cells), Pdgfrb (pericytes) and Acta2 (smooth muscle cells). Of note, the data processing and in silico EC selection (steps 4–10) was performed using algorithms implemented in the BIOMEX software (Taverna et al., 2020).
All raw sequencing data referred to in this study are available at ArrayExpress (ArrayExpress: E-MTAB-8077; (Kalucka et al., 2020).
Limitations
We acknowledge a number of limitations pertaining to the protocols detailed above. First, the protocols were established using 8-week-old male C57BL6/J mice, and therefore further adjustments might be needed to isolate ECs from different mouse strains (BALB/c, CD-1 or SCID), gender, or from mice at different developmental stages. Second, the protocols are optimized for blood vessel ECs and not for lymphatic ECs. Thus, further adjustments have to be made to specifically isolate lymphatic ECs. For additional information about isolation of lymphatic ECs from e.g., murine lymph nodes and murine embryos we refer to (Fujimoto et al., 2020, Crosswhite, 2018). Third, ECs isolated using the protocols described above were used for multiple transcriptomics approaches (e.g., bulk or single cell RNA sequencing). Additional optimizations should be performed in order to use isolated cells for other applications, e.g., in vitro cell culture. Additionally, estimated time in the protocols was established for isolation of one type of organ from up to 3 mice by one person at the time. When isolating from higher number of animals the estimated time (or number of required people) for each step may increase.
Of note, some protocols described above may slightly differ from the isolation protocols published in (Kalucka et al., 2020) due to further optimization and adjustments to improve EC isolation efficiency. Additionally, if multiple organs will be isolated from the same mouse, we suggest to have one person assigned per single organ isolation.
Troubleshooting
Problem 1
Low number of isolated ECs. A low number of isolated ECs may occur due to: i.) under- or over- digestion of the dissected tissue (steps 2b-k, 3b, 7b-e, 8a, 12 d, 12g, 16a, 16c, 16d); ii) poor enrichment with CD31 MicroBeads (Miltenyi Biotec, Cat#130-097-418) (steps 4, 9, 13, 17); or iii) loss of ECs during cell sorting (e.g.,: due to decreased cell viability or poor labeling with antibodies) (steps 5, 10, 14, 18).
Potential solution
To avoid extensive loss of ECs, we recommend to optimize the mechanical steps of digestion (vigorous shaking by hand, pipetting, cutting with scalpel blade, rotation speed) in order to obtain the maximum possible number of ECs from each tissue. Moreover, we recommend to optimize the volume of CD31 MicroBeads (Miltenyi Biotec, Cat#130-097-418) as well as the antibody concentrations according to the manufacturer's instructions, to avoid loss of ECs during magnetic- and cell sorting, respectively. If the problem persists, we advise to use and pool additional mice for the isolation of ECs of the particular organ. Additionally, other positive and negative selection technologies could be optimized and used, for example non-column based magnetic isolations (STEMCELL Technologies).
Problem 2
Low viability of isolated ECs. Low viability of the ECs could be caused by the extended time of the isolation procedure, extended digestion time and the number of manipulations on the cells (steps 2e, 2h, 2k, 3b, 4, 5, 8a, 9, 10,12d, 12g, 13, 14, 16c, 17, 18).
Potential solution
We recommend to process samples fast to reduce the time of isolation and to store cell solutions at 4°C or on ice during the isolation procedure. Optimal enzymatic digestion time is crucial for cell viability. If low EC viability persists during sorting, we advise to adjust the time of enzymatic digestion. Additionally, the concentration of the cell suspension should be adjusted when starting FACS for optimal flow rate to reduce sorting time.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Peter Carmeliet (peter.carmeliet@kuleuven.be), or technical contact, Joanna Kalucka (joanna.kalucka@aias.au.dk).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Original/source data for datatype in the paper is available in ArrayExpress: E-MTAB-8077. The published article by Kalucka et al. includes all datasets generated or analyzed during this study.
Acknowledgments
We acknowledge the help of A. Bouché and P. Vanwesemael for technical assistance. J.K., N.V.C., K.V., L.T., M.B., P.d.Z., and K.R. are supported by the Fonds voor Wetenschappelijk Onderzoek (FWO); J.K. by AIAS-CO-FUND II: GA: MSCA: 754513 and The Aarhus University Research Foundation; Lundbeckfonden: R307-2018-3667, Carlsberg Fonden: CF19-0687, and Steno Diabetes Center Aarhus (SDCA); L.-A.T. by University of Antwerp; V.G. by Strategisch Basisonderzoek Fonds voor Wetenschappelijk Onderzoek-Vlaanderen (SB-FWO); S.J.D. and K.D.F. by a Marie Curie-IEF Fellowship; and P.C. by Methusalem funding, FWO Vlaanderen, Foundation Against Cancer (2016-078), Kom op Tegen Kanker (Stand up to Cancer, Flemish Cancer Society), ERC Proof of Concept (ERC-713758), and an ERC Advanced Research Grant (ERC-743074). All the figures were created with BioRender.
Author contributions
L. Sokol, V.G., M.G.-C., N.V.C., and J.K. wrote the manuscript. The protocol for EC isolation from the heart was optimized by M.G.-C. and J.K., liver was optimized by J.K., colon was optimized by V.G., M.G.-C., and J.K., and small intestine was optimized by J.K., L. Sokol, and C.D.; J.K., N.V.C., L. Sokol, E.M., L-A.T., K.V., R.C., L.T., M.B., C.D., S.J.D., V.G., M.G.-C., P.d.Z., and K.D.F. participated in EC isolations. J.K, L.P.M.H.d.R., K.R., J.G., and L. Sokol performed scRNA-seq and bioinformatic analysis and result visualization. J.K. and M.P. performed flow cytometry. L.P.M.H.d.R., L.S., M.D., G.E., J.K., K.R., J.G., X.L., and P.C. provided advice and discussed results. J.K. coordinated optimization of protocols. P.C. conceptualized the study.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2021.100489.
Contributor Information
Joanna Kalucka, Email: joanna.kalucka@aias.au.dk.
Peter Carmeliet, Email: peter.carmeliet@kuleuven.vib.be.
Supplemental information
References
- Abraham G., Qiu Y., Inouye M. FlashPCA2: principal component analysis of Biobank-scale genotype datasets. Bioinformatics. 2017;33:2776–2778. doi: 10.1093/bioinformatics/btx299. [DOI] [PubMed] [Google Scholar]
- Crosswhite P. Isolation of LYVE-1+ endothelial cells from mouse embryos. Bio-protocol. 2018;8:e2962. doi: 10.21769/BioProtoc.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto N., He Y., D’ADDIO M., Tacconi C., Detmar M., Dieterich L.C. Single-cell mapping reveals new markers and functions of lymphatic endothelial cells in lymph nodes. bioRxiv. 2020 doi: 10.1371/journal.pbio.3000704. 2020.01.09.900241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalucka J., De Rooij L.P.M.H., Goveia J., Rohlenova K., Dumas S.J., Meta E., Conchinha N.V., Taverna F., Teuwen L.-A., Veys K. Single-cell transcriptome atlas of murine endothelial cells. Cell. 2020;180:764–779.e20. doi: 10.1016/j.cell.2020.01.015. [DOI] [PubMed] [Google Scholar]
- Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna F., Goveia J., Karakach T.K., Khan S., Rohlenova K., Treps L., Subramanian A., Schoonjans L., Dewerchin M., Eelen G., Carmeliet P. BIOMEX: an interactive workflow for (single cell) omics data interpretation and visualization. Nucleic Acids Res. 2020;48:W385–W394. doi: 10.1093/nar/gkaa332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Maaten L.J.P., Hinton G.E. Visualizing High-Dimensional Data Using t-SNE. J. Mach. Learn. Res. 2008;9:27. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original/source data for datatype in the paper is available in ArrayExpress: E-MTAB-8077. The published article by Kalucka et al. includes all datasets generated or analyzed during this study.