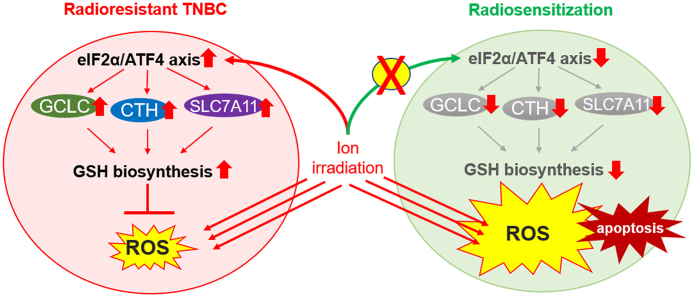
Abstract
Triple-negative breast cancer (TNBC) is the most aggressive breast cancer subtype. Radiotherapy is an effective option for the treatment of TNBC; however, acquired radioresistance is a major challenge to the modality. In this study, we show that the integrated stress response (ISR) is the most activated signaling pathway in radioresistant TNBC cells. The constitutive phosphorylation of eIF2α in radioresistant TNBC cells promotes the activation of ATF4 and elicits the transcription of genes implicated in glutathione biosynthesis, including GCLC, SLC7A11, and CTH, which increases the intracellular level of reduced glutathione (GSH) and the scavenging of reactive oxygen species (ROS) after irradiation (IR), leading to a radioresistant phenotype. The cascade is significantly up-regulated in human TNBC tissues and is associated with unfavorable survival in patients. Dephosphorylation of eIF2α increases IR-induced ROS accumulation in radioresistant TNBC cells by disrupting ATF4-mediated GSH biosynthesis and sensitizes them to IR in vitro and in vivo. These findings reveal ISR as a vital mechanism underlying TNBC radioresistance and propose the eIF2α/ATF4 axis as a novel therapeutic target for TNBC treatment.
Keywords: TNBC, Radioresistance, Integrated stress response, eIF2α, ATF4, GSH
Abbreviations: AARE, Amino acid response element; AMBIC, Ammonium bicarbonate; AP-1, Active protein-1 binding site; ARE, Antioxidant responsive element; CHIP, Chromatin immunoprecipitation; CRE, Cis-regulatory element; CTH, Cystathionine gamma-lyase; DEP, Differentially expressed proteins; HE, Hematoxylin and eosin; IF, Immunofluorescence; IR, Irradiation; ISR, Integrated stress response; OS, Overall survival; TSS, Transcription start site
Graphical abstract
Highlights
-
•
The eIF2α/ATF4 axis is constitutively activated in radioresistant TNBC.
-
•
Phosphorylated eIF2α increases the expression of ATF4 and GCLC at the translational level in TNBC.
-
•
The eIF2α/ATF4 axis activation causes radioresistance in TNBC by promoting GSH biosynthesis and ROS scavenging.
-
•
ATF4 promotes GSH biosynthesis in radioresistant TNBC by triggering the expression of GCLC, CTH, and SLC7A11.
-
•
Inhibition of the eIF2α/ATF4 axis can improve the sensitivity of TNBC to radiotherapy in vitro and in vivo.
1. Introduction
Triple-negative breast cancer (TNBC) is the most aggressive subtype of BC, which is associated with ~67% of BC deaths despite accounting for only 10–30% of all BC cases [1]. Due to the lack of estrogen receptor (ER), human epidermal receptor 2 (HER2), and progesterone receptor (PR), TNBC is inherently resistant to classic BC targeted therapies, including hormone therapy, making it the most challenging BC in the clinic.
Radiotherapy (RT) is an effective option for TNBC local control, especially in patients receiving breast-conserving surgery [2]. Compared with those without post-operational treatment, adjuvant RT decreased the 10-year recurrence rate of TNBC from 35% to 19% and significantly improved patients’ 15-year survival rate [3]. Nevertheless, TNBC is more likely to develop a radioresistant phenotype than other BC subtypes, resulting in a higher recurrence rate and a poor response to subsequent treatment [4,5]. So far, the regulatory mechanisms involved in TNBC radioresistance remain poorly characterized. There is also a lack of clinically effective radiosensitizers for the treatment of TNBC patients [6]. Therefore, elucidating the mechanism of TNBC radioresistance and identifying potential therapeutic targets may have a high clinical impact.
The integrated stress response (ISR) is initiated by the phosphorylation of eukaryotic Initiation Factor 2α (eIF2α) on serine 51 in response to various stressed conditions, including glucose deprivation, amino acid starvation, endoplasmic reticulum stress, and hypoxia [7]. The phosphorylated site inhibits the release of eIF2α from eIF2α-GDP/eIF2β complexes, which blocks the global protein synthesis while allows the translation of mRNAs with small open reading frames in their 5’ UTRs (uORFs). Several studies indicated that ISR activation plays a critical role in protecting cells from oxidative stress injury [[8], [9], [10]]. For example, the hyperphosphorylated eIF2α in cancer cells was shown to repress cytotoxics-induced accumulation of reactive oxygen species (ROS) and cause chemoresistance [11,12]. Although the effect was considered to be associated with the inhibition of reduced glutathione (GSH) depletion and the efflux of oxidative glutamate [13,14], the exact mechanism by which eIF2α phosphorylation contributes to therapeutic resistance is not fully understood, and the role of ISR in cancer radioresistance remains to be elucidated.
In this study, we demonstrate for the first time that the eIF2α/ATF4 axis plays a key role in the fine-tuning of the GSH biosynthesis gene transcriptional program involved in TNBC radioresistance. We show that constitutive eIF2α phosphorylation and ATF4 activation are required to maintain intracellular GSH homeostasis and redox balance in radioresistant TNBC under oxidative stress conditions. The high intratumoral expression of the axis-associated genes predicts a poor clinical prognosis in TNBC patients, whereas the eIF2α/ATF4 axis inhibition can restore the sensitivity of radioresistant TNBC to IR in vitro and in vivo. These findings reveal ISR as a vital mechanism contributing to cancer radioresistance and provide a rationale for exploring eIF2α/ATF4 signaling as a novel therapeutic target for TNBC treatment.
2. Materials and methods
2.1. Cell culture
Human TNBC cell lines (MDA-MB-231, -436, and -468) were purchased from the American Type Culture Collection (ATCC, VA, USA). MDA-MB-231, -436, and -468 cells were cultured in the Iscove's Modified Dulbecco's Medium (IMDM) (Gibco, VIC, Australia), and RPMI-1640 medium (Gibco), Dulbecco's modified Eagle's medium (DMEM) (Gibco), respectively, supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin-streptomycin solution (Gibco). All cell lines were maintained in a humidified atmosphere with 5% CO2 at 37 °C.
2.2. Establishment of radioresistant TNBC cell lines
Parental TNBC cells in 75 cm2 flasks were placed on a 5-cm motorized shelf with a distance to X-ray tube being 76 cm in the X-Rad 320 Biological Irradiator (Accela s.r.o., Prague, Czech Republic) and were irradiated at 0.8 Gy/min (min) dose rate at 320 kV and 12.5 mA. Each cell line received a total dose of 60 Gy IR (2 Gy per fraction × 5 fractions per week for 6 weeks). The radioresistant cells (MDA-MB-231-RR, −436-RR, and −468-RR) were derived from surviving clones after IR.
2.3. Colony formation assay
Exponentially growing cells were seeded in a 6-well plate at a density of 2 × 103 per well. They were treated with the indicated conditions and cultured for 9–14 days. Colonies (>50 cells) were stained with 0.5% crystal violet for 15 min at room temperature and were immediately counted and photographed.
2.4. Sample preparation for label-free proteomics
Cells were lysed using the lysis buffer consisting of 3% sodium deoxycholate, 6 M urea, and 1 × Halt™ protease and phosphatase inhibitor cocktail (Thermo Scientific, VIC, Australia) in 50 mM ammonium bicarbonate (AMBIC) buffer. After sonicating and agitating, samples were centrifuged (14,000×g for 15 min at 4 °C) and the supernatant was collected for BCA assay. Protein samples were reduced with 15 mM dithiothreitol at 60 °C for 30 min and then alkylated with 20 mM iodoacetamide at room temperature for 30 min in the dark. These samples were diluted with 50 mM AMBIC buffer six times before digestion which was performed with the Pierce™ Trypsin Protease (Thermo Scientific) at a trypsin:protein ratio of 1:50 (w/w) at 37 °C overnight. On the second day, formic acid (FA) was added to stop digestion, and the precipitates were removed by centrifugation. The supernatants were desalted and cleaned using the Pierce™ C18 Spin Tips (Thermo Scientific). Collected samples were dried using the SpeedVac centrifugal evaporator and then resolved in 0.1% FA to a final concentration of 1 μg/μL.
2.5. Label-free quantitative proteomics
Label-free proteomics was carried out using the high-performance liquid chromatography (HPLC) with tandem mass spectrometric (MS/MS) as described previously [15]. A detailed description of methods is included in Supplementary materials and methods.
2.6. Protein identification and quantitation
Peak picking of peptides was performed using the Progenesis QI (Waters, NSW, Australia). Ion abundances and features of peptides were aligned to a reference sample containing equal contributions of all samples. Peptide abundances were normalized against the total abundances and compared by one-way analysis of variance (ANOVA). The ms/ms spectra of peptides were exported to the Mascot Daemon v2.5.1 (Matrix Science, London, UK) for protein identification against the SwissProt database (downloaded Feb 2018; containing 556,568 sequences) and with the following parameter set: (i) taxonomy, homo sapiens (human); (ii) fixed modifications, carbamidomethyl DTT (C); (iii) variable modifications, cysteinyl (C), oxidation (C), oxidation (M), phospho (STY); (iv) enzyme, semiTrypsin; (v) maximum missed cleavages, 2; (vi) peptide charge 2+, 3+ and 4+; (vii) peptide tolerance, ± 4 ppm; (viii) ms/ms tolerance, 0.4 Da. False discovery rate (FDR) with q value set at <0.01 was used as a filter at both protein and peptide level, and only proteins with at least two unique peptides were included for the final report. Pathways enriched within the dataset were analyzed using the Ingenuity Pathways Analysis software (IPA®, QIAGEN, MD, USA). The core analysis was carried out using the default settings, with only direct relationships and experimentally observed confidence considered. The P-value for the correlation between identified proteins and a given canonical pathway was calculated by Fisher's exact test.
2.7. Western blot (WB)
Total protein was extracted with the RIPA lysis buffer containing 1 × Halt™ protease and phosphatase inhibitor cocktail (Thermo Scientific), while nuclear protein was extracted using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Scientific). WB was performed as described previously [16]. A detailed description of methods and antibodies (Abs) is included in Supplementary materials and methods.
2.8. Immunofluorescence (IF)
Dual-color staining was performed with primary and secondary Abs and propidium iodide (PI, 0.2 μg/mL), while four-color staining was achieved using rabbit primary Abs, the Zenon™ Tricolor Rabbit IgG Labeling Kit (Thermo Scientific), and 4′,6-diamidino-2-phenylindole (DAPI, 0.5 μg/mL). A detailed description of methods and Abs is included in Supplementary materials and methods.
2.9. ROS detection
ROS detection was performed with the CellROX™ green probe (Thermo Scientific). The probe can measure the oxidative stress caused by superoxide anion and hydroxyl radicals in live cells [17]. Cells were seeded in a 96-well plate at the density of 2.5 × 104 cells per well for 24 h and then treated with 4 Gy IR in the culture medium. After 24 h, the CellROX® Green Reagent was added into the cells with a final concentration of 5 μM and incubated for 30 min at 37 °C in the dark. Then cells were washed with 1 × PBS three times, and the fluorescence was measured at excitation/emission = 485/520 nm using the Infinite® M200 PRO microplate reader (Tecan, Männedorf, Switzerland).
2.10. Glutathione detection
GSH and oxidized glutathione (GSSG) detection were achieved using the GSH/GSSG Ratio Detection Assay Kit (Abcam, VIC, Australia) according to the specification. GSH and GSSG were labeled with 1 × Thiol Green solution and 1 × GSSG Probe solution, respectively, and were detected by the Infinite® M200 PRO microplate reader at excitation/emission of 490/520 nm.
2.11. Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated using the High Pure RNA Isolation Kit (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer's instruction. Reverse transcription was performed with iScript Advanced cDNA Synthesis Kit (BioRad, NSW, Australia) with 1 μg total RNA. The qRT-PCR was performed with PrimePCR SYBR® Green Assays (BioRad) and 2 μL cDNA on a CFX96™ Real-Time PCR System (BioRad). GAPDH was used as a reference, and the 2−ΔΔCT method was used for calculating the relative gene expression. Primers for qRT-PCR are listed in Supplementary materials and methods.
2.12. Polysome preparation
Polysomes were size-fractionated by a 10 mL sucrose density gradient made by 1 mL of each sucrose solution (bottom to top: 50%–5%). Briefly, cells were incubated in the medium containing 100 μg/mL cycloheximide (Sigma-Aldrich, NSW, Australia) for 10 min at 37 °C and were lysed in polysome lysis buffer containing 1% Triton™ X-100 (Sigma-Aldrich). The supernatant was collected by centrifugation and was added to the top of the gradient in an ultracentrifuge tube. The gradient was centrifuged for 2 h (36,000 rpm at 4 °C) using the SW 41 Ti Swinging-Bucket Rotor (Beckman Coulter Life Sciences, NSW, Australia). A total of twelve fractions were collected from the top to the bottom of the gradient using the BR-188 Density Gradient Fractionator (Brandel, MD, USA). All fractions were stored at −80 °C for RNA isolation and qRT-PCR.
2.13. Chromatin-immunoprecipitation (CHIP)
ChIP assay was performed using the Pierce Agarose ChIP Kit (Thermo Scientific). Briefly, cells were fixed with 1% formaldehyde and incubated with 1× glycine solution for 5 min at room temperature. Cell pellets were lysed in the Lysis Buffer 1 on ice for 10 min and sonicated for 15 s. For chromatin digestion, 10 U/μL micrococcal nuclease was used. Digested chromatin was incubated with anti-ATF4 (D4B8, CST, MA, USA), anti-Nrf2 (ab62352, Abcam), or anti-IgG (ab2410, Abcam) antibodies overnight at 4 °C and then incubated with protein A/G plus agarose for 1 h at 4 °C. The CHIPs were incubated with IP elution buffer supplemented with 6 μL of 5 M NaCl and 2 μL of 20 mg/mL Proteinase K for 40 min at 65 °C. The reverse crosslinking of ChIPs was performed with a DNA clean-up column and DNA binding buffer. The purified DNA was subject to qRT-PCR analysis, and the fold enrichment of a gene promoter was calculated by dividing the cycle threshold (Ct) value from anti-ATF4 or anti-Nrf2 group by the Ct value from the anti-IgG group. PCR primers targeting the core promoter region of a gene are listed in Supplementary materials and methods.
2.14. Gene overexpression
The pCMV6 vector and pCMV6-GADD34 plasmid were purchased from OriGene Technologies (Rockville, MD, USA). The pcDNA3 vector and pcDNA3-eIF2αS51A plasmid were gifts from David Ron (Addgene plasmid #21808 and #21810, MA, USA). Transient overexpression of GADD34 and eIF2αS51A was achieved by the Lipofectamine 3000 (Thermo Scientific) according to the manufacturer's instructions. Cells were incubated with the plasmid-lipid complex for 72 h before the following assays. For MDA-MB-231 and -436 cells, the average transfection efficiency is ~90% by flow cytometry. Stable overexpression of eIFα2S51A was achieved by lentiviral particles containing the pLenti-C-Myc-DDK-P2A-Puro-eIF2αS51A, assembled by OriGene Technologies. Briefly, cells were incubated with lentiviral particles for 18 h in the medium containing 8 μg/mL polybrene and then cultured in the lentiviral particle-free medium for another 72 h. Puromycin (1 μg/mL) was used to select clones with stable eIF2αS51A expression for in vivo experiments. The pLenti vector clone (PS100092V, OriGene Technologies) was used as a control.
2.15. Gene knockdown
ATF4 knockdown was performed using the Lipofectamine RNAiMAX transient transfection reagent (Thermo Scientific) and the combination of two Silencer® Select ATF4-specific siRNAs (Assay ID s1702 and s1704, Thermo Scientific), as per the manufacturer's protocol. Silencer™ Select Negative Control siRNA (Thermo Scientific) was used as a control. After transfection for 72 h, cells were tested for ATF4 expression and used for the following assays.
2.16. Cell apoptosis detection
Apoptosis detection was performed using the Annexin V-FITC Apoptosis Detection Kit (Abcam) on the LSR Fortessa™ X-20 flow cytometer (BD Bioscience, NSW, Australia) at the 530/30 and 610/20 bandpass filters according to the instructions.
2.17. Mouse xenograft experiments
Female athymic (nu/nu) BALB/c mice (5 weeks old, 18–20 g) were provided by Guangdong Medical Laboratory Animal Center (Guangzhou, China) and were housed in a specific pathogen-free (SPF) animal facility in the Experimental Animal Center of Sun Yat-sen University (Guangzhou, China). All procedures involving mice were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University (IACUC-2020-B0813). After one-week adaption, exponentially growing MDA-MB-231-RR cells (7 × 106) stably expressing eIF2αS51A or the control vector in 0.2 mL 1 × PBS were subcutaneously injected into the right flank of mice to develop xenograft tumors. Tumor size was monitored using a caliper and calculated applying the formula: length × width2 × 0.5. When the average tumor volume reached 100 mm3, mice were randomly divided into 4 groups with 6 mice per group: 1) vector; 2) eIF2αS51A; 3) vector + local IR; 4) eIF2αS51A + local IR. Local IR was given at 2 Gy per day for 5 consecutive days (10 Gy in total). A lead shield was used to ensure that only the right flank of each mouse was exposed to IR. All mice were euthanized 20 days after IR completion, and tumor samples were collected and stored at −80 °C or fixed in formalin, respectively, for the following assay.
2.18. Tissue microarray (TMA) analysis
TMA slides (Cat# BR1202 and BRN801a) were obtained from US Biomax (Rockville, MD, USA), and the pathological information is available at www.biomax.us. Immunohistochemical (IHC) staining of these paraffinized tissues was performed as described previously [16]. A detailed description of methods and Abs is included in Supplementary materials and methods.
2.19. Bioinformatic analysis
The annotation and RNA-sequencing data of 2509 BC samples in the METABRIC dataset were obtained using the cBioPortal (https://www.cbioportal.org). A total of 1904 BC samples with complete clinical information were sorted from the dataset, and a total of 298 TNBC samples were obtained from the dataset, as defined by the ‘negative’ in the fields of HER2 status, ER status, and PR status. Gene expression was normalized by GAPDH. The three-gene signature was constructed using GCLC, SLC7A11, and CTH, and the risk score of the samples was calculated by the sum of individual gene expression multiplied by its Cox regression coefficient. The cutoff value used to stratify samples into the high and low expression (risk score) groups of a gene (signature) was optimized by the X-Tile [18], and the Kaplan-Meier survival curves were generated accordingly. The log-rank test was used to compare the survival curves.
2.20. Statistical analysis
All data were analyzed using the Prism 9 (GraphPad Software Incorporated, USA). The correlation analysis between protein expressions was performed with the linear regression, and the correlation coefficient R was calculated. The comparison of the two groups was performed using unpaired Student's t-test, while the comparison of three or more groups was achieved using one-way ANOVA with Tukey's post hoc test. Statistical significance was defined by P < 0.05.
3. Results
3.1. The eIF2α/ATF4 axis is constitutively activated in radioresistant TNBC cells
The radioresistant TNBC cells were developed using fractional IR and confirmed by radiosensitivity assays (Supplementary Fig. 1A). These radioresistant cells also showed strong resistance to cisplatin, doxorubicin, and olaparib (Supplementary Fig. 1B-D). Morphologically, radioresistant TNBC cells were flat and swollen and were likely to grow into a more compact colony than parental cells (Supplementary Fig. 2A, B). This is consistent with the observation that radioresistant TNBC cells displayed a lower migrative and invasive potential (Supplementary Fig. 2C). Furthermore, the proliferation rate of radioresistant cells was lower than their parental cells (Supplementary Fig. 2D).
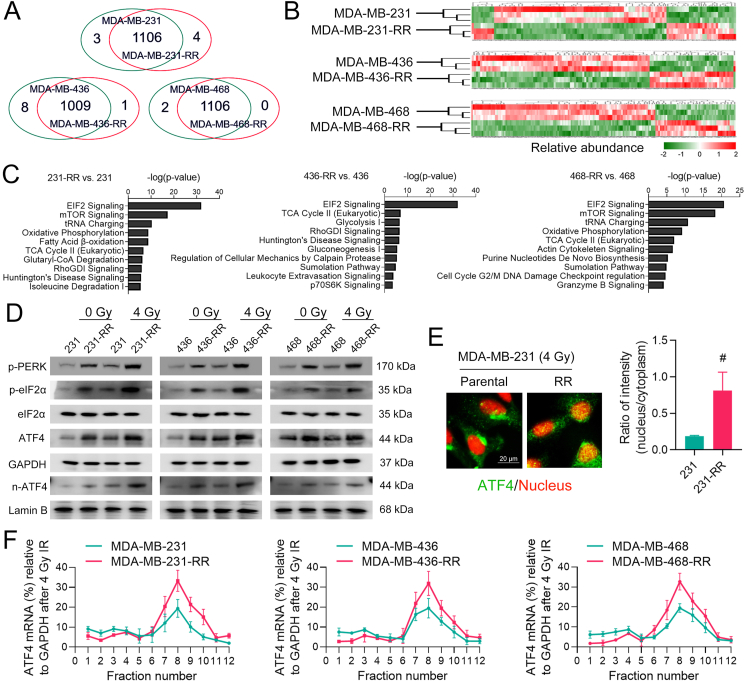
In this study, our first objective was to identify critical pathways participating in TNBC radioresistance via label-free proteomics. After MASCOT searching, a total of 1113, 1018, and 1108 proteins were identified from MDA-MB-231, -436, and -468 cell lines, respectively (Fig. 1A). A full list of these proteins is shown in Supplementary Table 1. The heatmap across differentially expressed proteins (DEPs, FDR-adjusted P < 0.05) shows that most proteins were down-regulated in radioresistant cells (Fig. 1B). To increase the confidence in pathway analysis and correct for multiple testing, only DEPs with log2 fold change ≥1 and ≤ -1were sent to IPA core analysis (Supplementary Fig. 3A, B). The top ten up-regulated and down-regulated pathways in radioresistant TNBC cells are shown in Fig. 1C and Supplementary Fig. 3C, respectively. Notably, the eIF2 signaling was among the most up-regulated pathway in radioresistant cell lines and was worthy of further investigation (Fig. 1C).
Fig. 1.
The eIF2α/ATF4 axis is constitutively activated in radioresistant TNBC cells. (A) Venn diagram shows the number of proteins identified from cell samples (FDR < 0.01 and unique peptide ≥ 2). The cross-section shows the number of common proteins between two cell lines, while the two sides show the number of uniquely expressed proteins. (B) Heatmap shows the hierarchical clustering of differentially expressed proteins (DEPs) (P < 0.05) between radioresistant and parental cells. Protein abundance is expressed using the standardized scale ranging from −2 to 2. High expression of a given protein is marked in red, while low expression is marked in green. White represents the intermediate expression. (C) Core functional analysis shows the top ten up-regulated canonical pathways in radioresistant cells. (D) Cells were treated with 0 or 4 Gy IR. The protein expression of p-PERK, p-eIF2α, eIF2α, ATF4, and n-ATF4 (nuclear ATF4) was determined by WB 24 h after IR. (E) The intracellular distribution of ATF4 (green) in MDA-MB-231 and -231-RR cells treated with 4 Gy IR was determined by IF 24 h after IR, and the nucleus was stained with red. Representative IF images are shown at 1000 × magnification. The nucleus/cytoplasm ratio of ATF4 fluorescent signal was compared. (F) The expression of ATF4 mRNAs in polysome fractions between radioresistant and parental TNBC cells treated with 4 Gy IR was determined by qRT-PCR 24 h after IR. #P < 0.05 vs. parental cells (n = 5). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
ISR initiated by eIF2α phosphorylation optimizes cellular responses to a wide range of stressors [7], implying that the eIF2 signaling may function in cancer radioresistance. To test our hypothesis, we first detected the status of eIF2α and found that the level of eIF2α phosphorylation in radioresistant TNBC cells was higher than that in parental cells, even after IR treatment (Fig. 1D). ATF4 is a crucial effector in response to ISR activation [10]. Increased protein expression of total ATF4 and nuclear ATF4 (n-ATF4) was also confirmed in radioresistant TNBC cells (Fig. 1D), indicating the activation of ATF4. This finding is further supported by the IF assay showing that ATF4 was highly accumulated in the nucleus of MDA-MB-231-RR cells treated with 4 Gy IR (Fig. 1E). Previous studies demonstrated that the translation of mRNAs containing an uORF could be selectively promoted during ISR [10], we thus performed the polysome fractionation assay to detect the alternation in the translational efficiency of ATF4 mRNA. The results show that the translation of ATF4 mRNA was accelerated in radioresistant cells as compared to parental cells after IR (Fig. 1F). These results suggest that the eIF2α/ATF4 axis is constitutively activated in radioresistant TNBC cells and may play an important role in regulating the cellular resistance to IR.
3.2. The eIF2α/ATF4 axis promotes GSH biosynthesis in radioresistant TNBC cells
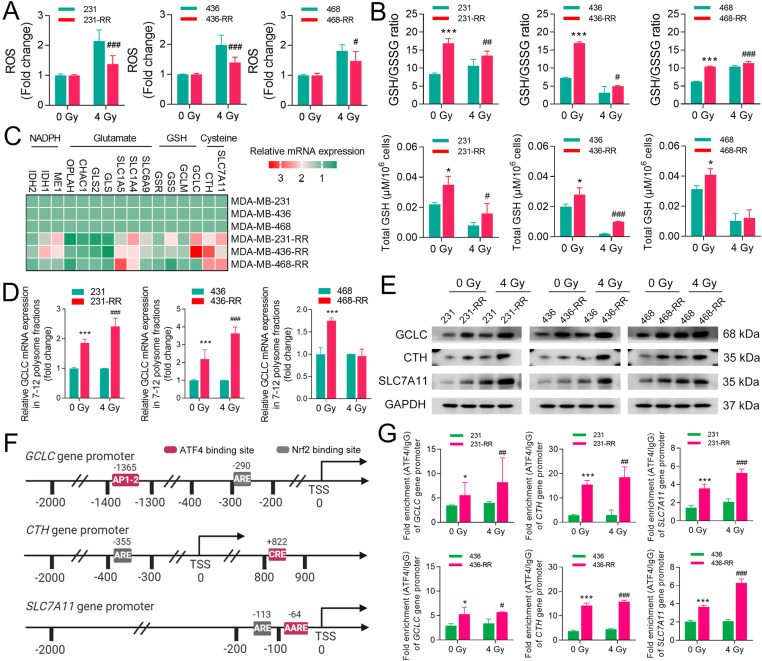
Intracellular accumulation of ROS is a key mechanism for IR-induced cell death [19]. Results from ROS detection show that there was a much lower ROS accumulation in radioresistant TNBC cells after IR (Fig. 2A). GSH is a direct and critical scavenger for ROS and may affect radiosensitivity [20]. We thus detected intracellular glutathione and found that radioresistant TNBC cells had a much higher GSH/GSSG ratio, as well as a higher level of total GSH, than parental cells in both IR and non-IR conditions (Fig. 2B). Inhibition of GSH synthesis sensitized these TNBC cells to IR (Supplementary Fig. 4), suggesting that GSH function is essential for maintaining the radioresistant phenotype of TNBC.
Fig. 2.
The eIF2α/ATF4 axis promotes GSH biosynthesis in radioresistant TNBC cells. TNBC cells were treated with 0 or 4 Gy IR. (A) The intracellular level of ROS was detected 24 h after IR. (B) Total GSH and GSH/GSSG ratio were detected after 24 h after IR. (C) Heatmap shows the relative mRNA expression of key genes involved in GSH metabolism between radioresistant and parental TNBC cells. High expression of a given mRNA is marked in red, while low expression is marked in green. Pink represents the intermediate expression. (D) The relative expression of polysome-associated GCLC mRNAs (fraction 7–12) was detected by qRT-PCR 24 h after IR. (E) The protein expression of GCLC, CTH, and SLC7A11 in TNBC cells was determined by WB 24 h after IR. (F) Schematic representation of the Nrf2-and ATF4-binding sites in human promoter regions of GCLC, CTH, and SLC7A11. The transcription start site (TSS) is assigned the 0 position. Antioxidant responsive element (ARE) is the consensus sequence for Nrf2 binding. Active protein-1 binding site (AP-1), cis-regulatory element (CRE), and amino acid response element (AARE) are the validated binding sites for ATF4 in human GCLC, CTH, and SLC7A11 genes, respectively. (G) The enrichment of core promoter regions of GCLC, CTH, and SLC7A11 with ATF4 or IgG antibody in TNBC cells was analyzed by ChIP-qPCR 24 h after IR. *P < 0.05 and ***P < 0.001 vs. parental cells without IR; #P < 0.05, ##P < 0.01, and ###P < 0.001 vs. parental cells treated with IR (n = 3). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To gain insights into the relationship between ISR and GSH, we analyzed the expression of key genes involved in glutathione metabolism [21]. We found that genes involved in cysteine uptake and synthesis [solute carrier family 7 member 11 (SLC7A11) and cystathionine-γ-lyase (CTH)], GSH synthesis [glutamate-cysteine ligase catalytic subunit (GCLC)], glutamate uptake [solute carrier family 1 member 4 (SLC1A4) and solute carrier family 1 member 5 (SLC1A5)], and NADPH generation [malic enzyme 1 (ME1) and isocitrate dehydrogenase (IDH1)] were up-regulated in radioresistant cells (Fig. 2C). Besides, the mRNAs of GCLC were shown to contain an uORF and hence may be translationally regulated by the ISR [13]. In this study, we found that the translational efficiency of GCLC mRNA was significantly increased in MDA-MB-231-RR and -436-RR cells regardless of IR (Fig. 2D). Subsequently, we confirmed that GCLC, CTH, and SLC7A11 were highly expressed in radioresistant cells at the protein level as compared to parental cells (Fig. 2E), demonstrating that GSH biosynthesis is promoted in radioresistant TNBC cells.
Both ATF4 and nuclear factor erythroid-2-related factor 2 (Nrf2) were identified as transcriptional activators for human GCLC [22,23], CTH [24,25], and SLC7A11 [14,26] (Fig. 2F), yet their roles in regulating TNBC radioresistance are poorly understood. To that end, we determined Nrf2 status in our cell models. However, we did not detect an up-regulation of total or nuclear Nrf2 protein in radioresistant TNBC cells compared with parental cells, though an increase in nuclear Nrf2 was observed, upon the IR, in both cell lines (Supplementary Fig. 5A). Next, we performed CHIP assays to detect the binding activity of ATF4 or Nrf2 to their core promoter regions. The results show that more ATF4 accumulated in the promoter regions of GCLC, CTH, and SLC7A11 in radioresistant TNBC cells at the baseline level compared with parental cells, and similar results were observed after 4 Gy IR (Fig. 2G). In contrast, no further enrichment of Nrf2 in the promoter regions of GCLC, SLC7A11, and CTH was found in radioresistant TNBC cells compared with parental cells, though it was stimulated by IR (Supplementary Fig. 5B). These results suggest that ATF4 may be associated with an acquired up-regulation of GSH in radioresistant TNBC cells.
3.3. The high expression of the eIF2α/ATF4 axis is found in human TNBC tissues and is associated with a poor prognosis
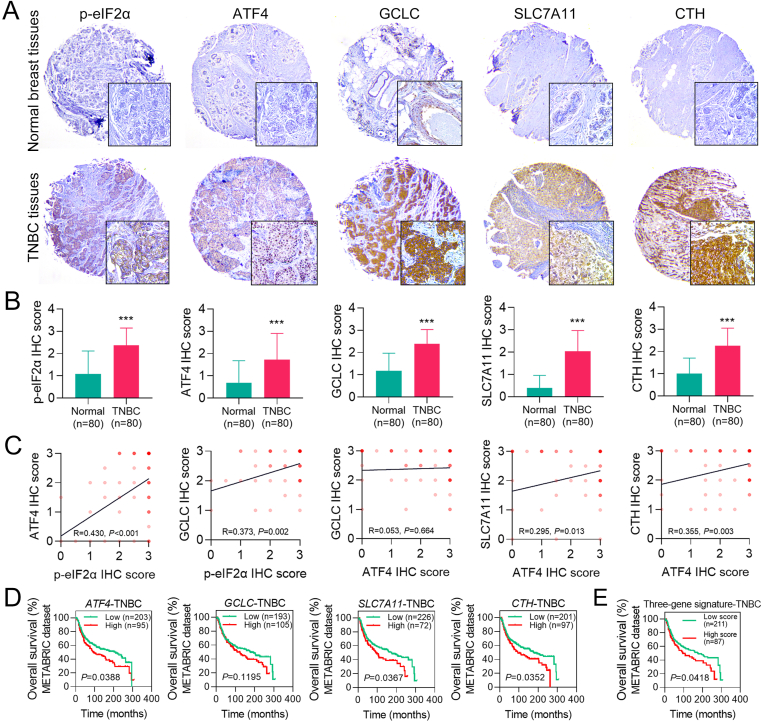
To further determine the importance of the eIF2α/ATF4 axis in TNBC, we analyzed the expression of the eIF2α/ATF4 signaling and its target genes in normal breast and TNBC tissues using human TMAs. The IHC results show that the protein expression of p-eIF2α, ATF4, GCLC, CTH, and SLC7A11 in TNBC tissues was much higher than that in normal breast tissues (Fig. 3A, B). To verify if the protein expression of GCLC, SLC7A11, and CTH is linked with the activation of the eIF2α/ATF4 axis in TNBC, the correlation analysis on protein expression was performed using the IHC score. The results confirmed a correlation between p-eIF2α and ATF4/GCLC, and the expression of ATF4 was shown to be associated with CTH and SLC7A11 but not with GCLC (Fig. 3C). Next, we explored the clinical significance of the ATF4 signaling using a TNBC patient cohort from the METABRIC database. We found that the high intratumoral expression of ATF4, SLC7A11, and CTH was associated with a lower overall survival (OS) rate in TNBC patients (Fig. 3D). Moreover, a higher risk score of the three-gene (GCLC, SLC7A11, and CTH) signature was correlated to a worse OS in the patients (Fig. 3E). These data suggest that the activation of ATF4 signaling predicts a poor prognosis in TNBC.
Fig. 3.
The high expression of the eIF2α/ATF4 axis is found in human TNBC tissues and is associated with a poor prognosis in TNBC patients. (A) The protein level of p-eIF2α, ATF4, GCLC, SLC7A11, and CTH in human normal breast and TNBC tissues was measured by IHC using human TMAs. Representative IHC images are shown at 40 × and 200 × magnification. Brown indicates protein staining, while blue represents the nucleus. (B) The average IHC scores of p-eIF2α, ATF4, GCLC, SLC7A11, and CTH between normal breast and TNBC tissues were compared. (C) Protein expression correlation in human TNBC tissues was determined using correlation analysis based on the IHC score. The positive R-value with a P-value less than 0.05 indicates a significantly positive correlation. (D) The association of intratumoral ATF4, GCLC, SLC7A11, or CTH expression with overall survival (OS) status in the patient cohort (n = 298) from the METABRIC dataset was analyzed using the Kaplan-Meier method and log-rank test. (E) A three-gene signature was established using GCLC, SLC7A11, and CTH. The association of the risk score of this gene signature with OS status in the patient cohort (n = 298) from the METABRIC dataset was analyzed using the Kaplan-Meier method and log-rank test. ***P < 0.001 vs. normal breast tissues (n = 80). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Dephosphorylation of eIF2α abolishes ATF4-mediated GSH biosynthesis in radioresistant TNBC cells
The eIF2αS51A (Ser-51-Ala mutation) is a dominant-negative eIF2α allele that can function as eIF2α but prevent the phosphorylation of eIF2α on Ser51, while GADD34 (growth arrest and DNA damage-inducible protein) can direct protein phosphatase 1 to dephosphorylate eIF2α. By overexpressing eIF2αS51A and GADD34 in MDA-MB-231-RR and -436-RR cells, respectively, we investigated the role of eIF2α in regulating ATF4 and downstream gene expression in TNBC.
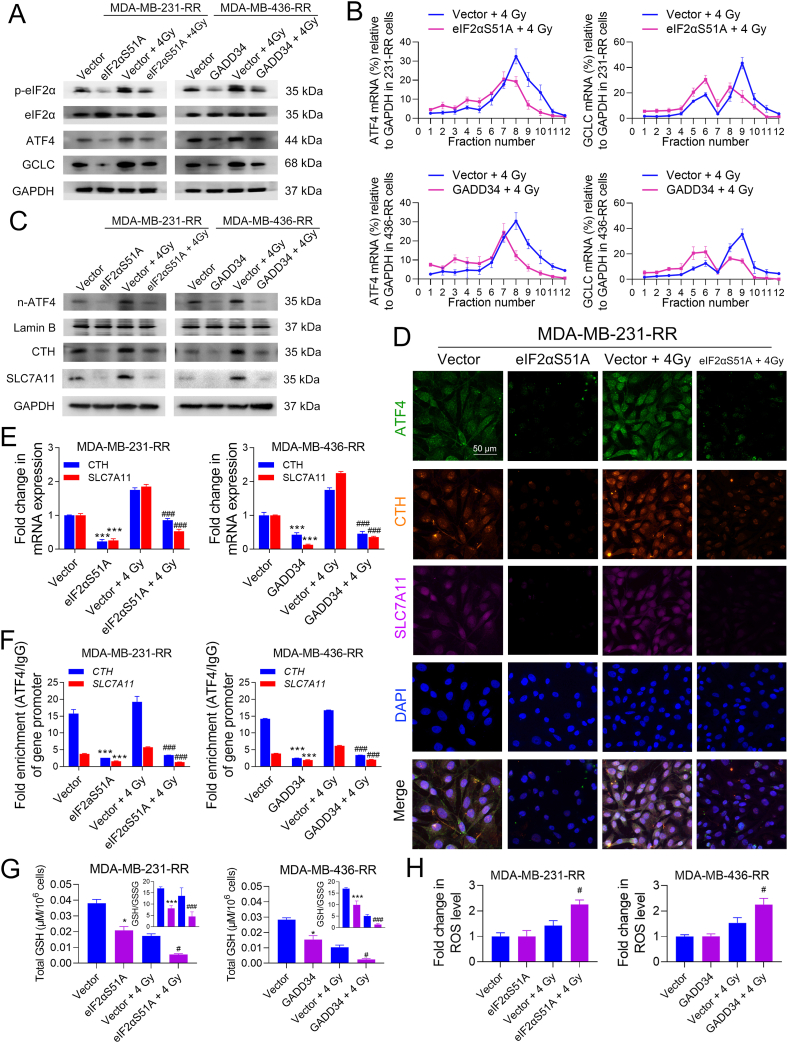
As shown in Fig. 4A, eIF2αS51A or GADD34 overexpression was sufficient to down-regulate p-eIF2α in radioresistant cells, as well as the protein level of ATF4 and GCLC (Fig. 4A). The mRNA analysis with enriched polysome fractions shows that ATF4 and GCLC mRNAs shifted from heavy ribopolysomes into light ones in eIF2αS51A-overexpressed MDA-MB-231-RR cells as compared to the vector cells, and the level of mRNAs associated with 7–12 polysomes was significantly decreased (Fig. 4B). Similar results were also obtained from GADD34-overexpressed MDA-MB-436-RR cells (Fig. 4B), suggesting that ATF4 and GCLC protein expression are, at least partially, regulated by eIF2α at the translational level. Furthermore, we found that the overexpression of eIF2αS51A or GADD34 reduced the baseline and IR-induced protein expression of n-ATF4, CTH, and SLC7A11 in radioresistant TNBC cells (Fig. 4C). Results from the IF assay show that eIF2αS51A overexpression decreased the protein expression of ATF4, CTH, and SLC7A11 in MDA-MB-231-RR cells treated with 0 or 4 Gy IR (Fig. 4D). Consistently, eIF2αS51A or GADD34 overexpression significantly down-regulated baseline and IR-induced mRNA expression of CTH and SLC7A11 in radioresistant TNBC cells (Fig. 4E). Also, they reduced ATF4 accumulation on the promoter regions of CTH and SLC7A11 (Fig. 4F). Glutathione detection shows that both total GSH and GSH/GSSG ratio were significantly decreased by eIF2α dephosphorylation in radioresistant TNBC cells treated with 0 or 4 Gy IR (Fig. 4G). Results from ROS detection show that eIF2αS51A or GADD34 overexpression significantly increased IR-induced ROS accumulation in radioresistant TNBC cells (Fig. 4H). Collectively, these data suggest that eIF2α phosphorylation is critical in regulating ATF4 transcriptional activation and GSH biosynthesis in radioresistant TNBC cells.
Fig. 4.
Dephosphorylation of eIF2α abolishes ATF4-mediated GSH biosynthesis in radioresistant TNBC cells. MDA-MB-231-RR and -436-RR cells were transfected with eIF2αS51A- and GADD34-overexpression plasmid, respectively, and then treated with 0 or 4 Gy IR. (A) The protein expression of p-eIF2α, eIF2α, ATF4, and GCLC was determined by WB 24 h after IR. (B) The expression of ATF4 and GCLC mRNAs in polysome fractions was detected by qRT-PCR 24 h after IR. (C) The protein expression of n-ATF4, CTH, and SLC7A11 was determined by WB 24 h after IR. (D) The protein level of ATF4, CTH, and SLC7A11 was detected by IF 24 h after IR. The nucleus was stained with blue. Representative IF images are shown at 400 × magnification. (E) The relative mRNA expression of CTH and SLC7A11 was detected by qRT-PCR 24 h after IR. (F) The enrichment of the core promoter regions of CTH and SLC7A11 genes with ATF4 or IgG antibody was analyzed by ChIP-qPCR 24 h after IR. (G) Total GSH and GSH/GSSG ratio were detected 24 h after IR. (H) The intracellular level of ROS was detected 24 h after IR. *P < 0.05 and ***P < 0.001 vs. vector group; #P < 0.05 and ###P < 0.001 vs. vector + IR group (n = 3). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. Dephosphorylation of eIF2α sensitizes radioresistant TNBC to IR in vitro and in vivo
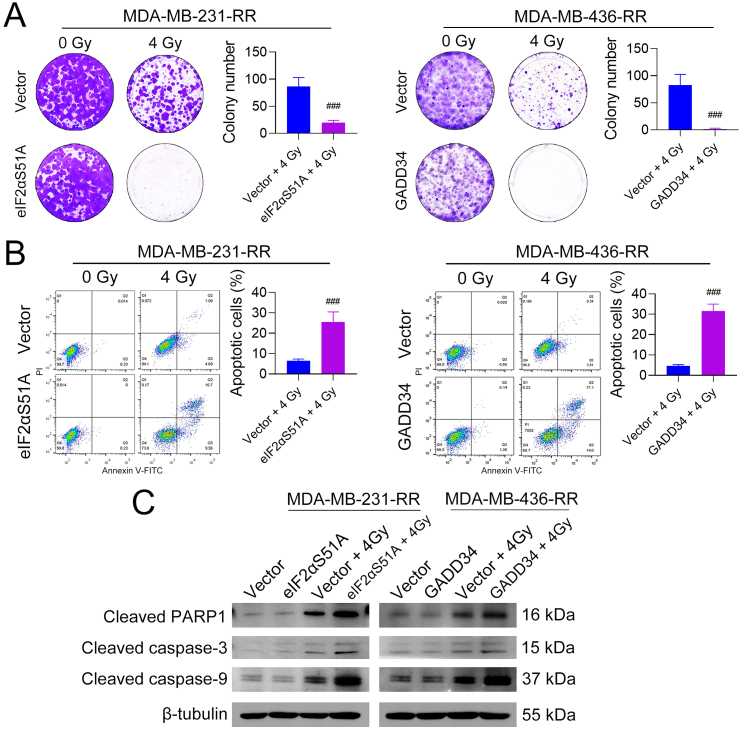
We next explored whether dephosphorylation of eIF2α could reverse the radioresistance of TNBC. Colony formation assays show that eIF2αS51A and GADD34 overexpression significantly decreased the survival rate of MDA-MB-231-RR and -436-RR cells treated with 4 Gy IR, respectively (Fig. 5A). Moreover, eIF2α dephosphorylation sensitized MDA-MB-231-RR and -436-RR cells to cisplatin, doxorubicin, and olaparib (Supplementary Fig. 6A), and a similar result was also obtained by directly inhibiting GSH synthesis (Supplementary Fig. 6B), suggesting that ISR might play a role in TNBC resistance to cytotoxics through regulating GSH. Flow cytometry analysis shows that eIF2αS51A or GADD34 overexpression dramatically increased the percentage of apoptotic cells caused by 4 Gy IR as compared to the vector group (Fig. 5B). To further validate these results, we also detected pro-apoptosis proteins and found that eIF2αS51A or GADD34 overexpression significantly increased the protein expression of cleaved poly (ADP-ribose) polymerase 1 (PARP1), caspase-3, and caspase-9 in radioresistant cells exposed to 4 Gy IR (Fig. 5C).
Fig. 5.
Dephosphorylation of eIF2α sensitizes radioresistant TNBC cells to IR. MDA-MB-231-RR and −436-RR cells were transfected with eIF2αS51A- and GADD34-overexpression plasmid, respectively, and then treated with 0 or 4 Gy IR. (A) Cells were cultured for colony formation after IR. Colonies were stained after 9–14 days and the number was compared. (B) Cell apoptosis was detected by flow cytometry 24 h after IR. (C) The protein expression of cleaved PARP1, cleaved caspase-3, and cleaved caspase-9 was detected by WB 24 h after IR. ###P < 0.001 vs. vector + IR group (n = 3).
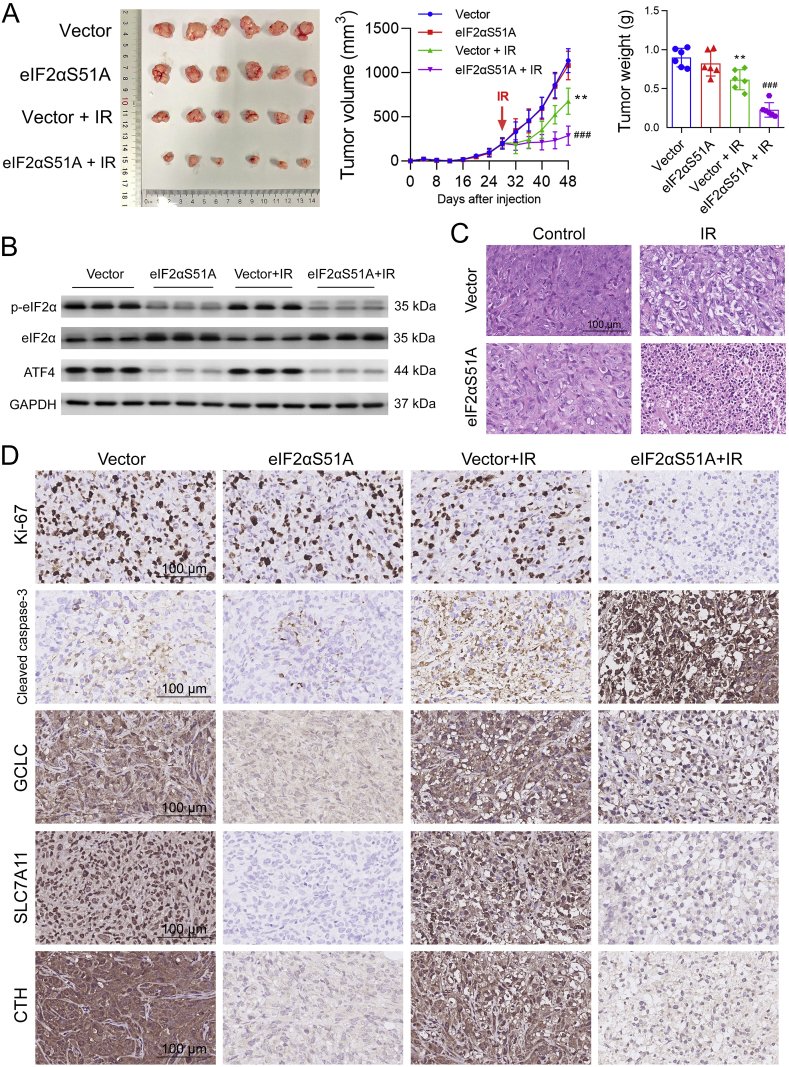
Subsequently, we further assessed the radiosensitizing effect of eIF2α dephosphorylation in mice bearing subcutaneous xenograft of MDA-MB-231-RR cells. We found that, compared with the vector group, eIF2αS51A overexpression exerted no impact on tumor growth but resulted in a greater shrinkage in tumor size and a significant decrease in tumor weight in combination with local IR (Fig. 6A). Thereafter, we confirmed that the eIF2α/ATF4 axis was inhibited in eIF2αS51A-overexpressed xenografts using WB analysis (Fig. 6B). Furthermore, we found that, compared with IR alone, combinatorial eIF2α dephosphorylation obviously increased tumor cell apoptosis, as shown by the HE staining (Fig. 6C). The eIF2α dephosphorylation also decreased intratumoral expression of Ki-67, GCLC, SLC7A11, and CTH and increased IR-induced expression of active caspase-3 (Fig. 6D). These results suggest that dephosphorylation of eIF2α can increase the radiosensitivity of TNBC in vitro and in vivo.
Fig. 6.
Dephosphorylation of eIF2α sensitizes radioresistant TNBC xenografts to IR in vivo. (A) Mice bearing tumor xenografts developed from vector- or eIF2αS51A-overexpressing MDA-MB-231-RR cells were treated with local IR (2 Gy per day for 5 days, 10 Gy in total) or sham radiation. Tumor size was monitored every 4 days, and the tumor weight was measured at the end of the experiment. (B) The protein expression of p-eIF2α, eIF2α, and ATF4 in tumor xenografts was compared by WB. (C) The histopathological examination of tumor xenograft tissues was performed using the HE stain. Representative HE staining images are shown at 200 × magnification. (D) The protein level of Ki-67, cleaved caspase-3, GCLC, SLC7A11, and CTH in tumor xenografts was detected by IHC. Representative IHC images are shown at 200 × magnification. **P < 0.05 vs. vector group; ###P < 0.001 vs. vector + local IR group (n = 6).
3.6. The ISR causes radioresistance in TNBC cells by promoting GSH biosynthesis
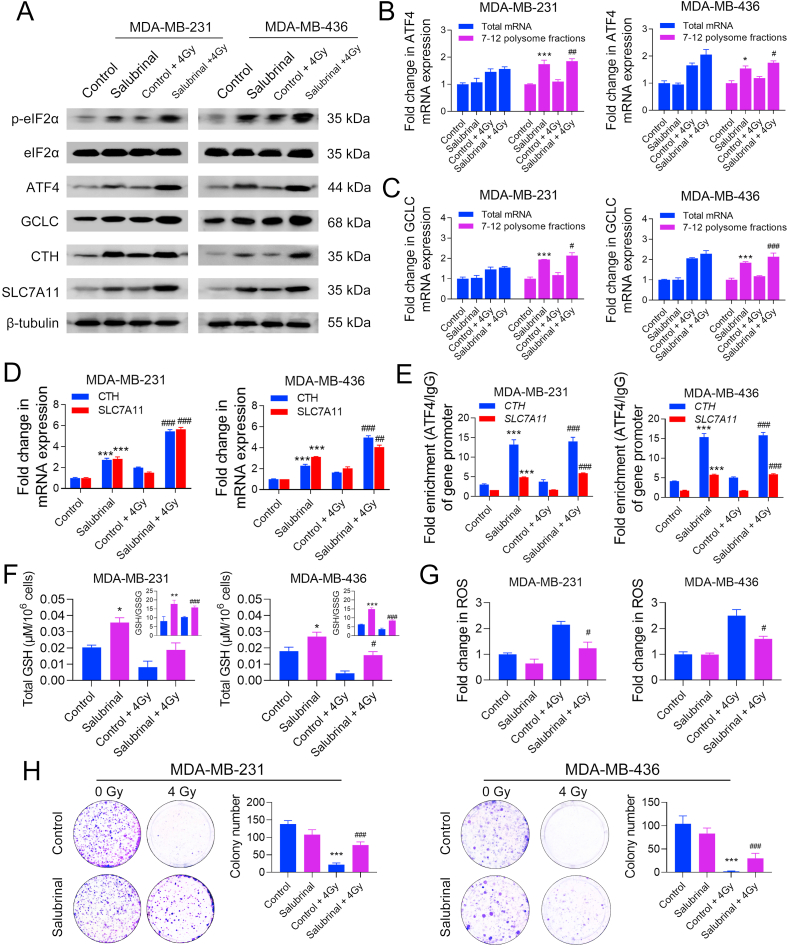
In this study, we further investigated the role of the ISR in promoting TNBC radioresistance by using salubrinal, a selective inhibitor of eIF2α phosphatase, to trigger eIF2α phosphorylation in parental MDA-MB-231 and -436 cells. As shown in Supplementary Fig. 7, treatment with 40 or 80 μM salubrinal for 24 h did not exert a cytotoxic effect on cell viability but significantly increased the eIF2α phosphorylation level in MDA-MB-231 cells; thus, a concentration of 40 μM was selected for the following study.
Through WB analysis, we found that treatment with 40 μM salubrinal significantly increased the protein expression of p-eIF2α, ATF4, GCLC, CTH, and SLC7A11 in MDA-MB-231 and -436 cells as compared to the control group (Fig. 7A). Polysome fractionation assays show that salubrinal markedly increased the translational efficiency of ATF4 and GCLC mRNAs in TNBC cells (Fig. 7B, C). Furthermore, the mRNA expression of CTH and SLC7A11 was also increased by salubrinal (Fig. 7D). This is consistent with the CHIP results showing that the binding of ATF4 to the core promoter regions of CTH and SLC7A11 was increased by salubrinal (Fig. 7E). Glutathione detection shows that treatment with 40 μM salubrinal could effectively up-regulate total GSH level and GSH/GSSG ratio and suppress IR-induced ROS accumulation in MDA-MB-231 and -436 cells (Fig. 7F, G). Compared with the control groups, treatment with salubrinal significantly increased the survival rate of MDA-MB-231 and -436 cells after IR (Fig. 7H), demonstrating that ISR can protect TNBC cells from IR-induced cell death by promoting GSH biosynthesis.
Fig. 7.
ISR causes radioresistance in TNBC cells by promoting GSH biosynthesis. MDA-MB-231 and -436 cells were treated with 0 or 40 μM salubrinal for 48 h and then treated with 0 or 4 Gy IR. (A) The protein expression of p-eIF2α, eIF2α, ATF4, CTH, and SLC7A11 was detected by WB 24 h after IR. (B–C) The relative expression of total mRNAs and polysome-associated (fraction 7–12) mRNAs of ATF4 and GCLC was detected by qRT-PCR 24 h after IR. (D) The mRNA expression of CTH and SLC7A11 was detected by qRT-PCR 24 h after IR. (E) The enrichment of the core promoter regions of CTH and SLC7A11 genes with ATF4 or IgG antibody was analyzed by ChIP-qPCR 24 h after IR. (F) Total GSH and GSH/GSSG ratio were determined 24 h after IR. (G) The intracellular level of ROS was detected 24 h after IR. (H) The radiosensitivity of TNBC cells was evaluated by colony formation assays. The colony numbers were compared and representative images are shown. *P < 0.05, **P < 0.01, and ***P < 0.001 vs. control group; #P < 0.05, ##P < 0.01, and ###P < 0.001 vs. control + IR group (n = 3).
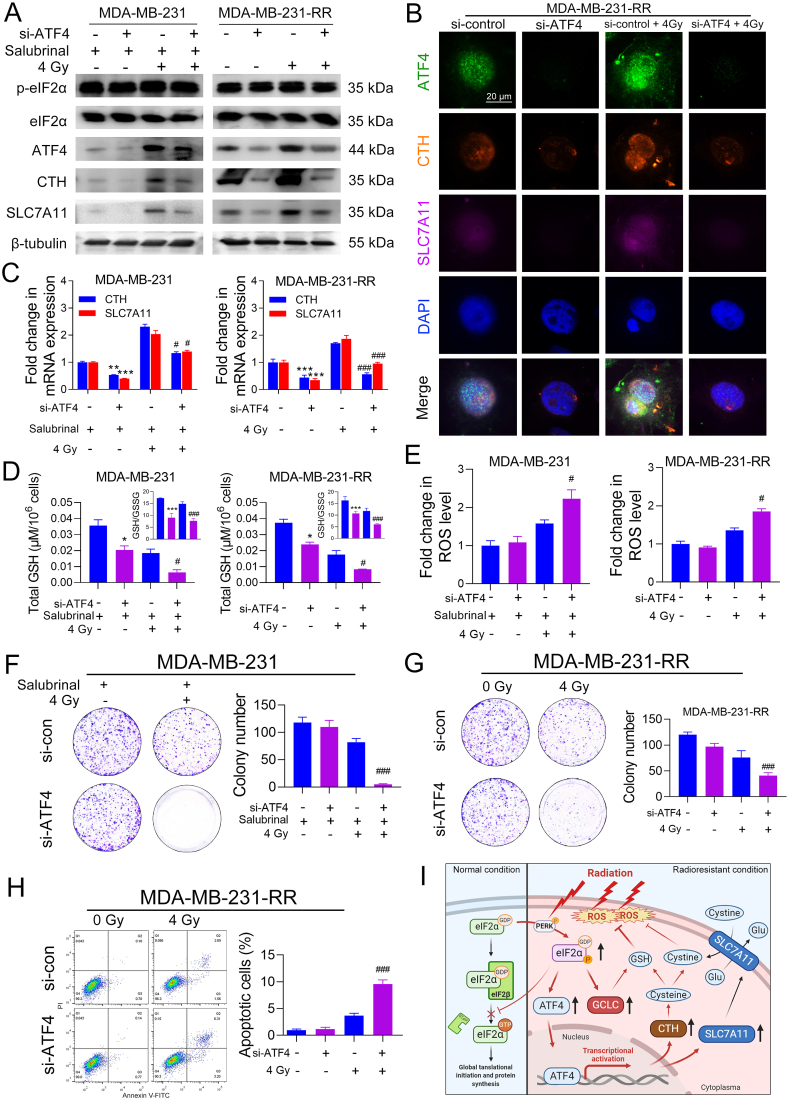
3.7. ATF4 mediates ISR-induced radioresistance in TNBC cells
To elucidate the role of ATF4 in ISR-promoted radioresistance in TNBC, we used ATF4-specific siRNAs (si-ATF4) to knock down the protein expression of ATF4 and this is performed in two ways. The one is that we knocked down ATF4 protein in MDA-MB-231 cells and then treated these cells with 40 μM salubrinal, followed by 0 or 4 Gy IR, and the other way is that we performed the knock-down assay in MDA-MB-231-RR cells and then directly treated these cells with 0 or 4 Gy IR.
As shown in Fig. 8A, si-ATF4 transfection significantly decreased the protein expression of CTH and SLC7A11 in salubrinal-treated MDA-MB-231 cells and MDA-MB-231-RR cells with or without IR. Consistent results were also observed from the IF assay in MDA-MB-231-RR cells (Fig. 8B). The qRT-PCR analysis shows that si-ATF4 markedly decreased the mRNA expression of CTH and SLC7A11 in salubrinal-treated MDA-MB-231 cells and MDA-MB-231-RR cells (Fig. 8C). Further investigation shows that ATF4 knockdown significantly reduced total GSH and GSH/GSSG ratio and increased IR-induced ROS accumulation in salubrinal-treated MDA-MB-231 cells and MDA-MB-231-RR cells (Fig. 8D, E). Furthermore, colony formation assays show that ATF4 knockdown significantly increased the sensitivity of salubrinal-treated MDA-MB-231 cells and MDA-MB-231-RR cells to IR (Fig. 8F, G). Consistently, flow cytometry analysis shows that si-ATF4 significantly increased the percentage of apoptotic cells in MDA-MB-231-RR cells after 4 Gy IR as compared to the si-control group (Fig. 8H). Besides, ATF4 knockdown also increased the sensitivity of MDA-MB-231-RR cells to cisplatin, doxorubicin, and olaparib (Supplementary Fig. 4C). These results suggest an essential role of ATF4 in mediating ISR-promoted GSH biosynthesis and radioresistance in TNBC.
Fig. 8.
ATF4 mediates ISR-induced radioresistance in TNBC cells. MDA-MB-231 cells were transfected with si-ATF4 or si-control for 72 h and then treated with 40 μM salubrinal for 48 h before 0 or 4 Gy IR. MDA-MB-231-RR cells were transfected with si-ATF4 or si-control for 72 h and then treated with 0 or 4 Gy IR. (A) The protein expression of p-eIF2α, eIF2α, ATF4, CTH, and SLC7A11 was detected by WB 24 h after IR. (B) The protein level of ATF4, CTH, and SLC7A11 in MDA-MB-231-RR cells was detected by IF 24 h after IR. The nucleus was stained with blue. Representative IF images are shown at 400 × magnification. (C) The relative mRNA expression of CTH and SLC7A11 was detected by qRT-PCR 24 h after IR. (D) Total GSH and GSH/GSSG ratio were determined 24 h after IR. (E) The intracellular level of ROS was detected 24 h after IR. (F–G) Cell radiosensitivity was evaluated using colony formation assays. The colony numbers were compared and representative images are shown. (H) Cell apoptosis was detected by flow cytometry 24 h after IR. *P < 0.05, **P < 0.01, and ***P < 0.001 vs. si-control group; #P < 0.05 and ###P < 0.001 vs. si-control + IR group (n = 3). (I) The schematic of the eIF2α/ATF4-mediated mechanism underlying TNBC radioresistance. In normal conditions, the eIF2β catalyzes the exchange of inactive eIF2α-GDP to active eIF2α-GTP, which promotes the release of eIF2β and the formation of a ternary complex. The process plays an important role in the initiation of 5′cap-dependent mRNA translation. In the radioresistant condition, constitutive phosphorylation of eIF2α at ser51 by activated PERK blocks the release of eIF2β and results in the global inhibition of 5′cap-dependent protein synthesis while selectively promotes the translation of uORF-containing mRNAs such as those of ATF4 and GCLC. This facilitates the transactivation of ATF4 and up-regulates the protein expression of GCLC, CTH, and SLC7A11, which increases GSH biosynthetic and ROS scavenging efficiency, leading to a radioresistant phenotype in TNBC cells. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Recurrent TNBC after RT leads to lethal distant metastasis and a poor response to follow-up treatment [4,5], as evidenced by the cross-resistance to conventional chemotherapy and novel targeted therapy, suggesting that radioresistance is a major challenge in TNBC treatment. Proteomics is a high-throughput screen method to investigate pathway enrichment in biological samples, which provides a direct profile of protein expression with signaling networks for disease biomarker discovery [27]. Using proteomics, we firstly demonstrated that ISR was initiated with the activation of the eIF2α/ATF4 axis and global inhibition of protein synthesis in radioresistant TNBC cells. The proposed role of the eIF2α/ATF4 axis in TNBC radioresistance is illustrated in Fig. 8I.
Cellular ISR constitutes a primary protective mechanism that allows stressed cells to survive by inhibiting the global translation and re-structuring cell transcriptome into recuperating from stress [28]. The protein kinase R-like endoplasmic reticulum kinase (PERK) is one of the most crucial sensors that trigger the ISR by phosphorylating eIF2α. Previous studies demonstrated that IR could induce the accumulation of misfolded or unfolded proteins in the endoplasmic reticulum and subsequently activate PERK [29,30]. Consistently, our results documented an up-regulation of p-PERK in radioresistant TNBC cells, suggesting that IR may induce ISR in TNBC cells through unfolded protein response. ATF4 is a member of the ATF/cyclic AMP response element-binding protein subfamily of the basic leucine zipper transcription factor superfamily [31], which regulates amino acid metabolism, resistance to oxidative stress, and cellular fate as a mediator to ISR [10]. The two uORFs in ATF4 mRNAs allow them to be translated by the 43S pre-initiation complex during ISR in an eIF2α-dependent manner [32], and consistent results were observed in our study. Further, we used salubrinal, a specific eIF2α phosphatase inhibitor that can mimic the ISR activation, to investigate the role of the ISR in TNBC radioresistance and found that salubrinal protected parental TNBC cells against IR-induced apoptosis, demonstrating that ISR is an important mechanism underlying TNBC radioresistance.
Although persistent and severe stress is likely to cause cell death, surviving cells may develop an adaptive oxidative stress resistance [33]. Activation of antioxidative signaling cascades has been proposed to explain the cancer adaption to stressful conditions, in our case, radiation, and escape from treatment-induced growth inhibition and apoptosis [34,35]. In this study, we showed evidence of an acquired enhancement in antioxidant capacity due to up-regulated GSH biosynthesis in radioresistant TNBC cells. These data are in accordance with a metabolite profiling study which revealed that GSH biosynthetic pathway might be a therapeutic target for TNBC [36]. Looking at downstream, GCLC is a rate-limiting GSH synthesis enzyme. SLC7A11 is an essential precursor for GSH synthesis, while CTH is responsible for the synthesis of cysteine which is an essential component for GSH. ATF4 can regulate the transcription of GCLC and SLC7A11 by binding to the AP-1 site [23] and amino acid response element [14], respectively, and regulate CTH by binding to a cis-regulatory intronic element [24]. Through CHIP-qRT-PCR analysis, we demonstrated that these genes were transcriptionally activated by ATF4 in TNBC radioresistance, highlighting the role of ATF4 as a mediator between ISR and enhanced GSH biosynthesis in TNBC cells.
The promoter regions of GCLC, SLC7A11, and CTH also contain an antioxidant response element (ARE) sequence, which makes them target genes of Nrf2 [22,25,26]. Similar to ATF4, Nrf2 regulates a variety of detoxification and cytoprotective enzymes by binding to the ARE region and plays a critical role in maintaining cellular redox homeostasis. The protective role of Nrf2 against cytotoxics-induced oxidative stress and apoptosis is well established [37]. Moreover, McDonald et al. [38] and Bailleul et al. [39] found that Nrf2 activation mediates radioresistance in mouse embryonic fibroblasts and human pancreatic cancer. Notably in our study, nuclear Nrf2 expression, as well as the transcriptional activity of Nrf2, was up-regulated in both radioresistant and parental TNBC cells in response to IR, implying a potential role of Nrf2 against IR-induced oxidative stress [40]. Although our evidence did not support the association of the Nrf2/ARE signaling with an acquired up-regulation of GSH biosynthesis in radioresistant TNBC cells, the role of Nrf2 in TNBC radioresistance still deserves further investigation.
Additionally, it is noteworthy to mention that there is a common molecular feature of the three TNBC cell lines used in this study that they all carry TP53 missense mutations. These mutations impair the tumor-suppressive nature of the p53 pathway due to the loss of control in cell proliferation and death and cause “gain-of-function” properties in BC, including pro-survival [41], metastasis [42], and eventually radioresistance [43], through activating various oncogenic signalings. As TP53 mutations are highly prevalent in TNBC patients (found in 80% of tumors) and may represent an important mechanism of inherent therapeutic resistance [44], further studies are warranted to elucidate the relationship between the mutant p53 and eIF2α/ATF4 axis in TNBC radioresistance.
In this study, we firstly showed a significant correlation of the eIF2α/ATF4 axis genes with a poor prognosis in TNBC patients. Further retrospective and prospective studies are thus needed to validate them as potential biomarkers for the prediction of patient outcomes. Additionally, the results from our in vivo assay highlight the therapeutic potential of eIF2α/ATF4 interference as a radiosensitizing strategy.
In conclusion, our results clearly demosntrate that the constitutive activation of ISR in TNBC cells induces an ATF4-mediated transcription of genes involved in GSH biosynthesis. This leads to an increase in intracellular GSH synthesis and ROS scavenging, protecting TNBC cells from IR-induced apoptosis. These findings provide a better understanding of the role of the eIF2α/ATF4 axis in TNBC therapeutic resistance and may enable new targeted therapeutic strategies for the treatment of recurrent TNBC.
Author contributions
Conception and design: Y.L and X.B; Acquisition of data: X.B; analysis and interpretation of data: X.B, J.N, J.B, Y.L; writing, review, and/or revision of the manuscript: X.B, J.N, J.B, Y.L, V.W; administrative, technical, or material support: P.G, Y.L, J.B, J.N, S.W, Y.Z.
Declaration of competing interest
The authors declare no competing financial interest.
Acknowledgments
The authors thank Prof. David Ron (Cambridge Institute for Medical Research, University of Cambridge) for providing the pcDNA-eIF2αS51A plasmid. We also thank Dr. Brendan Lee (Biological Resources Imaging Laboratory, UNSW Sydney) and Mr. Ken Hopper (Cancer Care Centre, St George Hospital, Sydney, Australia) for their expertise in cell irradiation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.101993.
Funding
This work was supported by the St George Cancer Care Centre Research Trust Fund, Prostate and Breast Cancer Foundation, Cancer Institute NSW Early Career Fellowship, and China Scholarship Council-University of New South Wales Ph.D. Scholarship.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Marra A., Viale G., Curigliano G. Recent advances in triple negative breast cancer: the immunotherapy era. BMC Med. 2019;17:90. doi: 10.1186/s12916-019-1326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moran M.S. Radiation therapy in the locoregional treatment of triple-negative breast cancer. Lancet Oncol. 2015;16:e113–e122. doi: 10.1016/S1470-2045(14)71104-0. [DOI] [PubMed] [Google Scholar]
- 3.Early Breast Cancer Trialists' Collaborative G, Darby S., McGale P., Correa C., Taylor C., Arriagada R. Effect of radiotherapy after breast-conserving surgery on 10-year recurrence and 15-year breast cancer death: meta-analysis of individual patient data for 10,801 women in 17 randomised trials. Lancet. 2011;378:1707–1716. doi: 10.1016/S0140-6736(11)61629-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen P.L., Taghian A.G., Katz M.S., Niemierko A., Abi Raad R.F., Boon W.L. Breast cancer subtype approximated by estrogen receptor, progesterone receptor, and HER-2 is associated with local and distant recurrence after breast-conserving therapy. J. Clin. Oncol. 2008;26:2373–2378. doi: 10.1200/JCO.2007.14.4287. [DOI] [PubMed] [Google Scholar]
- 5.Kyndi M., Sorensen F.B., Knudsen H., Overgaard M., Nielsen H.M., Overgaard J. Estrogen receptor, progesterone receptor, HER-2, and response to postmastectomy radiotherapy in high-risk breast cancer: the Danish Breast Cancer Cooperative Group. J. Clin. Oncol. 2008;26:1419–1426. doi: 10.1200/JCO.2007.14.5565. [DOI] [PubMed] [Google Scholar]
- 6.Speers C., Zhao S.G., Kothari V., Santola A., Liu M., Wilder-Romans K. Maternal embryonic leucine zipper kinase (MELK) as a novel mediator and biomarker of radioresistance in human breast cancer. Clin. Canc. Res. 2016;22:5864–5875. doi: 10.1158/1078-0432.CCR-15-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pakos-Zebrucka K., Koryga I., Mnich K., Ljujic M., Samali A., Gorman A.M. The integrated stress response. EMBO Rep. 2016;17:1374–1395. doi: 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Back S.H., Scheuner D., Han J., Song B., Ribick M., Wang J. Translation attenuation through eIF2alpha phosphorylation prevents oxidative stress and maintains the differentiated state in beta cells. Cell Metabol. 2009;10:13–26. doi: 10.1016/j.cmet.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu P.D., Jousse C., Marciniak S.J., Zhang Y., Novoa I., Scheuner D. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 2004;23:169–179. doi: 10.1038/sj.emboj.7600030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harding H.P., Zhang Y., Zeng H., Novoa I., Lu P.D., Calfon M. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 11.Chen L., He J., Zhou J., Xiao Z., Ding N., Duan Y. EIF2A promotes cell survival during paclitaxel treatment in vitro and in vivo. J. Cell Mol. Med. 2019;23:6060–6071. doi: 10.1111/jcmm.14469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palam L.R., Gore J., Craven K.E., Wilson J.L., Korc M. Integrated stress response is critical for gemcitabine resistance in pancreatic ductal adenocarcinoma. Cell Death Dis. 2015;6 doi: 10.1038/cddis.2015.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan S., Somia N., Maher P., Schubert D. Regulation of antioxidant metabolism by translation initiation factor 2alpha. J. Cell Biol. 2001;152:997–1006. doi: 10.1083/jcb.152.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewerenz J., Maher P. Basal levels of eIF2alpha phosphorylation determine cellular antioxidant status by regulating ATF4 and xCT expression. J. Biol. Chem. 2009;284:1106–1115. doi: 10.1074/jbc.M807325200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wasinger V.C., Yau Y., Duo X., Zeng M., Campbell B., Shin S. Low mass blood peptides discriminative of inflammatory bowel disease (ibd) severity: a quantitative proteomic perspective. Mol. Cell. Proteomics. 2016;15:256–265. doi: 10.1074/mcp.M115.055095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ni J., Cozzi P.J., Hao J.L., Beretov J., Chang L., Duan W. CD44 variant 6 is associated with prostate cancer metastasis and chemo-/radioresistance. Prostate. 2014;74:602–617. doi: 10.1002/pros.22775. [DOI] [PubMed] [Google Scholar]
- 17.Celeghini E.C.C., Alves M.B.R., de Arruda R.P., de Rezende G.M., Florez-Rodriguez S.A., de Sa Filho M.F. Efficiency of CellROX deep red((R)) and CellROX orange((R)) fluorescent probes in identifying reactive oxygen species in sperm samples from high and low fertility bulls. Anim. Biotechnol. 2021;32:77–83. doi: 10.1080/10495398.2019.1654485. [DOI] [PubMed] [Google Scholar]
- 18.Camp R.L., Dolled-Filhart M., Rimm D.L. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin. Canc. Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 19.Srinivas U.S., Tan B.W.Q., Vellayappan B.A., Jeyasekharan A.D. ROS and the DNA damage response in cancer. Redox Biol. 2019;25:101084. doi: 10.1016/j.redox.2018.101084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miura M., Sasaki T. Role of glutathione in the intrinsic radioresistance of cell lines from a mouse squamous cell carcinoma. Radiat. Res. 1991;126:229–236. [PubMed] [Google Scholar]
- 21.Bansal A., Simon M.C. Glutathione metabolism in cancer progression and treatment resistance. J. Cell Biol. 2018;217:2291–2298. doi: 10.1083/jcb.201804161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moinova H.R., Mulcahy R.T. An electrophile responsive element (EpRE) regulates beta-naphthoflavone induction of the human gamma-glutamylcysteine synthetase regulatory subunit gene. Constitutive expression is mediated by an adjacent AP-1 site. J. Biol. Chem. 1998;273:14683–14689. doi: 10.1074/jbc.273.24.14683. [DOI] [PubMed] [Google Scholar]
- 23.Liu C.W., Hua K.T., Li K.C., Kao H.F., Hong R.L., Ko J.Y. Histone methyltransferase G9a drives chemotherapy resistance by regulating the glutamate-cysteine ligase catalytic subunit in head and neck squamous cell carcinoma. Mol. Canc. Therapeut. 2017;16:1421–1434. doi: 10.1158/1535-7163.MCT-16-0567-T. [DOI] [PubMed] [Google Scholar]
- 24.Mistry R.K., Murray T.V., Prysyazhna O., Martin D., Burgoyne J.R., Santos C. Transcriptional regulation of cystathionine-gamma-lyase in endothelial cells by NADPH oxidase 4-dependent signaling. J. Biol. Chem. 2016;291:1774–1788. doi: 10.1074/jbc.M115.685578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lambertini E., Penolazzi L., Angelozzi M., Grassi F., Gambari L., Lisignoli G. The expression of cystathionine gamma-lyase is regulated by estrogen receptor alpha in human osteoblasts. Oncotarget. 2017;8:101686–101696. doi: 10.18632/oncotarget.21514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye P., Mimura J., Okada T., Sato H., Liu T., Maruyama A. Nrf2- and ATF4-dependent upregulation of xCT modulates the sensitivity of T24 bladder carcinoma cells to proteasome inhibition. Mol. Cell Biol. 2014;34:3421–3434. doi: 10.1128/MCB.00221-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang L., Graham P., Hao J., Bucci J., Malouf D., Gillatt D. Proteomics discovery of radioresistant cancer biomarkers for radiotherapy. Canc. Lett. 2015;369:289–297. doi: 10.1016/j.canlet.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 28.Adomavicius T., Guaita M., Zhou Y., Jennings M.D., Latif Z., Roseman A.M. The structural basis of translational control by eIF2 phosphorylation. Nat. Commun. 2019;10:2136. doi: 10.1038/s41467-019-10167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z., Lv Y., Zhao N., Guan G., Wang J. Protein kinase R-like ER kinase and its role in endoplasmic reticulum stress-decided cell fate. Cell Death Dis. 2015;6 doi: 10.1038/cddis.2015.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harding H.P., Zhang Y., Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 31.Ameri K., Harris A.L. Activating transcription factor 4. Int. J. Biochem. Cell Biol. 2008;40:14–21. doi: 10.1016/j.biocel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 32.Harding H.P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 33.Landriscina M., Maddalena F., Laudiero G., Esposito F. Adaptation to oxidative stress, chemoresistance, and cell survival. Antioxidants Redox Signal. 2009;11:2701–2716. doi: 10.1089/ars.2009.2692. [DOI] [PubMed] [Google Scholar]
- 34.Bump E.A., Brown J.M. Role of glutathione in the radiation response of mammalian cells in vitro and in vivo. Pharmacol. Ther. 1990;47:117–136. doi: 10.1016/0163-7258(90)90048-7. [DOI] [PubMed] [Google Scholar]
- 35.Snezhkina A.V., Kudryavtseva A.V., Kardymon O.L., Savvateeva M.V., Melnikova N.V., Krasnov G.S. ROS generation and antioxidant defense systems in normal and malignant cells. Oxid Med Cell Longev. 2019;2019:6175804. doi: 10.1155/2019/6175804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beatty A., Fink L.S., Singh T., Strigun A., Peter E., Ferrer C.M. Metabolite profiling reveals the glutathione biosynthetic pathway as a therapeutic target in triple-negative breast cancer. Mol. Canc. Therapeut. 2018;17:264. doi: 10.1158/1535-7163.MCT-17-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bai X., Chen Y., Hou X., Huang M., Jin J. Emerging role of NRF2 in chemoresistance by regulating drug-metabolizing enzymes and efflux transporters. Drug Metab. Rev. 2016;48:541–567. doi: 10.1080/03602532.2016.1197239. [DOI] [PubMed] [Google Scholar]
- 38.McDonald J.T., Kim K., Norris A.J., Vlashi E., Phillips T.M., Lagadec C. Ionizing radiation activates the Nrf2 antioxidant response. Canc. Res. 2010;70:8886–8895. doi: 10.1158/0008-5472.CAN-10-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailleul J., Yazal T., Sung D., Palomera D., Sehgal A., Dao A. Abstract C59: NRF2 drives metabolic reprogramming in irradiated pancreatic cancer cells and promotes radioresistance. Canc. Res. 2019;79:C59–C. [Google Scholar]
- 40.Yang Y., Yee D. Abstract 1207: Nrf2 suppression results in growth inhibition and enhances response to chemotherapy and radiation therapy in triple-negative breast cancer cells. Canc. Res. 2015;75:1207. [Google Scholar]
- 41.Lim L.Y., Vidnovic N., Ellisen L.W., Leong C.O. Mutant p53 mediates survival of breast cancer cells. Br. J. Canc. 2009;101:1606–1612. doi: 10.1038/sj.bjc.6605335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wellenstein M.D., Coffelt S.B., Duits D.E.M., van Miltenburg M.H., Slagter M., de Rink I. Loss of p53 triggers WNT-dependent systemic inflammation to drive breast cancer metastasis. Nature. 2019;572:538–542. doi: 10.1038/s41586-019-1450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jameel J.K., Rao V.S., Cawkwell L., Drew P.J. Radioresistance in carcinoma of the breast. Breast. 2004;13:452–460. doi: 10.1016/j.breast.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 44.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.