Abstract
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is the third highly pathogenic coronavirus to emerge in the human population in last two decades. SARS-CoV-2 spread from Wuhan, China, across the globe, causing an unprecedented public healthcare crisis. The virus showed remarkable age dependent pathology, with symptoms resembling common cold in most adults and children while causing more severe respiratory distress and significant mortality in older and frail humans. Even before the SARS-CoV-2 outbreak infectious diseases represented one of the major causes of death of older adults. Loss of immune function and reduced protection from infectious agents with age – immunosenescence - is a result of complex mechanisms affecting production and maintenance of immune cells as well as the initiation, maintenance and termination of properly directed immune responses. Here we briefly discuss the current knowledge on how this process affects age-dependent outcomes of SARS-CoV-2 infection.
KEYWORDS: Sars-CoV-2, COVID-19, aging, immunity, severity
SARS-CoV-2 virus and COVID-19 infection
In December 2019, a novel coronavirus was identified as cause of outbreak of severe respiratory illnesses in the city of Wuhan, China.1 The virus shared 79.6% sequence identity to SARS-CoV, which caused a small global outbreak in 2002, and was thus named SARS-CoV-2. Clinical disease caused by the virus was termed Coronavirus disease-19 (COVID-19). The virus spread globally and in March 2020 World Health Organization declared the outbreak a global pandemic.2 As of early April, 2021 the virus has infected more than 130 million people globally and caused >3 million deaths.
Of the four families of Coronaviridae (alpha, beta, gamma, and delta), all human corona viruses (CoV) belong to either alpha (229E, NL63) or beta (OC43, HKU1, SARS-CoV,MERS-CoV, and SARS-CoV-2) family,3 with the latter containing all three CoV highly pathogenic to humans. Four seasonal cold human CoVs (229E, NL63, OC43, and HKU1) account for 10% to 30% of upper respiratory tract infections4 manifested as common cold, although even these viruses can cause more severe symptoms in frail older subjects.5
CoV are single-strand RNA viruses with four structural proteins - S (spike), E (envelope), M (membrane), and N (nucleoprotein) - and multiple ORFs encoding non-structural and accessory proteins.6 Both SARS-CoV-1 and SARS-CoV-2 enter cells via the interaction between the viral spike protein and the host cell surface enzyme angiotensin-converting enzyme 2 (ACE2),1 , 7 although there is evidence for other cell surface molecules such as CD147 (Basignin)8 and the serine protease TMPRSS29 as coreceptors and/or entry co-factors. SARS-CoV-2 binds to ACE2 with 10-20 higher affinity than SARS-CoV-1, partially explaining the difference in infectivity.10 ACE2 catalyzes the hydrolysis of angiotensin II and is a critical regulator of renin-angiotensin system and downregulation of ACE2 via virus binding results in disruption of renin–angiotensin system.11 ACE2 has a protective role in lung,12 kidney and heart injury.13 Therefore at least a part of pathology could be a direct consequence of virus binding to the host ACE2 receptor. This is consistent with findings that injection of SARS-CoV-1 spike protein into mice worsens acute lung failure in vivo.7 ACE2 is expressed in multiple tissues and various epithelial cell types in the respiratory airway, with highest expression in nasal epithelial cells,14 pointing to these cells as possible loci of original infection. SARS-CoV-2 has a direct cytopathic effect on human airway epithelial cells.15 Viral shedding primarily occurs from upper respiratory tract but fecal shedding also occurs16 and is often used for early epidemiological detection.17
Person to person transmission is thought to occur mostly by droplets, although evidence supports the possibility of airborne transmission to a lesser extent.18 Fomite transmission, although possible, presents low risk in real life situations.19 The possibility of fecal-oral transmission also cannot be excluded.20 , 21 Pharyngeal virus shedding is highest before or early after onset of symptoms16 , 22 and the majority of transmission is estimated to occur from presymptomatic or asymptomatic subjects,23 explaining why traditional epidemiological measures were unsuccessful in stopping SARS-CoV-2 spread.
Incubation period is 5.7 days on average24 but SARS-CoV-2 infection remains completely or largely asymptomatic in 20-40% people.25 , 26 In symptomatic humans, disease severity ranges from mild flu-like disease to severe respiratory syndrome and death. Clinical picture is nonhomogeneous with most common symptoms being fever and cough.27 Disease follows a severe course in up to 20% of subjects with acute respiratory distress syndrome (ARDS) as the most frequent complication.28 Other complications include myocarditis29 and kidney injury.30 The vast majority of severe cases, hospitalizations and deaths (8 out of 10) occur in people above 65 years of age,31 and even people 50 and older exhibit sharp increases in hospitalization (4x) and mortality (10x) relative to those 18-29 years of age. The numbers for those >85 years of age are staggering – 13-fold more hospitalizations, and 630-fold higher likelihood of death than those 18-29 years old (cdc.gov/coronavirus). The elderly often present atypically31 with lower incidence of fever.32 COVID-19 shows remarkable age-specific outcomes with log-linear increase in infection fatality rate by age among individuals older than 30 years.33 , 34 Other than age, risk factors for severe disease include hypertension, obesity, smoking, type 2 diabetes and male sex.35 Of interest, age-related frailty assessed by a clinical scale was found to be associated with COVID-19 severity in a prospective cohort study of humans >60 years.36 Another study showed, disease outcomes were better predicted by frailty than either age or comorbidity.37 Frailty is a geriatric syndrome characterized by reduced energy levels, muscle loss and increased vulnerability associated with a hyperinflammable state and elevated levels of proinflammatory cytokines, particularly IL-6.38 , 39 This preexisting hyperinflammable state could be contributing to COVID-19 severity which is associated with overactivation of the innate immune system.40 Prevalence of frailty syndrome is markedly increased in persons above 80 years41 but physiological and functional changes are distinct from the usual age-related changes.42
To effectively protect older adults against SARS-CoV-2, one must dissect potential contributors to the above age-related susceptibility to COVID-19. Age related decreases in respiratory function have the potential to account for high incidence of respiratory symptoms among the elderly.43 However, chest computer tomography did not show increased lung damage in elderly despite increased disease severity,44 suggesting that lung aging by itself may not be the determining factor of severe COVID-19. Consistent with that, severity was associated with damage to other organs, mainly heart, liver and kidneys.35 On the other hand, hypercoagulopathy likely plays a role in organ damage and anticoagulant therapy has been both shown to reduce mortality45 and is widely used in suspected severe COVID-19 cases. SARS-CoV-2 is able to infect vascular endothelial cells which express high levels of ACE2.46 , 47 Resulting endothelial injury and inflammation induces a hypercoagulative state and increased thrombotic events.48 In addition to direct cytopathic effect of the virus, the downregulation of ACE2 and endothelial damage, there is abundant evidence that immune system dysregulation plays a major role in COVID-19 tissue injury.49, 50, 51, 52, 53 There is evidence that human leukocyte antigen (HLA) class I molecules play a role, as HLA-A*01:01 allele was associated with higher risk of severe COVID-19.54 A genome-wide association study performed on >2000 intensive care patients found 9 loci associated with severe COVID-19, out of which 5 were genes linked to the immune system, most notably low expression of interferon receptor gene IFNAR2 and high expression of chemotactic receptor CCR2.55 Immune correlates of disease severity or protection are studied intensively. However, even a year into the pandemic, we do not understand the precise role and importance of the immune aging in the pathogenesis and severity of COVID-19. At least one major obstacle to understanding how aged immune system alterations translate into higher risk of COVID-19 in older adults is lack of adequate (aged) animal models.56 SARS-Cov-2 virus causes upper respiratory tract infection in Syrian hamsters and ferrets, with mild clinical symptoms and transmission to cage mates.56 , 57 However, in both cases there is no resource for aged animals. Only mild clinical disease has been reported in non-human primates58 but more severe pneumonia and increased viral replication was observed in aged rhesus macaques59 highlighting the need for aged animals. Viral spike protein of both SARS-CoV-1 and 2 does not bind to mouse ACE260 so several transgenic mice expressing human ACE receptor have been developed61, 62, 63 but availability of aged animals is scarce. A different approach is to use mutagenesis to develop mouse adapted viral strains. A recombinant SARS-Cov-2 virus which can infect BALB/c mice was developed and showed age related pathogenesis.64 While the immune system is necessary for viral clearance and while severe cases show delayed viral clearance,65 immune hyperactivation is associated with pathogenesis. Innate immune responses are initiated after viral components, mostly ss and dsRNA, are recognized by pattern recognition receptors (PRR). Their activation induces type I interferon (IFN-I) responses that engender inflammatory cytokine cascades important for limiting viral spread. Initiation of these inflammatory signals leads to recruitment of immune cells to sites of infection starting with neutrophils. Antigen presenting cells, most notably dendritic cells, present viral peptides on MHC molecules to initiate T cell responses, whereas parallel activation of B cells by soluble virus epitopes initiate humoral (antibody) responses. Here, we will briefly outline age-related changes to these processes and how they might directly contribute to poor COVID-19 prognosis.
Age related changes of the immune system
Aging results in multiple measurable alterations in the innate and adaptive arms of immunity. Termed immunoscenescence, this process leads to a variable but often marked reduction of immune protection against infections that is deleterious to the health and wellbeing of a substantial fraction of older adults.66
Age-related defects in innate immunity can broadly be attributed to decreased phagocytic capacity and impaired/delayed migration, differentiation, and cytokine production by innate immune cells. Neutrophils display reduced cytokine signaling and effector molecule production in older adults. Defects in specific pattern recognition receptor (PRR) expression and signaling have been shown to partially account for the hampered responsiveness of old neutrophils to pathogens. These changes have been correlated with poor prognosis in bacterial infections including sepsis.67, 68, 69 Old macrophages also exhibit reduced migration and phagocytosis, that interestingly leads to reduced removal of dying inflammatory neutrophils in the lung of old mice during influenza infection, suggesting that similar mechanisms could feed into severe COVID-19 pathology.70 , 71 Old NK cells exhibit a more mature phenotype and have depressed cytokine secretion and cytotoxic potential. This could be due, in part, to alterations in the expression of activating and inhibitory receptors. In the ectromelia (mouse pox) model, these defects have been shown to explain increased viral susceptibility in old mice.72 , 73 Finally, old dendritic cells are less efficient at capturing and processing antigen, which leads to their reduced activation and consequent suboptimal activation of naïve T cells.74 , 75 All innate cells subsets exhibit more or less pronounced defects in migration, although it remains to be shown whether these defects occur due to underproduction or dysregulated production of chemokines directing their migration, or to the inability of cells themselves to appropriately respond to chemokine cues.76 Another possibility is increased production of negative regulators of chemotaxis such as prostaglandin D2.77
There are both quantitative and qualitative changes in B cell function with age. The absolute quantity of both bone-marrow resident B cell progenitors and their naïve daughter cells is reduced with age, leading to underproduction of new naïve B cells. Functionally, the formation and output of germinal center (GC) reactions in primary and secondary responses are both impaired in old age.78 Defects in the GC reaction is the result of age-related decline in function of both follicular dendritic cells (FDCs) and T cells, along with intrinsic defects within B cells themselves. Decreased FDC functionality is in part due to lower expression of Fc receptors, leading to impaired antigen capture and presentation, while defects in CD4 help may stem from decreased expression of CD40L, an important costimulatory molecule in GC reactions.79 , 80
Antibodies produced in old mice following B cell activation are of lower quality compared to those produced in adult mice. One apparent cause of this defect is the impaired production of the E2A gene-encoded E47 transcription factor.81 With age, dysregulation of the expression of the mRNA-degradation promoting protein ZFP36 increases with age leads to a higher turnover rate of E47 mRNA in older animals. This instability leads to under-induction of activation-induced cytidine deaminase (AID). Because AID plays a crucial role in both class-switch recombination and somatic hypermutation in activated B-cells, the culmination of these defects is the production of antibodies of inferior avidity and function in old mice.81 , 82
Multiple age-associated defects appear in the T cell compartment in advanced age. The earliest hallmark of T cell aging is thymic involution, marked by degeneration and atrophy of thymic stroma and a concordant and progressive reduction in naïve T cell output.83 While the homeostatic maintenance of naïve T cells in peripheral lymphoid organs becomes the dominant means of retaining the naïve T cell pool, this process also gradually weakens with aging.84 Eventually, in the last third of life this leads to reduced diversity of the T cell receptor (TCR) repertoire, and a relative (and in the presence of cytomegalovirus, absolute) accumulation of memory T cells, potentially producing holes in the T cell repertoire mobilized against a given epitope or pathogen.85 , 86
This general collapse of naïve T cell homeostasis is accompanied by decreased primary (new) T cell responses in magnitude and differentiation. This is likely a combination of reduced naïve T cell numbers (and perhaps diversity) and of cell-extrinsic defects in peripheral lymphoid organ structure,87 , 88 that fail to orchestrate coordinated and efficient movements and cell-cell communication in the course of primary responses.89 Studies of human blood have shown that CD8 T cell responses may be either more or differently impacted by aging relative to CD4 T cells.90 , 91 However, in both aged mice and humans, it has been demonstrated that naïve T cells are less functional following priming as measured by cytotoxic function, cytokine production, and proliferative capacity.92 , 93
Immunological features of COVID-19 in aged subjects
SARS-CoV-2 virus control seems to be directly related to COVID-19 severity, as virus load in severe cases was found to be higher and clearance delayed compared to mild cases.65 Even after adjusting for age and comorbidities higher viral load was predictive of mortality.94 Peak viral load was increased in aged humans suggesting that immune system in elderly is less able to counteract viral replication and spread.95 The initial step in counteracting viral spread is induction of anti-viral defenses in different cells by type I interferons. SARS-Cov-2 evades IFN-I response more efficiently than MERS and SARS-CoV-1 via its nsp1 and nsp6 proteins which suppress IFN-I signaling.96 Impaired IFN-I responses in white blood cells of severe COVID-19 patients were found to be associated with a persistent viremia and exacerbated inflammatory response.97 In contrast, bronchoalveolar lavage fluid of severe COVID-19 patients showed increased expression of IFN stimulated genes.98 Contradictory findings regarding IFN-I responses and COVID-19 severity might be explained by differences in sampling time and tissue sampled, and resolving them would be highly significant as recombinant IFN-I is currently investigated as potential COVID-19 treatment.99 Progression to severe clinical picture is associated with hyperactivation of the immune system manifested by increased levels of inflammatory cytokines in circulation, also called “cytokine storm”.40 Levels of C-reactive protein (CRP) and interleukin-6 (IL-6) were particularly strongly associated with increased mortality.100 Other cytokines elevated in severe cases included IL2, IL7, IL10, GSCF, IP10, MCP1, MIP1A, and TNFα.101 Basal IL-6 and TNF-α levels in circulation increase with age, a condition sometimes termed inflammaging,102 and increased IL-6 is in particular associated with age-related frailty and all cause mortality.38 , 103 Given that IL-6 levels predicted COVID-19 mortality, blocking IL-6 signaling was considered a promising therapeutic target; however, clinical trials of tocilizumab and sarilumab, monoclonal antibody directed against IL-6 receptor, have yielded mixed results on survival in hospitalized COVID-19 patients104 , 105 for reasons not understood at this time.
In addition to increased cytokine levels, multiple cellular immunity defects are associated with COVID-19 severity. Severe cases show decreased lymphocyte blood counts and increased neutrophil counts, such that the neutrophil to lymphocyte ratio emerged as an independent predictor of mortality.106 Autopsy samples from the lungs displayed neutrophil infiltration in pulmonary capillaries,107 raising the possibility that reduced removal of neutrophils by alveolar macrophages with aging, which contributes to severity of influenza in old mice, could be at play here too. Microvascular thrombi, containing neutrophil extracellular traps associated with platelets and fibrin were found in the lung, kidney, and heart post mortem.108 Activated neutrophils are known to contribute to large-vessel thrombosis109 and excessive reactive oxygen species production is also suspected to contribute to tissue damage in severe COVID-19.50 Previous research in rodent viral71 and bacterial110 models showed that aged animals displayed increased infiltration of neutrophils in lungs which contributed to pneumonia severity. This excessive neutrophil infiltration was associated with increased chemokine production by senescent epithelial cells71 and impaired toll like receptor activation.110 Lymphopenia in severe cases affected primarily T lymphocytes, particularly CD4+ and CD8+ T cells.111 Multiple reports showed a decrease in naïve CD4+ T cells in bloodstream of severe cases and increased expression of activation markers such as CD38 and HLA-DR.111, 112, 113 Although decreased naïve T cells in the blood were associated with severity most of these studies lacked older participants with moderate disease so some of the observed phenotypes might be features of aging by itself. Moderate cases were characterized by the presence of highly clonally expanded CD8+ in bronchoalveolar lavage fluid suggesting a strong T cell response is protective.114 Antigen specific CD4+ T cell responses correlated with SARS-CoV-2 specific IgG and IgA antibody titers.115 However, SARS-Cov-2 specific T cells responses have been detected in up to 40% healthy controls116, 117, 118 leading to hypotheses that cross reactive memory T cells from previous common cold CoV infection might be protective.119 Ex vivo peptide stimulation revealed a range of preexisting memory T cells that are cross-reactive between SARS-CoV-2 and the common cold coronaviruses. Cross-reactivity was associated with epitopes derived from SARS-CoV-2 spike, N, nsp8, nsp12, and nsp13 proteins.120 At the moment, it is unclear whether and how the presence and the exact specificity of these cross-reactive T cells affects disease severity.
While lymphopenia affected B cells to a lesser extent,121 there were pronounced oligoclonal expansions of plasmablasts in severe cases.122 Overall antibody titers were increased in severe cases and were not affected by age.123 , 124 However, in elderly subjects neutralizing antibody titers were less correlated with antigen specific CD4+ and CD8+ T cell responses, suggesting that potential lack of coordination in adaptive immune responses may contributes to disease severity.125
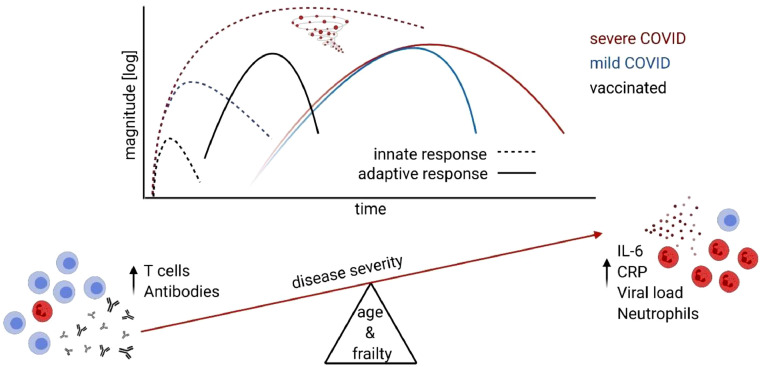
The protective ability of early adaptive immune responses is highlighted by development of multiple successful SARS-CoV-2 vaccines (Figure 1 ). While several vaccine candidates are still in clinical trials, two mRNA vaccines have been approved and are in mass use in the US as of December 2020.126 Both of these vaccines were shown to induce neutralizing antibodies and Th1 cell response127 , 128 and reduce the incidence of symptomatic and severe COVID-19 with high efficacy even in participants 65 years of age or older.129 It remains to be seen how broadly protective and durable these responses will be in older adults.
Figure 1.
Severe cases of COVID-19 are characterized by prolonged hyperactivation of innate immunity manifested by increased levels of inflammatory cytokines in circulation, also called “cytokine storm” as well as increased neutrophil count. This primarily occurs in aged and frail subjects. Development of successful vaccine shows that early adaptive immune response from T cells and neutralizing antibodies is protective and prevents the severe course of COVID-19.
In lieu of a conclusion…
We have learned an extraordinary amount of information about the virology, pathogenesis and immunology of SARS-COV-2 over the past year, with >100,000 papers containing COVID-19 and/or SARS-CoV-2 listed in PubMed and preprint servers in 2020 alone. However, we still very much lack a comprehensive picture of the disease and of virus pathogenesis, as well as of the interactions of SARS-CoV-2 with its (human) host. It is evident that COVID-19 severity is associated with delayed viral clearance, hyperactivtion of the innate immune system, increased antibody titers and T cell lymphopenia (Figure 1). At the moment, it is unclear how frailty and aging predispose for these phenotypes and increased severity. Below, we outline some of the most burning immunological questions, and hope that these and related questions will be answered with utmost urgency:
-
■
Are innate sensors specifically disabled in older adults to make them more vulnerable to COVID-19?
-
■
Are there aging-related cytokine dysfunctions similar to recently discovered type I IFN genetic defects that underlie severe COVID-19 in older adults? Do they kick in only when the older adaptive immune system cannot terminate infection on time?
-
■
Do low numbers of naïve T and B cells with aging predispose towards poor immunity and poor outcomes?
-
■
Is the reduced diversity of the T and B cell response with age linked to impaired immune responses?
-
■
Can the older immune system target all the key antigens of the virus, or is the virus more likely to slip by it?
-
■
Do the remaining CD4 and CD8 cells respond with correct and strong effector function?
-
■
Is the sum of immune defects sufficient to permit variant selection in in older adults?
-
■
Do prior coronavirus infections differentially shape the ability of the older immune system to respond to SARS-CoV-2?
-
■
Do older adults generate long-lived and protective memory responses?
-
■
Do T memory responses in older population target the same array of virus epitopes as in adults?
-
■
Are virus escape variants more likely to slip by older T memory (Tm) responses?
-
■
How well will older T cells respond to SARS-CoV-2 vaccination?
Footnotes
Supported in part by grants from the UArizona BIO5 Institute, the Elizabeth Bowman Endowed Professorship, the USPHS awards AG-020719, AG-057701 and AG-052359 and the contract CTR050053 from the Arizona Department of Health Service (Prime: US Department of Treasury) to J.N-Z., as well as by the CDC contract 75D30120C08379 (PI J. Burgess; sub-PI Nikolich).
References
- 1.Zhou P, Yang X-L, Wang X-G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qamar MA. COVID-19: a look into the modern age pandemic. J. Public Heal. 2020 doi: 10.1007/s10389-020-01294-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perlman S, Netland J. Coronaviruses post-SARS: Update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paules CI, Marston HD, Fauci AS. Coronavirus infections-more than just the common cold. JAMA - J Am Med Assoc. 2020;323:707–708. doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- 5.Greenberg SB. Update on human rhinovirus and coronavirus infections. Semin Respir Crit Care Med. 2016;37:555–571. doi: 10.1055/s-0036-1584797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luk HKH, Li X, Fung J, Lau SKP, Woo PC Y. Molecular epidemiology, evolution and phylogeny of SARS coronavirus. Infect Genet Evol. 2019;71:21–30. doi: 10.1016/j.meegid.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuba K., Imai Y, Rao S, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus – induced lung injury. Nat Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang K., et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct Target Ther. 2020;5:1–10. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inhibitor P., et al. 2020. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and is Blocked by a Clinically Proven Article SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and is Blocked by a Clinically Proven Protease Inhibitor; pp. 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wrapp D, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. bioRxiv. 2020;1263:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zoufaly A., et al. Human recombinant soluble ACE2 in severe COVID-19. Lancet Respir. Med. 2020;8:1154–1158. doi: 10.1016/S2213-2600(20)30418-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imai Y., et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crackower MA, Sarao R, Oliveira-dos-Santos AJ, Da Costa J, Zhang L. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417:822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 14.Sungnak W., et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu N., et al. Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells. Nat Commun. 2020;11:1–8. doi: 10.1038/s41467-020-17796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wölfel R., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 17.Gundy PM, Gerba CP, Pepper IL. Survival of coronaviruses in water and wastewater. Food Environ Virol. 2009;1:10–14. [Google Scholar]
- 18.Ng K., et al. COVID-19 and the risk to health care workers: a case report. Ann. Intern. Med. 2020;172:766–767. doi: 10.7326/L20-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mondelli MU, Colaneri M, Seminari EM, Baldanti F, Bruno R. Low risk of SARS-CoV-2 transmission by fomites in real-life conditions. Lancet Infect. Dis. 2020;3099:30678. doi: 10.1016/S1473-3099(20)30678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Widders A, Broom A, Broom J. SARS-CoV-2: The viral shedding vs infectivity dilemma. Infect Dis Heal. 2020;25:210–215. doi: 10.1016/j.idh.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Y, et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He X., et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 23.Johansson M.A., et al. SARS-CoV-2 transmission from people without COVID-19 symptoms key points + supplemental content. JAMA Netw Open. 2021;4 doi: 10.1001/jamanetworkopen.2020.35057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wassie GT, Azene AG, Bantie GM, Dessie G, Aragaw AM. Incubation period of severe acute respiratory syndrome novel Coronavirus 2 that causes Coronavirus disease 2019: a systematic review and meta-analysis. Curr Ther Res - Clin Exp. 2020;93 doi: 10.1016/j.curtheres.2020.100607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buitrago-Garcia D, et al. Occurrence and transmission potential of asymptomatic and presymptomatic SARSCoV-2 infections: A living systematic review and meta-analysis. PLoS Med. 2020;17:1–25. doi: 10.1371/journal.pmed.1003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oran D.P., Topol E.J. Prevalence of asymptomatic SARS-CoV-2 infection : a narrative review. Ann Intern Med. 2020;173:362–367. doi: 10.7326/M20-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guan W., et al. Clinical characteristics of Coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tzotzos SJ, Fischer B, Fischer H, Zeitlinger M. Incidence of ARDS and outcomes in hospitalized patients with COVID-19: a global literature survey. Crit Care. 2020;24:1–4. doi: 10.1186/s13054-020-03240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siripanthong B, et al. Recognizing COVID-19–related myocarditis: The possible pathophysiology and proposed guideline for diagnosis and management. Hear Rhythm. 2020;17:1463–1471. doi: 10.1016/j.hrthm.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nadim MK, et al. COVID-19-associated acute kidney injury: consensus report of the 25th Acute Disease Quality Initiative (ADQI) Workgroup. Nat Rev Nephrol. 2020;16:747–764. doi: 10.1038/s41581-020-00356-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lithander FE, et al. COVID-19 in older people: a rapid clinical review. Age Ageing. 2020;49:501–515. doi: 10.1093/ageing/afaa093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo T, et al. Clinical characteristics of elderly patients with COVID-19 in Hunan Province, China: a multicenter, retrospective study. Gerontology. 2020;66:467–475. doi: 10.1159/000508734. [DOI] [PubMed] [Google Scholar]
- 33.O'Driscoll M, et al. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2918-0. [DOI] [PubMed] [Google Scholar]
- 34.Yang W, et al. Estimating the infection-fatality risk of SARS-CoV-2 in New York City during the spring 2020 pandemic wave: a model-based analysis. Lancet Infect Dis. 2020;3099:1–10. doi: 10.1016/S1473-3099(20)30769-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolff D, Nee S, Sandy N, Michael H. Risk factors for Covid ‑ 19 severity and fatality : a structured literature review. Infection. 2020 doi: 10.1007/s15010-020-01509-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma Y, et al. The association between frailty and severe disease among COVID-19 patients aged over 60 years in China: A prospective cohort study. BMC Med. 2020;18:1–8. doi: 10.1186/s12916-020-01761-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hewitt J, et al. The effect of frailty on survival in patients with COVID-19 (COPE): a multicentre, European, observational cohort study. Lancet Public Heal. 2020;5:e444–e451. doi: 10.1016/S2468-2667(20)30146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soysal P, et al. Inflammation and frailty in the elderly: a systematic review and meta-analysis. Ageing Res Rev. 2016;31:1–8. doi: 10.1016/j.arr.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 39.Van Epps P., et al. Frailty has a stronger association with inflammation than age in older veterans. Immun Ageing. 2016;13:1–9. doi: 10.1186/s12979-016-0082-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ragab, D, Eldin, HS, Taeimah, M, Khattab, R. The COVID-19 cytokine storm ; what we know so far. 11, 1–4 (2020). [DOI] [PMC free article] [PubMed]
- 41.Collard RM, Boter H, Schoevers RA, Oude Voshaar RC. Prevalence of frailty in community-dwelling older persons: A systematic review. J Am Geriatr Soc. 2012;60:1487–1492. doi: 10.1111/j.1532-5415.2012.04054.x. [DOI] [PubMed] [Google Scholar]
- 42.Mohler MJ, Fain MJ, Wertheimer AM, Najafi B, Nikolich-Žugich J. The Frailty syndrome: clinical measurements and basic underpinnings in humans and animals. Exp Gerontol. 2014;54:6–13. doi: 10.1016/j.exger.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 43.Perrotta F, Corbi G, Mazzeo G, et al. COVID-19 and the elderly: insights into pathogenesis and clinical decision-making. Aging Clin Exp Res. 2020;32:1599–1608. doi: 10.1007/s40520-020-01631-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mori H, et al. Comparison of COVID-19 disease between young and elderly patients: hidden viral shedding of COVID-19. J Infect Chemother Off J Japan Soc Chemother. 2021;27:70–75. doi: 10.1016/j.jiac.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gómez-Mesa JE, Galindo-Coral S, Montes MC, Muñoz Martin AJ. Thrombosis and coagulopathy in COVID-19. Curr Probl Cardiol. 2021;46 doi: 10.1016/j.cpcardiol.2020.100742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nägele MP, Haubner B, Tanner FC, Ruschitzka F, Flammer AJ. Endothelial dysfunction in COVID-19: current findings and therapeutic implications. Atherosclerosis. 2020;314:58–62. doi: 10.1016/j.atherosclerosis.2020.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varga Z., et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klok F.A., et al. Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb Res. 2020;191:145–147. doi: 10.1016/j.thromres.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poonia B., Kottilil S. Immune correlates of COVID-19 control. Front Immunol. 2020;11:1–9. doi: 10.3389/fimmu.2020.569611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Massaad, C, Nuss, P, Benoliel, J-J, Becker, C. COmmEnT Tissue damage from neutrophil-induced oxidative stress in COVID-19. Nat Rev Immunol1–2 doi: 10.1038/s41577-020-0407-1 [DOI] [PMC free article] [PubMed]
- 51.Nienhold R, et al. Two distinct immunopathological profiles in autopsy lungs of COVID-19. Nat Commun. 2020;11:1–13. doi: 10.1038/s41467-020-18854-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schurink B., et al. Viral presence and immunopathology in patients with lethal COVID-19: a prospective autopsy cohort study. The Lancet Microbe. 2020;1:e290–e299. doi: 10.1016/S2666-5247(20)30144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edén A, et al. CSF biomarkers in patients with COVID-19 and neurologic symptoms: a case series. Neurology. 2021;96:e294–e300. doi: 10.1212/WNL.0000000000010977. [DOI] [PubMed] [Google Scholar]
- 54.Shkurnikov M, Nersisyan S, Jankevic T, et al. Association of HLA class I genotypes with severity of coronavirus disease-19. Front Immunol. 2021;12:423. doi: 10.3389/fimmu.2021.641900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pairo-Castineira E, et al. Genetic mechanisms of critical illness in Covid-19. Nature. 2020;591 doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 56.Muñoz-Fontela C, et al. Animal models for COVID-19. Nature. 2020;586:509–515. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi J, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science (80-.) 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rockx B., et al. Comparative pathogenesis of COVID-19, MERS and SARS in a non-human primate model. bioRxiv. 2020;1015:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu P., et al. Age-related rhesus macaque models of COVID-19. Anim Model Exp Med. 2020;3:93–97. doi: 10.1002/ame2.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel Coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS Coronavirus. J. Virol. 2020;94:1–9. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCray PB, Pewe L, Wohlford-Lenane C, et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome Coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tseng C-T K, Huang C, Newman P, et al. Severe acute respiratory syndrome Coronavirus infection of mice transgenic for the human angiotensin-converting enzyme 2 virus receptor. J Virol. 2007;81:1162–1173. doi: 10.1128/JVI.01702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bao L, Deng W, Huang B, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 64.Dinnon KH, Leist SR, Schäfer A, et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Y, Yan LM, Wan L, et al. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis. 2020;20:656–657. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nikolich-Žugich J. The twilight of immunity: emerging concepts in aging of the immune system. Nat Immunol. 2018;19:10–19. doi: 10.1038/s41590-017-0006-x. [DOI] [PubMed] [Google Scholar]
- 67.Hazeldine J, Lord JM. Innate immunesenescence: underlying mechanisms and clinical relevance. Biogerontology. 2015;16:187–201. doi: 10.1007/s10522-014-9514-3. [DOI] [PubMed] [Google Scholar]
- 68.Tseng CW, Kyme PA, Arruda A, et al. Innate immune dysfunctions in aged mice facilitate the systemic dissemination of methicillin-resistant S. aureus. PLoS One. 2012;7:1–9. doi: 10.1371/journal.pone.0041454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simell B, Vuorela A, Ekström N, et al. Aging reduces the functionality of anti-pneumococcal antibodies and the killing of Streptococcus pneumoniae by neutrophil phagocytosis. Vaccine. 2011;29:1929–1934. doi: 10.1016/j.vaccine.2010.12.121. [DOI] [PubMed] [Google Scholar]
- 70.Wong CK, Smith CA, Sakamoto K, Kaminski N, Koff JL, Goldstein DR. Aging impairs alveolar macrophage phagocytosis and increases influenza-induced mortality in mice. J Immunol. 2017;199:1060–1068. doi: 10.4049/jimmunol.1700397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kulkarni U, Zemans RL, Smith CA, Wood SC, Deng JC, Goldstein DR. Excessive neutrophil levels in the lung underlie the age-associated increase in influenza mortality. Mucosal Immunol. 2019;12:545–554. doi: 10.1038/s41385-018-0115-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Solana R, Tarazona R, Gayoso I, Lesur O, Dupuis G, Fulop T. Innate immunosenescence: effect of aging on cells and receptors of the innate immune system in humans. Semin. Immunol. 2012;24:331–341. doi: 10.1016/j.smim.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 73.Manser AR, Uhrberg M. Age-related changes in natural killer cell repertoires: impact on NK cell function and immune surveillance. Cancer Immunol Immunother. 2016;65:417–426. doi: 10.1007/s00262-015-1750-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chougnet CA, Thacker RI, Shehata HM, et al. Loss of phagocytic and antigen cross-presenting capacity in aging dendritic cells is associated with mitochondrial dysfunction. J Immunol. 2015;195:2624–2632. doi: 10.4049/jimmunol.1501006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li G, Smithey MJ, Rudd BD, Nikolich-Žugich J. Age-associated alterations in CD8α+ dendritic cells impair CD8 T-cell expansion in response to an intracellular bacterium. Aging Cell. 2012;11:968–977. doi: 10.1111/j.1474-9726.2012.00867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shaw AC, Goldstein DR, Montgomery RR. Age-dependent dysregulation of innate immunity. Nat Rev Immunol. 2013;13:875–887. doi: 10.1038/nri3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao J, Zhao J, Legge K, Perlman S. Age-related increases in PGD 2 expression impair respiratory DC migration, resulting in diminished T cell responses upon respiratory virus infection in mice. J Clin Invest. 2011;121:4921–4930. doi: 10.1172/JCI59777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ademokun A, Wu YC, Dunn-Walters D. The ageing B cell population: composition and function. Biogerontology. 2010;11:125–137. doi: 10.1007/s10522-009-9256-9. [DOI] [PubMed] [Google Scholar]
- 79.Zheng B, Han S, Takahashi Y, Kelsoe G. Immunosenescence and germinal center reaction. Immunol Rev. 1997;160:63–77. doi: 10.1111/j.1600-065x.1997.tb01028.x. [DOI] [PubMed] [Google Scholar]
- 80.Eaton SM, Burns EM, Kusser K, Randall TD, Haynes L. Age-related defects in CD4 T cell cognate helper function lead to reductions in humoral responses. J. Exp. Med. 2004;200:1613–1622. doi: 10.1084/jem.20041395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frasca D, Landin AM, Lechner SC, et al. Aging down-regulates the transcription factor E2A, activation-induced cytidine deaminase, and Ig class wwitch in human B cells. J Immunol. 2008;180:5283–5290. doi: 10.4049/jimmunol.180.8.5283. [DOI] [PubMed] [Google Scholar]
- 82.Crooke SN, Ovsyannikova IG, Poland GA, Kennedy RB. Immunosenescence and human vaccine immune responses. Immun Ageing. 2019;16:1–16. doi: 10.1186/s12979-019-0164-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nikolich-Žugich J. Aging of the T cell compartment in mice and humans: from no naive expectations to foggy memories. J Immunol. 2014;193:2622–2629. doi: 10.4049/jimmunol.1401174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.den Braber I., Mugwagwa T, Vrisekoop N, et al. Maintenance of peripheral baive T cells is sustained by thymus output in mice but not humans. Immunity. 2012;36:288–297. doi: 10.1016/j.immuni.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 85.Qi Q, Liu Y, Cheng Y, et al. Diversity and clonal selection in the human T-cell repertoire. 2014. doi:10.1073/pnas.1409155111 [DOI] [PMC free article] [PubMed]
- 86.Smithey MJ, Venturi V, Davenport MP, Buntzman A S, Vincent BG. Lifelong CMV infection improves immune defense in old mice by broadening the mobilized TCR repertoire against third-party infection. Proc Natl Acad Sci U S A. 2018;115:E6817–E6825. doi: 10.1073/pnas.1719451115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Becklund BR, Purton JF, Ramsey C, et al. The aged lymphoid tissue environment fails to support naïve T cell homeostasis. Sci Rep. 2016;6:30842. doi: 10.1038/srep30842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thompson HL, Smithey MJ, Uhrlaub JL, et al. Lymph nodes as barriers to T-cell rejuvenation in aging mice and nonhuman primates. Aging Cell. 2019;18:e12865. doi: 10.1111/acel.12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Richner JM, Gmyrek GB, Govero J, et al. Age-dependent cell trafficking defects in draining lymph nodes impair adaptive immunity and control of west nile virus infection. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wertheimer AM, Bennett MS, Park B, et al. Aging and cytomegalovirus infection differentially and jointly affect distinct circulating T cell subsets in humans. J Immunol. 2014;192:2143–2155. doi: 10.4049/jimmunol.1301721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Czesnikiewicz-Guzik M, Lee W-W, Cui D, et al. T cell subset-specific susceptibility to aging. Clin Immunol. 2008;127:107–118. doi: 10.1016/j.clim.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thompson, HL, Renkema, R, Smithey, MJ. Defective transcriptional programming of effector CD8 T cells in aged mice is cell-extrinsic and can be corrected by administration of IL-12 and IL-18. 10, 2–15 (2019). [DOI] [PMC free article] [PubMed]
- 93.Kim C, Jadhav RR, Gustafson CE, et al. Defects in antiviral T Cell responses inflicted by aging-associated miR-181a deficiency. Cell Rep. 2019;29:2202–2216.e5. doi: 10.1016/j.celrep.2019.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pujadas E, Chaudhry F, McBride R, et al. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir Med. 2020;8:e70. doi: 10.1016/S2213-2600(20)30354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.To KK-W, Tsang OT-Y, Leung W-S, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xia H., Cao Z, Xie X, et al. Article evasion of type I interferon by SARS-CoV-2 ll ll evasion of type I interferon by SARS-CoV-2. Cell Reports. 2020;33 doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hadjadj, J, Yatim, N, Barnabei, L, Corneau, A, Boussier, JE. (2, 6, 7). 724, 718–724 (2020).
- 98.Zhou Z, Ren L, Zhang L, Jin Q, Li M, Wang J. Heightened innate immune Responses in the Respiratory Tract of COVID-19 Patients ll ll Short Article Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe. 2020:883–890. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee, JS. The type I interferon response in COVID-19 : implications for treatment. Nat. Rev. Immunol.19–20 doi: 10.1038/s41577-020-00429-3. [DOI] [PMC free article] [PubMed]
- 100.Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID ‑ 19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020 doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Franceschi C, Campisi J. Chronic inflammation (Inflammaging) and its potential contribution to age-associated diseases. J Gerontol - Ser A Biol Sci Med Sci. 2014;69:S4–S9. doi: 10.1093/gerona/glu057. [DOI] [PubMed] [Google Scholar]
- 103.Puzianowska-Kuźnicka M., Owczarz M, Wieczorowska-Tobis K, et al. Interleukin-6 and C-reactive protein, successful aging, and mortality: the PolSenior study. Immun Ageing. 2016;13:21. doi: 10.1186/s12979-016-0076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Reiss WG, Pharm D, Kramer B, et al. Tocilizumab in Patients Hospitalized with Covid-19 Pneumonia. Immun Ageing. 2016;13:21. [Google Scholar]
- 105.Gordon AC, Mouncey PR, Al-beidh F, et al. Interleukin-6 receptor antagonists in critically Ill patients with Covid-19 – Writing Committee : corresponding author. medRxiv. 2021:1–12. doi: 10.1056/NEJMoa2100433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu Y, Du X, Chen J, et al. Neutrophil-to-lymphocyte ratio as an independent risk factor for mortality in hospitalized patients with COVID-19. J Infect. 2020;81:e6–e12. doi: 10.1016/j.jinf.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Barnes BJ, Adrover JM, Baxter-Stoltzfus A, et al. Targeting potential drivers of COVID-19: Neutrophil extracellular traps. J Exp Med. 2020;217:1–7. doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nicolai L, Leunig A, Brambs S, et al. Immunothrombotic dysregulation in COVID-19 pneumonia is associated with respiratory failure and coagulopathy. Circulation. 2020:1176–1189. doi: 10.1161/CIRCULATIONAHA.120.048488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Massberg S, Grahl L, Von Bruehl ML, et al. Reciprocal coupling of coagulation and innate immunity via neutrophil serine proteases. Nat Med. 2010;16:887–896. doi: 10.1038/nm.2184. [DOI] [PubMed] [Google Scholar]
- 110.Hinojosa E, Boyd AR, Orihuela C. Age-associated inflammation and Toll-like receptor dysfunction prime the lungs for pneumococcal pneumonia. J Infect Dis. 2009;200:546–554. doi: 10.1086/600870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen G., Wu D, Guo W, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mathew D., Giles JR, Baxter AE, et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rydyznski Moderbacher C., Ramirez SI, Dan JM, et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liao M, Liu Y, Yuan J, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 115.Grifoni A., Weiskopf D, Ramirez SI, et al. Article targets of T cell responses to SARS-CoV-2 Coronavirus in humans with COVID-19 disease and unexposed individuals ll article targets of T Cell responses to SARS-CoV-2 Coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Braun J, Loyal L, Frentsch M, et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 117.Grifoni A, Weiskopf D, Ramirez SI, et al. Targets of T Cell responses to SARS-CoV-2 Coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sekine T, Perez-Potti A, Rivera-Ballesteros O, et al. Robust T Cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Le Bert N, Tan AT, Kunasegaran K, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 120.Mateus J, Grifoni A, Tarke A, et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;94:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mathew, D. Deep immune profiling of COVID-19 patients reveals patient heterogeneity and distinct immunotypes with implications for therapeutic interventions. (2020). [DOI] [PMC free article] [PubMed]
- 122.Kuri-cervantes L, Pampena MB, Meng W, et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci Immunol. 2020;5:eabd7114. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lau EHY, Tsang OTY, Hui DSC, et al. Neutralizing antibody titres in SARS-CoV-2 infections. Nat Commun. 2021;12:1–7. doi: 10.1038/s41467-020-20247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ripperger T.J., Uhrlaub JL, Watanabe M, et al. Orthogonal SARS-CoV-2 serological assays enable surveillance of low-prevalence communities and reveal durable humoral immunity. Immunity. 2020;53:925–933.e4. doi: 10.1016/j.immuni.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sette A, Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021:1–20. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Banerji A, Wickner PG, Saff R, et al. mRNA vaccines to prevent COVID-19 disease and reported allergic reactions: current evidence and suggested approach. J Allergy Clin Immunol Pract. 2020 doi: 10.1016/j.jaip.2020.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Anderson EJ, Rouphael NG, Widge AT, et al. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N Engl J Med. 2020;383:2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sahin U, Muik A, Derhovanessian E, et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 129.Spector SA, Rouphael N, Creech CB, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]