Abstract
This article reviews the many and varied mass spectrometry based responses to the SARS-CoV2 coronavirus amidst a continuing global healthcare crisis. Although RT-PCR is the most prevalent molecular based surveillance approach, improvements in the detection sensitivities with mass spectrometry coupled to the rapid nature of analysis, the high molecular precision of measurements, opportunities for high sample throughput, and the potential for in-field testing, offer advantages for characterising the virus and studying the molecular pathways by which it infects host cells. The detection of biomarkers by MALDI-TOF mass spectrometry, studies of viral peptides using proteotyping strategies, targeted LC-MS analyses to identify abundant peptides in clinical specimens, the analysis of viral protein glycoforms, proteomics approaches to understand impacts of infection on host cells, and examinations of point-of-care breath analysis have all been explored. This review organises and illustrates these applications with reference to the many studies that have appeared in the literature since the outbreak. In this respect, those studies in which mass spectrometry has a major role are the focus, and only those which have peer-reviewed have been cited.
Keywords: Mass spectrometry, SARS-CoV2, Coronavirus, Analysis, Proteomics
1. Introduction
Just over a century from the 1918 influenza pandemic, warnings about a future viral pandemic have been realised with the emergence and spread of the SARS-CoV2 coronavirus. First detected in China in late 2019 [1], the virus rapidly spread throughout the world and has currently been associated with over 2.5 million deaths and some 113 million cases of infection [2], with Europe and the Americas particularly impacted. Beyond the global health emergency, the pandemic is estimated to have resulted in an economic cost exceeding $USD 10 trillion [3], resulting from a decrease in the global economy of some 5%, only matched by the depression early in the twentieth century and the two world wars.
A global scientific effort has presented a united front to contain, monitor and respond to the virus through the implementation of a range of analytical, both molecular and non-molecular, approaches. Chief among the technologies employed for the detection and surveillance of the virus has been reverse transcription–polymerase chain reaction (RT-PCR)-based analysis and sequencing [4,5]. RT-PCR, quantitative PCR, Nucleic Acid Sequence-Based Amplification (NASBA), Loop Mediated Isothermal Amplification (LAMP), and CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) approaches have all been applied where some have potential for point-of-care diagnosis at the bedside [5,6].
Mass spectrometry has long been used for the study and analysis of viruses [[7], [8], [9], [10]], though recent advances in instrumentation have offered improvements in sensitivity, resolution and mass accuracy, as well as the ability to analyse whole viruses and their interactions. Some 200 publications describing the application of mass spectrometry, in some form, to characterise the coronavirus [11] have appeared in the literature since the outbreak of the pandemic and this review attempts to organise and review the approaches against conventional methods. It also explores the possibility of a frontline approach for point-of-care diagnosis using a mass spectrometry platform technology.
2. Discussion
2.1. SARS-CoV2 coronavirus, its structure and components, by the numbers
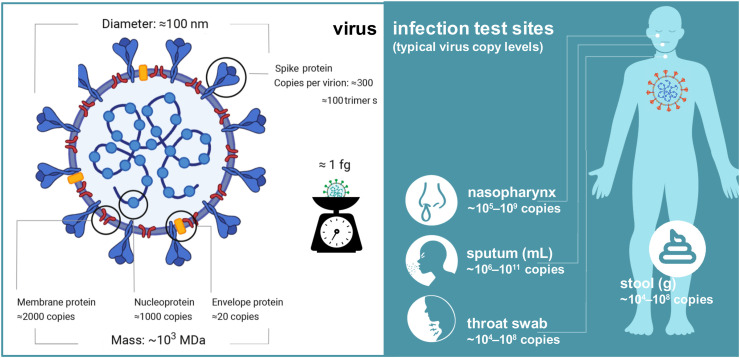
A brief review of the nature of the SARS-CoV2 coronavirus is worthy of note since the subsequent mass spectrometric analysis is dependent upon it. The virus is a beta-coronavirus whose genome is comprised of a single strand of RNA of some 30 kilobases in length. This contains 10 genes that encode 26 proteins, some from the cleavage of a polyprotein with proteases that are themselves part of that protein. In addition, an RNA polymerase and associated factors to copy the genome, a proof-reading exonuclease, and several other non-structural proteins are encoded [12]. The remaining genes code for structural components of the virus that comprise a surface spike (S) protein which binds host cell receptors [13], a nucleocapsid (N) protein that packages the genome [14] and two membrane-bound (M) proteins that include the envelope (E) protein (Fig. 1 ). These are present in some 300 (S), 1000 (N), 2000 (M) and 20 (E) copies per virion with a diameter of some 100 nm and a total mass of approximately 1 fg [15].
Fig. 1.
Anatomy of the structure of the SARS-CoV2 coronavirus particle showing the structural proteins, copy numbers, virion size and mass, and infection sites and copies per typical specimen [15,17]. All values are approximate only.
The structural proteins have molecular weights of 141.2 (S - consisting of two subunits S1 and S2), 45.6 (N), 25.1 (M), and 8.4 (E) kDa. respectively [16]. The high copy numbers for the membrane and envelope proteins aid their detection by mass spectrometry, as does the sheer size of the surface spike protein particularly when detected in a digested form [16]. The spike protein adopts the form of a trimer on the surface of the virus where its two subunits (S1 and S2), generated by cleavage of the S protein at residues 685–686 and each approximately 70 kDa in size, catalyse the attachment of the virus to the membrane of a host cell and facilitate its fusion respectively.
Also of importance to mass spectrometric analysis is the viral copy numbers per isolate, usually assessed based upon RNA quantitation. Viral loads vary widely from some 104-1011 copies per specimen or standardized volume (Fig. 1) [15,17]. These values, however, are approximate only and depend on the period since initial infection and the virus recovery procedures. Despite this, upper respiratory tract swabs typically contain the highest levels of virus and are the primary sources for downstream analysis by mass spectrometry.
2.2. Mass spectrometric analysis of virus components for diagnostics
By far the most common approach to detect viruses, and microorganisms in general, by mass spectrometry involves analysis of the component proteins. This can be accomplished directly after their release from a sample isolate as intact proteins or more often after digestion of the whole virus or component proteins. Proteins or their peptide components are then subjected to either MALDI or LC-ESI based analysis, or some combination of both. Different laboratories have their particular biases in this regard, in part associated with the mass spectrometer configurations at hand. In some cases, particularly within clinical lab settings, any released biomolecular component (DNA/RNA, protein, lipid, etc.) is used as a biomarker based upon its molecular weight alone without any attempt to identify the nature of that component.
2.2.1. Unidentified potential biomarker detection with MALDI-TOF
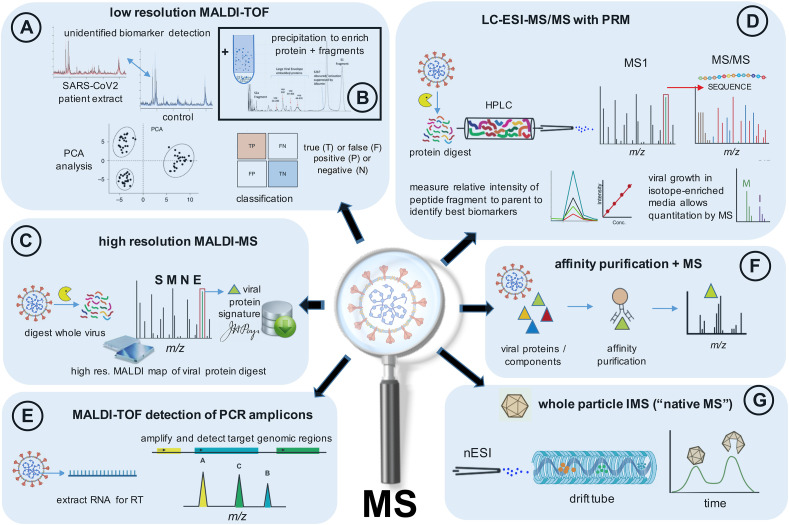
Among the first peer-reviewed papers to appear in the literature, the detection of SARS-CoV-2 in nasal swabs using MALDI-TOF MS aside conventional RT-PCR analysis was investigated [18]. In this study, MALDI mass spectra of nasal mucous secretion samples from three South American countries that had been confirmed either positive or negative for SARS-CoV-2 by RT-PCR. No report of the levels of virus in the secretion samples was made, but they can be expected to correspond to levels obtained from nasopharynx swabs (Fig. 1). A total of 362 specimens (comprising 211 positive and 151 negative) were directly subjected to MALDI-TOF analysis and an intensity comparison was made based on seven selected peaks of distinct m/z values, mostly between 3000 and 3500 with one peak each at m/z 7612 and 10,444. No effort to identify the biomarkers was made; rather a principal cluster analysis (PCA) of the seven selected peaks using a machine learning approach was able to identify the presence or absence of SARS-CoV2 (Fig. 2 A) based upon detection of the ions and their relative mean intensities with 7% false positives and 5% false negatives. When PCA was performed with combined samples from the three laboratories, the results for the positive samples and negative controls did not completely resolve, though they did so when those of each laboratory were handled independently. Demonstrating a high degree of variability of detected components in the specimens, the study noted that among the selected peaks, only the m/z 7612 component (unidentified) was common to all spectra across all laboratories. The most substantial mean intensity difference was exhibited by ions at m/z 3358 that was best used to differentiate the control group from the SARS-CoV2 positive group.
Fig. 2.
Schematic showing major mass spectrometry (MS) approaches/workflows used to characterise SARS-CoV2 biomarkers, the structural protein components, or proteolytic peptides thereof, or whole virus using MALDI and LC-ESI based techniques. Performance (sensitivities, resolution, dynamic ranges, etc.) depend on specific MS instrument configuration and operation. Refer to cited studies and datasets of Section 2.
A similar preliminary study of 311 patient specimens was conducted by Rocca et al. [19]. The authors used the intensities of 10 peaks taken from the MALDI-TOF spectra and applied a machine learning algorithm and three difference classification models to assess its performance in detecting positive and negative SARS-CoV2 samples directly from nasopharyngeal swabs samples. Samples which exhibited six peaks at 3372, 3442, 3465, 3488, 6347 and 10,836 Da were used to assess the absence of infection based upon their reduced intensity or absence in the positive samples. An overall accuracy of some 68% was reported (reflecting a positive prediction value of 60% and negative prediction value of 73.2% in the proportion of samples within cut-off parameters), but it was noted that none of the peaks found could be molecularly attributed to virus-specific proteins and were likely to some type of viral host components.
2.2.2. Viral protein and proteolytic peptide detection with MALDI-MS
Irrespective of the source of specimen, the removal of non-protein contaminants can improve mass spectrometric detection and the reliability of any molecular assay. The presence of abundant host molecules can mask and also suppress the ionisation of viral proteins at low abundance. Virus enrichment, and sample clean-up, substantially improves the detection of lower abundance viral protein signals and improves MALDI shot-to-shot and sample-to-sample reproducibility. Iles and co-workers [20] utilised the cold addition of acetone to precipitate viral protein from background host proteins and other contaminants where the pellet recovered after centrifugation was resuspended in a solubilisation buffer. Low resolution MALDI-TOF of the S-protein subunits and their fragments, S1 (at ~ m/z 79,000) and S2 (~m/z 62,000–72000) were detected together with other putatively identified viral-associated envelope protein fragments (ranging from ~ m/z 26,000–47000) at elevated intensities in saliva and gargle samples of infected patients [20] (Fig. 2B).
This, and the studies cited above, demonstrate the importance of higher resolution mass spectra to more confidently identify the components, particularly if intact viral proteins are to be identified. Furthermore, the establishment of a library of reference SARS-CoV2 viral protein mass spectra would enable one to perform a so-called biotyping experiment in which viral protein biomarkers could be assigned with more confidence. To circumvent this requirement, digested viral proteins can be detected at sufficient resolution to be identified on most MALDI-TOF based systems with reasonable reliability.
When high resolution mass accuracy is employed on a Fourier-transform based instrument (i.e. an ion cyclotron resonance (ICR) or Orbitrap), sufficient mass accuracy is obtained to allow viral peptides to be assigned unequivocally by mass alone (Fig. 2C). In one such MALDI based study [16], a whole virus digest, analyzed across a mass range of m/z 500–3000, detected peptides of the nucleocapsid, membrane, and spike proteins with a typical sequence coverage of some 27%, or equivalent to that found by LC-ESI-MS [21]. Mass accuracies exceeded 3 ppm and a study of nasopharyngeal specimens found detection limits were better than 105 copies (compare with Fig. 1) when using full scan mass spectra. Much lower limits are possible with selected ion monitoring. Importantly the ability to detect peptide ions unique to the virus by mass alone using high resolution mass spectrometry forms part of a proteotyping strategy applied to other respiratory viruses including influenza and parainfluenza [22].
2.2.3. Proteolytic peptide detection with LC-ESI-MS and tandem mass spectrometry
Nikolaev and co-workers [21] employed LC-ESI MS to detect peptides of the more abundant nucleocapsid protein within the combined muscosal swabs of SARS-CoV2 infected patients taken of the lower part of the nasopharynx and posterior wall of the oropharynx. In this study, viral protein was precipitated from cooled inactivated samples and either digested immediately with trypsin, or first reduced and alkylated before digestion, in express or standard protocols respectively. Tandem mass spectra (MS/MS) were recorded for 14 fully or partially digested peptides on a hybrid ion mobility Q-TOF instrument and their sequences were derived through a comparison of detected fragments with those predicted. These peptides were exclusively detected, but to differing degrees, in the SARS-CoV2 positive samples.
A number the same peptides were also detected in a similar study [23] of three gargle samples of SARS-CoV2 infected patients. The authors of this study identified unique nucleoprotein peptides originating in two out of three samples with viral loads estimated to be of the order of 105 to 106 RNA equivalents per μL of gargle solution (c.f. throat swab concentrations, Fig. 1) that were not detected in the lower concentration (~103 equivalents/μL) sample. Tandem LC-MS instruments provide a means to at least partially purify the sample during analysis but suffer from the drawback of a considerable analysis time (up to several hours) per sample [21], due to the LC run time and the need to wash and equilibrate the column after each run.
The use of a targeted strategy, in which selected peptide markers are detected together with their fragments in so-called parallel reaction monitoring (PRM), can help to reduce analysis times (Fig. 2D). Cazares et al. [24] employed such a strategy in mock infected samples to detect peptides of the SARS-CoV-2 spike and nucleocapsid proteins. Four proteolytic peptides, two each from the spike and nucleoprotein, were selected based upon their reproducible production following digestion, and low limits of detection. Two such peptides were detected in PRM experiments in the ~200–400 amol range following serial dilution of the samples. In such mock samples, this equated to the detection of virus at titre levels above some 2 × 105 pfu/mL (see Fig. 1).
Gouveia and co-workers [25] utilised a similar procedure to identify a shortlist of 14 peptides derived from the matrix, nucleocapsid and spike proteins in a cell cultured SARS-CoV-2 virus sample, two of which matched those selected in the work of Cazares [24]. In a proof-of-concept study [26], the same group acquired MS/MS spectra of peptides detected in two nasopharyngeal swabs but found only a small proportion of the peptide sequences could be mapped to microorganisms, raising concerns about false discovery rates. In simulated swabs containing specific quantities of SARS-CoV-2 virus, mixed with other nasal proteins, a single viral peptide of the nucleocapsid protein was reported at low ng or pfu level. An order of magnitude more material was needed to detect peptides from multiple proteins necessary for a more unequivocal analysis. Of nine positive clinical specimens, the authors detected virus peptides in two samples.
Another study by Singh and colleagues [27], who also employed PRM, selected two peptides from the spike and a replicase polyprotein from a shortlist of eight peptides from these and the nucleoprotein. A detection sensitivity of 90% (from 57 of 63 samples) and specificity of 100% in terms of RT-PCR confirmed positive samples was achieved. This study also reported the two peptides were detected in upper respiratory tract swabs of patients who have symptomatically recovered from SARS-CoV2 and had tested negative for RT-PCR analyses, demonstrating the potential of the MS approach to diagnosis asymptomatic SARS-CoV2 in patients. A larger study of close to 1000 specimens, employing the PRM strategy, detected peptides of SARS-CoV-2 nucleocapsid protein both qualitatively and quantitatively by incorporating 15N-labelled standards, in up to 84% of the positive cases with up to 97% specificity [28]. In this study, the use of a robotic sample handler enabled the analysis of 4 samples every 10 min.
2.2.4. Detection of DNA amplicons with mass spectrometry
A complementary strategy to the analysis of viral protein components is the implementation of mass spectrometry for the rapid detection of amplified polymerase chain reaction (PCR) products. This has been applied previously to a range of respiratory viruses [29,30] using ESI [29] and MALDI based instruments [30].
In the MALDI-TOF based study, serially diluted solutions of plasmids containing nearly the full-length sequence of target genes of human coronaviruses. The approach employed multiplex PCR, primer extension and MALDI-TOF identification of the amplicons. Virus was able to detect as low as 10 copies while the virus was detected in 22% (29/131) of clinical specimens using primers and extension probes specific to the RNA-dependent RNA polymerase (RdRp) and nucleocapsid (N) genes. The results were in accord with companion genomic analysis and PCR-sequencing.
In a more recent study, viral RNA was isolated and amplified from 44 nasopharyngeal or oropharyngeal swab specimens, that had tested either positive (22) or negative (22) for SARS-CoV-2 virus, and analysed by MALDI-TOF MS (Fig. 2E) [31]. The assay was designed to detect the following SARS-CoV-2 targets: viral nucleocapsid genes (N1-3 across bases 28,653–28760, 28,880–28978, 28,076–28190 respectively), and ORF1ab/nsp3 and ORF1ab/nsp10 across bases 3223–3335 and 13,342–13432 respectively. Five amplicons, with masses of between 5356 and 7010, were detected in two open reading frame (ORF1ab, and ORF1) and three nucleocapsid protein coding regions (N1, N2 and N3) that were unique to positive samples, while the detection of unextended primers were diagnostic of a negative sample. From this perspective, samples were identified as positive if two or more amplicon targets were detected and negative if less than two were detected. While the total run time for an exclusive RT-PCR analysis was considerably less than when combined with MS detection (some 80 versus 340 min), both required a similar hands on intervention time [31]. The MS-based method also has a fast turnaround time from sample to diagnosis and therefore is suitable for routine use.
2.2.5. Post-translational modification analysis; glycosylation and phosphorylation
In the case of the SARS-CoV2 coronavirus, a particular focus has been on the study of the glycosylation profile of the surface spike protein. The SARS-CoV-2 S glycoprotein contains 22 N-linked glycosylation sites per protomer and, experimentally, it has been confirmed that some 19 of them are glycosylated. These oligosaccharides contribute to spike protein folding, influence priming by host proteases, and regulate antibody recognition in response to the virus. They participate in viral entry into the host, proteolytic cleavage of viral proteins, and recognition and neutralization of the virus by the host's immune system.
To resolve the site-specific glycosylation of the SARS-CoV-2 S protein and visualize the glycoform heterogeneity across the protein surface, one study purified recombinant SARS-CoV-2 S-protein using size exclusion chromatography to ensure the presence of native-like trimeric protein [32]. This was then cleaved with three different proteases separately and the products analysed by LC-ESI-MS. The three proteases were selected to generate glycopeptides that contain a single N-linked glycan. A dispersion of oligomannose-type glycans was reported across both the S1 and S2 subunits. Whereas the glycan content (28%) was above that observed on typical host glycoproteins, it is lower than that for the envelope protein of HIV. This reduced glycan shield, it has been suggested [32], may be of benefit in the elicitation of neutralizing antibodies when developing immunotherapies.
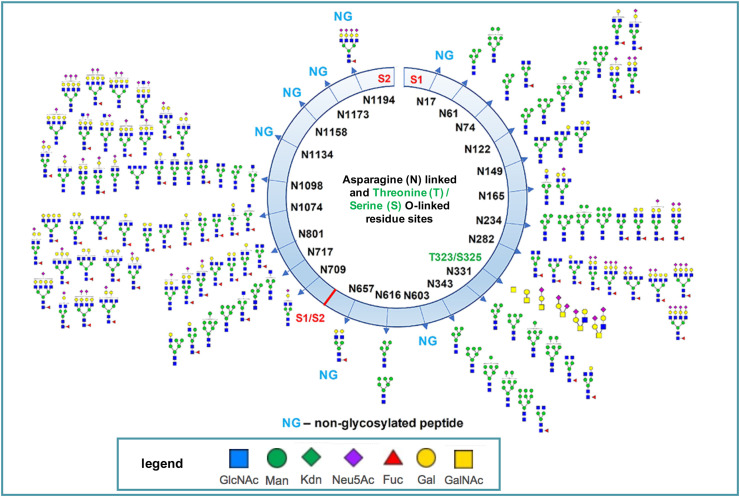
A second study [33], which expressed the two subunits separately, identified the glycan compositions at 17 out of the 22 predicted N-glycosylation sites and found the remaining five sites unoccupied. High mannose, hybrid and complex-type glycans across the N-glycosylation sites were observed (Fig. 3 ) though seven sites had little to no glycosylation. N-linked residues at positions 17, 603, 1134, 1158 and 1173 in S-protein were completely unglycosylated. Two highly sialylated-glycans at 234 and 282, adjacent to the RBD, may act as a determinant in virus binding with human Angiotensin-Converting Enzyme 2 (hACE2), a type I transmembrane metallocarboxypeptidase, that is attached to the cell membranes located in the lung which serves as the SARS-CoV2 virus' entry receptor.
Fig. 3.
Map of the N and O-linked glycosylation sites and heterogeneity of the S-protein determined by LC-ESI-MS/MS. The site of cleavage of the S-protein (at residues 685–686) to generate the S1 and S2 subunits is shown. Asparagine (N) linked and Threonine and Serine (T/S) O-linked sites are shown numbered according to the intact protein. Monosaccharide symbols follow the Symbol Nomenclature for Glycans (SNFG) system. Adapted from reference 31 with permission.
As well as the complex heterogeneity seen at N-glycosylation sites, the study also identified two unexpected O-glycosylation sites within the receptor-binding domain (RBD) of the S1 subunit at residues Thr323 and Ser325. N-acetyl-galactosamine and neuraminic acid glycoconjugates predominated at the former residue, while N-acetylhexosamine and neuraminic acid glyconjugates were detected at the latter. Although the function of these glycosylation sites remains unknown, it was suggested that they may play a role in shielding protein epitopes and aid immunoevasion.
Another recent study [34] has investigated post-translational modifications in both the SARS-CoV2 surface protein and hACE2. It provided additional structural details to study mechanisms underlying host attachment, immune response mediated by S protein and hACE2. All seven glycosylation sites in hACE2 were found to be completely occupied, mainly by complex N-glycans. However, this glycosylation did not directly contribute to the binding affinity between S-protein and hACE2 which was found to be impacted by additional post-translational modifications including multiple methylated sites in both proteins and hydroxylproline at multiple sites in hACE2.
A global phosphoproteomics survey of SARS-CoV-2 infection in Vero E6 cells [35] used LC-MS to reveal a rewiring of phosphorylation on host and viral proteins. Cells were infected with SARS-CoV-2 virus were harvested at various time points over a 24 h period. The proteins released upon lysing the cells were digested. Chromatographic separation and enrichment of phosphorylated peptides was followed by LC-MS/MS to identify the phosphorylated proteins against a database of host and SARS-CoV-2 protein sequences. Across a virus-host protein-protein interaction map of 332 human proteins that interact with SARS-CoV-2 viral proteins, 40 were found to undergo significantly different phosphorylation following infection versus an uninfected control. Among them several RNA-processing proteins were differentially phosphorylated during infection, including LARP1 and RRP9. Phosphorylation decreases on several sites in the La-motif related protein (LARP), an RNA-binding protein that regulates the translation of specific target mRNA species. This may consequently increase LARP1 affinity for other untranslated regions (UTRs), driving an inhibition of cell protein synthesis. These insights using proteomics datasets into how SARS-CoV-2 virus operate hijacks host cells provide opportunities to identify attractive targets for therapeutic intervention.
3. Interaction of the SARS-CoV2 virus with host cells; proteomic analysis of viral replication and inhibition
Host-cell interactions with viruses are the subject of much interest, and here too mass spectrometry has played a role. One study [36] used three glycoform models for the S-protein, based upon a mass spectrometric analysis of the glycosylation sites by LC-nESI-MS/MS, and molecular dynamic simulations to examine its interaction with the ACE2-RBD. Two glycans on ACE2 at N-linked residues 90 and 322 were predicted to form interactions with the S protein. The ACE2 glycan at position 90 was found to be close enough to the S-trimer surface to repeatedly form interactions, and the glycan arms interacted with multiple regions of the surface over the course of the simulations. Intermolecular glycan-glycan interactions were also observed repeatedly between the N-linked glycan at position 546 of ACE2 and those in the S protein at N-linked residues 74 and 165.
Proteomics strategies have examined host cell translation changes after infection with SARS-CoV-2. Bojkova et al. [37] infected human epithelial cells with SARS-CoV-2 and cultured the cells for 24 h, after which visible cytopathogenic effects were apparent, in a medium containing isotopically-enriched lysine and arginine. The authors used LC-MS/MS to monitor the levels and translation rates for five viral proteins, compared to a mock infection control, by summing the intensities of all peptide segments confirmed for each unique protein. To identify potential inhibitors of SARS-CoV-2 replication, they determined proteins with abundance trajectories that were similar to the detected viral proteins in order to study the pathways that are potentially important for virus amplification. The study further tested two translation inhibitors, ribavirin and NMS873, with different modes of action and found that these prevented viral replication.
Quantitative mass spectrometry, with and without the use of stable isotopes, has begun to reveal mechanisms underlying SARS-CoV-2 infection, including several key processes used by the virus to adapt their host. In a separate protocol, V'kovski and co-workers [38] adopted enzyme-catalysed biotin-labelling of proteins within the coronavirus replicase transcriptase complex (RTC) that likely contribute to the viral life cycle using affinity purification and identification of biotinylated proteins by mass spectrometry (Fig. 2F).
4. Metabolomics profiling of SARS-CoV2 infection
The metabolomics profiling of infected patients provides another means to better understand the underlying pathologic processes and pathways, and to identify potential diagnostic biomarkers. One study [39] adopted a targeted quantitative approach to analyse metabolites isolated from the blood plasma of infected patients using a combination of direct injection (DI) and reverse-phase LC-MS/MS. Up to 150 different endogenous metabolites including amino acids, acylcarnitines, biogenic amines and derivatives, uremic toxins, glycerophospholipids, sphingolipids, and sugars, were studied in SARS-CoV2 infected samples versus negative controls. Some derivatization and extraction of the analytes, and multiple reaction monitoring (MRM) was employed where isotopically-labelled and other internal standards were used for metabolite quantification purposes. A total of 183 plasma metabolites were detected using both DI-LC-MS/MS, and companion proton NMR, where the presence of eight were found to be the best indicators of positive coronavirus disease infection, with a particular signature being the increased levels of kynurenine.
Increased degradation of tryptophan, with a consequential increase in kynurenine, occurs during the immune response and is driven by the release of interferon-gamma from the activated T-cells upon virus infection. SARS-CoV2 T-cell activation causes an approximate 10-fold increase in plasma interferon-gamma in critically ill patients when compared with healthy subjects [39]. Although the presence of plasma kynurenine effectively discriminated infected patients from healthy control subjects, further specificity was provided by a measure of the arginine/kynurenine ratio where arginine was found to be significantly depressed in infected patients. Arginine is an amino acid precursor for nitric oxide which increases blood flow and oxygen to wounds. Thus arginine is essential for tissue repair and its depletion could potentially delay and/or compromise patient recovery.
4.1. Breath analysis and paperspray detection of metabolites for MS diagnostics
The desire for a rapid, cost effective and non-invasive molecular test of SARS-CoV2 viral infections has awakened the role of breath analysis [40,41]. The detection of volatile organic compounds (VOCs) by mass spectrometry has been active for several decades. However, relatively little work has attempted to diagnose viral infections using VOCs, since viruses hijack the host cell metabolism and, in so doing, do not produce their own metabolites [40].
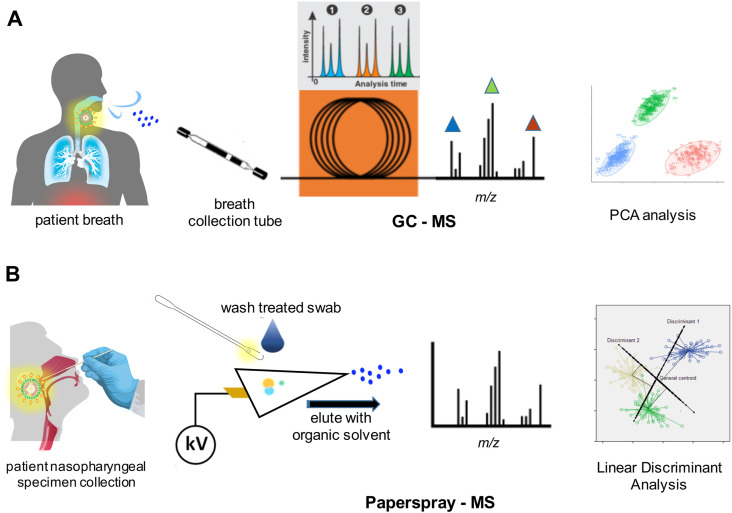
Various breath sampling devices are available that can be used by non-specialist staff. The key analytical aim is to detect elevated or reduced levels of VOCs at concentrations that are only a small percentage of exhaled carbon dioxide. GC-MS offers a sensitive and comparatively rapid approach with which to analyse breath samples (Fig. 4 A). A feasibility GC-ion mobility MS based study [42] involving ninety-eight patients (positive and negative to coronavirus, with some exhibiting other conditions including asthma in the negative cohort) at two different centres involved multivariate analysis of aldehydes (ethanal, octanal), ketones (acetone, butanone), and methanol that discriminated SARS-CoV2 infection from other conditions. An unidentified component with significant predictive power of infection severity was isolated in one group, while heptanal was identified as a biomarker in the second. Diagnosis was possible with 80% and 81.5% accuracy in the two groups based upon the MS results.
Fig. 4.
Schematics of (A) GC-MS analysis of volatiles in breath of infected patients and (B) nasopharyngeal swab specimens by paperspray mass spectrometry. Performance (sensitivities, resolution, dynamic ranges, etc.) depend on specific MS instrument configuration and operation. Refer to cited studies and datasets of Fig. 4.
Further studies, some published ahead of peer-review, note some caution in such diagnoses and suggest more work is needed to discriminate SARS-CoV2 infection from other respiratory virus infections as well as complications associated with patients who smoke. The future use of selected ion flow tube mass spectrometry (SIFT-MS) for SARS-CoV2 detection in real-time breath analysis also remains a possibility since the approach has advantages for the detection of volatile organic compounds (VOCs) of humid air, in the form of exhaled breath, without sample pre-treatment or separation.
An alternative that avoids complications with breath analysis, but still offers a rapid point-of-care test possibility uses paperspray mass spectrometry. Here analytic samples are deposited directly onto a paper or similar hydrophobic surface and eluted with solvent using an electrospray type format (Fig. 4B). DeSilva and colleagues [43] used a water proof Teslin substrate to examine the metabolite and lipid profile in mixed upper and lower respiratory tract swabs of infected patients employing positive and negative mass spectrometry analysis on a linear ion trap. Based on a comparison of relative peak intensities, nine down-regulated and twenty-two up-regulated metabolites were identified across 10 SARS-CoV2 infected samples employing a linear discriminant function analysis (LDA). Among the predominant changes in the swab samples, seventeen lipids were significantly elevated in abundance in the positive infection group. The authors reported, based on the statistical analysis, a 93.3% correlation with the results of a PCR classification.
5. Ion mobility mass spectrometry of virus components
Although the same mass resolution and accuracy is not achieved for the detection of whole virus particles (Fig. 2G) and their complexes, there is merit in such mass spectrometric based investigations. Ion mobility mass spectrometry offers an alternative to X-ray crystallography and cryo-electron microscopy in the study of virus assembly, composition, and heterogeneity as well as structural dynamics, despite its inability to provide the same level of structural detail of crystallographic and microscopic studies.
Ion mobility mass spectrometry has been applied to study the binding of the receptor-binding domain of SARS-CoV2 spike protein with the ACE-2 host cell receptor [44]. A combination of molecular modelling and IMS was used to investigate the role of heparin in destabilizing the RBD-ACE2 association.
The detection of both the monomeric (with a molecular mass of 33,795 Da) and homodimeric complex form of a protease of SARS-CoV2 virus has also shown in a preliminary communication [45] together with its dissociation constant. The unit, named the main protease, or Mpro, is a cysteine protease that cleaves the encoded polyproteins at eleven sites resulting in a complex of twelve non-structural proteins (nsp5-nsp16). However, the dissociation constant for SARS-CoV-2 Mpro complex was determined by serial sample dilution and ion mobility mass spectrometry (MS) to be 0.14 μM, or over an order of magnitude lower than that obtained by analytical ultracentrifugation, raising caution about the native aspects of such experiments. The binding of several candidate small molecule inhibitors was undertaken with a view to assess their ability to bind to the dimer of SARS-CoV-2 Mpro and inhibit the virus' ability to replicate.
6. Evolution of viral proteins – protein phylogenetics with mass spectral data
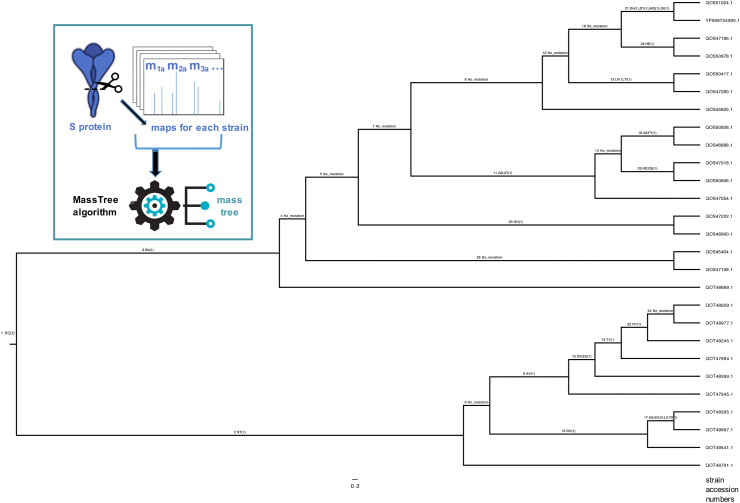
Phylogenetic studies of viral protein evolution are another area where mass spectrometry is beginning to be applied. Mutations in the SARS CoV-2 virus are now becoming more common as the world's population begins to be vaccinated. Those that help the virus to evade immune responses, vaccines and/or therapies are of most concern. It has been shown in a series of studies, which have recently been reviewed [46], that mass map profiles can be used to generate phylogenetic trees that are highly congruent with sequence based trees. Importantly, the sequence-free mass approach can determine most common amino acid mutations from a pairwise comparision of mass differences alone that, using a purpose built algorithm, are also displayed at branch nodes across the tree. These so-called mass trees [47] allow the evolution of the protein, and the virus strain from which they were derived, to be charted and followed by tracing non-synonymous mutations patterns along interconnected branches. Ancestral and descendant mutations can be studied in the context of the origins of antiviral resistance or other evolutionary events.
A recent application of the approach examined the evolution of the SARS-CoV2 S-protein [48]. This is the subject of particular interest given the impact of mutations on the virus' transmissibility and virulence. Areas within predicted epitopes of high antigenicity are of particular concern in terms of the effectiveness of a universal vaccine. Mass maps for this protein across 27 strains of the virus were used to build the mass tree shown (Fig. 5 box insert). Of the mutations shown on the tree (Fig. 5), the algorithm correctly assigned all but four mutations. These outliers were present in peptide segments with one or two other mutations, such that a comparison of their mass differences did not correspond to a detectable single point mutation based on mass alone.
Fig. 5.
Phylogenetic mass-based tree showing the evolution of SARS-CoV2 S-protein and associated single point mutations across 27 strains of the virus. Expand (zoom) to view labels.
7. Comparisons of RT-PCR versus mass spectrometric detection for viral diagnosis
While RT-PCR based analyses continue to be the “gold standard” for the molecular surveillance and characterisation of the SARS-CoV2 virus, the approach is not immune to false positive and false negative results [49,50]. In the real world, testing conditions are far from perfect, and accuracy suffers with higher false positive and negative rates. RT-PCR assays are typically complete within 2–4 h, but this is after the specimens have been processed for analysis [51], and detection limits down to some 10 copies of virus have been demonstrated [52] (Table 1 ). Though amplification allows for the generation of extra copies, PCR sequencing is also necessary to monitor ongoing mutations in the SARS-CoV-2 genome, in part to decide whether the primers and probes designed remain suitable for the detection of mutated virus strains.
Table 1.
Comparison of RT-PCR versus amplicon and viral peptide detection for SARS-CoV2 diagnosisa.
| Step/parameter | RT-PCR detection [5] | DNA amplicon detection by MS [30,31] | Viral peptide detection by MALDI-MS [16] | Viral peptide sequencing by LC-ESI-MS/MS [21] |
|---|---|---|---|---|
| Viral component recovery | RNA | RNA | protein | protein |
| Recovery time | minutes | minutes | 1–2 h | 1–2 h |
| Steps pre-analysis | reverse transcription, denaturation, annealing, amplificationb | reverse transcription, denaturation, annealing, amplificationb | proteolytic digestion with/without reduction/alkylation | proteolytic digestion with/without reduction/alkylation |
| Pre-analysis time | 30 min | 30 min | 4–14 hc | 4–14 hc |
| Detection time | 2–4 h | few minutes | few minutes | 30 min – 1 h |
| Detection limit (copies) | ~10 | >10–102 [30] | > 105d | 105–106 |
| Reliability/confidence | up to 95% | high (with multiple amplicons detected) | high (with multiple peptides detected) | high (with multiple peptides sequenced) |
| Analysis cost/sample (USD) | $10 | $10-50 | $100 | $250 |
| Instrument cost (USD) | $20 K+ | $100 K+ | $100–1000 K+ | $250–500 K+ |
All times and figures are approximate only and depend on specific protocols and equipment employed. Citations are to representative studies.
According to real-time RT-PCR detection of SARS-CoV-2 protocol, Institut Pasteur, Paris (https://www.who.int/docs/default-source/coronaviruse/real-time-rt-pcr-assays-for-the-detection-of-sars-cov-2-institut-pasteur-paris.pdf).
Improve using immobilized enzyme digestion to 1–2 h.
Improve by one or two orders of magnitude with selected ion monitoring (SIM).
By comparison the direct analysis of viral proteins, or their peptide counterparts, with mass spectrometry is most challenged by the limit of detection. Studies reported in this review have consistently detected virus using MALDI and LC-ESI-MS approaches down to some 105 copies (Table 1). Even with the use of selected ion or reaction monitoring, only a magnitude or two improvement in sensitivity can be expected without other advances in detection capability. Thus, at best, without further advances in mass spectrometry technology at least one order of magnitude more material is required over RT-PCR analysis.
Similar detection limits restrict the application of the DNA amplicon detection by mass spectrometry [30]. Thus when the number of copies is low, an MS-only based method may fail to detect the virus. Thus larger PCR volumes and/or more PCR cycles are required for the analysis of such samples. Like RT-PCR approaches, the MS-based strategy would not be able to detect new variants without prior knowledge from gene based sequencing. Where MS approaches do offer advantage is in the speed of analysis since a simple mass-only (MS1) analysis can be performed in minutes and with high sample throughout versus the hours necessary to perform PCR (Table 1).
Nonetheless, the wider application of mass spectrometry to studies of viral protein structures, their binding properties, affinities and function, in both qualitative and quantitative studies, far outreaches the applicability of PCR based methods. The rapid analysis times for the mass analysis stage are only encumbered by sample preparation and, where applicable, LC or other chromatographic or electrophoretic separation.
8. Conclusions and future outlook
The rapid and confident detection of a virus is a key requirement in an infectious disease outbreak such as that seen for the SARS-CoV2 pandemic. This detection needs to be both sensitive and be able to be performed by individuals with little training and expertise after appropriate inactivation of the virus [54]. Point of care tests are particularly attractive in this regard, either at a border entry point or elsewhere in field. The growing miniaturisation of mass spectrometer instruments and the potential of rapid swab based analysis by paperspray or portable MALDI based instruments are attractive in this regard. Paperspray or related technologies provide a rapid (seconds per sample) analytical technique with minimal to no sample preparation requirements, while MALDI strategies can be implemented with high sample throughput with minimal sample treatment and are more tolerant to ESI-based methods to salts and other contaminants. The recent coupling of solid-phase microextraction swabs to mass spectrometric analysis offer future potential for virus analysis [53]. The complementary nature of MS techniques to PCR-based methods allow the approaches to work hand-in-hand to accelerate management and responses to the SARS-CoV2 virus. MS based viral peptide detection strategies have already shown, in the case of multiple vaccine and other mixed strains, that it is possible to detect co-infections when sufficient mass resolution is achieved [22].
These analyses can be supported by more labour intensive laboratory based methods including GC and LC-MS to study disease prognosis, assess biomarkers for diagnosis and treatment, and evaluate the performance of vaccines and therapeutics. The impact of SARS-CoV2 infection on host cells and the effects of viral post-translational modifications such as glycosylation are already the focus of much interest employing multi-faceted mass spectrometry techniques. The use of MS datasets to study the evolution of the virus [48] including the identification of mutations that may limit detection by PCR or those that enable the virus to evade immune responses or challenge existing vaccines and/or therapies have great future potential. Without doubt, the expanded application of mass spectrometry and related “omics” strategies to better respond to virus outbreaks is sure to build on foundation studies [[7], [8], [9], [10],22] that predated the SARS-CoV2 pandemic. The growing reach of mass spectrometry into structural protein biology applications, using a range of approaches involving chemical and enzymatic treatments that have recently been compared side-by-side [55], should also aid in our understanding of the virus, its molecular machinery and dynamics.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus−infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization Coronavirus disease (COVID-19) weekly epidemiological and operational updates. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports
- 3.The Economist . seventh ed. 2021. What Is the Economic Cost of Covid-19? [Google Scholar]
- 4.Tahamtan A., Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev. Mol. Diagn. 2020;20:453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.da Silva S.J.R., Silva C.T.A.D., Guarines K.M., Mendes R.P.G., Pardee K., Kohl A., Pena L. Clinical and laboratory diagnosis of SARS-CoV-2, the virus causing COVID-19. ACS Infect. Dis. 2020;6:2319–2336. doi: 10.1021/acsinfecdis.0c00274. [DOI] [PubMed] [Google Scholar]
- 6.Islam K.U., Iqbal J. An update on molecular diagnostics for COVID-19. Front. Cell Infect. Microbiol. 2020;10:560616. doi: 10.3389/fcimb.2020.560616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trauger S.A., Junker T., Siuzdak G. Investigating viral proteins and intact viruses with mass spectrometry. Modern. Mass Spectrometry. 2003:265–282. [Google Scholar]
- 8.Downard K.M., Morrissey B., Schwahn A.B. Mass spectrometry analysis of the influenza virus. Mass Spectrom. Rev. 2009;28:35–49. doi: 10.1002/mas.20194. [DOI] [PubMed] [Google Scholar]
- 9.Ganova-Raeva L.M., Khudyakov Y.E. Application of mass spectrometry to molecular diagnostics of viral infections. Expert Rev. Mol. Diagn. 2013;13:377–388. doi: 10.1586/erm.13.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milewska A., Ner-Kluza J., Dabrowska A., Bodzon-Kulakowska A., Pyrc K., Suder P. Mass spectrometry in virological sciences. Mass Spectrom. Rev. 2020;39:499–522. doi: 10.1002/mas.21617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.NCBI Pubmed Central (PMC) search (23 February 2021) with terms “mass+spectrometry+coronavirus”. https://www.ncbi.nlm.nih.gov/pmc/
- 12.Yao H., Song Y., Chen Y., Wu N., Xu J., Sun C., Zhang J., Weng T., Zhang Z., Wu Z., Cheng L., Shi D., Lu X., Lei J., Crispin M., Shi Y., Li L., Li S. Molecular architecture of the SARS-CoV-2 virus. Cell. 2020;183:730–738. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F. Structure, function, and evolution of coronavirus spike. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye Q., West A.M.V., Silletti S., Corbett K.D. Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein. Protein Sci. 2020;29:1890–1901. doi: 10.1002/pro.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bar-On Y.M., Flamholz A., Phillips R., Milo R. SARS-CoV-2 (COVID-19) by the numbers. eLife. 2020;9 doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dollman N.L., Griffin J.H., Downard K.M. Detection, mapping, and proteotyping of SARS-CoV-2 coronavirus with high resolution mass spectrometry. ACS Infect. Dis. 2020;6:3269–3276. doi: 10.1021/acsinfecdis.0c00664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.L., Peiris M., Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nachtigall F.M., Pereira A., Trofymchuk O.S., Santos L.B. Detection of SARS-CoV-2 in nasal swabs using MALDI-MS. Nat. Biotechnol. 2020;38:1168–1173. doi: 10.1038/s41587-020-0644-7. [DOI] [PubMed] [Google Scholar]
- 19.Rocca M.F., Zintgraff J.C., Dattero M.E., Santos L.S., Ledesma M., Vay C., Prieto M., Benedetti E., Avaro M., Russo M., Nachtigall F.M., Baumeister E. A combined approach of MALDI-TOF mass spectrometry and multivariate analysis as a potential tool for the detection of SARS-CoV-2 virus in nasopharyngeal swabs. J. Virol. Methods. 2020;286:113991. doi: 10.1016/j.jviromet.2020.113991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iles R.K., Zmuidinaite R., Iles J.K., Carnell G., Sampson A., Heeney J.L. Development of a clinical MALDI-ToF mass spectrometry assay for SARS-CoV-2: rational design and multi-disciplinary team work. Diagnostics. 2020;10:746. doi: 10.3390/diagnostics10100746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikolaev E.N., Indeykina M.I., Brzhozovskiy A.G., Bugrova A.E., Kononikhin A.S., Starodubtseva N.L., Petrotchenko E.V., Kovalev G.I., Borchers C.H., Sukhikh G.T. Mass-spectrometric detection of SARS-CoV-2 virus in scrapings of the epithelium of the nasopharynx of infected patients via nucleocapsid N protein. J. Proteome Res. 2020;19:4393–4397. doi: 10.1021/acs.jproteome.0c00412. [DOI] [PubMed] [Google Scholar]
- 22.Downard K.M. Proteotyping for the rapid identification of influenza virus and other biopathogens. Chem. Soc. Rev. 2013;42:8584–8595. doi: 10.1039/c3cs60081e. [DOI] [PubMed] [Google Scholar]
- 23.Ihling C., Tänzler D., Hagemann S., Kehlen A., Hüttelmaier S., Arlt C., Sinz A. Mass spectrometric identification of SARS-CoV-2 proteins from gargle solution samples of COVID-19 patients. J. Proteome Res. 2020;19:4389–4392. doi: 10.1021/acs.jproteome.0c00280. [DOI] [PubMed] [Google Scholar]
- 24.Cazares L.H., Chaerkady R., Samuel Weng S.H., Boo C.C., Cimbro R., Hsu H.E., Rajan S., Dall'Acqua W., Clarke L., Ren K., McTamney P., Kallewaard-LeLay N., Ghaedi M., Ikeda Y., Hess S. Development of a parallel reaction monitoring mass spectrometry assay for the detection of SARS-CoV-2 spike glycoprotein and nucleoprotein. Anal. Chem. 2020;92:13813–13821. doi: 10.1021/acs.analchem.0c02288. [DOI] [PubMed] [Google Scholar]
- 25.Gouveia D., Grenga L., Gaillard J.C., Gallais F., Bellanger L., Pible O., Armengaud J. Shortlisting SARS-CoV-2 peptides for targeted studies from experimental data-dependent acquisition tandem mass spectrometry data. Proteomics. 2020;20 doi: 10.1002/pmic.202000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gouveia D., Miotello G., Gallais F., Gaillard J.C., Debroas S., Bellanger L., Lavigne J.P., Sotto A., Grenga L., Pible O., Armengaud J. Proteotyping SARS-CoV-2 virus from nasopharyngeal swabs: a proof-of-concept focused on a 3 Min mass spectrometry window. J. Proteome Res. 2020;19:4407–4416. doi: 10.1021/acs.jproteome.0c00535. [DOI] [PubMed] [Google Scholar]
- 27.Singh P., Chakraborty R., Marwal R., Radhakrishan V.S., Bhaskar A.K., Vashisht H., Dhar M.S., Pradhan S., Ranjan G., Imran M., Raj A., Sharma U., Singh P., Lall H., Dutta M., Garg P., Ray A., Dash D., Sivasubbu S., Gogia H., Madan P., Kabra S., Singh S.K., Agrawal A., Rakshit P., Kumar P., Sengupta S. A rapid and sensitive method to detect SARS-CoV-2 virus using targeted-mass spectrometry. J. Proteins Proteome. 2020;11:159–165. doi: 10.1007/s42485-020-00044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cardozo K.H.M., Lebkuchen A., Okai G.G., Schuch R.A., Viana L.G., Olive A.N., Lazari C.D.S., Fraga A.M., Granato C.F.H., Pintão M.C.T., Carvalho V.M. Establishing a mass spectrometry-based system for rapid detection of SARS-CoV-2 in large clinical sample cohorts. Nat. Commun. 2020;11:6201. doi: 10.1038/s41467-020-19925-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mengelle C., Mansuy J.M., Da Silva I., Guerin J.L., Izopet J. Evaluation of a polymerase chain reaction-electrospray ionization time-of-flight mass spectrometry for the detection and subtyping of influenza viruses in respiratory specimens. J. Clin. Virol. 2013;57:222–226. doi: 10.1016/j.jcv.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiu L., Zhang C., Wu Z., Peng J. Establishment and application of a universal coronavirus screening method using MALDI-TOF mass spectrometry. Front. Microbiol. 2017;8:1510. doi: 10.3389/fmicb.2017.01510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wandernoth P., Kriegsmann K., Groh-Mohanu C., Daeumer M., Gohl P., Harzer O., Kriegsmann M., Kriegsmann J. Detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by mass spectrometry. Viruses. 2020;12:849. doi: 10.3390/v12080849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shajahan A., Supekar N.T., Gleinich A.S., Azadi P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 2020;30:981–988. doi: 10.1093/glycob/cwaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Z., Ren K., Zhang X., Chen J., Jiang Z., Jiang J., Ji F., Ouyang X., Li L. Mass spectrometry analysis of newly emerging coronavirus HCoV-19 spike protein and human ACE2 reveals camouflaging glycans and unique post-translational modifications. Engineering (Beijing) 2020 doi: 10.1016/j.eng.2020.07.014. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouhaddou M., Memon D., Meyer B., White K.M., Rezelj V.V., Correa Marrero M., Polacco B.J., Melnyk J.E., Ulferts S., Kaake R.M., Batra J., Richards A.L., Stevenson E., Gordon D.E., Rojc A., Obernier K., Fabius J.M., Soucheray M., Miorin L., Moreno E., Koh C., Tran Q.D., Hardy A., Robinot R., Vallet T., Nilsson-Payant B.E., Hernandez-Armenta C., Dunham A., Weigang S., Knerr J., Modak M., Quintero D., Zhou Y., Dugourd A., Valdeolivas A., Patil T., Li Q., Hüttenhain R., Cakir M., Muralidharan M., Kim M., Jang G., Tutuncuoglu B., Hiatt J., Guo J.Z., Xu J., Bouhaddou S., Mathy C.J.P., Gaulton A., Manners E.J., Félix E., Shi Y., Goff M., Lim J.K., McBride T., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., De Wit E., Leach A.R., Kortemme T., Shoichet B., Ott M., Saez-Rodriguez J., tenOever B.R., Mullins R.D., Fischer E.R., Kochs G., Grosse R., García-Sastre A., Vignuzzi M., Johnson J.R., Shokat K.M., Swaney D.L., Beltrao P., Krogan N.J. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 2020;182:685–712. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao P., Praissman J.L., Grant O.C., Cai Y., Xiao T., Rosenbalm K.E., Aoki K., Kellman B.P., Bridger R., Barouch D.H., Brindley M.A., Lewis N.E., Tiemeyer M., Chen B., Woods R.J., Wells L. Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe. 2020;28:586–601. doi: 10.1016/j.chom.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S., Cinatl J., Münch C. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583:469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.V'kovski P., Steiner S., Thiel V. Proximity labeling for the identification of coronavirus-host protein interactions. Methods Mol. Biol. 2020;2203:187–204. doi: 10.1007/978-1-0716-0900-2_14. [DOI] [PubMed] [Google Scholar]
- 39.Fraser D.D., Slessarev M., Martin C.M., Daley M., Patel M.A., Miller M.R., Patterson E.K., O'Gorman D.B., Gill S.E., Wishart D.S., Mandal R., Cepinskas G. Metabolomics profiling of critically ill coronavirus disease 2019 patients: identification of diagnostic and prognostic biomarkers. Crit Care Explor. 2020;2 doi: 10.1097/CCE.0000000000000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gould O., Ratcliffe N., Król E., de Lacy Costello B. Breath analysis for detection of viral infection, the current position of the field. J. Breath Res. 2020;14 doi: 10.1088/1752-7163/ab9c32. [DOI] [PubMed] [Google Scholar]
- 41.Walker H.J., Burrell M.M. Could breath analysis by MS could be a solution to rapid, non-invasive testing for COVID-19? Bioanalysis. 2020;12(17):1213–1217. doi: 10.4155/bio-2020-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruszkiewicz D.M., Sanders D., O'Brien R., Hempel F., Reed M.J., Riepe A.C., Bailie K., Brodrick E., Darnley K., Ellerkmann R., Mueller O., Skarysz A., Truss M., Wortelmann T., Yordanov S., Thomas C.L.P., Schaaf B., Eddleston M. Diagnosis of COVID-19 by analysis of breath with gas chromatography-ion mobility spectrometry - a feasibility study. E-Clin. Med. 2020;29:100609. doi: 10.1016/j.eclinm.2020.100609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Silva I.W., Nayek S., Singh V., Reddy J., Granger J.K., Verbeck G.F. Paper spray mass spectrometry utilizing Teslin® substrate for rapid detection of lipid metabolite changes during COVID-19 infection. Analyst. 2020;145:5725–5732. doi: 10.1039/d0an01074j. [DOI] [PubMed] [Google Scholar]
- 44.Yang Y., Du Y., Kaltashov I.A. The utility of native MS for understanding the mechanism of action of repurposed therapeutics in COVID-19: heparin as a disruptor of the SARS-CoV-2 interaction with its host cell receptor. Anal. Chem. 2020;92:10930–10934. doi: 10.1021/acs.analchem.0c02449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El-Baba T.J., Lutomski C.A., Kantsadi A.L., Malla T.R., John T., Mikhailov V., Bolla J.R., Schofield C.J., Zitzmann N., Vakonakis I., Robinson C.V. Allosteric inhibition of the SARS-CoV-2 main protease: insights from mass spectrometry based assays. Angew Chem. Int. Ed. Engl. 2020;59:23544–23548. doi: 10.1002/anie.202010316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Downard K.M. Sequence-free phylogenetics with mass spectrometry. Mass Spectrom. Rev. 2020 doi: 10.1002/mas.21658. in press. Online November 10. [DOI] [PubMed] [Google Scholar]
- 47.Lun A.T., Swaminathan K., Wong J.W., Downard K.M. Mass trees: a new phylogenetic approach and algorithm to chart evolutionary history with mass spectrometry. Anal. Chem. 2013;85:5475–5482. doi: 10.1021/ac4005875. [DOI] [PubMed] [Google Scholar]
- 48.Mann C., Downard K.M. Evolution of SARS CoV-2 coronavirus surface protein investigated with mass spectrometry based phylogenetics. Anal. Lett. 2021;54 doi: 10.1080/00032719.2021.1928685. in press. [DOI] [Google Scholar]
- 49.Katz A.P., Civantos F.J., Sargi Z., Leibowitz J.M., Nicolli E.A., Weed D., Moskovitz A.E., Civantos A.M., Andrews D.M., Martinez O., Thomas G.R. False-positive reverse transcriptase polymerase chain reaction screening for SARS-CoV-2 in the setting of urgent head and neck surgery and otolaryngologic emergencies during the pandemic: clinical implications. Head Neck. 2020;42:1621–1628. doi: 10.1002/hed.26317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wikramaratna P.S., Paton R.S., Ghafari M., Lourenço Estimating the false-negative test probability of SARS-CoV-2 by RT-PCR. Euro Surveill. 2020;25:2000568. doi: 10.2807/1560-7917.ES.2020.25.50.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feng W., Newbigging A.M., Le C., Pang B., Peng H., Cao Y., Wu J., Abbas G., Song J., Wang D.B., Cui M., Tao J., Tyrrell D.L., Zhang X.E., Zhang H., Le X.C. Molecular diagnosis of COVID-19: challenges and research needs. Anal. Chem. 2020;92:10196–10209. doi: 10.1021/acs.analchem.0c02060. [DOI] [PubMed] [Google Scholar]
- 52.Fung B., Gopez A., Servellita V., Arevalo S., Ho C., Deucher A., Thornborrow E., Chiu C., Miller S. Direct comparison of SARS-CoV-2 analytical limits of detection across seven molecular assays. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.01535-20. e01535-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu L., Yuan Z.C., Li Z.M., Huang Z., Hu B. In vivo solid-phase microextraction swab sampling of environmental pollutants and drugs in human body for nano-electrospray ionization mass spectrometry analysis. Anal. Chim. Acta. 2020;1124:71–77. doi: 10.1016/j.aca.2020.05.022. [DOI] [PubMed] [Google Scholar]
- 54.Patterson E.I., Prince T., Anderson E.R., Casas-Sanchez A., Smith S.L., Cansado-Utrilla C., Solomon T., Griffiths M.J., Acosta-Serrano A., Turtle L., Hughes G.L. Methods of inactivation of SARS-CoV-2 for downstream biological assays. J. Infect. Dis. 2020;222:1462–1467. doi: 10.1093/infdis/jiaa507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maleknia S.D., Downard K.M. Mass spectrometry in structural proteomics: the case for radical probe protein footprinting. Trends Anal. Chem. 2019;110:293–302. [Google Scholar]