Abstract
Introduction
Pregnant women are increasingly being prescribed and abusing opioid drugs. As the primary communication organ between mother and conceptus, the placenta may be vulnerable to opioid effects but also holds the key to better understanding how these drugs affect long-term offspring health. We hypothesized that maternal treatment with oxycodone (OXY), the primary opioid at the center of the current crisis, would deleteriously affect placental structure and gene expression patterns.
Methods
Female mice were treated daily with 5 mg OXY/kg or saline solution (Control, CTL) for two weeks prior to breeding and until placenta were collected at embryonic age 12.5. A portion was fixed for histology, and the remainder was frozen for RNA isolation followed by RNAseq.
Results
Maternal OXY treatment reduced parietal trophoblast giant cell (pTGC) area and decreased the maternal blood vessel area within the labyrinth region. OXY exposure affected placental gene expression profiles in a sex dependent manner with female placenta showing up-regulation of many placental enriched genes, including Ceacam11, Ceacam14, Ceacam12, Ceacam13, Prl7b1, Prl2b1, Ctsq, and Tpbpa. In contrast, placenta of OXY exposed males had alteration of many ribosomal proteins. Weighted correlation network analysis revealed that in OXY females vs. CTL females, select modules correlated with placental histological changes induced by OXY. Such associations were lacking in the male OXY vs. CTL male comparison.
Discussion
Results suggest OXY exposure alters placental histology. In response to OXY exposure, female placenta responds by upregulating placental enriched transcripts that are either unchanged or downregulated in male placenta. Such changes may shield female offspring from developmental origins of health and disease-based diseases.
Keywords: Opioid, Drug Abuse, Analgesic, Trophoblast, Reproduction, DOHaD, Sexual Dimorphism, Placental Lactogens, Trophoblast Giant Cells
1. Introduction
Opioid drugs are increasingly prescribed analgesic agents to control pain. In 2016 alone, prescription opioid pain relievers were abused by approximately four percent of the United States population [1]. This abuse is considered one of the greatest non-infectious disease public health and economic challenges facing our nation [1]. Oxycodone (OxyContin) is at the epicenter of the current health crisis. Opioid use disorder (OUD), as it has come to be coined, is especially a problem in women of child-bearing age. A Substance Abuse and Mental Health Services Administration (SAMHSA) report revealed female treatment admissions for opioid pain relievers far outnumbered male admissions in all age categories [2].
The current rate of OUD during pregnancy is approximately 5.6 per 1000 live births [3]. One report suggested that more than 85% of pregnancies with women plagued with OUD were left untreated [4]. Pregnant women with OUD had delayed and reduced rates of prenatal care compared to women without substance use disorders [5]. Moreover, neonates prenatally exposed to opioids are at risk for neonatal abstinence syndrome (NAS, or also referred to as neonatal opioid withdrawal syndrome) [6]. Birth outcomes associated with maternal OUD include poor fetal growth, potential premature birth, low birthweight, and possible congenital defects [7, 8]. One study showed that such infants had elevated neonatal intensive care unit (NICU) admission rates and required longer hospitalization periods [5]. Thus, overall healthcare costs for women with OUD and their infants is substantially greater than those without this disease [5]. Even if infants born to mothers with OUD do not demonstrate outright health problems at birth, they may be at greater risk for later diseases due to developmental origin of health and disease (DOHaD) effects [9, 10]. Thus, it is important to understand how such drugs reach the fetus and how the conceptus responds when confronted with opioids, such as oxycodone.
The placenta is in direct contact with maternal tissue, and thus, anything circulating in the bloodstream has to go through the placenta to reach the fetus. While transient, the placenta has intimate contact with the underlying uterine tissue and assumes a variety of roles in nutrient, gas, and waste exchange. In eutherian mammals, it produces a range of hormones and cytokine factors that promote and buffer the needs of the fetus in the face of uterine challenges. Rodents and humans that have an invasive hemochorial type of placentation have syncytiotrophoblast cells that are involved in nutrient and gas exchange and are immersed in maternal blood [11]. This proximity though renders such placental cells susceptible to drugs circulating within the maternal blood stream. While the placenta has the ability to detoxify certain xenobiotic chemicals and thereby offer some protection for the fetus, as an organ that responds to hormones and other factors, it can also be affected by such exogenous compounds. It is also clear that how the placenta responds to environmental challenges is sexually dimorphic in nature [11-13].
Interestingly, the placenta has a tightly regulated opioid system that has been suggested to be integral in regulating placental factors regulating maternal recognition of pregnancy, such as hCG and placental lactogens [14-18]. Thus, maternal opioid treatment can tamper with this key regulatory pathway in the placenta. Only a handful of studies though have examined how opioids, and oxycodone in particular, affect the placenta or individual trophoblast cells [19-22]. The collective findings suggest cause for concern that opioids can disrupt placental steroid hormone production, human chorionic gonadotropin (hCG) production, and the expression of receptors essential for normal placental function.
To address some of the critical gaps in our knowledge, we sought to examine how maternal exposure to oxycodone affects the global transcriptome and histological structure of the mouse placenta, which have similar features and trophoblast cell-types as the human placenta [11-13]. Both species have an invasive and hemochorial form of placentation where the trophoblast cells are bathed in maternal blood. With its short gestational period, the mouse is also the ideal model for studying how various in utero environmental changes affect placental function. Our overarching hypothesis was that such maternal treatment would disrupt the genetic machinery and lead to associated architectural changes in the placenta. To test this hypothesis, female mice were exposed two weeks prior to breeding (periconceptional period) and up until the time the placenta were collected at embryonic age E12.5, which is considered mid-gestation and a period where we, and others, have shown that various in utero environmental changes have shown to affect placental structure and function [12, 23-26]. A high throughput RNA-seq method was used to examine the global transcriptomic changes affected by oxycodone in male and female conceptuses. Additionally, we considered whether such treatment might abolish normally sexually dimorphic differences in gene expression. Histomorphometrics were used to assess the various regions of the placenta, namely the labyrinth, spongiotrophoblast, and parietal trophoblast giant cell (pTGC) region. Weighted correlation network analysis (WGCNA) [27] was used to link gene expression changes and histological (phenotypic) changes, as we have done previously [28-30].
2. Materials and methods
2.1. Animals and treatments
All animal experiments were approved by the University of Missouri Animal Care and Use Committee (Protocol #9590) and conformed to the NIH Guidelines for the Care and Use of Laboratory Animals. Seven-week-old male and female CF1 mice were ordered from Envigo (Madison, WI). Females were habituated for one week prior to being placed on one of two treatments. Female mice were randomly assigned to be in the oxycodone (OXY, Catalogue # O1378; Sigma Chemical, St. Louis, MO) treatment or saline control (CTL) group, and each group initially included 15 females. At 8 weeks of age, the OXY group received 5 mg OXY/kg body weight in 0.9% saline with an average volume of 0.099 ml injected intraperitoneally (IP) daily for two weeks prior to breeding and until the placentas were collected at embryonic age (E) 12.5. During this time, the CTL group received comparable IP injections of 0.9% saline. Females were weighed weekly throughout the course of the experiment, and the dose of OXY was accordingly adjusted to maintain giving 5 mg/kg IP daily injection. This dose and route of administration (IP) was chosen based on previous studies that examined the effects of adult rodent exposure to OXY and verified that such concentrations mimics those for humans with OUD [31-33]. To date, most studies that have examined the direct effects of OXY have used either IP injections or added it to the water. While addition to the water would be less stressful, a concern with this approach is that there could then be variability in exposures across animals. No adverse health effects were noted in mice given OXY or saline alone IP injections. The treatments were initiated two weeks prior to breeding to encompass the periconceptional period, which may be important in pre-implantation embryo and initial placental development [12, 23]. Animals were provided food and water ad libitum, and fed AIN93G phytoestrogen-free diet (Envigo, Madison, WI) to reduce any exogenous hormone exposure.
2.2. Breeding and placental collections
After two weeks of being treated daily with either OXY or CTL, females were paired with potential CF1 breeder males and checked the next morning for a vaginal plug. When a vaginal plug was observed, this was considered E 0.5 days post-coitus. If no vaginal plug was observed in the morning, the males were placed in separate cages and then re-paired that evening. Females were continued on the respective treatments until they were humanely euthanized at E 12.5. At this time, each uterine horn was incised, and the uterine position of each conceptus was delineated. Fetal tissue was collected for PCR sexing. Half of the fetal placental tissue (as determined by the discoid morphology) where the underlying uterine tissue was dissected away was frozen in liquid nitrogen and the other half of the fetal placenta with underlying uterine tissue was fixed in 4% paraformaldehyde (PFA) for histological analyses. The fetal and placental collections and preservation were done as described previously [12, 23].
2.3. Fetal PCR sexing
To determine the sex of each conceptus, DNA was isolated with the DNeasy Blood & Tissue Kit (Catalogue #69504; Qiagen, Gaithersburg, MD) from fetal tissue. Polymerase Chain Reaction (PCR) amplification was then performed for Sry (Y-chromosome specific gene, forward primer: 5′TCATGAGACTGCCAACCACAG3′; reverse primer: 5′CATGACCACCACCACCACCAA3′) and myogenin (Myog- autosomal control gene; forward primer: 5′TTACGTCCATCGTGGACAGC3′; reverse primer: 5′TGGGCTGGGTGTTAGTCTTA3′), as detailed in [34]. Once the sex of the conceptuses was established, one male and one female placental pair from each litter and preferably in the middle of the uterine horns were selected for further analyses. For the CTL group, 10 female and 10 male placentas were tested, and for the OXY group, 11 female and 11 male placentas were tested. These placentas were chosen based on those that had an RNA integrity number (RIN) score > 7.0, as detailed below.
2.4. Placental histological analyses
Before embedding for histology, placenta tissue was marked with tattoo ink to aid in orientation. All attempts were made to obtain a cross section through all fetal placental layers. In those sections, where such layers were only partially included or missing, the tissue block was deparaffinized and re-orientated until such sections could be obtained. In those placentas where adequate orientation was not achieved, they were replaced with same sex placental samples from the same dam. Sections of placenta for histology were fixed overnight in a 4% PFA solution at 4 °C. They were then subjected to three washes in 1X phosphate buffered saline (PBS) before being placed in histology cassettes (Fisher Scientific, St. Louis. MO) and stored in 70% ethanol until being processed. Histological sections (4-5 μm thick) were cut at the IDEXX Co. Histology Laboratory (Columbia, MO) and stained with PAS-Hematoxylin (PAS-H). Histological slides were viewed, and images photographed under a Leica DM 5500B (Wetzlar, Germany) upright microscope with Leica DFC290 color digital camera at the University of Missouri Cytology Core Facility. Images taken from same tissue sections were stitched together by using Leica Application Suite software (V4.12, Leica). Morphometric analyses on the obtained images were performed with the Adobe Photoshop program (v2020, San Jose, CA). The labyrinth (LA), spongiotrophoblast (spongioTB), and parietal trophoblast giant cell (pTGC) areas were measured within each placental section and compared relative to each other to control for differences in size across sections examined. The number of apoptotic pTGC cells was also determined. Apoptosis of these cells was defined as having a shrunken and pyknotic nucleus with dense nuclear aggregations [35, 36].
For maternal blood vessel area measurement within the labyrinth (LA) region, a box measuring 500 × 500 pixels was placed over 5 to 7 random LA areas for each placenta section. Ten maternal blood vessels in this area were randomly selected, the edge of the blood vessel was outlined, and the area was measured with the Adobe Photoshop program (Adobe, San Jose, CA).
2.5. Placental RNA isolation
RNA was isolated from each of the selected placental samples with the Qiagen AllPrep DNA/RNA/miRNA Universal Kit (Catalogue #80224; Qiagen, Germantown, MD). The quantity and quality of the RNA was determined with a Nanodrop ND1000 spectrophotometer (Nanodrop Products, Wilmington, DE). The results were further confirmed by analyzing the RNA on the Fragment Analyzer (Advanced Analytical Technologies, Ankeny, IA). Only those samples that had an RIN score > 7.0 were selected for RNA sequencing (RNAseq).
2.6. Illumina TruSeq RNA library preparation and sequencing
Libraries were constructed per the manufacturer’s protocol with reagents supplied in Illumina’s TruSeq mRNA Stranded Library Preparation Kit and sequenced at the University of Missouri DNA Core Facility. In brief, poly-A containing mRNA was purified from total RNA (850 ng) using poly-T oligo beads, mRNA was fragmented, and double-stranded cDNA generated from cleaved RNA using random hexamers. cDNA then underwent end repair and adapter ligation followed by PCR amplification. Unique dual indexing was used to mitigate index hopping events known to be an issue on patterned flow cells with exclusion amplification chemistry.
The size and purity of the final library was determined using the Fragment Analyzer (Agilent Technologies, Inc.), quantified with the Qubit fluorometer by means of the Qubit dsDNA HS Assay kit (Invitrogen), and diluted according to Illumina’s standard sequencing protocol. Libraries were pooled and run on an Illumina NovaSeq 6000 sequencer using a paired end 50 bp read format on a S1flow cell to generate ~35 million paired reads per sample. The actual number of reads obtained for each sample is listed in Supplementary Table 1.
2.7. RNAseq data processing
The reads were trimmed with cutadapt (Version 2.8), and reference: DOI:10.14806/ej.17.1.200 for Illumina adapters, for ambiguous nucleotides (N’s), and for artificial poly-G [37] for reads whose 3' ends overlap with the adapter for a minimum of 3 bases with 90% identity. After trimming, reads with fewer than ten bases were discarded. The filtered trimmed reads were aligned to the reference mouse genome (GRCm38) by HISAT2 version 2.0.5 to achieve a high overall alignment (~97%) [38]. The aligned reads were further filtered to remove reads that mapped to the mitochondrial genome and those that aligned to pseudochromosomes. The number of reads that aligned to each protein-coding gene were counted with the featureCount tool from the Subread software suite Version 2.0.0 [39].
2.8. Differential gene expression analysis (DGEA): DESeq2
The raw read counts were used to carry out differential gene expression analysis (DGEA) by means of DESeq2 to study the effects of maternal oxycodone exposure on the placental transcriptome [40]. The genes with an average of less than 5 read counts in at least one group were filtered out before carrying out DGEA. Genes were considered upregulated if they had an absolute fold-change ≥ 1.5 and adjusted p-value ≤ 0.05.
2.9. Tissue-specific gene enrichment analysis
Tissue-specific gene enrichment analysis was determined by TissueEnrich [41]. We used the mouse ENCODE [42] dataset to carry out the enrichment analysis with default settings. Enrichments were considered significant if P was ≤ 0.01 and fold-change ≥ 2.
2.10. Functional enrichment analysis
Protein-protein interactions (PPI) for proteins encoded by DEG for the various comparisons: OXY females vs. CTL females, OXY males vs. CTL males, CTL males vs. CTL females, and OXY males vs. OXY females were determined with the STRING Database [43]. The PPI files generated with STRING were imported into the cytohubba app [44] in Cytoscape [45] to examine for the top 10 hub genes. Within this program, hub genes were determined with MCC analysis, as recommended [44]. For functional enrichment analysis, DEG were imported in g:GOSt (https://biit.cs.ut.ee/gprofiler/gost), and the significance threshold used was FDR ≤ 0.01 (calculated using the Benjamini-Hochberg method). The output included all significant GO terms and pathways (KEGG, Reactome, and WikiPathways).
2.11. qPCR validation
Eight placental enriched genes that were DE in OXY female vs. CTL female placenta were analyzed by qPCR analysis. Several of these were also considered hub genes for this comparison. Total RNA, which had been treated with DNase to remove any genomic DNA contamination, was reverse transcribed into cDNA by using the QuantiTect Reverse Transcription Kit (Catalogue #205310, Qiagen). The qPCR procedure was performed on the Bio-Rad CFX Connect Real-Time PCR Detection System (Bio-Rad). Primers were designed by using NCBI Primer-Blast online (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) and purchased from IDT technologies (Coralville, IA). Primer sequences and efficiencies for the genes examined are listed in Supplementary Table 2. The Bio-Rad SYBR Green Master Mix (Catalogue # 1725121, Hercules, CA) was used according to the manufacture’s protocol. The cycling conditions for qPCR were 1) 95°C for 5 min for polymerase activation 2) 40 cycles of; denaturation 15 seconds at 95 °C, annealing and extension 30 seconds at 56 °C, and 3) dissociation melt curve analysis of denaturation at 95°C, complete annealing at 56 °C, followed by a gradual increase in temperature up to 95°. Both ubiquitin C (Ubc) and glyceraldehyde 3-phosphate dehydrogenase (Gapdh) genes were used as internal reference controls. Ubc has been shown to be an appropriate reference control for the mouse placenta [46]. We included the two internal reference controls that showed strong similarity in Ct results to increase the strength and confidence in the comparisons.
2.12. Statistical analyses for placental histology
Fetal sex ratio data were analyzed by using the generalized linear model included in SAS version 9.4 (PROC GENMOD procedure, SAS Institute, Cary, NC). The data were distributed as a binomial and transformed by means of a logit-link function. The χ2 test was used to test deviation from a 1:1 ratio (i.e., a value of 0.5 for the fraction of male fetuses), as well as for differences in sex ratio between groups. The antilog of the logit and the antilog of the differences between logit estimates produced the odds and odds ratio, respectively. The χ2 test was also used to test for differences in pregnancy rates between CTL and OXY groups.
Data for dependent variables of maternal blood vessel size within the labyrinth region, pTGC apoptosis, ratios between LA, spongioTB, and pTGC areas in placenta were analyzed for normality by using the Wilk-Shapiro test (V9.4; SAS Analytics, Cary, NC). Maternal blood vessel size within the labyrinth region and ratio data were log transformed to approach a normal distribution. These data were then analyzed by the PROC MIXED procedure of SAS v9.4. Sources of variation considered were treatment, sex, and treatment × sex interaction with an individual mouse serving as the experimental unit to determine treatment effects. For all data, a P value of ≤0.05 was considered significant. All data are presented as actual means (). The error bars for all figures and reported data represent the standard error of the mean (SEM).
Gene expression data as determined by qPCR analyses were normalized by using combined average dCt values of the two housekeeping genes: Gapdh and Ubc and then analyzed based on treatment X sex interactions with the PROC GLM procedure of SAS. Graphs are based on 2−ΔΔCt values relative to CTL values for each sex with the mean value for these CTL groups set to 1 for graphing purposes. For all data, a p-value of ≤ 0.05 was considered significant. All data are presented as means ± standard error of the means (SEM).
2.13. Weighted correlation network analysis (WGCNA)
WGCNA describes correlation patterns among genes based on their gene expression data. The WGCNA package in R [27] was used to find unsigned weighted gene co-expression modules. The blockwiseModules function was run with a soft thresholding power of 18 to indicate the gene cluster. In order to identify significant gene modules, correlation scores between genes and traits were used to rank the gene cluster, which utilized the eigengene network methodology by using Pearson Correlation/Bicor. Trait data used for this analysis was LA to pTGC ratio, SpongioTB to pTGC ratio, pTGC apoptosis, and maternal blood vessel size in the labyrinth region. Genes within module eigengenes (ME) that were significant were further analyzed as detailed for functional enrichment analyses for DE genes.
3. Results
3.1. Pregnancy rate, number of conceptuses, and sex ratio of conceptuses
To examine whether maternal OXY exposure affected general pregnancy success rate, total fetal implantation, or led to skewing in conceptus sex ratio by E12.5, these parameters were compared between the two groups. No differences were detected in pregnancy success rate, number of conceptuses, or conceptus sex ratio at this stage of gestation (Supplementary Table 3). No fetal abnormalities were detected at E12.5 in either group.
3.2. Placental histological changes induced by maternal OXY treatment
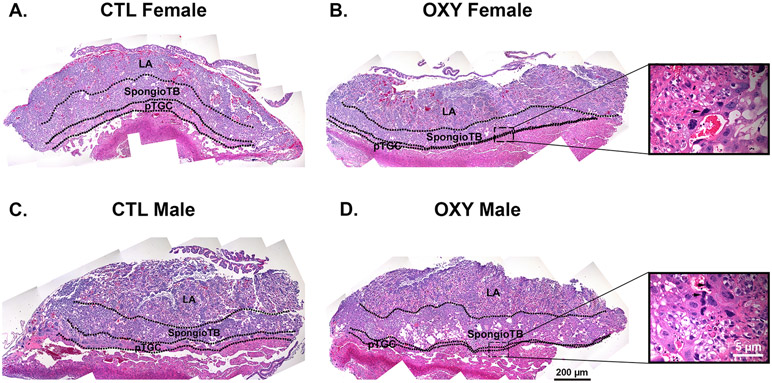
The placental histology from CTL females, OXY females, CTL males, and OXY males is shown in Fig. 1. As we, and others, have found other chemicals, such as bisphenol A (BPA) and bisphenol S (BPS) can affect the distribution of the various placental layers [23, 47, 48], we first sought to quantify the ratio of the LA to pTGC, LA to SpongioTB, and spongioTB to pTGC area. No differences were detected in the LA to spongioTB area. However, noticeable differences were observed in the LA to pTGC area and spongioTB to pTGC area in females and males. In CTL females and OXY females, the LA to pTGC ratio was 5.79 ± 0.56 vs. 19.70 ± 3.63, respectively (p < 0.001, Fig. 2A). In CTL males and OXY males, the LA to pTGC ratio was 8.23 ± 0.76 vs. 19.46 ± 2.5, respectively (p < 0.001, Fig. 2A). Comparison of the spongioTB to pTGC ratio in CTL females vs. OXY females was similarly elevated in the latter (2.55 ± 0.21 vs. 9.14 ± 1.48, respectively; p < 0.001; Fig. 2B). This same pattern change in this ratio was observed in CTL males vs. OXY males (3.25 ± 0.30 vs. 9.15 ± 1.46, respectively, p < 0.01; Fig. 2B). The collective findings suggest that in both the female and male placenta, maternal OXY treatment reduces either the differentiation of trophoblast cells allocated to the pTGC compartment and/or causes apoptosis of these cells.
Fig. 1.
Histology of the placenta in CTL and OXY exposed females and males. A) CTL female placenta. B) OXY female placenta. Inset shows apoptotic pTGC (arrow). C) CTL male placenta. D) OXY male placenta. Inset shows apoptotic pTGC designated (arrows). LA = labyrinth; SpongioTB = spongiotrophoblast; pTGC = parietal trophoblast giant cells.
Fig. 2.
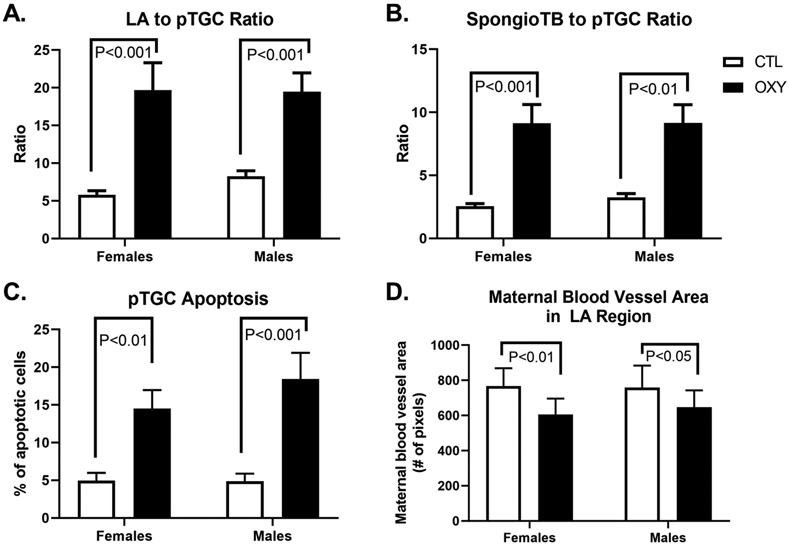
Quantification of placental histological changes. A) Labyrinth (LA) to parietal trophoblast giant cell (pTGC) area. B) Spongiotrophoblast (SpongioTB) to pTGC area. C) Number of apoptotic pTGC. D) Maternal blood vessel area in LA region. P value differences are designed in each of the graphs. For the CTL group, 10 females and 10 males were tested, and for the OXY group, 11 females and 11 males were tested.
We thus determined the number of apoptotic pTGC cells based on shrunken and pyknotic nuclei. pTGC apoptosis was increased in OXY females relative to CTL females (14.50 ± 2.47 vs. 4.94 ± 1.05, respectively; p < 0.01; Fig. 2C). Similarly, OXY males had greater number of apoptotic pTGC compared to CTL males (18.43 ± 3.48 vs. 4.87 ± 1.02; p < 0.001; Fig. 2C). Lastly, we examined the amount of maternal blood vessel penetration into the LA region as determined by pixel numbers. CTL females had greater area of the LA region occupied by maternal blood vessels relative to OXY females (786.3 ± 100.6 vs. 600.4 ± 91.7 pixels, respectively; p < 0.01; Fig. 2D). Similar differences were observed in CTL males compared to OXY males (758.7 ± 125.0 vs. 647.8 ± 95.3, p < 0.05; Fig. 2D). Higher magnification views of maternal blood vessels within the LA region for each group are shown in Supplementary Fig. 1.
3.2. Transcriptomic analyses of mouse placental tissues exposed to OXY or CTL
RNAseq was performed on placentas recovered from OXY treated or CTL mouse dams at E 12.5. The average number of paired-end reads was 40,484,781, the average percent of alignment was 95.64% and the average number of mapped paired-end reads was 38,725,330 (Supplementary Table 1).
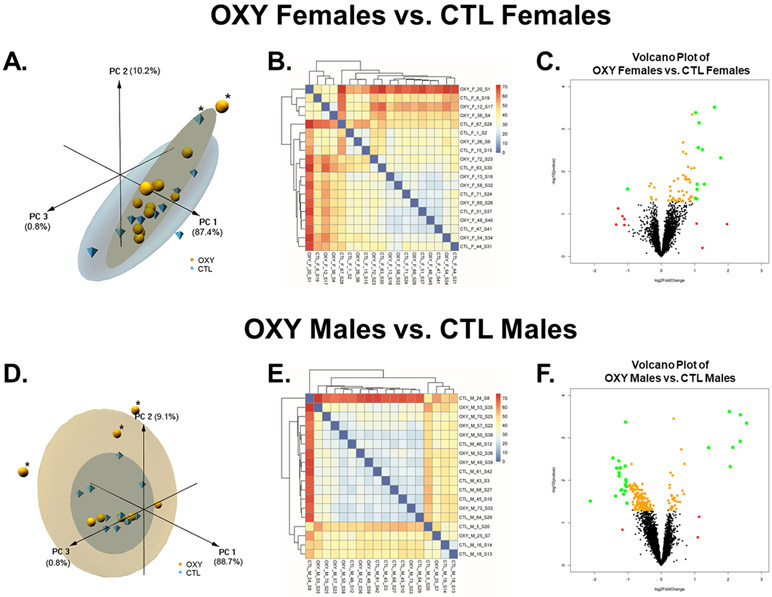
Effects of maternal OXY exposure based on fetal sex was first considered. Principal Component Analyses (PCA) of OXY female exposed vs. CTL female placentas revealed that the PC1, PC2, and PC3 axes accounted for 87.4, 10.2, and 0.8%, respectively, of the variation (Fig. 3A). This analyses also revealed that one OXY female and one CTL female placenta were outliers. These samples were thus not considered in subsequent analyses. While no clear separation was detected in the PCA and heatmap analyses based on all genes (Fig. 3A and 1B), volcano plot analyses done after outliers detailed above were removed, revealed select genes that were considered DE (Fig. 3C). Comparison of OXY male vs. CTL male placenta with PCA analyses revealed that the PC1, PC2, and PC3 axes accounted for 88.7, 9.1, and 0.8%, respectively, of the variation (Fig. 3D). This analyses also revealed that 3 OXY male placentas were considered outliers and removed from further analyses. No clear separation between OXY male vs. CTL male placentas was identified based on PCA or heatmap analyses with all identified genes (Fig. 3D and E). Volcano plot analyses revealed though select genes that were DE (Fig. 3F).
Fig. 3.
Effects of oxycodone on placental gene expression in female and male mouse conceptuses. A) 3D PCA plot of RNAseq results from placenta of OXY females vs. CTL females. B) Heatmap based on all transcripts identified in placenta of OXY females vs. CTL females d. C) Volcano plot analyses of gene transcripts identified in placenta of OXY females vs. CTL females. Transcripts represented by Orange: p < 0.05; Transcripts represented by Red: log2FoldChange < −1 or > 1; Transcripts represented by Green: p < 0.05 AND log2FoldChange < −1 or > 1. D) 3D PCA plot of RNAseq results from placenta of OXY males vs. CTL males. E) Heatmap based on all transcripts identified in placenta of OXY males vs. CTL males. F) Volcano plot analyses of gene transcripts identified in placenta of OXY males vs. CTL males. Transcripts represented by Orange: p < 0.05; Transcripts represented by Red: log2FoldChange < −1 or > 1; Transcripts represented by Green: p < 0.05 AND log2FoldChange < −1 or > 1. For the CTL group, 10 females and 10 males were initially tested, and for the OXY group, 11 females and 11 males were initially tested. However, the PCA analysis revealed that one OXY female and one CTL female placenta were outliers. These samples were thus not considered in subsequent analyses. Likewise, this analysis revealed that 3 OXY male placentas were outliers and removed from further RNAseq analyses.
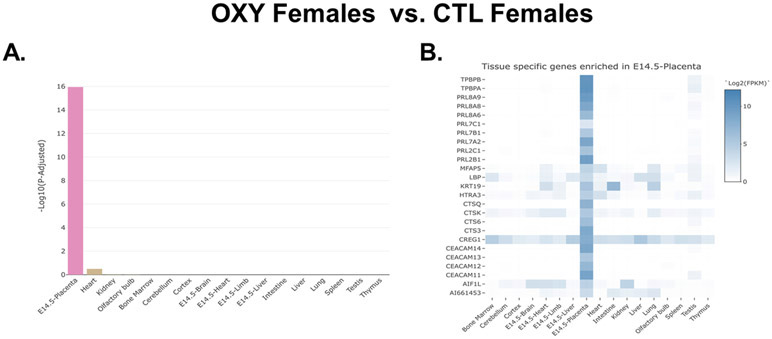
The full list of all genes identified based on OXY female vs. CTL female placenta is provided in Supplementary File 1. As shown in this file, there were genes DE based on p value. However, no genes were DE based on a false discovery rate (FDR). The reason for this finding is not clear as our mapped reads and % efficiency are considered more than sufficient for surveying eukaryotic transcriptomes [28, 49]. Moreover, even after excluding potential outliers, the data analyses still includes 10 OXY female and 9 CTL female placenta, and this number of replicates or even less has been sufficient in our previous placental transcriptome studies to detect gene expression differences [12, 23]. In examining those genes considered DE based on p values, we noticed that many of them are important for placental function, and thus decided to further investigate them. The top 25 DE genes based on p values in placentas of OXY females vs. CTL females are listed in Table 1. Next, we used the TissueEnrich program [42] to determine whether these DE genes had placenta-specific gene expression patterns. Indeed, this program revealed that DE genes based on OXY female vs. CTL female placenta were overwhelmingly confined to the placenta (Fig. 4A). Heatmap analysis with this program showed that various DE genes based on this comparison, including Tpbpb, Tpbpa, various prolactin genes, such Prl8a9, Prl8a8, Prl8a6, Prl7c1, and other Prl isoforms, Cts6q, Ceacam14, Ceacam13, Ceacam12, and Ceacam11, that are enriched in the mouse placenta (Fig. 4B). Most of these were upregulated in the placenta of OXY exposed females. It should be noted that TissueEnrich is a web tool that first determines if a gene has tissue-specific expression by comparing its expression across a large number of tissues. In order to do so, the tool must use consortium data that was generated across different tissues. For mouse, TissueEnrich includes data from the mouse ENCODE project, which only has data from E14.5 placenta. We believe this data can be used in this study for two reasons. First, E12.5 and E14.5 are both considered mid-gestation by which time a mature placenta has formed. Second, according to TissueEnrich, the genes shown in Fig. 4 have higher expression in E14.5 placenta compared to other mouse tissues, and from our data, we know that these genes are also expressed at E12.5.
Table 1.
Top 25 DE genes in placenta of OXY females vs. CTL females. Bolded and shaded genes are also considered hub genes.
| Gene ID | Gene Name | Fold change |
P value | Expression in OXY Females |
|---|---|---|---|---|
| Prl7b1 | prolactin family 7, subfamily b, member 1 (Prl7b1) | 3.015145 | 0.000305 | ↑ |
| Ipp | IAP promoted placental gene (Ipp) | 2.04192 | 0.000411 | ↑ |
| Prl8a6 | prolactin family 8, subfamily a, member 6 (Prl8a6) | 1.900212 | 0.000452 | ↑ |
| Prl8a8 | prolactin family 8, subfamily a, member 81 (Prl8a8) | 2.190423 | 0.00071 | ↑ |
| Psg-ps1 | pregnancy specific glycoprotein pseudogene 1 (Psg-ps1) | 1.575866 | 0.002067 | ↑ |
| Ceacam12 | carcinoembryonic antigen-related cell adhesion molecule 12 (Ceacam12) | 2.148957 | 0.00272 | ↑ |
| Prl7a2 | prolactin family 7, subfamily a, member 2 (Prl7a2) | 2.344508 | 0.003038 | ↑ |
| Rgs5 | regulator of G-protein signaling 5 (Rgs5) | 1.577403 | 0.00343 | ↑ |
| Creg1 | cellular repressor of E1A-stimulated genes 1 (Creg1) | 1.669612 | 0.003814 | ↑ |
| S100a8 | S100 calcium binding protein A8 (calgranulin A) (S100a8) | 3.434036 | 0.004753 | ↑ |
| Ceacam14 | carcinoembryonic antigen-related cell adhesion molecule 14 (Ceacam14) | 1.746187 | 0.007763 | ↑ |
| Prl7c1 | prolactin family 7, subfamily c, member 1 (Prl7c1) | 1.983514 | 0.008284 | ↑ |
| Tpbpb | trophoblast specific protein beta (Tpbpb) | 1.867087 | 0.008407 | ↑ |
| Prl8a9 | prolactin family8, subfamily a, member 9 (Prl8a9) | 1.57127 | 0.012147 | ↑ |
| Ceacam13 | carcinoembryonic antigen-related cell adhesion molecule 13 (Ceacam13) | 1.653706 | 0.012975 | ↑ |
| Igfbp7 | insulin-like growth factor binding protein 7 (Igfbp7) | 1.412147 | 0.014668 | ↑ |
| Ctsq | cathepsin Q (Ctsq) | 1.638126 | 0.014715 | ↑ |
| Ceacam11 | carcinoembryonic antigen-related cell adhesion molecule 11 (Ceacam11) | 1.740848 | 0.014965 | ↑ |
| Gpx3 | glutathione peroxidase 3 (Gpx3) | 1.677336 | 0.01554 | ↑ |
| Tpbpa | trophoblast specific protein alpha (Tpbpa) | 1.785004 | 0.015593 | ↑ |
| Far1 | fatty acyl CoA reductase 1 (Far1) | 1.300561 | 0.016593 | ↑ |
| Prl2c1 | prolactin family 2, subfamily c, member 1 (Prl2c1) | 1.816418 | 0.01796 | ↑ |
| Igsf8 | immunoglobulin superfamily, member 8 (Igsf8) | 0.805401 | 0.019183 | ↓ |
| Ctsk | cathepsin K (Ctsk) | 2.442033 | 0.019817 | ↑ |
| Fabp4 | fatty acid binding protein 4, adipocyte (Fabp4) | 2.105421 | 0.019899 | ↑ |
Fig. 4.
TissueEnrich results for OXY female vs. CTL female placentas. A) Bar plot distribution of the tissue enrichment of all DE genes. B) Heatmap analyses revealing those DE genes that are enriched in the placenta. This analysis is based on the results from 10 OXY females and 9 CTL females.
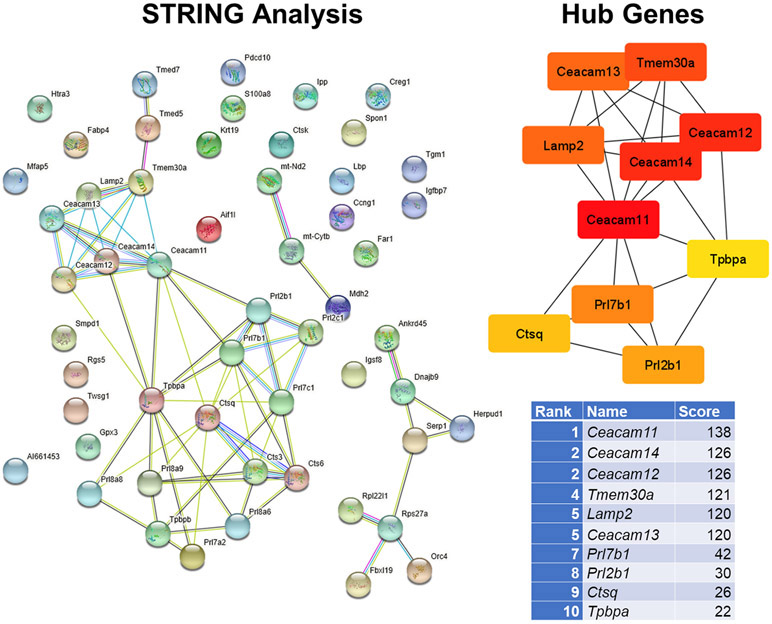
The STRING program was used to examine protein-protein-interactions based on the proteins encoded by the DE genes (Fig. 5). This information was then imported into the cytohubba app [44] in Cytoscape [45] to identify the top 10 hub genes. The STRING diagram reveals two distinct clusters of the above placental enriched genes (Fig. 4). Moreover, the top 10 hub genes based on OXY female vs. CTL female placentas were these same placental enriched genes, including Ceacam11, Ceacam14, Ceacam12, Ceacam13, Prl7b1, Prl2b1, Ctsq, and Tpbpa. Of the hub genes, 8 are enriched in the placenta as shown in Fig. 4. Seven of these hub genes are among the top 25 DE genes for the female group comparisons (Table 1).
Fig. 5.
STRING and hub gene analyses for OXY females vs. CTL females. PPI were determined by STRING analysis. The PPI files generated with STRING were imported into the cytohubba app [44] in Cytoscape [45] to examine for the top 10 hub genes. Within this program, hub genes were determined with MCC analysis, as recommended [44]. This analysis is based on the results from 10 OXY females and 9 CTL females.
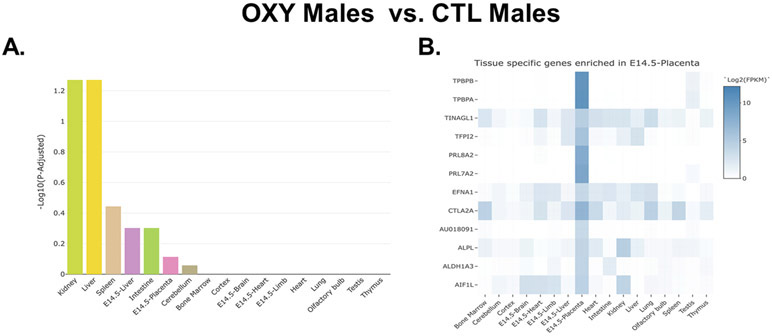
Genes identified based on the OXY male vs. CTL male comparisons were next considered. The full list of genes identified by this comparison are listed in Supplementary File 2. Several genes were considered DE based on p value, but as with the female comparison, none of these were considered significant based on an FDR. The top 25 DE based on p values are listed in Table 2. In contrast to the female group comparisons, the male group comparisons did not include many that appeared to be unique to the placenta. This finding was further supported when we analyzed this set of genes with the TissueEnrich program. Those that were DE in OXY males vs. CTL males were associated with the kidney and liver and less so with the placenta as further indicated by the fact that the enrichment values for the male comparison are less than those in the female comparison (Fig. 6A). Further examination revealed that were select genes enriched in the placenta, such as Tpbpb, Tpbpa, Prl8a2, and Prl7a2. Contrary to the female group comparisons, these placental enriched genes were downregulated in the placenta of OXY exposed males.
Table 2.
Top 25 DE genes in placenta of OXY males vs. CTL males. Genes that are bolded and shaded are also considered hub genes.
| Gene ID | Gene Name | Fold change |
P value | Expression in OXY males |
|---|---|---|---|---|
| Cubn | cubilin (intrinsic factor-cobalamin receptor) (Cubn) | 4.100144 | 0.000239 | ↑ |
| Apob | apolipoprotein B (Apob) | 5.172242 | 0.000285 | ↑ |
| Tpm3 | tropomyosin 3, gamma (Tpm3) | 1.292477 | 0.000352 | ↑ |
| Tfpi2 | tissue factor pathway inhibitor 2 (Tfpi2) | 0.476091 | 0.000424 | ↓ |
| Apoa4 | apolipoprotein A-IV (Apoa4) | 5.861951 | 0.000449 | ↑ |
| Afp | alpha fetoprotein (Afp) | 5.140322 | 0.001201 | ↑ |
| Ttr | transthyretin (Ttr) | 4.401619 | 0.001678 | ↑ |
| Lrp2 | low density lipoprotein receptor-related protein 2 (Lrp2) | 2.670474 | 0.001692 | ↑ |
| Ubc | ubiquitin C (Ubc) | 1.514529 | 0.001867 | ↑ |
| Usmg5 | upregulated during skeletal muscle growth 5 (Usmg5) | 0.366133 | 0.002957 | ↓ |
| Rps14 | ribosomal protein S14 (Rps14) | 0.41332 | 0.003472 | ↓ |
| Trf | transferrin (Trf) | 1.61551 | 0.004082 | ↑ |
| Prl7a2 | prolactin family 7, subfamily a, member 2 (Prl7a2) | 0.540139 | 0.00414 | ↓ |
| Col6a3 | collagen, type VI, alpha 3 (Col6a3) | 0.472914 | 0.004608 | ↓ |
| Apoa1 | apolipoprotein A-I (Apoa1) | 4.182753 | 0.004747 | ↑ |
| Rps27 | ribosomal protein S27 (Rps27) | 0.427968 | 0.005039 | ↓ |
| Rpl22l1 | ribosomal protein L22 like 1 (Rpl22l1) | 0.395386 | 0.005126 | ↓ |
| Rps27a | ribosomal protein S27A (Rps27a) | 0.422993 | 0.005161 | ↓ |
| Chchd1 | coiled-coil-helix-coiled-coil-helix domain containing 1 (Chchd1) | 0.41672 | 0.006623 | ↓ |
| Ass1 | argininosuccinate synthetase 1 (Ass1) | 1.21153 | 0.006628 | ↑ |
| Hspa8 | heat shock protein 8 (Hspa8) | 1.26608 | 0.007615 | ↑ |
| 1810022K09Rik | RIKEN cDNA 1810022K09 gene (1810022K09Rik) | 0.416109 | 0.007896 | ↓ |
| Rpl22 | ribosomal protein L22 (Rpl22) | 0.478678 | 0.009642 | ↓ |
| Fkbp3 | FK506 binding protein 3 (Fkbp3) | 0.48227 | 0.010581 | ↓ |
| Chchd2 | coiled-coil-helix-coiled-coil-helix domain containing 2 (Chchd2) | 0.571114 | 0.011822 | ↓ |
Fig. 6.
TissueEnrich results for OXY male vs. CTL male placentas. A) Bar plot distribution of the tissue enrichment of all DE genes. B) Heatmap analyses revealing those DE genes that are enriched in the placenta. This analysis is based on the results from 8 OXY males and 10 CTL males.
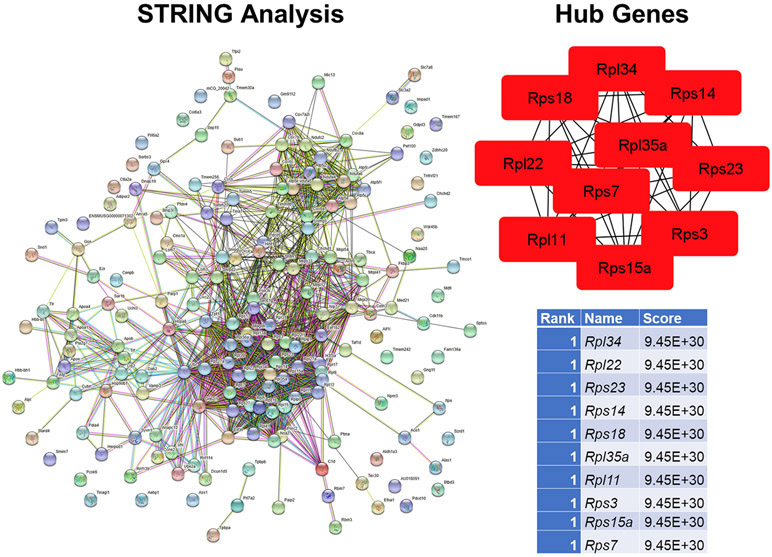
As with the female group comparisons, STRING and cytohubba analyses was done to examine for PPI and hub genes, respectively. The STRING diagram reveals huge clusters of genes encoding ribosomal protein subunits (Fig. 7A). All of the top 10 (equally tied for #1) are ribosomal protein forms, either Rps or Rpl forms (Fig. 7B). Of these Rps14 and Rpl22 are included in the top 25 DE genes based on the male group comparisons (Table 2). Both of these genes, along with other Rps and Rpl forms, show reduced expression in the placenta of male exposed to OXY.
Fig. 7.
STRING and hub gene analyses for OXY males vs. CTL males. PPI were determined by STRING analysis. The PPI files generated with STRING were imported into the cytohubba app [44] in Cytoscape [45] to examine for the top 10 hub genes. Within this program, hub genes were determined with MCC analysis, as recommended [44]. This analysis is based on the results from 8 OXY males and 10 CTL males.
To determine whether developmental exposure to OXY affected normal sexually dimorphic patterns in placental gene expression, transcript profiles of males and females within each group were compared. For the PCA analyses of CTL males vs. CTL females, the PC1, PC2, and PC3 axes accounted for 89.0, 8.1, and 1.1 % of the variation, respectively (Supplementary Fig. 2). While no differences were detected in the heatmap based on all genes identified in this comparison, DE genes were detected with volcano plot analyses (Supplementary Fig. 2). PCA comparison of OXY males vs. OXY females revealed that the PC1, PC2, and PC3 axes accounted for 88.6, 9.4, and 0.8% of the variation, respectively (Supplementary Fig. 2). While no differences were revealed with the heatmap for all genes identified in this comparison, the volcano plot identified select genes that were DE (Supplementary Fig. 2).
The full list of all genes identified based on CTL male vs. CTL female placenta is provided in Supplementary File 3. Several genes were identified to be DE based on FDR of ≤ 0.05. The top 25 DE genes are listed in Supplementary Table 4. Within this list, several of the genes were deemed hub genes, as detailed below, including Trf, Apoa1, Apoa4, Apob, Rbp4, Trf, Apoa2, Apoe, and Fga.
The full list of DE genes for OXY males vs. OXY female placenta is provided in Supplementary File 4. Similar to CTL male vs. CTL female placenta, several genes were DE based on a FDR ≤ 0.05. The top 25 DE genes are listed in Supplementary Table 5. Of these, only one gene, Rpl22, was considered a hub gene. Ogt is an X-linked chromosome gene that has previously been shown to demonstrate sexually dimorphic patterns of expression under maternal stress [26]. In the case of CTL males vs. CTL females, it is reduced in the latter based on P value but not FDR (Supplementary Table 4). In contrast, this gene is significantly downregulated in OXY males vs. OXY females based on P value and FDR (Supplementary Table 5).
When the DE genes based on CTL males vs. CTL females were analyzed with the TissueEnrich program, it revealed that the gene-sets were enriched for those associated with liver, placenta, kidney and intestine (Supplementary Fig. 3). Those enriched in the placenta included Tpbpb, Serpinb9e, Serpinbdc, Prl8a6, Prl7b1, Prl7a2, Ctsm, Cecam14, Cecam12, and Ceacam11 (Supplementary Fig. 3). TissueEnrich analyses of DE genes based on the OXY males vs. OXY female placental comparison reveals that they are enriched for transcripts associated with the placenta followed by heart (Supplementary Fig. 3). Further examination reveals that DE genes for this comparison contain more placental enriched ones that CTL males vs. CTL females, including Tpbpa, Prl8a9, Prl8a6, Prl7c1, Prl7a2, Prl2b1, Gzme, Ctla2b, Ceacam13, Cdh5, and Au018091 (Supplementary Fig. 3).
STRING analyses [50] of proteins encoded by the DE genes for CTL males vs. CTL females revealed one major cluster and two minor clusters with the proteins demonstrating limited PPI (Supplementary Fig. 4). Hub gene analyses based on the STRING analysis revealed that Apob, Trf, Apoa1, Apoe, Apoa2, Fga, Apoa4, Ttr, Rbp4, and Gpc3 are the top 10 hub genes (Supplementary Fig. 4). All of these genes were decreased in CTL males vs. CTL females.
Based on the proteins encoded for the DE genes with the OXY male vs. OXY female placenta, there are many interwoven and tight connections falling into two main clusters with the STRING analysis [50] (Supplementary Fig. 5). The top 10 hub genes based on this comparison and Fig. are all associated with ribosomal proteins, tied for the top rank, and include Rp111, Rps25, Rpl22, Rps3, Rpl35a, Rpl23, Rps8, Rpl31, Rps5, and Rpl34, all of which were decreased in OXY males relative to OXY females (Supplementary Fig. 5).
The g:Profiler web-based program was used to perform functional enrichment analyses on the DE gene-sets identified for the four comparisons detailed above. The detailed list for each is provided in Supplementary File 5. The top 10 GO terms for OXY females vs. CTL females are prolactin receptor binding, hormone activity, cytokine receptor binding, receptor ligand activity, signaling receptor activator activity, cysteine-type endopeptidase activity, long-chain fatty acid binding, cysteine-type peptidase activity, and signaling receptor binding (Supplementary Table 6). For OXY males vs. CTL males, the top ten G0 terms include structural constituents of ribosome, structural molecule activity, proton transmembrane transporter activity, RNA binding, rRNA binding, proton-transporting ATP synthase activity, rotational mechanism, oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor, and heme-copper terminal oxidase activity (Supplementary Table 7).
GO terms that were enriched for CTL males vs. CTL females include phosphatidylcholine-sterol O-acyltransferase activator activity, cholesterol transfer activity, sterol transfer activity, lipoprotein particle receptor binding, sterol transporter activity, lipid transfer activity, small molecule binding, alcohol binding, lipid transporter activity, and high-density lipoprotein particle receptor binding (Supplementary Table 8). Those for OXY males vs. OXY females include structural constituent of ribosome, structural molecule activity, proton transmembrane transporter activity, RNA binding, rRNA binding, proton-transporting ATP synthase activity, rotational mechanism, oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor, heme-copper terminal oxidase activity, cytochrome-c oxidase activity, and proton channel activity (Supplementary Table 9).
3.3. qPCR validation of placental enriched genes.
Eight placental enriched genes that were upregulated in OXY female vs. CTL female placenta based on RNAseq analyses were further validated with qPCR analyses. As shown in Fig. 4, several of these are also considered hub genes based on this comparison. Based on qPCR analyses, Ceacam11, Ceacam12, Ceacam14, Prl2b1, Prl7b1, and Tpbpb were upregulated in OXY female vs. CTL female placenta (Supplementary Fig. 6). These results match those obtained with RNAseq. Ceacam13 and Tpbpa, which RNA-seq showed both to be upregulated in OXY female placenta, only showed a trend towards upregulation with qPCR analyses (p= 0.07 and 0.1, respectively). With RNAseq analyses, Tpbpa and Tpbpb were downregulated in OXY male vs. CTL male placenta. Based on qPCR analyses, the same was the case for Tpbpa (p < 0.05) and Tpbpb tended to be lower (p = 0.1, Supplementary Fig. 6). qPCR and RNAseq analyses further revealed that none of these other placental enriched genes were DE in OXY male vs. CTL male placenta.
3.4. Integrative correlation analyses
WGCNA [27] was used to link genes identified with the various pairwise comparisons and placental morphometric analyses (LA to pTGC ratio, SpongioTB to pTGC ratio, apoptotic pTGC, and area of maternal blood vessels within the LA region). For these analyses, 8292 genes for female OXY vs. female CTL, 8355 gene for OXY male OXY vs. CTL male, 8298 genes for CTL male vs. CTL female comparison, and 6273 gene for OXY males vs. OXY females were used for gene module and placenta morphometrics correlation analyses. Individual modules are represented by different colors, and module eigengenes (ME) were then correlated with these histological measurements. Different color modules are defined as clusters of densely inter-connected genes as determined by the program [27]. The grey color module though includes genes that do not fit into the other modules.
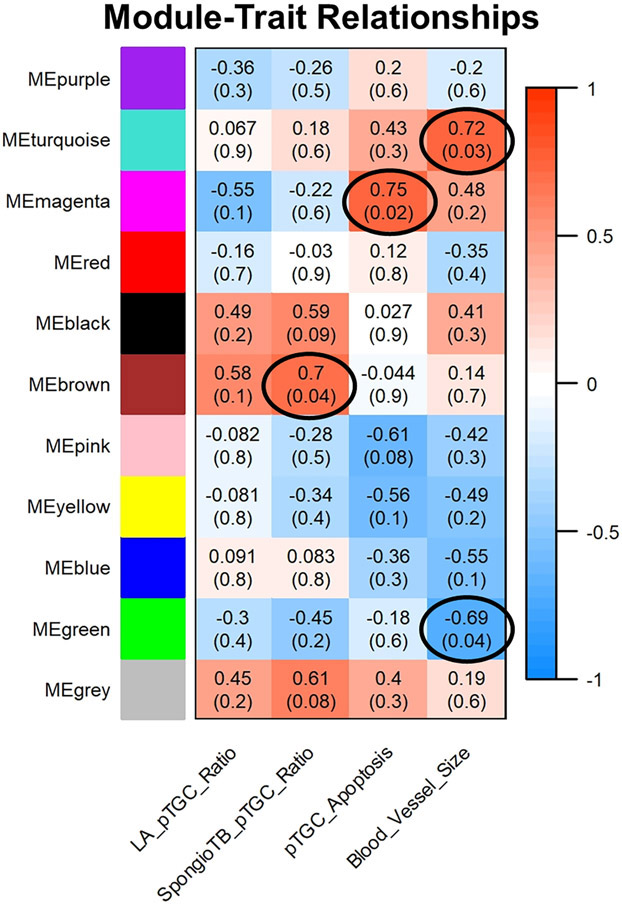
Supplementary Fig. 7 shows the resulting dendrogram and heatmap for the OXY female vs. CTL female placental comparison, which reveals clear separation into different color modules. As shown in Fig. 8, the ME Brown was positively associated with SpongioTB to pTGC ratio (r = 0.7; p = 0.04). ME Magenta was positively associated with pTGC apoptosis (r = 0.75; p = 0.02). ME turquoise was positively associated with maternal blood vessel area (r= 0.72; p = 0.03); whereas, ME Green was negatively correlated with this category (r = −0.69; p = 0.04). Genes within these four modules are listed in Supplementary File 6.
Fig. 8.
Relationship of WGCNA results and placental morphometric assessments for OXY female vs. CTL female comparison. Modules identified in Supplementary Fig. 5 were correlated with placental morphometric assessments (Fig. 2). Each row corresponds to a Module Eigengene (ME) and colors represent the correlation coefficient between the ME and one of the four placental histological measurements (LA to pTGC ratio; SpongioTB to pTGC ratio; pTGC apoptosis; Maternal blood vessel area in LA region). Numbers at the top of rows represents degree of correlation, and values with a negative integer indicate an inverse correlation with one of the four placental measurements. ME Brown was positively associated with SpongioTB to pTGC ratio (r = 0.7; p = 0.04). ME Magenta was positively associated with pTGC apoptosis (r = 0.75; p = 0.02). ME turquoise was positively associated with maternal blood vessel area (r= 0.72; p = 0.03); whereas, ME Green was negatively correlated with this category (r = −0.69; p = 0.04). A legend that ranges from blue to red shades (−1 to 1, respectively) is included to the right of the Figure. Genes within these four modules are listed in Supplementary File 6. This analysis is based on the results from 10 OXY females and 9 CTL females.
TissueEnrich analyses revealed that for ME Brown, ME Magenta, ME Turquoise, and ME Green for OXY females vs. CTL females, there is enrichment for placental-associated genes (Supplementary Figs. 8 and 9). For instance, ME Magenta for this comparison includes Prl8a2 and several Gzm forms that are enriched in the placenta (Supplementary Fig. 8D). ME Turquoise for this comparison includes Tpbpa, Prl7d1, Prl7a1, and Ceacam9 (Supplementary Fig. 9B). ME Green includes Prl6a1 and Prl2b1 (Supplementary Fig. 9D).
Hub genes for the ME Brown based on OXY females vs. CTL female placental comparison include Keap1, Fbxo10, Spsb1, Asb13, Ube2o, Nedd4l, Ube2a1, Traf7, Ube3b, and Cul7 (Supplementary Fig. 10). Those for ME Magenta with this comparison include Prf1, Serpinb6a, Fabp5, Gzmd, Gzme, Cdkn1b, Sepp2, Rab31, Gpx8, and B2m (Supplementary Fig. 10). Hub genes for ME Turquoise based on this comparison consist of Rps27a, Vprbp, Ube2z, Fbx119, Rnf126, Rlim, Anapc1, Anapc2, Anapc10, and Fzr1 (Supplementary Fig. 11). Those within the ME Green include Ap2a2, Snap91, Eps15, Arpc3, Itsn1, Arpc5, Ston2, Pacsin3, Col1a1, Col5a1.
WebGestalt based on KEGG and Reactome pathways was used to perform functional enrichment analyses for each of these ME in the OXY females vs. CTL females (Supplementary File 7). For ME Brown, likely enriched pathways are protein processing in endoplasmic reticulum, extracellular matrix organization, signaling by VEGF, RHO GTPases Activate WASPs and WAVEs, Asparagine N-linked glycosylation, N-glycan biosynthesis, regulation of actin cytoskeleton, focal adhesion, VEGFA-VEGFR2 pathway, and collagen biosynthesis and modifying enzymes (Supplementary File 7). Those for ME Magenta include Metabolism of angiotensinogen to angiotensin, regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs), peptide hormone metabolism, calcitonin-like ligand receptors, sphingolipid de novo biosynthesis, triglyceride catabolism, class B/2 (secretin family receptors), interferon signaling, interferon gamma signaling, and triglyceride metabolism (Supplementary File 7). Pathways likely enriched for ME Turquoise are metabolism of proteins, gene expression (transcription), RNA polymerase II transcription, metabolism of RNA, Huntington disease, ribosome, mRNA splicing, mRNA splicing- major pathway, translation, and oxidative phosphorylation (Supplementary File 7). Those for ME Green consist of transport of small molecules, vesicle-mediated transport, EPHD-mediated forward signaling, membrane trafficking, axon guidance, protein digestion and absorption, endocytosis, EPH-ephrin signaling, HDL remodeling, and regulation of PTEN gene transcription (Supplementary File 7).
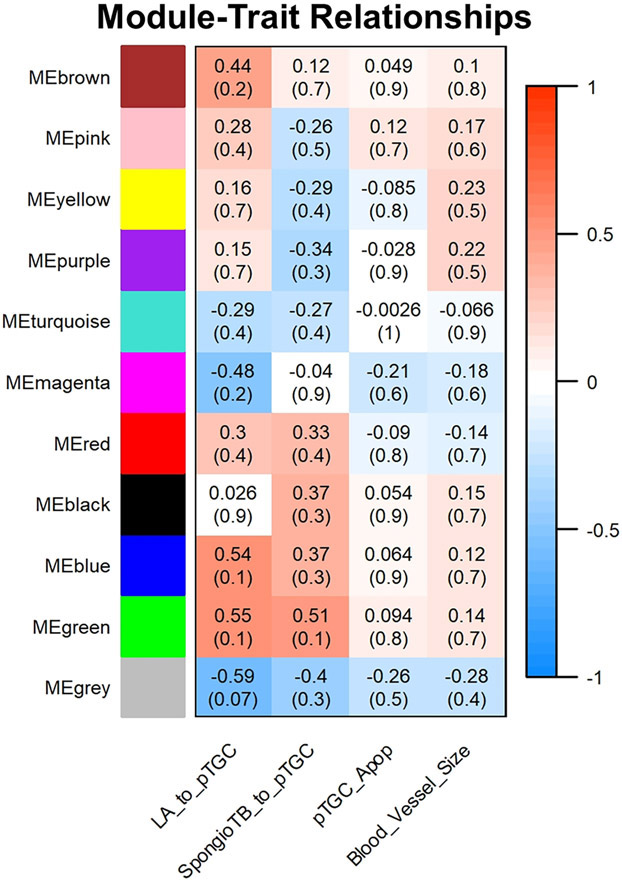
While the dendrogram and heatmap for OXY males vs. CTL males reveals clear separation into different color modules (Supplementary Fig. 12), none of these were significantly associated with any of the categories analyzed for the placental histology (Fig. 9). Accordingly, no follow-up analyses were done with any of these ME.
Fig. 9.
Relationship of WGCNA results and placental morphometric assessments for OXY male vs. CTL male comparison. Modules identified in Supplementary Fig. 8 were correlated with placental morphometric assessments (Fig. 2). Each row corresponds to a Module Eigengene (ME) and colors represent the correlation coefficient between the ME and one of the four placental histological measurements (LA to pTGC ratio; SpongioTB to pTGC ratio; pTGC apoptosis; Maternal blood vessel area in LA region). Numbers at the top of rows represents degree of correlation, and values with a negative integer indicate an inverse correlation with one of the four placental measurements. For this comparison, no ME correlated with any of these four placental measurements. A legend that ranges from blue to red shades (−1 to 1, respectively) is included to the right of the Figure. This analysis is based on the results from 8 OXY males and 10 CTL males.
The dendrogram and heatmap analyses for CTL males vs. CTL females demonstrate clear color module separation (Supplementary Fig. 13). Based on the module trait relationship diagram, ME purple was negatively associated with LA to pTGC ratio (r = −0.53; p = 0.02, Supplementary Fig. 14). ME Blue, Red and Grey were positively associated with SpongioTB to pTGC ratio (ME Blue: r = 0.57; p = 0.01; ME Red: r = 0.48; p = 0.05; ME Grey: r = 0.48; p = 0.04). The genes within each of these color modules is provided in Supplementary File 8. As ME Grey consists of a random assortment of genes that do not fit into other color modules, this ME was not considered in further analyses.
Genes within all three of these modules (ME Purple, Blue, and Red) for CTL male vs. CTL female were primarily enriched for placenta-associated genes (Supplementary Fig. 15). The ME Purple included Dmrtc1b, which is exclusively expressed by the placenta (Supplementary Fig. 15B). Placenta-specific transcripts in ME Red for this comparison include Psg27, Psg23, Psg19, Psg16, and Ceacam5 (Supplementary Fig. 15D).
Supplementary Fig. 16 provides information on the top hub genes for the above three ME. Top hub genes for ME Purple include Polr2e, Mrps6, Mrps18a, Canx, Eci2, Hmgcl, Gtf2h4, Kdm6a, Copb1, and Gpr137b. The top hub genes for ME Blue are Fbxw7, Fbxo32, Keap1, Asb13, Gan, Kctd6, Fbxo22, Fbxo10, Ube2s, and Nedd4l. ME Red top hub genes are placental enriched genes that consist of Psg17, Psg18, Psg19, Psg28, Psg23, Psg29, Ceacam5, Psg21, Psg27, and Psg25.
WebGestalt based on KEGG and Reactome pathways was used to perform functional enrichment analyses for each of these ME in the CTL males vs. CTL females (Supplementary File 9). For ME purple, presumptive functionally enriched pathways include peroxisome, metabolism, peroxisomal protein import, protein localization, formation of TC-NER pre-incision complex, beta-oxidation of very long chain fatty acids, formation of RNA Pol II elongation complex, RNA polymerase II transcription elongation, gap-filling DNA repair synthesis and ligation in TC-NER, dual inclusion of TC-NER. Those for ME Blue include small cell lung cancer, laminin interactions, pathways in cancer, metabolism of proteins, focal adhesion, protein processing in endoplasmic reticulum, cell junction organization, cell-cell communication, signaling by receptor tyrosine kinases, and asparagine N-linked glycosylation. Pathways likely enriched for ME Red include protein processing in endoplasmic reticulum, N-glycan biosynthesis, asparagine N-linked glycosylation, transport to the Golgi and subsequent modification, extracellular matrix organization, glycosaminoglycan degradation, vesicle-mediated transport, plasma lipoprotein assembly, remodeling, and clearance, collagen biosynthesis and modifying enzymes, and glycosaminoglycan metabolism.
Supplementary Fig. 17 shows the dendrogram and heatmap analyses for OXY males vs. OXY females with clear color module separation evident. While ME Grey was inversely associated with LA to pTGC ratio (r = −0.53; p = 0.03), this ME is considered non-specific in that it includes all genes that do not fit into the color modules (Supplementary Fig. 18). As such, no follow-up analyses were performed on this non-specific ME. The genes within this module are included for reference in Supplementary File 10.
4. Discussion
The goals of the current studies were to determine the effects of maternal exposure to oxycodone on the placental structure and global transcriptome profile. We also sought to integrate the histological changes with transcripts identified in the various comparisons to determine which sets of genes might relate to pathophysiological changes in the placenta.
Our findings reveal that maternal oxycodone exposure reduced the area of pTGC relative to both the LA and SpongioTB area. We also found greater number of apoptotic pTGC in this group. Thus, it might be speculated that OXY reductions in the pTGC area could be due to both reduced allocation of progenitor trophoblast stem cells to differentiate into pTGC and apoptosis of those that have differentiated into these cells. These possibilities merit further evaluation. We and other researchers have seen similar reductions in these cells when mouse dams are exposed to the endocrine disrupting chemicals (EDC), BPA or BPS [23, 47, 48]. These placental cells directly contact the underlying uterine myometrium, and thus, they are likely vulnerable to toxicants and drugs in her bloodstream. We, and others, have also shown that the pTGC likely either uptake neurotransmitters, including serotonin and dopamine, from the underlying uterine tissue or may directly synthesize such compounds [23, 51, 52]. In so doing, the pTGC may affect fetal brain development through the placenta-brain-axis [13].
Diethylstilbestrol (DES), which was administered to pregnant mothers in the 1940s-1970s under the mistaken notion that it prevented miscarriage, reduces the amount of rough endoplasmic reticulum (RER) in these cells [53]. Perfluorooctanoic acid (PFOA) exposure in mice causes reductions in pTGC [54]. In contrast, hyperosmolar stress results in increased differentiation of progenitor cells to pTGC [55]. Based on current results and these other data, it is likely that OXY acts directly on pTGC.
Direct effects of OXY on the placenta are further supported by the fact that the placenta contains an endogenous opioid regulatory system that has been suggested to be integral in regulating placental factors regulating maternal recognition of pregnancy, such as hCG and placental lactogens [14-18]. Opioid receptors, including μ forms that would be activated by OXY, are expressed in the human and rodent placenta [56, 57].
The other major histological finding was that maternal OXY treatment reduced the area of the LA region occupied by maternal blood vessels. As the primary source of oxygen and nutrients for the fetus and removal of fetal waste material [58], such changes may compromise the placenta and correspondingly fetal development. It is not clear though if such reductions are due to reduced angiogenesis and arborization of maternal blood vessels or remodeling defects within the labyrinth region. Similarly, the pyrethroid insecticide, fenvalerate (FEN), causes reductions in the blood sinusoid area in the placental labyrinth layer [58]. The histological changes in the pTGC region and maternal blood vessels within the LA region were observed to the same extent in male and female placenta exposed to OXY. However, gene expression patterns showed sexually dimorphic differences in response to OXY exposure.
The placenta of female conceptuses exposed to OXY had prominent enrichment of placental enriched genes that were also considered hub genes, including Ceacam11, Ceacam14, Ceacam12, Ceacam13, Prl7b1, Prl2b1, Ctsq, and Tpbpa. CEACAMs, including CEACAM11, and pregnancy specific glycoproteins (PSGs) are members of the carcinoembryonic antigen (CEA) family, which is a large group of evolutionarily and structurally divergent glycoproteins that are differentially expressed in trophoblast cells during placental development [59]. Such placental molecules might be important in suppressing a maternal immune attack against the hetero-allotypic conceptus that is only 50% genetically related [59]. Prolactin family paralog/placental lactogen expression by the placenta may regulate maternal physiological responses, including uterine decidualization [60]. Such hormones may also act in a paracrine manner to provide potential protection of the placenta against maternal stressors. For instance, mice with genetic deletion of Prl7b1 show normal placentation under basal conditions [61]. However, when subjected to maternal hypoxia, wild-type (WT) mice showed adaptations to this stressor that were absent in Prl7b1 null placentas [61]. In line with the upregulation of several Prl forms in the female placenta exposed to OXY, functional enrichment analysis revealed that the primary pathways presumably altered in this group relative to CTL females are prolactin receptor binding, hormone activity, cytokine receptor binding, receptor ligand activity, and signaling receptor activator activity. The surge in Prl and other placental-enriched transcripts in the placenta of OXY exposed females might be a mechanism to confer additional protection. Consistent with this possibility, functional enrichment analysis revealed that positive regulation of cell cycle proliferation was also associated with DE genes identified in the OXY female vs. CTL female placenta (Supplementary File 5). DE genes within this category included Prl7B1, Prl8A6, Prl8A8, Prl7A2, Prl7C1, Prl8A9, Prl2C1, Fabp4, Tgm1, Pdcd10, and Prl2B1. For this comparison and OXY male vs. CTL male placenta, there was surprisingly no enrichment of apoptotic pathways.
Tpbpa and Tpbpb were also up-regulated in OXY female relative to CTL female placenta with the former considered a hub gene. Tpbpa expression is initiated in the ectoplacental cone between embryonic days (E) 7.5 and 8.5 and is then evident in the junctional zone. Tpbpa-positive cells are precursors of various trophoblast subtypes, including TGCs and glycogen trophoblast cells [62]. Tpbpb also localizes to the junctional zone in the mouse placenta [63]. Female placenta exposed to OXY may increase the expression for both genes in attempts to compensate for reductions in pTGCs. If such is the case, it will be of interest to examine additional gestational timepoints to determine if at later stages in pregnancy, the number of pTGCs is similar in OXY and CTL female placenta.
Comparison of gene expression profiles in OXY males vs. CTL males reveals a completely different pattern of expression relative to counterpart females. In males, only a few of the DE transcripts is considered to be placental enriched, Tpbpb, Tpbpa, Prl8a2, and Prl7a2. These placental enriched genes were downregulated in the placenta of OXY exposed males. As Tpbpa is associated with progenitor cells for TGCs, it is possible downregulation of this gene at e12.5 may manifest in later reductions of pTGCs at later gestational timepoints. Hub genes that were downregulated in male OXY placenta include various Rps and Rpl forms. Decreased expression of genes encoding ribosomal proteins may compromise protein translation of other transcripts. It remains to be determined whether these sexually dimorphic patterns in placental gene expression in response to OXY exposure may later affect offspring health, such as through the placenta-brain-axis. Currently, we are testing various behavioral domains in male and female mouse offspring derived from other dams exposed to OXY relative to controls. Our prediction based on the placental expression profiles is that male offspring developmentally exposed to OXY will show greater behavioral disturbances in terms of cognitive impairments, increased anxiogenic and stereotypic behaviors, and reduced socio-communication; whereas, such deficits will be reduced or even absent in their female siblings. We, and others, have shown that by mounting robust placental gene expression changes in response to maternal challenges, including high fat diet or stress, females demonstrate later resistance to behavioral changes relative to male counterparts [12, 26, 64-66].
Past work reveals that the placenta is a sexually dimorphic organ, which could be due to epigenetic factors, escape from X inactivation, steroid hormone influence, and other factors [11-13, 67]. Examination of the signature gene expression patterns between male and female placenta within the same group shows that OXY exposure alters such sex differences. Hub genes that are altered in CTL males vs. CTL females center around those encoding apolipoproteins that associate with high density lipid proteins, such as Apob, Apoa1, Apoe, Apoa2, and Apoa4. All of these are decreased in CTL males relative to CTL females. Two other hub genes in this comparison are transferrin (Trf) and transthyretin (Ttr), which are also decreased in CTL males. Transferrin is important in iron binding, and transthyretin transports vitamin A and thyroxine. It is not clear if these above sex differences in placental gene expression patterns confers some inherent advantages for females in terms of nutrient and hormone transport. In a human trophoblast cell line, BeWo, APOA1 stimulates an increase in placental lactogen 1 [68] .
Comparison of OXY males vs. OXY females reveals a completely different picture in terms of hub genes and overall expression pattern. All of the hub genes for this comparison are various Rpl and Rps forms. All of which are decreased in male OXY vs. female OXY placenta. This change is assuredly due to the marked suppression of these genes in male OXY exposed placenta, which is not observed in females. O-linked-N-acetylglucosamine transferase (Ogt) is an X-linked and nutrient-sensor gene that is reduced in CTL males vs. CTL females, as has been reported by others [26, 69]. However, maternal OXY exposure enhanced these sex differences. Similar findings have been reported in relation to maternal stress [26]. It remains to be determined how such sex differences in placental gene expression under basal conditions and in response to maternal OXY exposure affect later offspring health outcomes, especially neurobehavioral responses.
Lastly, we used WGCNA analyses to interlink the histopathological changes in the placenta to genes identified in the various comparisons. In the female OXY vs. female CTL placenta, ME Brown positively correlated with spongioTB to pTGC ratio, and the primary pathways likely enriched with this gene-set include extracellular matrix reorganization, VEGF signaling, and regulation of actin bioskeleton, and collagen biosynthesis and modifying enzymes. Such remodeling may stimulate differentiation of spongioTB over pTGCs. ME Magenta positively correlated with pTGC apoptosis, and within this ME, overly-represented pathways include peptide hormone signaling, interferon signaling, interferon gamma signaling, and triglyceride metabolism and catabolism. Interferon gamma signaling has been previously implicated in promoting trophoblast apoptosis [70, 71].
It is not clear why none of the ME significantly correlated with these parameters for OXY males vs. OXY females. For the sex comparisons, only ME in the CTL males vs. CTL females showed significant associations. In examining the pathways enriched in the three modules, ME Purple, ME Blue, and ME Red that correlated with one or more of the histological parameters, Asparagine N-linked glycosylation, was enriched in both ME Blue and ME Red that were positively linked with spongioTB to pTGC ratio. Asparagine-linked-glycosylation is important in placental specific proteins from other species, including pregnancy associated glycoproteins in pigs and cattle [72, 73] and hCG in humans [74]. Conceivably, this biochemical alteration may affect the potency of such placental-specific hormones and factors, and thereby potential paracrine effects within trophoblast cells.
Current data suggest that the pTGC are especially vulnerable to the effects of maternal OXY exposure. While the current RNA-seq approach with whole placental tissue may shed some light on how maternal OXY exposure induces changes in the pTGC and other trophoblast cells, it is not possible to determine which cells gave rise to such gene expression differences. To delineate further gene expression changes to specific trophoblast cells, future studies will employ the recently developed Visium Spatial Transcriptomics (ST) technology [75, 76]. This approach provides quantitative mRNA profiles of cells in intact tissue and may be useful to examine how gene expression changes in different trophoblast cell lineages relate to and possibly influence each other and the underlying uterine tissue. This technology though was not available at the time the current studies were initiated. It will be of interest to compare results with this recently introduced method to current results. Underlying mechanisms leading to pTGC apoptosis also warrants further examination. A spatial transcriptomics approach will assumingly be more sensitive and provide clues as to underlying gene expression changes and pathways leading to such cell-specific disruptions. In a separate study, we are currently examining how maternal OXY treatment affects offspring neurobehavioral outcomes. Such neural effects may be direct in origin or through the placenta-brain axis [13]. This work will be the subject of a future manuscript.
In conclusion, current results indicate that maternal OXY exposure reduces the pTGC area but increases pTGC apoptosis in E12.5 placenta of both males and females. This treatment, however, leads to sex-dependent differences in placental gene expression with females demonstrating a more robust response to this environmental challenge. Several placental enriched genes were increased in the placenta of OXY exposed females; whereas, placental associated genes were downregulated in males, along with several ribosomal protein associated genes. Exposure to OXY also disrupted normal sex differences in placental gene expression patterns. Integrative correlation analyses also revealed that in OXY females vs. CTL females, select ME are associated with histological measurements related to pTGC area and apoptosis. It remains to be determined whether such genes and associated pathways in these ME affects pTGC differentiation and apoptosis. Gene expression changes in the placenta of OXY exposed females may buffer the offspring from later DOHaD-associated diseases, which is the subject of ongoing studies.
Supplementary Material
Supplementary Fig. 1. Higher magnification region of LA region for each group to show maternal blood vessels (designated with arrows) in each group. For maternal blood vessel area measurement within the labyrinth (LA) region, a box measuring 500 × 500 pixels was placed over 5 to 7 random LA areas for each placenta section. Ten maternal blood vessels in this area were randomly selected, the edge of the blood vessel was outlined, and the area was measured with the Adobe Photoshop program (Adobe, San Jose, CA).
Supplementary Fig. 2. Sex differences in placental gene expression CTL and OXY exposed conceptuses. A) 3D PCA plot of RNAseq results from placenta of CTL males vs. CTL females. B) Heatmap based on all transcripts identified in placenta of CTL males vs. CTL females. C) Volcano plot analyses of gene transcripts identified in placenta of CTL males vs. CTL females. Transcripts represented by Orange: p < 0.05; Transcripts represented by Red: log2FoldChange < −1 or > 1; Transcripts represented by Green: p < 0.05 AND log2FoldChange < −1 or > 1. D) 3D PCA plot of RNAseq results from placenta of OXY males vs. OXY females. E) Heatmap based on all transcripts identified in placenta of OXY males vs. OXY females. F) Volcano plot analyses of gene transcripts identified in placenta of OXY males vs. OXY females. Transcripts represented by Orange: p < 0.05; Transcripts represented by Red: log2FoldChange < −1 or > 1; Transcripts represented by Green: p < 0.05 AND log2FoldChange < −1 or > 1. These analyses are based on the results from 10 CTL males, 9 CTL females, 10 OXY females, and 8 OXY males.
Supplementary Fig. 3. TissueEnrich results for CTL male vs. CTL female placenta and OXY male vs. OXY female placenta. A) Bar plot distribution of the tissue enrichment of all DE genes for CTL male vs. CTL female comparison. B) Heatmap analyses revealing those DE genes for CTL male vs. CTL female comparison that are enriched in the placenta. C) Bar plot distribution of the tissue enrichment of all DE genes for OXT male vs. OXY female comparison. D) Heatmap analyses revealing those DE genes for OXY male vs. OXY female comparison that are enriched in the placenta. These analyses are based on the results from 10 CTL males and 9 CTL females.
Supplementary Fig. 4. STRING and hub gene analyses for CTL males vs. CTL females. PPI were determined by STRING analysis. The PPI files generated with STRING were imported into the cytohubba app [44] in Cytoscape [45] to examine for the top 10 hub genes. Within this program, hub genes were determined with MCC analysis, as recommended [44]. These analyses are based on the results from 10 CTL males and 9 CTL females.
Supplementary Fig. 5. STRING and hub gene analyses for OXY males vs. OXY females. PPI were determined by STRING analysis. The PPI files generated with STRING were imported into the cytohubba app [44] in Cytoscape [45] to examine for the top 10 hub genes. Within this program, hub genes were determined with MCC analysis, as recommended [44]. These analyses are based on the results from 8 OXY males and 10 OXY females.
Supplementary Fig. 6. qPCR results for placental enriched genes. Ceacam11, Ceacam12, Ceacam14, Prl2b1, Prl7b1, and Tpbpb were upregulated in OXY female vs. CTL female placenta. Ceacam13 and Tpbpa only showed a trend towards upregulation with these analyses (p= 0.07 and 0.1, respectively). Tpbpa was significantly reduced in OXY male vs. CTL male placenta (p < 0.05), and Tpbpb tended to be lower for this comparison (p = 0.1). None of the other placental enriched genes tested with qPCR analyses were DE in OXY male vs. CTL male placenta.
Supplementary Fig. 7. Dendrogram and heatmap of WGCNA results for OXY female vs. CTL female placental comparison. As detailed in [27], the heatmap represents the Topological Overlap Matrix (TOM) for all analyzed genes. Lighter colors in the heatmap depict low overlap and darker red colors indicate greater overlap. Along the diagonal, blocks of darker colors represent the different color modules. The gene dendrogram and module assignment are shown at the left and top of the image. This analysis is based on the results from 10 OXY females and 9 CTL females.
Supplementary Fig. 8. TissueEnrich results for ME Brown and ME Magenta for OXY female vs. CTL female placental comparison A) Bar plot distribution of the tissue enrichment of genes in ME Brown for this comparison. B) Heatmap analyses of genes within ME Brown for this comparison. C) Bar plot distribution of the tissue enrichment of genes within ME Magenta for this comparison. D) Heatmap analyses of genes within ME Magenta for this comparison. This analysis is based on the results from 10 OXY females and 9 CTL females.
Supplementary Fig. 9. TissueEnrich results for ME Turquoise and ME Green for OXY female vs. CTL female placental comparison A) Bar plot distribution of the tissue enrichment of genes in ME Turquoise for this comparison. B) Heatmap analyses of genes within ME Turquoise for this comparison. C) Bar plot distribution of the tissue enrichment of genes within ME Green for this comparison. D) Heatmap analyses of genes within ME Green for this comparison. This analysis is based on the results from 10 OXY females and 9 CTL females.
Supplementary Fig. 10. Hub genes for ME Brown and ME Magenta for OXY female vs. CTL female placental comparison. This analysis is based on the results from 10 OXY females and 9 CTL females.
Supplementary Fig. 11. Hub genes for ME Turquoise and ME green for OXY female vs. CTL female placental comparison. This analysis is based on the results from 10 OXY females and 9 CTL females.
Supplementary Fig. 12. Dendrogram and heatmap of WGCNA results for OXY male vs. CTL male placental comparison. As detailed in [27], the heatmap represents the Topological Overlap Matrix (TOM) for all analyzed genes. Lighter colors in the heatmap depict low overlap and darker red colors indicate greater overlap. Along the diagonal, blocks of darker colors represent the different color modules. The gene dendrogram and module assignment are shown at the left and top of the image. This analysis is based on the results from 8 OXY males and 10 CTL males.
Supplementary Fig. 13. Dendrogram and heatmap of WGCNA results for CTL male vs. CTL female placental comparison. As detailed in [27], the heatmap represents the Topological Overlap Matrix (TOM) for all analyzed genes. Lighter colors in the heatmap depict low overlap and darker red colors indicate greater overlap. Along the diagonal, blocks of darker colors represent the different color modules. The gene dendrogram and module assignment are shown at the left and top of the image. This analysis is based on the results from 10 CTL males and 9 CTL females.
Supplementary Fig. 14. Relationship of WGCNA results and placental morphometric assessments for CTL male vs. CTL female placental comparison. Modules identified in Supplementary Fig. 13 were correlated with placental morphometric assessments (Fig. 2). Each row corresponds to a Module Eigengene (ME) and colors represent the correlation coefficient between the ME and one of the four placental histological measurements (LA to pTGC ratio; SpongioTB to pTGC ratio; pTGC apoptosis; Maternal blood vessel area in LA region). Numbers at the top of rows represents degree of correlation, and values with a negative integer indicate an inverse correlation with one of the four placental measurements. ME purple was negatively associated with LA to pTGC ratio (r = −0.53; p = 0.02). ME Blue, Red and Grey were positively associated with SpongioTB to pTGC ratio (ME Blue: r = 0.57; p = 0.01; ME Red: r = 0.48; p = 0.05; ME Grey: r = 0.48; p = 0.04). A legend that ranges from blue to red shades (−1 to 1, respectively) is included to the right of the Figures. The genes within each of these color modules is provided in Supplementary File 8. This analysis is based on the results from 10 CTL males and 9 CTL females.
Supplementary Fig. 15. TissueEnrich results for ME Purple, ME Blue, and ME Red for CTL male vs. CTL female placental comparison A) Bar plot distribution of the tissue enrichment of genes in ME Purple for this comparison. B) Heatmap analyses of genes within ME Purple for this comparison. C) Bar plot distribution of the tissue enrichment of genes within ME Blue for this comparison. D) Heatmap analyses of genes within ME Blue for this comparison. C) Bar plot distribution of the tissue enrichment of genes within ME Red for this comparison. D) Heatmap analyses of genes within ME Red for this comparison. This analysis is based on the results from 10 CTL males and 9 CTL females.
Supplementary Fig. 16. Hub genes for ME Purple, ME Blue, and ME Red for CTL male vs. CTL female placental comparison. This analysis is based on the results from 10 CTL males and 9 CTL females.
Supplementary Fig. 17. Dendrogram and heatmap of WGCNA results for OXY male vs. OXY female placental comparison. As detailed in [27], the heatmap represents the Topological Overlap Matrix (TOM) for all analyzed genes. Lighter colors in the heatmap depict low overlap and darker red colors indicate greater overlap. Along the diagonal, blocks of darker colors represent the different color modules. The gene dendrogram and module assignment are shown at the left and top of the image. This analysis is based on the results from 8 OXY males and 10 OXY females.
Supplementary Fig. 18. Relationship of WGCNA results and placental morphometric assessments for OXY male vs. OXY female placental comparison. Modules identified in Supplementary Fig. 17 were correlated with placental morphometric assessments (Fig. 2). Each row corresponds to a Module Eigengene (ME) and colors represent the correlation coefficient between the ME and one of the four placental histological measurements (LA to pTGC ratio; SpongioTB to pTGC ratio; pTGC apoptosis; Maternal blood vessel area in LA region). Numbers at the top of rows represents degree of correlation, and values with a negative integer indicate an inverse correlation with one of the four placental measurements. ME Grey was inversely associated with LA to pTGC ratio (r = −0.53; p = 0.03). A legend that ranges from blue to red shades (−1 to 1, respectively) is included to the right of the Figure. The genes within this module are included for reference in Supplementary File 10. This analysis is based on the results from 8 OXY males and 10 OXY females.
Acknowledgements
We appreciate all of the assistance from the undergraduate students who helped in taking care of the mice, Dr. Peter Langfelder for assisting with the WGCNA analyses, and Dr. Scott Givan, bioinformaticians, and computer scientists at the University of Missouri who assisted with the RNAseq analyses. Dr. Cheryl Rosenfeld is supported by NIEHS 1R01ES025547. Dr. Geetu Tuteja is supported by NICHD RHD096083A.
Footnotes
Conflict of interest statement
The authors have no conflicts of interests to declare.
Data access availability
RNA sequence data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE152172.
References
- [1].Reinhart M, Scarpati LM, Kirson NY, Patton C, Shak N, Erensen JG, The economic burden of abuse of prescription opioids: A systematic literature review from 2012 to 2017, Appl Health Econ Health Policy 16(5) (2018) 609–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].SAMHSA., Behavioral health barometer: United States, 2015 Report. , Rockville, MD, 2015. [PubMed] [Google Scholar]
- [3].Patrick SW, Schumacher RE, Benneyworth BD, Krans EE, McAllister JM, Davis MM, Neonatal abstinence syndrome and associated health care expenditures: United States, 2000-2009, Jama 307(18) (2012) 1934–40. [DOI] [PubMed] [Google Scholar]
- [4].Heil SH, Jones HE, Arria A, Kaltenbach K, Coyle M, Fischer G, Stine S, Selby P, Martin PR, Unintended pregnancy in opioid-abusing women, J Subst Abuse Treat 40(2) (2011) 199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Clemans-Cope L, Lynch V, Howell E, Hill I, Holla N, Morgan J, Johnson P, Cross-Barnet C, Thompson JA, Pregnant women with opioid use disorder and their infants in three state Medicaid programs in 2013-2016, Drug Alcohol Depend 195 (2019) 156–163. [DOI] [PubMed] [Google Scholar]
- [6].Jones HE, Kaltenbach K, Benjamin T, Wachman EM, O'Grady KE, Prenatal opioid exposure, neonatal abstinence syndrome/neonatal opioid withdrawal syndrome, and later child development research: shortcomings and solutions, J Addict Med 13(2) (2019) 90–92. [DOI] [PubMed] [Google Scholar]
- [7].C. https://www.cdc.gov/pregnancy/opioids/basics.html, Basics about opioid use during oregnancy, (2019).
- [8].Yazdy MM, Desai RJ, Brogly SB, Prescription opioids in pregnancy and birth outcomes: a review of the literature, J Pediatr Genet 4(2) (2015) 56–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Grandjean P, Barouki R, Bellinger DC, Casteleyn L, Chadwick LH, Cordier S, Etzel RA, Gray KA, Ha EH, Junien C, Karagas M, Kawamoto T, Paige Lawrence B, Perera FP, Prins GS, Puga A, Rosenfeld CS, Sherr DH, Sly PD, Suk W, Sun Q, Toppari J, van den Hazel P, Walker CL, Heindel JJ, Life-long implications of developmental exposure to environmental stressors: new perspectives, Endocrinology 156(10) (2015) 3408–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Rosenfeld CS, The Epigenome and Developmental Origns of Health and Disease, Elsevier; (2015). [Google Scholar]
- [11].Rosenfeld CS, Sex-specific placental responses in fetal development, Endocrinology 156(10) (2015) 3422–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mao J, Zhang X, Sieli PT, Falduto MT, Torres KE, Rosenfeld CS, Contrasting effects of different maternal diets on sexually dimorphic gene expression in the murine placenta, Proc Natl Acad Sci USA 107(12) (2010) 5557–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rosenfeld CS, The placenta-brain-axis J Neurosci Res (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ahmed MS, Cemerikic B, Agbas A, Properties and functions of human placental opioid system, Life Sci 50(2) (1992) 83–97. [DOI] [PubMed] [Google Scholar]
- [15].Cemerikic B, Cheng J, Agbas A, Ahmed MS, Opioids regulate the release of human chorionic gonadotropin hormone from trophoblast tissue, Life Sci 49(11) (1991) 813–24. [DOI] [PubMed] [Google Scholar]
- [16].Cemerikic B, Schabbing R, Ahmed MS, Selectivity and potency of opioid peptides in regulating human chorionic gonadotropin release from term trophoblast tissue, Peptides 13(5) (1992) 897–903. [DOI] [PubMed] [Google Scholar]
- [17].Cemerikic B, Zamah R, Ahmed MS, Opioids regulation of human chorionic gonadotropin release from trophoblast tissue is mediated by gonadotropin releasing hormone, J Pharmacol Exp Ther 268(2) (1994) 971–7. [PubMed] [Google Scholar]
- [18].Petit A, Gallo-Payet N, Bellabarba D, Lehoux JG, Belisle S, The modulation of placental lactogen release by opioids: a role for extracellular calcium, Mol Cell Endocrinol 90(2) (1993) 165–70. [DOI] [PubMed] [Google Scholar]
- [19].Cemerikic B, Genbacev O, Sulovic V, Beaconsfield R, Effect of morphine on hCG release by first trimester human trophoblast in vitro, Life Sci 42(18) (1988) 1773–9. [DOI] [PubMed] [Google Scholar]
- [20].Zharikova OL, Deshmukh SV, Kumar M, Vargas R, Nanovskaya TN, Hankins GD, Ahmed MS, The effect of opiates on the activity of human placental aromatase/CYP19, Biochem Pharmacol 73(2) (2007) 279–86. [DOI] [PubMed] [Google Scholar]
- [21].Serra AE, Lemon LS, Mokhtari NB, Parks WT, Catov JM, Venkataramanan R, Caritis SN, Delayed villous maturation in term placentas exposed to opioid maintenance therapy: a retrospective cohort study, Am J Obstet Gynecol 216(4) (2017) 418.e1–418.e5. [DOI] [PubMed] [Google Scholar]
- [22].Neradugomma NK, Liao MZ, Mao Q, Buprenorphine, Norbuprenorphine, R-Methadone, and S-Methadone Upregulate BCRP/ABCG2 Expression by Activating Aryl Hydrocarbon Receptor in Human Placental Trophoblasts, Mol Pharmacol 91(3) (2017) 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mao J, Jain A, Denslow ND, Nouri MZ, Chen S, Wang T, Zhu N, Koh J, Sarma SJ, Sumner BW, Lei Z, Sumner LW, Bivens NJ, Roberts RM, Tuteja G, Rosenfeld CS, Bisphenol A and bisphenol S disruptions of the mouse placenta and potential effects on the placenta-brain axis, Proc Natl Acad Sci USA 117(9) (2020) 4642–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gabory A, Ferry L, Fajardy I, Jouneau L, Gothie JD, Vige A, Fleur C, Mayeur S, Gallou-Kabani C, Gross MS, Attig L, Vambergue A, Lesage J, Reusens B, Vieau D, Remacle C, Jais JP, Junien C, Maternal diets trigger sex-specific divergent trajectories of gene expression and epigenetic systems in mouse placenta, PloS One 7(11) (2012) e47986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gallou-Kabani C, Gabory A, Tost J, Karimi M, Mayeur S, Lesage J, Boudadi E, Gross MS, Taurelle J, Vige A, Breton C, Reusens B, Remacle C, Vieau D, Ekstrom TJ, Jais JP, Junien C, Sex- and diet-specific changes of imprinted gene expression and DNA methylation in mouse placenta under a high-fat diet, PloS One 5(12) (2010) e14398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Howerton CL, Morgan CP, Fischer DB, Bale TL, O-GlcNAc transferase (OGT) as a placental biomarker of maternal stress and reprogramming of CNS gene transcription in development, Proc Natl Acad Sci USA 110(13) (2013) 5169–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Langfelder P, Horvath S, WGCNA: an R package for weighted correlation network analysis, BMC Bioinformatics 9 (2008) 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mesa AM, Mao J, Nanjappa MK, Medrano TI, Tevosian S, Yu F, Kinkade J, Lyu Z, Liu Y, Joshi T, Wang D, Rosenfeld CS, Cooke PS, Mice lacking uterine enhancer of zeste homolog 2 have transcriptomic changes associated with uterine epithelial proliferation, Physiol Genomics 52(2) (2020) 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Manshack LK, Conard CM, Bryan SJ, Deem SL, Holliday DK, Bivens NJ, Givan SA, Rosenfeld CS, Transcriptomic alterations in the brain of painted turtles (Chrysemys picta) developmentally exposed to bisphenol A or ethinyl estradiol, Physiol Genomics 49(4) (2017) 201–215. [DOI] [PubMed] [Google Scholar]
- [30].Rosenfeld CS, Hekman JP, Johnson JL, Lyu Z, Ortega MT, Joshi T, Mao J, Vladimirova AV, Gulevich RG, Kharlamova AV, Acland GM, Hecht EE, Wang X, Clark AG, Trut LN, Behura SK, Kukekova AV, Hypothalamic transcriptome of tame and aggressive silver foxes (Vulpes vulpes) identifies gene expression differences shared across brain regions, Genes Brain Beh 19(1) (2020) e12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Szumlinski KK, Coelho MA, Tran T, Stailey N, Lieberman D, Gabriella I, Swauncy I, Brewin LW, Ferdousian S, Who is HOT and who is LOT? Detailed characterization of prescription opioid-induced changes in behavior between 129P3/J and 129S1/SvlmJ mouse substrains, Genes Brain Beh (2019) e12609. [DOI] [PubMed] [Google Scholar]
- [32].Zhang Y, Picetti R, Butelman ER, Schlussman SD, Ho A, Kreek MJ, Behavioral and neurochemical changes induced by oxycodone differ between adolescent and adult mice, Neuropsychopharmacol 34(4) (2009) 912–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Liu YL, Liang JH, Yan LD, Su RB, Wu CF, Gong ZH, Effects of l-tetrahydropalmatine on locomotor sensitization to oxycodone in mice, Acta Pharmacol Sin 26(5) (2005) 533–8. [DOI] [PubMed] [Google Scholar]
- [34].Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R, Male development of chromosomally female mice transgenic for Sry, Nature 351(6322) (1991) 117–21. [DOI] [PubMed] [Google Scholar]
- [35].Velayuthaprabhu S, Matsubayashi H, Sugi T, Nakamura M, Ohnishi Y, Ogura T, Archunan G, Expression of apoptosis in placenta of experimental antiphospholipid syndrome mouse, Am J Reprod Immunol 69(5) (2013) 486–94. [DOI] [PubMed] [Google Scholar]
- [36].Velayuthaprabhu S, Matsubayashi H, Sugi T, Nakamura M, Ohnishi Y, Ogura T, Tomiyama T, Archunan G, A unique preliminary study on placental apoptosis in mice with passive immunization of anti-phosphatidylethanolamine antibodies and anti-factor XII antibodies, Am J Reprod Immunol 66(5) (2011) 373–84. [DOI] [PubMed] [Google Scholar]
- [37].Martin M, Cutadapt removes adapter sequences from high-throughput sequencing reads, EMBnet.journal; Vol 17, No 1: Next Generation Sequencing Data AnalysisDO - 10.14806/ej.17.1.200 (2011). [DOI] [Google Scholar]
- [38].Kim D, Langmead B, Salzberg SL, HISAT: a fast spliced aligner with low memory requirements, Nat Methods 12(4) (2015) 357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Liao Y, Smyth GK, Shi W, featureCounts: an efficient general purpose program for assigning sequence reads to genomic features, Bioinformatics 30(7) (2014) 923–30. [DOI] [PubMed] [Google Scholar]
- [40].Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol 15(12) (2014) 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Jain A, Tuteja G, TissueEnrich: Tissue-specific gene enrichment analysis, Bioinformatics 35(11) (2019) 1966–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, Ren B, A map of the cis-regulatory sequences in the mouse genome, Nature 488(7409) (2012) 116–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C, STRING v10: protein-protein interaction networks, integrated over the tree of life, Nucleic Acids Res 43(Database issue) (2015) D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Chin C-H, Chen S-H, Wu H-H, Ho C-W, Ko M-T, Lin C-Y, Cytohubba: Identifying hub objects and sub-networks from complex interactome, BMC Syst. Biol 8(4) (2014) S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T, Cytoscape: a software environment for integrated models of biomolecular interaction networks, Genome Res. 13(11) (2003) 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Solano ME, Thiele K, Kowal MK, Arck PC, Identification of suitable reference genes in the mouse placenta, Placenta 39 (2016) 7–15. [DOI] [PubMed] [Google Scholar]
- [47].Tait S, Tassinari R, Maranghi F, Mantovani A, Bisphenol A affects placental layers morphology and angiogenesis during early pregnancy phase in mice, J Appl Toxicol 35(11) (2015) 1278–91. [DOI] [PubMed] [Google Scholar]
- [48].Tachibana T, Wakimoto Y, Nakamuta N, Phichitraslip T, Wakitani S, Kusakabe K, Hondo E, Kiso Y, Effects of bisphenol A (BPA) on placentation and survival of the neonates in mice, J Reprod Dev 53(3) (2007) 509–14. [DOI] [PubMed] [Google Scholar]
- [49].Johnson SA, Spollen WG, Manshack LK, Bivens NJ, Givan SA, Rosenfeld CS, Hypothalamic transcriptomic alterations in male and female California mice (Peromyscus californicus) developmentally exposed to bisphenol A or ethinyl estradiol, Physiol Rep 5(3) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, STRING v10: protein–protein interaction networks, integrated over the tree of life, Nucleic Acids Res. 43(D1) (2014) D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yavarone MS, Shuey DL, Sadler TW, Lauder JM, Serotonin uptake in the ectoplacental cone and placenta of the mouse, Placenta 14(2) (1993) 149–61. [DOI] [PubMed] [Google Scholar]
- [52].Cote F, Fligny C, Bayard E, Launay JM, Gershon MD, Mallet J, Vodjdani G, Maternal serotonin is crucial for murine embryonic development, Proc Natl Acad Sci USA 104(1) (2007) 329–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Nagao T, Kagawa N, Saito Y, Komada M, Developmental effects of oral exposure to diethylstilbestrol on mouse placenta, J Appl Toxicol 33(11) (2013) 1213–21. [DOI] [PubMed] [Google Scholar]
- [54].Suh CH, Cho NK, Lee CK, Lee CH, Kim DH, Kim JH, Son BC, Lee JT, Perfluorooctanoic acid-induced inhibition of placental prolactin-family hormone and fetal growth retardation in mice, Mol Cell Endocrinol 337(1-2) (2011) 7–15. [DOI] [PubMed] [Google Scholar]
- [55].Liu J, Xu W, Sun T, Wang F, Puscheck E, Brigstock D, Wang QT, Davis R, Rappolee DA, Hyperosmolar stress induces global mRNA responses in placental trophoblast stem cells that emulate early post-implantation differentiation, Placenta 30(1) (2009) 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Mantione KJ, Angert RM, Cadet P, Kream RM, Stefano GB, Identification of a μ opiate receptor signaling mechanism in human placenta, Med Sci Monit 16(11) (2010) Br347–52. [PubMed] [Google Scholar]
- [57].Zhu Y, Pintar JE, Expression of opioid receptors and ligands in pregnant mouse uterus and placenta, Biol Reprod 59(4) (1998) 925–32. [DOI] [PubMed] [Google Scholar]
- [58].Guo C, Yang Y, Shi MX, Wang B, Liu JJ, Xu DX, Meng XH, Critical time window of fenvalerate-induced fetal intrauterine growth restriction in mice, Ecotoxicol Environ Saf 172 (2019) 186–193. [DOI] [PubMed] [Google Scholar]
- [59].Finkenzeller D, Fischer B, Lutz S, Schrewe H, Shimizu T, Zimmermann W, Carcinoembryonic antigen-related cell adhesion molecule 10 expressed specifically early in pregnancy in the decidua is dispensable for normal murine development, Mol Cell Biol 23(1) (2003) 272–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Simmons DG, Rawn S, Davies A, Hughes M, Cross JC, Spatial and temporal expression of the 23 murine Prolactin/Placental Lactogen-related genes is not associated with their position in the locus, BMC Genomics 9 (2008) 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Bu P, Alam SM, Dhakal P, Vivian JL, Soares MJ, A prolactin family paralog regulates placental adaptations to a physiological stressor, Biol Reprod 94(5) (2016) 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Hu D, Cross JC, Ablation of Tpbpa-positive trophoblast precursors leads to defects in maternal spiral artery remodeling in the mouse placenta, Dev Biol 358(1) (2011) 231–9. [DOI] [PubMed] [Google Scholar]
- [63].Henke C, Ruebner M, Faschingbauer F, Stolt CC, Schaefer N, Lang N, Beckmann MW, Strissel PL, Strick R, Regulation of murine placentogenesis by the retroviral genes Syncytin-A, Syncytin-B and Peg10, Differentiation 85(4-5) (2013) 150–60. [DOI] [PubMed] [Google Scholar]
- [64].Bale TL, The placenta and neurodevelopment: sex differences in prenatal vulnerability, Dialogues Clin Neurosci 18(4) (2016) 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Gabory A, Ferry L, Fajardy I, Jouneau L, Gothié JD, Vigé A, Fleur C, Mayeur S, Gallou-Kabani C, Gross MS, Attig L, Vambergue A, Lesage J, Reusens B, Vieau D, Remacle C, Jais JP, Junien C, Maternal diets trigger sex-specific divergent trajectories of gene expression and epigenetic systems in mouse placenta, PloS One 7(11) (2012) e47986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Gallou-Kabani C, Gabory A, Tost J, Karimi M, Mayeur S, Lesage J, Boudadi E, Gross MS, Taurelle J, Vigé A, Breton C, Reusens B, Remacle C, Vieau D, Ekström TJ, Jais JP, Junien C, Sex- and diet-specific changes of imprinted gene expression and DNA methylation in mouse placenta under a high-fat diet, PloS One 5(12) (2010) e14398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Gabory A, Roseboom TJ, Moore T, Moore LG, Junien C, Placental contribution to the origins of sexual dimorphism in health and diseases: sex chromosomes and epigenetics, Biol Sex Differ 4(1) (2013) 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Handwerger S, Myers S, Richards R, Richardson B, Turzai L, Moeykins C, Meyer T, Anantharamahiah GM, Apolipoprotein A-I stimulates placental lactogen expression by human trophoblast cells, Endocrinology 136(12) (1995) 5555–60. [DOI] [PubMed] [Google Scholar]
- [69].Pantaleon M, Steane SE, McMahon K, Cuffe JSM, Moritz KM, Placental O-GlcNAc-transferase expression and interactions with the glucocorticoid receptor are sex specific and regulated by maternal corticosterone exposure in mice, Sci Reports 7(1) (2017) 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Lash GE, Otun HA, Innes BA, Kirkley M, De Oliveira L, Searle RF, Robson SC, Bulmer JN, Interferon-gamma inhibits extravillous trophoblast cell invasion by a mechanism that involves both changes in apoptosis and protease levels, FASEB J 20(14) (2006) 2512–8. [DOI] [PubMed] [Google Scholar]
- [71].Zhang L, Zhao M, Jiao F, Xu X, Liu X, Jiang Y, Zhang H, Ou X, Hu X, Interferon gamma is involved in apoptosis of trophoblast cells at the maternal-fetal interface following Toxoplasma gondii infection, Int J Infect Dis 30 (2015) 10–6. [DOI] [PubMed] [Google Scholar]
- [72].Klisch K, Leiser R, In bovine binucleate trophoblast giant cells, pregnancy-associated glycoproteins and placental prolactin-related protein-I are conjugated to asparagine-linked N-acetylgalactosaminyl glycans, Histochem Cell Biol 119(3) (2003) 211–7. [DOI] [PubMed] [Google Scholar]
- [73].Szafranska B, Majewska M, Panasiewicz G, N-glycodiversity of the Pregnancy-Associated Glycoprotein family (PAG) produced in vitro by trophoblast and trophectoderm explants during implantation, placentation and advanced pregnancy in the pig, Reprod Biol 4(1) (2004) 67–89. [PubMed] [Google Scholar]
- [74].Bielinska M, Matzuk MM, Boime I, Site-specific processing of the N-linked oligosaccharides of the human chorionic gonadotropin alpha subunit, J Biol Chem 264(29) (1989) 17113–8. [PubMed] [Google Scholar]
- [75].Berglund E, Maaskola J, Schultz N, Friedrich S, Marklund M, Bergenstrahle J, Tarish F, Tanoglidi A, Vickovic S, Larsson L, Salmen F, Ogris C, Wallenborg K, Lagergren J, Stahl P, Sonnhammer E, Helleday T, Lundeberg J, Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity, Nat Commun 9(1) (2018) 2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Maniatis S, Aijo T, Vickovic S, Braine C, Kang K, Mollbrink A, Fagegaltier D, Andrusivova Z, Saarenpaa S, Saiz-Castro G, Cuevas M, Watters A, Lundeberg J, Bonneau R, Phatnani H, Spatiotemporal dynamics of molecular pathology in amyotrophic lateral sclerosis, Science 364(6435) (2019) 89–93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Higher magnification region of LA region for each group to show maternal blood vessels (designated with arrows) in each group. For maternal blood vessel area measurement within the labyrinth (LA) region, a box measuring 500 × 500 pixels was placed over 5 to 7 random LA areas for each placenta section. Ten maternal blood vessels in this area were randomly selected, the edge of the blood vessel was outlined, and the area was measured with the Adobe Photoshop program (Adobe, San Jose, CA).
Supplementary Fig. 2. Sex differences in placental gene expression CTL and OXY exposed conceptuses. A) 3D PCA plot of RNAseq results from placenta of CTL males vs. CTL females. B) Heatmap based on all transcripts identified in placenta of CTL males vs. CTL females. C) Volcano plot analyses of gene transcripts identified in placenta of CTL males vs. CTL females. Transcripts represented by Orange: p < 0.05; Transcripts represented by Red: log2FoldChange < −1 or > 1; Transcripts represented by Green: p < 0.05 AND log2FoldChange < −1 or > 1. D) 3D PCA plot of RNAseq results from placenta of OXY males vs. OXY females. E) Heatmap based on all transcripts identified in placenta of OXY males vs. OXY females. F) Volcano plot analyses of gene transcripts identified in placenta of OXY males vs. OXY females. Transcripts represented by Orange: p < 0.05; Transcripts represented by Red: log2FoldChange < −1 or > 1; Transcripts represented by Green: p < 0.05 AND log2FoldChange < −1 or > 1. These analyses are based on the results from 10 CTL males, 9 CTL females, 10 OXY females, and 8 OXY males.
Supplementary Fig. 3. TissueEnrich results for CTL male vs. CTL female placenta and OXY male vs. OXY female placenta. A) Bar plot distribution of the tissue enrichment of all DE genes for CTL male vs. CTL female comparison. B) Heatmap analyses revealing those DE genes for CTL male vs. CTL female comparison that are enriched in the placenta. C) Bar plot distribution of the tissue enrichment of all DE genes for OXT male vs. OXY female comparison. D) Heatmap analyses revealing those DE genes for OXY male vs. OXY female comparison that are enriched in the placenta. These analyses are based on the results from 10 CTL males and 9 CTL females.
Supplementary Fig. 4. STRING and hub gene analyses for CTL males vs. CTL females. PPI were determined by STRING analysis. The PPI files generated with STRING were imported into the cytohubba app [44] in Cytoscape [45] to examine for the top 10 hub genes. Within this program, hub genes were determined with MCC analysis, as recommended [44]. These analyses are based on the results from 10 CTL males and 9 CTL females.
Supplementary Fig. 5. STRING and hub gene analyses for OXY males vs. OXY females. PPI were determined by STRING analysis. The PPI files generated with STRING were imported into the cytohubba app [44] in Cytoscape [45] to examine for the top 10 hub genes. Within this program, hub genes were determined with MCC analysis, as recommended [44]. These analyses are based on the results from 8 OXY males and 10 OXY females.
Supplementary Fig. 6. qPCR results for placental enriched genes. Ceacam11, Ceacam12, Ceacam14, Prl2b1, Prl7b1, and Tpbpb were upregulated in OXY female vs. CTL female placenta. Ceacam13 and Tpbpa only showed a trend towards upregulation with these analyses (p= 0.07 and 0.1, respectively). Tpbpa was significantly reduced in OXY male vs. CTL male placenta (p < 0.05), and Tpbpb tended to be lower for this comparison (p = 0.1). None of the other placental enriched genes tested with qPCR analyses were DE in OXY male vs. CTL male placenta.
Supplementary Fig. 7. Dendrogram and heatmap of WGCNA results for OXY female vs. CTL female placental comparison. As detailed in [27], the heatmap represents the Topological Overlap Matrix (TOM) for all analyzed genes. Lighter colors in the heatmap depict low overlap and darker red colors indicate greater overlap. Along the diagonal, blocks of darker colors represent the different color modules. The gene dendrogram and module assignment are shown at the left and top of the image. This analysis is based on the results from 10 OXY females and 9 CTL females.
Supplementary Fig. 8. TissueEnrich results for ME Brown and ME Magenta for OXY female vs. CTL female placental comparison A) Bar plot distribution of the tissue enrichment of genes in ME Brown for this comparison. B) Heatmap analyses of genes within ME Brown for this comparison. C) Bar plot distribution of the tissue enrichment of genes within ME Magenta for this comparison. D) Heatmap analyses of genes within ME Magenta for this comparison. This analysis is based on the results from 10 OXY females and 9 CTL females.
Supplementary Fig. 9. TissueEnrich results for ME Turquoise and ME Green for OXY female vs. CTL female placental comparison A) Bar plot distribution of the tissue enrichment of genes in ME Turquoise for this comparison. B) Heatmap analyses of genes within ME Turquoise for this comparison. C) Bar plot distribution of the tissue enrichment of genes within ME Green for this comparison. D) Heatmap analyses of genes within ME Green for this comparison. This analysis is based on the results from 10 OXY females and 9 CTL females.
Supplementary Fig. 10. Hub genes for ME Brown and ME Magenta for OXY female vs. CTL female placental comparison. This analysis is based on the results from 10 OXY females and 9 CTL females.
Supplementary Fig. 11. Hub genes for ME Turquoise and ME green for OXY female vs. CTL female placental comparison. This analysis is based on the results from 10 OXY females and 9 CTL females.
Supplementary Fig. 12. Dendrogram and heatmap of WGCNA results for OXY male vs. CTL male placental comparison. As detailed in [27], the heatmap represents the Topological Overlap Matrix (TOM) for all analyzed genes. Lighter colors in the heatmap depict low overlap and darker red colors indicate greater overlap. Along the diagonal, blocks of darker colors represent the different color modules. The gene dendrogram and module assignment are shown at the left and top of the image. This analysis is based on the results from 8 OXY males and 10 CTL males.
Supplementary Fig. 13. Dendrogram and heatmap of WGCNA results for CTL male vs. CTL female placental comparison. As detailed in [27], the heatmap represents the Topological Overlap Matrix (TOM) for all analyzed genes. Lighter colors in the heatmap depict low overlap and darker red colors indicate greater overlap. Along the diagonal, blocks of darker colors represent the different color modules. The gene dendrogram and module assignment are shown at the left and top of the image. This analysis is based on the results from 10 CTL males and 9 CTL females.
Supplementary Fig. 14. Relationship of WGCNA results and placental morphometric assessments for CTL male vs. CTL female placental comparison. Modules identified in Supplementary Fig. 13 were correlated with placental morphometric assessments (Fig. 2). Each row corresponds to a Module Eigengene (ME) and colors represent the correlation coefficient between the ME and one of the four placental histological measurements (LA to pTGC ratio; SpongioTB to pTGC ratio; pTGC apoptosis; Maternal blood vessel area in LA region). Numbers at the top of rows represents degree of correlation, and values with a negative integer indicate an inverse correlation with one of the four placental measurements. ME purple was negatively associated with LA to pTGC ratio (r = −0.53; p = 0.02). ME Blue, Red and Grey were positively associated with SpongioTB to pTGC ratio (ME Blue: r = 0.57; p = 0.01; ME Red: r = 0.48; p = 0.05; ME Grey: r = 0.48; p = 0.04). A legend that ranges from blue to red shades (−1 to 1, respectively) is included to the right of the Figures. The genes within each of these color modules is provided in Supplementary File 8. This analysis is based on the results from 10 CTL males and 9 CTL females.
Supplementary Fig. 15. TissueEnrich results for ME Purple, ME Blue, and ME Red for CTL male vs. CTL female placental comparison A) Bar plot distribution of the tissue enrichment of genes in ME Purple for this comparison. B) Heatmap analyses of genes within ME Purple for this comparison. C) Bar plot distribution of the tissue enrichment of genes within ME Blue for this comparison. D) Heatmap analyses of genes within ME Blue for this comparison. C) Bar plot distribution of the tissue enrichment of genes within ME Red for this comparison. D) Heatmap analyses of genes within ME Red for this comparison. This analysis is based on the results from 10 CTL males and 9 CTL females.
Supplementary Fig. 16. Hub genes for ME Purple, ME Blue, and ME Red for CTL male vs. CTL female placental comparison. This analysis is based on the results from 10 CTL males and 9 CTL females.
Supplementary Fig. 17. Dendrogram and heatmap of WGCNA results for OXY male vs. OXY female placental comparison. As detailed in [27], the heatmap represents the Topological Overlap Matrix (TOM) for all analyzed genes. Lighter colors in the heatmap depict low overlap and darker red colors indicate greater overlap. Along the diagonal, blocks of darker colors represent the different color modules. The gene dendrogram and module assignment are shown at the left and top of the image. This analysis is based on the results from 8 OXY males and 10 OXY females.
Supplementary Fig. 18. Relationship of WGCNA results and placental morphometric assessments for OXY male vs. OXY female placental comparison. Modules identified in Supplementary Fig. 17 were correlated with placental morphometric assessments (Fig. 2). Each row corresponds to a Module Eigengene (ME) and colors represent the correlation coefficient between the ME and one of the four placental histological measurements (LA to pTGC ratio; SpongioTB to pTGC ratio; pTGC apoptosis; Maternal blood vessel area in LA region). Numbers at the top of rows represents degree of correlation, and values with a negative integer indicate an inverse correlation with one of the four placental measurements. ME Grey was inversely associated with LA to pTGC ratio (r = −0.53; p = 0.03). A legend that ranges from blue to red shades (−1 to 1, respectively) is included to the right of the Figure. The genes within this module are included for reference in Supplementary File 10. This analysis is based on the results from 8 OXY males and 10 OXY females.