Abstract
Osteoporosis (OP) results from an imbalance between bone formation, which is regulated by osteoblasts, and bone resorption, which is mediated by osteoclasts. MicroRNA-22-3p (miR-22-3p) expression is decreased during the process of osteoclast differentiation and p38α mitogen-activated protein kinase (MAPK)14 promotes the proliferation and differentiation of osteoclast progenitors. However, whether miR-22-3p could target MAPK14 to regulate the progression of OP remains unknown, which was the aim of the present study. CD14+ PBMCs were used for the establishment of osteoclastic differentiation in vitro. In the present study, reverse transcription quantitative PCR was used to determine the mRNA expression of MAPK14, tartrate resistant acid phosphatase (TRAP), nuclear factor of activated T-cells (NFATC1) and cathepsin K (CTSK). Western blotting was applied to determine the protein expression of MAPK14, TRAP, NFATC1, CTSK, p-p65 and p65. Dual luciferase reporter assay was applied to confirm the relation between miR-22-3p and MAPK14. Cell Counting Kit-8 assay and flow cytometry assays were used to determine the cell proliferation and cell apoptosis, respectively. The results demonstrated that miR-22-3p expression was lower while MAPK14 expression was higher in the serum from patients with OP compared with healthy volunteers. Furthermore, miR-22-3p expression was negatively correlated with MAPK14 expression in patients with OP. In addition, miR-22-3p expression was decreased and MAPK14 expression was increased during the progression of CD14+peripheral blood mononuclear cells (PBMCs) osteoclastic differentiation in a time-dependent manner. Furthermore, miR-22-3p inhibited the proliferation and differentiation and promoted the apoptosis of CD14+PBMCs by targeting MAPK14. In summary, the findings from the present study suggested that miR-22-3p may serve a potential therapeutic role in patients with OP.
Keywords: osteoporosis, microRNA-22-3p, mitogen-activated protein kinase 14
Introduction
Osteoporosis (OP) remains the most common progressive skeletal disease. This disease limits the activity of patients (1) and decreases the quality of life in elders and postmenopausal women (2). The modeling and remodeling of bone is a dynamic metabolic process primarily mediated by the osteoblasts, which form new bone by secretion of bone matrix and acceleration of calcium (Ca2+) deposition, and the osteoclasts, which resorb old bone by resolving mineralized bone matrix (3,4). In case of imbalance between osteoblastic bone formation and osteoclastic bone resorption (5), which can lead to upregulated bone resorption, downregulated bone formation or both (6,7), OP can occur.
MicroRNAs (miRNAs) are a subclass of non-coding RNAs of 19-25 nucleotides in length, which regulate the expression of target genes at post-transcriptional level (8). Previous studies have reported the importance of miRNAs in the regulation of skeletal development, bone formation and homeostasis (9,10). For example, miR-125b, miR-29a and miR-378 have been demonstrated to be involved in osteoblastic differentiation (11-13). Furthermore, miR-21, miR-155 and miR-223 have been found to be implicated in osteoclastic differentiation (14,15). In 2015, miR-22-3p expression was found to be decreased in the bone of patients who suffered a bone fracture (16). In 2016, miR-22-3p was discovered to be downregulated during the progression of osteoclastic differentiation (17). In 2018, miR-22-3p expression was reported to be decreased in the bone of patients with OP (18). In 2020, extracellular vesicle-encapsulated miR-22-3p from bone marrow mesenchymal stem cells were found to induce osteogenic differentiation via Fat mass- and obesity-associated gene inhibition (19). However, the underlying mechanisms of miR-22-3p during the process of osteoclast differentiation have not yet been reported, which was explored in the present study.
Materials and methods
Clinical samples
A total of 30 healthy volunteers (mean age, 59.79±6.53 years; age range, 47-68 years) and 30 postmenopausal women with OP (mean age, 60.23±7.15 years; age range, 49-69 years) were included in the present study. Blood samples (5 ml) were collected from each participant for the determination of miR-22-3p and p38α mitogen-activated protein kinase (MAPK)14 expression. This study was approved by the Ethics Committee of Chongqing Public Health Medical Center (approval no. CMC20180106). Each participant provided a signed informed consent prior to the beginning of the study.
Bone mineral density (BMD) measurement
The BMD of each participant was determined at the lumbar spine (L1-L4) and the left femoral neck through dual-energy X-ray absorptiometry (DXA) using a Hologic 4500 bone densitometer (Hologic), according to the World Health Organization criteria (20). Each scan was handled by a single technician and repeated thrice with repositioning between each scan. The precision of the machine, which is presented as percentage coefficient of variation (CV%), varied between subregions. CV% for BMD of the lumbar spine (L1-L4) was <1%, whereas CV% for BMD of the left femoral was <2%.
Isolation and incubation of CD14+peripheral blood mononuclear cells (PBMCs)
Osteoclasts are primary bone-resorbing cells which can form from precursor fusion, and CD14+PBMCs are early progenitors for osteoclasts (21). Furthermore, osteoclast formation can occur in CD14+PBMCs with the stimulation of macrophage colony stimulating factor (M-CSF) and receptor activator of nuclear factor-κB ligand (RANKL) (22). Subsequently, the present study used CD14+PBMCs for the study of OP in vitro. PBMCs were extracted as previously described (22). CD14+PBMCs were purified using CD14 antibody-coated magnetic cell sorting MicroBeads (Miltenyi Biotec GmbH). When CD14+PBMCs reached 90% purity by flow cytometry, they were seeded into 48-well plates at the density of 2.5x105 cells/well. CD14+PBMCs were incubated in α-MEM (Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), penicillin (50 IU/ml) and streptomycin (50 mg/ml) (Gibco; Thermo Fisher Scientific, Inc.) and placed in an incubator with 5% CO2 at 37˚C for 2 h.
Osteoclastic differentiation
For the establishment of osteoclastic differentiation in vitro, CD14+PBMC initial complete culture medium was changed for α-MEM containing M-CSF (25 ng/ml; R&D Systems, Inc.) and RANKL (25 ng/ml; R&D Systems, Inc.). The complete culture medium containing differentiation factors, 10% FBS, penicillin (50 IU/ml) and streptomycin (50 mg/ml) was refreshed every three days and floating cells were removed. After incubation for six days, CD14+PBMCs were collected for subsequent experiments.
Cell transfection
For the overexpression of miR-22-3p, CD14+PBMCs (2x104 cells/ml; 200 µl) were seeded into 48-well plates and transfected with miR-NC mimic (sense, 5'-UUCUCCGAACGUGUCACGU-3' and antisense, 5'-ACGUGACACGUUCGGAGAA-3') or miR-22-3p mimic (sense, 5'-AAAAGCUGCCAGUUGAAGAACUGU-3' and antisense, 5'-ACAGUUCUUCAACUGGCAGCUUUU-3') (50 nmol/l; Guangzhou Ribobio Co., Ltd.) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After transfection for 48 h, CD14+PBMCs were subsequently treated with M-CSF and RANKL as aforementioned and collected for subsequent experiments.
To overexpress MAPK14, CD14+PBMCs (2x104 cells/ml; 200 µl) were seeded into 48-well plates and transfected with pcDNA3.1 or pcDNA3.1-MAPK14 (2 µg; Sangon Biotech Co., Ltd.) using Lipofectamine 2000. After transfection for 48 h, CD14+PBMCs were subsequently treated with M-CSF and RANKL as aforementioned and collected for subsequent experiments. At 48 h after the transfection, cells were used for subsequent experiments.
Dual luciferase reporter assay
The binding site between miR-22-3p and MAPK14 was predicted by TargetScan 7.1 (http://www.targetscan.org/vert_71/). To verify the interaction between miR-22-3p and MAPK14, CD14+PBMCs (1x105 cells/ml) were seeded into 24-well plates and co-transfected with wild type (WT) or mutant (MUT) MAPK14 3'-UTR (400 ng) which was cloned into psiCHECK2 (Promega Corporation) and miR-NC mimic or miR-22-3p mimic (20 nmol/l) using Lipofectamine 2000. After incubation for 48 h, CD14+PBMCs were collected for the determination of luciferase activity with the dual-luciferase reporter assay (Promega Corporation). The relative luciferase activity was normalized to Renilla luciferase activity (Promega corporation).
Cell proliferation assay
CD14+PBMC proliferation was determined using Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.). CD14+PBMCs were seeded into 96-well (1x103/well) and incubated for 0, 24, 48 or 72 h. CCK-8 solution (10 µl) was added into each well and incubated for 1 h at 37˚C. Absorbance at 450 nm was determined on a microplate reader.
Cell apoptosis assay
CD14+PBMC cell apoptosis was determined using the Annexin V-FITC/propidium iodide (PI) Apoptosis Detection kit (Sigma-Aldrich; Merck KGaA) on a flow cytometer (BD Biosciences). Briefly, CD14+PBMCs were collected by after centrifugation at 1,000 x g for 5 min at 4˚C and resuspended in 500 µl binding buffer. Subsequently, CD14+PBMCs were stained with Annexin V-FITC (100 µl) for 15 min and PI (10 µl) for 5 min in the dark. The cell apoptotic rate was quantified on a flow cytometry and analyzed by CellQuest version 3.3 (BD Biosciences).
RNA isolation from serum and cells
Serum samples were obtained from whole blood by centrifugation at 2,000 x g for 15 min at room temperature after being incubated at room temperature for 30 min. For detection of miRNA, total RNA was isolated from serum (200 µl) of participants or CD14+PBMCs using miRNeasy Serum/Plasma kit (Qiagen GmbH). For the detection of mRNA, total RNA was isolated from serum of participants or CD14+PBMCs using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.).
Reverse transcription quantitative (RT-q)PCR
Complementary DNA (cDNA) was obtained from miRNA and mRNA using TaqMan MicroRNA Reverse Transcription kit (Thermo Fisher Scientific, Inc.) and First Strand cDNA Synthesis Kit (Takara Bio, Inc.), respectively. RT-qPCR reactions were performed as follows: 95˚C for 30 sec, followed by 40 cycles at 95˚C for 5 sec and at 60˚C for 20 sec using SYBR Green qPCR assay kit (Takara Bio, Inc.) on an ABI 7500 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The relative expression levels of miR-22-3p and of MAPK14, tartrate resistant acid phosphatase (TRAP), nuclear factor of activated T-cells (NFATC1) and cathepsin K (CTSK) were normalized to endogenous controls U6 or GAPDH, respectively, and were expressed as 2-∆∆Cq (23). The following primer sequences were used: GAPDH forward, 5'-GCACCGTCAAGGCTGAG AAC-3' and reverse, 5'-TGGTGAAGACGCCAGTGGA-3'; U6 forward, 5'-GTGCTCGCTTCGGCAGCACAT-3' and reverse, 5'-AATATGGAACGCTTCACGAAT-3'; CTSK forward, 5'-TCCGCAATCCTTACCGAATA-3' and reverse, 5'-AACTTGAACACCCACATCCTG-3'; NFATC1 forward, 5'-TACCAGCGTTTCACCTACCT-3' and reverse, 5'-CCCTTTTCCTTTCCTTTTCA-3'; TRAP forward, 5'-AGACATCAATGACAAGAGGT-3' and reverse, 5'-AAG TGCAGGCGGTAGAAAGG-3'; miR-22-3p forward, 5'-AAGCTGCCAGTTGAAGAACTGT-3' and reverse, 5'-ACAGTTCTTCAACTGGCAGCTT-3'; MAPK14 forward, 5'-GAAAAGGGTCTTCTTGGCAGCTT-3' and reverse, 5'-AAGCTGCCAAGAAGACCCTTTTC-3'.
Western blotting
CD14+PBMCs were lysed on ice using RIPA (Sigma-Aldrich; Merck KGaA) containing PMSF and protease inhibitors (Roche Diagnostics GmbH). Protein concentration was determined by a BCA kit (Thermo Fisher Scientific, Inc.). Proteins (20 µg per lane) were separated by 10% SDS-PAGE and transferred onto PVDF membranes (EMD Millipore). Membranes were blocked with 5% bovine serum albumin (Beyotime Institute of Biotechnology) at room temperature for 1 h and were incubated with primary antibodies against GAPDH (1:1,000; cat. no. 5174; Cell Signaling Technology, Inc.), MAPK14 (1:1,000; cat. no. 8690; Cell Signaling Technology, Inc.), TRAP (1:1,000; cat. no. ab65854; Abcam), NFATC1 (1:1,000; cat. no. ab25916; Abcam), CTSK (1:1,000; cat. no. ab37259; Abcam), p-p65 (1:1,000; cat. no. 3033; Cell Signaling Technology, Inc.) and p65 (1:1,000; cat. no. 8242; Cell Signaling Technology, Inc.) at 4˚C overnight. Membranes were then incubated with anti-rabbit (1:2,000; cat. no. 7074) or anti-mouse HRP-conjugated secondary antibody (1:2,000; cat. no. 7076; both from Cell Signaling Technology, Inc.) at room temperature for 1 h. Enhanced chemiluminescence reagent (Bio-Rad Laboratories, Inc.) was used to detect the signal on the membrane. Densitometry analysis was performed using Image J software (v1.50; National Institutes of Health).
Statistical analyses
Statistical analyses were performed with the GraphPad Prism 6.0 (GraphPad Software, Inc.). Data were expressed as the means ± standard deviation. Unpaired student's t-test was used to analyze differences between two groups from CD14+PBMCs and participants. One-way ANOVA followed by Tukey's post hoc test was used to analyze differences among multiple groups. Pearson correlation analysis was used to analyze the relationship between miR-22-3p and MAPK14 expression. P<0.05 was considered to indicate a statistically significant difference.
Results
Clinicopathological characteristics of participants
The clinicopathological characteristics of participants are presented in Table I. There was no significant difference between the mean age of postmenopausal women with OP and healthy volunteers (60.23±7.15 years vs. 59.79±6.53 years, respectively). The body mass indexes in the two groups were also not different (24.23±3.09 vs. 23.59±4.14 in postmenopausal women with OP and healthy volunteers, respectively). The mean femoral neck BMD and lumbar spine BMD (0.62±0.13 and 0.61±0.09, respectively) of patients with postmenopausal OP were significantly lower compared with those of healthy volunteers (0.89±0.12 and 0.85±0.16, respectively). The comparison of BMD was not adjusted for age and BMI according to a previous study (24).
Table I.
Clinicopathological characteristics of participants.
| Variable | Healthy volunteers | Patients with OP | P-value |
|---|---|---|---|
| Age, years | 59.79±6.53 | 60.23±7.15 | >0.05 |
| BMI, kg/m2 | 23.59±4.14 | 24.23±3.09 | >0.05 |
| Femoral neck BMD, g/cm2 | 0.89±0.12 | 0.62±0.13 | <0.001 |
| Lumbar spine BMD, g/cm2 | 0.85±0.16 | 0.61±0.09 | <0.001 |
miR-22-3p expression level in participants and CD14+PBMCs
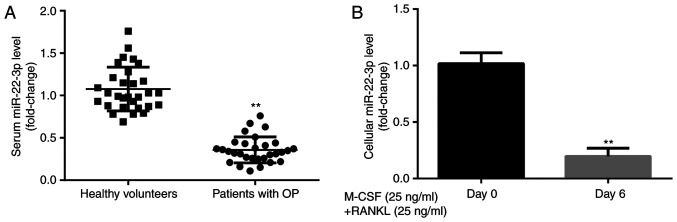
To explore the effects of miR-22-3p during the development of OP, miR-22-3p expression level was evaluated in healthy volunteers and patients with postmenopausal OP. The results from RT-qPCR demonstrated that miR-22-3p expression was significantly lower in patients with postmenopausal OP compared with healthy volunteers (Fig. 1A). Furthermore, miR-22-3p expression level was significantly lower in CD14+PBMCs treated with M-CSF and RANKL for 6 days compared with those untreated (Fig. 1B). These results suggested that miR-22-3p may be involved in the development of OP.
Figure 1.
miR-22-3p expression level is decreased in serum and CD14+PBMCs from patients with OP. miR-22-3p expression was lower in (A) patients with postmenopausal OP compared with healthy volunteers and (B) CD14+PBMCs treated with M-CSF and RANKL for 6 days compared with untreated cells. **P<0.01. OP, osteoporosis; miR, microRNA; PBMCs, peripheral blood mononuclear cells; M-CSF, macrophage colony stimulating factor; RANKL, receptor activator of nuclear factor-κB ligand.
miR-22-3p targets the expression of MAPK14
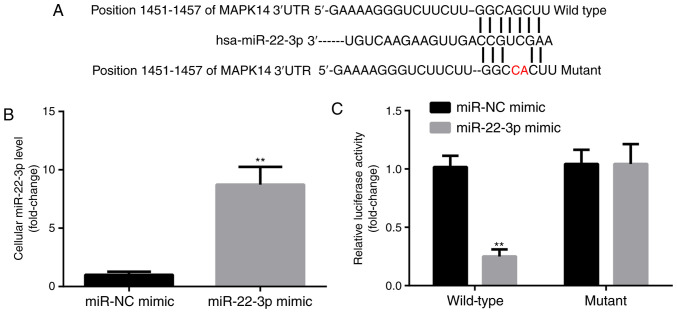
MAPK14 was predicted to be a target gene for miR-22-3p by TargetScan. The predicted pairing between MAPK14 3'UTR and miR-22-3p is presented in Fig. 2A. In addition, compared with miR-NC mimic group, miR-22-3p mimic significantly increased miR-22-3p expression level in CD14+PBMCs (Fig. 2B). Subsequently, the interaction between miR-22-3p and MAPK14 in CD14+PBMCs was detected by dual luciferase reporter assay. Compared with miR-NC mimic group, miR-22-3p mimic significantly decreased the luciferase activity of the WT-MAPK14 but not MUT-MAPK14 (Fig. 2C). These findings confirmed the potential interaction between miR-22-3p and MAPK14.
Figure 2.
miR-22-3p targets the expression of MAPK14. (A) Binding sites between MAPK14 and miR-22-3p. (B) miR-22-3p mimic was successfully transfected into CD14+PBMCs. (C) miR-22-3p targeted WT-MAPK14. **P<0.01. miR, microRNA; PBMCs, peripheral blood mononuclear cells; NC, negative control.
MAPK14 expression in participants and CD14+PBMCs
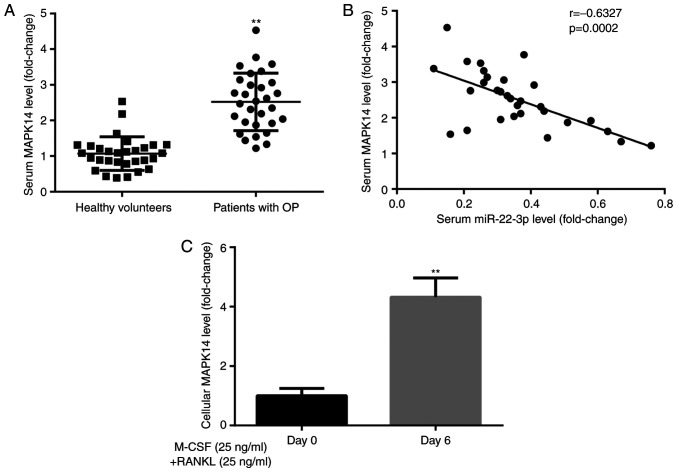
To explore the effects of MAPK14 during the development of OP, MAPK14 expression was evaluated in healthy volunteers and patients with postmenopausal OP. The results from RT-qPCR demonstrated that MAPK14 expression was significantly higher in patients with postmenopausal OP compared with healthy volunteers (Fig. 3A). Furthermore, Pearson correlation analysis demonstrated that miR-22-3p and MAPK14 expression levels were negatively correlated in patients with postmenopausal OP (Fig. 3B).
Figure 3.
MAPK14 expression level is increased in serum and CD14+PBMCs from patients with OP.(A) MAPK14 expression was higher in patients with postmenopausal OP compared with healthy volunteers. (B) Negative correlation between miR-22-3p and MAPK14 expression. (C) MAPK14 expression was increased in CD14+PBMCs treated with M-CSF and RANKL for 6 days compared with untreated cells. **P<0.01. OP, osteoporosis; miR, microRNA; PBMCs, peripheral blood mononuclear cells; M-CSF, macrophage colony stimulating factor; RANKL, receptor activator of nuclear factor-κB ligand.
In addition, results from RT-qPCR showed that MAPK14 expression level was significantly higher in CD14+PBMCs treated with M-CSF and RANKL for 6 days compared with those untreated (Fig. 3C). These findings suggested that MAPK14 may be implicated in the development of OP.
miR-22-3p mimic inhibits CD14+PBMC proliferation by targeting MAPK14
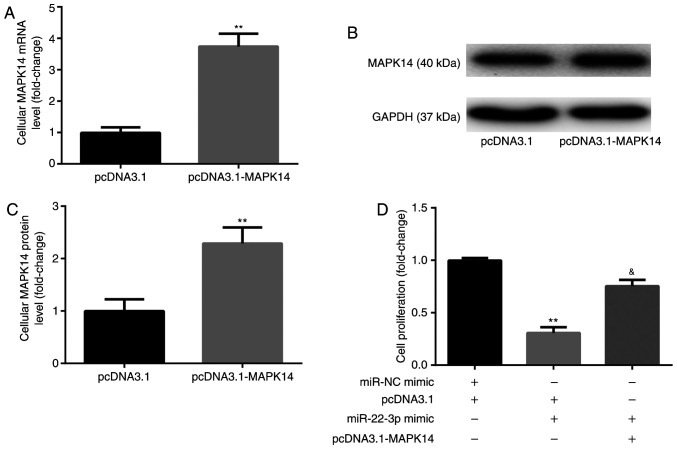
Prior to the assessment of cell proliferation, CD14+PBMCs were transfected with pcDNA3.1-MAPK14 to overexpress MAPK14. The results from RT-qPCR and western blotting demonstrated that the mRNA (Fig. 4A) and protein (Fig. 4B and C) expression of MAPK14 was significantly increased following transfection with pcDNA3.1-MAPK14 in CD14+PBMCs compared with pcDNA3.1 group, which confirmed the successful transfection of pcDNA3.1-MAPK14 into CD14+PBMCs.
Figure 4.
miR-22-3p mimic inhibits CD14+PBMC proliferation by targeting MAPK14. (A-C) pcDNA3.1-MAPK14 was successfully transfected in CD14+PBMCs. (D) pcDNA3.1-MAPK14 reversed the effects of miR-22-3p mimic on CD14+PBMCs proliferation. **P<0.01 vs. pcDNA3.1 or miR-NC mimic + pcDNA3.1. &P<0.05 vs. miR-22-3p mimic + pcDNA3.1. OP, osteoporosis; miR, microRNA; PBMCs, peripheral blood mononuclear cells; M-CSF, macrophage colony stimulating factor; RANKL, receptor activator of nuclear factor-κB ligand; NC, negative control.
The results from CCK-8 showed that at day 6 of incubation with M-CSF and RANKL, miR-22-3p mimic significantly decreased CD14+PBMC proliferation compared with miR-NC mimic + pcDNA3.1 group, which was significantly rescued following co-transfection with pcDNA3.1-MAPK14 (Fig. 4D).
miR-22-3p mimic promotes CD14+PBMC apoptosis by targeting MAPK14
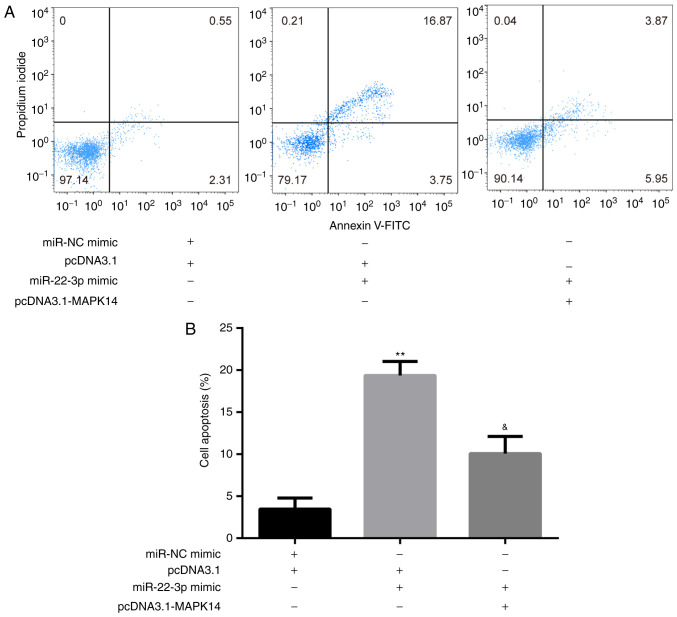
The results from flow cytometry showed that at day 6 of incubation of M-CSF and RANKL, miR-22-3p mimic significantly increased CD14+PBMC apoptosis compared with miR-NC mimic + pcDNA3.1 group, which was significantly rescued following co-transfection with pcDNA3.1-MAPK14 (Fig. 5A and B).
Figure 5.
miR-22-3p mimic promotes CD14+PBMCs apoptosis by targeting MAPK14. (A and B) pcDNA3.1-MAPK14 reversed miR-22-3p mimic-induced CD14+PBMC apoptosis. **P<0.01 vs. pcDNA3.1 or miR-NC mimic + pcDNA3.1. &P<0.05 vs. miR-22-3p mimic + pcDNA3.1. miR, microRNA; PBMCs, peripheral blood mononuclear cells; NC, negative control.
miR-22-3p mimic inhibits CD14+PBMC differentiation by targeting MAPK14
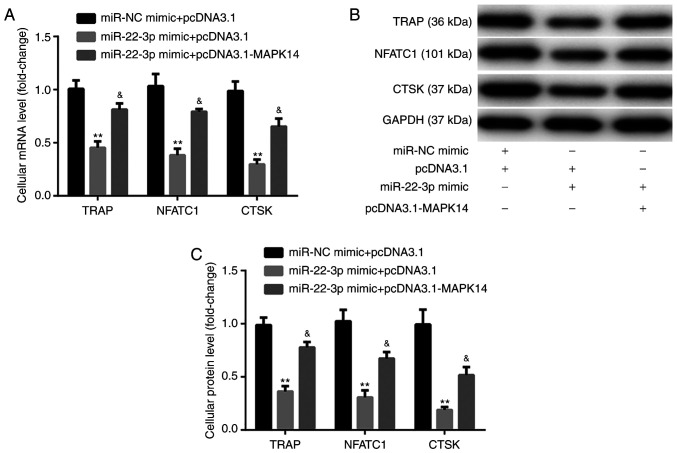
The results from RT-qPCR and western blotting exhibited that at day 6 after incubation with M-CSF and RANKL, miR-22-3p mimic significantly decreased the mRNA and protein expression of the osteoclast markers, including TRAP, NFATC1, and CTSK, compared with miR-NC mimic + pcDNA3.1 group. These observations were reversed following co-transfection with pcDNA3.1-MAPK14 (Fig. 6A-C). These findings indicated that miR-22-3p mimic may attenuate OP via reducing the osteoclastic proliferation and differentiation, while inducing osteoclastic apoptosis by targeting MAPK14.
Figure 6.
miR-22-3p mimic inhibits CD14+PBMC differentiation by targeting MAPK14. (A-C) pcDNA3.1-MAPK14 reversed miR-22-3p mimic-induced CD14+PBMCs decreased differentiation. **P<0.01 vs. pcDNA3.1 or miR-NC mimic + pcDNA3.1. &P<0.05 vs. miR-22-3p mimic + pcDNA3.1. miR, microRNA; PBMCs, peripheral blood mononuclear cells; NC, negative control; TRAP, tartrate resistant acid phosphatase; NFATC1, nuclear factor of activated T-cells; CTSK, cathepsin K.
miR-22-3p mimic inhibits NF-κB activity by targeting MAPK14
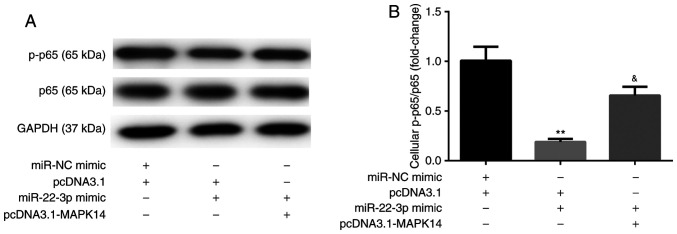
The results from western blotting showed that, at day 6 after incubation with M-CSF and RANKL, miR-22-3p mimic decreased the protein expression of NF-κB p-p65 compared with miR-NC mimic + pcDNA3.1 group. These observations were reversed following co-transfection with pcDNA3.1-MAPK14 (Fig. 7A and B). These results suggested that miR-22-3p mimic may attenuate OP by targeting MAPK14 and inactivating NF-κB p65.
Figure 7.
miR-22-3p mimic inactivates NF-κB by targeting MAPK14. (A and B) pcDNA3.1-MAPK14 reversed miR-22-3p mimic-induced inactivation of NF-κB p65 in CD14+PBMCs **P<0.01 vs. pcDNA3.1 or miR-NC mimic + pcDNA3.1. &P<0.05 vs. miR-22-3p mimic + pcDNA3.1. miR, microRNA; PBMCs, peripheral blood mononuclear cells; NC, negative control; p, phosphorylated.
Discussion
Abnormal expression of miRNAs has been shown to be associated with the process of OP (16), and certain miRNAs are implicated in multiple biological processes, including bone remolding (25). For example, estrogen can downregulate miR-21 biogenesis and upregulate its target gene Fas ligand, inhibiting therefore osteoclastogenesis and inducing osteoclastic apoptosis (26,27). Furthermore, miR-223, of which expression is decreased during RAW264.7 osteoclast differentiation, represses osteoclastogenesis by targeting nuclear factor I A (28,29). In addition, Linc02349 can induce the osteogenesis of human umbilical cord-derived stem cells by sponging miR-25-3p and miR-33b-5p (30). The present study aimed therefore to determine the effect of miR-22-3p on OP in osteoclast cells. It was demonstrated that miR-22-3p inactivated p38/NF-κB pathway by MAPK14 inhibition, thus inhibiting osteoclastic proliferation and differentiation, while promoting osteoclastic apoptosis.
Originally, the present study demonstrated that miR-22-3p expression was lower in patients with postmenopausal OP and CD14+PBMCs treated with M-CSF and RANKL for 6 days compared with healthy volunteers and untreated CD14+PBMCs, respectively. These findings suggested the involvement of miR-22-3p in the development of OP, which was consistent with a previous study reporting miR-22-3p downregulation during osteoclast differentiation (17). Subsequently, the target genes for miR-22-3p had to be investigated.
In the present study, MAPK14 was verified as a target of miR-22-3p in CD14+PBMCs. The p38 MAPKs represent a class of four paralogous mammalian genes, including p38α/MAPK14, p38β/MAPK11, p38γ/MAPK12 and p38δ/MAPK13, which together with ERK1/2 and ERK5 and the c-Jun N-terminal kinases, belong to a wider family of serine-threonine and tyrosine kinases that regulate numerous cellular processes (31). Activated p38 MAPKs can phosphorylate and activate 200-300 downstream targets (32). In addition, p38 MAPK14/11 controls the entry into primitive endoderm differentiation during the development of preimplantation mouse embryo (33). Furthermore, p38α MAPK can promote bone loss. For example, specific p38α inhibitors can prevent bone loss in postmenopausal OP (34), p38α MAPK promotes bone loss induced by ovariectomy via increasing RANKL in osteoblast lineage cells (35), and p38α MAPK induces the proliferation and differentiation of osteoclast progenitors as well as bone remodeling (36). Similarly, the present study demonstrated that MAPK14 expression was higher in patients with postmenopausal OP and CD14+PBMCs treated with M-CSF and RANKL for 6 days compared with healthy volunteers and untreated CD14+PBMCs, respectively.
Subsequently, the present study showed that miR-22-3p mimic significantly decreased osteoclastic proliferation and induced osteoclastic apoptosis by targeting MAPK14. Furthermore, the expression of the osteoclast markers TRAP, NFATC1, and CTSK (37) was significantly decreased by miR-22-3p mimic, which was rescued following MAPK14 overexpression. These results demonstrated the involvement of miR-22-3p and MAPK14 in osteoclastic proliferation, apoptosis and differentiation. Although a previous study reported that miR-22-3p targets MAPK14 in Huntington's disease (38), the present study verified for the first time their relationship in OP, which is of great importance. However, the effects of miR-22-3p and MAPK14 in the regulation of the potential signaling pathways are unknown.
As functional cytokines during osteoclastic differentiation in mammals, M-CSF and RANKL can activate NF-κB pathway, which in turn induces the osteoclastic survival and differentiation (39,40). The present study reported that miR-22-3p mimic could inactivate NF-κB p65 by targeting MAPK14.
The current study presented the following limitations: i) Data on resorptive activity and images of osteoclast maturation were lacking; ii) in vivo experiments were lacking; and iii) miRNA-22-3p downregulation experiments to verify the present findings were missing. Future investigation will address these limitations.
In summary, the present study demonstrated that miR-22-3p mimic could attenuate OP via reducing osteoclastic proliferation and differentiation, while inducing osteoclastic apoptosis by targeting MAPK14 and inactivating NF-κB. These findings may provide a potential therapeutic target for patients with OP.
Acknowledgements
Not applicable.
Funding Statement
Funding: The present study was supported by the Chongqing Medical Research Program (grant no. zy201602149).
Availability of materials and methods
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XJ, MY and WH performed the experiments and data analysis. SC conceived the project, supervised the experiments and data analysis, and prepared the manuscript. XJ, MY, WH and SC confirmed the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Chongqing Public Health Medical Center (approval no. CMC20180106). Each participant provided signed informed consent prior to the beginning of the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that there have no competing interests.
References
- 1.Cosman F, de Beur SJ, LeBoff MS, Lewiecki EM, Tanner B, Randall S, Lindsay R. Erratum to: Clinician's guide to prevention and treatment of osteoporosis. Osteoporos Int. 2015;26:2045–2047. doi: 10.1007/s00198-015-3037-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguyen BN, Hoshino H, Togawa D, Matsuyama Y. Cortical thickness index of the proximal femur: A radiographic parameter for preliminary assessment of bone mineral density and osteoporosis status in the age 50 years and over population. Clin Orthop Surg. 2018;10:149–156. doi: 10.4055/cios.2018.10.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiao W, Wang Y, Pacios S, Li S, Graves DT. Cellular and molecular aspects of bone remodeling. Front Oral Biol. 2016;18:9–16. doi: 10.1159/000351895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karsenty G. Transcriptional control of skeletogenesis. Annu Rev Genomics Hum Genet. 2008;9:183–196. doi: 10.1146/annurev.genom.9.081307.164437. [DOI] [PubMed] [Google Scholar]
- 5.Teitelbaum SL. Bone resorption by osteoclasts. Science. 2000;289:1504–1508. doi: 10.1126/science.289.5484.1504. [DOI] [PubMed] [Google Scholar]
- 6.Manolagas SC. From estrogen-centric to aging and oxidative stress: A revised perspective of the pathogenesis of osteoporosis. Endocr Rev. 2010;31:266–300. doi: 10.1210/er.2009-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rachner TD, Khosla S, Hofbauer LC. Osteoporosis: Now and the future. Lancet. 2011;377:1276–1287. doi: 10.1016/S0140-6736(10)62349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gámez B, Rodriguez-Carballo E, Ventura F. MicroRNAs and post-transcriptional regulation of skeletal development. J Mol Endocrinol. 2014;52:R179–R197. doi: 10.1530/JME-13-0294. [DOI] [PubMed] [Google Scholar]
- 10.Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, Zhang Y. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol. 2012;8:212–227. doi: 10.1038/nrendo.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizuno Y, Yagi K, Tokuzawa Y, Kanesaki-Yatsuka Y, Suda T, Katagiri T, Fukuda T, Maruyama M, Okuda A, Amemiya T, et al. miR-125b inhibits osteoblastic differentiation by down-regulation of cell proliferation. Biochem Biophys Res Commun. 2008;368:267–272. doi: 10.1016/j.bbrc.2008.01.073. [DOI] [PubMed] [Google Scholar]
- 12.Wang FS, Chuang PC, Lin CL, Chen MW, Ke HJ, Chang YH, Chen YS, Wu SL, Ko JY. MicroRNA-29a protects against glucocorticoid-induced bone loss and fragility in rats by orchestrating bone acquisition and resorption. Arthritis Rheum. 2013;65:1530–1540. doi: 10.1002/art.37948. [DOI] [PubMed] [Google Scholar]
- 13.Kahai S, Lee SC, Lee DY, Yang J, Li M, Wang CH, Jiang Z, Zhang Y, Peng C, Yang BB. MicroRNA miR-378 regulates nephronectin expression modulating osteoblast differentiation by targeting GalNT-7. PLoS One. 2009;4(e7535) doi: 10.1371/journal.pone.0007535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia Z, Chen C, Chen P, Xie H, Luo X. MicroRNAs and their roles in osteoclast differentiation. Front Med. 2011;5:414–419. doi: 10.1007/s11684-011-0168-0. [DOI] [PubMed] [Google Scholar]
- 15.van Wijnen AJ, van de Peppel J, van Leeuwen JP, Lian JB, Stein GS, Westendorf JJ, Oursler MJ, Im HJ, Taipaleenmäki H, Hesse E, et al. MicroRNA functions in osteogenesis and dysfunctions in osteoporosis. Curr Osteoporos Rep. 2013;11:72–82. doi: 10.1007/s11914-013-0143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weilner S, Skalicky S, Salzer B, Keider V, Wagner M, Hildner F, Gabriel C, Dovjak P, Pietschmann P, Grillari-Voglauer R, et al. Differentially circulating miRNAs after recent osteoporotic fractures can influence osteogenic differentiation. Bone. 2015;79:43–51. doi: 10.1016/j.bone.2015.05.027. [DOI] [PubMed] [Google Scholar]
- 17.Ma Y, Shan Z, Ma J, Wang Q, Chu J, Xu P, Qin A, Fan S. Validation of downregulated microRNAs during osteoclast formation and osteoporosis progression. Mol Med Rep. 2016;13:2273–2280. doi: 10.3892/mmr.2016.4765. [DOI] [PubMed] [Google Scholar]
- 18.Mäkitie RE, Hackl M, Niinimäki R, Kakko S, Grillari J, Mäkitie O. Altered microRNA profile in osteoporosis caused by impaired WNT signaling. J Clin Endocrinol Metab. 2018;103:1985–1996. doi: 10.1210/jc.2017-02585. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Wang Y, Zhao H, Han X, Zhao T, Qu P, Li G, Wang W. Extracellular vesicle-encapsulated miR-22-3p from bone marrow mesenchymal stem cell promotes osteogenic differentiation via FTO inhibition. Stem Cell Res Ther. 2020;11(227) doi: 10.1186/s13287-020-01707-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Assessment of fracture risk and its application to screening for postmenopausal osteoporosis. Report of a WHO Study Group. World Health Organ Tech Rep Ser. 1994;843:1–129. doi: 10.1007/BF01622200. World Health Organization. [DOI] [PubMed] [Google Scholar]
- 21.Hemingway F, Cheng X, Knowles HJ, Estrada FM, Gordon S, Athanasou NA. In vitro generation of mature human osteoclasts. Calcif Tissue Int. 2011;89:389–395. doi: 10.1007/s00223-011-9530-0. [DOI] [PubMed] [Google Scholar]
- 22.Sørensen MG, Henriksen K, Schaller S, Henriksen DB, Nielsen FC, Dziegiel MH, Karsdal MA. Characterization of osteoclasts derived from CD14+ monocytes isolated from peripheral blood. J Bone Miner Metab. 2007;25:36–45. doi: 10.1007/s00774-006-0725-9. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Cheng P, Chen C, He HB, Hu R, Zhou HD, Xie H, Zhu W, Dai RC, Wu XP, Liao EY, et al. miR-148a regulates osteoclastogenesis by targeting V-maf musculoaponeurotic fibrosarcoma oncogene homolog B. J Bone Miner Res. 2013;28:1180–1190. doi: 10.1002/jbmr.1845. [DOI] [PubMed] [Google Scholar]
- 25.Jing D, Hao J, Shen Y, Tang G, Li ML, Huang SH, Zhao ZH. The role of microRNAs in bone remodeling. Int J Oral Sci. 2015;7:131–143. doi: 10.1038/ijos.2015.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.García Palacios V, Robinson LJ, Borysenko CW, Lehmann T, Kalla SE, Blair HC. Negative regulation of RANKL-induced osteoclastic differentiation in RAW264.7 cells by estrogen and phytoestrogens. J Biol Chem. 2005;280:13720–13727. doi: 10.1074/jbc.M410995200. [DOI] [PubMed] [Google Scholar]
- 27.Sugatani T, Hruska KA. Down-regulation of miR-21 biogenesis by estrogen action contributes to osteoclastic apoptosis. J Cell Biochem. 2013;114:1217–1222. doi: 10.1002/jcb.24471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kagiya T, Nakamura S. Expression profiling of microRNAs in RAW264.7 cells treated with a combination of tumor necrosis factor alpha and RANKL during osteoclast differentiation. J Periodontal Res. 2013;48:373–385. doi: 10.1111/jre.12017. [DOI] [PubMed] [Google Scholar]
- 29.Shibuya H, Nakasa T, Adachi N, Nagata Y, Ishikawa M, Deie M, Suzuki O, Ochi M. Overexpression of microRNA-223 in rheumatoid arthritis synovium controls osteoclast differentiation. Mod Rheumatol. 2013;23:674–685. doi: 10.1007/s10165-012-0710-1. [DOI] [PubMed] [Google Scholar]
- 30.Cao L, Liu W, Zhong Y, Zhang Y, Gao D, He T, Liu Y, Zou Z, Mo Y, Peng S, et al. Linc02349 promotes osteogenesis of human umbilical cord-derived stem cells by acting as a competing endogenous RNA for miR-25-3p and miR-33b-5p. Cell Prolif. 2020;53(e12814) doi: 10.1111/cpr.12814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev. 2011;75:50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cuadrado A, Nebreda AR. Mechanisms and functions of p38 MAPK signalling. Biochem J. 2010;429:403–417. doi: 10.1042/BJ20100323. [DOI] [PubMed] [Google Scholar]
- 33.Thamodaran V, Bruce AW. p38 (Mapk14/11) occupies a regulatory node governing entry into primitive endoderm differentiation during preimplantation mouse embryo development. Open Biol. 2016;6(160190) doi: 10.1098/rsob.160190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caverzasio J, Higgins L, Ammann P. Prevention of trabecular bone loss induced by estrogen deficiency by a selective p38alpha inhibitor. J Bone Miner Res. 2008;23:1389–1397. doi: 10.1359/jbmr.080410. [DOI] [PubMed] [Google Scholar]
- 35.Thouverey C, Caverzasio J. Ablation of p38α MAPK signaling in osteoblast lineage cells protects mice from bone loss induced by estrogen deficiency. Endocrinology. 2015;156:4377–4387. doi: 10.1210/en.2015-1669. [DOI] [PubMed] [Google Scholar]
- 36.Cong Q, Jia H, Li P, Qiu S, Yeh J, Wang Y, Zhang ZL, Ao J, Li B, Liu H. p38α MAPK regulates proliferation and differentiation of osteoclast progenitors and bone remodeling in an aging-dependent manner. Sci Rep. 2017;7(45964) doi: 10.1038/srep45964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 38.Jovicic A, Zaldivar Jolissaint JF, Moser R, Silva Santos MF, Luthi-Carter R. MicroRNA-22 (miR-22) overexpression is neuroprotective via general anti-apoptotic effects and may also target specific Huntington's disease-related mechanisms. PLoS One. 2013;8(e54222) doi: 10.1371/journal.pone.0054222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwon M, Kim JM, Lee K, Park SY, Lim HS, Kim T, Jeong D. Synchronized cell cycle arrest promotes osteoclast differentiation. Int J Mol Sci. 2016;17(1292) doi: 10.3390/ijms17081292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boyce BF. Advances in osteoclast biology reveal potential new drug targets and new roles for osteoclasts. J Bone Miner Res. 2013;28:711–722. doi: 10.1002/jbmr.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.