Abstract
MicroRNA (miR)-770-5p expression is increased in patients with type 2 diabetes mellitus (T2DM) compared with healthy controls; however, the roles and molecular mechanism underlying miR-770-5p in T2DM are not completely understood. In the present study, the reverse transcription-quantitative PCR (RT-qPCR) results indicated that miR-770-5p expression was significantly increased and Bcl-2 associated athanogene 5 (BAG5) expression was significantly decreased in the serum of patients with T2DM compared with healthy volunteers. TargetScan and a dual luciferase reporter gene system were used to predict and verify BAG5 as a target gene of miR-770-5p. Additionally, the RT-qPCR results demonstrated that miR-770-5p expression was significantly increased and BAG5 expression was significantly decreased in uric acid (UA)-treated Min6 cells compared with control cells. Min6 cells were transfected with miR-770-5p inhibitor and BAG5-small interfering (si)RNA to alter expression levels. The results indicated that miR-770-5p negatively regulated BAG5. The effect of miR-770-5p knockdown on UA-induced pancreatic β-cell damage and dysfunction was subsequently assessed. Min6 cells were transfected with miR-770-5p inhibitor or miR-770-5p inhibitor + BAG5-siRNA for 48 h, followed by treatment with or without 5 mg/dl UA for 24 h. Cell viability, apoptosis, apoptosis-related factor expression levels and insulin secretion were assessed. The results demonstrated that UA treatment significantly reduced cell viability, increased cell apoptosis and reduced insulin secretion in Min6 cells compared with the control group. miR-770-5p inhibitor significantly attenuated UA-induced injury and dysfunction of Min6 cells, whereas BAG5 knockdown abolished the protective effects of miR-770-5p inhibitor on UA-damaged Min6 cells. In conclusion, miR-770-5p was highly expressed in the serum of patients with T2DM compared with healthy volunteers. In UA-treated pancreatic β-cells, compared with the inhibitor control group, miR-770-5p knockdown regulated the expression of apoptosis-related genes, increased cell viability, inhibited cell apoptosis and increased insulin secretion by targeting BAG5. Therefore, the present study suggested that miR-770-5p inhibitor may serve a protective role in T2DM.
Keywords: microRNA-770-5p, Bcl-2 associated athanogene 5, type 2 diabetes, pancreatic β-cells
Introduction
Diabetes, whose global prevalence in adults has risen from 4.7% in 1980 to 8.5% in 2014, is currently one of the most prevalent non-communicable diseases threatening global human health (1). At present, the etiology and pathogenesis of type 2 diabetes mellitus (T2DM) are not completely understood. T2DM is characterized by insulin resistance and pancreatic β-cell hypofunction (2), and is often accompanied by a variety of complications, such as kidney disease, fundus disease and neuropathy, that cause damage to various organs and tissues in the body (3,4).
MicroRNAs (miRNAs/miRs) are single-stranded, endogenous non-coding RNAs that are 18-22 nucleotides in length, which regulate gene expression at the post-transcriptional level to exert their biological effects (5,6). miRNAs serve important roles in physiological alterations in the human body, and during the occurrence and development of various diseases, such as cancer, cardiovascular disease and diabetes (7).
Alterations in serum miRNA expression levels occur earlier compared with target genes, they are relatively stable in body fluids, and they display intercellular shuttle ability and tissue-specific specificity; therefore, miRNAs are well suited as biomarkers for the early detection and diagnosis of diseases (8). It has been reported that miRNAs serve important roles in the development of islet cell failure in patients with T2DM (9).
miR-770-5p is a form of mature miR-770and the high evolutionary conservation of miR-770-5p suggests that it may be of particular biological significance (10). Several studies have demonstrated that miR-770 is clinically significant and serves a crucial role in the carcinogenesis and progression of different types of cancer, such as non-small cell lung, breast, ovarian and gastric cancer, as well as hepatocellular carcinoma (11-15). Moreover, previous studies have indicated the significant role of miR-770-5p in diabetic nephropathy and gestational diabetes mellitus (16-18), suggesting that miR-770-5p serves a key role in the regulation of cell function (11-18). It has also been reported that compared with healthy volunteers, miR-770-5p expression was significantly increased in patients with T2DM (9); however, the mechanism underlying miR-770-5p in patients with T2DM is not completely understood. As the function of islet cells serves critical roles in T2DM development (19,20), the aim of the present study was to investigate the expression of miR-770-5p in patients with T2DM, as well as its role in pancreatic β-cells to provide insight into potential therapeutic targets for T2DM.
Materials and methods
Clinical samples
Peripheral blood samples from 20 patients with T2DM (14 male patients and 8 female patients; age, 32-61 years) and 20 healthy subjects (14 male patients and 8 female patients; age, 29-64 years) were collected from the First Affiliated Huai'an People's Hospital of Nanjing Medical University (Huai'an, China) between May 2017 and September 2018. T2DM was diagnosed according to the World Health Organization criteria (21) and was defined as a fasting plasma glucose of ≥7 mmol/l (126 mg/dl), a 2-h glucose of ≥11.1 mmol/l (200 mg/dl) in the 75-g Oral Glucose Tolerance Test and/or HbA1c ≥6.5%. The inclusion criteria were as follows: body mass index <40 and no history of diagnosis of diabetes. The exclusion criteria were as follows: i) Type 1 diabetes; ii) other types of diabetes; iii) severe infections; iv) acute cerebrovascular disease; or iv) those who had recently undergone surgery. There were no significant differences in age and sex between patients with T2DM and healthy volunteers. The present study was approved by the Ethics Committee of The First Affiliated Huai'an People's Hospital of Nanjing Medical University (approval no. KY-P-2017-009-01). Written informed consent was obtained from each patient prior to sample collection. Blood samples were immediately frozen and stored at -80˚C. To detect the expression levels of miR-770-5p and BAG5 mRNA in patients with T2DM, serum was isolated from the peripheral blood samples by centrifugation at 1,500 x g for 10 min at 4˚C.
Cell culture and treatment
Min6 cells (a mouse insulinoma cell line; American Type Culture Collection) were cultured in 5 mM glucose DMEM (Hyclone; Cytiva) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 50 mmol/l β-mercaptoethanol (Sigma-Aldrich; Merck KGaA), 100 U/ml penicillin and 0.1 mg/ml streptomycin (Sigma-Aldrich; Merck KGaA) at 37˚C with 5% CO2.
Uric acid (UA) was prepared as previously reported (22). Briefly, UA sodium salt (10-100 g/ml; Sigma-Aldrich; Merck KGaA) was added to prewarmed EBM-2 medium (Lonza Group, Ltd.; 37˚C). The mixture was agitated at 37˚C for 30 min and passed through a sterile 0.22-µm filter.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from cells or serum samples using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Total RNA (1 µg) was reverse transcribed into cDNA using the PrimeScript RT reagent kit (Takara Bio, Inc.) according the manufacturer's protocol. Subsequently, qPCR was performed using SYBR® Premix Ex Taq™ (Takara Bio, Inc.). The primer sequences used for qPCR were as follows: U6 forward, 5'-GCT TCGGCAGCACATATACTAAAAT-3' and reverse, 5'-CGCT TCACGAATTTGCGTGTCAT-3'; GAPDH forward, 5'-CTTT GGTATCGTGGAAGGACTC-3' and reverse, 5'-GTAGAGGC AGGGATGATGTTCT-3'; miR-770-5p forward, 5'-CCAGTAC CACGTGTCAG-3' and reverse, 5'-GAACATGTCTGCGTAT CTC-3'; BAG5 forward, 5'-TGTCCCCGGGTTTAGGGGTG TTC-3' and reverse, 5'-TTCACAAGCACTGTCCCGCCC-3'; Bcl-2 forward, 5'-TTGGATCAGGGAGTTGGAAG-3' and reverse, 5'-TGTCCCTACCAACCAGAAGG-3'; and Bax forward, 5'-CGTCCACCAAGAAGCTGAGCG-3 and reverse 5'-CGTCCACCAAGAAGCTGAGCG-3'. The following thermocycling conditions were used for qPCR: Initial denaturation at 95˚C for 10 min; followed by 35 cycles of 95˚C for 15 sec and 55˚C for 40 sec. miRNA and mRNA expression levels were quantified using the 2-ΔΔCq method (23) and normalized to the internal reference genes U6 and GAPDH, respectively.
Dual luciferase reporter assay
TargetScan (version 7.2; targetscan.org/vert_72) was used to predict the potential targets of miR-770-5p, and the binding sites between miR-770-5p and BAG5 were identified. To investigate the association between miR-770-5p and BAG5, a dual luciferase reporter assay was performed. The binding site for miR-770-5p in the 3'-untraslated region (UTR) of BAG5 was constructed using synthetic oligonucleotides (Beijing Augct DNA-Syn Biotechnology Co., Ltd.) and cloned into the pmirGLO Dual-Luciferase expression vector (Promega Corporation) to generate wild-type (WT)-BAG5 (5'-UAGUUGGAGGAUGA AUACUGGAG-3'). The mutated 3'-UTR sequence of BAG5 was cloned into the pmirGLO Dual-Luciferase expression vector to generate mutated (MUT)-BAG5 (5'-UAGUUGGAG GAUGAAACUACCAG-3'). Min6 cells (5x104) cultured in 24-well plates were co-transfected with 100 nM miR-770-5p mimic (5'-UCCAGUACCACGUGUCAGGGCCA-3') or 100 nM mimic control (5'-UCGCUUGGUGCAGGUCGG GAA-3') and 1 ng WT-BAG5 or 1 ng MUT-BAG5 reporter plasmids using the Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. At 48 h post-transfection, cells were harvested, and a dual luciferase assay system (Promega Corporation) was used to detect luciferase activities according to the manufacturer's protocol. Luciferase activity was normalized to that of Renilla luciferase activity.
Western blotting
Western blotting was used to measure protein expression levels. Briefly, cells were lysed using RIPA lysis buffer (Shanghai Yeasen Biotechnology Co., Ltd.) and centrifuged at 12,000 x g at 4˚C for 5 min. Protein concentrations were determined using a bicinchoninic acid assay. Total protein (40 µg per lane) was separated via 10% SDS-PAGE and transferred to PVDF membranes (Invitrogen; Thermo Fisher Scientific, Inc.). Following blocking in 5% skimmed dry milk at room temperature for 1 h, the membranes were incubated overnight at 4˚C with primary antibodies targeted against the following: Caspase-3 (cat. no. 14220; 1:1,000; Cell Signaling Technology, Inc.), cleaved caspase-3 (cat. no. 9664; 1:1,000; Cell Signaling Technology, Inc.), Bcl-2 (cat. no. 3498; 1:1,000; Cell Signaling Technology, Inc.), Bax (cat. no. 2772; 1:1,000; Cell Signaling Technology, Inc.), BAG5 (cat. no. ab97660; 1:1,000; Abcam) and β-actin (cat. no. 4970; 1:1,000; Cell Signaling Technology, Inc.). Subsequently, the membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibodies (cat. no. ab205718; 1:2,000; Abcam) for 1 h at room temperature. Protein bands were visualized using an ECL chemiluminescence kit (Thermo Fisher Scientific, Inc.). Protein expression was semi-quantified using Gel-Pro Analyzer densitometry software (version 6.3; Media Cybernetics, Inc.) with β-actin as the loading control.
Cell transfection and treatment
Min6 cells were transfected with 100 nM miR-770-5p inhibitor (5'-UGGCCCUGACACG UGGUACUGGA-3'; Shanghai GenePharma Co., Ltd.), 100 nM inhibitor control (5'-CAGUACUUUUGUGUAGUACAA-3'; Shanghai GeneChem Co., Ltd.), 2 µM control-small interfering (si)RNA (cat. no. sc-37007; Santa Cruz Biotechnology, Inc.), 2 µM BAG5-siRNA (cat. no. sc-72605; Santa Cruz Biotechnology, Inc.) or 100 nM miR-770-5p inhibitor + 2 µM BAG5-siRNA using Lipofectamine® 3000 reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. At 48 h post-transfection, transfection efficiency was determined via RT-qPCR.
After transfection, cells were treated with 5 mg/dl UA at 37˚C for 24 h and then subjected to subsequent experiments. Cells without any treatment were considered as the control.
MTT assay
Min6 cell viability was measured using an MTT assay (Beyotime Institute of Biotechnology). Min6 cells were seeded (5x103 cells/well) into 96-well plates and incubated overnight in DMEM supplemented with 10% FBS. Following treatment and transfection, 20 µl MTT solution (5 mg/ml in distilled water) was added to each well and incubated for 4 h at 37˚C. The medium was removed and 150 µl DMSO was added to dissolve purple formazan. The optical density was measured at a wavelength of 490 nm using a POLARstar OPTIMA multifunctional micro-plate reader (BMG Labtech GmbH).
Apoptosis analysis
Min6 cell apoptosis was measured by performing flow cytometry. Min6 cells (106) were digested using 0.2% trypsin, followed by washing with pre-cooled PBS. Cells were stained using an Annexin V-FITC/PI kit (cat. no. 70-AP101-100; Multisciences (Lianke) Biotech Co., Ltd.) according to the manufacturer's protocol. In brief, Min6 cells were stained with 5 µl Annexin V-FITC and 10 µl PI at room temperature for 5 min in the dark. Apoptotic cells (early and late apoptosis) were analyzed using a FACSCalibur flow cytometer (BD Biosciences) and CellQuest software version 5.1 (BD Biosciences).
Glucose-stimulated insulin secretion assay
Min6 cells were incubated in glucose-free Krebs Ringer bicarbonate (KRB) buffer (115 mM NaCl, 4.7 mM KCl, 1.2 mM MgSO4, 1.2 mM KH2PO4, 20 mM NaHCO3, 16 mM HEPES and 2.56 mM CaCl2; Sigma-Aldrich; Merck KGaA) supplemented with 0.2% BSA (Sigma-Aldrich; Merck KGaA) at 37˚C for 1 h. Subsequently, cells were treated with KRB buffer with basal glucose (3.3 mM) or high concentration glucose (16.7 mM; Sigma-Aldrich; Merck KGaA) at 37˚C for 1 h. The supernatants were collected by centrifugation at 4˚C for 10 min at 500 x g, and subsequently insulin concentrations were determined using an Insulin ELISA kit (cat. no. PI602; Beyotime Institute of Biotechnology) according to the manufacturer's protocol.
Statistical analysis
Statistical analyses were performed using SPSS software (version 20.0; IBM Corp.). Comparisons between two groups were analyzed using a unpaired Student's t-test. Comparisons among multiple groups were analyzed using one-way ANOVA followed by Tukey's post hoc test. Data are presented as the mean ± SD of three independent experiments. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-770-5p is highly expressed in the serum of patients with T2DM
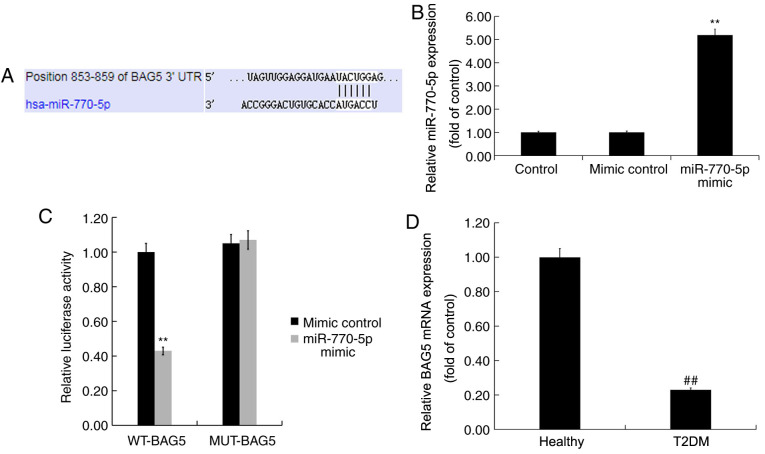
RT-qPCR was performed to detect the expression levels of miR-770-5p in the serum of patients with T2DM and healthy volunteers. miR-770-5p expression levels in the serum of patients with T2DM were significantly higher compared with healthy volunteers (Fig. 1).
Figure 1.
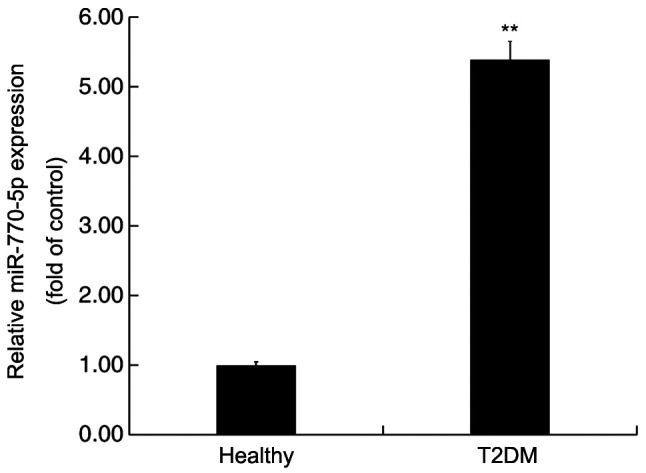
miR-770-5p expression levels in the peripheral blood of patients with T2DM. Reverse transcription-quantitative PCR was performed to detect the expression levels of miR-770-5p in the peripheral blood of patients with T2DM and healthy volunteers. **P<0.01 vs. healthy. miR, microRNA; T2DM, type 2 diabetes mellitus.
BAG5 is a target gene of miR-770-5p
TargetScan was used to predict the possible targets of miR-770-5p. miR-770-5p had numerous potential target genes, including BAG5 (Fig. 2A). As a member of the BAG protein family, which has been reported to enhance cell proliferation and survival (24), BAG5 is involved in several diseases via regulation of cell proliferation and gene expression (25-28). Islet β-cell destruction and apoptosis serve an important role in the development of DM (29). However, the role of BAG5 in T2DM, and the relationship between miR-770-5p and BAG5 are not completely understood. Thus, BAG5 was selected for further examination in the present study. The dual luciferase reporter assay is a well-established experimental method for identifying whether a miRNA directly binds to its target gene (30,31). Therefore, to determine whether miR-770-5p directly targeted BAG5, a dual luciferase reporter assay was used. Compared with the mimic control group, miR-770-5p mimic significantly increased miR-770-5p expression levels in Min6 cells (Fig. 2B). Subsequently, Min6 cells were co-transfected with WT-BAG5 or MUT-BAG5 and miR-770-5p mimic or mimic control. The results indicated that miR-770-5p mimic- + WT-BAG5-transfected cells displayed significantly reduced levels of luciferase activity compared with mimic control- + WT-BAG5-transfected cells (Fig. 2C). By contrast, the luciferase activities of miR-770-5p mimic- + MUT-BAG5-transfected cells were not significantly altered compared with mimic control- + MUT-BAG5-transfected cells, suggesting that miR-770-5p directly targeted the 3'-UTR of BAG5.
Figure 2.
BAG5 is a target gene of miR-770-5p. (A) TargetScan was used to predict the binding sites between BAG5 and miR-770-5p. (B) Transfection efficiency of miR-770-5p mimic. (C) Dual luciferase reporter assays were performed to verify the interaction between BAG5 and miR-770-5p. (D) Reverse transcription-quantitative PCR was performed to detect BAG5 expression levels in the peripheral blood of patients with T2DM and healthy volunteers. **P<0.01 vs. mimic control; ##P<0.01 vs. healthy. BAG5, Bcl-2 associated athanogene 5; miR, microRNA; T2DM, type 2 diabetes mellitus; WT, wild-type; MUT, mutant; UTR, untranslated region.
RT-qPCR was performed to detect the mRNA expression levels of BAG5 in the serum of patients with T2DM and healthy volunteers. BAG5 mRNA expression levels in the serum of patients with T2DM were significantly decreased compared with healthy volunteers (Fig. 2D).
UA treatment upregulates miR-770-5p expression and downregulates BAG5 expression in Min6 cells
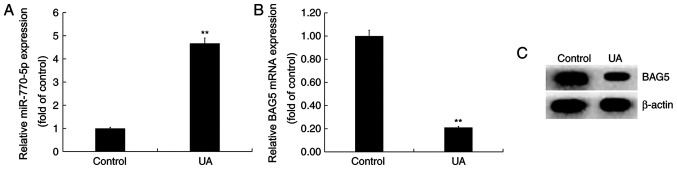
Min6 cells were treated with UA (5 mg/dl) for 24 h. Compared with the control group, miR-770-5p expression was significantly increased in the UA treatment group (Fig. 3A). By contrast, the mRNA and protein expression levels of BAG5 were decreased in UA-treated Min6 cells compared with control cells (Fig. 3B and C).
Figure 3.
Effect of UA treatment on miR-770-5p and BAG5 expression levels in Min6 cells. Min6 cells were treated with UA (5 mg/dl) for 24 h. (A) miR-770-5p expression levels were detected via RT-qPCR. BAG5 (B) mRNA and (C) protein expression levels were determined via RT-qPCR and western blotting, respectively. **P<0.01 vs. control group. UA, uric acid; miR, microRNA; BAG5, Bcl-2 associated athanogene 5; RT-qPCR, reverse transcription-quantitative PCR.
miR-770-5p negatively regulates BAG5 expression in Min6 cells
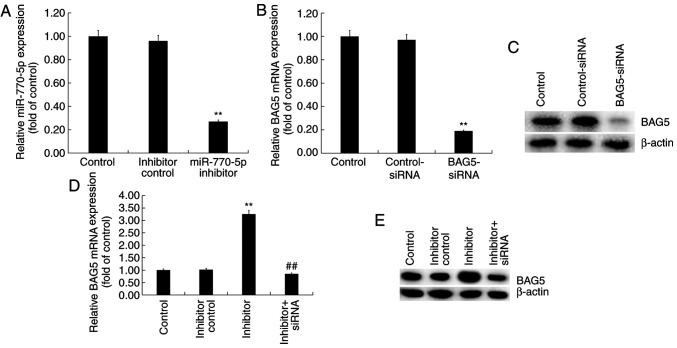
Min6 cells were transfected with control-siRNA, BAG5-siRNA, miR-770-5p inhibitor, inhibitor control or BAG5-siRNA + miR-770-5p inhibitor for 48 h. The RT-qPCR results demonstrated that miR-770-5p inhibitor significantly decreased miR-770-5p expression levels in Min6 cells compared with inhibitor control (Fig. 4A). BAG5-siRNA markedly reduced BAG5 mRNA and protein expression levels in Min6 cells compared with control-siRNA (Fig. 4B and C). Moreover, compared with the inhibitor control group, miR-770-5p inhibitor increased the mRNA (Fig. 4D) and protein (Fig. 4E) expression levels of BAG5 in Min6 cells, which was reversed by co-transfection with BAG5-siRNA.
Figure 4.
Regulatory effect of miR-770-5p on BAG5 expression in Min6 cells. Min6 cells were transfected with control-siRNA, BAG5-siRNA, inhibitor control, miR-770-5p inhibitor or BAG5-siRNA + miR-770-5p inhibitor for 48 h. (A) Transfection efficiency of miR-770-5p inhibitor. The transfection efficiency of BAG5-siRNA was assessed via (B) RT-qPCR and (C) western blotting. BAG5 (D) mRNA and (E) protein expression levels were determined via RT-qPCR and western blotting, respectively. **P<0.01 vs. control; ##P<0.01 vs. inhibitor. miR, microRNA; BAG5, Bcl-2 associated athanogene 5; siRNA, small interfering RNA; RT-qPCR, reverse transcription-quantitative PCR.
miR-770-5p inhibitor alleviates UA-induced injury and dysfunction in Min6 cells
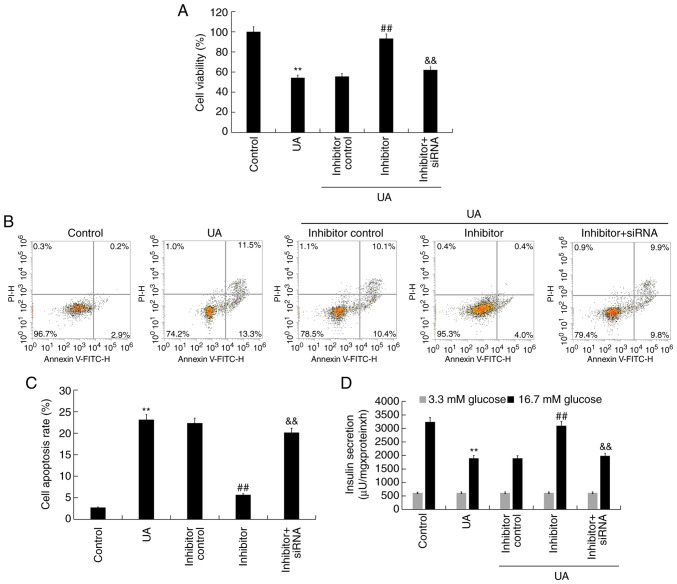
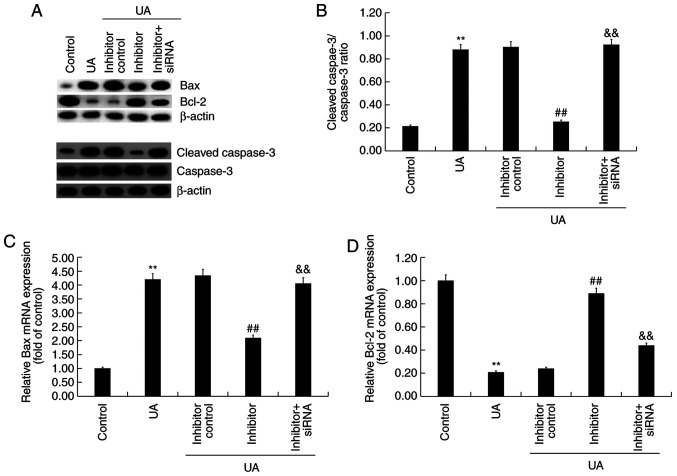
To explore the effect of miR-770-5p knockdown on UA-induced Min6 cell injury and dysfunction, Min6 cells were transfected with inhibitor control, miR-770-5p inhibitor, or miR-770-5p inhibitor + BAG5-siRNA for 48 h, and subsequently treated with UA (5 mg/dl) for 24 h. Cell viability, apoptosis and insulin secretion were assessed. Compared with UA treatment alone, miR-770-5p inhibitor significantly increased cell viability (Fig. 5A) and decreased cell apoptosis (Fig. 5B and C) in UA-treated Min6 cells. Similarly, miR-770-5p inhibitor significantly increased insulin secretion from UA- and glucose-treated Min6 cells compared with UA treatment alone (Fig. 5D). Compared with the control group, UA treatment markedly increased Bax and cleaved-caspase-3 protein expression levels, reduced Bcl-2 protein expression levels and upregulated the ratio of cleaved-caspase-3/caspase-3 in Min6 cells (Fig. 6A and B). miR-770-5p inhibitor markedly decreased Bax and cleaved-caspase-3 protein expression levels, increased Bcl-2 protein expression levels, and downregulated the ratio of cleaved-caspase-3/caspase-3 in UA-treated Min6 cells compared with the UA + inhibitor control treatment group. Moreover, compared with the control group, UA treatment significantly increased Bax mRNA expression levels and decreased Bcl-2 mRNA expression levels in Min6 cells (Fig. 6C and D). However, miR-770-5p inhibitor significantly decreased Bax expression levels and increased Bcl-2 mRNA expression levels in UA-treated Min6 cells compared with UA treatment alone. Furthermore, miR-770-5p inhibitor-mediated effects on UA-treated Min6 cells were significantly reversed by co-transfection with BAG5-siRNA.
Figure 5.
Effect of UA and miR-770-5p inhibitor on Min6 cells. Following transfection, Min6 cells were treated with UA (5 mg/dl) for 24 h. (A) Cell viability was detected by performing an MTT assay. Cell apoptosis was (B) determined by flow cytometry and (C) quantified. (D) A glucose-stimulated insulin secretion assay was performed to detect the insulin secretion. **P<0.01 vs. control; ##P<0.01 vs. UA; &&P<0.01 vs. inhibitor. UA, uric acid; miR, microRNA; siRNA, small interfering RNA.
Figure 6.
Effect of UA and miR-770-5p inhibitor on apoptosis-related gene expression levels in Min6 cells. Following transfection, Min6 cells were treated with UA (5 mg/dl) for 24 h. (A) Western blotting was performed to measure the protein expression levels of Bax, Bcl-2, cleaved-caspase-3 and caspase-3. (B) The ratio of cleaved-caspase-3/caspase-3. Reverse transcription-quantitative PCR were performed to measure (C) Bax and (D) Bcl-2 mRNA expression levels. **P<0.01 vs. control; ##P<0.01 vs. UA; &&P<0.01 vs. inhibitor. UA, uric acid; miR, microRNA; siRNA, small interfering RNA.
Discussion
According to relevant epidemiological statistics, the number of new cases of diabetes mellitus (DM) worldwide is increasing on a yearly basis (32). Although multiple pathogenic mechanisms underlying DM complications have been identified, the clinical results achieved are limited and the exact pathogenesis is not completely understood. With continuous advances in molecular biotechnology, the discovery of miRNAs has provided a novel insight into the investigation of DM (33). An increasing number of studies have demonstrated that miRNAs serve important roles in the pathological processes underlying DM (9,33,34). In the present study, the expression of miR-770-5p and its target gene BAG5 in patients with T2DM was assessed. miR-770-5p expression was significantly increased in the serum of patients with T2DM compared with healthy volunteers. By contrast, BAG5 expression was significantly decreased in the serum of patients with T2DM compared with healthy volunteers.
The relationship between T2DM and serum UA levels has been investigated (35). A number of studies have reported that the risk of T2DM was greatly increased under high UA conditions (36,37). Enhanced UA levels not only resulted in insulin resistance but also negatively affected pancreatic β-cell survival and insulin release (38). The Min6 cell line has been used to investigate pancreatic β-cell function in vitro (38,39). In the present study, UA-treated Min6 cells were used to explore the effect of miR-770-5p on UA-induced pancreatic β-cell damage and dysfunction.
Islet β-cell destruction, apoptosis and dedifferentiation can result in the loss of islet β-cells, which decreases insulin secretion resulting in blood sugar level disorders, ultimately leading to diabetes and the associated complications (29). miRNAs destruct multiple target genes, such as C-C chemokine receptor type 7 and CD247, in islet β-cells (40). Apoptosis refers to the autonomous and orderly death of cells under the regulation of genes in order to maintain the homeostasis of the internal environment (41). Islet β-cell apoptosis is a primary cause of DM and islet β-cell function damage is also closely associated with DM. Islet β-cell function damage mechanisms may be associated with oxidative stress, glucose toxicity, lipotoxicity and hypoxia. Inflammation caused by these mechanisms may initially maintain and repair the function of pancreatic islet β-cell, but persistent inflammatory reactions can cause damage and permanent destruction to islet β-cells (42,43). Several miRNAs have been reported to be involved in islet β-cell apoptosis, including miR-23a-3p, miR-23b-3p and miR-149-5p. These miRNAs are associated with type 1 diabetes mellitus via downregulating Bcl-2 expression, which promotes islet β-cell apoptosis (44). miR-770-5p, which has been studied in several types of cancer, such as non-small cell lung, breast, ovarian and gastric cancer, as well as hepatocellular carcinoma (11-15), was reported to serve key roles in diabetic nephropathy and gestational diabetes mellitus (16-18). A recent study indicated that compared with healthy volunteers, miR-770-5p expression was upregulated in patients with gestational diabetes mellitus, and it can regulate INS-1 insulinoma cell proliferation, apoptosis and insulin secretion by targeting tumor protein p53 regulated inhibitor of apoptosis 1(18). However, the roles and mechanism underlying miR-770-5p in pancreatic β-cells are not completely understood.
In the present study, BAG5 was identified as a direct target of miR-770-5p. BAG5 expression was significantly downregulated in patients with T2DM and UA-treated Min6 cells compared with healthy volunteers and control cells, respectively. BAG5 is a member of the BAG protein family, which enhance cell proliferation and survival (24). Previous studies have demonstrated that BAG5 is involved in several diseases, including cancer and Alzheimer's disease, via regulation of apoptosis and gene expression (25-28). Therefore, it was hypothesized that miR-770-5p may serve an important role in T2DM via regulating islet β-cell apoptosis by targeting BAG5. The effects of miRNA-770-5p on islet β-cell damage and dysfunction were determined by assessing cell viability using an MTT assay, detecting apoptosis via flow cytometry, and measuring the expression levels of apoptosis-associated genes and proteins via RT-qPCR and western blotting, respectively. The results demonstrated that UA treatment reduced cell viability, promoted apoptosis, increased the cleaved-caspase-3/caspase-3 ratio and Bax expression levels, and decreased Bcl-2 expression levels in Min6 cells compared with the control group. However, UA treatment-mediated effects were significantly attenuated by transfection with miR-770-5p inhibitor. Moreover, miR-770-5p-mediated effects on UA-treated Min6 cells were significantly reversed by co-transfection with BAG5-siRNA. The aforementioned results were consistent with a previous study that reported that miRNAs are involved in islet β-cell destruction and apoptosis (44).
Insulin is a protein hormone secreted by islet β-cells that are stimulated by endogenous or exogenous substances (45). Insulin serves an important role in the balance of blood sugar levels in the human body, whereby insufficiency results in hyperglycemia, DM and DM complications (46). Previous studies have reported that miRNAs can regulate the synthesis and secretion of insulin (47,48) and regulate the blood sugar balance in the human body (49). miRNAs not only promote insulin synthesis and secretion, but also serve a negative regulatory role in insulin synthesis and secretion (50-52). In the present study, the effects of miR-770-5p on insulin secretion were determined. The results demonstrated that compared with the control group, UA treatment significantly reduced insulin secretion in Min6 cells, which was significantly reversed by transfection with miR-770-5p inhibitor. However, miR-770-5p inhibitor-mediated effects on insulin secretion in UA-treated Min6 cells were significantly reversed by co-transfection with BAG5-siRNA. A key limitation of the present study was that a UA + miR-770-5p inhibitor + control-siRNA group was not included.
In conclusion, the results of the present study demonstrated that miR-770-5p expression was significantly upregulated in patients with T2DM compared with healthy volunteers. Compared with the inhibitor control group, miR-770-5p knockdown increased cell viability, reduced cell apoptosis, regulated the expression of apoptosis-related genes and increased insulin secretion in UA-treated pancreatic β-cells by targeting BAG5, thus suppressing the development and/or progression of T2DM. The results of the present study provided novel insight into potential prevention and treatment strategies for T2DM. However, the present study was only a preliminary study of the role of miR-770-5p in T2DM. Therefore, in vivo studies are required to confirm the role of miR-770-5p in T2DM. Additionally, the relationship between miR-770-5p and BAG5 expression levels and the clinicopathological characteristics of patients with T2DM should be investigated in future studies.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
MW contributed to designing the study, collecting, analyzing and interpreting the data, and preparing the manuscript. JW, TJ and KZ contributed to collecting, analyzing and interpreting the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of The First Affiliated Huai'an People's Hospital of Nanjing Medical University (approval no. KY-P-2017-009-01). Written informed consent was obtained from each patient prior to sample collection.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kanter JE, Bornfeldt KE. Impact of Diabetes Mellitus. Arterioscler Thromb Vasc Biol. 2016;36:1049–1053. doi: 10.1016/j.jdiacomp.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashcroft FM, Rorsman P. Diabetes mellitus and the β cell: The last ten years. Cell. 2012;148:1160–1171. doi: 10.1016/j.cell.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wright E Jr, Scism-Bacon JL, Glass LC. Oxidative stress in type 2 diabetes: The role of fasting and postprandial glycaemia. Int J Clin Pract. 2006;60:308–314. doi: 10.1111/j.1368-5031.2006.00825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oelze M, Schuhmacher S, Daiber A. Organic nitrates and nitrate resistance in diabetes: The role of vascular dysfunction and oxidative stress with emphasis on antioxidant properties of pentaerithrityl tetranitrate. Exp Diabetes Res. 2010;2010(213176) doi: 10.1155/2010/213176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 6.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 7.Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: Shared themes amid diversity. Nat Rev Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camussi G, Deregibus MC, Bruno S, Cantaluppi V, Biancone L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 2010;78:838–848. doi: 10.1038/ki.2010.278. [DOI] [PubMed] [Google Scholar]
- 9.Wang C, Wan S, Yang T, Niu D, Zhang A, Yang C, Cai J, Wu J, Song J, Zhang CY, et al. Increased serum microRNAs are closely associated with the presence of microvascular complications in type 2 diabetes mellitus. Sci Rep. 2016;6(20032) doi: 10.1038/srep20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo J, Han J, Liu J, Wang S. MicroRNA-770-5p contributes to podocyte injury via targeting E2F3 in diabetic nephropathy. Braz J Med Biol Res. 2020;53(e9360) doi: 10.1590/1414-431x20209360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Liang Y, Sang Y, Song X, Zhang H, Liu Y, Jiang L, Yang Q. miR-770 suppresses the chemo-resistance and metastasis of triple negative breast cancer via direct targeting of STMN1. Cell Death Dis. 2018;9(14) doi: 10.1038/s41419-017-0030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, Yang Y, Zhang X. miR-770 inhibits tumorigenesis and EMT by targeting JMJD6 and regulating WNT/β-catenin pathway in non-small cell lung cancer. Life Sci. 2017;188:163–171. doi: 10.1016/j.lfs.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Wu WJ, Shi J, Hu G, Yu X, Lu H, Yang ML, Liu B, Wu ZX. Wnt/β-catenin signaling inhibits FBXW7 expression by upregulation of microRNA-770 in hepatocellular carcinoma. Tumour Biol. 2016;37:6045–6051. doi: 10.1007/s13277-015-4452-5. [DOI] [PubMed] [Google Scholar]
- 14.Guo W, Dong Z, Liu S, Qiao Y, Kuang G, Guo Y, Shen S, Liang J. Promoter hypermethylation-mediated downregulation of miR-770 and its host gene MEG3, a long non-coding RNA, in the development of gastric cardia adenocarcinoma. Mol Carcinog. 2017;56:1924–1934. doi: 10.1002/mc.22650. [DOI] [PubMed] [Google Scholar]
- 15.Zhao H, Yu X, Ding Y, Zhao J, Wang G, Wu X, Jiang J, Peng C, Guo GZ, Cui S. miR-770-5p inhibits cisplatin chemoresistance in human ovarian cancer by targeting ERCC2. Oncotarget. 2016;7:53254–53268. doi: 10.18632/oncotarget.10736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Li H. miR-770-5p facilitates podocyte apoptosis and inflammation in diabetic nephropathy by targeting TIMP3. Biosci Rep. 2020;40(BSR20193653) doi: 10.1042/BSR20193653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang SZ, Qiu XJ, Dong SS, Zhou LN, Zhu Y, Wang MD, Jin LW. MicroRNA-770-5p is involved in the development of diabetic nephropathy through regulating podocyte apoptosis by targeting TP53 regulated inhibitor of apoptosis 1. Eur Rev Med Pharmacol Sci. 2019;23:1248–1256. doi: 10.26355/eurrev_201902_17018. [DOI] [PubMed] [Google Scholar]
- 18.Zhang YL, Chen XQ. Dysregulation of microRNA-770-5p influences pancreatic-β-cell function by targeting TP53 regulated inhibitor of apoptosis 1 in gestational diabetes mellitus. Eur Rev Med Pharmacol Sci. 2020;24:793–801. doi: 10.26355/eurrev_202001_20062. [DOI] [PubMed] [Google Scholar]
- 19.Böni-Schnetzler M, Meier DT. Islet inflammation in type 2 diabetes. Semin Immunopathol. 2019;41:501–513. doi: 10.1007/s00281-019-00745-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mezza T, Cinti F, Cefalo CMA, Pontecorvi A, Kulkarni RN, Giaccari A. β-Cell Fate in Human Insulin Resistance and Type 2 Diabetes: A Perspective on Islet Plasticity. Diabetes. 2019;68:1121–1129. doi: 10.2337/db18-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: Diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1998;15:539–553. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 22.Kanellis J, Watanabe S, Li JH, Kang DH, Li P, Nakagawa T, Wamsley A, Sheikh-Hamad D, Lan HY, Feng L, et al. Uric acid stimulates monocyte chemoattractant protein-1 production in vascular smooth muscle cells via mitogen-activated protein kinase and cyclooxygenase-2. Hypertension. 2003;41:1287–1293. doi: 10.1161/01.HYP.0000072820.07472.3B. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Townsend PA, Cutress RI, Sharp A, Brimmell M, Packham G. BAG-1: A multifunctional regulator of cell growth and survival. Biochim Biophys Acta. 2003;1603:83–98. doi: 10.1016/s0304-419x(03)00002-7. [DOI] [PubMed] [Google Scholar]
- 25.Bruchmann A, Roller C, Walther TV, Schäfer G, Lehmusvaara S, Visakorpi T, Klocker H, Cato AC, Maddalo D. Bcl-2 associated athanogene 5 (Bag5) is overexpressed in prostate cancer and inhibits ER-stress induced apoptosis. BMC Cancer. 2013;13(96) doi: 10.1186/1471-2407-13-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo K, Li L, Yin G, Zi X, Liu L. Bag5 protects neuronal cells from amyloid β-induced cell death. J Mol Neurosci. 2015;55:815–820. doi: 10.1007/s12031-014-0433-1. [DOI] [PubMed] [Google Scholar]
- 27.Gupta MK, Tahrir FG, Knezevic T, White MK, Gordon J, Cheung JY, Khalili K, Feldman AM. GRP78 Interacting Partner Bag5 Responds to ER Stress and Protects Cardiomyocytes From ER Stress-Induced Apoptosis. J Cell Biochem. 2016;117:1813–1821. doi: 10.1002/jcb.25481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Guo J, Fei E, Mu Y, He S, Che X, Tan J, Xia K, Zhang Z, Wang G, et al. BAG5 protects against mitochondrial oxidative damage through regulating PINK1 degradation. PLoS One. 2014;9(e86276) doi: 10.1371/journal.pone.0086276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue M, Jackson CJ. Activated protein C and its potential applications in prevention of islet β-cell damage and diabetes. Vitam Horm. 2014;95:323–363. doi: 10.1016/B978-0-12-800174-5.00013-2. [DOI] [PubMed] [Google Scholar]
- 30.Liu Q, Axtell MJ. Quantitating plant microRNA-mediated target repression using a dual-luciferase transient expression system. Methods Mol Biol. 2015;1284:287–303. doi: 10.1007/978-1-4939-2444-8_14. [DOI] [PubMed] [Google Scholar]
- 31.Wang Z, Zhu Z, Lin Z, Luo Y, Liang Z, Zhang C, Chen J, Peng P. miR-429 suppresses cell proliferation, migration and invasion in nasopharyngeal carcinoma by downregulation of TLN1. Cancer Cell Int. 2019;19(115) doi: 10.1186/s12935-019-0831-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: Estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27:1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 33.Tiwari J, Gupta G, de Jesus Andreoli Pinto T, Sharma R, Pabreja K, Matta Y, Arora N, Mishra A, Sharma R, Dua K. Role of microRNAs (miRNAs) in the pathophysiology of diabetes mellitus. Panminerva Med. 2018;60:25–28. doi: 10.23736/S0031-0808.17.03382-1. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Sun X, Icli B, Feinberg MW. Emerging Roles for MicroRNAs in Diabetic Microvascular Disease: Novel Targets for Therapy. Endocr Rev. 2017;38:145–168. doi: 10.1210/er.2016-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Behradmanesh S, Horestani MK, Baradaran A, Nasri H. Association of serum uric acid with proteinuria in type 2 diabetic patients. J Res Med Sci. 2013;18:44–46. [PMC free article] [PubMed] [Google Scholar]
- 36.Ye X, Cao Y, Gao F, Yang Q, Zhang Q, Fu X, Li J, Xue Y. Elevated serum uric acid levels are independent risk factors for diabetic foot ulcer in female Chinese patients with type 2 diabetes. J Diabetes. 2014;6:42–47. doi: 10.1111/1753-0407.12079. [DOI] [PubMed] [Google Scholar]
- 37.Viazzi F, Piscitelli P, Giorda C, Ceriello A, Genovese S, Russo G, Guida P, Fioretto P, De Cosmo S, Pontremoli R. Metabolic syndrome, serum uric acid and renal risk in patients with T2D. PLoS One. 2017;12(e0176058) doi: 10.1371/journal.pone.0176058. AMD-Annals Study Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding Y, Shi X, Shuai X, Xu Y, Liu Y, Liang X, Wei D, Su D. Luteolin prevents uric acid-induced pancreatic β-cell dysfunction. J Biomed Res. 2014;28:292–298. doi: 10.7555/JBR.28.20130170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adeyemo AA, Zaghloul NA, Chen G, Doumatey AP, Leitch CC, Hostelley TL, Nesmith JE, Zhou J, Bentley AR, Shriner D, et al. South Africa Zulu Type 2 Diabetes Case-Control Study: ZRANB3 is an African-specific type 2 diabetes locus associated with beta-cell mass and insulin response. Nat Commun. 2019;10(3195) doi: 10.1038/s41467-019-10967-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fornari TA, Donate PB, Assis AF, Macedo C, Sakamoto-Hojo ET, Donadi EA, Passos GA. Comprehensive Survey of miRNA-mRNA Interactions Reveals That Ccr7 and Cd247 (CD3 zeta) are Posttranscriptionally Controlled in Pancreas Infiltrating T Lymphocytes of Non-Obese Diabetic (NOD) Mice. PLoS One. 2015;10(e0142688) doi: 10.1371/journal.pone.0142688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elmore S. Apoptosis: A review of programmed cell death. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerber PA, Rutter GA. The Role of Oxidative Stress and Hypoxia in Pancreatic Beta-Cell Dysfunction in Diabetes Mellitus. Antioxid Redox Signal. 2017;26:501–518. doi: 10.1089/ars.2016.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu Z, Gilbert ER, Liu D. Regulation of insulin synthesis and secretion and pancreatic Beta-cell dysfunction in diabetes. Curr Diabetes Rev. 2013;9:25–53. [PMC free article] [PubMed] [Google Scholar]
- 44.Grieco FA, Sebastiani G, Juan-Mateu J, Villate O, Marroqui L, Ladrière L, Tugay K, Regazzi R, Bugliani M, Marchetti P, et al. MicroRNAs miR-23a-3p, miR-23b-3p, and miR-149-5p Regulate the Expression of Proapoptotic BH3-Only Proteins DP5 and PUMA in Human Pancreatic β-Cells. Diabetes. 2017;66:100–112. doi: 10.2337/db16-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao X, Mohan R, Özcan S, Tang X. MicroRNA-30d induces insulin transcription factor MafA and insulin production by targeting mitogen-activated protein 4 kinase 4 (MAP4K4) in pancreatic β-cells. J Biol Chem. 2012;287:31155–31164. doi: 10.1074/jbc.M112.362632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schroeder IS, Kania G, Blyszczuk P, Wobus AM. Insulin-producing cells. Methods Enzymol. 2006;418:315–333. doi: 10.1016/S0076-6879(06)18019-2. [DOI] [PubMed] [Google Scholar]
- 47.Ramzy A, Mojibian M, Kieffer TJ. Insulin-Deficient Mouse β-Cells Do Not Fully Maturebut Can Be Remedied Through Insulin Replacementby Islet Transplantation. Endocrinology. 2018;159:83–102. doi: 10.1210/en.2017-00263. [DOI] [PubMed] [Google Scholar]
- 48.Melkman-Zehavi T, Oren R, Kredo-Russo S, Shapira T, Mandelbaum AD, Rivkin N, Nir T, Lennox KA, Behlke MA, Dor Y, et al. miRNAs control insulin content in pancreatic β-cells via downregulation of transcriptional repressors. EMBO J. 2011;30:835–845. doi: 10.1038/emboj.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guay C, Regazzi R. MicroRNAs and the functional β cell mass: For better or worse. Diabetes Metab. 2015;41:369–377. doi: 10.1016/j.diabet.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 50.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 51.Fred RG, Mehrabi S, Adams CM, Welsh N. PTB and TIAR binding to insulin mRNA 3'- and 5'UTRs; implications for insulin biosynthesis and messenger stability. Heliyon. 2016;2(e00159) doi: 10.1016/j.heliyon.2016.e00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Setyowati Karolina D, Sepramaniam S, Tan HZ, Armugam A, Jeyaseelan K. miR-25 and miR-92a regulate insulin I biosynthesis in rats. RNA Biol. 2013;10:1365–1378. doi: 10.4161/rna.25557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.